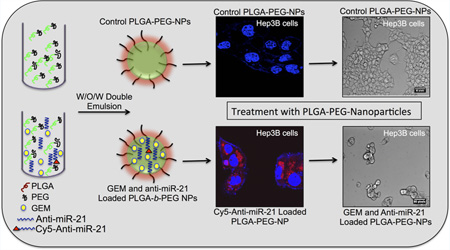
Abstract
Hepatocellular carcinoma (HCC) is highly prevalent, and the third most common cause of cancer-associated deaths worldwide. HCC tumors respond poorly to chemotherapeutic anticancer agents due to inherent and acquired drug resistance, and low drug permeability. Targeted drug delivery systems with significant improvement in therapeutic efficiency are needed for successful HCC therapy. Here, we report the results of a technique optimized for the synthesis and formulation of antisense-miRNA-21 and gemcitabine (GEM) co-encapsulated PEGylated-PLGA nanoparticles (NPs) and their in vitro therapeutic efficacy in human HCC (Hep3B and HepG2) cells. Water-in-oil-in-water (w/o/ w) double emulsion method was used to coload antisense-miRNA-21 and GEM in PEGylated-PLGA-NPs. The cellular uptake of NPs displayed time dependent increase of NPs concentration inside the cells. Cell viability analyses in HCC (Hep3B and HepG2) cells treated with antisense-miRNA-21 and GEM co-encapsulated NPs demonstrated a nanoparticle concentration dependent decrease in cell proliferation, and the maximum therapeutic efficiency was attained in cells treated with nanoparticles co-encapsulated with antisense-miRNA-21 and GEM. Flow cytometry analysis showed that control NPs and antisense-miRNA-21-loaded NPs are not cytotoxic to both HCC cell lines, whereas treatment with free GEM and GEM-loaded NPs resulted in ~9% and ~15% apoptosis, respectively. Cell cycle status analysis of both cell lines treated with free GEM or NPs loaded with GEM or antisense-miRNA-21 displayed a significant cell cycle arrest at the S-phase. Cellular pathway analysis indicated that Bcl2 expression was significantly upregulated in GEM treated cells, and as expected, PTEN expression was noticeably upregulated in cells treated with antisense-miRNA-21. In summary, we successfully synthesized PEGylated-PLGA nanoparticles co- encapsulated with antisense-miRNA-21 and GEM. These co-encapsulated nanoparticles revealed increased treatment efficacy in HCC cells, compared to cells treated with either antisense-miRNA-21- or GEM-loaded NPs at equal concentration, indicating that down-regulation of endogenous miRNA-21 function can reduce HCC cell viability and proliferation in response to GEM treatment.
Keywords: PLGA nanoparticles, gemcitabine, drug delivery, antisense-miRNA-21, miRNA-21, Bcl2, PTEN, hepatocellular carcinoma
Graphical abstract
INTRODUCTION
The World Health Organization reports that one of the world’s leading causes of death is associated with cancer.1,2 Hepatocellular carcinoma (HCC) is the third most common cause of cancer-associated deaths worldwide, next to stomach and lung cancers.3–5 HCC is a primary malignancy of the liver cells (hepatocytes) that, in most cases, develops due to the risk factors such as chronic Hepatitis B and C infection, alcohol abuse, hemochromatosis, or exposure to several chemicals.4,5 HCC poorly responds to chemotherapeutics owing to the obstacles such as intrinsic and acquired drug resistance and low permeability of drugs.6,7 Existing treatment approaches involve surgical resection or liver transplantation, but tumor recurrence and metastasis remain common problems.8 Improved treatment strategies including new chemotherapeutics and superior drug delivery systems are required for effective HCC treatment.9,10 Several nanoparticle (NP)-mediated treatment strategies have been reported for better delivery of chemotherapeutics to HCC cells.11,12 The advantages of NP-mediated drug delivery methods include their ability to modify the pharmacological effect, pharmacokinetics, bioavailability, bio-distribution, extended circulation time of drugs, and drug targeting to the site of action.13,14
Gemcitabine (GEM; 2′-deoxy-2′,2′-difluorocytidine) is currently used as a chemotherapeutic drug for treating several kinds of cancers, including pancreatic cancer,15 nonsmall cell lung carcinoma, ovarian cancer, and breast cancer.16 GEM is also a promising chemotherapeutic agent for treating HCC, when administered either by itself or in combination with other chemotherapeutics.17 After intravenous injection of GEM, it undergoes rapid enzymatic degradation in systemic circulation and causes various adverse effects, including hair loss, fever, fatigue, nausea, and vomiting. Various drug delivery systems have been developed using polymer nanoparticles to overcome adverse effects associated with GEM.16,18–24 GEM-loaded poly(lactide)-co-glycolide-block-poly(ethylene glycol) (PLGA-b-PEG-NH2) nanoparticles have been prepared by adopting water-in-oil-in-water (w/o/w) double emulsion method with 35% encapsulation efficiency, demonstrating significant cytotoxic effect in MIA PaCa-2 pancreatic cancer cells.18 GEM-loaded chitosan NPs prepared using coacervation method developed by Arias et al. produced a substantial improvement in antitumor activity against a subcutaneous tumor graft of L1210 mouse lymphocytic leukemia cells in mice compared to free gemcitabine treatment.23 A more precise drug delivery system is required to increase the delivery of active GEM to tumors and to achieve enhanced antitumor effect.
MicroRNAs (miRNAs or miRs) are a family of small noncoding RNAs of 18–24 nucleotides endogenously expressed in cells and are involved in the regulation of gene expression in cells.16,25–27 MiRNAs trigger translational repression through interactions with the 3′-untranslated region of mRNA regulating gene expression. These miRNAs play a significant role in regulating genes involved in developmental timing. Moreover, several miRNAs have been shown to be regulating genes involved in various cellular processes during tumorigenesis.28 Numerous research results have indicated that miRNAs expression is dysregulated in human HCC.29–31 The overexpression of some miRNAs promotes cancer development by stimulating growth signaling of cancer cells; these miRNAs are called oncomiRs.16 Acquisition of drug resistance or chemotherapy resistance is another common problem in HCC patients, and altered expression of miRNAs has been shown to increase drug resistance in cisplatin treated HCC.32 These results indicate that regulating the function of oncomiRs that are overexpressed in cancer or miRNAs responsible for drug resistance can improve cancer therapy. Targeting miRs is a promising new method in the development of a novel class of anticancer therapeutics. Small chemically modified antisense oligonucleotides, complementary to the mature miRs sequences, are developed for blocking the function of endogenous miRs, called miR inhibitors or anti-miRs or antisense-miRs.33,34 Anti-miRs prevent miRNA activity by irreversibly binding to the target miRNAs. MiRNA-21 is one of the first miRNAs identified in mammalian cells, termed as oncomiR, which has been connected with a wide variety of cancers, including HCC.35–37 MiR-21 up-regulation promoted HCC cell proliferation through repression of mitogen-activated protein kinase 3, favoring angiogenesis and invasion and reducing cell death.37,38 MiR-21 inhibition in HCC cell lines increased the expression of PTEN (phosphatase and tensin homologue) tumor suppressor protein, decreasing the cell proliferation, migration, and invasion.36 Unfortunately, the synthetic naked anti-miRs are unstable in a nuclease rich serum and plasma environment.39 Effective shielding agents are required for the delivery of these nucleic acids in vivo. Numerous delivery methods have been developed to protect nucleic acids from degradation.16,39–43
PLGA is a biocompatible and biodegradable copolymer that has been the most attractive and effective polymeric drug delivery carrier reported for clinical applications.44,45 Biodegradable PLGA-b-PEG NPs or PEGylated PLGA NPs have been widely studied for the purpose of anticancer drug delivery.16,40 PEGylation of the PLGA has been shown to improve the NP’s circulation time in vivo and tumor uptake through the enhanced permeability and retention (EPR) effect.39,46 Furthermore, PEGylation protects NPs from the immune recognition and increases bioavailability.16 PEGylated PLGA NPs composed of a hydrophobic PLGA core and encircled by a hydrophilic PEG layer are one of the best-controlled release systems for targeted drug delivery.47
To the best of our knowledge, combinational treatment of HCC by antisense-miRNA-21 and GEM NPs has not been previously reported. Here, we report the synthesis of PEGylated-PLGA NPs co-encapsulated with antisense-miRNA-21 and GEM and their antiproliferative and cytotoxic effects in HCC (Hep3B and HepG2) cell lines.
