Abstract
Aim: The treatment of psoriasis remains elusive, underscoring the need for identifying novel disease targets and mechanism-based therapeutic approaches. We recently reported that the PI3K/Akt/mTOR pathway that is frequently deregulated in many malignancies is also clinically relevant for psoriasis. We also provided rationale for developing delphinidin (Del), a dietary antioxidant for the management of psoriasis. This study utilized high-throughput biophysical and biochemical approaches and in vitro and in vivo models to identify molecular targets regulated by Del in psoriasis.
Results: A kinome-level screen and Kds analyses against a panel of 102 human kinase targets showed that Del binds to three lipid (PIK3CG, PIK3C2B, and PIK3CA) and six serine/threonine (PIM1, PIM3, mTOR, S6K1, PLK2, and AURKB) kinases, five of which belong to the PI3K/Akt/mTOR pathway. Surface plasmon resonance and in silico molecular modeling corroborated Del's direct interactions with three PI3Ks (α/c2β/γ), mTOR, and p70S6K. Del treatment of interleukin-22 or TPA-stimulated normal human epidermal keratinocytes (NHEKs) significantly inhibited proliferation, activation of PI3K/Akt/mTOR components, and secretion of proinflammatory cytokines and chemokines. To establish the in vivo relevance of these findings, an imiquimod (IMQ)-induced Balb/c mouse psoriasis-like skin model was employed. Topical treatment of Del significantly decreased (i) hyperproliferation and epidermal thickness, (ii) skin infiltration by immune cells, (iii) psoriasis-related cytokines/chemokines, (iv) PI3K/Akt/mTOR pathway activation, and (v) increased differentiation when compared with controls.
Innovation and Conclusion: Our observation that Del inhibits key kinases involved in psoriasis pathogenesis and alleviates IMQ-induced murine psoriasis-like disease suggests a novel PI3K/AKT/mTOR pathway modulator that could be developed to treat psoriasis. Antioxid. Redox Signal. 26, 49–69.
Keywords: : delphinidin, psoriasis, imiquimod model of inflammation, antioxidants
Introduction
Psoriasis is a common and currently incurable chronic inflammatory skin disease that affects an estimated 125 million people worldwide (26, 32). The cause of psoriasis is incompletely understood, but the disease is characterized by sharply demarcated, erythematous scaly skin plaques resulting from epidermal keratinocyte hyperproliferation, aberrant differentiation, parakeratosis, immune cell infiltration, and angiogenesis (32). Psoriasis is also associated with significant decline in quality of life and an increased risk of arthritis and cardiovascular disease. Current treatment options are mainly palliative and involve corticosteroids, UV light therapy, and immune-modulating therapy.
Innovation.
This study provides the first evidence for a novel mechanism and target of the small-molecule dietary antioxidant, delphinidin, against chronic inflammatory skin condition, psoriasis. Delphinidin inhibits key kinases involved in psoriasis pathogenesis and alleviates imiquimod (IMQ)-induced murine psoriasis-like disease, suggesting a novel PI3K/AKT/mTOR pathway modulator for psoriasis. Unlike other rapalogs, Del binds mammalian target of rapamycin (mTOR) with a much higher affinity and does not require the formation of a ternary complex for its activity. Our observations provide a preclinical rationale for developing Del as an antipsoriatic drug, alone or in combination with other known drugs, as a powerful strategy for treatment of psoriasis.
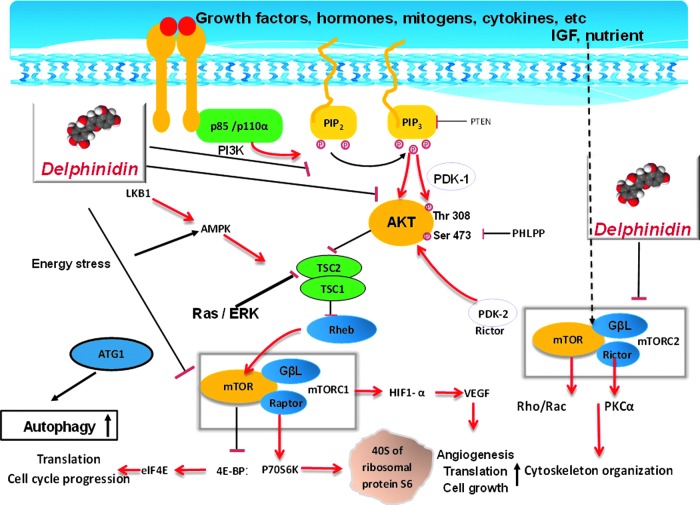
The phospho-inositol-3 kinase (PI3K)/protein kinase B (Akt) and mammalian target of rapamycin (mTOR) signal transduction pathway is centrally implicated in the regulation of a variety of important physiologic functions, such as metabolism, protein synthesis, cell growth, survival, cell cycle, angiogenesis, and apoptosis (1, 5, 24, 28). The pathway is frequently deregulated in diverse malignancies (10, 24) and recent data suggest its clinical relevance in inflammatory diseases, including psoriasis (7, 22). The PI3K/Akt/mTOR pathway is tightly regulated through feedback loops, in part, via mTOR with linkage through Akt (see diagram in Fig. 10).
FIG. 10.
Schematic illustration of the effect of delphinidin on PI3K/Akt/mTOR signaling. Simultaneously targeting both PI3K/Akt and mTOR has the potential to inhibit both upstream and downstream signaling in the pathway, resulting in decrease in cell and tissue growth, angiogenesis, and normalization of tissue architecture. Delphindin by inhibiting the PI3K/Akt pathway also keeps the mTOR pathway in check and results in inhibition of cell survival and growth. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
mTOR exists in two functionally distinct protein complexes, mTORC1 and mTORC2. mTORC1 phosphorylates the p70S6 kinase (p70S6K), which in turn phosphorylates the S6 ribosomal protein and 4E-BP1, leading to protein translation (21). mTORC2 functions in the feedback loop to activate Akt by phosphorylation on serine 473 (36), which in turn activates mTORC1 via phosphorylation of TSC2 and PRAS40, thus promoting keratinocyte hyperproliferation and inhibiting differentiation (22). Because the PI3K/Akt/mTOR pathway is hyperactivated both in human and murine psoriasis, it is an attractive antipsoriatic drug target (7, 16, 22).
Rapamycin and its analogs are the best-known allosteric inhibitors of the PI3K/Akt/mTOR pathway and are being used for treating several types of cancers (31). These allosteric inhibitors, in complex with FKBP12, target the FKB domain of mTOR (11) and partially inhibit mTOR through binding to mTORC1, but not mTORC2 (48). However, inhibition of mTORC1 is not sufficient to achieve a broad and multifactorial therapeutic effect owing to failure to inhibit mTORC2 and resistance to this treatment has been reported. This resistance has been partially ascribed to a feedback loop that triggers Akt activation via p70S6K inhibition (33, 37, 44).
The great similarity between the mTOR and the class I PI3K catalytic domains has enabled the development of novel PI3K/mTOR kinase inhibitors that can suppress mTORC1 and mTORC2 or concurrently inhibit mTOR and PI3K kinase activities, thereby attenuating Akt activation as observed in phase I clinical trials in patients with various types of cancers (39). Moreover, initial evidence from clinical data suggests that mTOR inhibitors may improve therapeutic benefit for psoriasis (17), thus there is an urgent need to develop novel mTOR-based targets and mechanism-based strategies to improve treatment outcomes (3, 13, 23). We and others have shown aberrant activation of the PI3K/Akt/mTOR components in inflamed skin lesions of both human psoriasis and a Toll-like receptor-7/8 ligand imiquimod (IMQ)-induced murine psoriasis-like skin model compared with healthy skin (7, 22).
We reported that delphinidin [3, 5, 7, 3′-, 4′-, 5′-hexahydroxyflavylium] (referred to herein as Del), a potent antioxidant found abundantly in pigmented fruits and vegetables, has proapoptotic, antiproliferative, anti-inflammatory, and prodifferentiation effects (6, 9, 34). Although Del induces mutifactorial effects, precise understanding of its targets in the biological system remains unknown, necessitating the exploration of its molecular mechanisms and targets, as well as its usefulness for treating psoriasis.
In this study, we report the identification of Del as a novel specific inhibitor of both lipid (PI3Ks) and serine/threonine (mTOR/p70S6K) kinases. This interaction counteracts the S6K-1/IRS-1 feedback loop in the hyperproliferative psoriasis-like mouse model. Combined kinome-level screen, binding constant (Kd), surface plasmon resonance (SPR), and in silico molecular docking analyses revealed strong interaction affinity between Del and PI3Ks (α, 2Cβ, and γ), mTOR, and p70S6K, but not Akt. Additionally, Del inhibits activation of these kinases in vitro in cultured normal human epidermal keratinocytes (NHEKs). Furthermore, with an in vivo mouse model, we provide evidence that topical application of Del significantly alleviates IMQ-induced psoriasis-like skin lesions in Balb/c mice.
Results
Del binds to several lipid and serine/threonine kinases
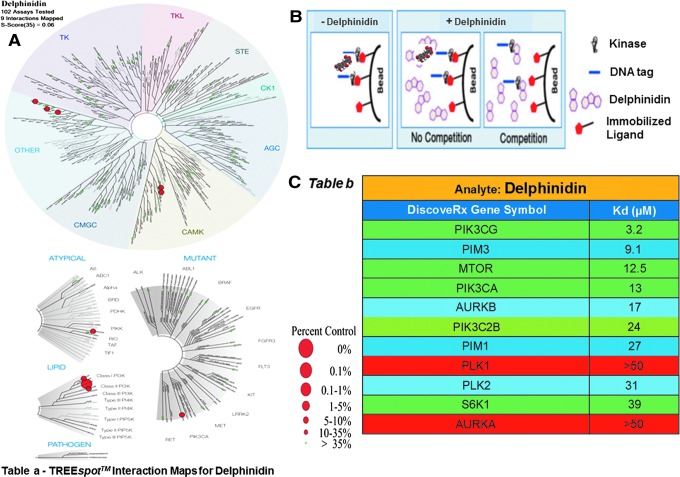
We measured the interaction between Del (50 μM) screened against a panel of 102 human kinase targets, using an active site-directed quantitative competitive ligand binding assay platform (KINOMEscan, DiscoveRx, CA) (24–26). Del was found to bind to several lipid (PIK3CG, PIK3C2B, and PIK3CA) and serine/threonine (PIM1, PIM3, AURKA, AURKB, mTOR, S6K1, PLK1, and PLK2) kinases (Fig. 1A–TREE spot image). Subsequent quantitative Kd analyses confirmed these interactions for all but two kinases (AURKA and PLK1) (Fig. 1B). Del displayed highest affinity for PIK3CG (3.2 μM) and PIM3 (9.1 μM) and modest affinity (10–40 μM) for other kinases (Fig. 1C- Table and Supplementary Fig. S1A, B; Supplementary Data are available online at www.liebertpub.com/ars).
FIG. 1.
