Abstract
Rice blast, caused by the fungal pathogen Magnaporthe oryzae, is a major constraint to rice production worldwide. In this study, we developed monogenic near-isogenic lines (NILs) NILPi9, NILPizt, and NILPi54 carrying genes Pi9, Pizt, and Pi54, respectively, by marker assisted backcross breeding using 07GY31 as the japonica genetic background with good agronomic traits. Polygene pyramid lines (PPLs) PPLPi9+Pi54 combining Pi9 with Pi54, and PPLPizt+Pi54 combining Pizt with Pi54 were then developed using corresponding NILs with genetic background recovery rates of more than 97%. Compared to 07GY31, the above NILs and PPLs exhibited significantly enhanced resistance frequencies (RFs) for both leaf and panicle blasts. RFs of both PPLs for leaf blast were somewhat higher than those of their own parental NILs, respectively, and PPLPizt+Pi54 exhibited higher RF for panicle blast than NILPizt and NILPi54 (P < 0.001), hinting an additive effect on the resistance. However, PPLPi9+Pi54 exhibited lower RF for panicle blast than NILPi9 (P < 0.001), failing to realize an additive effect. PPLPizt+Pi54 showed higher resistant level for panicle blast and better additive effects on the resistance than PPLPi9+Pi54. It was suggested that major R genes interacted with each other in a way more complex than additive effect in determining panicle blast resistance levels. Genotyping by sequencing analysis and extreme-phenotype genome-wide association study further confirmed the above results. Moreover, data showed that pyramiding multiple resistance genes did not affect the performance of basic agronomic traits. So the way to enhance levels of leaf and panicle blast resistances for rice breeding in this study is effective and may serve as a reference for breeders.
Key Message: Resistant levels of rice blast is resulted from different combinations of major R genes, PPLPizt+Pi54 showed higher resistant level and better additive effects on the panicle blast resistance than PPLPi9+Pi54.
Keywords: Japonica rice, blast resistance, polygene pyramid lines
Introduction
Rice (Oryza sativa) is a staple food crop for more than 50% of the world’s population. Rice blast is caused by Magnaporthe oryzae, a fungus that infects all parts of rice plant but causes the greatest losses when necks and panicles are infected, it has been leading to severe yield losses worldwide and threatening global food security (Liu et al., 2014). Using host resistance has been proven to be the most effective and economical method to control rice blast (Fukuoka et al., 2009). So far, 102 rice blast R genes have been identified (Su et al., 2015; Vasudevan et al., 2015; Zheng et al., 2016). Among them, 27 genes have been cloned: Pib, Pb1, Pita, Pi9, Pi2, Pizt, Pid2, Pi33, Pii, Pi36, Pi37, Pikm, Pit, Pi5, Pid3, Pid3-A4, Pi54, Pish, Pik, Pikp, Pia, PiCO39, Pi25, Pi1, Pi21, P50 and Pi65(t) (Liu et al., 2014; Su et al., 2015; Zheng et al., 2016). The majority of rice blast R genes are associated with a HR according to the gene-for-gene concept, and race specificity is the key feature of this R gene-mediated disease resistance (Fukuoka et al., 2009). Due to highly frequent variation in the M. oryzae population (Bonman, 1992), durable resistance of new rice varieties simply with only a major R gene could be lost quickly, especially when such a variety is grown in large areas (McDonald and Linde, 2002). Therefore, to acquire a durable and broad-spectrum resistant variety, pyramiding multiple R genes into a current rice variety is an important and practicable breeding strategy on controlling blast disease (Hittalmani et al., 2000; Fukuoka et al., 2012). However, with so many available blast R genes, methods to pyramid R genes and actual resistance levels of each R gene are still unknown.
Pi9, at the Piz locus, producing broad-spectrum blast resistance was cloned from chromosome 6 of Oryza minuta, a tetraploid wild species of the Oryza genus (Zhou et al., 2006). Pizt, a multiple allele of Pi9, was isolated from rice cultivar Toride 1 (Mackill and Bonman, 1992). Pi9 and Pizt belong to the NBS-LRR class of R genes (Qu et al., 2006). The NBS-LRR class encodes a receptor-like kinase (Chen et al., 2006). Pi54 (formerly known as Pi-kh) was first identified in the indica rice cultivar HR22 (Kiyosawa and Murty, 1969), and was cloned from the indica rice cultivar Tetep (Sharma et al., 2005). Another donor variety of Pi54 is K3 (Xu et al., 2008). Pi54 belongs to the CC-NBS-LRR class of R genes and expresses a protein that can activate several downstream related pathways against pathogen attack. The above R genes confer broad-spectrum resistance to indica rice blast isolates (Ballini et al., 2008; Rai et al., 2011; Khanna et al., 2015). However, under japonica genetic background, actual levels of blast resistances of Pi9, Pizt, and Pi54 had not been reported. Here, we developed monogenic NILs NILPi9, NILPizt, NILPi54, and PPLs PPLPi9+Pi54, and PPLPizt+Pi54, respectively, by MABB using 07GY31 as the japonica genetic background. This study will not only report an effective way to enhance levels of leaf and panicle blast resistances in rice plants during rice breeding, but also provide important information that levels of panicle blast resistance are actually resulted from different combinations of major R genes, major R genes would not just produce a simple additive effect, gene reaction would also happen.
Materials and Methods
Development of NILPi9, NILPizt, NILPi54, PPLPi9+Pi54, and PPLPi9+Pizt
Using 75-1-127, Toride 1, and K3 as donor parents of genes Pi9, Pizt, and Pi54, respectively, and 07GY31, a blast-susceptible japonica variety, as the recurrent parent, F1 progenies of 75-1-127/07GY31, Toride 1/07GY31, and K3/07GY31 were produced and backcrossed, resulting in three BC1F1 populations, respectively (Figure 1). Markers closely linked with Pi9, Pizt, and Pi54, respectively, were then used to check targeted genes among the above BC1F1 populations. Twenty plants with the targeted gene from each BC1F1 population were selected to backcross with the recurrent parent, up to BC5F1. During the development, no selection for agronomic traits was carried out. After selfing the BC5F1 and identification of corresponding targeted gene among each BC5F2 population, 10 plants with homozygous genotype of the targeted gene were selected randomly from each segregating population. BC5F3 seeds were then harvested individually from each selected BC5F2 plant, constituting NILPi9, NILPiztt, and NILPi54, each featured a blast resistant phenotype.
