Abstract
Saffron is one of the highly exotic spices known for traditional values and antiquity. It is used for home décor besides serving as a colorant flavor and is widely known for medicinal value. Over the last few years, saffron has garnered a lot of interest due to its anti-cancer, anti-mutagenic, anti-oxidant and immunomodulatory properties. Integration of systems biology approaches with wide applications of saffron remains a growing challenge as new techniques and methods advance. Keeping in view of the dearth of a review summarizing the omics and systems biology of saffron, we bring an outline on advancements in integrating omic technologies, the medicinal plant has seen in recent times.
Key Words: Genomics, Systems Biology, Medicinal value, Therapeutics
Introduction
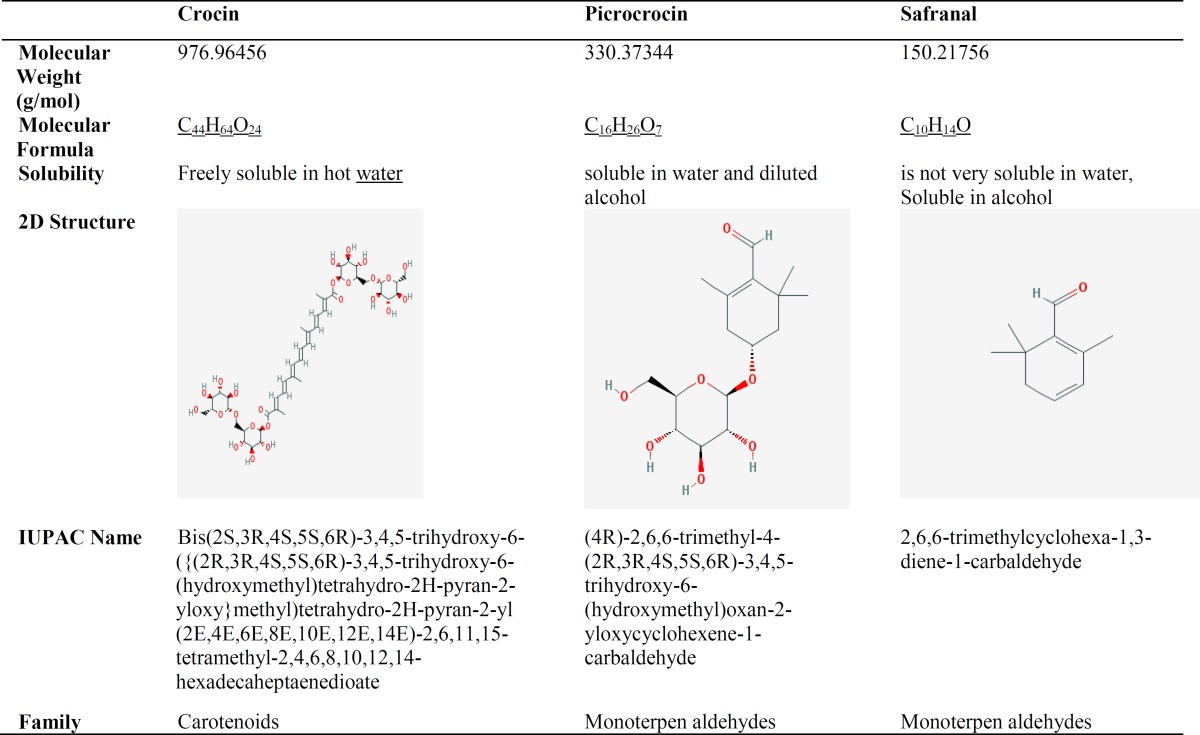
Saffron (Crocus sativus L.) is the most expensive spice and a profitable crop of the world, which is well known for its color, taste and medicinal value. The crop is being cultivated in many countries since ancient times with Iran producing approximately 95 % of the world’s saffron (Mosavi and Bathaie). It is characterized by its long, red stigmas, which contain natural carotenoid compounds such as crocin - responsible for the color(Singla and Bhat, 2011 ▶), crocetin and picrocrocin - responsible for the bitter taste along with other compounds like kaempferols (Carmona et al., 2007 ▶) and saffranal - responsible for the flavor (Rezaee and Hosseinzadeh, 2013 ▶).
