Abstract
G. pentaphyllum (Gynostemma pentaphyllum), a creeping herbaceous perennial with many important medicinal properties, is widely distributed in Asia. Gypenosides (triterpenoid saponins), the main effective components of G. pentaphyllum, are well studied. FPS (farnesyl pyrophosphate synthase), SS (squalene synthase), and SE (squalene epoxidase) are the main enzymes involved in the synthesis of triterpenoid saponins. Considering the important medicinal functions of G. pentaphyllum, it is necessary to investigate the transcriptomic information of G. pentaphyllum to facilitate future studies of transcriptional regulation. After sequencing G. pentaphyllum, we obtained 50,654,708 unigenes. Next, we used RPKM (reads per kilobases per million reads) to calculate expression of the unigenes and we performed comparison of our data to that contained in five common databases to annotate different aspects of the unigenes. Finally, we noticed that FPS, SS, and SE showed differential expression of enzymes in DESeq. Leaves showed the highest expression of FPS, SS, and SE relative to the other two tissues. Our research provides transcriptomic information of G. pentaphyllum in its natural environment and we found consistency in unigene expression, enzymes expression (FPS, SS, and SE), and the distribution of gypenosides content in G. pentaphyllum. Our results will enable future related studies of G. pentaphyllum.
1. Introduction
Gynostemma pentaphyllum (Thunb.) Makino is a kind of creeping herbaceous perennial that is distributed in Asia. Gynostemma pentaphyllum (G. pentaphyllum) grows in many places of China, including Guangxi, Guangdong, Fujian, Guizhou, Yunnan, Hubei, Anhui, Hebei, Jiangsu, Henan, Shandong, Sichuan, and Shanxi and in Taiwan. G. pentaphyllum also grows in neighboring countries such as Bangladesh, India, Indonesia, Japan, Republic of Korea, and Malaysia (data was obtained from the Checklist of South China Botanical Garden) [1]. Gypenosides (triterpenoid saponins), the major effective components of G. pentaphyllum, have various bioactivities that explain the extensive application of G. pentaphyllum in natural medicines [2–13]. For instance, the gypenosides exhibit a hypoglycemic effect by increasing the secretion of insulin [11–13]. Other functions like anticancer function, anti-inflammatory function, antianxiety function, blood fat-reducing, liver cells-protecting, neuroprotection, and immunoprotection have also been reported [2–10].
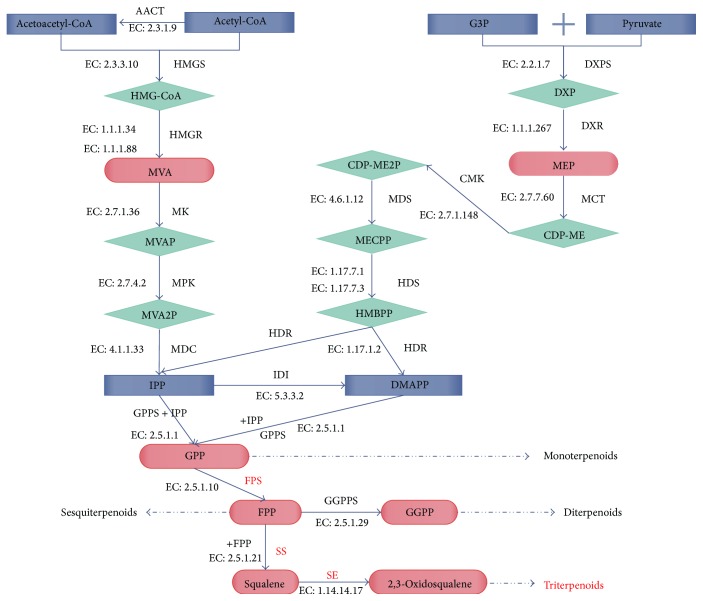
Gypenosides are secondary metabolites in the synthesis pathway of triterpenoids. Mevalonate or isoprenoid are the precursors in the beginning of the pathway of triterpenoid synthesis, which we refer to as the MVA (mevalonic acid) or MEP (methylerythritol phosphate) pathway (Figure 1) [14–16]. The synthesis pathway of the triterpenoids can be decomposed into three parts: (1) the synthesis of IPP (isopentenyl pyrophosphate) or DMAPP (dimethylallyl pyrophosphate); (2) the synthesis and cyclization of the squalene; and (3) the functionalization reaction that proceeds with complexity of the squalene (Figure 1). FPS (farnesyl pyrophosphate synthase), SS (squalene synthase), and SE (squalene epoxidase) were previously identified as the main enzymes involved in the synthesis of triterpenoid saponins [17–20]. FPS, SS, and SE are required for the synthesis and cyclization of the squalene that combines two sesquiterpenoids into one triterpenoid (C15 + C15 = C30) [16, 21]. After this step, triterpenoids can be transformed into many isoform types like protosteryl type (chair-chair-chair-boat conformations), dammarenyl type (chair-chair-chair-boat conformations), cadinyl type (chair-chair-chair-boat conformations), and hopene and tetrahymanol (chair-chair-chair-chair conformations or chair-chair-chair-boat conformations) [21].
Figure 1.
Triterpenoid synthesis pathway. Note: AACT: acetyl-CoA C-acetyltransferase; HMGS: hydroxymethylglutaryl-CoA synthase; HMG-CoA: hydroxymethylglutaryl-CoA; HMGR: hydroxymethylglutaryl-CoA reductase; MVA: mevalonate; MK: mevalonate kinase; MVAP: mevalonate phosphate; MPK: mevalonate phosphate kinase; MVAPP: mevalonate diphosphate; MDC: mevalonate diphosphate decarboxylase; G3P: D-glyceraldehyde-3-phosphate acetaldehydetransferase; DXPS: 1-deoxy-D-xylulose-5-phosphate synthase; DXP: 1-deoxy-D-xylulose-5-phosphate; DXR: 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MEP: 2-C-methyl-D-erythritol 4-phosphate; MCT: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CDP-ME: 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; CDP-ME2P: 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; MECPP: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HDS: l-hydroxy-2-methyl-butenyl-4-diphosphate synthase; HMBPP: l-hydroxy-2-methyl-2-butenyl-4-diphosphate; HDR: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IPP: isopentenyl-PP; IDI: isopentenyl-diphosphate delta-isomerase; DMAPP: dimethylallyl-PP; GPS: geranyl-diphosphate synthase; GPS: geranyl-diphosphate synthase; GPPS: geranylgeranyl diphosphate synthase; GPP: geranylgeranyl diphosphate; FPS: farnesyl diphosphate synthase; GGPPS: geranylgeranyl diphosphate synthase; GGPP: geranylgeranyl diphosphate; SS: squalene synthase; and SE: squalene epoxidase.