FIGURE 1.
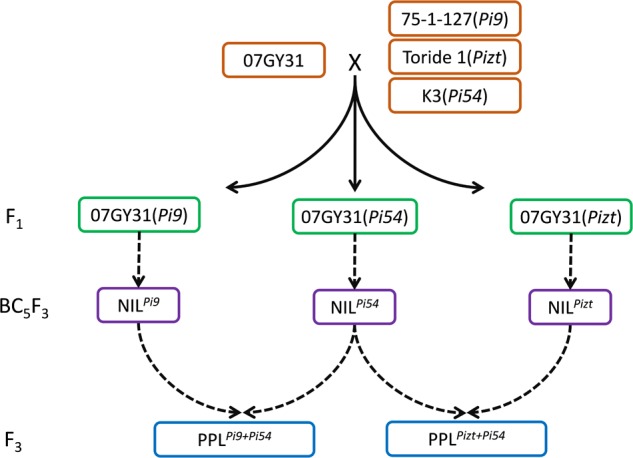
Development of NILPi9, NILPizt, NILPi54, PPLPi9+Pi54, and PPLPizt+Pi54 MAS, marker-assisted selection; NIL, near-isogenic line; PPL, polygene pyramid line.
To further enhance levels of blast resistance, PPLs combining Pi9 with Pi54 and PPLs combining Pizt with Pi54 were then developed by MAS (Figure 1). Crossing NILPi9 with NILPi54 produced F1 with heterozygous genotypes at Pi9 and Pi54 loci. After self-pollination, 211 plants from the resulted F2 segregating population were used to select lines with both homozygous genotypes at Pi9 and Pi54 loci, and nine double homozygous plants were acquired. After determination of genetic background recovery rates, a plant showed the highest genetic background recovery rate (98.52%) was found and named PPLPi9+Pi54. Its seeds were then harvested to plant for the evaluation of both leaf and panicle blast resistances. The development process of PPLs combining Pizt with Pi54 was similar to that of PPLPi9+Pi54, resulting in a PPL plant, named PPLPizt+Pi54, with a genetic background recovery rate of 98.67%. Its seeds were also harvested to plant for the evaluation of both leaf and panicle blast resistances.
To verify whether major-R-gene combination could affect levels of blast resistances, PPLPizt+Pi54 and PPLPi9+Pi54 were selected to cross with 07GY31, respectively. Their F1 progenies were subjected to selfing and hence two F2 segregating populations were produced, respectively. F3 seeds from each F2 single plant were individually harvested and used to plant and determine panicle blast resistance levels which would indirectly reflect panicle blast resistance levels of their parent F2 plants. All plants were grown in the Yangzhou Wantou experimental fields at the Lixiahe Agricultural Research Institute of Jiangsu Province (119°42′ E, 32°39′ N) and in Sanya City in Hainan Province (110°02′E, 18°48′ N).
Evaluation of Seedling Leaf and Panicle Blast Resistances for NILs and PPLs
For pathogen collection for the determination of both leaf and panicle blast resistances, M. oryzae (rice blast pathogen) isolates were obtained from diseased panicles by single-spore isolation. In total, 112 physiological races of M. oryzae were collected, from Anhui, Jiangsu, Hunan, Hubei, Hainan, Henan, and Guangdong, for the leaf blast resistance determination for the above NILs, PPLs, 07GY31, Tetep, Zhenglong 13, Sifeng 43, Dongnong 363, Kanto 51, Hejiang 18, and LTH. Wu et al.’s (2015) method was referred to for leaf blast resistance evaluation. 7 days after inoculation, disease reaction of plant leaves was recorded in accordance with standard methods (Bonman, 1992). Test fields (test nurseries) were located at the experimental fields at the Lixiahe Agricultural Research Institute of Jiangsu Province (119°42′ E, 32°39′N). Resistance levels of lines were measured with RFs. RFs meaning proportions of plants showing resistant phenotype, were calculated according to the following formula: RF = R/(R + S) × 100%, where R is the total number of plants showing resistant phenotype, while S is the total number of plants showing sensitive phenotype. For the determination of panicle blast resistance for the above NILs, PPLs, 07GY31, in total, 64 M. oryzae isolates from Anhui, Guangdong, Henan, Hubei, Hainan, Jiangsu, and Zhejiang were utilized. 192 experiment plots were involved in a randomized block design to realize three biological repeats, and 30 plants were grown in each plot. All plants were grown under natural field conditions. One booting panicle was selected for each plant based on the principle that the distance between pulvini of flag leaf and penultimate leaf is 4 cm and was hence injected with 1 mL conidial suspension at a concentration of 5 × 104 conidia/mL (Puri et al., 2009). Panicle blast resistance evaluation was based on the severity of symptoms in infected panicles 30 days after heading according to Titone et al.’s (2015) methods.
All NILs, PPLs, and 07GY31 were also subjected to the determination of blast resistance in blast nurseries in two hot spot fields (one in Xinyi City in Jiangsu Province and the other in Hefei City in Anhui Province). Field rows were involved in a CRBD, and produced three replications. For each line or variety, 60 plants were planted in five rows, meaning 12 plants per row. The evaluation of leaf blast and panicle blast severity in each of the NILs and the recurrent parent was performed using a 0–9 ordinal scale ([IRRI], 2002), where 0–1 = highly resistant, 2–3 = resistant, 4 = mildly resistant, 5–6 = mildly susceptible, 7 = susceptible and 8–9 = highly susceptible, resistant level reflected as RPR according to the following formula: RPR = (HR + R + MR)/60 × 100%, where HR is the total number of plants showing high resistant phenotype, while R is the total number of plants showing resistant phenotype and MR is the total number of plants showing mildly resistant phenotype. RPR were determined 30 days after heading. For further verifying results from the comparison of panicle blast resistance levels of NILs and PPLs, three fields surrounded by virgin land were selected from Jiangsu and Anhui, respectively.