Apart from vitamins and minerals, the flower and perianth are sources of a variety of chemicals such as anthocyanins (derivatives of delphinidin and petunidin), carotenoids (zeaxanthin, lycopene and α- and β-carotenes), volatile compounds, viz. flavonol glycosides (kaempferol, rhamnopyranoside, rutin, quercetin, etc.) and phenolics (vanillic, syringic, gallic, caffeic and salicylic acids) that have been relatively used for medicinal purposes. For example, crocin (C44H64O24), the diester which is formed from the disaccharidegentiobiose and the dicarboxylic acidcrocetin (C20H24O4), has found universal acceptance as a phytotherapeutic drug (Frusciante et al., 2014 ▶). Saffron, as a functional spice (Kyriakoudi et al., 2015 ▶), has been used as a flavor since ancient times. Its stigmas have considerable amount of riboflavin (vitamin B2) (Schmidt et al., 2007 ▶), which also contributes to the yellow color along with the highly water soluble compound,crocin (Tsatsaroni and Liakopoulou-Kyriakides, 1995 ▶). About 50,000 years ago, saffron-based pigment was used in home décor and cave art for wall paintings in Iraq (Bathaie et al., 2014 ▶; Zargari, 1990 ▶). Despite its high cost, saffron is also used as a fabric dye. There is an increasing demand for natural plants materials and their essential oils for cosmetic purposes, such as the aqueous, ethanolic and methanolic extracts from saffron petals (Formisanoet al., 2008 ▶). Historical studies of its uses in the ancient times show that saffron was used as a perfume and as gifts in various countries like Greece and ancient Persia (Abrishami, 1997 ▶; Leffingwell, 2002 ▶).
Of late, an increasing number of studies have explored the therapeutic effects and health benefits of saffron extracts and/or its components (apocarotenoids) against an array of diseases. Saffron finds applications as an anti-depressant (Hosseinzadeh et al., 2004 ▶), anticancer (Amin et al., 2011 ▶), hypnotic (Hosseinzadeh and Noraie, 2009 ▶), anti-inflammatory (Poma et al., 2012 ▶), hepatoprotective (Omidi et al., 2014 ▶), anti-tumor (Abdullaev, 2004 ▶; Festuccia et al., 2014 ▶); aphrodisiac (Hosseinzadeh et al., 2008 ▶) agent and as a treatment for memory diseases (Ghadrdoost et al., 2011 ▶) and skin disorders (Tabassum and Hamdani, 2014 ▶). Saffron stigmas possess antioxidant and free-radical scavenging activities as its metabolites prevent lipid peroxidation and human platelet aggregation (Jessie and Krishnakantha, 2005 ▶). A study on crocetin and saffranal demonstrated that the former is more effective in inhibiting free-radical formation in male Swiss albino mice aged 6–8 weeks (Hamid et al., 2009 ▶). This crocetin effect on level of lipid peroxidation and marker enzymes in lung cancer suggests crocetin as a potent anti-tumor agent. The anti-cancer activity of saffron and crocin usually results in cell cycle arrest. In the aforementioned studies, it has been observed that some anti-tumor drugs used in the treatment of cancer show genotoxicity. In addition, reports indicated the anti-genotoxic, anti-oxidant and chemo-preventive potential of saffron against well-known anti-tumor drugs like cisplatin (CIS), cyclophosphamide (CPH), doxorubicin (DOX) and mitomycin-C (MMC) using comet assay (Chahine et al., 2015 ▶). Furthermore, saffron as an antidepressant has been used as a promising natural alternative for the treatment of mild-to-moderate depression; however, it is essential to determine the optimal dosages and duration of this treatment. In this regard, the anti-tumor effect of saffron on skin cancer was reported (Mathews-Roth, 1982 ▶).
Aqueous saffron was shown to suppress oxidative stress in dimethylbenz[a] anthracene (DMBA) -induced skin carcinoma in mice when treated early (Das et al., 2010 ▶). Owing to its anti-oxidant properties, it can protect the central nervous system from oxidative lesion and improve learning power. Recent reviews have discussed randomized controlled trials on the effectiveness of saffron on psychological and behavioral outcomes; current human clinical evidence recommends the use of saffron for treatment of a range of pathologies, including Alzheimer's disease, age-related macular degeneration and cardiac ischaemia (Hausenblas et al., 2015 ▶; Broadhead et al., 2015 ▶). Nonetheless, the complete list of applications of saffron is beyond the scope of this review.