Like the ginsenosides, gypenosides (triterpenoids) in G. pentaphyllum have various and vital applications in medicine and health [22]. However, gypenosides showed much higher heterogeneity when compared with ginsenosides and more than 169 kinds of gypenosides were found in G. pentaphyllum [23–28]. In other words, more than five times the number of triterpenoid saponins was found in G. pentaphyllum relative to Panax ginseng (P. ginseng). It was interesting that G. pentaphyllum has such diversity in triterpenoids compared to other plants [26]. This is likely related to different expression of genes and enzymes involved in the synthesis pathway of triterpenoids. Nowadays, transcriptomic sequencing (RNA sequencing) is a more and more popular tool to explore transcriptomic process [29–37], because RNA sequencing has several advantages relative to DNA sequencing like lower fee, higher efficiency, more advanced features, and so forth [36–39]. In 2011, Sathiyamoorthy Subramaniyam analyzed the transcriptome of G. pentaphyllum related to the synthesis pathway of triterpenoids. However, this article had two important limitations. First, the samples used for sequencing only included two tissues (leaves and roots) and G. pentaphyllum that was sampled and sequenced was planted in water and not in its natural environment [32]. This is important because G. pentaphyllum exhibits great phenotypic diversity in different environments because of its strong adaptability [33, 40–44]. Additionally, although the author showed sequencing data in the paper, no association analysis among unigenes expression, enzyme expression, or the distribution of gypenosides content of G. pentaphyllum was determined. In 2015, Zhao et al. identified EST-SSR makers by analyzing the sequencing data of two species of Gynostemma (Cucurbitaceae) [33]. In that article, the tissues were natural and complete, but the three tissues (young leaves, flowers, and immature seeds) from each kind of G. pentaphyllum were mixed up together to extract RNA for constructing cDNA to sequence. In other words, the sequencing data and related information in that article were a mixed result and these results could not be classified by tissues. Therefore, to address this deficiency of knowledge, we collected G. pentaphyllum in natural environment and sequenced its transcriptome separately by Illumina's NextSeq 500.
2. Materials and Methods
2.1. Sample Collection and Preparation
G. pentaphyllum used for sequencing was planted in the Medicinal Plant Garden of Guangxi Traditional Chinese Medical University, Nanning City, Guangxi Autonomous Region, China. In July 2015, we harvested G. pentaphyllum after identification by Mr. Yilin Zhu (Guangxi Traditional Chinese Medical University). Fibrous roots, leaves, and stems were separately collected and cleaned and removed of impurities like soil (biological repeat of collections of each tissue was three times) (Figure S1–S5 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/7840914). Finally, the samples were saved in cryotubes and submerged in liquid nitrogen immediately.
2.2. Illumina Sequencing
The plant tissue sample of G. pentaphyllum was sent to Personalbio Company (Shanghai City, China) for transcriptome sequencing using Next-Generation Sequencing (NGS) technology based on the sequencing platform of Illumina's NextSeq 500. First, the mRNA was cleaved into little segments after treatment with chemical reagents and high temperature. Next, the segments were used to construct a cDNA library that was sequenced by paired end (PE) reads.
2.3. Unigene Assembly
Trinity (r20140717, k-mer 25 bp) professional software was used to assemble the RNA sequence [45]. First, high-quality sequences were constructed into a short-sequence library with length of k-mer. Next, primary contig sequences were obtained by the extension of the short-sequence library using overlaps with a k-mer-1 length. Next, primary contig sequences were categorized by their overlaps and categorical contigs were constructed into the De Bruijn graph. Based on the recognition rate of reads in each category, transcript sequences were restored by the contigs. After assemblage by Trinity, BLAST (version 2.2.30+) was used to compare the assembled sequences with reference sequences in NCBI (National Center for Biotechnology Information) nonredundant protein (NR) sequences to determine the best comparison results. Finally, sequences with the same gi number were classified as the same unigene and the longest sequence was regarded as the representative sequence of that unigene [46].
2.4. Analysis of Unigene Expression
RPKM (reads per kilobases per million reads) was used to calculated unigene expression of G. pentaphyllum and the calculation method of RPKM is described below [47]. Before we calculated the unigene expression, we need to process the read count of unigenes with Bowtie 2 (2.2.4, default setting) [48]. The RPKM density distribution generally reflected the pattern of gene expression. Typically, unigenes with medium expression cover the majority of the area under the curve (AUC) in the density distribution of RPKM (as drawn with the density function of software R). Oppositely, unigenes with higher or lower expression occupy the minority of AUC. DESeq (version 1.18.0) software was used to analyze the differential expression of unigenes in our study [49]. The expression of unigenes was compared by the fold change (fold change > 2) and its significance (p value < 0.05). The final result was displayed by Venn diagram.
| (1) |
2.5. Functional Annotation
After categorization, unigenes were annotated for functions using five databases: NCBI nonredundant protein (NR) sequences, Gene Ontology (GO) [50, 51], Kyoto Encyclopedia of Genes and Genome (KEGG) [52, 53], evolutionary genealogy of genes: Nonsupervised Orthologous Groups (eggNOG), [54] and Swiss-Prot [55, 56].
2.6. Analysis of the Distribution of Gypenosides Content in G. pentaphyllum
First, dried powder of the sample (about 15 mg) was mixed with 10 mL of extraction solvent (ethanol containing 5.0% pure water) and was processed by a continued supersonic treatment for 30 minutes. Second, the mixed solvent was evaporated to dryness using a rotary evaporator. The dry gypenosides were redissolved in 5.0 mL hot water (50°C). Third, this solution was applied to a chromatography column containing D101 macroporous resin and allowed to stand for 20 minutes. Fourth, pure water was used to rinse unbound material from the macroporous resin, while extraction solvent (ethanol containing 50% pure water) was used to remove the gypenosides form column. Fifth, the solvent of gypenosides was brought up to 5.0 mL and processed with a color reaction by vanillin. We then detected the absorbance of the solvent (after color reaction) by UV-Vis Spectrophotometer at a wavelength of 584 nm. Finally, we used Panaxadiol (C30H52O3) as a standard sample to calculate the actual content of gypenosides in samples by the standard curve method.