RESULTS AND DISCUSSION
Synthesis and Characterization of PEGylated-PLGA NPs Co-encapsulated with Antisense-miRNA-21 and GEM
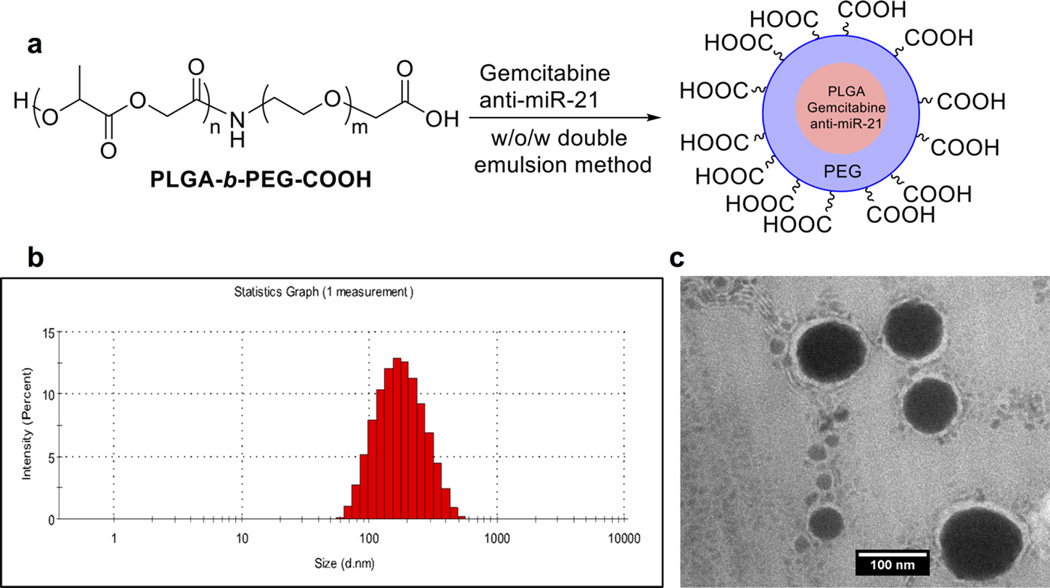
Owing to the highly hydrophilic nature of antisense-miRNA-21 and GEM, we have formulated PEGylated-PLGA NPs loaded with antisense-miRNA-21 and GEM using the w/ o/w double emulsion method (Figure 1a). We developed an optimal procedure to load a higher concentration GEM using dimethyl sulfoxide (DMSO) as a cosolvent to dissolve GEM with PLGA-b-PEG copolymer (PLGA, MW 7000–17000; PEG, MW 3400) and hydrophobic Span 80, and hydrophilic poly(vinyl alcohol) (PVA) as NP stabilizers for the first and second emulsions, respectively. We used spermidine as a counterion for encapsulating antisense-miRNA-21.39 Hydrodynamic sizes of NPs determined using dynamic light scattering (DLS) were in the range of 150–230 nm (Figure 1b). The size and morphology of NPs were further confirmed by transmission electron microscopy (TEM) and displayed uniform spherical shape with sizes in the range of 100–200 nm (Figure 1c).
Figure 1.
Synthesis and characterization of PEGylated-PLGA-NPs: (a) Formulation of gemcitabine and antisense-miRNA-21 co-encapsulated PEGylated-PLGA-NPs; (b) hydrodynamic particle size of NPs prepared by w/o/w double emulsion method by dynamic light scattering (DLS); (c) TEM images of PEGylated-PLGA-NPs.
Identification of Optimal Method To Co-load Anti-sense-miRNA-21 and GEM in PEGylated-PLGA NPs
After experimenting with several formulations, we optimized GEM loading in PEGylated-PLGA- NPs by employing the w/o/w method using 1% Span 80 and 1% PVA as stabilizers (Table 1). We were able to achieve only 20–24% encapsulation efficiency (ee) using a previously described double emulsion method (Table 1, entry 1).39 We assumed that lower ee that resulted from a previously described procedure may be due to the leakage of water-soluble GEM into the aqueous layer. Hence, we decided to use a cosolvent method by dissolving GEM in DMSO and premixing it with PLGA-b-PEG copolymer in dichloromethane. The ee slightly increased (24–31%) when DMSO was utilized as a cosolvent (Table 1, entries 2 and 3). We also tried a emulsion-diffusion evaporation method41 and achieved only 27% ee (Table 1, entry 4). Changing the surfactant to 2% PVA for the first emulsion and 1% PVA for the second emulsion resulted in 28% ee (Table 1, entry 5). However, the higher ee (42%) was achieved using 1% Span 80 for the first emulsion and 1% PVA for the second emulsion (Table 1, entry 6). Substituting for water as GEM solvent, instead of utilizing the cosolvent method, resulted in only 24% ee (Table 1, entry 7). These results signify the importance of premixing GEM with PLGA-b-PEG polymer. The best GEM encapsulation method (Table 1, entry 6) for loading antisense-miRNA-21 resulted in 66% ee of antisense-miRNA-21 (Table 1, entry 8), which is close to the optimal ee for loading antisense-miRNA-21.39 Coloading of antisense-miRNA-21 and GEM resulted in 64% and 40% ee for antisense-miRNA-21 and GEM, respectively (Table 1, entry 9).
Table 1.
Optimization of Antisense-miRNA-21 and Gemcitabine Loading in PEGylated-PLGA NPs
| S no. | method | surfactant for emulsification |
gemcitabine solvent | mean size (nm)a |
PDIa | ζ potentiala | encapsulation efficiency gemcitabine/antisense-miRNA-21 (%) |
|---|---|---|---|---|---|---|---|
| 1 | w/o/w | 3% Span 80, 1% Tween 80 |
water | 148.4 | 0.163 | −20.4 | 20.4/− |
| 2 | w/o/w | 1% PVA | DCM/DMSO (10:1) |
195.5 | 0.082 | −8.6 | 24.4/− |
| 3 | w/o/w | 1% Span 80, 1% PVA | DCM/DMSO (10:1) |
230.8 | 0.242 | −10.9 | 31.2/− |
| 4 | EDE | 2% PVA | DCM/DMSO (10:1) |
170.4 | 0.320 | −7.5 | 27.4/− |
| 5 | w/o/w | 2% PVA, 1% PVA | DCM/DMSO (10:1) |
189.9 | 0.206 | −9.4 | 28/− |
| 6 | w/o/w | 1% Span 80, 1% PVA | DCM/DMSO (10:1) |
204.9 | 0.155 | −14.7 | 42/− |
| 7 | w/o/w | 1% Span 80, 1% PVA | water | 232.6 | 0.191 | −12.9 | 24.0/− |
| 8 | w/o/w | 1% Span 80, 1% PVA | DCM/DMSO (10:1) |
215.1 | 0.189 | −22.6 | −/68.3 |
| 9 | w/o/w | 1% Span 80, 1% PVA | DCM/DMSO (10:1) |
224.9 | 0.145 | −18.8 | 40/64.2 |
Note: average of three DLS measurements.
For cell culture studies, we used the identified optimal methods shown in Table 1 for making different NPs (GEM, entry 6; antisense-miRNA-21, entry 8; coloading, entry 9). We prepared control NPs, antisense-miRNA-21, and GEM individually loaded, as well as co-encapsulated NPs, and characterized for their hydrodynamic size, polydispersity index (PDI), and ζ potential (Table 2, entries 1–4). These NPs possessed hydrodynamic sizes between 195 and 215 nm, with 0.140–0.190 PDI and ζ potential of −14 to −23 mV. Control NPs and GEM-loaded NPs had a ζ potential of around −15 mV (Table 2, entries 1 and 2). As anticipated, ζ potential was increased to −23 mV when NPs were co-encapsulated with 10 nmol of antisense-miRNA-21 (Table 2, entry 3), owing to the highly anionic nature of antisense-miRNAs. However, ζ potential decreased to −19 mV when they were loaded with 5 nmol of antisense-miRNA-21 (Table 2, entry 4). This result indicates that ζ potential varies with the concentrations of microRNA loaded in NPs.
Table 2.
Optimized PEGylated-PLGA NPs Used in the Cell Culture Studies
| gemcitabine/antisense-miRNA-21 |
|||||||
|---|---|---|---|---|---|---|---|
| S no. | PEGylated-PLGA-NPs | mean Size (nm)a |
PDIa | ζ potential | EE (%)a | loading (%) |
antisense-miRNA molecules/NP |
| 1 | control NPs | 198.4 ± 18.4 | 0.187 | −15.7 ± 3.1 | |||
| 2 | gemcitabine | 201.8 ± 15.1 | 0.155 | −14.7 ± 2.6 | 42 ± 6.2/− | 2.1/− | |
| 3 | antisense-miRNA-21 | 205.1 ± 18.1 | 0.189 | −22.6 ± 2.9 | −/64.8 ± 9.6 | −/0.45 | 1680 ± 185 |
| 4 | gemcitabine and antisense-miRNA-21 |
212.6 ± 16.3 | 0.148 | −18.8 ± 2.4 | 40 ± 6.3/62.7 ± 8.6 | 2.0/0.43 | 1630 ± 176 |
Note: average from three experiments.