Challenges on genomics, transcriptomics, metabolomics and proteomics
Genomics
Saffron is a perennial sterile plant reproducing only vegetatively using the corms. Although a lot of work has been carried out using tissue culture and hybridization (Rubio-Moraga et al., 2014 ▶; Mir et al., 2015 ▶), propagation through corms offers no or little genetic variation in the form of somatic mutations, segregation distortions, transversions, etc., which neither of them combining in a population nor bringing heritable changes due to its sterility (Agayev et al., 2009 ▶). The triploid saffron (2n=3x=24) is known to be a probable progeny of C. cartwrightianus, which contributes to two of the three genomes, while the other parental lineages remain unclear (Fernández, 2004 ▶). Detailed intraspecific chromosome variations with respect to geographical area, classification, complex cytology and morphological characteristics (corm tunics, leaves, flowers, etc.) of the Crocus sativusseries have been reported (Saxena, 2010 ▶). The morphogenetic architecture of saffron is still an issue of debate because various studies report contradicting molecular results. However, many studies identify variations in phenotypic and phytochemical traits due to the epigenetic changes urging the immediate need for developing molecular markers to identify these variations at molecular level, which can be further exploited for the improvement of saffron (Mir et al., 2015 ▶). Also, 27 SSRs markers were evaluated on eight Iranian-cultivated saffron ecotypes and 29 wild alleles to assess the molecular variability and discriminating capacity of these markers regarding their effectiveness in establishing genetic relationships in these Crocus ecotypes (Nemati et al., 2014 ▶). More recent reports analyzed 112 accessions using Factorial Correspondence Analysis (individual level) of Amplified Fragment Length Polymorphism (AFLP) and methyl-sensitive AFLP to search for variations at the genetic and epigenetic (cytosine methylation) levels (Busconi et al., 2015 ▶). These studies indicated the presence of high epigenetic variability (33.57 % polymorphic peaks and 28 types of effective epigenotypes). Efforts are underway to prevent genetic erosion and induce genetic variability in order to develop superior varieties of saffron throughout the world.
Transcriptomics
‘Saffron omics’, an initiative of the European Cooperation in Science and Technology (COST), aims to strengthen collaborative research on developing 'omic' approaches in defining the structural organization of saffron genome, DNA fingerprinting to protect the quality and improve the genetic, chemical fingerprinting, proteomics, transcriptomics and metabolomics of this crop (http://www.saffronomics.org/). Currently, there are 6,768 saffron ESTs available at (http://www.ncbi.nlm.nih.gov/nucest/?term=%22Saffron%22), since the first set of 6,603 high quality ESTs from cDNA library of a saffron stigma were produced by D'Agostino et al., (2007) ▶ (available at http://www.saffrongenes.org). Transcriptomic and genomic studies onsaffron have received much lesser attention when compared to its potential applications in therapeutics and phytochemistry. This is probably due to the low or almost null genetic variability attributed to sterile triploid and vegetative propagation (Piqueras et al., 1999 ▶). The ESTs identified till date correspond to floral development (four MIKC type-II MADS-box cDNAs) (Tsaftaris et al., 2011 ▶), markers to detect adulteration in traded saffron (Bar-MCA analysis (Jiang et al., 2014 ▶), AS-PCR and SCAR (Shen et al., 2007 ▶; Torelli et al., 2014 ▶), environmental and pathogenic stresses (Husaini, 2014 ▶) and developmental pathways (Álvarez-Ortí et al., 2004 ▶). Similar transcriptomic studies on saffron have led to the dissection biosynthetic pathways of carotenoids (Castillo et al., 2005 ▶) and flavonoids for characterization of glucosyltransferase (Moraga et al., 2009 ▶). Additionally, deep transcriptomics analysis has identified carotenoid cleavage dioxygenase (CCD2), a novel dioxygenase which catalyzes the first step of crocin biosynthesis originating from carotenoid zeaxanthin (Frusciante et al., 2014 ▶). Although bioinformatics tools have been applied for the prediction and regulation of signaling pathways, a stringent validation using in vitro experiments should not be given a miss. One such immune-perspective model was deliberated with TGFβ (Kahlem and Newfeld, 2009 ▶) where successful applications of both fine-scale and network-scale informatics approaches for understanding signaling pathways were reviewed. Similarly, T and B-cell epitopes of Iranian saffron profiling were predicted using bioinformatics tools (Saffari et al., 2008 ▶).