DNA Extraction and Genotyping
For MAS during each generation, three-week-old rice leaves were individually collected from NIL and PPL plants, and immediately frozen in liquid nitrogen and stored at -80°C for future DNA extraction. Genomic DNA was rapidly extracted by the TPS method for future molecular marker analysis (Rogers and Bendich, 1985). PCR amplification was conducted according to the workflow described by Xiao et al. (2016). Molecular markers closely linked with major Pi genes Pizt, Pi9, and Pi54, respectively, (Supplementary Table 1) were used for MAS during the development of each NIL population and each PPL population.
For GBS analysis, genomic DNA was extracted from 100 mg (fresh weight) of three-week-old rice leaf tissue, using the DNA Secure Plant Kit (Qiagen, USA) and following manufacturer’s instruction. Each extracted genomic DNA was qualitatively estimated by electrophoresis on 1% agarose gels, quantitatively measured by Biophotometer Plus (Eppedorf, Germany), and diluted to100 ng/μL with TE buffer. Diluted genomic DNA was then stored at -20°C.
Genome Alignment and Variant Calling
Genotyping by sequencing was conducted by the Illumina HiSeqTM 2000 system generating 90 bp paired-end read. Raw reads with a mapping quality score of less than 20 were discarded. BWA v0.5.9 (Li and Durbin, 2010) was used to map raw paired-end reads to the japonica Os-Nipponbare-Reference-IRGSP-1.0 genome assembly (International Rice Genome Sequencing Project1.01). Alignment files were then input into the GATK V1.2 (McKenna et al., 2010) to identify SNPs. Multiple SNP calling was performed using the GATK Unified Genotyper caller. SNPs with quality scores of >20 and coverage of between two and twice the mean coverage of all accessions were selected. If a SNP was called at the same position in more than one accession, it would be retained. XP-GWAS (Yang et al., 2015)2 was then conducted to find out blast-resistance-related QTLs based on the acquired SNP genotypes.
Evaluation of Agronomic Traits for NILs
To check whether introduction of a major Pi gene could affect agronomic traits and select NILs with good agronomic traits, two test sites, one located at the Lixiahe Agricultural Research Institute of Jiangsu Province and the other in Anhui Province, were used to evaluate basic yield-related traits including period from HD to date of 50% flowering, PH, PN, GNP, SR, 1000-GW and YPP, of NILs. The evaluation was performed based on a CRBD with two replications. Each line involving 120 plants was planted in 10 rows. Planting methods were the same as the above. Agronomic traits were measured according to the Standard Evaluation System for Rice (International Rice Research Institute [IRRI], 2002). Five single plants between the second and the sixth rows in each plot were taken for measurements of agronomic traits.
Result
NILPizt, NILPi54, and NILPi9 Exhibited Significantly Higher RFs for Both Leaf and Panicle Blasts than 07GY31
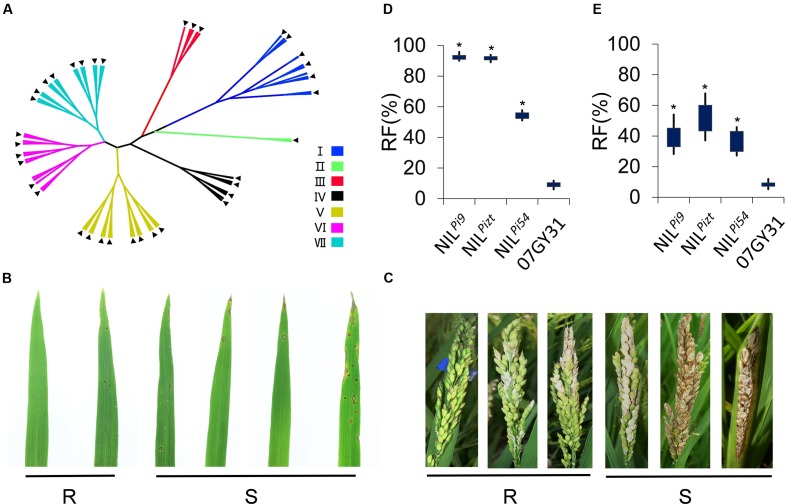
Genetic background recovery rates of NILPi9, NILPi54, and NILPizt were found to be 98.25, 97.33, and 97.82%, respectively (Supplementary Table 2). These NILs were then subjected to both inoculations of leaf and panicle blasts for the determination of resistances. Rice blast pathogens were collected from 7 regions of China, namely, Jiangsu, Hunan, Anhui, Hubei, Sanya, Henan, and Guangdong. Totally 112 physiological races were involved. Based on resistance responses of varieties Tetep, Zhenlong13, Sifeng4, Dongnong363, Guandong51, Hejiang18, and LTH to the races, we divided these races into seven types, namely, typed I, II, III, IV, V, VI, and VII (Figure 2A). Plants rated 0–3 in the identification of resistance to either leaf or panicle blast were considered to be resistant to the corresponding blast and each got an “R,” while plants rated 4–5 each got an “S” (Figures 2B,C). All the 112 races were involved in the identification of resistance to leaf blast. Finally, we found RFs of NILPizt, NILPi54, and NILPi9 were significantly enhanced for leaf blast (P < 0.001), compared to that of 07GY31. NILPizt and NILPi9 exhibited significantly higher RFs than NILPi54 (P < 0.001) (Figure 2D). Data indicated that resistance levels of NILPizt, NILPi54, and NILPi9 for panicle blast were also significantly enhanced, compared to that of 07GY31, based on 64 selected to identify resistance responses. Moreover, NILPizt and NILPi9 exhibited significantly higher RF than NILPi54 (Figures 2D,E).
FIGURE 2.
Evaluation of leaf and panicle blast resistances for NILs and PPLs.
(A) Seven groups classification of 112 physiological races based on the resistance responses of varieties Tetep, Zhenlong13, Sifeng4, Dongnong363, Kanto 51, Hejiang18, and LTH; (B) Classification of leaf blast resistance; (C) Classification of panicle blast resistance; (D) Resistance levels of NILPizt, NILPi54, NILPi9, and 07GY31 for leaf blast; (E) Resistance levels of NILPizt, NILPi54, NILPi9, and 07GY31for panicle blast. R: Resistant; S: Susceptible. ∗Statistically significant at P < 0.001 level;  :64 selected physiological races for the determination of panicle blast resistance.