In Vitro Drug Release Studies of PEGylated-PLGA NPs Loaded with GEM
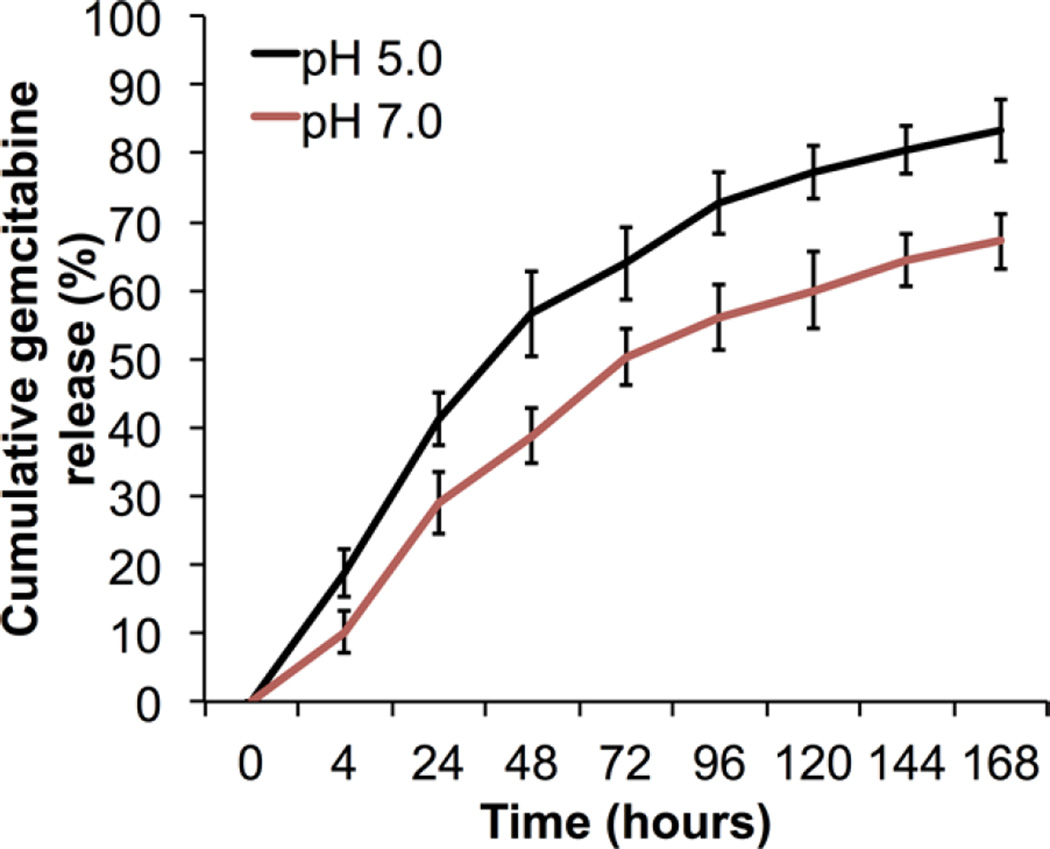
Slow and sustained release properties of drug delivery agents are essential for minimizing the negative side effects of anticancer drugs. Hydrophobic PLGA degrades slowly through hydrolysis of its ester bonds in water, while releasing encapsulated drugs and its monomers lactic acid and glycolic acid inside the cells.16 In our previous study we have shown that antisense-miRNA-21 and antisense-miRNA-10b co-encapsulated in PEGylated-PLGA NPs displayed significant stability for more than a week, even in cell culture medium.39 In this study after optimizing the GEM loading into NPs, we performed in vitro drug release studies (Figure 2). We have collected released GEM over time and cumulatively calculated the GEM percentage. These GEM-loaded NPs showed an initial burst release of 19% and 41% at pH 5.0 and 10% and 29% at pH 7.0 measured after 4 and 24 h, respectively. Subsequently, GEM was released in a sustained manner with 57% and 39% release after 48 h, 64% and 50% after 72 h, and 73% and 56% after 96 h at pH 5.0 and pH 7.0, respectively. At the later time points, a smaller amount of GEM was released gradually with 83% and 67% release after 7 days at pH 5.0 and pH 7.0, respectively. These results demonstrated a higher release of GEM from NPs at pH 5.0, compared to pH 7.0 (Figure 2). Khaira et al. reported that GEM loaded in starch NPs showed fast drug release properties with nearly 60% burst release of GEM after 10 h and 80% in 24 h.48 However, GEM loaded in PLGA NPs prepared with Pluronic F68 surfactant displayed an initial burst release (40%), followed by slow and sustained release (80%) up to 96 h at pH 7.4.20 The PEGylated-PLGA-NPs formulated by us with a combination of hydrophobic surfactant Span 80 and hydrophilic surfactant PVA slowly released GEM over time in a sustained manner for up to 83% in 7 days (Figure 2). This result indicates the usefulness of PEGylated-PLGA-NPs for anticancer drug delivery to show a sustained therapeutic effect. In addition, slow and sustained release of GEM from PEGylated-PLGA-NPs provides the optimal condition to release active GEM over time by avoiding its enzymatic degradation inside the cells and also by decreasing negative side effects, which further highlights the need of lower doses of GEM administration for clinical patient care.
Figure 2.
In vitro GEM release studies of GEM-loaded PEGylated-PLGA-NPs for 7 days.
Cell Uptake Studies of Cy5-Conjugated Antisense-miRNA-21 and GEM Co-encapsulated NPs in Hep3B Cells
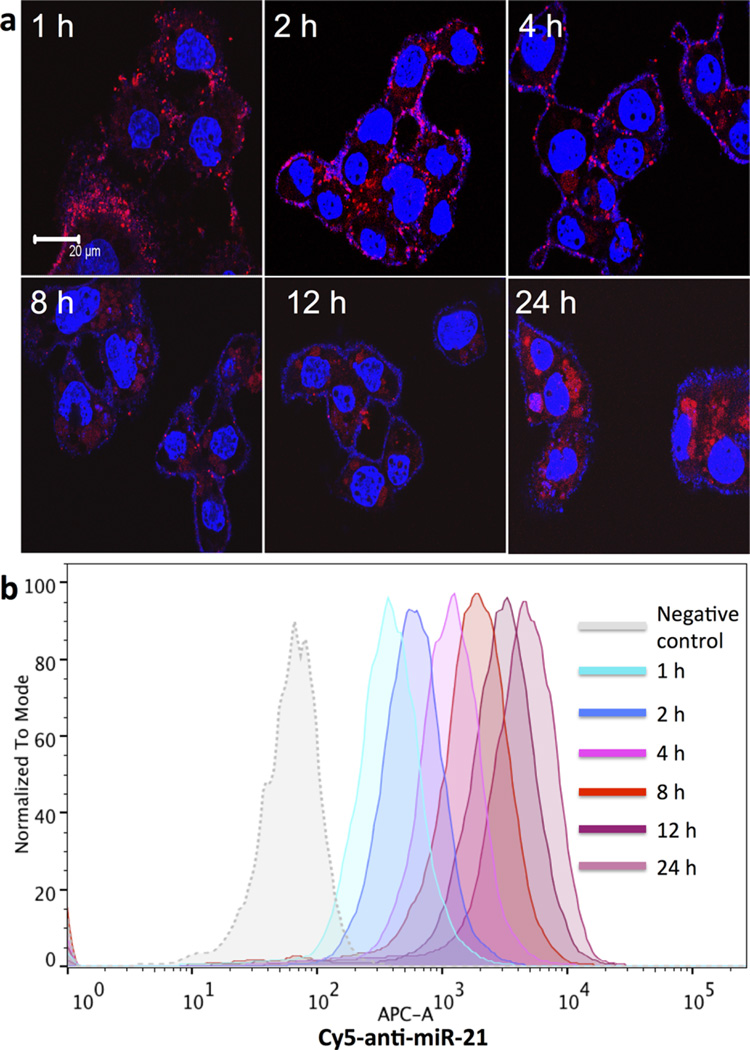
Antisense-miRNA-21 with 10% Cy5-conjugated anti-sense-miRNA-21 and GEM coloaded NPs was used to track the cellular uptake and to monitor the intracellular delivery of the NPs in Hep3B cells. We used confocal fluorescent microscopy and fluorescence-activated cell sorting (FACS) analysis to measure uptake at different time points (Figure 3). The microscopic images revealed time dependent increase in uptake of NPs. The NPs cellular entry was detected 1 h after initial treatment, with ring formation over the surface of the cells. NPs entered cell cytoplasm after 2–4 h, indicated by the reduced Cy5 NPs ring intensity. The intracellular uptake of NPs increased over time at 6–8 h, and a large percentage of NPs was detected inside the cells at 24 h (Figure 3a). Similarly, quantitative FACS analysis of cells treated at the same concentration of NPs assessed over time revealed a significant level of Cy5-conjugated antisense-miRNA-21-loaded PEGylated-PLGA NPs uptake (measured by Cy5 fluorescence) 1 h after initial treatment compared to untreated cells (negative control), and increased over time (Figure 3b).