Metabolomics
Metabolome is a unique collection of cellular working parts that are associated with the expression of the sequenced genomes in all living organism including bacteria, plant, animal, etc. In the recent past, metabolomic analysis proved to be an incipient tool for functional gene annotation and characterization, particularly for those genes involved in regulatory pathways. Metabolomic studies aid in identifying substrates and products of enzymes without the need for going through heterologous expression systems (Beale and Sussman, 2011 ▶). Saffron metabolomics has provided an unbiased, comprehensive qualitative and quantitative overview of its metabolites such as crocetin esters, picrocrocin, safranal, etc., elucidating their association with therapeutic and aesthetic properties (Ordoudi et al., 2015 ▶). Previous studies have identified more than 160 volatile compounds (see Table 1) using chromatography combined with spectroscopy (UV, IR, NMR) and mass spectrometry (MS) techniques (Assimiadiset al., 1998; Calsteren et al., 1997 ▶). Metabolite fingerprinting obtained using 1H NMR spectra and chemo-metrics was reported for the authentication of both Iranian and Italian saffron (Cagliani et al., 2015 ▶; Yilmaz et al., 2010 ▶). The insights to the structural variations in crocetin esters and picrocrocin and differentiation of sugars bound to them using 1H NMR method were well documented (Ordoudi et al., 2015 ▶; Ordoudi and Tsimidou, 2004 ▶). While 1H NMR serves as a potent tool to control saffron quality deterioration, it offers specific advantages to characterize secondary metabolites. Simultaneous identification and quantification of metabolites is necessary to understand the dynamics of the metabolome in analyzing fluxes and pathways associated with saffron. However, the major challenge remains in finding variations in biochemical pathways and metabolic networks that might correlate with the physiological and developmental phenotype of a cell and tissue.
Table 1.
Chemical properties of saffron metabolites (Source: Pubchem andWikipedia
Proteomics
Identification of proteins and prediction of their structure from the amino acid sequence are challenges for researchers across different biological disciplines. Complete understanding of the biological role of proteins requires knowledge of their structure and function (Pieper et al., 2006 ▶). Although proteomics studies hold promise in characterization of both known and unknown proteins, to date, only 312 protein sequence entries are reported in GenBank: (http://www.ncbi.nlm.nih.gov/protein/?term=Saffron). Protein information provides the possibility of predicting three-dimensional structure. Proteomic analysis carried out earlier identified differentially accumulated proteins in somatic embryos of saffron, which provide insights into underlying molecular mechanisms (Sharifi et al., 2012 ▶). In addition, dearth of validated structure information for a majority of plant proteins is a major hindrance to functional annotation, evolutionary analyses and building interaction networks (Pentony et al., 2012 ▶). Although there are a plethora of tools available for the prediction and visualization of secondary and tertiary structures, detailed analyses were limited to a few selected plant gene families. For instance, UniProt hosts mere 98 protein entries for C.sativus, of which only 5 have been reviewed; leaving a huge scope for both in silico and in vitro studies. Presently, there is a demand for atomic-level structural refinements that can generate 3D models for use in drug screening and inferring biochemical function for these saffron proteins, especially when large template structures become available Furthermore, three crystal structures are available in protein data bank (PDB) (http://www.rcsb. org/pdb/explore/explore.do?structureId=3U8E ) which can bridge that demand in finding insights into the above-mentioned mechanisms.
A need for integrated Systems Biology approaches
Over the last few years, saffron has garnered a lot of interest due to its therapeutic potential (Naghshineh et al., 2015 ▶). Integration of systems biology approaches in drug discovery has tremendous application in investigating the drug–target interaction mechanisms and in identifying novel targets in a network context (Vandamme et al., 2014 ▶; Harrold et al., 2013 ▶).