:64 selected physiological races for the determination of panicle blast resistance.
PPLPizt+Pi54Exhibited Higher RF for Panicle Blast than PPLPi9+Pi54
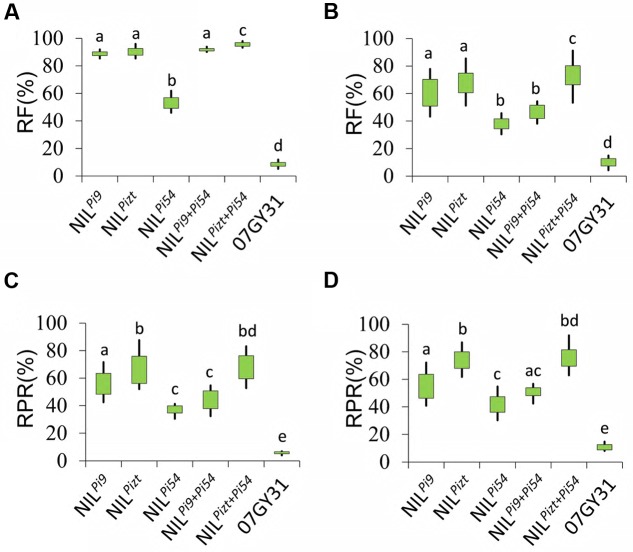
Pyramiding multiple major resistance genes is one of the main methods to enhance variety resistance. NILPizt and NILPi9 were then crossed with NILPi54, respectively. Finally, two polygene pyramid lines, PPLPizt+Pi54 and PPLPi9+Pi54, were developed using MAS. Data showed that RFs of all the PPLs for leaf blast were somewhat higher than those of their own parental NILs, respectively, (Figures 3A,B), and PPLPizt+Pi54 exhibited higher RF for panicle blast than NILPizt and NILPi54 (P < 0.001), hinting an additive effect on the resistance. However, PPLPi9+Pi54 exhibited lower RF for panicle blast than NILPi9 (P < 0.001), failing to realize an additive effect. For further verification, two fields surrounded by virgin land were selected for the determination of resistance to panicle blast from Anhui and Jiangsu, respectively. Interestingly, PPLPizt+Pi54 showed higher resistant level for panicle blast and better additive effects on the resistance than PPLPi9+Pi54 (Figures 3C,D). In these fields, PPLPi9+Pi54 still showed significantly lower resistant level for panicle blast than NILPi9 (P < 0.001), meaning major R genes interacted with each other in a way more complex than additive effect in determining panicle blast resistance levels.
FIGURE 3.
Blast resistance comparison between NILs and PPLs. (A) Resistance frequencies of NILs, PPLs, and 07GY31 for leaf blast; (B) Resistance frequencies of NILs, PPLs, and 07GY31 for panicle blast; (C) Resistance levels of NILPizt, NILPi54, NILPi9, and 07GY31for panicle blast; (C) Resistance levels of NILs, PPLs, and 07GY31 for the disease spectrum at Anhui test site; (D) Resistance levels of NILs, PPLs, and 07GY31 for the disease spectrum at Jiangsu test site. Entries with different letters were statistically significantly different at P < 0.001 level; RPR; RF.
Levels of Panicle Blast Resistance Were Determined by Major Gene Combinations
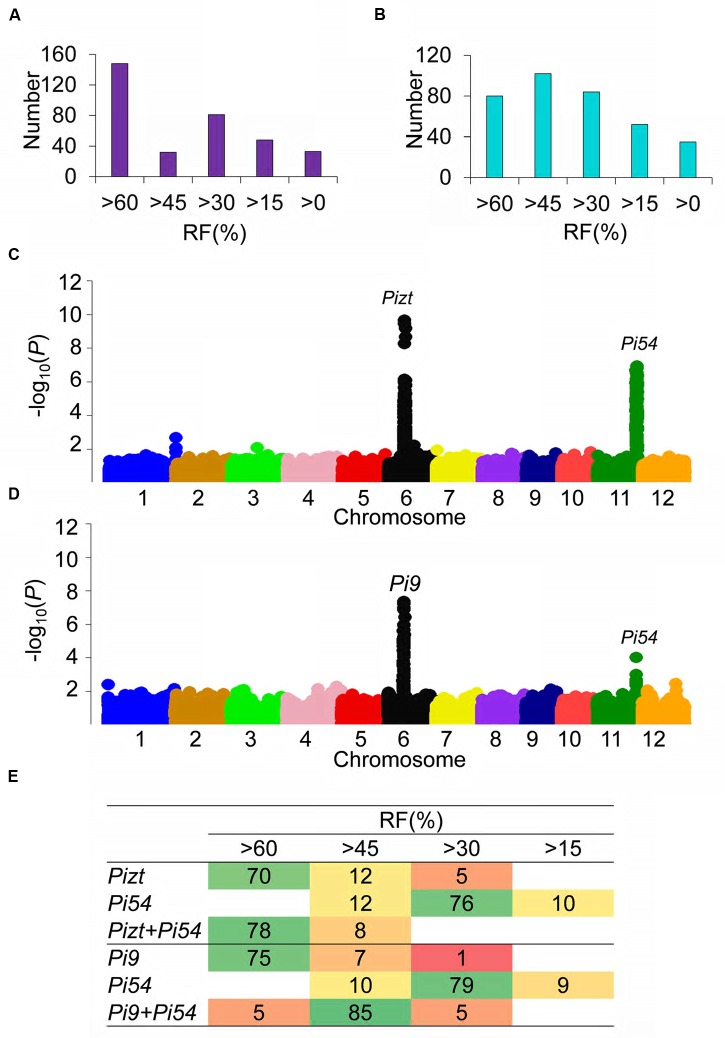
For further verifying the above results, three physiological races were selected from each of the following 7 regions: Jiangsu, Hunan, Anhui, Hubei, Henan, Guangdong, and Sanya. These 21 physiological races were involved in determining the resistance level of each F2 single-plant in each F2 segregating population (Figure 4A). Panicle blast resistance of each F2 single-plant was reflected by RFs of its 20 offspring F3 plants. F2 segregating population of PPLPi9+Pi54/07GY31 was constituted by 342 single plants, and F2 segregating population of PPLPizt+Pi54/07GY31 was constituted by 353 single plants. 120 single F2 plants with RFs of either more than 60% or less than 15% were selected from each F2 segregating population to construct resistant and susceptible pools; each pool contained 60 single plants. Each mixed pool was then subjected to GBS. Raw reads of each pool were individually assembled using the Os-Nipponbare-Reference-IRGSP-1.0 genome assembly as the reference genome. A sequence with a total length of 3.3 Gb, an average sequencing depth of 5.8× and a coverage of 91.2% over the reference genome, and totally 49,366 SNPs were used to perform QTL mapping for each F2 segregating population.