Figure 3.
Time dependent cellular uptake of gemcitabine and antisense-miRNA-21 co-encapsulated PEGylated-PLGA-NPs in Hep3B cells. (a) Confocal microscopic images (red, Cy5-fluorescence from co-encapsulated Cy5-antisense-miRNA-21; blue, DAPI nuclear stain). (b) FACS analysis of Cy5-antisense-miRNA-21 co-encapsulated NPs treated Hep3B cells.
Antiproliferative Effect of Free GEM, GEM NPs, Antisense-miRNA-21 NPs, and Antisense-miRNA-21-GEM Co-encapsulated NPs in HCC (Hep3B and HepG2) Cells
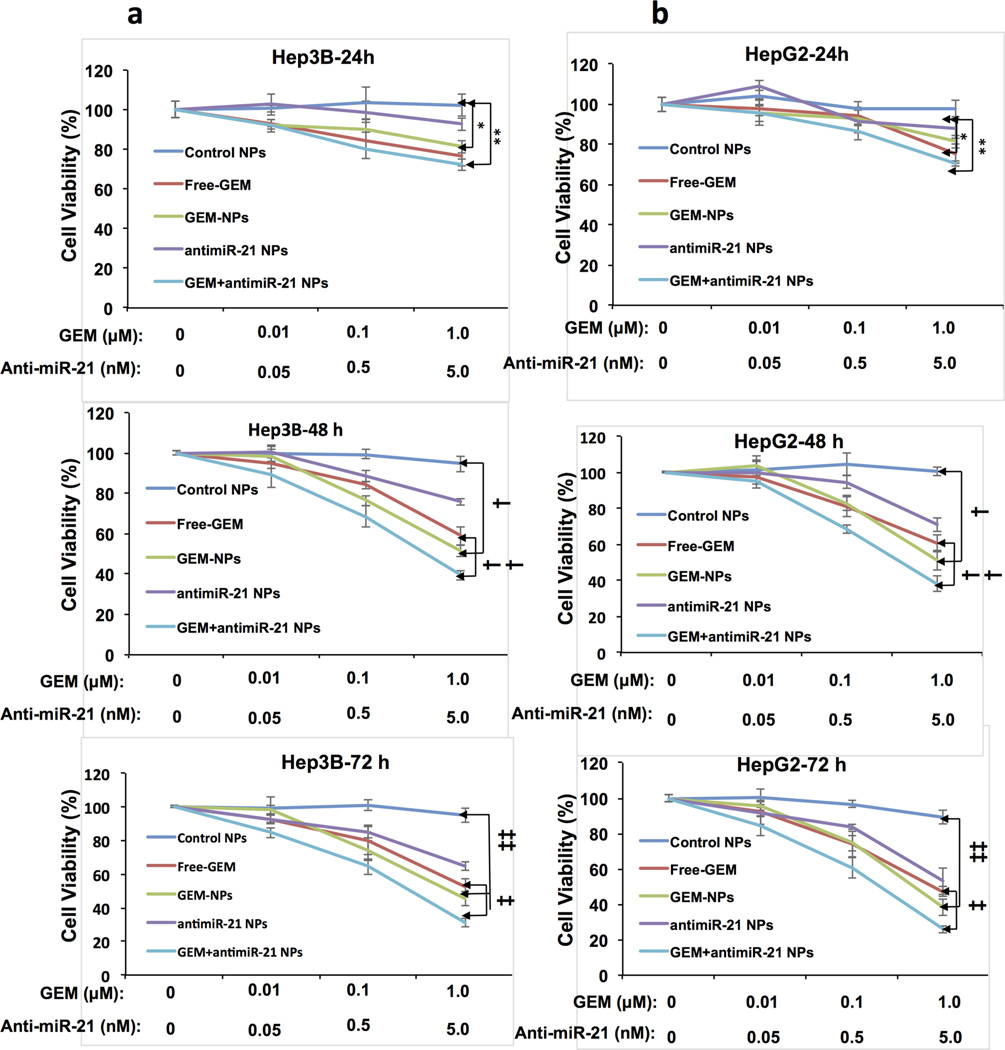
Antiproliferative effects of GEM have been predominantly studied in pancreatic cancer cells.21,49,50 However, in 2008 Matsumoto et al. reported an antiproliferative effect of free GEM in both Hep3B and HepG2 cells and found a dose and time dependent cell growth arrest when applied at increasing doses between100 nM and 1 mM.51 We studied the antiproliferative effect of antisense-miRNA-21- and GEM-loaded NPs in HCC cells. Hep3B and HepG2 cells were seeded in 96-well cell culture plates (10000 cells/well in 100 µL of DMEM containing 2% FBS) and the cells were further incubated for 24 h. The next day, cells were treated with free GEM, GEM NPs, antisense-miRNA-21 NPs, and antisense-miRNA-21 and GEM co-encapsulated PEGylated-PLGA-NPs, as well as with control NPs at several concentrations, and were further incubated for different time points (24–72 h) at 37 °C in 5% CO2. After each time point, the cells were assessed by MTT assay. Results indicated that both HCC cell lines exhibited dose dependent (10 nM to 1 µM) reduction in cell viability with respect to GEM after 24–72 h post-treatments (Figure 4). Antisense-miRNA-21 and GEM co-encapsulated NPs treated cells exhibited lower cell viability compared to free GEM and GEM NPs treated cells in a dose dependent manner, in both cell lines, at 24 and 48 h of treatment (p < 0.01 antisense-miRNA-21-GEM NPs vs free GEM and GEM NPs at 1 µM; Figure 4); as anticipated, control NPs did not result in any considerable toxicity. Interestingly, antisense-miRNA-21-loaded NPs produced a significant antiproliferative effect in both cell lines (p < 0.01, antisense-miRNA-21 NPs vs control NPs at 1 µM; Figure 4). However, NPs loaded with antisense-miRNA-21 alone did not cause any cell death, as revealed by FACS analysis (Figure 5). This indicates that at lower concentrations antisense-miRNA-21 acts as a cytostatic rather than a cytotoxic agent. Overall, our results suggest that co-delivery of antisense-miRNA-21 and GEM increased the therapeutic efficacy against HCC cells. The peak plasma concentration of GEM reported in pancreatic cancer patients is 100 µM.51 Our dose studies suggest that lower concentrations of antisense-miRNA-21 and GEM inhibit HCC cell proliferation. This indicates that our therapeutic approach is more efficient for further in vivo studies, including those intended for applications in humans.
Figure 4.
Cell viability evaluations of HCC cells after treatment with free GEM, GEM/antisense-miRNA-21, individually, and co-encapsulated in PEGylated-PLGA-NPs by MTT assay: Dose response in Hep3B (a) and HepG2 (b) cells treated with free GEM, GEM-loaded NPs, antisense-miRNA-21-loaded NPs, and GEM-antisense-miRNA-21 co-encapsulated NPs by MTT assay [24 h, (*) p < 0.01 control NPs vs GEM NPs, and (**) p < 0.01 control NPs vs GEM-antisense-miRNA-21 NPs at 1 µM; 48 h, (†) p < 0.01 control NPs vs GEM NPs at 1 µM, and (††) p < 0.01 GEM-antisense-miRNA-21 NPs vs free GEM at 1 µM; 72 h, (‡) p < 0.01 GEM-antisense-miRNA-21 NPs vs free GEM at 1 µM, and (‡‡) p < 0.01 control NPs vs GEM NPs at 1 µM].
Figure 5.