Similarly, few studies on saffron have applied target deconvolution, reverse screening, modelling and docking for retrospective identification of molecular targets and functional components (Nithya and Shakthisekharan 2015 ▶; Bhattacharjee et al., 2012 ▶). Systems biology approaches have been applied to the non-therapeutic aspects of saffron, such as building complete metabolic pathways of the bioactive compounds, spatial-temporal expression of genes involved in clonal propagation and quantification of factors. A striking example of one such analysis was demonstrated recently (Zeraatkar et al., 2015 ▶), wherein a three-dimensional geometrical model of saffron flower was generated for the first time, using reverse engineering and laser scanning technology. The mechanical behavior of the flower could play an important role in the design of post-harvesting machinery and process. Predicting biological functions and metabolic pathways was linked to the construction of protein interaction networks (PIN) (Guan and Kiss-Toth, 2008 ▶; Wetei et al., 2013 ▶).
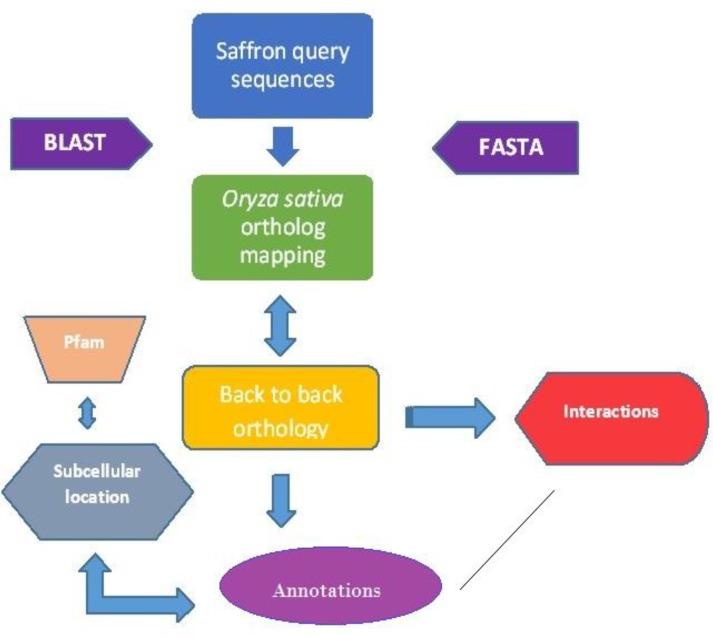
In silico molecular dynamics and docking approach have been employed to investigate interactions between secondary metabolites of saffron (safranal, crocetin and dimethylcrocetin) and transport proteins such as β-lactoglobulin, could be valuable factors in controlling their transport to biological sites (Sahihi 2015 ▶). Reports on saffron have often highlighted the need for refining bioinformatics tools available with transcriptomic and genomic data (Fernandez and Gomez Gomez, 2005 ▶; Husaini et al., 2009 ▶; Gomez Gomez et al., 2009 ▶) but little has been done in this direction. Towards this end, the current section focuses on in silico approaches to build a protein interaction network of candidates involved in crocetin biosynthesis pathway. We have worked on a case study with 35 saffron protein sequences selected as a query to search for orthologs (Oryza sativa as reference).
The annotation scores, based on the features are taken as per our former annotation approach (Suravajhala and Sundararajan, 2012 ▶). Sequence similarity searches were done on local FASTA (http://fasta.bioch .virginia.edu) and using BLASTp(http://blast.ncbi.nlm.nih.gov/Blast.cgi) tool against non-redundant protein sequences of Oryza sativa.