FIGURE 4.
Extreme-phenotype genome -wide association study of QTL loci related to panicle blast resistance. (A) Panicle blast resistance classification in the F2 segregating population of PPLPizt+Pi54/07GY31; (B) Panicle blast resistance classification in the F2 segregating population of PPLPi9+Pi54/07GY31; (C) XP-GWAS of panicle-blast-resistance-related loci in the F2 segregating population of PPLPizt+Pi54/07GY31; (D) XP-GWAS of panicle-blast-resistance-related loci in the F2 segregating population of PPLPi9+Pi54/07GY31. (E) Distributions of Pi9, Pizt, and Pi54 in the F2 segregating population of PPLPizt+Pi54/07GY31 and the F2 segregating population of PPLPi9+Pi54/07GY31. Numbers in the table of Figure 4E is the number of plants with Pi9, Pizt, Pi54, Pizt+Pi54, Pi9+Pi54.
As shown in Figure 4B, two strong peaks were detected for the F2 segregating population of PPLPizt+Pi54/07GY31, indicating that only two major genes would affect the resistance to panicle blast in this population. One peak was found to be associated with the range from 10.1 to 11.29 Mb on chromosome 6, and this range completely covered gene Pizt (P = 2.26 × 10-10); and the other peak was found to be associated with the range from 24.98 to 25.96 Mb on chromosome 11, this range completely covered gene Pi54 (P = 1.19 × 10-9) (Figure 4C). The above indicated that Pizt and Pi54 should be the only two genes determining panicle blast resistance in the F2 segregating population of PPLPizt+Pi54/07GY31.
We further used tightly linked markers to scan the genome of each plant and work out the distributions of Pizt and Pi54 in the F2 segregating population of PPLPizt+Pi54/07GY31. As shown in Figure 4E, RFs of more than 60% mainly involved plants with mono-gene Pizt, and plants pyramiding both Pizt and Pi54, while RFs varying from 30 to 45% mainly involved plants with mono-gene Pi54. These distributions indicated that pyramiding both Pizt and Pi54 significantly enhanced panicle blast resistance levels.
Interestingly, in the F2 segregating population of PPLPi9+Pi54/07GY31, only one strong peak (P = 4.46 × 10-8) was detected (Figure 4D). This peak was found to be associated with the range from 10.09 to 10.47 Mb on chromosome 6, and this range completely covered gene Pi9. Besides this strong peak, only one weak peak (P = 8.12 × 10-3) was detected, this peak was associated with the range from 24.87 to 25.99 Mb on chromosome 11, and this range completely covered gene Pi54.
Tightly linked markers were also used to scan the genome of each plant and work out the distributions of Pi9 and Pi54 in the F2 segregating population of PPLPi9+Pi54/07GY31. Different from the F2 segregating population of PPLPizt+Pi54/07GY31, RFs of more than 60% in the F2 segregating population of PPLPi9+Pi54/07GY31 mainly involved plants with mono-gene Pi9, while RFs ranging from 40 to 60% mainly involved plants pyramiding both Pi9 and Pi54 (Figure 4E). PPLPizt+Pi54 showed higher resistant level for panicle blast and better additive effects on the resistance than PPLPi9+Pi54. Therefore, we confirmed that rice plant resistance to panicle blast was directly determined by the combination of resistance genes.
Introduction of Resistance Gene(s) Had No Effects on Basic Agronomic Traits
NILs, PPLs, and 07GY31 were subjected to the phenotyping of the following basic agronomic traits: growth period, 1000-grain weight, effective panicles per plant, grain number per panicle, seed setting rate, and yield. Phenotypic data did not indicate any significant difference between other lines and 07GY31, except the chalkiness of NILPiz and PPLPizt+Pi54 was higher than 07GY31. It was suggested that significant enhancement of rice blast resistance levels did not affect levels of else agronomic traits, since genetic background recovery rates of the above NILs were high (Table 1).
Table 1.
Performance of basic agronomic traits in NILs and PPLs.
| Plant height(cm) | Heading days(d) | Panicle number | Grain number per panicle | Seeding rate (%) | 1,000 grain weight (g) | Yield (kg) | Head rice rate | Chalkiness rate | Amylose content | |
|---|---|---|---|---|---|---|---|---|---|---|
| NILPizt | 100.45 ± 3.04a | 98 ± 1.41a | 12.15 ± 0.64a | 129.25 ± 6.15a | 91.85 ± 0.78a | 25.15 ± 0.36a | 551.2 ± 7.07a | 74.15 ± 0.78a | 31.85 ± 0.78b | 16.9 ± 0.85a |
| NILPi54 | 99.2 ± 1.84a | 97.5 ± 0.71a | 12.25 ± 1.34a | 132.85 ± 7.42a | 92.85 ± 0.92a | 25.45 ± 0.35a | 561.5 ± 12.02a | 73.15 ± 0.92a | 22.65 ± 4.17a | 17.45 ± 1.49a |
| NILPi9 | 100.35 ± 1.49a | 96.5 ± 0.701a | 12.65 ± 0.21a | 124.70 ± 3.25a | 91.95 ± 1.06a | 25.2 ± 0.85a | 561.5 ± 7.78a | 74.35 ± 0.21a | 24.45 ± 0.92a | 16.35 ± 0.78a |
| PPLPizt+Pi54 | 99.5 ± 3.39a | 97.5 ± 0.71a | 11.95 ± 0.35a | 130.75 ± 7.71a | 91.7 ± 1.27a | 25.45 ± 0.49a | 562 ± 19.79a | 74.15 ± 0.49a | 35.45 ± 4.17b | 16.75 ± 0.78a |
| PPLPi9+Pi54 | 101.35 ± 1.48a | 98.2 ± 0.6a | 12.65 ± 1.49a | 125.65 ± 4.31a | 92.4 ± 1.13a | 25.1 ± 0.57a | 553 ± 4.24a | 74.35 ± 0.35a | 26.2 ± 1.27a | 16.15 ± 0.79a |
| 07GY31(CK) | 99.45 ± 0.78a | 98.2 ± 0.61a | 11.75 ± 0.49a | 128.65 ± 2.62a | 92.15 ± 0.35a | 25.05 ± 0.35a | 556 ± 8.49a | 73.55 ± 0.92a | 24.75 ± 4.45a | 16.1 ± 1.42a |
Entries with different letters were statistically significantly different at P < 0.001 level.