3. Result
3.1. Overview of the Sequencing and Assembly
The mRNA of G. pentaphyllum was cut into many small segments to construct a cDNA library. After sequencing the cDNA library, we obtained 352,999,296 original reads. However, this set of original reads also contained a lot of adapters and low-quality sequences, so 103,500,643 reads were filtered out leaving 249,488,643 clean reads of high quality. Next, 1,119,964 contigs with a total length of 249,488,543 bp were assembled by the overlaps of the original reads and we used these contigs to restore the transcriptome sequences. In the next step, we harvested 159,858,904 transcriptome sequences and used BLAST (Basic Local Alignment Search Tool) for all transcriptome sequences in the NR database. The transcriptome sequence with the highest score in BLAST was saved and the transcriptome sequences with same gi number were categorized as coming from the same unigene. Finally, we obtained 50,654,708 unigenes with a mean length of 755 bp. The overview of sequences is presented in Table 1.
Table 1.
Overview of the sequencing and assembly.
| Categories | Description | Number |
|---|---|---|
| Total reads | Total number of reads (RAW) | 356,311,342 |
| Number of clean reads | 352,999,296 | |
| Contigs | Total length (bp) | 249,488,543 |
| Sequence number | 1,119,964 | |
| Max. length (bp) | 13,870 | |
| Mean length (bp) | 223 | |
| GC% | 48 | |
| Transcriptome | Total length (bp) | 159,858,904 |
| Sequence number | 319,480 | |
| Max. length (bp) | 11,670 | |
| Mean length (bp) | 500 | |
| GC% | 46 | |
| Unigenes |
Total length (bp) | 50,654,708 |
| Sequence number | 67,068 | |
| Max. length (bp) | 11,670 | |
| Mean length (bp) | 755 | |
| GC% | 44 |
3.2. Result of Annotation
We used five databases, NR, GO, KEGG, eggNOG, and Swiss-Port, to annotate unigenes for functions (Figure S6–S9). The overview of annotation is listed in Table 2. The result of each annotation is provided in the support file. Generally speaking, eggNOG showed an identification rate of 96.57% and KEGG displayed the lowest identification rate of 8.77% when comparing unigenes with the reference sequences. Based on GO annotation, the unigenes were categorized into different categories based on different functions and GO Slim displayed general characteristic about the distribution of the unigenes. Then, we used the eggNOG database to explore the biological function of protein in more detail because eggNOG classifies different protein sequences into a more detailed directory. Similarly, Swiss-Port was also used for annotation of the protein sequences, and Swiss-Port builds on eggNOG and provided more detailed structural information about the protein. Finally, KEGG is the last but most important database we used to annotate enzymes, since the KEGG pathway annotation showed us the network of the intermolecular reaction. This allows determination of the enzymes that are located in the synthesis pathway of triterpenoid saponins.
Table 2.
Overview of annotation.
| Annotation in database | Unigene number | Percentage (%) |
|---|---|---|
| NR | 67,068 | 100 |
| GO | 40,623 | 61 |
| KO | 5,884 | 9 |
| eggNOG | 64,768 | 97 |
| Swiss-Prot | 55,429 | 83 |
| In all databases | 5,031 | 7.5 |
3.3. Expression of Unigenes
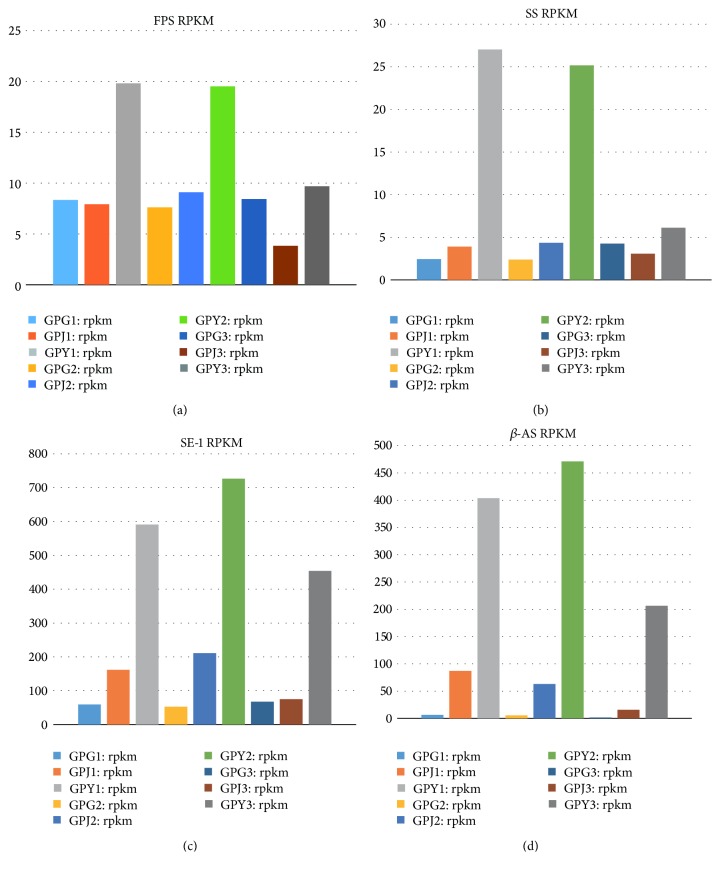
RPKM is a normalization method to calculate gene expression and we used the density distribution of the RPKM to show the expression of unigenes. The map of the density distribution showed that our unigenes expression conformed to standards because unigenes with mid-range expression occupied the majority of AUC (area under the curve) and unigenes with lower or higher expression were the minority of AUC (Figure 2). In the density distribution, unigenes in fibrous roots showed much higher expression than in the stems and leaves. Although stems and leaves showed similar unigene expression, stems had slightly higher unigene expression than the leaves. We analyzed the result of expression of the unigenes of FPS, SS, SE, and β-AS (beta-amyrin synthase) in Table 3. Noticeably, the unigenes that encoded FPS, SS, SE, and β-AS showed the highest expression in leaves and the lowest expression in fibrous roots. In the unigene expression of β-AS, the leaves showed almost 125 times higher expression than in the fibrous roots. The unigenes of FPS, SS, SE, and β-AS showed higher expression in the stems than in the fibrous roots. Based on this sequencing data, we detected the differential expression of unigenes using the software DESeq. We obtained the upregulated and downregulated unigenes in the pairwise comparison among the data from the fibrous roots, stems, and leaves (Table 4). We also determined the unigenes that showed differential expression in all samples. We used Venn diagram function in software R to describe the general distribution of unigenes with differential expression (Figure 3). Combining the data in Table 4 and Figure 3, we concluded that 10832 unigenes showed differential expression. Additionally, 699 unigenes displayed differential expression in all samples, while 6512 unigenes showed differential expression in the pairwise comparison of samples.