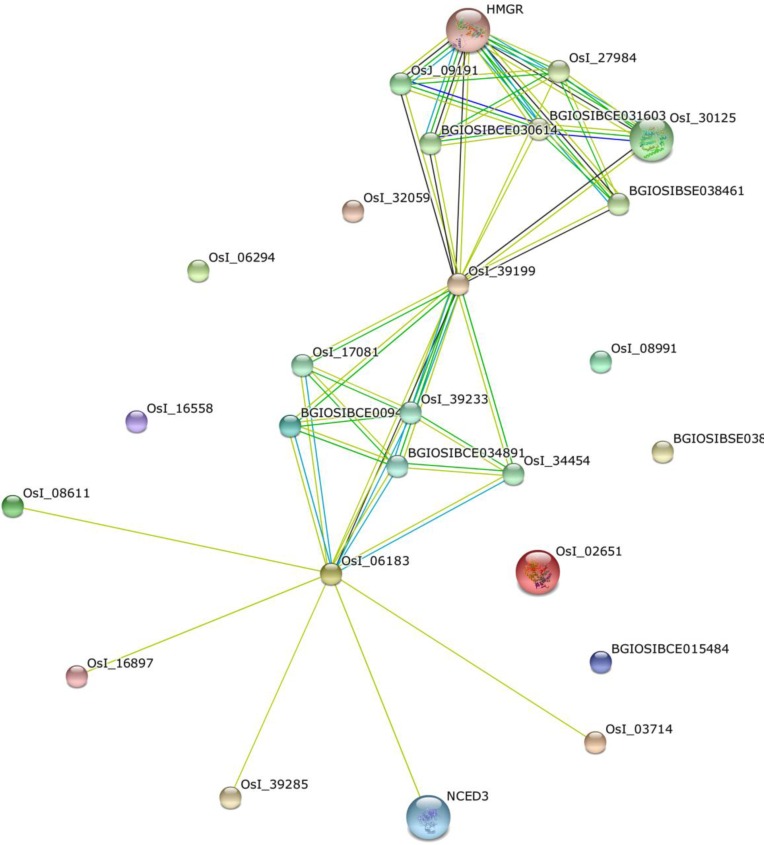
Further characterization involving Pfam score, orthology inference, functional linkages, back-to-back orthology, subcellular location and protein associations were considered from known databases and visualizers (Figure 1). Each protein was given a value of 1 if the protein matched the classifier; else 0 was rendered (Table 2). Although only 10 out of the 35 query proteins selected have orthologs in Oryza, classification scoring approach revealed 15 crocetin-related proteins to have functional protein associations (Table 2). These were visualized by a protein interaction network (Figure 2) where in, interologs of three genes, viz. HMGR (putative 3-hydroxy-3-methylglutaryl-CoA reductase), lycopene cyclase and phytoene synthase are known to be co-expressed. These candidates that are derived from the methods employed in this analysis are concurrent with earlier reports which focused on transcriptome and metabolome experiments. It would be interesting to exploit pull-down assays and computational biology tools which could enhance our knowledge of the carotenoid biosynthetic pathway and establish other key protein interacting partners.
Figure 1.
A flowchart of tools used for annotation methodology employed in obtaining the 35 protein sequences
Table 2.
A case study with six-point classification scoring strategy for identification of 35 protein candidate sequences. Table 2a: Identification of protein families and orthologous sequences for the query sequences from saffron
|
Organism
|
Classification 1
|
Classification 2
|
||||||
|---|---|---|---|---|---|---|---|---|
| Saffron | Protein family scores | Orthology | ||||||
| Accession | Nomenclature | Identity | E value | Score | Score | Accession | Organism | Score |
| AIF76151.1 | UDP-glucosyltransferase UGT85U2, partial | UDPGT | 2.70E-31 | 108.7 | 1 | EAY87581.1 | Oryza sativa | 0 |
| AIF76152.1 | UDP-glucosyltransferase UGT85U1 | UDPGT | 6.00E-32 | 110.8 | 1 | EAY87581.1 | Oryza sativa | 0 |
| Q84KG5.1 | Carotenoid 9,10(9',10')-cleavage dioxygenase | RPE65 | 5.60E-146 | 486.9 | 1 | ABA99624.2 | Oryza sativa | 0 |
| AIF27228.1 | carotenoid cleavage dioxygenase 7 | RPE65 | 2.50E-96 | 323.1 | 1 | EAY95081.1 | Oryza sativa | 0 |
| AIF27229.1 | carotenoid cleavage dioxygenase 8a | RPE65 | 2.10E-103 | 346.5 | 1 | NP_001044229.2 | Oryza sativa | 0 |
| AIF27230.1 | carotenoid cleavage dioxygenase 8b | RPE65 | 2.90E-103 | 346 | 1 | NP_001044229.2 | Oryza sativa | 0 |
| AIG94929.1 | carotenoid cleavage dioxygenase 2 | RPE65 | 7.40E-134 | 446.9 | 1 | ABA99624.2 | Oryza sativa | 0 |
| CAI60776.1 | phytoene synthase, partial | SQS_PSY | 3.60E-35 | 121.5 | 1 | AAK07734.1 | Oryza sativa | 1 |