Small molecule–kinase interaction maps and binding constants for Del. (A) Dendrograms of the human kinase family showing kinase interactions with Del and selectivity against different members of the kinase family. The interaction map is produced from TREEspot™ profile visualization software (www.kinomescan.com) with permission from DiscoveryRx (http://treespot.discoverx.com). The target kinases identified to bind in the primary screen are indicated by red spheres. The radii of the spheres correspond to Del inhibitory potency (Kd) at 50 μM inhibitor (Del) concentration, where larger circles indicate higher affinity binding. The green spheres represent kinases not found to bind (Complete results of all 104 kinase screens against Del showing different binding modes of different kinase families can be found in Supplementary Fig. S1A). (B) Schematics of the KINOMEscan™ model, which is based on a competition binding assay that quantitatively measures the ability of a compound to compete with an immobilized active site-directed ligand. The assay is performed by combining three components: DNA-tagged kinase; immobilized ligand; and a test compound (Del). The ability of the test compound to compete with the immobilized ligand is measured via quantitative PCR of the DNA tag. (C) Table, showing/depicting matrix of binding constants (Kds) of a single screen at 50 μM Del concentration, shows 9 of the 11 identified Kds ≥1 μM (high to modest affinity) and only 2 without binding as indicated for the different kinases identified from primary screen. (Del's detailed analysis curve fitting data can be found in Supplementary Fig. S1B). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Del physically interacts and binds with the PI3Ks, mTOR, and p70S6K kinases
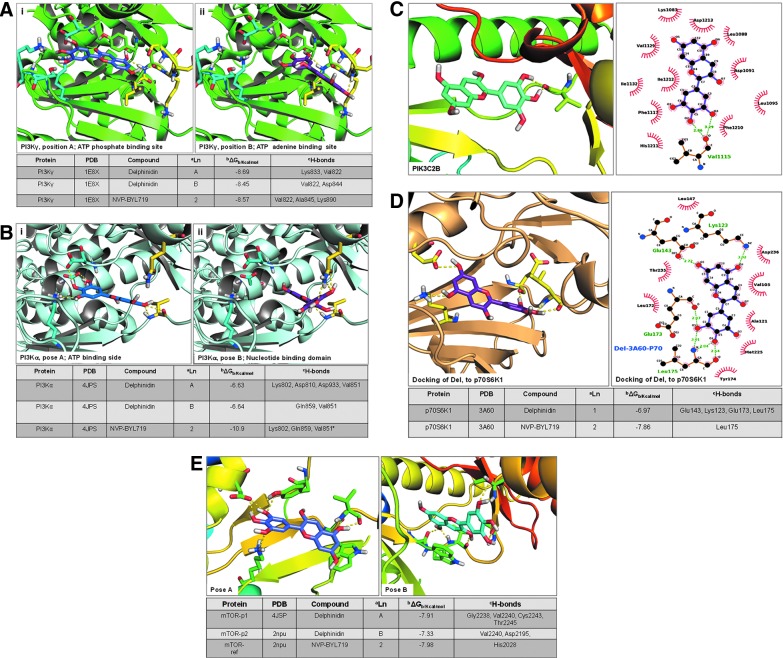
We conducted intensive in silico molecular docking analysis to determine the actual interaction of Del with individual PI3K isoforms (α, 2cβ, and γ), mTOR, and p70S6K kinases, including Akt. Autodoc4 analysis predicted physical interaction and revealed strong binding of Del with PI3Ks (PIK3CA, PIK3C2B, and PIK3CG, with binding energy of −6.64, −7.82, and −8.69 Kcal/mol, respectively) (Fig. 2). For PI3K-γ (gamma) kinase (PDB code 1E8X), Del clusters in two preferred sites, the ATP phosphate binding site A and the ATP adenine binding site B (Fig. 2A). In position A, the hydroxyls of the phenyl ring chelate to Lys 833 in the lysine/aspartate-rich phosphate binding site, with the remaining hydroxyls chelated near the nucleotide binding region, and the hydrogen bonds are made with the Val 822 backbone amide and carboxyl (Fig. 2Ai). In position B, one of the three phenyl-OH chelates with a backbone amide of Val 882 and a terminal hydroxyl chelates with the backbone amide of Asp 844 in the nucleotide loop (Fig. 2Aii). The largest docking cluster was found to be in site A, with an average binding energy of −8.69 Kcal/mol, and site B has an average cluster of −8.45 Kcal/mol.
FIG. 2.
Predicted docking mode of Del with PI3K/Akt and mTOR pathway components identified in Kinome-level primary screens. Predictive Autodock4 docking computational model view of the (A, i,ii) PI3K-gamma kinase (PDB code 1E8X)-Del complex. Del docking clusters in two preferred sites, the ATP phosphate binding site A (blue ligand, cyan side chains) or the ATP adenine binding site B (purple ligand, yellow side chains). For Del in A (blue carbons), the hydroxyls of the phenyl ring chelate to Lys 833 in the lysine/aspartate-rich phosphate binding site, with the remaining hydroxyls chelated near the nucleotide binding region (yellow residues). Hydrogen bonds are made with the Val 822 backbone amide and carboxyl. For Del in B (purple carbons), one of the three phenyl OH chelates with a backbone amide of Val 882 and a terminal hydroxyl chelating with the backbone amide of Asp 844 in the nucleotide loop (yellow residues). Hydrogen bonding is given as yellow lines. The largest docking cluster is in site A, with an average binding energy of −8.69 Kcal/mol. The site B has a clustering average of −8.45 Kcal/mol. (B, i, ii) PI3K alpha kinase (PDB code 4JPS)-Del complex. The dockings cluster in two sites, as they do in the PI3K gamma. Site A, is in the ATP phosphate binding site (blue ligand, cyan side chains), and site B prefers the nucleotide binding domain (purple ligand, yellow side chains). In site A, the three hydroxyls hydrogen bond to the ligands that bind phosphate, Lys 802, Asp 810, and Asp 933. The nucleotide end in site A forms H bonds with Val 851 backbone atoms. For site B (purple ligand, yellow side chains), the ligand fits in a reverse direction, with the three hydroxyls hydrogen bonding with Gln 859 side chain and the backbone of Val 851. Hydrogen bonding is given as yellow lines. The free energy of binding for the two clusters A and B is −6.63 Kcal/mol and −6.64 Kcal/mol. (C) PI3KC2B (PI3KC2 beta kinase) target homology fitted from PI3K-delta (PDB code 4XE0) crystal structure (40) using the Swiss-Model Repository database found a binding site very close in overall fit to the PI3K-gamma binding site, but contains several different amino acids. Autodock4 protein-ligand docking of PI3KC2-beta-Del complex showed that Del binds and clusters at a much lower energy of −7.82 Kcal/mol compared with the PI3K-gamma target. This is a big difference and agrees with the chemistry, which shows lower binding. The pymol and ligplot figures show one backbone carbonyl involved in the docking Val 1115. (Complete results of Autodock4 docking of Del docked in the AKT molecule can be found in Supplementary Fig. S2A, B). (D) P70 kinase (PDB code 3A60)-Del complex. Del (cyan) docks at only one cluster of docking poses with a mean binding energy of −6.97 Kcal/mol. At p70 kinase kinase site, the three hydroxyls of Del bind to the backbone in the nucleotide binding site, while the other end of Del uses two hydroxyls to bridge the Glu 123 and the Lys 123 in the phosphate end of the binding pocket. The neighborhood of the ligand is given in the ligand plot to the left. (E) Docking of mTOR (PDB code 2NPU)-Del complex and of mTOR-4JSP (PDB code 4JSP)-Del complexes. Del docks in two preferred binding positions of nearly equal energy clusters, A at −7.91 Kcal/mol and B at −7.33 Kcal/mol. Using Pymol PyMOL Molecular Graphics System, Version 1.7.6 (Schrodinger, LLC), the cartoon format displays both docked Site A (blue ligand), the best binding docks in the preferred phosphate binding site using the Lys-Asp pair in the active site that chelates the phosphate in ATP, and Site B (green ligand), which excludes these residues and binds in the backbone nucleotide loop. The reference NVP molecule docks at a calculated energy of −7.98 Kcal/mol. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Del docking clusters in two sites (A and B) in the PI3K-α (alpha) kinase (PDB code 4JPS) as it does in the PI3K-γ (Fig. 2B). In site A (ATP phosphate binding site), the three hydroxyls hydrogen bond to the ligands that bind phosphate, Lys 802, Asp 810, and Asp 933 and form H-bonds to Val 851 backbone atoms at the nucleotide end (Fig. 2Bi, and Table). In site B (nucleotide binding domain), the ligand fits in a reverse direction, with the three hydroxyls hydrogen bonding to Gln 859 side chain and the backbone of Val 851(Fig. 2Bii, and Table). The free binding energy of clusters A and B is −6.63 Kcal/mol and −6.64 Kcal/mol, respectively, and agrees with the chemistry being much lower than in the gamma and NVP-ligand (−10.9 Kcal/mol) docking.
For the design of PI3K-C2β(beta) target, the three-dimensional structure of PI3K-C2β was homology fitted to the crystal structure of PI3K-δ(delta) (PDB code 4XE0) (40) using the Swiss-Model repository web site (http://swissmodel.expasy.org). A 32.09% sequence similarity over approximately 1500 amino acids was found as the best sequence fit in the Swiss-Model database. The binding site in the target is very close in overall fit to the PI3K-γ binding site, but contains several different amino acids. Despite the targets' overall similarity, Autodock4 protein-ligand (PI3K-C2β-Del) docking predicted that Del only binds at an energy of −7.82 Kcal/mol and forms a cluster at this energy, which is much lower than the PI3K-γ target (Fig. 2C), whereas the reference NVP ligand binds at energy of −9.34 Kcal/mol.
This is a big difference and agrees with the chemistry, which shows lower binding. If fact, PI3K-C2β presents only one backbone carbonyl involved in binding Val 1115, as opposed to the multiple hydrogen bonds in the reference molecule (data not shown). Because Del was not found to bind Akt, a downstream target of PI3K, we further utilized two different crystal structures (PDB codes 1UNQ and 3D0E) (20, 29) to investigate possible interactions missed by autodocking. Both 1 UNQ and 3D0E dockings confirmed less affinity binding of Del with Akt and had the best calculated binding energy of −6.15 (1UNQ) and −7.73(3D0E) Kcal/mol, respectively, and thus concurring with the binding assay (For details, see Supplementary Fig. S2A).
In the initial screen, Del was also found to bind p70S6K, a downstream target of the PI3K/Akt/mTOR pathway responsible for protein translation and cell growth. To confirm this observation, Del was docked in the P70 kinase site (PDB code 3A60). Only one cluster of docking pose with a mean binding energy of −6.97 Kcal/mol was observed. At one end in the kinase site, the three hydroxyl groups of Del bind to the backbone in the nucleotide binding site, while the other end utilizes two hydroxyls to bridge Glu 143 and Lys 123 in the phosphate end of the Akt binding pocket (Fig. 2D).
To investigate possible Del-mTOR binding sites, we used the 4JSP crystal structure (PDB code 4JSP) (18). Del docked at two preferred binding positions of nearly equal energy, A at −7.91 Kcal/mol and B at −7.33 Kcal/mol, in each of the two clusters displayed (Fig. 2E). Site A shares the best binding, where Del docks in the preferred phosphate binding site using the Lys-Asp pair in the active site that chelates the phosphate in ATP. Site B excludes these residues and binds in the backbone nucleotide loop. Numerous hydrogen bonds are made to the mTOR molecule in these sites.
Del interferes with mTOR and physically interacts within the FRB binding site
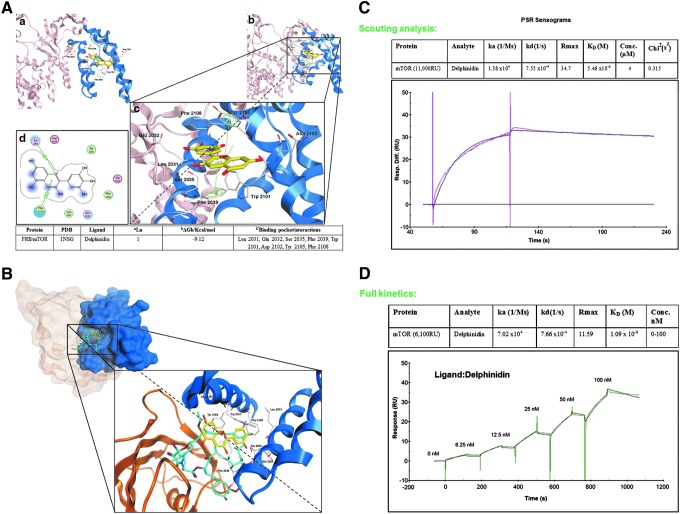
The mTOR complex is a central hub of the PI3K/Akt/mTOR signal transduction pathway and represents a major promising therapeutic target for clinical trials with druggable agents, such as rapamycin and rapalogs. To further characterize the putative interaction of Del with mTOR at the molecular level, we computationally examined its binding within the rapamycin binding site, FRB (residues 2019–2112) of the mTOR protein. Accordingly, docking experiments were performed using only the FRB from the three-dimensional structure of FKBP12-rapamycin-FRB ternary complex (at 2.2 Å resolution, PDB code: 1NSG) (25). As demonstrated from SPR studies (Fig. 3C), Del and rapamycin behave differently with respect to their binding affinity. These compounds are different in shape and Del does not need FKBP12 to interact with mTOR.
FIG. 3.