Figure 2.
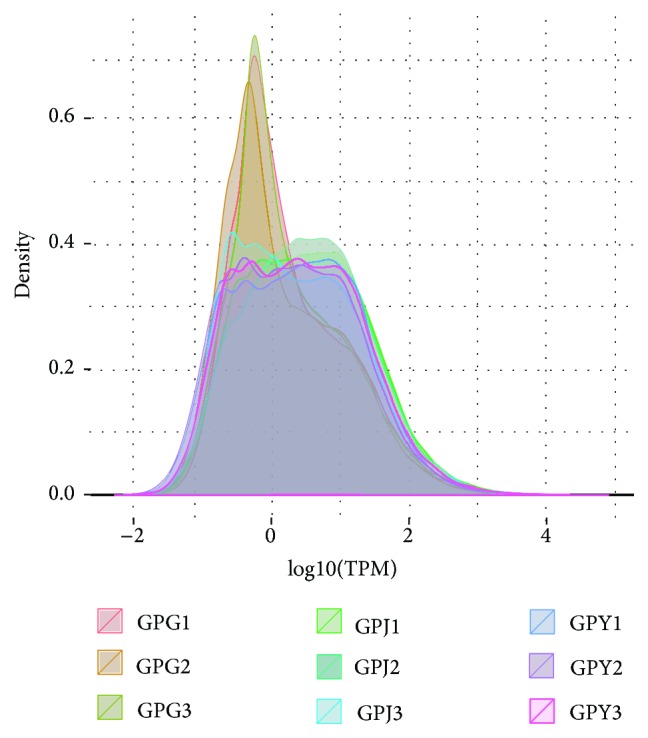
Density distribution of RPKM. Note: GPG: fibrous roots; GPJ: stems; GPY: leaves; RPKM: reads per kilobases per million reads.
Table 3.
RPKM of unigenes (FPS, SS, SE, and β-AS) in samples.
| Enzyme | Unigene ID | RPKM of sample 1 | RPKM of sample 2 | RPKM of sample 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fibrous roots | Stems | Leaves | Fibrous roots | Stems | Leaves | Fibrous roots | Stems | Leaves | ||
| FPS | c118250_g1_i1 | 8.35 | 7.93 | 19.81 | 7.61 | 9.11 | 19.53 | 8.44 | 3.84 | 9.7 |
| SS | c136108_g1_i1 | 2.44 | 3.91 | 27.03 | 2.39 | 4.36 | 25.18 | 4.26 | 3.06 | 6.12 |
| SE-1 | c127030_g2_i1 | 59.23 | 161.95 | 591.15 | 53.05 | 211.1 | 726.75 | 67.54 | 74.84 | 454.22 |
| SE-2 | c113536_g2_i1 | 0.79 | 0.95 | 5.66 | 0.26 | 0.94 | 2.87 | 0.6 | 0.52 | 2.7 |
| β-AS | c70785_g1_i1 | 6.49 | 86.98 | 403.79 | 5.5 | 62.95 | 471.36 | 1.66 | 15.79 | 206.31 |
Note: FPS: farnesyl pyrophosphate synthase; SS: squalene synthase: SE: squalene epoxidase; β-AS: beta-amyrin synthase.
Table 4.
Overview of the upregulated and downregulated unigenes.
| Case | Control | Upregulated unigenes | Downregulated unigenes | Total DE unigenes | |||
|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | ||
| GPG | GPJ | 3476 | 5.18 | 1759 | 2.62 | 5235 | 7.81 |
| GPG | GPY | 5590 | 8.33 | 3089 | 4.61 | 8679 | 12.94 |
| GPJ | GPY | 2938 | 4.38 | 1890 | 2.82 | 4828 | 7.2 |
Note: GPG: fibrous roots of G. pentaphyllum; GPJ: stems of G. pentaphyllum; GPY: leaves of G. pentaphyllum; DE: differential expression.
Figure 3.
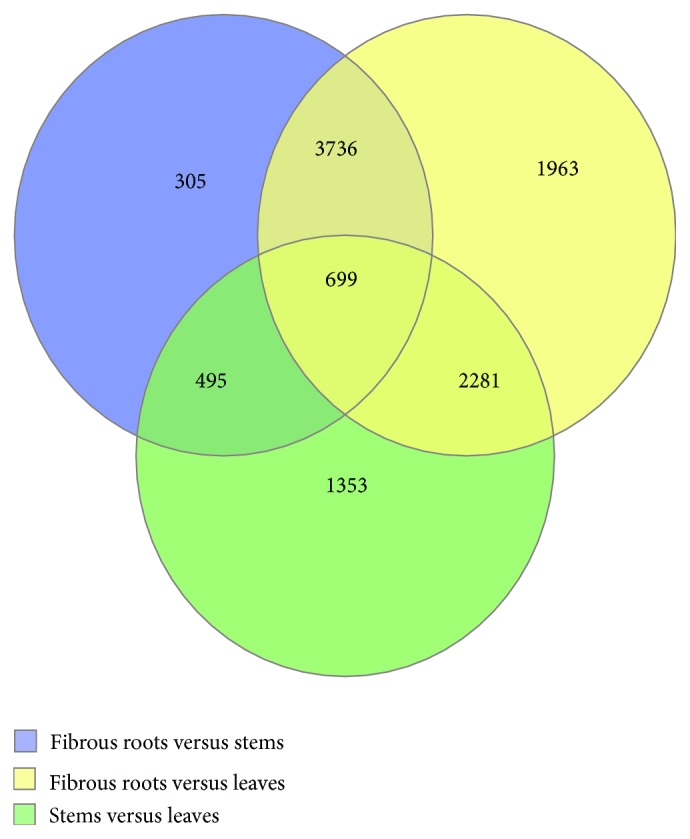
Venn diagram of differential expression of unigenes.
3.4. Content Distribution of Gypenosides in G. pentaphyllum
A UV-Vis Spectrophotometer was used to detect the content distribution of gypenosides in G. pentaphyllum. Leaves had the highest content (3.189%) of gypenosides of all samples (Table 5), and the content of gypenosides in stems (0.365%) or fibrous roots (0.172%) was much lower than the leaves. A correlation coefficient (R 2) of 0.996 in the standard curve indicates that our result was accurate and reliable (Figure S10).
Table 5.
