Summary
Severe chronic hepatic injury can induce complex reparative processes. Ductular reaction and the appearance of small hepatocytes are standard components of this response, which is thought to have both adverse (e.g. fibrosis, carcinogenesis) and beneficial (regeneration) consequences. This complex tissue reaction is regulated by orchestrated cytokine action. We have investigated the influence of the tyrosine kinase inhibitor imatinib on a regenerative process. Ductular reaction was induced in mice by the widely used choline‐deficient ethionine‐supplemented diet (CDE). Test animals were treated daily with imatinib. After 6 weeks of treatment, imatinib successfully reduced the extent of ductular reaction and fibrosis in the CDE model. Furthermore, the number of small hepatocytes increased, and these cells had high proliferative activity, were positive for hepatocyte nuclear factor 4 and expressed high levels of albumin and peroxisome proliferator‐activated receptor alpha. The overall functional zonality of the hepatic parenchyma (cytochrome P450 2E1 and glucose 6 phosphatase activity; endogenous biotin content) was maintained. The expression of platelet‐derived growth factor receptor beta, which is the major target of imatinib, was downregulated. The anti‐fibrotic activity of imatinib has already been reported in several experimental models. Additionally, in the CDE model imatinib was able to enhance regeneration and preserve the functional arrangement of hepatic lobules. These results suggest that imatinib might promote the recovery of the liver following parenchymal injury through the inhibition of platelet‐derived growth factor receptor beta.
Keywords: choline‐deficient ethionine‐supplemented diet, ductular reaction, imatinib, liver fibrosis, liver regeneration, PDGFR‐β
Introduction
The liver has an enormous capacity to regenerate following various types of injury. Obviously, the most efficient mechanism is the compensatory hyperplasia of the hepatocytes, studied mostly following surgical or chemical partial hepatectomy (Riehle et al. 2011). However, there are well‐known backup mechanisms such as the enlargement, hypertrophy of hepatocytes (Nagy et al. 2001; Miyaoka et al. 2012) and the regeneration that has been observed following chronic toxic injury (Itoh & Miyajima 2014; Michalopoulos & Khan 2015). This latter pathway of regeneration has been the subject of very intense investigation over for many years. There was almost complete agreement that the ‘new’ hepatocytes derive from the hepatic stem/progenitor cells, which are amplified through the so‐called ductular reaction. Recently, the origin and the precursor role of the ductular reaction have been intensely debated. (Schaub et al. 2014; Tarlow et al. 2014a,b; Yanger et al. 2014). Nevertheless, there is no doubt about the existence of a reserve regenerative mechanism in the liver (Boulter et al. 2013; Grompe 2014; Itoh & Miyajima 2014; Michalopoulos & Khan 2015). Whatever the origin of this regeneration is, it is mostly associated with an intense ductular reaction. There are good evidences even in human livers that this reserve mechanism could result in complete recovery of hepatic functions (Fujita et al. 2000; Quaglia et al. 2008). There are three major problems concerning the outcome of this regenerative process: (i) it is often not efficient enough ‐ liver transplantation is frequently required in fulminant hepatic failure, even if the explanted liver is rich in ductular reaction (Demetris et al. 1996; Quaglia et al. 2008); (ii) the ductular reaction, whatever its role is in the regenerative process, could result in hepatic fibrosis or even cirrhosis (Williams et al. 2014) and (iii) the ductular reaction could promote carcinogenesis (Alison et al. 2009). The hepatic ductules are closely associated with activated myofibroblasts, and these cells are thought to provide the necessary microenvironment or niche (Kordes & Häussinger 2013) for the ductular cells. Thus, myofibroblasts are key players of liver regeneration, but they are also responsible for the deposition of large amounts of extracellular matrix (ECM) material resulting in hepatic fibrosis. The optimization of the reparative process by promoting regeneration and ameliorating the unfavourable consequences remains a great challenge.
It is well known that the activation of hepatic myofibroblasts is driven by several growth factors, among others by platelet‐derived growth factor (PDGF) (Pinzani 2002). This growth factor is acting through a tyrosine kinase receptor, which can be efficiently inhibited by the clinically widely used drug imatinib (Grimminger et al. 2010). Indeed, imatinib efficiently attenuated the progression of hepatic fibrosis in several experimental models (Neef et al. 2006; Knight et al. 2008; El‐Agamy et al. 2011; Kim et al. 2012; Kuo et al. 2012). We have decided to study the impact of imatinib treatment on one of the most widely used experimental models of chronic hepatic injury/regeneration in mouse liver, induced by choline‐deficient ethionine‐supplemented diet (CDE) (Lenzi et al. 1992). Imatinib efficiently reduced the extent of fibrosis and ductular reaction and also maintained the metabolic zonation of the hepatic lobules. In addition, the number of small hepatocytes increased significantly. They proliferated and highly expressed hepatocyte‐specific genes. All these changes were assessed as signs of accelerated regeneration due to the effect of imatinib.
Methods
Animal experiments
All experiments were conducted on 8‐week‐old male C57Bl/6 mice kept under standard conditions. Ductular reaction was induced with choline‐deficient diet containing 0.5% ethionine (CDE; Altromin, Lage, Germany). Animals in Group 1 (n = 8) received CDE ad libitum for 6 weeks; Group 2 (n = 9) received imatinib treatment (25 mg/kg/day, per os; Glivec, Novartis, Basel, Switzerland) besides CDE. Each animal was given 200 mg/kg bromodeoxyuridine (BrdU) intraperitoneally 1 h before termination. After sacrificing the animals, samples from the liver were taken and fixed in formalin for histological examination and the rest were snap‐frozen in liquid nitrogen.