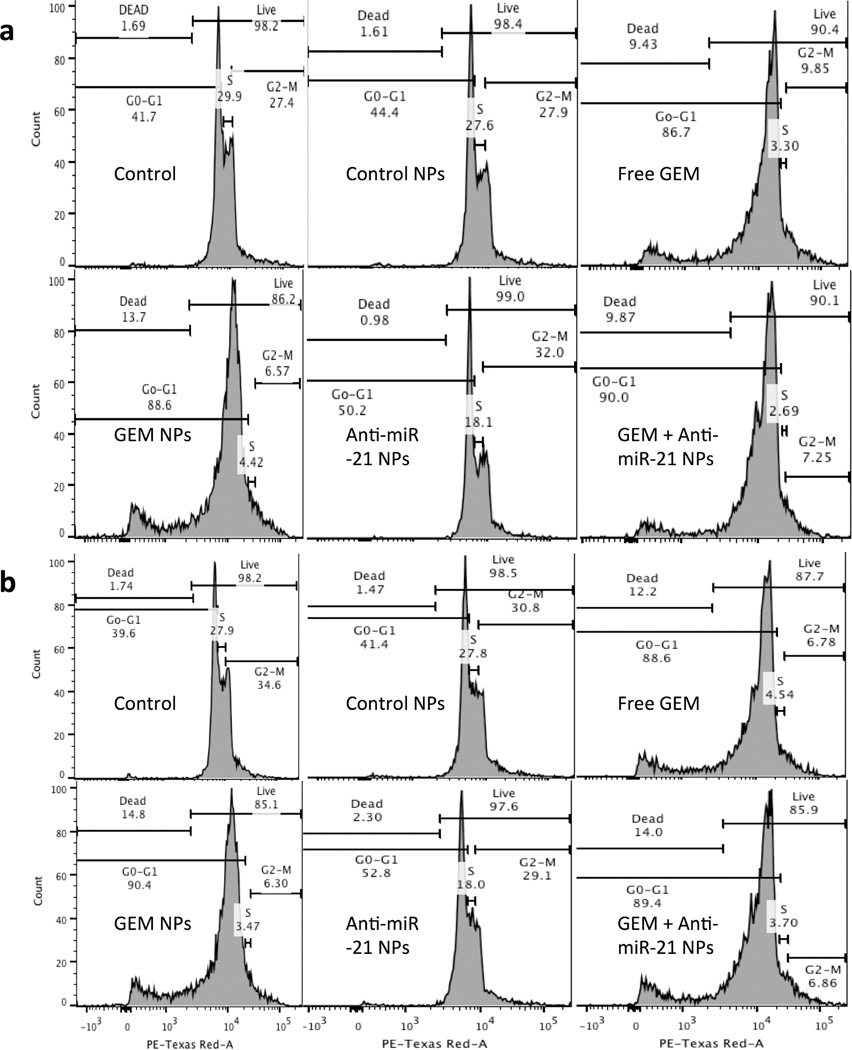
Flow cytometry (FACS) analysis of HEP3B (a) and HepG2 (b) cells treated with free GEM, GEM NPs, antisense-miRNA-21 NPs, and GEM-antisense-miRNA-21 NPs.
Cytotoxicity Evaluation of Antisense-miRNA-21- and GEM-Loaded NPs in HCC Cells
After successfully examining the antiproliferative effect of antisense-miRNA-21-GEM NPs, we further studied cytotoxicity of antisense-miRNA-21- and GEM-loaded NPs in HCC cells and analyzed the NPs’ cytotoxicity by flow cytometry. Hep3B and HepG2 cells were seeded in 12-well tissue culture plates (50000 cells/well, in 1 mL of DMEM containing 10% FBS in each well) and incubated for 24 h. The next day, cells were washed with PBS and supplemented with fresh medium (DMEM/2% FBS). Subsequently, cells were treated with control NPs, free GEM, GEM NPs, antisense-miRNA-21 NPs, or antisense-miRNA-21–GEM co-encapsulated NPs (GEM, 3 µM; antisense-miRNA-21, 15 nM) and further incubated for 48 h at 37 °C in 5% CO2. The cells were then collected and analyzed for treatment induced cell death and cell cycle status, after staining with propidium iodide (PI) by flow cytometry. We used logarithmic scale to analyze the live and dead cell populations and a linear scale to assess cell cycle status (Figure 5a,b, Table 3, and Supporting Information Figure S1). Results indicated that antisense-miRNA-21-loaded NPs and control NPs were not cytotoxic, as indicated by the absence of an apoptotic cell population in both cell lines (Figure 5a,b and Table 3). However, apoptotic populations were observed in free GEM and GEM NPs treated cells. Free GEM, GEM NPs, and antisense-miRNA-21–GEM-loaded NPs treatments resulted in 9%, 12%, and 14% and 15%, 10%, 14% apoptotic cells, respectively, in Hep3B and HepG2 cells. (Figure 5a,b and Table 3). Cell cycle status analysis of both HCC cell lines indicated that untreated control cells and control NPs treated cells displayed similar results. Whereas, a significant increase in the G0/G1 phase with a concomitant decrease in the G2/M phase, and a very high level reduction in S phase cells was observed in cells treated with free GEM and GEM NPs, antisense-miRNA-21 NPs, and antisense-miRNA-21–GEM co-encapsulated NPs. A reduction in S phase cells indicated that cell growth was arrested due to treatment. Especially, antisense-miRNA-21-loaded NPs treated cells showed a decrease in S phase cells with a slight increase in the G0-G1 phase, but not the G2-M phase, compared to untreated control cells (Supporting Information Figure S1, Figure 5a,b, and Table 3). This indicates that antisense-miRNA-21 alone can arrest growth of HCC cells but is not cytotoxic. Also, FACS results demonstrated that both HCC (Hep3B and HepG2) cell lines showed similar trends in response to antisense-miRNA–GEM combination treatment (Figure 5a,b and Table 3). They also suggest that GEM inhibits the growth of both HCC cell lines at lower doses compared to higher doses reported in previous studies.51 Our results are consistent with previous reports, where a significant level of cell cycle arrest was observed in Hep3B cells at the G0/G1 phase after the GEM treatment.51
Table 3.
Cell Cycle Analysis of HCC Cells by FACS
| Hep3B |
HepG2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| apoptotic cells (%) |
live cells (%) |
G0/G1 phase (%) |
S phase (%) |
G2/M phase (%) |
apoptotic cells (%) |
live cells (%) |
G0/G1 phase (%) |
S phase (%) |
G2/M phase (%) |
|
| control cells | 1.69 | 98.2 | 41.7 | 29.9 | 27.4 | 1.74 | 98.2 | 39.6 | 27.9 | 34.6 |
| control NPs | 1.61 | 98.4 | 44.4 | 27.6 | 27.9 | 1.47 | 98.5 | 41.4 | 27.8 | 30.8 |
| free GEM | 9.43 | 90.4 | 86.7 | 3.3 | 9.85 | 12.2 | 87.7 | 88.6 | 4.54 | 6.78 |
| GEM NPs | 13.7 | 86.2 | 88.6 | 4.42 | 6.57 | 14.8 | 85.1 | 90.4 | 3.47 | 6.30 |
| antisense-miRNA-21 NPs | 0.98 | 99.0 | 50.2 | 18.1 | 32.0 | 2.30 | 97.6 | 52.8 | 18.0 | 29.1 |
| antisense-miRNA-21 + GEM NPs | 9.87 | 90.1 | 90.0 | 2.69 | 7.25 | 14.0 | 85.9 | 89.4 | 3.70 | 6.86 |
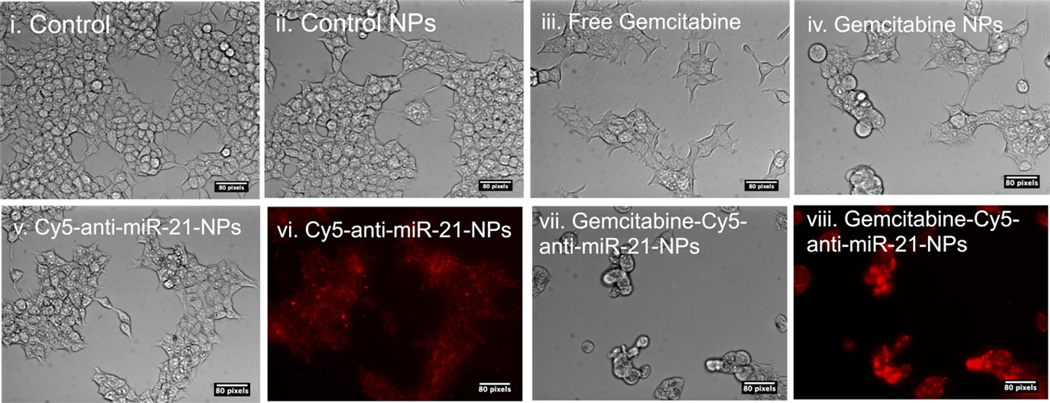
We have also tested the cell proliferation and the cellular entry of NPs in HCC cells by phase contrast and fluorescent microscopic imaging techniques. The microscopic images of Hep3B cells demonstrated a clear decrease in the concentration of cells when cells were treated with free GEM, GEM NPs, antisense-miRNA-21 NPs, and antisense-miRNA-21–GEM (10% Cy5-conjuagted antisense-miRNA-21) co-encapsulated NPs in comparison to untreated and control NPs (Figure 6). Moreover, antisense-miRNA–21-GEM (10% Cy5-conjuagted antisense-miRNA-21) co-encapsulated NPs treated cells showed a substantial collection of Cy5-antisense-miRNA-21 NPs inside the cells, as imaged by fluorescent microscope (Figure 6vi,viii).
Figure 6.
Fluorescent microscopic images of Hep3B cells which are treated with GEM and antisense-miRNA-21-loaded NPs for 72 h (i−v, vii); cell density of Hep3B cells by bright field imaging (vi, viii); Cy5-fluorescent signal from Hep3B cells.
