Abstract
Using the amber suppression approach, Nε-(4-azidobenzoxycarbonyl)-δ, ε-dehydrolysine, an allysine precursor is genetically encoded in E. coli. Its genetic incorporation followed by two sequential biocompatible reactions allows convenient synthesis of proteins with site-specific lysine dimethylation. Using this approach, dimethyl-histone H3 and p53 proteins have been synthesized and used successfully to probe functions of epigenetic enzymes including histone demethylase LSD1 and histone acetyltransferase Tip60. We confirmed that LSD1 is catalytically active toward H3K4me2 and H3K9me2 but inert toward H3K36me2 and methylation at p53 K372 directly activates Tip60 for its catalyzed acetylation at p53 K120.
Keywords: lysine dimethylation, dimethyllysine, amber suppression, genetic code expansion, allysine
Graphical abstract
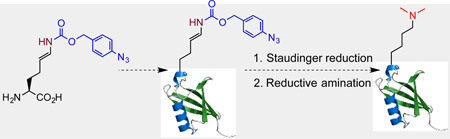
An allysine precursor, Nε-(4-azidobenzoxycarbonyl)-δ, ε-dehydrolysine is genetically encoded. Its incorporation followed by Staudinger reduction and reductive amination allows the synthesis of proteins with site-specific lysine dimethylation.
Protein lysine methylation is a reversible posttranslational modification that was originally discovered in histones but occurs also in many non-histone proteins.[1] There are three levels of lysine methylation, namely mono-, di-, and trimethylation that coordinate with other histone modifications to regulate chromatin-based transcriptional control and shape inheritable epigenetic programs in the eukaryotes.[2] Besides its epigenetic roles of chromatin regulation, lysine methylation also serves critical functions in regulating activities of transcription factors such as p53 and NF-κB.[3] Proteins with site-specific lysine methylation can be potentially synthesized by incubating target proteins with histone methyltransferases (HMTs). However, not all lysine methylation sites have their corresponding HMTs identified. In addition, the promiscuity of HMTs and the three levels of methylation add to high heterogeneity to finally methylated proteins, which makes it difficult to separate homogenous proteins with site-specific lysine methylation. Alternatively, native chemical ligation and expressed protein ligation, two generally applied chemical methods can be used for the synthesis of proteins with three lysine methylation types.[4] However, both methods suffer from limitations such as the requirement of a cysteine for the ligation process and the obstacle to install lysine methylation in the middle of a protein. Several groups have developed approaches that combine the amber suppression-based mutagenesis approach and photo- and chemical-based cleavage for the synthesis of proteins with lysine monomethylation. In these approaches, protected Nε-methyl-lysines are genetically incorporated into proteins and then deprotected byphoto- and chemical-based cleavage of the protection groups.[5] However, a similar method has not been developed for the synthesis of proteins with lysine dimethylation and lysine trimethylation. Chin and coworkers previously described a multi-step strategy for the synthesis of histones with lysine dimethylation that involves the genetic incorporation of a protected lysine at a designated lysine site of a histone, global protection of all other lysine residues and N-terminal amine in the expressed histone, the removal of the protection group from the genetic encoded modified lysine to recover lysine at the designated site, reductive alkylation with formaldehyde to install lysine dimethylation at the designated site, and the final removal of the global protection group to afford a dimethyl-histone.[6] Although elegant, this approach cannot be applied to proteins that are sensitive to denaturing conditions used for global protection and deprotection of lysine residues and N-terminal amine. Its incompatibility with cysteine that was not present in the original model histone is also a concern.