Ethical approval statement
The animal study protocols were conducted according to National Institute of Health (NIH) guidelines for animal care and were approved by the Animal Care and Use Committee of Semmelweis University (Nr: KA‐1771).
Morphological analysis
Immunohistochemistry
Frozen sections were fixed in methanol for 10 min and incubated in room temperature for 1 h with the primary antibodies (Table S1), then with appropriate secondary antibodies (Jackson Immunoresearch, West Grove, PA) and fluorescent dyes (Table S1).
Morphometric analysis
The area occupied by ductular reaction or myofibroblasts was measured on three images from each liver, which were captured from frozen sections immunostained for cytokeratin‐19 (CK‐19), desmin and PDGFR‐β with a Bio‐Rad confocal system (MRC 1024; Bio‐Rad, Richmond, CA), using a 10 × objective. The area percentage was determined with manual thresholding using the ImageJ 1.49k program (NIH, Bethesda, MD). The ratio of small and large hepatocytes and their proliferative activity was determined on frozen sections immunostained for β‐catenin and BrdU, and nuclei were outlined by 4′,6‐diamidino‐2‐phenylindole (DAPI). Sections were scanned with the Pannoramic 250 Flash scanner (3DHistech, Budapest, Hungary). On each section, hepatocytes in the 200 μm proximity of three portal areas were circumscribed manually using the Pannoramic Viewer 1.15.4 (3DHistech). Only those cells were counted, where the nucleus was distinguishable. For each cell, an estimated diameter was calculated by the program. The border between small and large hepatocytes was set at 22 μm. The BrdU labelling index of 500 pericentral large hepatocytes was also determined.
From each liver, three images from Picro‐sirius red‐stained sections were captured with a Zeiss Axioskop 2 plus microscope (Zeiss, Oberkochen, Germany) mounted with an Olympus PD50 digital camera (Olympus, Tokyo, Japan), using a 5 × objective. The area occupied by fibrotic tissue was measured using the Quick PhotoMicro 2.2 (Promicra, Prague, Czech Republic) software.
Histopathological analysis
Zonality of the liver lobules
To examine the distribution of endogenous biotin and cytochrome P450 2E1 (CYP2E1) isoenzyme, Streptavidin‐TRITC and CYP2E1 labelling was performed on frozen sections. Glucose‐6‐phosphatase (G6Pase) enzyme histochemistry was made on frozen sections as described before (Teutsch 1981). Sections were scanned with the Pannoramic 250 Flash scanner (3DHistech).
Oil red O staining
Frozen sections were fixed in 4% paraformaldehyde, rinsed in 60% isopropanol and then stained with oil red O working solution (60% oil red O stock solution, 40% distilled water) for 10 min. After rinsing with 60% isopropanol, haematoxylin background staining was performed. Sections were scanned with the Pannoramic 250 Flash scanner (3DHistech).
Quantitative Real‐Time Polymerase Chain Reaction
Microdissected samples
Frozen sections made from the livers of Group 2 animals were fixed in methanol, stained with RNase‐free haematoxylin and dried at room temperature. Laser microdissection of small and large hepatocytes was performed by using the PALM MicroBeam system (Zeiss). At least 100.000 μm2 area containing small or large hepatocytes was collected. Total RNA was isolated by the RNA Aqueous Micro Kit (cat. no. AM 1931; Life Technologies, Carlsbad, CA). The total amount of isolated RNA was used for reverse transcription.
Whole liver samples
Frozen sections from the livers of groups 1 and 2 mice were collected in lysis buffer. Total RNA was isolated with TRIzol (cat. no. 15596–018; Life Technologies). RNA concentration was measured by NanoDrop 1000 (Thermo Fisher); 1 μg RNA per sample was converted into cDNA.
A high‐capacity cDNA reverse transcription kit (cat. no. 4368814; Life Technologies) was used for cDNA synthesis as recommended by the supplier. PCR was performed by the ABI Prism 7300 sequence detection system, using Life Technologies TaqMan gene expression assays (Table S2) according to the manufacturer's instructions. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used as endogenous control. All samples were run in triplicate, in a 20 μl reaction volume. Results were obtained as threshold cycle (CT) values. Expression levels were calculated using the ΔCT method. The values were calculated as the mean values of three independent measurements, and the expression levels of mRNA in all samples were defined as a ratio to GAPDH expression. The average fold change compared with the control group was calculated using ΔΔCT method from RT‐PCR data.
Statistical analysis
Statistical analysis was performed with GraphPad Instat 3.06 (GraphPad Software, La Jolla, CA). Data are presented as means ± standard error (SEM). Deviation from Gaussian distribution was tested using Kolmogorov–Smirnov's method. Means were compared with Student's or Welch's t‐test according to the result of the F‐probe. Results were considered significant at P ≤ 0.05.