Immunoblot Analysis of Cells Treated with Antisense-miRNA-21- and GEM-Loaded Nanoparticles in HCC Cells
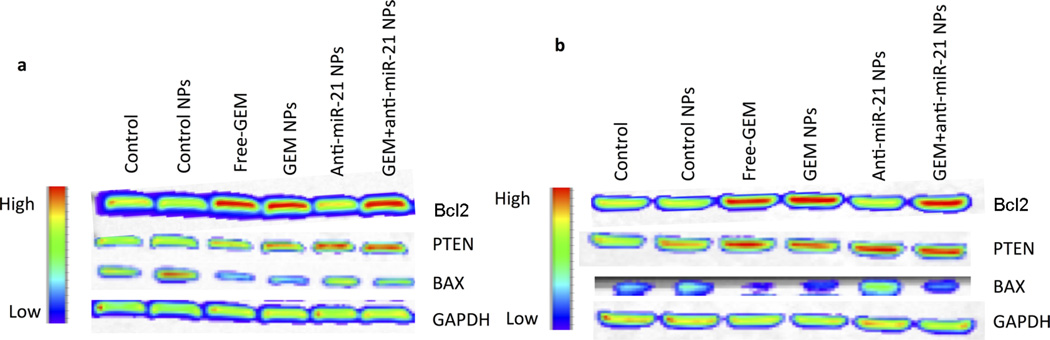
We also assessed the target proteins expression in HCC cells treated with antisense-miRNA-21 NPs (15 nM), free GEM (3 µM), antisense-miRNA-21 (15 nM), and GEM (3 µM) individually and co-encapsulated in NPs to further examine the pathways associated with the cytotoxic and antiproliferative effects (Figure 7 and Figure 8). The cells were incubated with NPs for 48 h, and the performed immunoblot analysis using antibodies detected PTEN, apoptotic protein B-cell lymphoma 2 (Bcl2), and Bcl2-associated X protein (BAX) (Figure 7). Results showed an increase in Bcl2 protein expression in both Hep3B and HepG2 cells treated with GEM-loaded NPs and free GEM, compared to control nanoparticle treated cells (Figure 7a,b). In contrast, a decrease in BAX protein expression was noticed in cells treated with GEM-loaded NPs and free GEM, compared to control nanoparticle treated cells (Figure 7a,b). However, a low level of BAX was found in cells at both treatment conditions, compared to Bcl2. The cells treated with nanoparticles loaded only with antisense-miRNA-21 displayed no effect on both Bcl2 and BAX expression levels, whereas NPs loaded with antisense-miRNA-21 by itself or in combination with antisense-miRNA-21–GEM treated cells displayed an increase in miRNA-21 target proteins expression (PTEN), compared to untreated cells (Figure 7a,b). In contrast, antisense-miRNA-21–GEM co-encapsulated NPs showed substantial up-regulation of Bcl2 expression in treated cells, indicating that cotreatment with GEM can up-regulate Bcl2 expression even in the presence of antisense-miRNA-21. PTEN is a prospective target of miRNA-21; hence, inhibition of miRNA-21 function in cells showed an increased expression of PTEN in HCC cells.36 Furthermore, our results demonstrate that inhibiting miRNA-21 with antisense-miRNA-21 increased PTEN expression in both HCC cell lines used for the study.
Figure 7.
Immunoblot analysis of Hep3B (a) and HepG2 (b) cells treated with free GEM and GEM and antisense-miRNA-21 individually and co-encapsulated NPs. (a, b) Optical chemiluminescence images showing Bcl2, PTEN, BAX, and GAPDH proteins stained with respective antibodies and imaged by optical CCD camera.
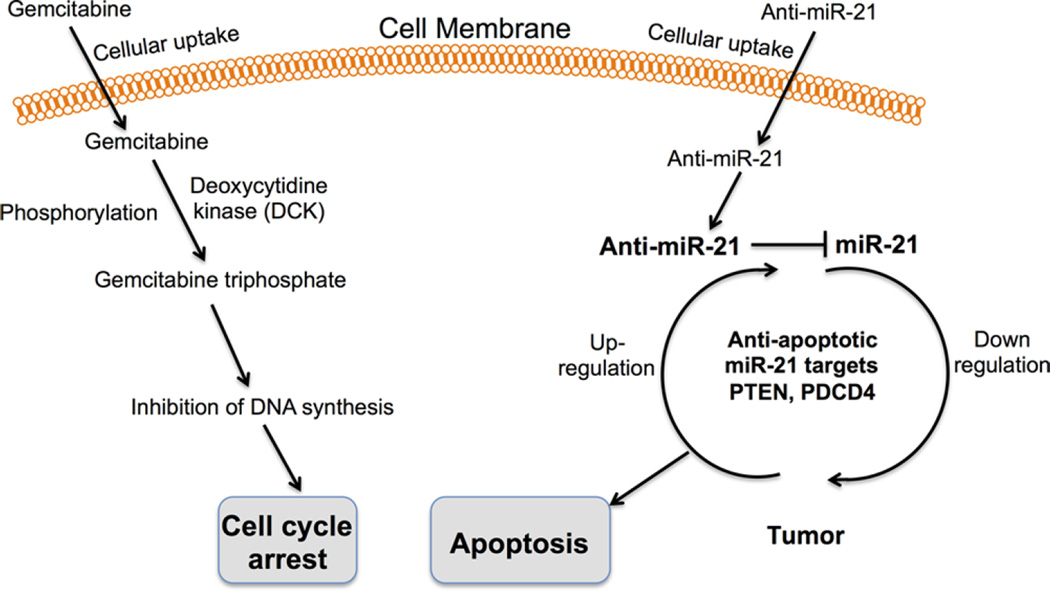
Figure 8.
Schematic illustrations of cellular mechanisms that regulate gemcitabine- and antisense-miRNA-21-mediated cell cycle arrest and apoptosis.
CONCLUSION
In conclusion, in this study we have developed an optimal formulation procedure for making PEGylated-PLGA-NPs co-encapsulated with antisense-miRNA-21 and GEM. Cell viability analysis demonstrated that antisense-miRNA-21 and GEM co-encapsulated NPs can improve treatment efficiency in HCC cells in comparison to NPs treated with equal concentrations of individually loaded antisense-miRNA-21 and GEM NPs. In addition, the delivery antisense-miRNA-21 effectively blocked endogenous miRNA-21 function and increased the level of target protein PTEN expression in cells. These results indicate that down-regulation of endogenous miRNA-21 function with antisense-miRNA-21 can reduce HCC cell proliferation by up-regulating miRNA-21 target proteins levels (Figure 8).
EXPERIMENTAL SECTION
Materials
All chemicals and reagents used in this study were purchased from standard commercial suppliers unless otherwise noted. The complete list of materials are given in the Supporting Information.
Methods
PLGA-b-PEG-COOH Co-polymer Synthesis
PLGA–PEG copolymer was synthesized using the procedure described in our previous work.41 In brief, to a solution of PLGA (MW 7000–17000) in dry dichloromethane (CH2Cl2), EDC and NHS were added and the reaction was continued for 4 h at room temperature (RT). The reaction mixture was poured into cold MeOH/Et2O) (1:1). The precipitated PLGA-NHS was centrifuged; supernatant was decanted, and the precipitate was dried under vacuum. The dried PLGA-NHS ester was solubilized in dry chloroform and mixed with heterobifunctional NH2–PEG–COOH (MW 3400) and diisopropylethylamine. The reaction was continued at RT for 24 h, and the reaction mixture was poured into cold MeOH/Et2O (1:1), washed twice, and dried under vacuum, which afforded the PLGA-b-PEG-COOH (yield, 74%; characterized by 1H NMR and MALDI-TOF).
Formulation and Characterization of PEGylated-PLGA-NPs Loaded with Antisense-miRNA-21 and GEM
The syntheses of PEGylated-PLGA-NPs loaded with the antisense-miRNA-21 and GEM were optimized using the procedure described in our previous work with some modifications.41 We used our optimized cosolvent w/ o/w method for making NPs and for studying various cell cultures. Antisense-miRNA-21 (0.5 nmol of Cy5-conjugated antisense-miRNA-21 + 4.5 nmol of antisense-miRNA-21) was mixed with spermidine (N/P ratio, 15:1) in DNase/RNAase free water (0.3 mL). A 10 mg amount of PLGA-b-PEG copolymer and 1% SPAN 80 (w/v) were dissolved in 1 mL of dichloromethane. GEM (0.5 mg) in 0.1 mL of DMSO was added to the above organic solution and mixed until the solution became clear and homogeneous. Then, spermidine-antisense-miRNA-21 complex was added to the above organic solution dropwise with slight stirring and sonicated at 40% amplitude for 60 s in an ice bath using a sonic dismembrator, which resulted in the formation of a first emulsion. The first emulsion was then added dropwise to 5 mL of 1% PVA (w/v) in autoclaved double distilled water with slight stirring and sonicated at 40% amplitude for 60 s in an ice bath, resulting in the formation of a secondary emulsion. It was then stirred for 3 h at RT, in order to vaporize the organic solvent and to strengthen the NPs. The strengthened NPs were sterilized by filtration (Whatman, 0.45 µm Puradisc 25 syringe filter, PES, sterile) and centrifuged by Amicon Ultra-15 centrifugal filter units (100000 DA, MWCO) and washed three times with DNase/RNase-free water. The NPs assessed for their size using DLS and loaded GEM and antisense-miRNA-21 concentrations using spectroscopy and gel electrophoresis were used for further studies. The control NPs and GEM-loaded NPs were formulated using a similar protocol with the elimination of the antisense-miRNA-21-spermidine step.