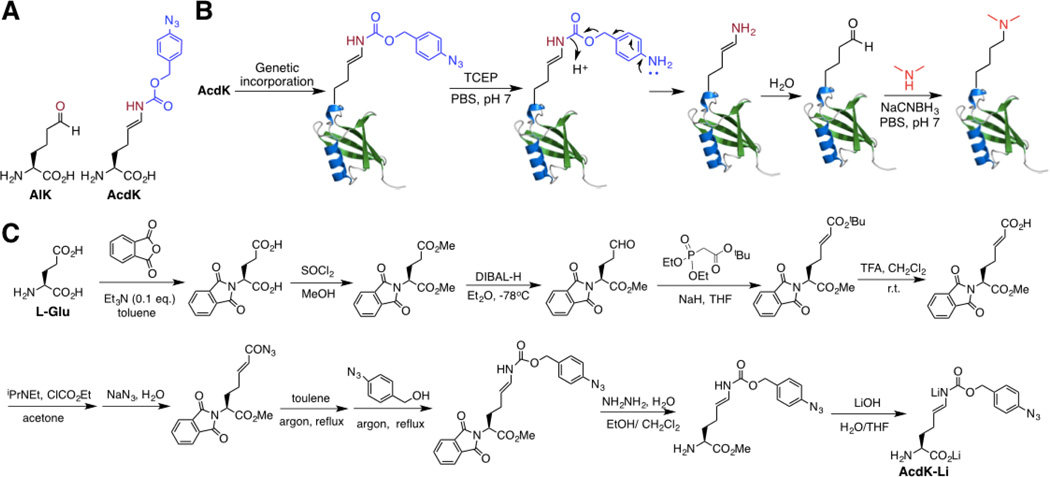
In order to site-specifically install lysine dimethylation in proteins, we envisioned that allysine (AlK, Figure 1A), a naturally occurring derivative of lysine in elastin and collagen[7] can be genetically encoded using the amber suppression mutagenesis approach and then undergo reductive amination with dimethylamine for the installation of site-specific lysine dimethylation in proteins. Given the concern of the cellular toxicity from its side chain aliphatic aldehyde, AlK was not directly used. Instead a precursor amino acid, Nε-(4-azidobenzoxycarbonyl)-δ, ε-dehydrolysine (AcdK, Figure 1A) that shields the side chain aldehyde was designed. AcdK has an azidobenzoxycarbonyl moiety whose reduction with a phosphine will trigger a self cleavage process to release δ, ε-dehydrolysine.[8] δ, ε-Dehydrolysine doesn’t stably exist in water and hydrolyzes instantaneously to form AlK. By genetically incorporating AcdK into proteins followed by Staudinger reduction with tris-(2-carboxyethyl)phosphine (TCEP) to recover AlK and then reductive amination with dimethyllysine in the presence of NaCNBH3, proteins with site-specific lysine dimethylation can be potentially synthesized (Figure 1B). Since both Staudinger reduction and reductive amination can be carried out in mild physiological conditions, this approach can be generally applied for the synthesis of proteins with site-specific lysine dimethylation as well as the synthesis of proteins with site-specific lysine monomethylation by simply changing dimethylamine to methylamine in the reductive amination step. A synthetic route of AcdK shown in Figure 1C that starts with L-glutamate and finishes as dilithium salt of AcdK was designed and successfully tested. Although the overall synthesis involves 9 steps, gram quantities of AcdK have been routinely produced.
Figure 1. The application and synthesis of AcdK.
(A) Structures of AlK and AcdK. (B) A diagram that illustrates the genetic incorporation of AcdK followed by Staudinger reduction, self-cleavage of the para-aminobenzyloxycarbonyl group, enamine hydrolysis, and reductive amination to generate a dimethyllysine in a protein. (C) The synthesis of AcdK. It starts with L-glutamate and finishes with a dilithium salt of AcdK.