Content distribution of gypenosides in G. pentaphyllum.
| Sample | Content of gypenosides | |||
|---|---|---|---|---|
| Number 1 | NUmber 2 | Number 3 | Average | |
| Fibrous roots | 0.1725% | 0.1731% | 0.1731% | 0.1729% |
| Stems | 0.3645% | 0.3657% | 0.3682% | 0.3662% |
| Leaves | 3.1887% | 3.1912% | 3.1931% | 3.1910% |
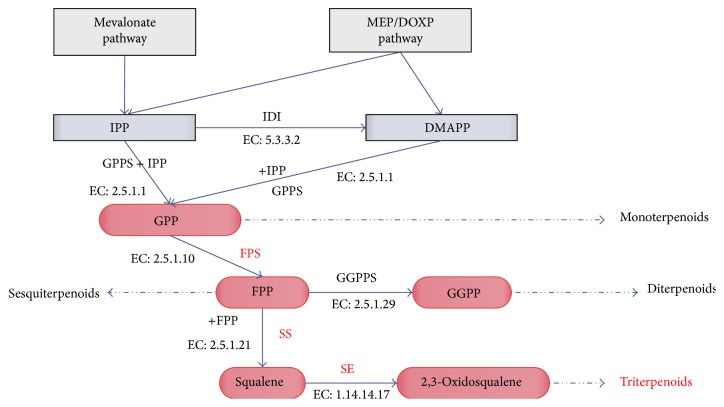
3.5. Expression of Enzymes in the Synthesis Pathway of Triterpenoids
Based on the KEGG Pathway, a more detailed result about the enzymes expression in the triterpenoids synthesis was obtained. We categorized related enzymes into three parts according to the synthesis pathway of the triterpenoids: (1) enzymes involved in the synthesis of IPP or DMAPP, (2) enzymes involved in the synthesis and cyclization of squalene, and (3) enzymes in the squalene functionalization reaction (Figure 1). We focused on FPS, SS, and SE because these three enzymes play an important role in the synthesis and cyclization of triterpenoids (Figure 4) [17–19]. The results are shown in Table 6 and Figure 5. We found three noticeable results. First, in the comparison of FPS, only one comparison between the fibrous roots and leaves was found. Leaves showed higher expression of FPS compared to the fibrous roots. Second, in the comparison of SS, two comparisons were found and the result showed that leaves had higher expression than the fibrous roots and stems. Third, in the comparison of SE, leaves showed higher expression than the stems and fibrous roots. The fibrous roots showed higher expression of SE than the stems.
Figure 4.
FPS, SS, and SE in the synthesis pathway of triterpenoids. Note: IPP: isopentenyl-PP; IDI: isopentenyl-diphosphate delta-isomerase; DMAPP: dimethylallyl-PP; GPS: geranyl-diphosphate synthase; GPPS: geranylgeranyl diphosphate synthase; GPP: geranylgeranyl diphosphate; FPS: farnesyl diphosphate synthase; GGPPS: geranylgeranyl diphosphate synthase; GGPP: geranylgeranyl diphosphate; SS: squalene synthase; and SE: squalene epoxidase.
Table 6.
Enzyme expression of FPS, SS, SE, and β-AS.
| Full name | Unigene ID | Short name | EC number | G versus J | G versus Y | J versus Y |
|---|---|---|---|---|---|---|
| Farnesyl diphosphate synthase | c118250_g1_i1 | FPS | EC: 2.5.1.1/2.5.1.10 | GPY up | ||
| Squalene synthase | c136108_g1_i1 | SS | EC: 2.5.1.21 | GPY up | GPY up | |
| Squalene epoxidase | c127030_g2_i1 | SE | EC: 1.14.13.132 | GPJ up | GPY up | GPY up |
| GPY up | GPY up | |||||
| β-amyrin synthase | c70785_g1_i1 | β-AS | EC: 5.4.99.39 | GPJ up | GPY up | GPY up |
Note: GPG or G: fibrous roots of G. pentaphyllum; GPJ or J: stems of G. pentaphyllum; GPY or Y: leaves of G. pentaphyllum.
Figure 5.
RPKM of FPS, SS, SE, and β-AS. Note: (a) RPKM of FPS; (b) RPKM of SS; (c) RPKM of SE-1; (d) RPKM of β-AS; GPG: fibrous roots; GPJ: stems; GPY: leaves; RPKM: reads per kilobases per million reads.
4. Discussion
G. pentaphyllum is a creeping herbaceous perennial with medicinal properties used in traditional Chinese medicine. Triterpenoid saponins, the main effective components of G. pentaphyllum, have been widely studied [2–13, 23, 24]. In this study, we obtained transcriptome information using RNA sequencing, a technique that exhibits higher efficiency and is less expensive than DNA sequencing [38]. We analyzed the associations of unigene expression, enzyme (the output of the unigenes) expression, and the content distribution of gypenosides (enzyme output) after functional annotation (Table 2), RPKM calculating (Table 3), and measurement of gypenosides content (Table 5).
We found that the expressions of unigenes and enzymes were positively associated with the distribution of gypenosides content. Generally speaking, unigenes and enzyme expression (FPS, SS, and SE) in the samples (fibrous roots, stems, and leaves) determined to the distribution of gypenosides content. Higher expression of unigenes and enzymes (encoded by unigenes) caused the higher content of enzymes' production (gypenosides), and the lower expression of unigenes and enzymes caused less enzyme production. This consistent result could facilitate future studies of other secondary metabolites in G. pentaphyllum. Since G. pentaphyllum has wide applications for health and medicine, it is essential to identify the secondary metabolites with various and vital medicinal functions.
One interesting finding was the observed differences between general expression and individual expression of unigenes (Figure 2 and Table 3). A group of specific unigenes (FPS, SS, and SE) located in a special pathway like triterpenoids synthesis could increase their expression to a much higher level as required for a special physiological activity like triterpenoid synthesis. This obvious difference in expression may result from changes in the regulation of transcription. It is currently a hot topic to study the differential expression of the transcriptome and the regulation of transcription of plants in response to stresses of a special environment or other genetic factors [57–63]. Another interesting observation was that the transcriptome sequences of G. pentaphyllum determined in our study showed high similarity to the transcriptome sequences related to bitterness in cucumber as reported previously [64]. We downloaded the available mRNA sequences of related enzymes from that article and blasted them against the unigene sequences of G. pentaphyllum. The BLAST result was surprising, as all thirteen available sequences related to bitterness in that article showed a high degree of similarity to the specific unigene sequences of G. pentaphyllum. This high similarity predicted that genes related to the biosynthesis, regulation, and domestication of bitterness in cucumber may also be present in G. pentaphyllum. G. pentaphyllum also has two tastes (sweet and bitter) and this difference of taste may be caused as in cucumber. The taste of G. pentaphyllum from bitter to sweet predicts that G. pentaphyllum may change in response to domestication and contain a similar mutation. A further genetic exploration of the domestication of G. pentaphyllum may provide understanding of its changed taste.