In Vitro Drug Release Studies
GEM release studies from PEGylated-PLGA-NPs were carried out at pH 5.0 and pH 7.0 in a dialysis setting. GEM encapsulated NPs were placed into the dialysis cassettes (cellulose membrane, MW cutoff 10000 kDa) and allowed to immerse in a beaker containing acidified water (pH of the water was adjusted to 5.0 using 0.1 N HCl) and ultrapure water (pH 7.0), in 37 °C incubator. At predetermined time intervals, a known volume of sample was collected from the outside solution chamber and replaced with an equal volume of fresh respective pH adjusted water. All the collected samples were concentrated and measured for GEM concentration by the UV absorbance at 268 nm using an Agilent Cary 60 UV–vis spectrophotometer.
Drug-Loaded NP Dose Dependent Therapeutic Analysis in Cells by MTT Assay
To study cell proliferation and viability in response to the dose of antisense-miRNA-21- and GEM-loaded NP treatment, Hep3B and HepG2 (10000 cells/well) cells were plated in phenol red free DMEM containing 2% FBS in 96-well tissue culture plates (100 µL/well) and incubated for 24 h at 37 °C and 5% CO2. After 24 h, the cells were treated with free GEM and various NPs at several concentrations, in triplicates for each concentration, for 24, 48, and 72 h at 37 °C and 5% CO2. Cell viability was measured by following a standard procedure (Life Technologies, Carlsbad, CA, USA). In brief, 12 mM MTT stock solution in phenol red free media was added after aspirating the medium. After 3 h of incubation, medium was carefully removed and 50 µL of DMSO was added to dissolve the metabolically reduced MTT derived formazan crystals by incubating the plate at 37 °C for 20 min. The absorbance (abs) was measured using microplate-reader at 540 nm. The relative cell viability in different treatment conditions was estimated and compared to control cells.
Apoptosis Analysis by Flow Cytometry
After treatment with various NPs, live and dead HCC (Hep3B and HepG2) cells were collected by trypsinization. The cells were fixed in 70% ethanol and stored at −20 °C. For flow cytometry measurements, ethanol was removed after centrifugation; the pelleted cells were washed with PBS and stained with PI (15 nM) containing RNase A (1 µg/mL) by incubating for 15 min. The PI stained cells was measured for dead or apoptotic cells by FACS, and the data were analyzed using FlowJo software.
Cellular Uptake Studies of Cy5-Conjugated Antisense-miRNA-21 and GEM Co-encapsulated NPs by Confocal Microscopy and FACS Analysis
To examine cellular uptake, Hep3B cells were grown on glass coverslips in 6-well plates for 24 h and were incubated with Cy5-conjugated antisense-miRNA-21 and GEM co-encapsulated NPs for 1 to 24 h at 37 °C and 5% CO2 by adding NPs at each time point. After incubation, cells were fixed in 4% PFA in PBS for 15 min, washed in PBS, blocked in 1% BSA/PBS for 1 h, permeabilized with 100% methanol for 20 min, and counterstained in 3 µM DAPI for 2 min, rinsed in PBS, mounted on glass slides, and imaged with a Leica SP2 AOBS confocal microscope. Similarly, to examine cellular uptake of NPs quantitatively by flow cytometry, Hep3B cells grown in 12-well culture plates for 24 h were incubated with Cy5-conjugated antisense-miRNA-21 and GEM co-encapsulated NPs for different time periods (1–24 h) at 37 °C with 5% CO2. The cells were collected after incubation for different time points and used for FACS analysis. The cell pellets were suspended in 0.3 mL of PBS and analyzed by flow cytometry by counting 10000 events, and the data were analyzed by FlowJo software.
Supplementary Material
Acknowledgments
We thank Dr. Sanjiv Sam Gambhir (Chair, Radiology, Stanford University), Dr. Ai Leen Koh, (Stanford Nano Shared Facilities) for her help in TEM imaging, and the Canary Center for providing facilities and support. This work in part was supported by NIH Grants R01CA161091 (to R.P) and R21EB022298 (to R.P. and J.K.W).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.6b08153.
Figure S1 showing PI-staining-based FACS analysis results and supporting materials and methods (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Boyle P, Levin B, editors. World Cancer Report 2008. Geneva, Switzerland: World Health Organization Press; 2008. [Google Scholar]
- 2.Stewart BW, Kleihues P. World Cancer Report 2003. Geneva, Switzerland: World Health Organization Press; 2003. [Google Scholar]
- 3.Bosch FX, Ribes J, Borras J. Epidemiology of Primary Liver Cancer. Semin. Liver Dis. 1999;19(3):271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 4.Finn RS. Development of Molecularly Targeted Therapies in Hepatocellular Carcinoma: Where Do We Go Now? Clin. Cancer Res. 2010;16(2):390–397. doi: 10.1158/1078-0432.CCR-09-2084. [DOI] [PubMed] [Google Scholar]
- 5.Davis GL, Dempster J, Meler JD, Orr DW, Walberg MW, Brown B, Berger BD, O’Connor JK, Goldstein RM. Hepatocellular Carcinoma: Management of an Increasingly Common Problem. Proc. (Bayl Univ Med. Cent) 2008;21(3):266–280. doi: 10.1080/08998280.2008.11928410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song XR, Zheng Y, He G, Yang L, Luo YF, He ZY, Li SZ, Li JM, Yu S, Luo X, Hou SX, Wei YQ. Development of PLGA Nanoparticles Simultaneously Loaded With Vincristine and Verapamil for Treatment of Hepatocellular Carcinoma. J. Pharm. Sci. 2010;99(12):4874–4879. doi: 10.1002/jps.22200. [DOI] [PubMed] [Google Scholar]
- 7.Huang M, Liu G. The Study of Innate Drug Resistance of Human Hepatocellular Carcinoma Bel7402 Cell Line. Cancer Lett. 1998;135(1):97–105. doi: 10.1016/s0304-3835(98)00280-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Shi ZL, Yang X, Yin ZF. Targeting of Circulating Hepatocellular Carcinoma Cells to Prevent Postoperative Recurrence and Metastasis. World J. Gastroenterol. 2014;20(1):142–147. doi: 10.3748/wjg.v20.i1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz M, Roayaie S, Konstadoulakis M. Strategies for the Management of Hepatocellular Carcinoma. Nat. Clin. Pract. Oncol. 2007;4(7):424–432. doi: 10.1038/ncponc0844. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Ye L, Pan J, Long M, Zhao Z, Yang H, Tian J, Wen Y, Dong S, Guan J, Luo B. Antitumoural Hydroxyapatite Nanoparticles-Mediated Hepatoma-Targeted Trans-Arterial Embolization Gene Therapy: In Vitro and In Vivo Studies. Liver Int. 2012;32(6):998–1007. doi: 10.1111/j.1478-3231.2012.02761.x. [DOI] [PubMed] [Google Scholar]
- 11.Mishra N, Yadav NP, Rai VK, Sinha P, Yadav KS, Jain S, Arora S. Efficient Hepatic Delivery of Drugs: Novel Strategies and Their Significance. BioMed Res. Int. 2013;2013:382184. doi: 10.1155/2013/382184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma P, Pandita A, Murthy RS. Concepts and Strategies for the Site Specific Delivery of Nanocarrier Based Delivery Systems for Treating Hepatocellular Carcinoma. Curr. Drug Delivery. 2013 Nov 24; [Epub ahead of print], PMID: 24266510. [PubMed] [Google Scholar]
- 13.Paulmurugan R, Bhethanabotla R, Mishra K, Devulapally R, Foygel K, Sekar TV, Ananta JS, Massoud TF, Joy A. Folate Receptor Targeted Polymeric Micellar Nanocarriers for Delivery of Orlistat as a Repurposed Drug against Triple Negative Breast Cancer. Mol. Cancer Ther. 2016;15(2):221–231. doi: 10.1158/1535-7163.MCT-15-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper DL, Conder CM, Harirforoosh S. Nanoparticles in Drug Delivery: Mechanism of Action, Formulation and Clinical Application Towards Reduction in Drug-Associated Nephrotoxicity. Expert Opin. Drug Delivery. 2014;11(10):1661–1680. doi: 10.1517/17425247.2014.938046. [DOI] [PubMed] [Google Scholar]