Discussion
Rice blast is one of the most destructive diseases. Based on related rice developmental stages, this disease is divided into leaf blast and panicle blast. Panicle blast is highly concentrated on by breeders and geneticists, because it is directly related to paddy rice production safety and rice quality. Pyramiding rice blast resistance genes has been becoming an effective strategy to develop a new variety with long lasting resistance. Currently, several broad-spectrum resistance genes such as Pi9, Pi2, Pizt, and Pi54, have already been cloned, but their distribution frequencies in elite parents of Chinese indica and japonica rice were low (Huang et al., 2015; Wu et al., 2015; Tian et al., 2016). Moreover, research in levels of resistances to leaf and panicle blasts of rice plants pyramiding either Pi9 or Pizt, and Pi54 hasn’t yet been reported. This study introduced Pi9, Pi54, and Pizt into 07GY31, respectively. Resulted monogenic NILs showed significantly enhanced levels of resistances to leaf and panicle blasts. Further, gene pyramiding was conducted in order to realize the enhancement of resistance levels. Resulted lines pyramiding either Pizt or Pi9, and Pi54 exhibited an additive effect on the resistance to leaf blast, compared to corresponding NILs. However, resistance levels of panicle blast were overall lower than those of leaf blast, similar to result reported by Wu et al. (2016). It might be because the pathogenetic process of panicle blast involved the development process of spike types and the rice maturation process, and ultimately, levels of resistance to panicle blast were comprehensive responses of plants to panicle-neck blast, grain blast and spike-neck blast, while leaf blast was simpler. Therefore, overall symptoms of panicle blast were prone to be more serious. Moreover, seedling leaf and young panicle are two different organs at different developmental stages; organ-specific genes might also affect plant resistances. For example, miR156 was specifically highly expressed in young panicles of rice, and existed along with the whole development process of young panicles (Wang et al., 2010). Coincidentally, in wheat, expression levels of miR156 had been reported to be related to levels of resistance to wheat powdery mildew caused by a fungal pathogen (Xin et al., 2010, and Bej and Basak, 2014). In additional, OsRac1, WRKY19, OsGF14b, and MAPK3/6 have been identified as downstream genes of R genes participate in resistance regulation (Liu et al., 2014). Liu et al. (2016) also reported low correlation between the levels of leaf and panicle blast resistance observed in the field, and provided that WRKY71 up-regulates the expression of OsGF14b by combining with the promoter of OsGF14b, which resulted in the level of panicle blast resistance is enhanced while the level of seedling leaf blast resistance is lowered. Therefore, it’s suggesting that organ-specifically expressed genes and downstream-regulating genes play an important role on blast resistant difference between seedling and panicle.
In this study, we also observed that lines pyramiding both Pizt and Pi54 showed an additive effect on resistance to panicle blast, while lines pyramiding both Pi9 and Pi54 exhibited lower resistance levels than Pi9 monogenic lines. Similar phenomena had been reported by Wang et al. (1994), negative interaction wherein some combinations of R genes actually cause low resistance, lines pyramiding both Piz5 and Pizt showed lower resistance levels than Piz5 monogenic lines in natural disease nurseries in IRRI (Los Banos, Laguna, Philippines) and India (Hittalmani et al., 2000). Therefore, pyramiding resistance genes did not always lead to enhanced resistance levels (Tabien et al., 2000), different combinations of resistance genes might directly resulted in various resistance levels. It is a pity that no follow-up validation studies have been reported. In this study, we had already located panicle blast resistance QTLs of the F2 segregating population of PPLPizt+Pi54/07GY31 and the F2 segregating population of PPLPi9+Pi54/07GY31. Data confirmed that PPLPizt+Pi54 were more resistant to panicle blast than PPLsPi9+Pi54. Therefore, it’s suggest that differences in molecular immune mechanisms existing between Pi9 and Pizt. Rice blast major resistance genes express proteins belong to the NOD-like receptor NLR family (nucleotide-binding domain leucine-rich repeat containing). NLRs can activate downstream gene expression programs after recognizing pathogens and hence initiate plant immune responses (Orbach et al., 2000; Qu et al., 2006; Hayashi and Yoshida, 2009). Therefore, genes that are downstream of and interact with major rice blast resistance genes also can affect plant resistance levels. Kawano et al.’s (2010) research mentioned a domain that is in Pi9 and called NB-ARC. They pointed out that OsRac1 initiated immune reactions after interacting with the NB-ARC domain of Pi9, but failed to combine with Pizt. Therefore, though Pi9 and Pizt exist as multiple alleles at Piz locus, their resistance mechanisms may have already been differentiated. Wu et al. (2016) reported NILs each combining the genetic background of indica rice variety Yangdao6 with one allele (Pizt, Pi2, Pigm, Pi40, Pi9, or Piz) at Piz locus, exhibited significantly different levels of panicle blast resistance. This result is similar to our result that resistance levels of NILPi9 and PPLPi9+Pi54 were more significantly different than those of NILPizt and PPLPizt+Pi54. Therefore, different major resistance genes may determine different molecular immune pathways, and which major resistance genes to select and pyramid becomes the key question during designing molecular breeding.