In silico molecular modeling of the interaction between Del in complex with mTOR in the FRKB12 domain. (A) Hypothetical model of FRB (blue) domain of mTOR protein in complex with Del (yellow). Predicted disposition of the best energy Del docking pose within the putative FRB binding pocket. (a–c) In silico docking studies were performed using MOE and 1NSG as the starting receptor docking site; Table at the bottom: summary of docking results for the top-ranked pose of Del bound to target protein mTOR. aLigand = top-ranked pose number; bΔGb = final score of free binding energy (ΔGbinding) for a representative conformer (energy values are expressed in kcal/mol); cBinding pocket/interactions: van der Waals, hydrophobic, cation–arene, and arene–arene stacking. Data were obtained from experiments performed twice with similar results. (d) Two-dimensional interaction map for Del onto the FRB mTOR protein binding site: amino acid residues (circles) are shown as follows: (a) hydrophobic residues (green interior), (b) polar residues (light purple), (c) basic residues (blue ring), and (d) acidic residues (red ring). Differences in solvent accessible surface area for Del ligand and for protein residues are plotted as a blue smud and a violet halo, respectively. The border of the binding pocket is marked by a dashed gray line. Dashed arrows denote tight interactions, with green bis-arene rings within a dashed line that denotes possible arene–arene stacking, while green arene-positive charge indicates possible cation–arene interaction. The interactions are summarized in the table. (The hypothetical model of Del (yellow) in complex with FRB (blue) domain of the full mTOR protein can be found in Supplementary Fig. S3). (B). Comparison between Del (yellow) and rapamycin (cyan) in complex with FK506-binding protein (FKBP12, light maroon) and the FKBP12-rapamycin binding (FRB) domain (blue) of FKBP12-rapamycin-associated protein. Graphical representation of the predicted disposition of Del and rapamycin docking pose within the FRB binding pocket. A model interaction site was obtained by superposing the Del and rapamycin within the binding site. This model was first built by manual docking, and energy minimization and visual inspection were performed by the MOE program. (C). Representative sensorgram showing initial scouting analysis between Del and mTOR. Del was flowed at 4 μM concentration of the analyte over the immobilized mTOR. The experiment was performed using a Biacore T-200 system. Chi-square (χ2) analysis was carried out between the actual sensorgram (magenta line) and the sensorgram generated from the BIAnalysis software (black line) to determine the accuracy of the analysis. A binding behavior was observed with KD = 5.46 × 10−8 M. χ2 value below 1 is highly significant (highly accurate). RU, response unit. (D). Sensorgram showing full kinetics between Del and mTOR. mTOR was directly immobilized in the sensor chip by amine coupling and Del was flowed over the protein-coated chip at different analyte concentrations (from highest to lowest, 100, 50, 25, 12.5, 6.25, 0 nM), with single-cycle kinetics. ka and kd or steady-state kinetics were used for determining the KD. Green line represents the best fitting of the association–dissociation processes with a 1:1 model. Data for affinity evaluation were obtained, that is, KD = 1.09 × 10−8 M, from a concentration-dependent binding curve for the interaction of increasing amounts of Del with constant concentration of mTOR. RU, response unit. Data shown are representative of three independent experiments. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Thus, we explored the binding between Del and mTOR, without FKBP12, assuming that Del binds within the FRB domain, which is inserted within the kinase N-lobe. Del docking suggested a nice fitting within the FRB protein (Fig. 3), and top-ranked energy scoring poses showed similar binding modes and tight affinity within the FRB amino acid pocket (Fig. 3A, a–c). Calculated free binding energy (ΔGb) for these conformations resulted around −9 kcal/mol (Fig. 3A and Table), indicating a ligand–receptor affinity better than the reported values for rapamycin (−6 kcal/mol) (2).
We further examined the hypothetical binding modes and interaction within the amino acid pocket that includes the following residues: Leu 2031, Glu 2032, Ser 2035, Phe 2039, Trp 2101, Asp 2102, Tyr 2105, and Phe 2108 (Fig. 3A, c–d, and Table). The most favorable conformation placed the oxonium ion of the chromenylium ring in close proximity to the Tyr 2135, involving strong cation–arene interaction (Fig. 3A, d). The pyrylium ring also established tight arene–arene stacking with the phenyl group of Phe 2039. Other amino acid residues involved in van der Waals and hydrophobic interactions with Del were located near the pocket. Particularly, the gallic moiety of Del was placed in close proximity to Phe 2108, Leu 2031, Glu 2032, and Ser 2035.
To display the difference in shape between Del and rapamycin and their disposition within the binding pocket, on the basis of the X-ray crystal structure of the FKBP12-rapamycin-FRB ternary complex used for docking experiments (1NSG), a model interaction site with a superposition of Del and rapamycin was built by manual docking. Energy minimization and visual inspection were performed by the MOE program (Fig. 3B). These insights suggest that with respect to rapamycin, Del would not be able to sterically block FRB and FKBP12 protein interactions, thus presumably acting by a different mechanism. It is noteworthy that Del interferes with the Ser2035, which is physiologically involved in regulation of phosphorylation of the key Thr389 of S6K1 protein, which in part could explain its mechanistic profile (Fig. 3B).
Finally, using SPR assay, we assessed the ability of Del to bind mTOR protein by real-time in vitro binding. Preliminary scouting analysis revealed a binding diagnostic behavior, and results indicated good affinity in nanomolar concentration range (KD = 5.46 × 10−8 M). Indeed, sensorgrams revealed slow association and dissociation processes (Fig. 3C). Maximum level of response unit (RU) increase was observed, that is, 11,000 RUs for scouting experiment. Full kinetics performed at different Del concentrations confirmed very strong affinity for mTOR in the nanomolar range (Fig. 3D) when fitting the curves with a 1:1 binding kinetic model provided, KD = 1.09 × 10−8 M.
Del treatment of NHEKs suppresses IL-22-induced proliferation and activation of PI3K/Akt and mTOR pathway components
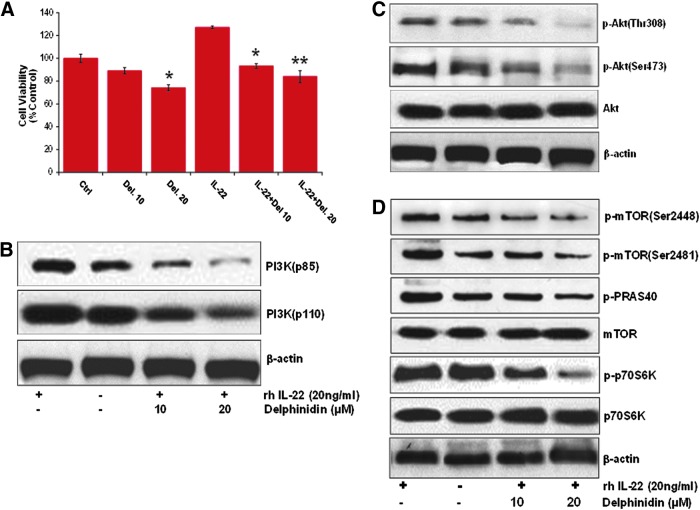
Since kinome-level screen, in silico molecular docking, and SPR studies revealed that Del physically interacts and strongly binds PI3K isoforms, mTOR, and p70S6K1 kinases, we investigated the biological significance of such interactions in cultured NHEKs. Using cell viability assay and Western blotting, we tested whether Del may provide growth inhibitory and chemotherapeutic effects against IL-22-induced proliferation and activation of PI3K/Akt/mTOR pathway targets (30). Cells were pretreated or not with Del (10 and 20 μM) and activated with or without rhIL-22 (20 ngml−1) for 48 h. Del treatment significantly inhibited IL-22-induced proliferation in a dose-dependent manner (Fig. 4A), resulting in 15% ± 2.4% and 26% ± 3.1% (p < 0.05) decrease of viable cells (Fig. 4A). Del (20 μM) also significantly decreased the number of viable cells in the absence of IL-22. Because keratinocyte hyperproliferation is a key ex vivo characteristic of psoriatic cell phenotype (27), these data suggest that Del can reduce the hyperproliferative and activated keratinocyte phenotype induced by IL-22.
FIG. 4.
Effect of Del treatment on IL-22-induced NHEK growth and expression and activation of PI3K/Akt and mTOR pathway targets in vitro. (A) Effect of Del on normal human epidermal keratinocyte (NHEK) growth and viability. NHEKs were pretreated with Del (10–20 μM) for 6 h before stimulation with rhIL-22 and further cultured for a total of 48 h with Del, and cell count of viable cells was determined by the MTT assay. Del treatment significantly decreased viability of IL-22-stimulated NHEKs. The data are expressed as the percentage of cell viability and represent the mean ± SEM of three experiments in which each treatment was performed in multiple wells. *denotes p < 0.05; **denotes p < 0.01. Effect of Del on (B) IL-22-induced increases in protein expression of PI3K (p85 and p110). (C) Effect of Del on phosphorylation of Akt. (D) Phosphorylation of mTOR and targets. The cells were pretreated with Del (10–20 μM; 6 h), followed by IL-22, for a total of 48 h and then harvested. Total cell lysates were prepared and 20 μg proteins was subjected to SDS-PAGE, followed by immunoblot analysis and chemiluminescence detection. Equal loading of protein was confirmed by stripping the immunoblot and reprobing it for β-actin. The immunoblots shown here are representative of three independent experiments with similar results. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To test whether Del exerts its activity by inhibiting PI3K/Akt/mTOR activity in NHEKs, we treated IL-22-stimulated NHEKs with/without 10 and 20 μM of Del and examined several key PI3K/Akt/mTOR signaling molecules known to be deregulated in human psoriasis (7, 22). Western blot results showed that IL-22 induced phosphorylation of PI3K (Fig. 4B), Akt at Ser473 (Fig. 4C), and mTOR at Ser2481 (Fig. 4D), which are all linked to mTOR activity. Del dose-dependently suppressed the IL-22-induced activation of PI3Ks (p110 and p85) and phosphorylation of Akt (both Ser473 and Thr308), mTOR (both Ser2448 and Ser2481), and downstream PRAS40 and p70S6K (Thr389) (Fig. 4B–D).
Del treatment inhibits TPA-induced NHEK secretion of proinflammatory cytokines and human neutrophil chemotactic activity in vitro in 2D cultures
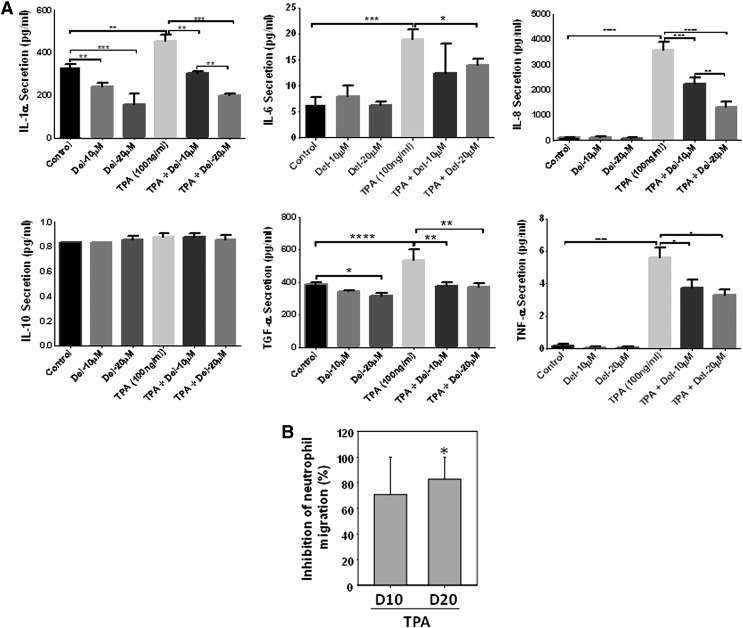
We next examined the effect of Del (10 and 20 μM) treatment on the production of TPA (100 ng/ml)-induced proinflammatory cytokines/chemokines, IL-1α, IL-6, IL-8, TNF-α, TGF-α, and IL-10, using a procarta-based multiplex immunoassay and further analyzed the neutrophil chemotactic activity of supernatants from control, stimulated, and Del-treated cells. As shown (Fig. 5A), Del treatment significantly and dose-dependently inhibited TPA-induced secretion of the proinflammatory cytokines, IL-1α, IL-6, IL-8, TNF-α, and TGF-α, a profibrotic mediator. However, both TPA and Del did not affect the production of the anti-inflammatory cytokine, IL-10 (Fig. 5A). Importantly, in agreement with the decrease of IL-8 production by Del-treated keratinocytes (Fig. 5A), the conditioned medium from TPA-activated and Del-treated keratinocyte cultures decreased neutrophil migration on fibronectin by ∼80% compared with migration induced by the supernatant medium from TPA-activated keratinocytes (Fig. 5B). This suggests that Del inhibits keratinocyte-produced chemoattractant activity for neutrophils.
FIG. 5.
Effect of Del treatment on TPA-induced NHEK secretion of proinflammatory cytokines and human neutrophil chemotactic activity in vitro. (A) NHEKs were cotreated with/without Del (10–20 μM) and/or TPA and cytokine production in supernatants was analyzed by Procarta 6-plex mix-matched immunoassay. Del (10 and 20 μM) dose-dependently inhibited TPA-induced NHEK secretion of the indicated cytokines. (B) Chemotactic activity for circulating human neutrophils using supernatants from TPA-stimulated NHEKs treated or not with Del (10 and 20 μM). Percentage inhibition of neutrophil migration using supernatant from Del-treated cells (10 and 20 μM) compared with TPA only is shown. (A, B) Data are mean ± SEM. *denotes p < 0.05; **denotes p < 0.01; and ***denotes p < 0.001, ****denotes p < 0.0001. For neutrophil chemotaxis in (B), graph is for three different keratinocyte-conditioned media and three different neutrophil donors. * indicates that migration is significantly inhibited by Del. For (A), results are representative of three independent experiments.