| CAI60777.1 | lycopene cyclase, partial [Crocus sativus] | Lycopene_cycl | 7.00E-39 | 133.8 | 1 | BAD16478.1 | Oryza sativa | 1 |
| CAC95133.1 | putative neoxanthin cleavage enzyme, partial | RPE65 | 5.90E-44 | 150.4 | 1 | EAZ24320.1 | Oryza sativa | 0 |
| ACD62475.1 | carotenoid cleavage dioxygenase 2 | RPE65 | 3.10E-129 | 431.6 | 1 | ABA99624.2 | Oryza sativa | 0 |
| ACD62476.1 | chromoplast carotenoid cleavage dioxygenase 4a | RPE65 | 1.20E-106 | 357.2 | 1 | EAZ24320.1 | Oryza sativa | 0 |
| ACD62477.1 | chromoplast carotenoid cleavage dioxygenase 4b | RPE65 | 9.10E-107 | 357.5 | 1 | EAZ24320.1 | Oryza sativa | 0 |
| ACM66950.1 | flavonoid glucosyltransferase | UDPGT | 1.90E-21 | 76.2 | 1 | BAD15509.1 | Oryza sativa | 0 |
| CAD33262.1 | zeaxanthin cleavage oxygenase | RPE65 | 2.80E-83 | 280.1 | 1 | EAZ24320.1 | Oryza sativa | 1 |
| CAC79592.1 | crocetindialdehyde | RPE65 | 5.60E-146 | 486.9 | 1 | ABA99624.2 | Oryza sativa | 0 |
| CAD33258.1 | betaine aldehyde dehydrogenase, partial | Aldedh | 3.40E-21 | 75.1 | 1 | AGP76273.1 | Oryza sativa | 1 |
| CAD70567.1 | aldehyde dehydrogenase | Aldedh | 3.90E-174 | 579.3 | 1 | NP_001043454.1 | Oryza sativa | 0 |
| CAC95134.1 | putative 3-hydroxy-3-methylglutaryl-CoA reductase, partial | HMG-CoA_red | 6.00E-64 | 216 | 1 | NP_001062221.1 | Oryza sativa | 1 |
| CAC95130.2 | beta-carotene hydroxylase | FA_hydroxylase | 3.40E-11 | 43.5 | 1 | EEC74425.1 | Oryza sativa | 1 |
| CAI79433.1 | beta-carotene hydroxylase enzyme, partial | FA_hydroxylase | 4.20E-07 | 30.3 | 1 | EEC74425.1 | Oryza sativa | 1 |
| CAI79451.1 | beta-carotene hydroxylase enzyme, partial | FA_hydroxylase | 4.20E-07 | 30.3 | 1 | EEC74425.1 | Oryza sativa | 1 |
| CAI79462.1 | beta-carotene hydroxylase enzyme, partial | FA_hydroxylase | no | no | 0 | NP_001053640.1 | Oryza sativa | 0 |
| ACD44928.1 | plastid 9-cis-epoxycarotenoid dioxygenase | RPE65 | 2.30E-134 | 448.6 | 1 | NP_001050765.1 | Oryza sativa | 0 |
| ADA82242.1 | lycopene beta cyclase | Lycopene_cycl | 5.40E-125 | 417.1 | 1 | BAD16478.1 | Oryza sativa | 0 |
| Q84K96.1 | CsZCD | RPE65 | 2.80E-83 | 280.1 | 1 | EAZ24320.1 | Oryza sativa | 1 |
| AEO50759.1 | CCD4c | RPE65 | 3.70E-117 | 391.8 | 1 | EAZ24320.1 | Oryza sativa | 0 |
| AAT84408.1 | beta carotene hydroxylase | FA_hydroxylase | 5.30E-07 | 30 | 1 | EEC74425.1 | Oryza sativa | 1 |
| AAQ56280.1 | glucosyltransferase-like protein | UDPGT | 1.20E-16 | 60.3 | 1 | NP_001053256.1 | Oryza sativa | 0 |
| AAP94878.1 | glucosyltransferase 2 | UDPGT | 4.60E-30 | 104.6 | 1 | NP_001063685.1 | Oryza sativa | 0 |
| Q6WFW1.1 | Crocetin glucosyltransferase 3 | UDPGT | 1.20E-16 | 60.3 | 1 | NP_001053256.1 | Oryza sativa | 0 |
| Q6X1C0.1 | Crocetin glucosyltransferase 2 | UDPGT | 4.60E-30 | 104.6 | 1 | NP_001063685.1 | Oryza sativa | 0 |