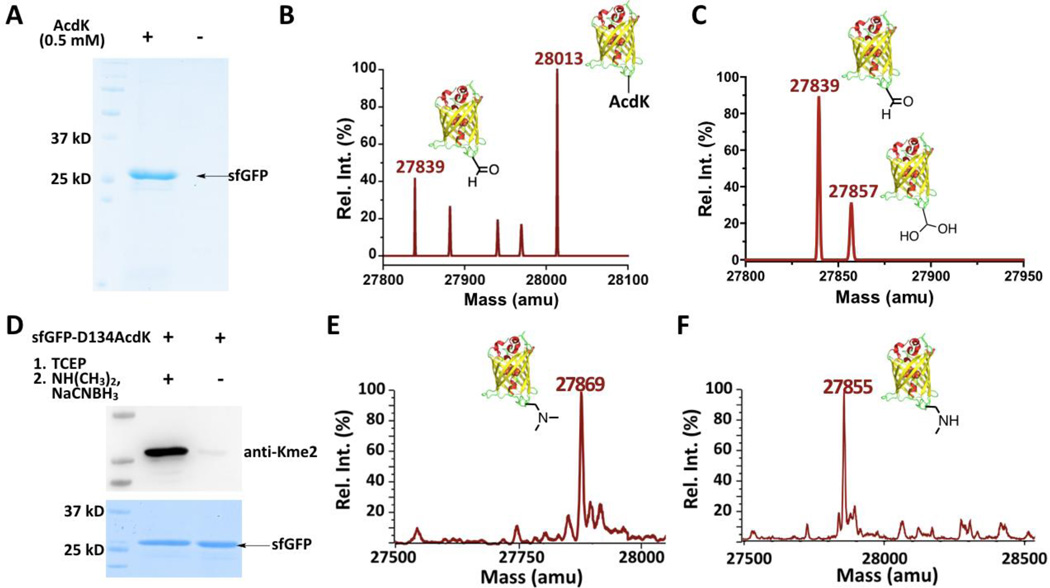
In order to use amber codon to genetically encode AcdK in E. coli, we employed an engineered pyrrolysyl-tRNA synthetase (PylRS)-tRNAPyl system.[9] Wild type and mutant PylRS-tRNAPyl pairs have been widely used for the genetic incorporation of a large number of lysine and phenylalanine derivatives into proteins at amber mutation sites in a series of cell strains.[10] A PylRS gene library in which codons for four active site residues, Y306, L309, C348, and Y384 of PylRS were randomized was first constructed. A broadly adopted and double-sieved selection protocol[10a] was applied for the selection of PylRS mutants that charge tRNAPyl with AcdK. The mutant, together with AcdK and tRNAPyl, that displays the best amber suppression efficiency in E. coli has mutations as L309T/C348G/Y384F and is coined as AcdKRS (Supplementary Figure 1). A plasmid pEVOL-AcdKRS that contains genes encoding AcdKRS and tRNAPyl was then constructed. This plasmid and another plasmid pBAD-sfGFP-D134TAG that contains a gene encoding superfolder green fluorescent protein (sfGFP) with an amber mutation at D134 and a C-terminal 6×His tag were used to cotransform E. coli BL21(DE3) cells. Growing the transformed cells in the presence of AcdK afforded full-length sfGFP (sfGFP-D134AcdK), which was markedly contrasted with non-detectable full-length sfGFP expressed in the absence of AcdK (Figure 2A). In the presence of 0.5 mM AcdK, 7 mg/L sfGFP-D134AcdK was expressed. This observation demonstrates the high selectivity of AcdKRS toward AcdK. The electrospray ionization mass spectrometry (ESI-MS) analysis of the purified sfGFP-D134AcdK displayed two major peaks (Figure 2B). One major peak at 28013 Da agrees well with the theoretically molecular weight of sfGFP-D134AcdK (28013 Da). The other major peak at 27839 Da implies partial reduction of AcdK in sfGFP-D134AcdK to AlK (the theoretic mass of sfGFP-D134AlK is 27839 Da). This was confirmed via labeling the purified sfGFP-D134AcdK readily with a hydroxylamine-conjugating fluorescein dye (Supplementary Figure 2; the original sfGFP fluorescence was quenched by denaturing the protein at the boiling temperature). Since no reducing reagents were provided during the purification process, this partial reduction is possibly due to reactions with endogenous thiol-containing reagents such as glutathione and H2S in E. coli cells.