Although this study was not the first report of the transcriptome information for G. pentaphyllum using sequencing, we corrected the limitations of the previous study and enriched the analysis of triterpenoid synthesis of G. pentaphyllum. G. pentaphyllum used for analysis in our study was planted in a natural environment and three common kinds of plant tissues (fibrous roots, stems, and leaves) were used to provide tissue samples for sequencing. Together with the sequencing, an exploration of the distribution of gypenosides content was performed to confirm the analysis of enzymes and unigenes. We found a positive association of unigene expression, enzyme expression, and the distribution of gypenosides. Our study will facilitate more genetic studies examining the regulation of transcription and the change of bitterness in G. pentaphyllum.
5. Conclusions
To provide more complete and high-quality transcriptional information of natural G. pentaphyllum, we used RNA sequencing technology to sequence the transcriptome of G. pentaphyllum. We found a positive association of unigene expression, enzyme expression, and the distribution of gypenosides. Our results will enable future related studies of G. pentaphyllum.
Supplementary Material
Figure S1. G. pentaphyllum (Sample) 01. Description: This is the picture of intact G. pentaphyllum in our study.
Figure S2. G. pentaphyllum (Sample) 02. Description: This is the picture of intact G. pentaphyllum in our study.
Figure S3. Leaves (Sample). Description: This is the picture of leaves sample of G. pentaphyllum in our study.
Figure S4. Stems (Sample). Description: This is the picture of stems sample of G. pentaphyllum in our study.
Figure S5. Fibrous Roots (Sample). Description: This is the picture of fibrous roots sample of G. pentaphyllum in our study.
Figure S6. The general result of annotation. Abbreviations: NR: Nonredundant protein sequences; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genome; eggNOG: Evolutionary genealogy of genes: Nonsupervised Orthologous Groups.
Figure S7. The result of GO Slim. Abbreviation: GO Slim: Cut-down versions of the GO ontologies.
Figure S8. The result of eggNOG annotation.
Figure S9. The result of KEGG annotation.
Figure S10. The standard curve of absorbance.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 31260069). The authors thank Mr. Yilin Zhu (Guangxi Traditional Chinese Medical University) for the identification of samples.
Abbreviations
- G. pentaphyllum:
Gynostemma pentaphyllum (Thunb.) Makino
- FPS:
Farnesyl pyrophosphate synthase
- SS:
Squalene synthase
- SE:
Squalene epoxidase
- RPKM:
Reads per kilobases per million reads
- NR:
Nonredundant protein sequences
- GO:
Gene Ontology
- eggNOG:
Evolutionary genealogy of genes: Nonsupervised Orthologous Groups
- KEGG:
Kyoto Encyclopedia of Genes and Genome
- MVA:
Mevalonic acid
- MEP:
Methylerythritol phosphate
- IPP:
Isopentenyl pyrophosphate
- DMAPP:
Dimethylallyl pyrophosphate
- P. ginseng:
Panax ginseng
- BLAST:
Basic Local Alignment Search Tool
- GO Slim:
Cut-down versions of the GO ontologies
- AUC:
Area under the curve
- β-AS:
Beta-amyrin synthase
- UV-Vis:
Ultraviolet-visible
- NGS:
Next-Generation Sequencing
- PE:
Paired end
- NCBI:
National Center for Biotechnology Information
- CDS:
Coding sequence
- RefSeq:
Reference sequences
- PDB:
Protein database
- EMBL:
European Molecular Biology Laboratory
- DDBJ:
DNA Data Bank of Japan
- KO:
KEGG Ortholog.
Disclosure
Qicong Chen is first author.
Data Access
The data and materials were listed in the supplementary information.
Competing Interests
The authors declare that they have no competing interests.
Authors' Contributions
Yaosheng Wu, Chengtong Ma, and Jieying Qian prepared the sample of G. pentaphyllum for sequencing in Guangxi Traditional Chinese Medical University. All authors worked together to analyze the sequencing data but Qicong Chen was responsible for the specific analysis of sequencing data. Yaosheng Wu provided the guidelines to detect the distribution of gypenosides content in G. pentaphyllum and the guidelines to write this report. Qicong Chen detected the distribution of gypenosides content in G. pentaphyllum and wrote this manuscript. All authors read and approved the final manuscript.
References
- 1.Checklist of South China Botanical Garden. http://www.efloras.org/florataxon.aspx?flora_id=610&taxon_id=200022642.