Topical Del treatment reduces IMQ-induced psoriasis-like skin inflammation in mice
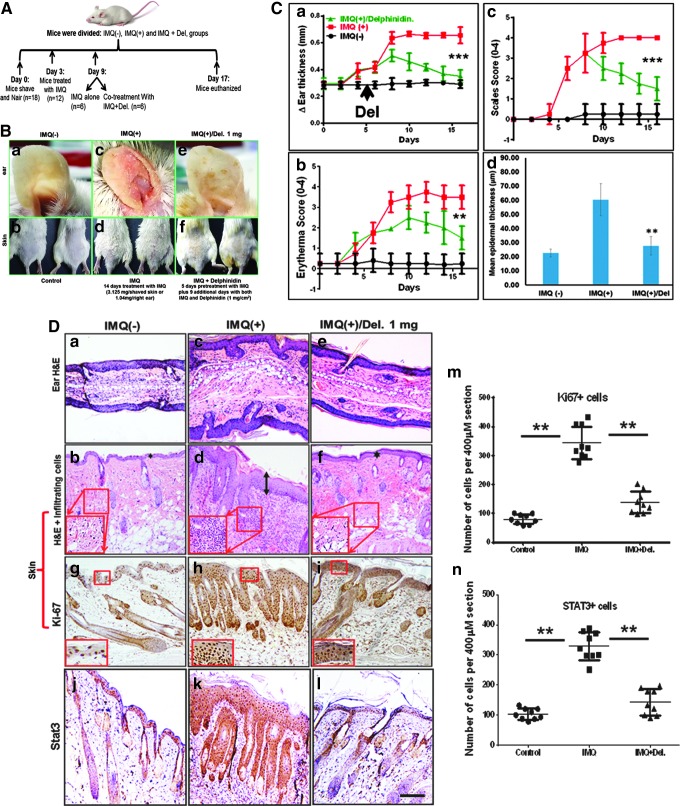
To establish antipsoriatic activity of Del in vivo, we designed a preclinical proof-of-concept study using an IMQ-induced Balb/c mouse model. This model displays many features characteristic of human psoriasis as previously described (45) and is schematically summarized in (Fig. 6A). At days 6–14 of the study, mice topically treated with 5% IMQ cream alone (n = 6) showed macroscopically prominent inflammatory, scaly, and thickened areas on the ears and trunk skin (Fig. 6B, c, d). In contrast, mice topically treated with control cream (Vaseline) (n = 6) did not show any skin abnormalities (Fig. 6B, a, b). We observed that Del treatment significantly suppressed psoriasis-like symptoms, including swelling, inflammation (erythema), and scaling compared with IMQ-treated mice (Fig. 6C, a–c).
FIG. 6.
Effect of Del treatment on pathological markers of IMQ-induced psoriasiform lesions in Balb/c mouse skin. Treatment with IMQ induces phenotypic skin changes resembling human psoriasis, and topical application of Del significantly alleviates these changes, including inhibition of epidermal hyperproliferation, decreases acanthosis, and reduces the severity of IMQ-induced psoriasis-like skin disease in Balb/c mice. Shown are macroscopic phenotypical presentation and histological analysis of inflamed mouse ear and back skin after 14 days of topical treatment with IMQ with or without cotreatment with Del. (A) Schematic representation of the treatment and experimental setup protocol before and after IMQ-induced lesion and/or Del treatment. Breifly, 6–8-week-old mice were topically treated for 14 consecutive days with 5% IMQ-containing cream (Aldara; 3.125 (skin−1) and 1.63 mg ear−1 day-1) or IMQ cream alone for 5 days, followed by cotreatment with IMQ cream and Del (1 mg cm−2 day−1) for further 9 days or control Vaseline cream. Fourteen days after the mice were sacrificed, samples were taken for further analyses. (c, d–m) Macroscopic views of ear (a, c, e) and back skin (b, d, f) from mice after 14 days. (B) Morphological analysis of inflamed ear and skin treated with IMQ alone (c, d), combination of IMQ and Del (e, f) compared with control (a, b). IMQ treatment induces inflammation and increased scales in ear and skin (c, d), Del treatment of inflamed IMQ-induced skin lesions markedly reduced both inflammation and scaling (e, f) to almost near normal phenotype observed in vehicle-treated control ear and skin (a, b). (C) Quantitative histological/severity assessment of changes in (a) ear thickness (measured in millimeters), (b) inflammation score, and (c) scales score throughout the 14-day experimental period, as well as (d) epidermal thickness after 14-day treatment in mice treated with vehicle control cream (Vaseline), compared with IMQ-alone (IMQ+) and IMQ and Del (IMQ+Del) therapy. Measure of epidermal thickness of the ear and back skin of mice shows that Del is effective in reducing IMQ-induced increases in epidermal thickness (d). For (a–c), each data point represents the mean of 6 random mice per group per time point measured. **denotes p < 0.01, ***denotes p < 0.001. (D) Hematoxylin and eosin (H&E) staining of mouse ear (a, c, e) and skin (b, d, f) of control (a, b), IMQ alone treated (c, d), and cotreated with IMQ and Del (e, f). (c, d) IMQ alone treated (middle panels) reflecting the hallmarks of psoriasis with abnormally thickened epidermis, parakeratosis (nucleated keratinocytes in the stratum corneum), thickened keratinized upper layer (hyperkeratosis (double-headed arrow), increased epidermal rete ridges, and increased immune cell influx (insets in H&E) compared with control (a, b). Data show that in addition to effectively reducing epidermal thickness, Del also reduces infiltration of immune cells in skin (e, f, and insets). Immunohistochemical staining for epidermal proliferation marker Ki67 (g–I, m) and Stat3 (j–l, n) (dark brown staining) expression shows increases in IMQ-treated section (h, k) and decrease of these markers after 9 days of consecutive treatment with Del. (m, n) Quantitative data are representative of the control group vs. IMQ-treated group vs. Del+IMQ-treated group. Symbols represent sections analyzed, and horizontal lines indicate averages of groups. **p ≤ 0.01. (magnification, × 200). IMQ, imiquimod. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Del treatment normalizes the epidermal architecture and promotes differentiation in inflamed psoriasis-like skin in mice
To further characterize the efficacy of topical Del treatment in reducing psoriasis-like disease, histological analyses were performed on ear and skin sections obtained from different treatment groups. Histopathological analysis revealed psoriasis-like phenotype in IMQ alone-treated mice, including acanthosis, epidermal rete ridge projections into the dermis, and microabscesses (Fig. 6D, a–f) and (Fig. 8b). In contrast, Del treatment led to a clear reduction in histological psoriasis-like features (Fig. 6D). More so, computer-assisted image analysis revealed that the average changes in ear thickness time course (p < 0.001; Fig. 6C, a, D, a, c, e) and epidermal thickness (p < 0.01; Fig. 6B, d, D, b, d, f) were significantly reduced after treatment with Del.
FIG. 8.
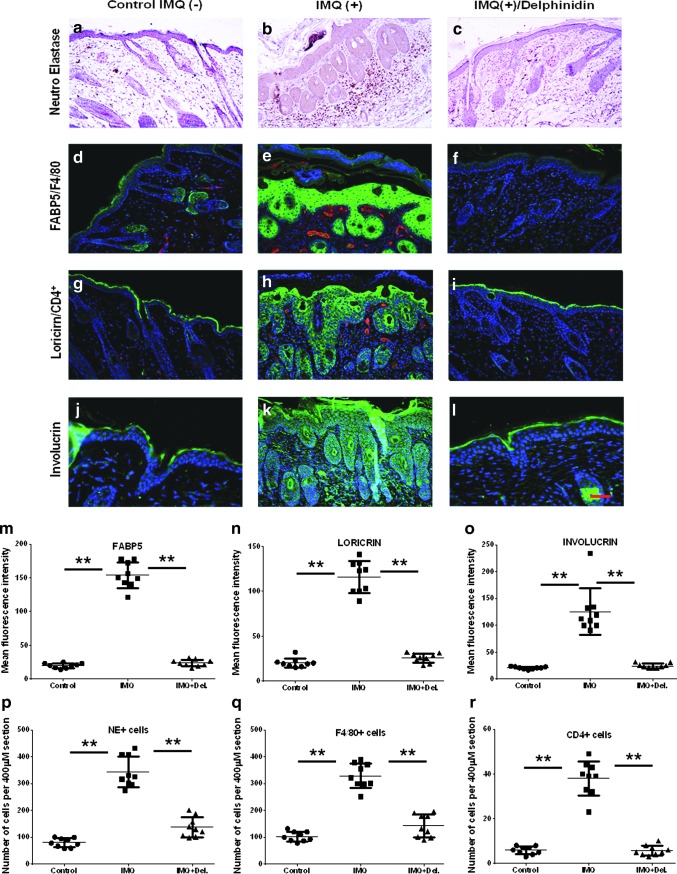
Effect of Del treatment on protein expression of infiltrating immune cells and psoriasis-related differentiation markers in IMQ-treated mouse skin. Immunolabeling and histological analysis of Balb/c mouse back skin sections after treatment with vehicle control cream (IMQ-, left panels), IMQ (IMQ+; middle panel), or IMQ and Del (IMQ+Del; right panel) using specific antibodies (magnification, × 200). (c) Immunolabeling of IMQ-induced mouse skin demonstrating granulocytic infiltrates into the skin and abnormal expression of differentiation markers. Immunohistochemical labeling for neutrophil elastase-positive (NE+) neutrophils in mouse skin shows an increase in infiltrates after IMQ treatment (b), which was significantly decreased to near normal control skin values (a) after Del treatment (c). IMQ application induces increases in epidermal microabscesses (b, e, h) as well as increases in the numbers of typical inflammatory cell infiltrates of (b) neutrophils in the epidermis and dermis and induction of microabscesses (brown esterase staining), (e) macrophages in the dermis (F4/80-staining, red), and (h) intraepidermal/dermal T cells (CD4 staining, red). Application of Del significantly reduced IMQ-induced skin infiltration of (f) macrophages in the dermis and (i) CD4+ T cells. Immunofluorescence staining of differentiation-related proteins in mouse skin sections after IMQ cream application (e, h, k) or IMQ and Del (f, i, l) coapplication. Staining of (e) FABP-5, (h) loricrin, and (k) involucrin in mice treated with IMQ alone shows a much abnormally broader staining pattern in several keratinocyte layers. Application of Del in combination with IMQ shows significantly increased and more focally expressed (f) FABP-5, (i) loricrin, and (l) involucrin, all restricted largely to the upper granular layer, as typically observed in normal epidermis (d, g, J), respectively. Nuclei counterstained blue with DAPI (4′,6-diamidino-2- phenylindole) (magnification, × 200). Quantitative immunostaining assessment of changes in expression of differentiation markers (m–o) and of infiltrating immune cell (p–r) markers in IMQ (e, h, k) and IMQ+Del (f, i, l) compared with control (d, g, j) mouse skin. Each data point represents the mean of seven random fields from four high-power magnification cryosections per mouse from six mice per group. Data are taken from at least two independent experiments. **denotes p < 0.01. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To further investigate the effect of Del treatment, we analyzed the expression of proliferation markers as well as the expression of Stat3, a known therapeutic target of psoriasis. Del treatment of IMQ-induced lesions significantly reduced the expression of both Ki-67(p < 0.01, Fig. 6D, g–i, m) and Stat3 (p < 0.01, Fig. 6D, j–l, n) to a staining pattern much more similar to that observed in uninflamed epidermis of control mouse skin (Fig. 6D, g, j, m, n). This was distinct from that of inflamed IMQ-treated mouse skin (Fig. 6D, h, k, m, n) and implied reduced proliferative activity of keratinocytes after Del treatment.
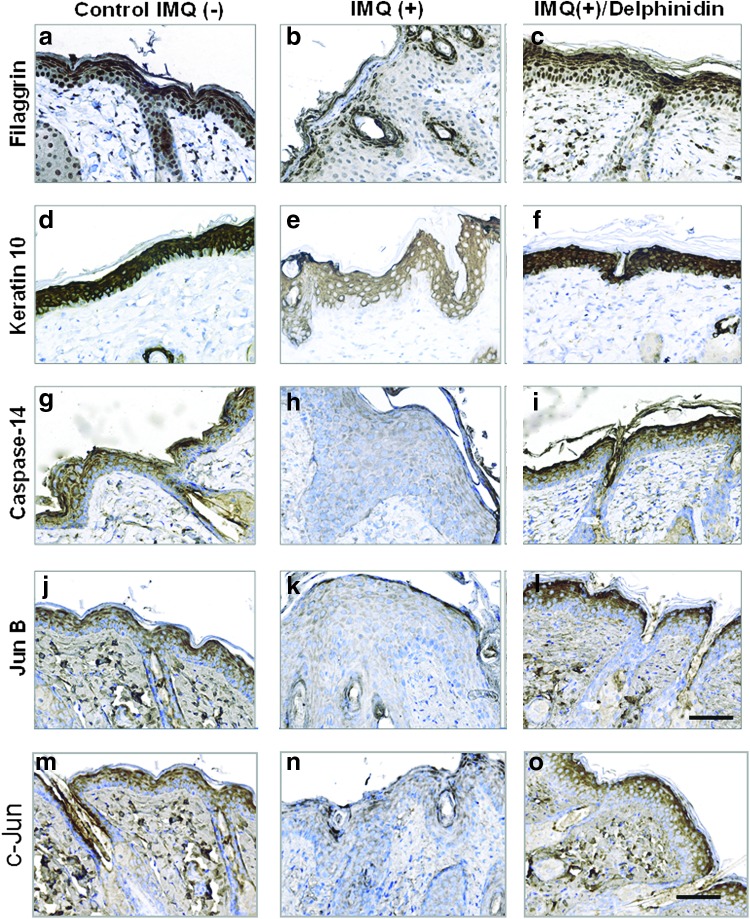
Additionally, epidermal cornification markers, keratin-10, loricrin, and involucrin, which are normally expressed in the suprabasal layers of epidermis (Fig. 7d) and (Fig. 8g, j [green staining]), displayed an abnormally broad staining pattern in several keratinocyte layers (Figs. 7e and 8h, k) of IMQ-induced skin lesions. By contrast, Del treatment of IMQ-induced skin lesions significantly normalized the expression of these markers, which are normally restricted largely to the upper granular layers (Figs. 7f and 8I, l, n, o).