During MABB period, backcrossing is one of the most common practices for removing donor parent chromosomes both linked and unlinked to the target gene. However, large linkage drags always found in high backcross generation. A fragment with 6.4 Mb around the blast R gene Pi33 from a wild rice was found in IR64 introgression lines (Ballini et al., 2007). And 11.6 Mb of chromosomal fragment around Pita from donor (Tetep) was also identified in BC5F2 individuals (Jia, 2009). Recently, some study also reported that most agriculture traits of NILs with high background recover rate were similar with recurrent parents, but some relating to grain quality traits (gel consistency, amylose content, etc.), heading date and plant height could be altered (Jiang et al., 2015; Khanna et al., 2015), it’s suggest that some genes or QTLs from the linkage drags influence on traits under receptor genetic background. Similar observation also present in our study, all NILs and PPLs that the genetic background recovery rates of are over 97% failed to exhibit significantly lower levels of yield trait than 07GY31, except the chalkiness rate of NILPizt and PPLPizt+Pi54 is higher than 07GY31. Since Toride 1, the donor of Pizt gene, possess higher chalkiness rate than 07GY31, some fragments impressing chalkiness as linkage drags introduced into 07GY31 during the MABB process. Anyway, the goal of MABB is recover phenotypically similar if not better improved lines than that of the receptor parent, with all desired plant type and grain quality, in short span of selection time. In our study, PPLPizt+Pi54 exhibited higher levels of resistances to leaf and panicle blasts than PPLPi9+Pi54. These results are already enough to provide a theoretical basis for deciding which major resistance genes to select and pyramid for developing a highly resistant variety. PPLPizt+Pi54 showed the highest levels of resistances and the highest yield in the multiple-environment trial. Potential material for future blast resistance breeding is already available.
Author Contributions
NX, YW, AL, and CL participated in the study conception and design. LY, CP, and YC contributed to DNA extraction and molecular marker identification. GL, YL, XZ, ZW, and ZD contributed to data analysis. NX wrote the manuscript. AL and LZ critically revised the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Key Studying and Developing Project of Jiangsu Province for Modern Agriculture (BE2015336), Natural Science Foundation of Jiangsu Province (BK20151318), National Natural Science Foundation of China (31401365), National Modern Agricultural Industry Technology System Special Fund (CARS-01-45), the Priority Academic Program Development of Jiangsu Higher Education Institutions and International Atomic Energy Agency (12228/RO).
Abbreviations
- BWA
Burrows-Wheeler Aligner
- CC-NBS-LRR
coiled coil-nucleotide binding site-leucine rich repeats
- CRBD
completely randomized block design
- GATK
Genome Analysis Toolkit
- GBS
genotyping by sequencing
- GNP
grain number per plant
- GW
1000-grain weight
- GWAS
genome-wide association study
- HD
heading day
- HR
hypersensitive response
- IRRI
International Rice Research Institute
- LTH
Lijiangxintuanheigu
- MABB
marker assisted backcross breeding
- MAS
marker associated selection
- NB-ARC
nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4
- NBS-LRR
nucleotide binding site-leucine rich repeat
- NILs
near-isogenic lines
- NLR
NOD-like receptor
- NOD
nucleotide-binding oligomerization domain
- PH
plant height
- PN
panicle number per plant
- PPLs
polygene pyramid lines
- RFs
resistance frequencies
- RPR
resistant panicle rate
- SNPs
single nucleotide polymorphisms
- SR
seeding rate
- XP-GWAS
extreme-phenotype genome-wide association study
- YPP
yield per plant
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01918/full#supplementary-material
References
- Ballini E., Berruyer R., Morel J. B., Lebrun M. H., Notteghem J. L., Tharreau D. (2007). Modern elite rice varieties of the ‘Green Revolution’ have retained a large introgression from wild rice around the Pi33 rice blast resistance locus. New Phytol. 175 340–350. 10.1111/j.1469-8137.2007.02105.x [DOI] [PubMed] [Google Scholar]
- Ballini E., Morel J. B., Droc G., Price A., Courtois B., Notteghem J. L., et al. (2008). A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21 859–868. 10.1094/MPMI-21-7-0859 [DOI] [PubMed] [Google Scholar]
- Bej S., Basak J. (2014). MicroRNAs: the potential biomarkers in plant stress response. Am. J. Plant Sci. 5 748–759. 10.1007/s00438-014-0953-y [DOI] [Google Scholar]
- Bonman J. M. (1992). Durable resistance to rice blast disease-environmental influences. Euphytica 63 115–123. 10.1007/BF00023917 [DOI] [Google Scholar]
- Chen X. W., Shang J. J., Chen D. X., Lei C. L., Zhai W. X., Liu G. Z., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46 229–233. 10.1111/j.1365-313X.2006.02739.x [DOI] [PubMed] [Google Scholar]
- Fukuoka S., Mizobuchi R., Saka N., Ivan S., Matsumoto T., Okuno K., et al. (2012). A multiple gene complex on rice chromosome 4 is involved in durable resistance to rice blast. Theor. Appl. Genet. 125 551–559. 10.1007/s00122-012-1852-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., Ebana K., et al. (2009). Loss of function of a prolinecontaining protein confers durable disease resistance in rice. Science 325 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Yoshida H. (2009). Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 57 413–425. 10.1111/j.1365-313X.2008.03694.x [DOI] [PubMed] [Google Scholar]
- Hittalmani S., Parco A., Mew T., Zeigler R. S., Huang N. (2000). Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100 1121–1128. 10.1007/s001220051395 [DOI] [Google Scholar]
- Huang X. H., Yang S. H., Gong J. Y., Zhao Y., Feng Q., Gong H., et al. (2015). Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6: 6258 10.1038/ncomms7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Research Institute [IRRI] (2002). Standard Evaluation System for Rice (SES), 4 Edn Los Banos: International Rice Research Institute (IRRI), 15–16. [Google Scholar]
- Jia Y. (2009). Artificial introgression of a large chromosome fragment around the rice blast resistance gene Pi-ta in backcross progeny and several elite rice cultivars. Heredity 103 333–339. 10.1038/hdy.2009.95 [DOI] [PubMed] [Google Scholar]