Results
CDE model
Imatinib treatment facilitates the appearance of small hepatocytes
The mice were kept on CDE for 6 weeks and were sacrificed. Histological examinations revealed important changes in the liver after imatinib treatment. The most striking difference was the appearance of small hepatocytes en masse in the periportal zone. These small hepatocytes were not as sharply separated from the neighbouring parenchyma as the ‘foci’ in hepatocarcinogenesis models, but they could be easily distinguished from the polymorphic, large hepatocytes. Small hepatocytes were present in the control livers (only CDE) periportally as well, but they did not form large confluent fields. To quantify the difference between the imatinib‐treated and control animals, the cell membrane was visualized by β‐catenin immunostaining (Figure 1). By means of morphometric analysis, the estimated diameter of individual periportal hepatocytes was demonstrated on a histogram (Figure 2) and an obvious shift towards smaller cell size could be observed following imatinib treatment. When the border between small and large hepatocytes was set at a diameter of 22 μm, the ratio of small hepatocytes was significantly higher in the imatinib‐treated animals (Table 1). A morphometric approach was also applied following CK‐19 (Figure 1a,b) and desmin (Figure 3a,b) immunostaining to analyse the extent of ductular reaction and the area occupied by myofibroblasts. Both values were significantly lower in the imatinib‐treated animals (Table 1). PDGFR is a well‐known target of imatinib (Grimminger et al. 2010), and hepatic myofibroblasts express mostly the β subtype of this receptor (Kocabayoglu et al. 2015). PDGFR‐β was highly expressed in the liver of CDE‐fed mice, but the expression was sharply reduced by imatinib treatment (Figure 3c,d, Table 1). The extent of fibrosis was evaluated by morphometric analysis of Picro‐sirius red‐stained sections. This parameter was also significantly lower following imatinib treatment (Figure 3e,f; Table 1). These morphological observations were completely supported by the quantitative real‐time PCR examination of the steady‐state mRNA level of the RNA samples isolated from tissue slices. The relative mRNA expression levels of CK‐19, desmin, smooth muscle actin (SMA) and PDGFR‐β were significantly lower in the imatinib‐treated mice (Figure 4a). Imatinib induced a significant downregulation of CK‐19, desmin, SMA and PDGFR‐β mRNA expression levels; among them, the decrease in PDGFR‐β expression level was the most striking (Figure 4b).
Figure 1.
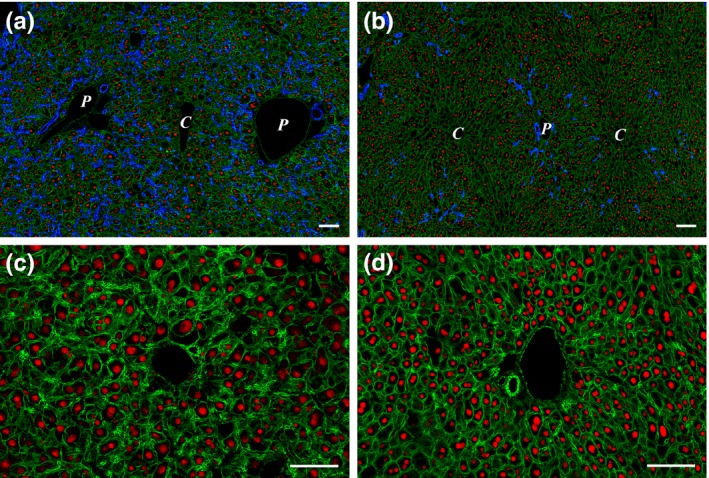
(a and b) Effect of imatinib on ductular reaction and hepatocytes in CDE‐fed mice. (a) A great number of ductules (CK‐19, blue) extend into the parenchyma from the portal areas in the CDE control animal (Group 1). (b) In the imatinib‐treated liver, there is a significant decrease in the amount of ductular reaction, while an increased number of small hepatocytes appear in the periportal areas. Cell membranes are visualized by β‐catenin (green), and hepatocyte nuclei are labelled with HNF‐4 (red). P stands for portal vein, and C stands for central vein. (c and d) Higher magnification of a portal area of a Group 1 (CDE; c) and a Group 2 (CDE+imatinib; d) animal. Sections are labelled with β‐catenin (green) and HNF‐4 (red). Note the high number of small hepatocytes surrounding the portal vein and the overall increase in the number of hepatocytes in the imatinib‐treated animal. Scale bar for Figure 1: 100 μm.
Figure 2.
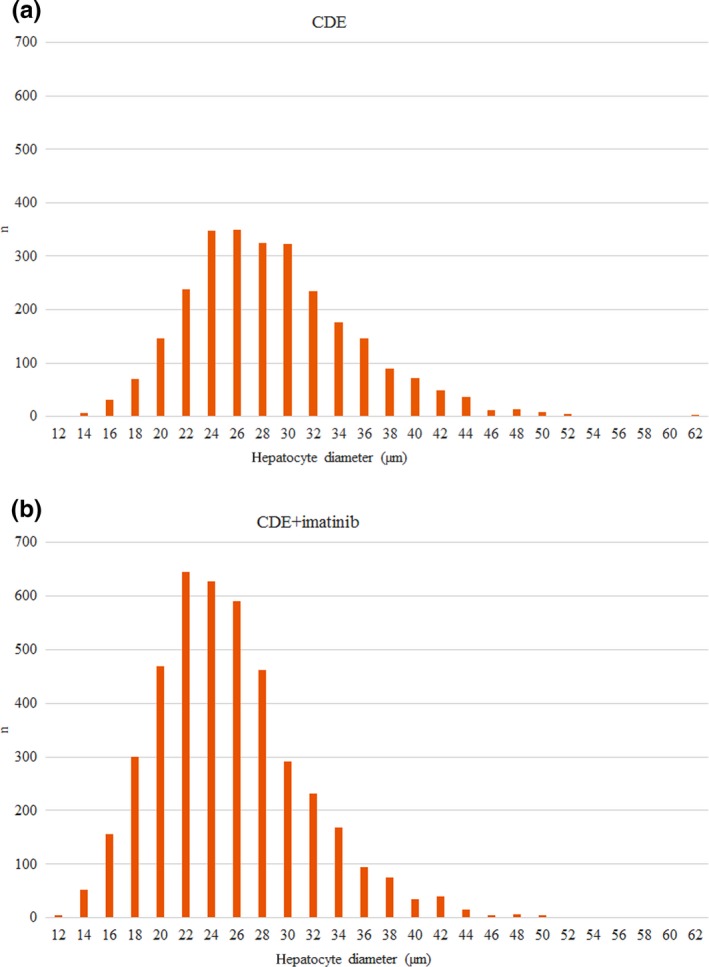
Estimated diameters of periportal hepatocytes in the 200 μm proximity of portal tracts in Group 1 (CDE; a) and Group 2 (CDE+imatinib; b). Note the shift towards smaller cell size in the imatinib‐treated group.