- 15.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, Nelson R, Dorr FA, Stephens CD, Von Hoff DD. Improvements in Survival and Clinical Benefit with Gemcitabine as First-Line Therapy for Patients with Advanced Pancreas Cancer: A Randomized Trial. J. Clin. Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 16.Devulapally R, Paulmurugan R. Polymer Nanoparticles for Drug and Small Silencing RNA Delivery to Treat Cancers of Different Phenotypes. Wiley Interdiscip Rev. Nanomed Nanobiotechnol. 2014;6(1):40–60. doi: 10.1002/wnan.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaaban S, Negm A, Ibrahim EE, Elrazak AA. Chemotherapeutic Agents for The Treatment of Hepatocellular Carcinoma: Efficacy and Mode Of Action. Oncol. Rev. 2014;8(1):246. doi: 10.4081/oncol.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwal S, Gupta S, Pabla D, Murthy RS. Gemcitabine-Loaded PLGA-PEG Immunonanoparticles for Targeted Chemotherapy of Pancreatic Cancer. Cancer Nanotechnol. 2013;4(6):145–157. doi: 10.1007/s12645-013-0046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Banderas L, Saez-Fernandez E, Holgado MA, Duran-Lobato MM, Prados JC, Melguizo C, Arias JL. Biocompatible Gemcitabine-Based Nanomedicine Engineered by Flow Focusing for Efficient Antitumor Activity. Int. J. Pharm. 2013;443(1–2):103–109. doi: 10.1016/j.ijpharm.2012.12.048. [DOI] [PubMed] [Google Scholar]
- 20.Kulhari H, Pooja D, Kota R, Reddy TS, Tabor RF, Shukla R, Adams DJ, Sistla R, Bansal V. Cyclic RGDfK Peptide Functionalized Polymeric Nanocarriers for Targeting Gemcitabine to Ovarian Cancer Cells. Mol. Pharmaceutics. 2016;13(5):1491–1500. doi: 10.1021/acs.molpharmaceut.5b00935. [DOI] [PubMed] [Google Scholar]
- 21.Arya G, Vandana M, Acharya S, Sahoo SK. Enhanced Antiproliferative Activity of Herceptin (HER2)-Conjugated Gemcitabine-Loaded Chitosan Nanoparticle in Pancreatic Cancer Therapy. Nanomedicine. 2011;7(6):859–870. doi: 10.1016/j.nano.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Joshi G, Kumar A, Sawant K. Enhanced Bioavailability and Intestinal Uptake of Gemcitabine HCl Loaded PLGA Nanoparticles after Oral Delivery. Eur. J. Pharm. Sci. 2014;60:80–89. doi: 10.1016/j.ejps.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Arias JL, Reddy LH, Couvreur P. Superior Preclinical Efficacy of Gemcitabine Developed as Chitosan Nanoparticulate System. Biomacromolecules. 2011;12(1):97–104. doi: 10.1021/bm101044h. [DOI] [PubMed] [Google Scholar]
- 24.Arias JL, Reddy LH, Couvreur P. Polymeric Nanoparticulate System Augmented the Anticancer Therapeutic Efficacy of Gemcitabine. J. Drug Target. 2009;17(8):586–598. doi: 10.1080/10611860903105739. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammond SM. MicroRNAs as Oncogenes. Curr. Opin. Genet. Dev. 2006;16(1):4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton AJ, Baulcombe DC. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Lee DY, Ben-David Y. The Roles of MicroRNAs in Tumorigenesis and Angiogenesis. Int. J. Physiol Pathophysiol Pharmacol. 2011;3(2):140–155. [PMC free article] [PubMed] [Google Scholar]
- 29.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of MiR-122 in the Rodent and Human Hepatocellular Carcinomas. J. Cell. Biochem. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, Lee CG. Profiling MicroRNA expression in Hepatocellular Carcinoma Reveals Micro-RNA-224 Up-Regulation and Apoptosis Inhibitor-5 as a MicroRNA-224-Specific Target. J. Biol. Chem. 2008;283(19):13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 31.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a Target of MiR-122a, a MicroRNA Frequently Down-Regulated in Human Hepatocellular Carcinoma. Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 32.Qin J, Luo M, Qian H, Chen W. Upregulated MiR-182 Increases Drug Resistance in Cisplatin-Treated HCC Cell by Regulating TP53INP1. Gene. 2014;538(2):342–347. doi: 10.1016/j.gene.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 33.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of MicroRNAs In Vivo with ‘Antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 34.Ross JS, Carlson JA, Brock G. MiRNA: The New Gene Silencer. Am. J. Clin. Pathol. 2007;128(5):830–836. doi: 10.1309/2JK279BU2G743MWJ. [DOI] [PubMed] [Google Scholar]
- 35.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of Novel Genes Coding for Small Expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 36.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA Involvement in Hepatocellular Carcinoma. J. Cell Mol. Med. 2008;12(6A):2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, Liu X. MicroRNA-21 Promotes Hepatocellular Carcinoma HepG2 Cell Proliferation Through Repression of Mitogen-Activated Protein Kinase-Kinase 3. BMC Cancer. 2013;13:469. doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devulapally R, Sekar NM, Sekar TV, Foygel K, Massoud TF, Willmann JK, Paulmurugan R. Polymer Nanoparticles Mediated Codelivery of AntimiR-10b and AntimiR-21 for Achieving Triple Negative Breast Cancer Therapy. ACS Nano. 2015;9(3):2290–2302. doi: 10.1021/nn507465d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang TY, Choe JW, Pu K, Devulapally R, Bachawal S, Machtaler S, Chowdhury SM, Luong R, Tian L, Khuri-Yakub B, Rao J, Paulmurugan R, Willmann JK. Ultrasound-Guided Delivery of MicroRNA Loaded Nanoparticles into Cancer. J. Controlled Release. 2015;203:99–108. doi: 10.1016/j.jconrel.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devulapally R, Sekar TV, Paulmurugan R. Formulation of Anti-miR-21 and 4-Hydroxytamoxifen Co-loaded Biodegradable Polymer Nanoparticles and Their Antiproliferative Effect on Breast Cancer Cells. Mol. Pharmaceutics. 2015;12(6):2080–2092. doi: 10.1021/mp500852s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa PM, Cardoso AL, Custodia C, Cunha P, Pereira de Almeida L, Pedroso de Lima MC. MiRNA-21 Silencing Mediated by Tumor-Targeted Nanoparticles Combined with Sunitinib: A New Multimodal Gene Therapy Approach for Glioblastoma. J. Controlled Release. 2015;207:31–39. doi: 10.1016/j.jconrel.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Bhargava-Shah A, Foygel K, Devulapally R, Paulmurugan R. Orlistat and Antisense-microRNA Loaded PLGA-PEG Nanoparticles for Enhanced Triple Negative Breast Cancer Therapy. Nanomedicine (London, U. K.) 2016;11(3):235–247. doi: 10.2217/nnm.15.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamaly N, Yameen B, Wu J, Farokhzad OC. Degradable Controlled-Release Polymers and Polymeric Nanoparticles: Mechanisms of Controlling Drug Release. Chem. Rev. 2016;116(4):2602–2663. doi: 10.1021/acs.chemrev.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hines DJ, Kaplan DL. Poly(Lactic-co-Glycolic) Acid-Controlled-Release Systems: Experimental and Modeling Insights. Crit. Rev. Ther. Drug Carrier Syst. 2013;30(3):257–276. doi: 10.1615/critrevtherdrugcarriersyst.2013006475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betancourt T, Byrne JD, Sunaryo N, Crowder SW, Kadapakkam M, Patel S, Casciato S, Brannon-Peppas L. PEGylation Strategies for Active Targeting of PLA/PLGA Nanoparticles. J. Biomed. Mater. Res., Part A. 2009;91A(1):263–276. doi: 10.1002/jbm.a.32247. [DOI] [PubMed] [Google Scholar]
- 47.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted Polymeric Therapeutic Nanoparticles: Design, Development and Clinical Translation. Chem. Soc. Rev. 2012;41(7):2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khaira R, Sharma J, Saini V. Development and Characterization of Nanoparticles for the Delivery of Gemcitabine Hydrochloride. Sci. World J. 2014;2014:560962. doi: 10.1155/2014/560962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teague A, Lim KH, Wang-Gillam A. Advanced Pancreatic Adenocarcinoma: A Review of Current Treatment Strategies and Developing Therapies. Ther. Adv. Med. Oncol. 2015;7(2):68–84. doi: 10.1177/1758834014564775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aggarwal S, Yadav S, Gupta S. EGFR Targeted PLGA Nanoparticles Using Gemcitabine for Treatment of Pancreatic Cancer. J. Biomed. Nanotechnol. 2011;7(1):137–138. doi: 10.1166/jbn.2011.1238. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto K, Nagahara T, Okano J-i, Murawaki Y. The Growth Inhibition of Hepatocellular and Cholangiocellular Carcinoma Cells by Gemcitabine and the Roles of Extracellular Signal-Regulated and Checkpoint Kinases. Oncol. Rep. 2008;20(4):863–872. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.