- Jiang J. F., Mou T. M., Yu H. H., Zhou F. S. (2015). Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance using Pi2 gene. Rice 8: 11 10.1186/s12284-015-0048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., et al. (2010). Activation of a Rac GTPase by the hNLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7 362–375. 10.1016/j.chom.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Khanna A., Sharma V., Ellur R. K., Shikari A. B., Singh U. D., Prakash G., et al. (2015). Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor. Appl. Genet. 128 1243–1259. 10.1007/s00122-015-2502-4 [DOI] [PubMed] [Google Scholar]
- Kiyosawa S., Murty V. V. S. (1969). The inheritance of blast-resistance in india rice variety, HR-22. Jpn. J. Breed. 19 269–276. 10.1270/jsbbs1951.19.269 [DOI] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yang J. Y., Zhang S. H., Zhao J. L., Feng A. Q., Yang T. F., et al. (2016). OsGF14e positively regulates panicle blast resistance in rice. Biochem. Biophys. Res. Communi. 471 247–252. 10.1016/j.bbrc.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Liu W. D., Liu J. L., Leach J. E., Wang G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52 213–241. 10.1146/annurev-phyto-102313-045926 [DOI] [PubMed] [Google Scholar]
- Mackill D. J., Bonman J. M. (1992). Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82 746–749. 10.1094/Phyto-82-746 [DOI] [Google Scholar]
- McDonald B. A., Linde C. (2002). The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124 163–180. 10.1023/A:1015678432355 [DOI] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: a map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Farrall L., Sweigard J. A., Chumley F. G., Valent B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12 2019–2032. 10.2307/3871102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri K. D., Shrestha S. M., Chhetri G. B. K., Joshi K. D. (2009). Leaf and neck blast resistance reaction in tropical rice lines under green house condition. Euphytica 165 523–532. 10.1007/s10681-008-9771-9 [DOI] [Google Scholar]
- Qu S. H., Liu G. F., Zhou B., Bellizzi M., Zeng L., Dai L. Y., et al. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172 1901–1914. 10.1534/genetics.105.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A. K., Kumar S. P., Gupta S. K., Gautam N., Singh N. K., Sharma T. R. (2011). Functional complementation of rice blast resistance gene Pi-kh(Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J. Plant Biochem. Biotechnol. 20 55–65. 10.1007/s13562-010-0026-1 [DOI] [Google Scholar]
- Rogers S. O., Bendich A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5 69–76. 10.1007/BF00020088 [DOI] [PubMed] [Google Scholar]
- Sharma T. R., Madhav M. S., Singh B. K., Shanker P., Jana T. K., Dalal V., et al. (2005). High-resolution mapping, cloning and molecular characterization of the Pi-k(h) gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genomics 274 569–578. 10.1007/s00438-005-0035-2 [DOI] [PubMed] [Google Scholar]
- Su J., Wang W. J., Han J. L., Chen S., Wang C. Y., Zeng L. X., et al. (2015). Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor. Appl. Genet. 128 2213–2225. 10.1007/s00122-015-2579-9 [DOI] [PubMed] [Google Scholar]
- Tabien R. E., Li Z., Paterson A. H., Marchetti M. A., Stansel J. W., Pinson S. R. M., et al. (2000). Mapping of four major rice blast resistance genes from ‘Lemont’ and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 101 1215–1225. 10.1007/s001220051600 [DOI] [Google Scholar]
- Tian D., Chen Z., Chen Z., Zhou Y., Wang Z., Wang F., et al. (2016). Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice 9: 19 10.1186/s12284-016-0091-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titone P., Mongiano G., Tamborini L. (2015). Resistance to neck blast caused by Pyricularia oryzae in Italian rice cultivars. Eur. J. Plant Pathol. 142 49–59. 10.1007/s10658-014-0588-1 [DOI] [Google Scholar]
- Vasudevan K., Gruissem W., Bhullar N. K. (2015). Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 5:15678 10.1038/srep15678.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Mackill D. J., Bonman J. M., McCouch S. R., Champoux M. C., Nelson R. J. (1994). RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xie W. B., Chen Y., Tang W. J., Yang J. Y., Ye R. J., et al. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61 752–766. 10.1111/j.1365-313X.2009.04100.x [DOI] [PubMed] [Google Scholar]
- Wu Y., Xiao N., Yu L., Pan C., Li Y., Zhang X., et al. (2015). Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PLoS ONE 10:e0126130 10.1371/journal.pone.0126130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yu L., Pan C., Dai Z., Li Y., Xiao N., et al. (2016). Development of near-isogenic lines with different alleles of Piz locus and analysis of their breeding effect under Yangdao 6 background. Mol. Breed. 36 1–12. 10.1007/s11032-016-0433-7 [DOI] [Google Scholar]
- Xiao N., Huang W. N., Li A. H., Gao Y., Li Y. H., Pan C. H., et al. (2016). Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Thero. Appl. Genet. 128 173–185. 10.1007/s00122-014-2420-x [DOI] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Xie C., Peng H., Ni Z., et al. (2010). Diverse set of MicroRNAs are responsive to powdery mildew infection and heat stress in wheat N (Triticumaestivum L.). BMC Plant Biol. 10:123 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Hayashi N., Wang C. T., Kato H., Fujimura T., Kawasaki S. (2008). Efficient authentic fine mapping of the rice blast resistance gene Pik-h in the Pik cluster, using new Pik-h-differentiating isolates. Mol. Breed. 22 289–299. 10.1094/PHYTO-99-8-0900 [DOI] [Google Scholar]
- Yang J. L., Jiang H. Y., Yeh C. T., Yu J. M., Jeddeloh J. A., Nettleton D., et al. (2015). Extreme-phenotype genome-wide association study (XP-GWAS): a method for identifying trait-associated variants by sequencing pools of individuals selected from a diversity panel. Plant J. 84 587–596. 10.1111/tpj.13029 [DOI] [PubMed] [Google Scholar]
- Zheng W., Wang Y., Wang L., Ma Z., Zhao J., Wang P., et al. (2016). Genetic mapping and molecular marker development for Pi65(t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor. Appl. Genet. 129 1035–1044. 10.1007/s00122-016-2681-7 [DOI] [PubMed] [Google Scholar]
- Zhou B., Qu S. H., Liu G. F., Dolan M., Sakai H., Lu G. D., et al. (2006). The eight amino-acid differences within three leucine-rich repeats between Pi2 and pita resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 19 1216–1228. 10.1094/MPMI-19-1216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.