Table 1.
Summary of the results
| Group 1 (CDE) | Group 2 (CDE+imatinib) | |
|---|---|---|
| Relative liver weight (%) | 4.63 ± 0.11 | 5.4 ± 0.43 |
| CK‐19 (%) | 8.97 ± 0.7* | 5.21 ± 1.03* |
| Desmin (%) | 18.01 ± 0.61* | 14.5 ± 0.67* |
| PDGFR‐β (%) | 9.66 ± 2.23* | 3.73 ± 3.09* |
| Picro‐sirius red (%) | 8.99 ± 0.4* | 3.85 ± 0.48* |
| Ratio of periportal small and large hepatocytes (%) | 18.61 ± 2.15* | 36.64 ± 5.07* |
| Proliferative activity of periportal small hepatocytes (%) | 3.93 ± 0.27* , ** | 8.32 ± 1.18* , *** |
| Proliferative activity of pericentral large hepatocytes (%) | 1.44 ± 0.16* , ** | 4.01 ± 0.56* , *** |
Those results are marked with, *where there was a significant difference between Group 1 (CDE) and Group 2 (CDE + imatinib). There was a significant difference between the proliferative activity of periportal small and pericentral large hepatocytes in both Group 1 (CDE; **) and Group 2 (CDE+imatinib; ***). Data are presented as means ± SEM. P ≤ 0.05. CDE, choline‐deficient ethionine‐supplemented diet; CK‐19, cytokeratin‐19; PDGFR‐β, platelet‐derived growth factor β.
Figure 3.
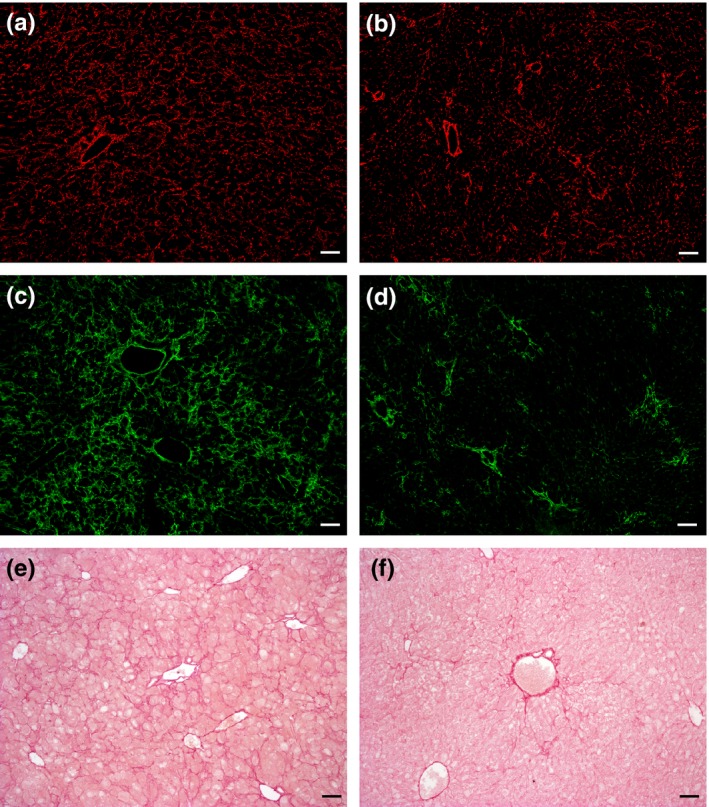
Effect of imatinib on myofibroblasts and the extent of fibrosis in CDE‐fed mice. Desmin (a and b) or PDGFR‐β (c and d) labelled frozen and Picro‐sirius red‐stained (e and f) paraffin‐embedded sections of Group 1 (CDE; a, c and e) and Group 2 (CDE+imatinib; b, d and f) animals. Imatinib treatment results in the reduction in the area occupied by desmin and PDFGR‐β‐positive myofibroblasts along with the reduction in the extent of fibrosis. Scale bar for Figure 3: 100 μm.
Figure 4.