- 2.Piao X., Wu Q., Yang J., Park S. Y., Chen D., Liu H. Dammarane-type saponins from heat-processed gynostemma pentaphyllum show fortified activity against A549 cells. Archives of Pharmacal Research. 2013;36(7):874–879. doi: 10.1007/s12272-013-0086-6. [DOI] [PubMed] [Google Scholar]
- 3.Piao X.-L., Xing S.-F., Lou C.-X., Chen D.-J. Novel dammarane saponins from Gynostemma pentaphyllum and their cytotoxic activities against HepG2 cells. Bioorganic and Medicinal Chemistry Letters. 2014;24(20):4831–4833. doi: 10.1016/j.bmcl.2014.08.059. [DOI] [PubMed] [Google Scholar]
- 4.Yang F., Shi H., Zhang X., Yu L. Two novel anti-inflammatory 21-nordammarane saponins from tetraploid jiaogulan (Gynostemma pentaphyllum) Journal of Agricultural and Food Chemistry. 2013;61(51):12646–12652. doi: 10.1021/jf404726z. [DOI] [PubMed] [Google Scholar]
- 5.Yang F., Shi H., Zhang X., Yang H., Zhou Q., Yu L. L. Two new saponins from tetraploid jiaogulan (Gynostemma pentaphyllum), and their anti-inflammatory and α-glucosidase inhibitory activities. Food Chemistry. 2013;141(4):3606–3613. doi: 10.1016/j.foodchem.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Müller C., Gardemann A., Keilhoff G., Peter D., Wiswedel I., Schild L. Prevention of free fatty acid-induced lipid accumulation, oxidative stress, and cell death in primary hepatocyte cultures by a Gynostemma pentaphyllum extract. Phytomedicine. 2012;19(5):395–401. doi: 10.1016/j.phymed.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Wang M., Wang F., Wang Y., et al. Metabonomics study of the therapeutic mechanism of Gynostemma pentaphyllum and atorvastatin for hyperlipidemia in rats. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0078731.e78731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi H. S., Park M. S., Kim S. H., Hwang B. Y., Lee C. K., Lee M. K. Neuroprotective effects of herbal ethanol extracts from gynostemma pentaphyllum in the 6-hydroxydopamine-lesioned rat model of parkinson's disease. Molecules. 2010;15(4):2814–2824. doi: 10.3390/molecules15042814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi H. S., Zhao T. T., Shin K. S., et al. Anxiolytic effects of herbal ethanol extract from gynostemma pentaphyllum in mice after exposure to chronic stress. Molecules. 2013;18(4):4342–4356. doi: 10.3390/molecules18044342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Im S.-A., Choi H. S., Choi S. O., et al. Restoration of electric footshock-induced immunosuppression in mice by gynostemma pentaphyllum components. Molecules. 2012;17(7):7695–7708. doi: 10.3390/molecules17077695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X.-S., Bi X.-L., Wan X., et al. Protein tyrosine phosphatase 1B inhibitory effect by dammarane-type triterpenes from hydrolyzate of total Gynostemma pentaphyllum saponins. Bioorganic & Medicinal Chemistry Letters. 2013;23(1):297–300. doi: 10.1016/j.bmcl.2012.10.097. [DOI] [PubMed] [Google Scholar]
- 12.Gao D., Zhao M., Qi X., et al. Hypoglycemic effect of Gynostemma pentaphyllum saponins by enhancing the Nrf2 signaling pathway in STZ-inducing diabetic rats. Archives of Pharmacal Research. 2016;39(2):221–230. doi: 10.1007/s12272-014-0441-2. [DOI] [PubMed] [Google Scholar]
- 13.Lokman E. F., Gu H. F., Wan Mohamud W. N., Östenson C.-G. Evaluation of antidiabetic effects of the traditional medicinal plant Gynostemma pentaphyllum and the possible mechanisms of insulin release. Evidence-Based Complementary and Alternative Medicine. 2015;2015:7. doi: 10.1155/2015/120572.120572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman J. D., Chappell J. Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Critical Reviews in Biochemistry and Molecular Biology. 1999;34(2):95–106. doi: 10.1080/10409239991209228. [DOI] [PubMed] [Google Scholar]
- 15.Haralampidis K., Trojanowska M., Osbourn A. E. Biosynthesis of triterpenoid saponins in plants. Advances in Biochemical Engineering/Biotechnology. 2002;75:31–49. doi: 10.1007/3-540-44604-4_2. [DOI] [PubMed] [Google Scholar]
- 16.Terpentoid backbone biosynthesis. http://www.genome.jp/kegg-bin/show_pathway?14607215648484/ko00900.args.
- 17.Kim Y.-K., Kim Y. B., Uddin M. R., Lee S., Kim S.-U., Park S. U. Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase. ACS Synthetic Biology. 2014;3(10):773–779. doi: 10.1021/sb400194g. [DOI] [PubMed] [Google Scholar]
- 18.Seo J. W., Jeong J. H., Shin C. G., et al. Overexpression of squalene synthase in Eleutherococcus senticosus increases phytosterol and triterpene accumulation. Phytochemistry. 2005;66(8):869–877. doi: 10.1016/j.phytochem.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Rasbery J. M. Genetic and biochemical characterization of the squalene epoxidase gene family in Arabidopsis thaliana 2007. http://hdl.handle.net/1911/20636.
- 20.He F., Zhu Y., He M., Zhang Y. Molecular cloning and characterization of the gene encoding squalene epoxidase in Panax notoginseng. DNA Sequence—Journal of DNA Sequencing and Mapping. 2008;19(3):270–273. doi: 10.1080/10425170701575026. [DOI] [PubMed] [Google Scholar]
- 21. Sesquiterpenoid and Triterpenoid Biosynthesis, http://www.genome.jp/kegg-bin/show_pathway?ko00909.
- 22. Pharmacopoeia of People's Republic of China, Beijing Chemical Industry Press: National Pharmacopoeia Committee, 1997.
- 23.Hu Y., Ip F. C. F., Fu G., Pang H., Ye W., Ip N. Y. Dammarane saponins from Gynostemma pentaphyllum . Phytochemistry. 2010;71(10):1149–1157. doi: 10.1016/j.phytochem.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Ky P. T., Huong P. T., My T. K., et al. Dammarane-type saponins from Gynostemma pentaphyllum . Phytochemistry. 2010;71(8-9):994–1001. doi: 10.1016/j.phytochem.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., Zhang W., Ji Y.-P., Zhao Y., Wang C.-G., Hu J.-F. Gynostemosides A-E, megastigmane glycosides from Gynostemma pentaphyllum . Phytochemistry. 2010;71(5-6):693–700. doi: 10.1016/j.phytochem.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Kim J. H., Han Y. N. Dammarane-type saponins from Gynostemma pentaphyllum . Phytochemistry. 2011;72(11-12):1453–1459. doi: 10.1016/j.phytochem.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Shi L., Cao J. Q., Shi S. M., Zhao Y. Q. Triterpenoid saponins from Gynostemma pentaphyllum. Journal of Asian Natural Products Research. 2011;13(2):168–177. doi: 10.1080/10286020.2010.547029. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X. S., Cao J. Q., Zhao C., Wang X. D., Wu X. J., Zhao Y. Q. Novel dammarane-type triterpenes isolated from hydrolyzate of total Gynostemma pentaphyllum saponins. Bioorganic & Medicinal Chemistry Letters. 2015;25(16):3095–3099. doi: 10.1016/j.bmcl.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Blanca J., Cañizares J., Roig C., Ziarsolo P., Nuez F., Picó B. Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae) BMC Genomics. 2011;12, article 104 doi: 10.1186/1471-2164-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyun T. K., Rim Y., Jang H., et al. De novo transcriptome sequencing of Momordica cochinchinensis to identify genes involved in the carotenoid biosynthesis. Plant Molecular Biology. 2012;79(4-5):413–427. doi: 10.1007/s11103-012-9919-9. [DOI] [PubMed] [Google Scholar]