FIG. 7.
Effect of Del treatment on protein expression of epidermal differentiation and AP-1 factor proteins in IMQ-treated mouse skin. Immunohistochemical analysis of differentiation and AP-1 factor protein expression in the epidermis of normal (a, d, g, j, m), IMQ-induced (b, e, h, k, n), and IMQ-induced and Del-treated (c, f, i, l) mouse skin. Normal epidermis shows a strong epidermal expression of differentiation proteins (a) filaggrin, (d) keratin-10, (g) caspase-14, and AP-1 factor proteins (j) JunB and (m) c-Jun (intense dark brown staining). IMQ application on the skin thickens the epidermis and decreases the expression showing only very weak and abnormally diffused staining patterns of filaggrin (b), keratin-10 (e), caspase-14 (h), JunB (k), and c-Jun (n) compared with vehicle-treated control epidermis (a, d, g, j, m). This IMQ-induced psoriasis-like skin lesion closely resembles clinical features observed in human psoriatic plaques. Thus, different layers of the epidermis show a strong reduction of protein expression per unit area in psoriasis-like skin (b, e, h, k, n). Del-treated IMQ-induced epidermis shows a thinner epidermis with an upregulated and normalized expression pattern of the differentiation-related protein markers, (c) filaggrin, (f) keratin-10, (i) caspase-14, (l) JunB, and (o) c-Jun. (magnification, × 400). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Importantly, as shown in Figure 7, IMQ treatment downregulated the expression of critically important differentiation-related proteins, including caspase-14, filaggrin, keratin10, and AP-1 factors (JunB and c-Jun), showing only weak and abnormally broad expression patterns (Fig. 7b, e, h, k, n) compared with control epidermis (Fig. 7a, d, g, j, m). By contrast, an upregulation of the expression of psoriasis-associated epidermal fatty acid-binding protein 5 (FABP5), a disease marker often overexpressed in lesioned psoriatic skin (7), was observed in all epidermal keratinocyte layers of IMQ-induced skin lesions (Fig. 8e, m).
Analysis of skin sections revealed that Del treatment of IMQ-induced skin lesion resulted in upregulation and normalization of caspase-14, filaggrin, keratin-10, JunB, and c-Jun (Fig. 7c, f, I, l, o) with expression confined to the differentiating layers, as typically observed in normal epidermis (Fig. 7a, d, g, j, m), but significantly higher than in IMQ alone-treated sections (Fig. 7b, e, h, k, n). Del treatment also normalized the protein expression of FABP5 to the upper granular layer similar to what is observed in normal epidermis (Fig. 8d, f, m) and less than the expression in IMQ alone-treated sections (Fig. 8e, m). Thus, treatment with Del normalized the epidermal skin architecture and enhanced differentiation in this IMQ-induced mouse model of psoriasis.
Del treatment inhibits inflammatory immune cell infiltration into inflamed psoriasis-like skin in mice
To characterize the immune cell infiltrates in the skin, we analyzed by quantitative immunostaining the expression markers of neutrophil, neutrophil elastase, (Fig. 8a–c), macrophage, F4/80 (Fig. 8d–f), and CD4+ T cells (Fig. 8g–i)(red staining) (Fig. 8p–r). The inflamed IMQ-treated mouse skin contained increased numbers of dermal and epidermal NE+-neutrophils and F4/80+ leukocytes (Fig. 8b, e). In contrast, quantitative analyses indicated that the overall number of NE+-neutrophilic granulocytes and F4/80+-macrophages was similar to control mice and in Del-treated inflamed skin (Fig. 8p, q) and was significantly reduced compared with IMQ-treated mice without Del (Fig. 8a, d, f, c, p, q). Similar to the pathophysiology seen in human psoriatic plaques; the number of dermal CD4+ T lymphocytes was increased in IMQ alone-treated mice (Fig. 8h) compared with control mice (Fig. 8g), and fewer and significantly reduced dermal and intraepidermal CD4+ T lymphocytes were detected in Del-treated inflamed skin sections (Fig. 8i, r). Thus, topical application of Del inhibits and reverses the inflammatory infiltrate typically associated with psoriasis.
Del treatment ameliorates IMQ-induced alteration of inflammatory immune molecules and PI3K/Akt/mTOR targets in mice
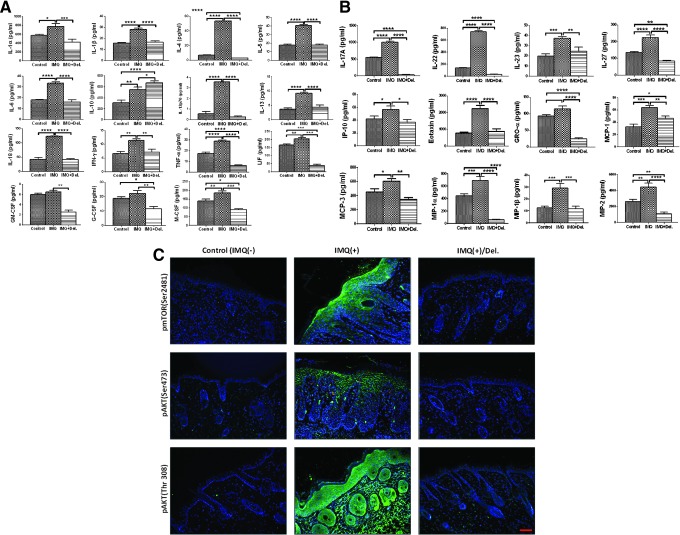
We examined the pathophysiological relevance of Del treatment in the inflammatory phenotype observed in IMQ-induced lesions, using a multiplex immunoassay. Quantitative analysis of protein expression of several immune molecules in skin lysates of IMQ-induced and Del-treated IMQ-induced lesions compared with normal control in Balb/c mouse skin revealed significant differences across the three groups in a large proportion of the 36 analytes examined. Post hoc t-tests revealed prominent increases in protein expression levels of 27 pro- and anti-inflammatory cytokines and chemokines in inflamed skin compared with normal control skin, including IL-1α, IL-1β, IL-4, IL-5, IL-6, IL-12p70, IL-13, IL-17A, IL-18, IL-22, IL-23, IL-27, IL-22, TNF-α, IFN-γ, GM-CSF, GRO-α, IP-10, MCP-1, MCP-3, MIP-1α, MIP-1β, MIP-2, Eotaxin, G-CSF, M-CSF, and LIF (Fig. 9A, B). However, no significant alterations in expression levels of IL-2, IL-3, IL-9, IL-15/IL-15R, IL-28, IL-31, IFN-α, RANTES, and ENA-78/LIX/CXCL5 were observed in this chronic phase model (Supplementary Fig. S4).
FIG. 9.
Del treatment regulates the expression levels of signature inflammatory mediators induced by topical IMQ treatment in Balb/c mice. Procarta multiplex immunoassay of mouse skin protein for cytokines and chemokines reported to be involved in aspects of psoriasis in vehicle-treated control (IMQ−; left), IMQ-treated (IMQ+; middle), and after treatment with Del and IMQ (IMQ+Del; right) of each bar graphs. Briefly, total skin lysates were isolated from the mouse back skin from each group and Procarta 36-multiplex immunoassay was performed to evaluate the expression levels of pro- and anti-inflammatory cytokines and chemokines. Comparison of levels of these immune analytes in controls IMQ-, IMQ+, and IMQ+Del groups: (A) Cytokines that have proinflammatory or anti-inflammatory, Th1, Th2, and activators of granulocytes and/or monocyte/macrophage potential that is elevated in IMQ and downregulated in IMQ+Del-treated (with exception of IL-10, which continually increased) vs. vehicle-treated controls. (B) Th17/Th22/Treg cytokines, type 1 and type 2, and granulocytes and/or monocyte/macrophage chemokines that are elevated in IMQ+ and are decreased in IMQ+Del-treated vs. vehicle-treated controls. (C) Immunostaining showed that Del downregulated the phosphorylation of mTOR and targets (green), blue represents nuclear staining by DAPI. Data show mean ± SEM for each cytokine and chemokine. Levels of immune molecules were largely increased in IMQ alone-treated mice and decreased in the Del+IMQ-treated mice compared with those of vehicle-treated controls, with the exception of IL10 (p < 0.0001), which was increased. Statistical significance is displayed for all the samples and only cytokines or chemokines meeting significance criteria (p < 0.05) post hoc analyses are represented (control group vs. IMQ-treated group or Del- treated and IMQ-treated groups). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 indicate p-values from two-sample t-test comparisons. (Complete results of inflammatory mediators not altered by Del or IMQ treatments can be found in Supplementary Fig. S4). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Interestingly, the protein expression was significantly reduced in Del-treated inflamed lesions (Fig. 9A, B), except for the anti-inflammatory cytokine IL-10 expression, which continued to increase, indicating an overall reduced inflammation upon Del treatment. Additionally, MIP-1α and MIP-1β (Fig. 9B), chemokines providing a strong T-cell chemotactic effect, and MIP-2 (Fig. 9B), a strong chemoattractant for neutrophils and T cells produced mainly by keratinocytes, were found to be highly expressed in IMQ alone-treated, but were significantly reduced in Del-treated, skin lesions (Fig. 9B), in agreement with Del's anti-inflammatory effect.
Furthermore, quantitative immunostaining analysis and Western blot evaluation of the effect of Del on PI3Ks and mTOR protein targets after 14 days showed a marked decrease in the expression of phospho-S6 and the phospho-Akt (Ser473) after Del treatment (Fig. 9C). These observations suggest that IMQ-induced psoriasis-like skin lesion development was suppressed by Del through inhibition of the PI3K/Akt/mTOR pathway and multiple psoriasiform disease markers.
Discussion
The present study employs pathway target discovery and therapeutic approaches to characterize and identify kinases as novel molecular targets of the antioxidant Del in the treatment of psoriasis. Kinase inhibitors are a class of therapeutic drugs that inhibit multiple targets, thus valuation of their biological consequences is important vis-à-vis their observed mode of interaction with human kinome, therapeutic efficacy, and safety (14, 15, 46). Using a quantitative kinome-level screen and thermodynamic Kds, we first identified and confirmed that of the 11 putative kinases Del binds to, 9 candidate targets belong to the lipid and serine threonine kinases. Interestingly, 5 of the 9 targets belonged to the PI3K/Akt/mTOR pathway.
To computationally model the interaction of Del, we employed both the autodock4 and MOE in silico molecular modeling methods to predict interactions and affinities for the identified PI3K/Akt/mTOR targets. Del favorably interacted with all investigated targets except Akt. Because in vitro cell-free binding or enzymatic activities do not exactly mirror activities observed in cells, knowledge of kinase inhibitor interaction patterns could aid interpretation of the observed preclinical and clinical activity.
To understand the impact of Del binding interactions in a biologically relevant context, we followed-up these studies using recombinant proteins in cell culture assays and in vivo disease models. Docking and SPR data revealed that Del strongly interacts and binds mTOR within the 1360–2549 amino acid residues, FRB domain (FKBP12-rapamycin binding domain (FRB), which is often inhibited by allosteric inhibitors, including rapamycin. Interestingly, the SPR affinity data of recombinant mTOR protein appeared about three orders of magnitude lower (i.e., in terms of absolute value) than those derived from experimental data obtained from TREESpot™. Therefore, we hypothesize that Del behaves as a specific and stable binder with mTOR, but differs mechanistically from rapamycin.
In fact, Banaszynski et al. (2) reported that rapamycin binds with moderate affinity to the 100 amino acids (Glu2015 to Gln2114) in the mTOR FRB domain with an SPR rapamycin-FRB interaction equilibrium binding, KD = 26 ± 0.8 μM, compared with FKBP12-rapamycin complex, which binds 2000-fold more tightly (KD = 12 ± 0.8 nM) to FRB than rapamycin alone. Therefore, while rapamycin essentially acts by simultaneously modulating interaction at the FKBP-FRB interface, playing a critical role in stabilizing the ternary complex, our results indicate that Del effectively binds to mTOR protein directly (KD = 10.9 nM) without the need of any counterpart. Furthermore, mTOR ATP binding affinity demonstrates that Del has relatively similar mTOR-regulating potential as does NVP-BYL719, the potent referenced PI3K inhibitor, and therefore can be a prospective compound for such studies.