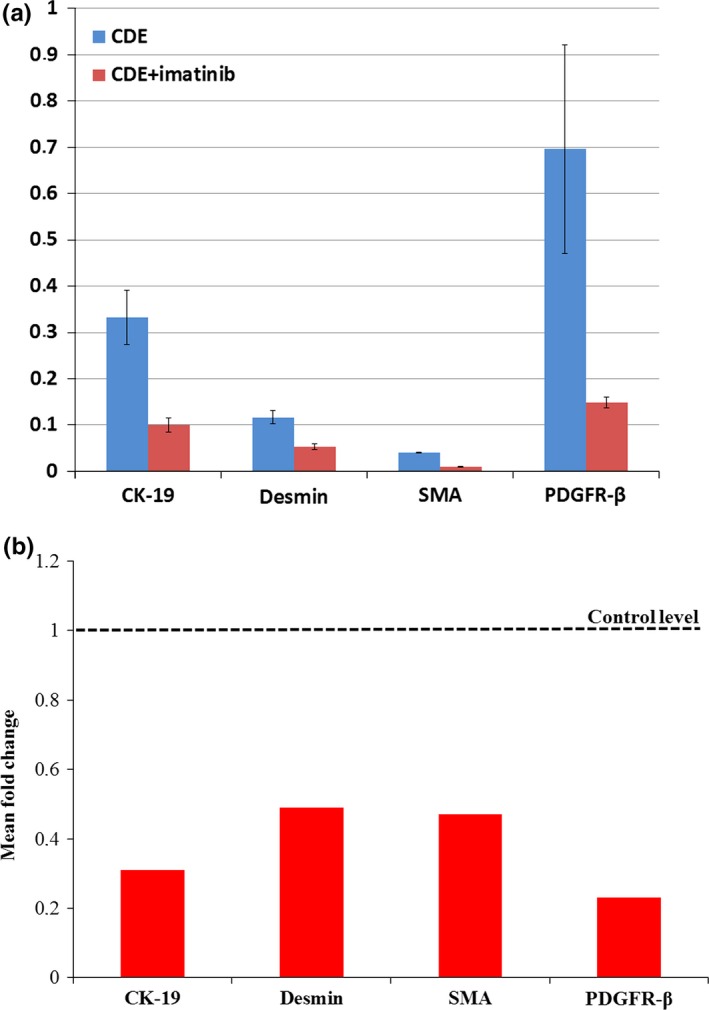
(a) QRT‐PCR analysis of CK‐19, desmin, SMA and PDGFR‐β mRNA in whole liver of Group 1 (CDE) and Group 2 (CDE+imatinib) animals. The relative expression levels of CK‐19, desmin, SMA and PDGFR‐β were determined by comparing with GAPDH expression level (100%). Bars represent 2−Δ CT values ± SEM. (b) Expression of CK‐19, desmin, SMA and PDGFR‐β in the imatinib‐treated group compared with the CDE control group. The sample from the control group is indicated by the dotted line, while treated samples are defined by red bars. Bars represent 2−ΔΔ CT values calculated by ΔΔCT method, and data are represented as means.
The relative liver weight of the imatinib‐treated animals was slightly higher than the weight of the controls, but the difference was statistically not significant (Table 1).
Characterization of small hepatocytes
The nuclear hepatocyte nuclear factor 4 (HNF‐4) staining of the small cells supported their hepatocytic commitment. The BrdU incorporation rate was significantly higher in the periportally located small hepatocytes than in the pericentral large hepatocytes in both groups. Furthermore, the small hepatocytes in the imatinib‐treated animals had higher proliferative rate than the small hepatocytes in the controls (Table 1). Quantitative real‐time PCR analysis of cyclin D1 expression of microdissected samples also supported the high proliferative activity of the small hepatocytes (2.4% in small hepatocytes vs. 1.335% in large hepatocytes). The steady‐state level of albumin and peroxisome proliferator‐activated receptor alpha (PPARα) mRNAs was higher or comparable in the small hepatocytes than in the large ones (albumin: 1922% vs. 1535.5%; PPARα: 3.65% vs. 4.95%).
Structural characterization of the liver
To get a ‘functional map’ of the liver parenchyma, we investigated the distribution of three zonal parameters. G6Pase activity and endogenous biotin content of hepatocytes are higher in the periportal cells, while CYP2E1 shows preferential pericentral expression in normal mouse liver (Figure S1). This zonality was almost completely lost in the mice fed with CDE diet (Figure 5a,c). However, it was well preserved, when imatinib was also administered (Figure 5b,d). Similar results were obtained with G6Pase enzyme histochemistry (data not shown). On the contrary, fatty change was more advanced following imatinib treatment (Figure 5e,f).
Figure 5.