[11] Given that AcdK is used as a precursor to install AlK in a protein, this partial conversion of AcdK to AlK is not a concern but a benefit. The purified sfGFP-D134AcdK was later converted quantitatively to sfGFP-D134AlK by reacting with TCEP (Figure 2C). The minor peak at 27857 Da in Figure 2C represents the hydrated form of sfGFP-D134AlK (the theoretic mass: 27857 Da). Partial hydration of an aliphatic aldehyde in water is a general observation. Given its reversible nature, this hydration is not expected to interfere with the following reductive amination. SfGFP-D134AlK was subsequently converted to sfGFP with dimethyllysine (Kme2) installed at D134 (sfGFP-D134Kme2) by incubating with dimethylamine and NaCNBH3. The installation of Kme2 was confirmed by both Western blot and ESI-MS analyses (Figure 2D–E; the theoretic molecular weight of sfGFP-D134Kme2 is 27868 Da). The Kme2 site was further confirmed with the tandem MS (MS/MS) analysis of trypsinized sfGFP-D134Km2 fragments (Supplementary Figure 3). We also tested the synthesis of sfGFP with methyllysine (Kme) installed at D134 (sfGFP-D134Kme) by reacting sfGFP-D134AlK with methylamine and NaCNBH3. The finally synthesized sfGFP-D134Kme displayed an ESI-MS peak at 27855 Da that agrees well with the theoretical mass (27854 Da) (Figure 2F). The formation of Kme in sfGFP-D134Kme cannot be independently confirmed with the Western blot analysis due to the lack of a commercial pan anti-Kme antibody. For both Staudinger reduction and reductive amination, reactions were carried out in the PBS buffer at pH 7 and room temperature, no protein aggregation and quenching of sfGFP fluorescence were observed, indicating that both reactions are compatible with preserving proteins in their native forms.
Figure 2. The genetic incorporation of AcdK into a model protein sfGFP for the lysine di- and monomethylation installation.
(A) The site-specific incorporation of AcdK into sfGFP at its D134 position to produce sfGFP-D134AcdK. Cells were transformed with two plasmids containing genes for AcdKRS, tRNAPyl, and sfGFP with an amber mutation at its D134 position and grown in LB media supplemented with or without 0.5 mM AcdK. (B–C) The ESI-MS spectra of purified sfGFP-S2AcdK (B) and its TCEP-reduced product (C). (D) The selective installation of Kme2 at the D134 position of sfGFP. SfGEP-D134AcdK was reduced with 5 mM TCEP in PBS buffer at pH 7 for 2 h followed by the reaction with 100 mM dimethylamine and 5 mM NaCNBH3 in PBS buffer at pH 7 for 8 h for the installation of Kme2 to form sfGEP-D134Kme2. The original sfGEP-D134AcdK was used as a control in the Western blot and SDS-PAGE analysis. The top panel shows a western blot probed by anti-Kme2 and the bottom panel shows a Coomassie blue stained gel for two proteins. (E–F) The ESI-MS spectraof sfGFP-D134Kme2 (E) and sfGFP-D134Kme (F).