In NHEK cultures, Del inhibited IL-22-induced expression of PI3Ks and phosphorylation of mTOR and effector molecules, including the substrates of mTORC1; p70S6K (Thr389) and mTORC2; and Akt (Ser473 and Thr308), respectively, resulting in growth inhibition. Treatment with IMQ produces psoriasis-like dermatitis in both humans and mice (19, 45) with lesions showing several parallels, including marked acanthosis and hyperkeratosis with focal parakeratosis, elongated rete ridges, dermal capillary dilation, and marked diffuse infiltration of mixed inflammatory cells, chiefly lymphocytes (45), making this an excellent preclinical model of psoriasis. Topical application of Del to IMQ-induced skin lesions normalized the epidermal architecture, reduced inflammation, and activated PI3K/Akt/mTOR signaling and strongly improved psoriatic clinical phenotype. Moreover, application of Del reduced immune cell infiltrates and the expression of immune mediators, including proinflammatory (Th1, Th2, Th17), activators of mononuclear cells, cytokines and chemokines known to be critical in human psoriasis pathophysiology (27). Therefore, Del treatment blocked epidermal hyperplasia, decreased immune cell infiltration, and led to normalization of the IL-23/T-helper-17 (Th17) and Th1/Th2 axis and consequently to an amelioration of the psoriatic phenotype.
We previously established prodifferentiation/anti-inflammatory effects of Del (6, 9, 34) and with others reported overexpression and clinical implication of PI3K/Akt/mTOR signaling in human psoriasis and murine psoriasis-like disease (7, 22). The epidermal growth factor receptor is an upstream activator of PI3K, which has been observed to be overexpressed in psoriasis, and which upon ligand binding generates phosphatidylinositol (3–5) P3, which activates Akt (8).
Akt activation correlates with disease progression in hyperproliferative diseases, such as cancer, human psoriasis, and in murine psoriasis-like disease (7, 22, 42). In this study, we demonstrate that Del treatment in vitro and in vivo in a preclinical IMQ-induced disease model inhibits the overexpression of PI3Ks and phosphorylation of mTORC1/mTORC2 and their targets, p70S6K and Akt (both at Thr308 and Ser473), respectively, including PRAS40, resulting in significant amelioration of psoriasis-like disease.
Intriguingly, Del's inhibition of Akt (Ser473) activation presents an advantage over the FDA-approved chemotherapeutic agent rapamycin and its derivatives (rapalogs) since Akt is the only known direct target of mTORC2, which is rapamycin insensitive, and its phosphorylation at Ser473 by mTORC2 ensures full activation (39) (Fig. 10). Moreover, the two mTOR complexes are linked through Akt since mTORC2-activated Akt activates mTORC1 through phosphorylation of PRAS40 and TSC2. However, allosteric inhibitors (rapalogs) specific for mTORC1 activate Akt (Ser473) in cancer due to abrogation of a negative feedback loop dependent on IGF-IR/insulin receptor substrate 1 (IRS-1) and involving the S6K-mediated suppression of upstream signaling (4, 12, 28). Thus, Del inhibition of phospho-Akt (Ser473) may result from the inhibition of mTORC2 and PI3K and/or mTORC1/2 and PI3K. In corollary with our findings, earlier studies have indicated that blockade of PI3Kδ and/or PI3Kγ is a possible treatment option for psoriasis and other IL-17-driven diseases (35). Furthermore, we also observed that Del binds other cell cycle and growth regulatory kinases, including PIM1, PIM3, AURKB, and PLK2, which are deregulated in several hyperproliferative diseases. To the best of our knowledge, the status of these kinases in psoriasis remains unexplored, suggesting a need for further investigation of their involvement in psoriasis. Our data provide a comprehensive description of the molecular mechanism and PI3K/Akt/mTOR pathway targeted by a small-molecule Del for the treatment of psoriasis.
Taken together, our study identified Del as a novel and potent dual PI3K/mTOR inhibitor, which possesses antipsoriatic activity in vitro in cell-free and cell culture, as well as in vivo in preclinical IMQ-induced psoriasis-like disease in Balb/c mice. Our findings strongly suggest that therapeutic intervention with Del as a PI3K/mTOR kinase inhibitor could provide substantial insights to improve the discovery and development of Del and other kinase inhibitors for psoriasis treatment. In addition, our observations also underscore the importance of testing Del in combination with known and conventional chemotherapeutic drugs.
Materials and Methods
Reagents
Del (Del), [3,5,7,3′-,4′-,5′-hexahydroxyflavylium], 12-O-tetradecanoyl-phorbol-13-acetate (TPA), and 3- (4,5-dimethylthiazol-2yl)-2,5- diphenyltetrazolium bromide (MTT) were from (Sigma–Aldrich). All stock solutions were diluted in DMSO and stored at −20°C; recombinant human IL-22 (rhIL-22) (R&D System), FRAP1 (mTOR), Recombinant Human Protein with GST-tag at N-terminal (1360–2549 aa, 163.9 kDa (Life Technologies), and plasma fibronectin (Calbiochem) were used. Details of antibodies used are in Supplementary Table S1: Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit secondary antibodies rabbit and Prolonged® Gold Anti-fade reagent containing 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). The BCA Protein Assay Kit (Pierce) and Novex precast Tris-glycine gels (Invitrogen or Bio-Rad) were used.
Competitive kinase binding assays
Del was screened against a panel of 102 kinase targets (Supplementary Table S2) using the KINOMEscan (DiscoveRx Corporation) quantitative ligand binding platform. KINOMEscan, an active site-directed competitive binding assay that quantitatively measures the interaction between a test compound and a kinase target, and the experiment does not require ATP, was performed as previously described (14, 15, 46), and furthermore assay conditions were optimized to ensure measurement of true thermodynamic Kds. In brief, DNA-tagged kinases were expressed using T7 phage display or using transiently transfected HEK-293 cells. Binding reactions were assembled by combining DNA-tagged kinase, streptavidin-coated magnetic beads loaded with kinase capture ligand, and test compound (50 μM Del) in 1 × binding buffer (16% SeaBlock, 0.32 × PBS, 0.02%BSA, 0.04% Tween 20, 0.004% sodium azide, 7.9 mM DTT). All reactions were performed in polypropylene 384-well plates in a final volume of 0.02 ml. The assay plates were incubated at room temperature with shaking for 1 h. After 1-h incubation, the affinity beads were subsequently washed (1 × PBS, 0.05% Tween 20) and eluted as earlier described (15, 43). The kinase concentration in the eluates was quantitated by qPCR.
Subsequent Kd analysis was performed for all interactions for which Del displayed evidence of binding (less than 35% of control) from the 50 μM primary screen. Kds were determined using 11-point threefold serial dilutions of Del starting at 50 μM and a DMSO control. Binding constants (Kds) were calculated with a standard dose–response curve using the Hill equation:
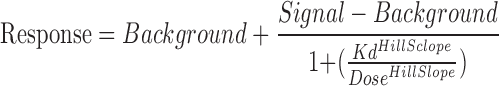 |
The hill slope was held to −1. Curves were fitted using a nonlinear least square fit with the Levenberg–Marquardt algorithm.
In silico computational molecular modeling study
Ligand docking studies were performed using two docking systems, the Autodock4 (Scripps Institute) and Molecular Operating Environment (MOE 2009.10; Chemical Computing Group, Inc.) software. Protein-ligand blind docking of Del to the targets was initially performed using Autodock4. The receptor site was prepared and Del was modeled with Sybyl (Certara Co., www.tripos.com) using the crystal structures of the targets, and some of the data obtained were also redocked using MOE as described below.
In brief, the docking model used was based on crystal structures with PDB entries as follows: PI3K-α (PDB code 4JPS) (18), PI3K-γ (PDB code 1E8X) (38), PI3K-C2β adapted from PI3K-δ (PDB code 4XE0) (40), Akt (PDB codes 1UNQ and 3D0E) (20, 29), p70S6K (PDB code 3A60) (41), and mTOR (PDB codes 2NPU and 4JSP) (18, 47), respectively, from the Protein Data Bank (www.pdb.org). Each target was individually docked and calculation was performed by setting docking grid sizes large enough to include the entire target molecule's crystal structure as well as the binding sites and Del. The results are displayed using Pymol (Schrodinger). For docking studies between Del and FRB domain of mTOR, the MOE 2009.10 software was utilized. The FRB domain (residues 2019–2112) of the mTOR protein of the three-dimensional structure of FKBP12-rapamycin-FRB ternary complex (at 2.2 Å resolution) used for docking experiments was downloaded from the PDB Data Bank (www.rcsb.org - PDB code: 1NSG) (25). Only for representation purpose, the truncated ∼1500 aa mTOR protein bound to full-length human mLST8 has been obtained by implementing the FRB portion from 1NSG X-ray crystal with the solved mTOR structure retrieved from 4JSV (47). Before docking, the protein structure was properly protonated using the Protonate 3D option and was geometrically optimized and minimized by employing MMFF94x force field with a generalized Born solvation model and RMSD gradient, 0.05 kcal mol-1 Å-1. The binding site was broadly defined based on residue proximity to the binding conformation of the ligand within the respective target protein. Rigid receptor-flexible ligand docking calculation was performed using the docking simulation feature MOE-dock by setting grid sizes that included FRB. The triangle matcher was used as placement method to generate docking poses, and the London ΔGbinding scoring function that estimates the free energy of binding (in kilocalories per mole, kcal/mol) was used to rank hit docking pose candidates. Only the best scored poses generated in the docking experiments were retained and examined with MOE.
SPR binding assays
Binding experiments were performed using a Biacore T-200 instrument (Biacore) at 25°C. The FRAP1 (mTOR) Recombinant Human Protein with GST-tag at N-terminal (1360–2549 aa, 163.9 kDa) was used, and 11000 and 6100 RUs (response units, for scouting and full kinetics, respectively) were directly immobilized on the N-ethyl-N-(3- dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide-activated flow cell2 and cell4 of the CM5 chip (sensor surface; GE certified) in water, respectively. The flow rate used for capturing the ligand is 5 μl/min. The unoccupied sites were blocked with 1 M ethanolamine. The analyte Del (Sigma-Aldrich) was then injected in solution that flowed over the chip surface at single analyte concentration. Binding of analyte to the immobilized protein was monitored in real time to obtain on (ka) and off (kd) rates. The equilibrium constant (KD) was calculated from the observed ka and kd or by steady-state kinetics, as for the small molecules. Stocks were prepared in 100% DMSO, and further dilutions were made in assay buffer containing 10 mM HEPES buffer (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.05% P20 (polyoxyethylenesorbitan), and 5% DMSO. First, scouting analysis was conducted to acquire preliminary estimates of the affinity between ligand and protein. Initial scouting analysis was performed at 4 μM of the analyte (Del) to determine yes/no binding. Full kinetics analysis was performed using Del that flowed over the surface at concentrations from 100 nM to 0 (run serial dilutions 100, 50, 25, 12.5, 6.25, 0 nM), with single cycle kinetics without regeneration, and a flow rate of 30 μl/min.
Keratinocyte isolation, culture conditions, treatment with TPA and IL-22, and cytokine detection
NHEKs were isolated from neonatal foreskin and adult skin biopsies, and primary cultures were established as previously described (9). Cells were cultured in CELLnTEC progenitor cell culture medium (ZenBio) supplemented with penicillin (100 U/ml), 100 μg/ml streptomycin (100 μg/ml), and amphotericin (100 μg/ml) (Life Technologies). The cells were maintained under standard cell culture conditions at 5% CO2 and 37°C in a humidified atmosphere incubator. Each vial of frozen cells was thawed and maintained for about 2 months (≈8 passages). Cells were cultured and treated with various agents, including rhIL-22 and TPA, in the presence/absence of different concentrations of Del as detailed below. For IL-22 stimulation, near-confluent NHEKs were treated with/without different concentrations of Del (0, 10, and 20 μM) for 8 h and then cotreated with or without rhIL-22 (20 ngml−1) for further 40 h (for a total of 48 h of exposure to Del). Cells were harvested and lysates were prepared for immunoblot analysis as described below.
For 12-O-tetradecanoyl-phorbol-13-acetate (TPA) stimulation, near-confluent NHEKs were pretreated with/without different concentrations of Del (10 and 20 μM) for 18 h and were then cotreated with/without 100 nM/ml TPA for the final 8 h (total of 24 h of exposure to Del). Cell culture supernatants were collected, centrifuged, and suspensions were stored at −80°C and subjected to a single freeze–thaw cycle for Procarta™ multiplex immunoassay analysis of keratinocyte-secreted proinflammatory cytokines and for human peripheral blood mononuclear cell-derived neutrophil chemotactic activity in vitro. The supernatant concentrations of the following cytokines, IL-1α, IL-6, IL-8 (CXCL8), IL-10, TNF-α, and TGF-α, were measured using a mix-matched 6-Plex Procarta immunoassay (Affymetric, eBiosciences) following the manufacturer's protocol as detailed below.