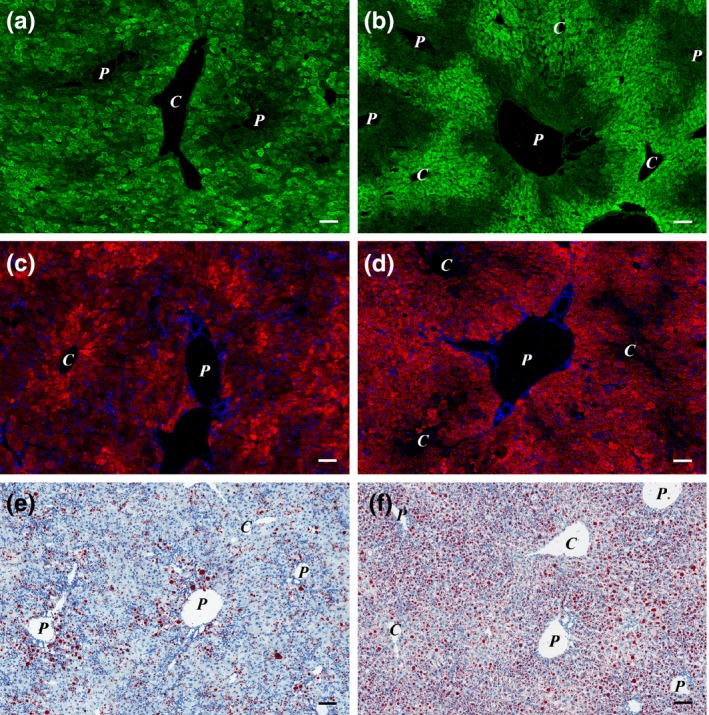
(a–d) Imatinib treatment preserves zonality of the liver in CDE‐fed mice. (a and b) CYP2E1 (green); (c and d) Streptavidin‐TRITC (red) and DAPI (blue) labelled; (e and f) oil red O stained frozen sections of Group 1 (CDE; a, c and e) and Group 2 (CDE+imatinib; b, d and f) animals. The pericentral expression of CYP2E1 (green) and the periportal distribution of endogenous biotin (red) can be observed in the imatinib‐treated animals (b and d), while the normal distribution of these two markers is lost in the mice fed with CDE only (a and c). The degree of steatosis increased in the imatinib‐treated group (f; Group 2). P stands for portal vein, and C stands for central vein. Scale bar for Figure 5: 100 μm.
Discussion
We have investigated the influence of the tyrosine kinase inhibitor imatinib on one of the most widely studied experimental models of chronic hepatic injury/regeneration in mice. Imatinib enhanced the emergence of small hepatocytes; at the same time, the ductular reaction and extent of fibrosis shrinked; and the metabolic zonality of the hepatic parenchyma was preserved. The small hepatocytes showed high proliferative activity and their nuclei contained HNF‐4 – a nuclear factor responsible for terminal differentiation of hepatocytes (Nagy et al. 1994) – and expressed highly hepatocyte‐specific genes: albumin and PPARα. In our interpretation, these results mean that imatinib accelerated the regeneration in liver chronically damaged by the CDE diet.
Imatinib, a small‐molecule tyrosine kinase inhibitor, has been designed originally for the treatment of chronic myelogenous leukaemia, but it turned out to inhibit several tyrosine kinase receptors efficiently, including PDGFR (Hantschel et al. 2008). After all, it was not surprising that in addition to the oncological applications it suppressed fibrogenesis in several experimental systems (Neef et al. 2006; Knight et al. 2008; El‐Agamy et al. 2011; Kim et al. 2012; Kuo et al. 2012) including the CDE model of hepatic fibrosis. We can confirm the anti‐fibrotic effect of imatinib in our experimental model, but in addition, the regeneration of the hepatic parenchyma was also more efficient.
Ductular reaction has been at the centre of studies investigating liver regeneration following chronic liver damage. Its biological role and function is still debated (Riehle et al. 2011; Itoh & Miyajima 2014; Michalopoulos & Khan 2015). The ductules can contribute to liver regeneration by differentiation into hepatocytes, even if this does not happen in all experimental models. There is, however, a general agreement that ductular reactions have unfavourable consequences as well, namely they can cause fibrosis and promote carcinogenesis (Alison et al. 2009; Williams et al. 2014). Our results suggest that by targeting myofibroblasts imatinib can diminish one of the adverse effects, fibrogenesis, without hindering regeneration. In fact, in our experiment the regeneration was even accelerated. Espanol‐Suner et al. (2012) were also able to facilitate regeneration by the prostaglandin I2 analogue iloprost, a drug inhibiting ECM and laminin deposition in the CDE model. These seem to be contradictory results, because it has been assumed that myofibroblasts were required for the growth of ductular reaction, which could be the precursor of hepatocytes. However, the hepatocytic but not ductular origin of regeneration has recently been demonstrated convincingly in the CDE model (Schaub et al. 2014; Tarlow et al. 2014a,b; Yanger et al. 2014). Furthermore, detailed analysis by Van Hul et al. (2009) revealed that CDE first activates myofibroblasts and initiates fibrosis, and the ductular reaction develops later. Knight et al. (2008) reported decreased tumour rate in a chronic CDE experiment if imatinib was administered in parallel. Thus, imatinib seems to promote regeneration while decreasing the adverse by‐products of ductular reaction. The cellular origin of the regenerating small hepatocytes was not addressed in our experiments. The decreased intensity of ductular reaction, together with the emergence of small hepatocytes, indirectly suggests their ductular origin. Regardless of whether they derive from hepatocytes or the ductular reaction, myofibroblasts seem to play a key role in the regulation of chronic injury‐induced tissue reaction. Imatinib influences all these processes through the inhibition of PDGFR‐β, one of the major receptors driving hepatic myofibroblasts. It has been reported in other experimental systems that, besides functional inhibition, small‐molecule tyrosine kinase inhibitors can decrease the expression of this receptor (Liu et al. 2011). We observed this downregulation at both mRNA and protein levels. The decrease in PDGFR‐β was greater than the reduction in the myofibroblast markers desmin and SMA; thus, this is an absolute downregulation, not only the consequence of decreased numbers of expressing cells. Kim et al. (2012) explained the anti‐fibrotic activity of imatinib by inducing cellular senescence and the so‐called senescence‐associated phenotype (Coppé et al. 2010) in myofibroblasts. Interleukin 6 (IL‐6) secretion was increased as part of this well‐orchestrated reaction. IL‐6 is a well‐known regenerative cytokine for the liver (Cressman et al. 1996). Unfortunately, myofibroblasts cannot be microdissected. We were not able to demonstrate upregulated IL‐6 mRNA expression in our RNA samples isolated from whole liver tissue (data not shown). This can be explained by the ‘dilution’ of myofibroblast RNA in whole liver samples, so the participation of this mechanism in the imatinib‐induced accelerated regeneration cannot be excluded.