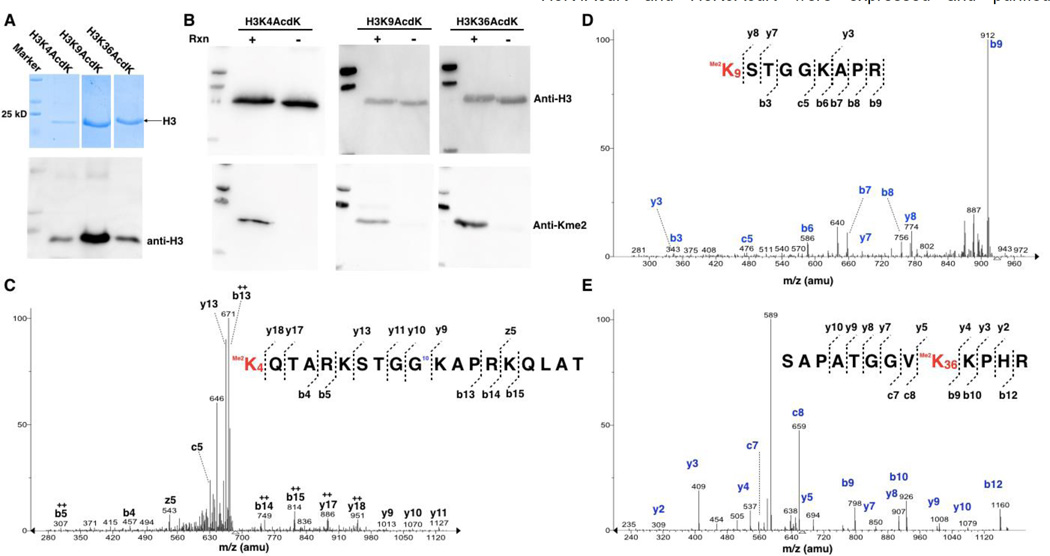
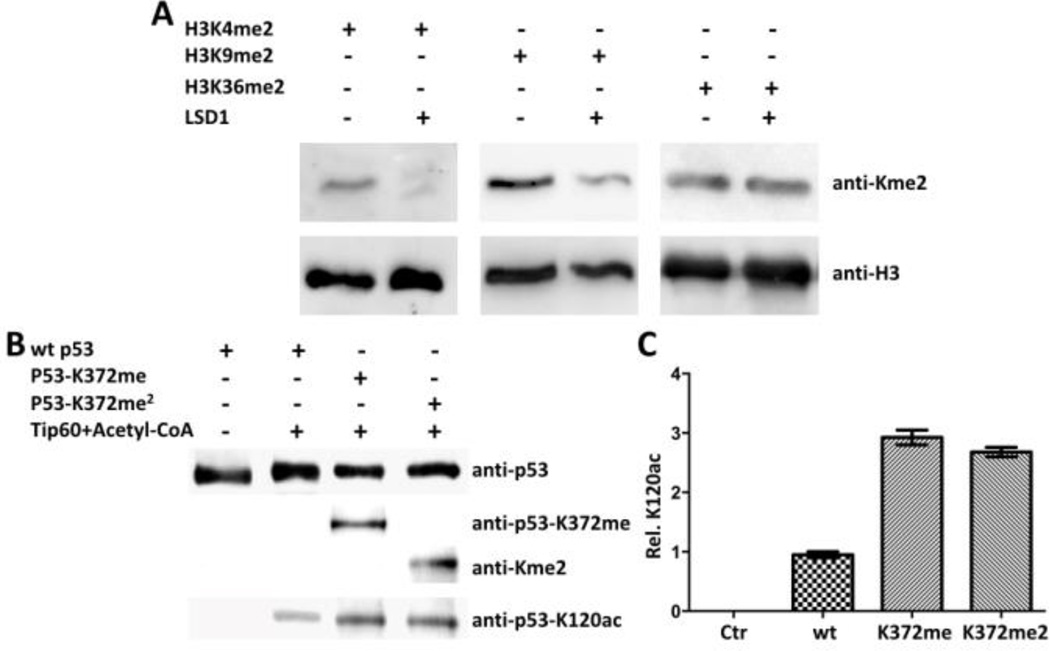
After the demonstration of the designed method for the synthesis of sfGFP installed site-specifically with Kme2, we next moved to synthesize three dimethyl-histone H3 variants and used them to probe substrate specificities of LSD1. LSD1 is a FAD-dependent histone demethylase that selectively targets H3 at K4 and K9 positions to remove their mono- and dimethylation but is inert toward other methylated H3 lysines.[12] In order to synthesize H3K4me2 and H3K9me2, two precursor proteins H3K4AcdK and H3K9AcdK were expressed and purified similarly as sfGFP-D134AcdK (Figure 3A). H3K36AcdK was synthesized in parallel as a control. All three proteins were then processed with Staudinger reduction and reductive amination to make H3K4me2, H3K9me2, and H3K36me2, respectively. The formation of the three dimethyl-H3 variants was confirmed with their detection by the pan anti-Kme2 antibody in the Western blot analysis (Figure 3B) and the Kme2 installation sites were further confirmed with the tandem MS analysis of trypsinized fragments of three histone proteins (Figure 3C–E). The installation of Kme2 at H3K4 was also independently confirmed with Western blotting by anti-H3K4me2 (Supplementary Figure 5). We also proceeded to synthesize H3K4me and H3K9me. However, commercial pan anti-Kme2/Kme antibodies from both Abcam and PTM Biolabs failed to recognize them although they detected dimethyl-H3 variants successfully (Supplementary Figure 6). The three synthesized dimethyl-H3 variants were then used as substrates of LSD1 to test its demethylation activities. As expected, LSD1 actively removed dimethylation at K4 and K9 but was inert toward dimethylation at K36 of H3 (Figure 4A). To show that the designed method can be applied to large proteins with multiple cysteine residues, we chose to work on p53. P53 is a tumor antigen and a transcription factor with 393 amino acids. Its mutations are associated with 60% of tumors. P53 undergoes methylation at K372 that activates its transcription activity.[13] It has been speculated that this activating role is achieved through the recruitment of Tip60, a histone acetyltransferase that has a methyllysine-binding chromodomain and acetylates p53 at K120.[14] We determined to confirm this via using p53 proteins that are site-specifically installed with lysine mono- and dimethylation. P53 with AcdK incorporated at K372 was first expressed in E. coli similarly as sfGFP-D134AcdK (Supplementary Figure 7). P53 typically has very low expression yields in E. coli.