Human neutrophil preparation and in vitro chemotaxis activity assay
For keratinocyte-conditioned medium preparation, supernatant samples from TPA-activated keratinocytes were obtained as above such that KS = keratinocyte supernatant and KSD = Del-treated keratinocyte supernatant.
For preparation of single-cell suspensions of neutrophil, peripheral blood was obtained from three different subjects under the study protocol approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board. Informed written consent was obtained from subjects before participation. Heparinized blood was centrifuged through Percoll (1.090 g/ml), red blood cells were lysed from the granulocyte-containing pellet, and neutrophils were purified by positive selection using anti-CD16 immunomagnetic beads (AutoMac system; Miltenyi Biotec, Inc.). Neutrophils typically >92% pure (≈5% of eosinophil) were resuspended into RPMI and 10% FBS at 2 × 106 cells/ml. For the migration or chemotaxis assay, 200 μl of plasma fibronectin at 10 μg/ml was added into polycarbonate filter membrane inserts from transwell chambers (3 μm pore size; Costar #3415). Chambers were incubated at 37°C for 2 h, washed with PBS, and dried overnight. Two hundred microliters (200 μl) of neutrophils (2 × 106cells/ml) was added into the insert and 400 μl of keratinocyte-conditioned medium diluted by four in RPMI 10% FBS was added into the bottom chamber. Conditioned medium from keratinocytes activated with TPA and treated or not with Del 10 or 20 μM (D10 or D20) was tested. As control, medium only with no keratinocytes (no KS) was included. Plates were then incubated at 37°C for 1 h, and migrated cells in the bottom chamber were counted using trypan blue, a hemacytometer, and a microscope. Typically ∼50% of the neutrophils have migrated to TPA-activated supernatants at the bottom chamber in 1 h. The % of migration inhibition was calculated with the following formula:
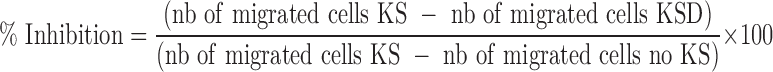 |
Determination of cell viability by MTT assay
The effect of Del on the viability of cells prestimulated with or without IL-22 was determined by the MTT assay as detailed below. Briefly, NHEKs were seeded at a density of 2 × 104 cells/well in 24-well poly(D)lysine (0.1 mg/ml; Sigma-Aldrich) precoated plates in 1 ml complete culture medium and incubated at 37°C and 5% CO2. At 80% confluence, cells were treated for 6 h with/without Del 0–20 μM, after which rhIL-22 (20 ng/ml) was added and further cultured. At 48 h of incubation, medium was removed and cells were rinsed with PBS and incubated for 3 h with 300 μl of MTT solution (0.5 mg/ml). The MTT solution was removed and formazan crystals were then solubilized in DMSO (300 μl) by shaking, and absorbance was spectrophotometrically recorded at 570 nm on a BioTek microplate reader (Bio-TEK Instruments, Inc.). Each experiment was repeated three times with similar results. The effect of Del on inhibition of IL-22-stimulated growth was assessed as percentage of cell viability in which untreated controls were considered as 100% viable.
Cell and skin tissue protein lysate extraction and Western blot analysis
NHEKs and mouse skin tissue lysates were prepared and immunoblotting was performed as previously described (7, 8). Briefly, following various treatments of NHEKs, cells were washed with cold PBS (10 mM, pH 7.4), followed by incubation in ice-cold 1 × lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid, 1 mM ethylenediaminetetraacetic acid, 20 mM NaF, 100 mM Na3VO4, 0.5% NP-40, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, pH 7.4) with freshly added protease inhibitor cocktail Set III (Calbiochem). Plates were placed on ice for 30 min, and cells were scraped and lysate collected in a microfuge tube and passed through a 22.5-gauge needle of syringe to break up the cell aggregates. The lysate was cleared by centrifugation at 14,000 g for 15 min at 4°C, and the supernatant (total cell lysate) was aliquoted and stored frozen at −80°C for further use in immunoassays to avoid additional freeze/thaw cycles for further analysis. The protein concentration was determined by BCA protein assay kit according to the manufacturer's protocol.
For mouse skin, epidermal tissue samples obtained from control and IMQ-treated mouse biopsies were lysed by homogenizing and ultrasonicating in ice-cold Cell Lysis Buffer (# EPX-99999-000; affymetrix, eBioscience) freshly supplemented with 1 mM PMSF and protease inhibitor cocktail Set III (Calbiochem La Jolla, CA), and tissue lysates were cleared by centrifugation at 14,000 g for 30 min at 4°C, quantified, and normalized as above and described earlier (7). Immunoblotting was conducted as previously reported (7). In brief, 10–20 μg of cell or tissue proteins was resolved with 4–12% SDS-PAGE and transferred onto nitrocellulose membranes, which were blocked with 7% nonfat milk/TBS-T or 5% bovine serum albumin (BSA) and hybridized with specific primary antibodies overnight at 4°C, followed by 2 h of hybridization with specified HRP-conjugated secondary antibody. The membrane-bound complexes were visualized using an enhanced chemiluminescence reagent (ECL kit; GE Healthcare) and autoradiographed using Gel-Doc automatic imager (Bio-Rad Laboratories) as described (8). Densitometric measurements of immunoblotted bands were determined using ImageJ digitalized software (National Institutes of Health).
Mice, IMQ-induced psoriasis-like dermatitis model, and treatments
Six- to eight-week-old Balb/c mice (Harlan laboratories) were maintained under specific pathogen-free conditions and fed standard chow diet. All animal experiments were performed in compliance with approved protocols and based on the guidelines established by the University of Wisconsin-Madison Institutional Animal Care and Use Committee. As previously described, psoriasis-like skin inflammation was induced following the model by Van der Fits et al. (45), with slight modification as described previously (7). In brief, the back skin of mice was shaved with an electric clipper (B. Braun Vet Care) and then treated with Nair, a depilatory cream to remove residual hair. After 48 h of resting, mice were initially divided into two different (control (n = 6) and IMQ-treated (n = 12)) groups. While control mice received a control cream (Vaseline; Walgreens Pharmacy), the IMQ-treated mice received on the right ear and the shaved back skin a daily topical dose of 20 and 62.5 mg of 5% IMQ cream (Aldara, 3M Pharmaceuticals), a Toll-like receptor-7/8 ligand, respectively, for 5 consecutive days to induce the disease before further splitting into different groups. On day 5, the IMQ-treated group was further divided into two different (IMQ(+) alone-treated (n = 6) and IMQ(+)-Del-treated groups, and both groups continued to receive the same booster dose of IMQ for a total of 14 consecutive days to achieve optimal chronic inflammation. However, from day 6, the control group continually received control cream and the IMQ(+) alone group continually received IMQ alone, while the IMQ(+)-Del-treated group first received topical application of Del (1 mg/cm2 of shaved skin area) and application of IMQ cream. The ear and skin were topically treated with delphinidin dissolved in DMSO and administered as 100 μl (on shaved back skin; 1 mg/cm2) and 20 μl (10 μl on each side of the right ear) daily from days 6–15 for IMQ. When coadministering drugs, a 3-h interval was allowed between applications and delphinidin was applied before IMQ. All mice were monitored daily and mouse ear thickness, erythema, and scaling were measured every alternate day throughout the entire 14 days of experiment. The concentration of Del used in this study was chosen based on our previous published protocol (34). Back and ear redness (erythema), presence of scales (scaling), and hardness of the skin were scored using a semiquantitative PASI scoring system from 0 to 4 based on their external physical appearance: 0 = no skin abnormalities, 1 = slight, 2 = moderate, 3 = marked, and 4 = severe. In addition, mouse ear skin thickening was assessed by measuring thickness using an Electronic Digital Caliper (Fisher Scientific, Pittsburgh, PA). After a total of 14 days or 9 consecutive days of Del treatment, all mice were euthanized and skin tissues were freshly harvested as earlier described (7, 35) for histological and biochemical analyses.
Histology, immunohistochemistry, and immunofluorescence analyses
Paraffin sections from the inflamed skin lesions of IMQ-induced, Del-treated+IMQ-induced and matched control mouse skin tissues were deparaffinized, processed for immunofluorescence and immunohistochemical staining, and images generated, processed, and analyzed essentially as previously described (7). Control immunostaining utilizing isotype controls were used. The quantification for antigen staining (number of brown, green, or red pixels) was performed using ImageJ software (National Institutes of Health) and the Nuance technology software with the average value of at least five fields for each mouse (7).
Human and mouse ProcartaPlex™ multiplex cytokine and chemokine immunoassays
The changes in multiple dysregulated immune mediator responses in this chronic phase, IMQ-induced murine psoriasis-like skin lesions, TPA-induced NHEK secretion of proinflammatory cytokines, and the role of Del intervention were examined using a mix-matched human 6-plex and a mouse 36-plex Procarta multiplex immunoassay technique (Affymetrix/eBioscience) according to the manufacturer's instructions. The Procarta multiplex technique blends sandwich immunoassay with fluorescent bead-based technology. Polystyrene microspheres (5.5-μm diameter) are each filled with a specific red/infrared fluorescent dye mixture (with 100 possible mix combinations). Each bead is also coated with specific biomarker reagents (such as antibodies against IL-17) in a combination that facilitates the quantification of up to 500 different analytes in a single microtiter well. Individual cytokines/chemokines were identified and classified by their bead color using red laser excitation, and their levels quantified via green laser excitation. This assay includes a wide range of cytokines and chemokines that reflect key processes relating to systemic activation of inflammatory signaling pathways involved in autoimmunity and anti-inflammatory responses. Both the human and mouse assays were performed according to the manufacturer's recommendations (Procarta multiplex cytokine and/or chemokine kits; Affymetrix/eBioscience). Utilizing a 36-plex multiplex magnetic bead-based immunoassay kit, mouse skin tissue lysate concentrations of the following immune molecules or analytes were determined: interleukin (IL)-1 family—IL-1α, IL-1β, and IL-18; IL/γ chain family—IL-2, IL-4, and IL-15/IL-15R; IL/β chain family—IL-3, IL-5, and GM-CSF (CSF2); IL-6 (gp130) family—IL-6, IL-31, and LIF; IL-12p70; IL-10; IL-9; IL-17A; IL-22; IL-23; IL-27; IL-13; IFNα2 (IFNα); IFNγ; IL-28; TNFα; CC chemokines—CCL2 (MCP-1), CCL-3 [macrophage inflammatory protein-1α (MIP-1α)], CCL4 (MIP-1β), CCL5 [regulated on activation, normal T cell expressed and secreted (RANTES)], CCL7 (MCP-3), and CCL-11 (eotaxin); CXC chemokines—CXCL-1 [growth-regulated oncogene a (GROα)], CXCL-2 (MIP-2), CXCL-5 [epithelial-derived neutrophil-activating peptide 78 (ENA-78)], and CXCL-10 (IP-10); and growth/cellular factors-M-CSF (CSF1) and G-CSF(CSF3) (granulocyte colony-stimulating factor; CSF3) (Customized Procarta immunoassay, Affymetrix/eBiosciences). Similarly, a customized mix-matched Human Procarta 6-plex magnetic bead-based immunoassay kit, consisting of IL-1α, IL-6, CXCL8 (IL-8), IL-10, TNF-α, and TGF-α) (Affymetrix/eBiosciences), was utilized to determine the concentration of TPA-induced and/or Del-treated NHEK secretion of immune molecules in culture supernatant. Cell culture supernatants, mouse skin lysates, reagents, standards, and protocols were prepared according to the manufacturer's instructions. Briefly, an aliquot of 25 μl (tissue lysates for mouse 36-Plex) or 50 μl (NHEK culture supernatant for Human 6-Plex) of each sample was added to each well of a 96-well black side/transparent bottom filter plate, initially preloaded with a panel of mix/matched magnetic bead-based anticytokine or chemokine antibodies covalently linked to unique polystyrene beads following the manufacturer's instructions. All skin lysates or cell culture supernatant samples were run in triplicate along with serial standards (7-point dilutions), added at this time with buffer controls. Next, an aliquot of 50 μl diluent was added, and the plate was incubated for 120 min at room temperature in the dark at 500 rpm. Plates were placed on a handheld magnet, followed by aspiration of each well content, and were washed three times with at least 150 μl of washing buffer to remove unbound antigen sample. After the last wash, the plate was blotted on absorbent paper towels to remove any residual buffer. An aliquot of 25 μl of biotinylated detection antibody mixture was added to each well, and the plate was sealed and incubated for 30 min at room temperature under 500 rpm. Buffer was aspirated and washing performed as above, and an aliquot of 50 μl (mouse kit) or 25 μl (human cell culture supernatant kit) Streptavidin-PE (SA-PE) enzyme working reagent was added to each well and incubated for 30 min under slow shaking at room temperature. A final aspiration and washing cycle was performed, followed by the addition of 120 μl reading buffer per well, and incubated for 5 min in the dark at room temperature with shaking and stored at 4°C overnight. The 96-well plate was then transferred to the xMAP Luminex reader (Luminex) for quantitative analysis. Mean fluorescence intensities of analyte-specific immunoassay bead sets were detected by a flow-based Luminex 3D suspension array system (Luminex). Optical density and protein content were detected in each well within 30 min, at 450 nm, and corrected at 570 nm. Digital images of the bead array were captured after laser excitation using a Pixel CCD camera xMAP technique (Luminex) and were processed on a computer workstation. Cytokine/chemokine concentrations were determined by importing into Procarta Plex Multiplex Analyst software v.1.0 (Affymetrix/eBioscience) and analyzed to obtain standard curves derived from the known reference concentrations supplied by the manufacturer and to quantify cytokine/chemokine levels. A five-parameter model was used to calculate final concentrations by interpolation, and values are expressed in picograms per milliliter (pgml−1). Concentrations obtained below the sensitivity limit of detection (LOD) of the method were recoded to the midpoint between zero and the LOD for that analyte for statistical comparisons and cytokine/chemokine concentrations were calculated accordingly.