The hepatocytes are specialized in the liver parenchyma to function with maximal efficiency. Their metabolic function depends on their position along the porto‐central axis of the hepatic lobule. The result of this specialization is the zonation of the liver lobule (Jungermann & Katz 1989). The disruption of zonation results in severe metabolic dysfunction (Sekine et al. 2006). During the CDE‐induced fibrosis and parenchymal injury, the zonal distribution of three different functional markers almost completely disappeared. Imatinib proved to be beneficial in this aspect as well because the zonality was preserved in the treated mice.
Besides all these favourable effects, steatosis was also enhanced following imatinib treatment. Hepatic toxicity, including fatty change in imatinib administration, has already been reported (Nassar et al. 2010), but this side effect might be reduced by optimalization of the dosage or schedule of treatment.
In conclusion, imatinib reduced fibrogenesis but accelerated liver regeneration by enhancing the emergence of small hepatocytes and preserving the functional zonation of liver parenchyma in the CDE model of chronic hepatic injury. This result may reveal a new paradigm for the treatment of chronic liver diseases.
Conflict of interest
The authors declare that there are no conflicts of interest.
Funding source
This study was funded by the Hungarian Scientific Research Fund (OTKA K116301 and PD109201) and János Bolyai Research Scholarship of the Hungarian Academy of Sciences. This research was also supported by the European Union and the State of Hungary and cofinanced by the European Social Fund in the framework of TÁMOP 4.2.4. A/1‐11‐1‐2012‐0001 ‘National Excellence Program’.
Supporting information
Figure S1. Distribution of CYP2E1 and endogenous biotin on frozen sections made from the liver of untreated 8 weeks old C57Bl/6 mouse. A: CYP2E1 (green) shows pericentral expression. B: Streptavidin‐TRITC (red) and DAPI (blue) labeled section. Endogenous biotin shows preferentially periportal localization. Scale bar for Figure S1: 100 µm.
Table S1. Primary antibodies, fluorescent dyes used for the immunohistochemical studies.
Table S2. Gene expression assays used for quantitative real‐time PCR.
References
- Alison M.R., Islam S. & Lin S. (2009) Stem cells in liver regeneration, fibrosis and cancer: the good, the bad and the ugly. J. Pathol. 217, 282–298. [DOI] [PubMed] [Google Scholar]
- Boulter L., Lu W.Y. & Forbes S.J. (2013) Differentiation of progenitors in the liver: a matter of local choice. J. Clin. Invest. 123, 1867–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Desprez P.Y., Krtolica A. & Campisi J. (2010) The senescence‐associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman D.E., Greenbaum L.E., DeAngelis R.A. et al (1996) Liver failure and defective hepatocyte regeneration in interleukin‐6‐deficient mice. Science 274, 1379–1383. [DOI] [PubMed] [Google Scholar]
- Demetris A.J., Seaberg E.C., Wennerberg A., Ionellie J. & Michalopoulos G. (1996) Ductular reaction after submassive necrosis in humans. Special emphasis on analysis of ductular hepatocytes. Am. J. Pathol. 149, 439–448. [PMC free article] [PubMed] [Google Scholar]
- El‐Agamy D.S., Shebl A.M. & Said S.A. (2011) Prevention and treatment of Schistosoma mansoni‐induced liver fibrosis in mice. Inflammopharmacology 19, 307–316. [DOI] [PubMed] [Google Scholar]
- Espanol‐Suner R., Carpentier R., Van Hul N. et al (2012) Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology 143, 1564–1575. [DOI] [PubMed] [Google Scholar]
- Fujita M., Furukawa H., Hattori M., Todo S., Ishida Y. & Nagashima K. (2000) Sequential observation of liver cell regeneration after massive hepatic necrosis in auxiliary partial orthotopic liver transplantation. Mod. Pathol. 13, 152–157. [DOI] [PubMed] [Google Scholar]
- Grimminger F., Schermuly R.T. & Ghofrani H.A. (2010) Targeting non‐malignant disorders with tyrosine kinase inhibitors. Nat. Rev. Drug Discov. 9, 956–970. [DOI] [PubMed] [Google Scholar]
- Grompe M. (2014) Liver stem cells, where art thou? Cell Stem Cell 15, 257–258. [DOI] [PubMed] [Google Scholar]
- Hantschel O., Rix U. & Superti‐Furga G. (2008) Target spectrum of BCR‐ABL inhibitors imatinib, nilotinib, and dasatinib. Leuk. Lymphoma 49, 615–619. [DOI] [PubMed] [Google Scholar]
- Itoh T. & Miyajima A. (2014) Liver regeneration by stem/progenitor cells. Hepatology 59, 1617–1626. [DOI] [PubMed] [Google Scholar]
- Jungermann K. & Katz N. (1989) Functional specialization of different hepatocyte populations. Physiol. Rev. 69, 708–764. [DOI] [PubMed] [Google Scholar]
- Kim Y., Fiel M.I., Albanis E. et al (2012) Anti‐fibrotic activity and enhanced interleukin‐6 production by hepatic stellate cells in response to imatinib mesylate. Liver Int. 32, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B., Tirnitz‐Parker J.E. & Olynyk J.K. (2008) C‐kit inhibition by imatinib mesylate attenuates progenitor cell expansion and inhibits liver tumor formation in mice. Gastroenterology 135, 969–979. [DOI] [PubMed] [Google Scholar]