[15] Therefore sfGFP with a C-terminal 6×His tag was fused to the C-terminus of p53 that also had a N-terminal GST tag for boosting up its expression in E. coli and easy separation from cell lysate. The expressed protein then underwent Staudinger reduction and reductive amination to generate p53 with mono- and dimethylation at K372 (p53-K372me and p53-K372me2), respectively. As shown in Figure 4B, the formation of p53-K372me and p53-K372me2 was conformed with the Western blot analysis using anti-p53-K372me and pan anti-Kme2 antibodies. A wild type p53 was also expressed as a control. All three proteins were then reacted with Tip60 in the presence of acetyl-CoA and subsequently probed by an anti-p53-K120ac antibody. As shown in the bottom panel in Figure 4B, Tip60 catalyzed K120 acetylation of all three proteins however K372 methylation significantly enhanced the reaction. Both mono- and dimethylation at K372 improved K120 acetylation by 2.5 to 3 folds (Figure 4C). This is the first evidence to show that K372 methylation of p53 directly recruits Tip60 to p53 for its acetylation at K120.
Figure 3. The synthesis of H3K4me2, H4K9me2, and H3K36me2.
(A) SDS-PAGE and western blot analyses of purified AcdK-containing H3 variants. Two plasmids pEVOL-AcdKRS and petDeut-1-H3 with a gene coding H3 with an amber mutation were used to co-transform E. coli BL21(DE3) cells. The transformed cells were grown in LB medium to OD 0.8. H3 expression was induced with the addition of 0.2% arabinose, 1 mM IPTG, and 2 mM AcdK. The expressed H3 variants were purified using Ni-NTA resins. (B) The Western blotting analysis of AcdK-containing H3 variants after their Staudinger reduction with TCEP and reductive amination with dimethylamine. Reaction conditions are ss same as shown in Figure 2D. (C–E) Tandem mass spectrometry analysis of Kme2-containing and trypsinized fragments of H3K4me2, H3K9me2, and H3K36me2.
Figure 4. Probing functions of epigenetic enzymes with synthetic dimethyl-H3 and p53 proteins.
(A) LSD1 activities on three dimethyl-H3 variants. 1 µg H3K4me2, H3K9me2, and H3K36me2 were incubated with 0.1 µg for 8 h before they were analyzed in SDS-PAGE and probed by anti-Kme2 and anti-H3 antibodies in western blotting. (B) Methylation at p53 K372 activates Tip60-catalyzed acetylation at p53 K120. 0.1 µg wild type (wt) p53, p53-K372me, and p53-K372me2 was incubated with 0.01 µg Tip60 for 2 h before they were analyzed by SDS-PAGE and probed with antibodies such as anti-p53-K372me, anti-Kme2, and anti-p53-K120ac in western blotting. (C) Quantified activation effects of p53 K372 methylation on p53 K120 acetylation by Tip60.