- 31.Sangwan R. S., Tripathi S., Singh J., Narnoliya L. K., Sangwan N. S. De novo sequencing and assembly of Centella asiatica leaf transcriptome for mapping of structural, functional and regulatory genes with special reference to secondary metabolism. Gene. 2013;525(1):58–76. doi: 10.1016/j.gene.2013.04.057. [DOI] [PubMed] [Google Scholar]
- 32.Subramaniyam S., Mathiyalagan R., Gyo I. J., Bum-Soo L., Sungyoung L., Chun Y. D. Transcriptome profiling and insilico analysis of Gynostemma pentaphyllum using a next generation sequencer. Plant Cell Reports. 2011;30(11):2075–2083. doi: 10.1007/s00299-011-1114-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y.-M., Zhou T., Li Z.-H., Zhao G.-F. Characterization of global transcriptome using illumina paired-end sequencing and development of EST-SSR markers in two species of Gynostemma (Cucurbitaceae) Molecules. 2015;20(12):21214–21231. doi: 10.3390/molecules201219758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X., Xu H., Ma X., Zhan R., Chen W. Triterpenoid saponin biosynthetic pathway profiling and candidate gene mining of the Ilex asprella root using RNA-Seq. International Journal of Molecular Sciences. 2014;15(4):5970–5987. doi: 10.3390/ijms15045970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin J. A., Wang Z. Next-generation transcriptome assembly. Nature Reviews Genetics. 2011;12(10):671–82. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 36.Nagalakshmi U., Waern K., Snyder M., et al. RNA-Seq: a method for comprehensive transcriptome analysis. In: Frederick M. A., editor. Current Protocols in Molecular Biology. chapter 4. 2010. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabbani B., Nakaoka H., Akhondzadeh S., Tekin M., Mahdieh N. Next generation sequencing: implications in personalized medicine and pharmacogenomics. Molecular BioSystems. 2016;12(6):1818–1830. doi: 10.1039/c6mb00115g. [DOI] [PubMed] [Google Scholar]
- 39. Next Generation Sequencing (NGS), https://en.wikibooks.org/wiki/Next_Generation_Sequencing_(NGS)
- 40.Wang C., Zhang H., Qian Z.-Q., Zhao G.-F. Genetic differentiation in endangered Gynostemma pentaphyllum (Thunb.) Makino based on ISSR polymorphism and its implications for conservation. Biochemical Systematics and Ecology. 2008;36(9):699–705. doi: 10.1016/j.bse.2008.07.004. [DOI] [Google Scholar]
- 41.Ming H. W., Cheng Z. Z. Effects of external support on the foraging behavior and reproductive strategies in Gynostemma pentaphylum populations. Acta Ecologica Sinica. 2001;21(1):47–50. [Google Scholar]
- 42.Zhou J., Wu Y. S., Zhao R. Q., et al. Molecular authentication of Gynostemma pentaphyllum through development and application of random amplification polymorphic DNA sequence-characterized amplified region marker. Genetics and Molecular Research. 2015;14(4):16204–16214. doi: 10.4238/2015.december.8.10. [DOI] [PubMed] [Google Scholar]
- 43.Xinfen G., Chen S. K., Gu Z. J., Zhao J. Z. A chromosomal study on the genus Gynostemma (Cucurbitaceae) Acta Botanica Yunnanica. 1995;17:312–316. [Google Scholar]
- 44. Gynostemma pentaphyllum, https://en.wikipedia.org/wiki/Gynostemma_pentaphyllum.
- 45.Haas B. J., Papanicolaou A., Yassour M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Ammar A., Elouedi Z., Lingras P. Foundations of Intelligent Systems: 20th International Symposium, ISMIS 2012, Macau, China, December 4–7, 2012. Proceedings. Vol. 7661. Berlin, Germany: Springer; 2012. RPKM: the rough possibilistic K-modes; pp. 81–86. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 48.Langmead B., Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10, article R106) doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gene Ontology, http://geneontology.org/
- 51.Ashburner M., Ball C., Ja Botstein D., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2015;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minoru K., Susumu G., Shuichi K., Yasushi O., Masahiro H. The KEGG resource for deciphering the genome. Nucleic Acids Research. 2004;32(22):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kyoto Encyclopedia of Genes and Genome, http://www.kegg.jp/
- 54. Evolutionary genealogy of genes: Non-supervised Orthologous Groups, http://eggnogdb.embl.de/#/app/home. [DOI] [PMC free article] [PubMed]
- 55.Powell S., Forslund K., Szklarczyk D., et al. EggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Research. 2014;42(1):D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swiss-Prot, http://web.expasy.org/docs/swiss-prot_guideline.html.
- 57.Qi B., Yang Y., Yin Y., Xu M., Li H. De novo sequencing, assembly, and analysis of the Taxodium “Zhongshansa” roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biology. 2014;14, article no. 201 doi: 10.1186/s12870-014-0201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang T., Hao L., Yao S., Zhao Y., Lu W., Xiao K. TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiology and Biochemistry. 2016;104:99–113. doi: 10.1016/j.plaphy.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 59.Jin W., Wang H., Li M., et al. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.) Plant Biotechnology Journal. 2016;14(11):2120–2133. doi: 10.1111/pbi.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai S., Tuan P. A., Saito T., et al. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta. 2016;244(3):573–586. doi: 10.1007/s00425-016-2524-4. [DOI] [PubMed] [Google Scholar]
- 61.Zhu Q., Li B., Mu S., et al. TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco. BMC Genomics. 2013;14(1, article 806) doi: 10.1186/1471-2164-14-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan F.-Q., Tu H., Liang W.-J., et al. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biology. 2015;15, article 89 doi: 10.1186/s12870-015-0450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu T., Zhu S., Tang Q., Tang S. Genome-wide transcriptomic profiling of ramie (Boehmeria nivea L. Gaud) in response to cadmium stress. Gene. 2015;558(1):131–137. doi: 10.1016/j.gene.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 64.Shang Y., Ma Y., Zhou Y., et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346(6213):1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. G. pentaphyllum (Sample) 01. Description: This is the picture of intact G. pentaphyllum in our study.
Figure S2. G. pentaphyllum (Sample) 02. Description: This is the picture of intact G. pentaphyllum in our study.
Figure S3. Leaves (Sample). Description: This is the picture of leaves sample of G. pentaphyllum in our study.
Figure S4. Stems (Sample). Description: This is the picture of stems sample of G. pentaphyllum in our study.
Figure S5. Fibrous Roots (Sample). Description: This is the picture of fibrous roots sample of G. pentaphyllum in our study.
Figure S6. The general result of annotation. Abbreviations: NR: Nonredundant protein sequences; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genome; eggNOG: Evolutionary genealogy of genes: Nonsupervised Orthologous Groups.
Figure S7. The result of GO Slim. Abbreviation: GO Slim: Cut-down versions of the GO ontologies.
Figure S8. The result of eggNOG annotation.
Figure S9. The result of KEGG annotation.
Figure S10. The standard curve of absorbance.