Statistical analyses
All statistical analyses were carried out with GraphPad prism version 6.1 (San Diego) and p-values <0.05 were considered significant. All quantitative data are expressed as mean ± SD, and significant differences were determined by the Student t-test or one-way ANOVA with Bonferroni post hoc test. Aggregate cytokine data were referenced against standards established with lysates or supernatants from control groups, which were analyzed concurrently with the study samples.
Supplementary Material
Abbreviations Used
- Akt/PKB
protein kinase B
- Del
delphindin generic name: chemical name: (2R,3R)-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-1(2H)-benzopyran-3,5,7-triol 3-(3,4,5-trihydroxybenzoate)
- DMSO
dimethyl sulfoxide
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GM-CSF
granulocyte–monocyte colony-stimulating factor
- HRP
horseradish peroxidase
- IMQ
imiquimod
- LOD
limit of detection
- MOE
molecular operating environment
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- MTT
3- (4,5-dimethylthiazol-2yl)-2,5- diphenyltetrazolium bromide
- NHEKs
normal human epidermal keratinocytes
- PBS
phosphate-buffered saline
- PBST
0.1% Tween 20 in PBS
- PDB
protein data bank
- PI3K
phosphoinositide 3-kinase
- PRAS40
proline-rich Akt substrate of 40 kDa
- Rap
rapamycin
- rhIL-22
recombinant human interleukin-22
- SPR
surface plasmon resonance
- TPA
12-O-tetradecanoyl-phorbol-13-acetate
- TSC2
tuberous sclerosis complex-2
Acknowledgments
This work was supported, in part, by the US PHS NIAMS Grants R01AR059742 (to H.M.) and the American Skin Association (ASA) grant Award (to J.C.C.). M.S. gratefully acknowledges the Fondazione Banco di Sardegna for partial financial support. The authors thank Dr. Andrea Brancale and Dr Nicolino Pala for the use of software for molecular modeling calculation and Dr. Babatunde S. Abiola for assistance in cell culture work.
Author Disclosure Statement
The authors declare no conflicts of interest exist.
References
- 1.Aksamitiene E, Kiyatkin A, and Kholodenko BN. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans 40: 139–146, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Banaszynski LA, Liu CW, and Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc 127: 4715–4721, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Bonner MY, Karlsson I, Rodolfo M, Arnold RS, Vergani E, and Arbiser JL. Honokiol bis-dichloroacetate (Honokiol DCA) demonstrates activity in vemurafenib-resistant melanoma in vivo. Oncotarget 7: 12857–12868, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck E, Eyzaguirre A, Rosenfeld-Franklin M, Thomson S, Mulvihill M, Barr S, Brown E, O'Connor M, Yao Y, Pachter J, Miglarese M, Epstein D, Iwata KK, Haley JD, Gibson NW, and Ji QS. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res 68: 8322–8332, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Cantley LC. The phosphoinositide 3-kinase pathway. Science 296: 1655–1657, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Chamcheu JC, Afaq F, Syed DN, Siddiqui IA, Adhami VM, Khan N, Singh S, Boylan BT, Wood GS, and Mukhtar H. Delphinidin, a dietary antioxidant, induces human epidermal keratinocyte differentiation but not apoptosis: studies in submerged and three-dimensional epidermal equivalent models. Exp Dermatol 22: 342–348, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamcheu JC, Chaves-Rodriquez MI, Adhami VM, Siddiqui IA, Wood GS, Longley BJ, and Mukhtar H. Upregulation of PI3K/AKT/mTOR, FABP5 and PPARbeta/delta in Human Psoriasis and Imiquimod-induced Murine Psoriasiform Dermatitis Model. Acta Derm Venereol, 96: 854–856, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamcheu JC, Navsaria H, Pihl-Lundin I, Liovic M, Vahlquist A, and Torma H. Chemical chaperones protect epidermolysis bullosa simplex keratinocytes from heat stress-induced keratin aggregation: involvement of heat shock proteins and MAP kinases. J Invest Dermatol 131: 1684–1691, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Chamcheu JC, Pal HC, Siddiqui IA, Adhami VM, Ayehunie S, Boylan BT, Noubissi FK, Khan N, Syed DN, Elmets CA, Wood GS, Afaq F, and Mukhtar H. Prodifferentiation, anti-inflammatory and antiproliferative effects of delphinidin, a dietary anthocyanidin, in a full-thickness three-dimensional reconstituted human skin model of psoriasis. Skin Pharmacol Physiol 28: 177–188, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen SJ, Nakahara T, Takahara M, Kido M, Dugu L, Uchi H, Takeuchi S, Tu YT, Moroi Y, and Furue M. Activation of the mammalian target of rapamycin signalling pathway in epidermal tumours and its correlation with cyclin-dependent kinase 2. Br J Dermatol 160: 442–445, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Choi J, Chen J, Schreiber SL, and Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273: 239–242, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, Liau L, Martin N, Becker D, Bergsneider M, Lai A, Green R, Oglesby T, Koleto M, Trent J, Horvath S, Mischel PS, Mellinghoff IK, and Sawyers CL. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med 5: e8, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa A, Bonner MY, and Arbiser JL. Use of Polyphenolic Compounds in Dermatologic Oncology. Am J Clin Dermatol, 17: 369–385, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, and Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29: 1046–1051, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, and Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 23: 329–336, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Faivre S, Kroemer G, and Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov 5: 671–688, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Frigerio E, Colombo MD, Franchi C, Altomare A, Garutti C, and Altomare GF. Severe psoriasis treated with a new macrolide: everolimus. Br J Dermatol 156: 372–374, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Furet P, Guagnano V, Fairhurst RA, Imbach-Weese P, Bruce I, Knapp M, Fritsch C, Blasco F, Blanz J, Aichholz R, Hamon J, Fabbro D, and Caravatti G. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett 23: 3741–3748, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Gilliet M, Conrad C, Geiges M, Cozzio A, Thurlimann W, Burg G, Nestle FO, and Dummer R. Psoriasis triggered by toll-like receptor 7 agonist imiquimod in the presence of dermal plasmacytoid dendritic cell precursors. Arch Dermatol 140: 1490–1495, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Heerding DA, Rhodes N, Leber JD, Clark TJ, Keenan RM, Lafrance LV, Li M, Safonov IG, Takata DT, Venslavsky JW, Yamashita DS, Choudhry AE, Copeland RA, Lai Z, Schaber MD, Tummino PJ, Strum SL, Wood ER, Duckett DR, Eberwein D, Knick VB, Lansing TJ, McConnell RT, Zhang S, Minthorn EA, Concha NO, Warren GL, and Kumar R. Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase. J Med Chem 51: 5663–5679, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, and Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T, Lin X, Meng X, and Lin M. Phosphoinositide-3 kinase/protein kinase-b/mammalian target of rapamycin pathway in psoriasis pathogenesis. A potential therapeutic target? Acta Derm Venereol 94: 371–379, 2014 [DOI] [PubMed] [Google Scholar]
- 23.Karlsson I, Zhou X, Thomas R, Smith AT, Bonner MY, Bakshi P, Banga AK, Bowen JP, Qabaja G, Ford SL, Ballard MD, Petersen KS, Li X, Chen G, Ogretmen B, Zhang J, Watkins EB, Arnold RS, and Arbiser JL. Solenopsin A and analogs exhibit ceramide-like biological activity. Vasc Cell 7: 5, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M. and Sabatini DM. mTOR signaling in growth control and disease. Cell 149: 274–293, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J, Choi J, and Clardy J. Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr 55: 736–744, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Lowes MA, Bowcock AM, and Krueger JG. Pathogenesis and therapy of psoriasis. Nature 445: 866–873, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Lowes MA, Suarez-Farinas M, and Krueger JG. Immunology of psoriasis. Annu Rev Immunol 32: 227–255, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markman B, Dienstmann R, and Tabernero J. Targeting the PI3K/Akt/mTOR pathway—beyond rapalogs. Oncotarget 1: 530–543, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DM. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem J 375: 531–538, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra A, Raychaudhuri SK, and Raychaudhuri SP. IL-22 induced cell proliferation is regulated by PI3K/Akt/mTOR signaling cascade. Cytokine 60: 38–42, 2012 [DOI] [PubMed] [Google Scholar]
- 31.Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons JJ, Wu H, and Sawyers CL. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A 98: 10314–10319, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nestle FO, Kaplan DH, and Barker J. Psoriasis. N Engl J Med 361: 496–509, 2009 [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, and Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66: 1500–1508, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal HC, Chamcheu JC, Adhami VM, Wood GS, Elmets CA, Mukhtar H, and Afaq F. Topical application of delphinidin reduces psoriasiform lesions in the flaky skin mouse model by inducing epidermal differentiation and inhibiting inflammation. Br J Dermatol 172: 354–364, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roller A, Perino A, Dapavo P, Soro E, Okkenhaug K, Hirsch E, and Ji H. Blockade of phosphatidylinositol 3-kinase PI3Kdelta or PI3Kgamma reduces IL-17 and ameliorates imiquimod-induced psoriasis-like dermatitis. J Immunol 189: 4612–4620, 2012 [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Shaw RJ. and Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441: 424–430, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Shi G, Blaszczyk J, Ji X, and Yan H. Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: synthesis and biochemical and crystallographic studies. J Med Chem 44: 1364–1371, 2001 [DOI] [PubMed] [Google Scholar]
- 39.Soares HP, Ming M, Mellon M, Young SH, Han L, Sinnet-Smith J, and Rozengurt E. Dual PI3K/mTOR inhibitors induce rapid over-activation of the MEK/ERK pathway in human pancreatic cancer cells through suppression of mTORC2. Mol Cancer Ther 14: 1014–1023, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somoza JR, Koditek D, Villasenor AG, Novikov N, Wong MH, Liclican A, Xing W, Lagpacan L, Wang R, Schultz BE, Papalia GA, Samuel D, Lad L, and McGrath ME. Structural, biochemical, and biophysical characterization of idelalisib binding to phosphoinositide 3-kinase delta. J Biol Chem 290: 8439–8446, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunami T, Byrne N, Diehl RE, Funabashi K, Hall DL, Ikuta M, Patel SB, Shipman JM, Smith RF, Takahashi I, Zugay-Murphy J, Iwasawa Y, Lumb KJ, Munshi SK, and Sharma S. Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation. J Biol Chem 285: 4587–4594, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syed DN, Adhami VM, Khan MI, and Mukhtar H. Inhibition of Akt/mTOR signaling by the dietary flavonoid fisetin. Anticancer Agents Med Chem 13: 995–1001, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Syed DN, Chamcheu JC, Khan MI, Sechi M, Lall RK, Adhami VM, and Mukhtar H. Fisetin inhibits human melanoma cell growth through direct binding to p70S6K and mTOR: findings from 3-D melanoma skin equivalents and computational modeling. Biochem Pharmacol 89: 349–360, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, and Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205, 2004 [DOI] [PubMed] [Google Scholar]
- 45.van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, and Lubberts E. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol 182: 5836–5845, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP, and Treiber DK. Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol 17: 1241–1249, 2010 [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, and Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature 497: 217–223, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu K, Shi C, Toral-Barza L, Lucas J, Shor B, Kim JE, Zhang WG, Mahoney R, Gaydos C, Tardio L, Kim SK, Conant R, Curran K, Kaplan J, Verheijen J, Ayral-Kaloustian S, Mansour TS, Abraham RT, Zask A, and Gibbons JJ. Beyond rapalog therapy: preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Res 70: 621–631, 2010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.