In summary, a method that allows expedient synthesis of proteins with site-specific lysine dimethylation has been successfully demonstrated. The method combines the amber suppression-based mutagenesis with two biocompatible organic reactions to install site-specific lysine dimethylation in proteins. Given its specificity, biocompatibility, and simplicity, it can be generally applied to synthesize proteins with site-specific lysine dimethylation for their functional investigation. Besides proteins with lysine dimethylation, we showed that the same method could be applied to synthesize proteins with lysine monomethylation. Other posttranslational lysine alkylation types that can be potentially installed site-specifically into proteins using the presented method include the enzymatic deoxyhypusine and hypusine formation in eukaryotic eIF5A and metabolic lysine glycation subtypes such as carboxymethylation.[16] Given that allysine itself naturally occurs in elastin and collagen for their crosslinking,[7] the application of our currently reported method in studying the elastin and collagen fibril formation is also anticipated.
Experimental Section
For details of synthesis and characterization of AcdK, selection of AcdKRS, expression of proteins and their treatment to install mono- and dimethylation, and assay conditions of LSD1 and Tip60, please see the supporting information.
Supplementary Material
Acknowledgments
Support of this work was provided from National Institute of Health (grant CA161158), National Science Foundation (grant CHE-1148684), Welch Foundation (grant A-1715), and the National Natural Science Fundation of China (grants 21402199 and 21502192). We thank Dr. Yohannes H. Reznom at the Laboratory for Biology Mass Spectrometry and Dr. Larry Dangott at the Protein Chemistry Lab of Texas A&M University for characterizing our proteins with mass spectrometry.
References
- 1.Hamamoto R, Saloura V, Nakamura Y. Nat Rev Cancer. 2015;15:110–124. doi: 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- 2.Greer EL, Shi Y. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]; b Lu T, Stark GR. Cancer Res. 2015;75:3692–3695. doi: 10.1158/0008-5472.CAN-15-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Chatterjee C, Muir TW. J Biol Chem. 2010;285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nguyen UT, Bittova L, Muller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW. Nat Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. J Am Chem Soc. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]; b Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. Mol Biosyst. 2010;6:1557–1560. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]; c Groff D, Chen PR, Peters FB, Schultz PG. Chembiochem. 2010;11:1066–1068. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ai HW, Lee JW, Schultz PG. Chem Commun (Camb) 2010;46:5506–5508. doi: 10.1039/c0cc00108b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen DP, Garcia Alai MM, Virdee S, Chin JW. Chem Biol. 2010;17:1072–1076. doi: 10.1016/j.chembiol.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi M, Sricholpech M. Essays Biochem. 2012;52:113–133. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Chen PR. Nat Chem Biol. 2016;12:129–137. doi: 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 10.a Neumann H, Peak-Chew SY, Chin JW. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]; b Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, Schultz PG. Angew Chem Int Ed Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, Fu X, Chang Z, Chen PR. Nat Chem Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]; d Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, Pai PJ, Russell DH, Liu WR. Mol Biosyst. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]; e Wan W, Tharp JM, Liu WR. Biochim Biophys Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Staros JV, Bayley H, Standring DN, Knowles JR. Biochem Biophys Res Commun. 1978;80:568–572. doi: 10.1016/0006-291x(78)91606-6. [DOI] [PubMed] [Google Scholar]; b Lippert AR, New EJ, Chang CJ. J Am Chem Soc. 2011;133:10078–10080. doi: 10.1021/ja203661j. [DOI] [PubMed] [Google Scholar]; c Peng H, Cheng Y, Dai C, King AL, Predmore BL, Lefer DJ, Wang B. Angew Chem Int Ed Engl. 2011;50:9672–9675. doi: 10.1002/anie.201104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]; b Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 13.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Tang Y, Luo J, Zhang W, Gu W. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 15.a Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T. Proc Natl Acad Sci U S A. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Midgley CA, Fisher CJ, Bartek J, Vojtesek B, Lane D, Barnes DM. J Cell Sci. 1992;101(Pt 1):183–189. doi: 10.1242/jcs.101.1.183. [DOI] [PubMed] [Google Scholar]
- 16.a Park MH. J Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Delgado-Andrade C. Food Funct. 2016;7:46–57. doi: 10.1039/c5fo00918a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.