- Kocabayoglu P., Lade A., Lee Y.A. et al (2015) β‐PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury but not carcinogenesis. J. Hepatol. 63, 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes C. & Häussinger D. (2013) Hepatic stem cell niches. J. Clin. Invest. 123, 1874–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo W.L., Yu M.C., Lee J.F., Tsai C.N., Chen T.C. & Chen M.F. (2012) Imatinib mesylate improves liver regeneration and attenuates liver fibrogenesis in CCl4‐treated mice. J. Gastrointest. Surg. 16, 361–369. [DOI] [PubMed] [Google Scholar]
- Lenzi R., Liu M.H., Tarsetti F. et al (1992) Histogenesis of bile duct‐like cells proliferating during ethionine hepatocarcinogenesis. Evidence for a biliary epithelial nature of oval cells. Lab. Invest. 66, 390–402. [PubMed] [Google Scholar]
- Liu Y., Wang Z., Kwong S.Q. et al (2011) Inhibition of PDGF, TGF‐β and Abl signaling and reduction of liver fibrosis by small molecule Bcr‐Abl tyrosine kinase antagonist Nilotinib. J. Hepatol. 55, 612–625. [DOI] [PubMed] [Google Scholar]
- Michalopoulos G.K. & Khan Z. (2015) Liver stem cells: experimental findings and implications for human liver disease. Gastroenterology 149, 876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Ebato K., Kato H., Arakawa S., Shimizu S. & Miyajima A. (2012) Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166–1175. [DOI] [PubMed] [Google Scholar]
- Nagy P., Bisgaard H.C. & Thorgeirsson S.S. (1994) Expression of hepatic transcription factors during liver development and oval cell differentiation. J. Cell Biol. 126, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P., Teramoto T., Factor V.M. et al (2001) Reconstitution of liver mass via cellular hypertrophy in the rat. Hepatology 33, 339–345. [DOI] [PubMed] [Google Scholar]
- Nassar I., Pasupati T., Judson J.P. & Segarra I. (2010) Histopathological study of the hepatic and renal toxicity associated with the co‐administration of imatinib and acetaminophen in a preclinical mouse model. Malays. J. Pathol. 32, 1–11. [PubMed] [Google Scholar]
- Neef M., Ledermann M., Saegesser H. et al (2006) Oral imatinib treatment reduces early fibrogenesis but does not prevent progression in the long term. J. Hepatol. 44, 167–175. [DOI] [PubMed] [Google Scholar]
- Pinzani M. (2002) PDGF and signal transduction in hepatic stellate cells. Front Biosci. 7, d1720–d1726. [DOI] [PubMed] [Google Scholar]
- Quaglia A., Portmann B.C., Knisely A.S. et al (2008) Auxiliary transplantation for acute liver failure: histopathological study of native liver regeneration. Liver Transpl. 14, 1437–1448. [DOI] [PubMed] [Google Scholar]
- Riehle K.J., Dan Y.Y., Campbell J.S. & Fausto N. (2011) New concepts in liver regeneration. J. Gastroenterol. Hepatol. 26(Suppl 1), 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J.R., Malato Y., Gormond C. & Willenbring H. (2014) Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 8, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S., Lan B.Y., Bedolli M., Feng S. & Hebrok M. (2006) Liver specific loss of beta‐catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology 43, 817–825. [DOI] [PubMed] [Google Scholar]
- Tarlow B.D., Finegold M.J. & Grompe M. (2014a) Clonal tracing of Sox 9 + liver progenitors in mouse oval cell injury. Hepatology 60, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow B.D., Pelz C., Naugler W.E. et al (2014b) Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutsch H.F. (1981) Chemomorphology of liver parenchyma. Qualitative histochemical distribution patterns and quantitative sinusoidal profiles of G6Pase, G6PDH and malic enzyme activity and of glycogen content. Prog. Histochem. Cytochem. 14, 1–92. [PubMed] [Google Scholar]
- Van Hul N.K.M., Abarca‐Quinones I., Sempoux C., Horsmans Y. & Leclercq I.A. (2009) Relation between liver progenitor cell expansion and extra cellular matrix deposition in a CDE‐induced murine model of chronic liver injury. Hepatology 49, 1625–1635. [DOI] [PubMed] [Google Scholar]
- Williams M.J., Clouston A.D. & Forbes S.J. (2014) Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology 146, 349–356. [DOI] [PubMed] [Google Scholar]
- Yanger K., Knigin D., Zong Y. et al (2014) Adult hepatocytes are generated by self‐duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Distribution of CYP2E1 and endogenous biotin on frozen sections made from the liver of untreated 8 weeks old C57Bl/6 mouse. A: CYP2E1 (green) shows pericentral expression. B: Streptavidin‐TRITC (red) and DAPI (blue) labeled section. Endogenous biotin shows preferentially periportal localization. Scale bar for Figure S1: 100 µm.
Table S1. Primary antibodies, fluorescent dyes used for the immunohistochemical studies.
Table S2. Gene expression assays used for quantitative real‐time PCR.