| CCG85331.1 | glucosyltransferase | UDPGT | 7.80E-23 | 80.7 | 1 | NP_001059726.1 | Oryza sativa | 0 |
| AAP94878.1 | glucosyltransferase 2 | UDPGT | 4.60E-30 | 104.6 | 1 | NP_001063685.1 | Oryza sativa | 0 |
| CAC95132.1 | putative neoxanthin cleavage enzyme, partial | RPE65 | 1.10E-41 | 142.9 | 1 | ABA99624.2 | Oryza sativa | 1 |
| CAC95131.1 | putative neoxanthin cleavage enzyme, partial | RPE65 | 9.80E-57 | 192.5 | 1 | ABA99624.2 | Oryza sativa | 1 |
The first classifier (a) starts with determing if the query proteins have defined domains and is given a score of 1 if there are Pfam matches. Pfam is a protein family feature wherein we could check the presence of domains and motif family members associated with the proteins. It is followed by identifying orthologs for the queries in Oryza with both BLAST and stand alone FASTA. A score of 1 is given only if the queries satisfy the sequence similarity evalutioncriteon in both of the tools
Figure 2.
A putative protein-protein interaction map of peers of HMG Co-A reductase (HMGR) of rice interologs in saffron.Interologs are orthologous set of interacting proteins in other organisms, here saffron. The PIN was constructed using STRING database (Szklarczyk D et al. 2015) with the potential candidate proteins from the six-point scoring schema (Table 2c) as queries searched against Oryza as reference organism. The nodes represent the proteins which are connected by edges in the form of lines
Table 2b.
Association studies for the query sequences
| Query |
Classification 3
|
1+2+3
|
||||
|---|---|---|---|---|---|---|
| GO/association studies |
|
|||||
| Accession | Gene | Annotation | Accessions | Score | ||
| AIF76151.1 | NA | 0 | 1 | |||
| AIF76152.1 | NA | 0 | 1 | |||
| Q84KG5.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 2 | |
| AIF27228.1 | NA | 0 | 1 | |||
| AIF27229.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 2 | |
| AIF27230.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 2 | |
| AIG94929.1 | NA | 0 | 1 | |||
| CAI60776.1 | NA | 0 | 2 | |||
| CAI60777.1 | NA | 0 | 2 | |||
| CAC95133.1 | NA | 0 | 1 | |||
| ACD62475.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 2 | |
| ACD62476.1 | NA | 0 | 1 | |||
| ACD62477.1 | NA | 0 | 1 | |||
| ACM66950.1 | NA | 0 | 1 | |||
| CAD33262.1 | NA | 0 | 2 | |||
| CAC79592.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 2 | |
| CAD33258.1 | NA | 0 | 2 | |||
| CAD70567.1 | NA | 0 | 1 | |||
| CAC95134.1 | NA | 0 | 2 | |||
| CAC95130.2 | NA | 0 | 2 | |||
| CAI79433.1 | NA | 0 | 2 | |||
| CAI79451.1 | NA | 0 | 2 | |||
| CAI79462.1 | NA | 0 | 0 | |||
| ACD44928.1 | NA | 0 | 1 | |||
| ADA82242.1 | NA | 0 | 1 | |||
| Q84K96.1 | NA | 0 | 2 | |||
| AEO50759.1 | NA | 0 | 1 | |||
| AAT84408.1 | NA | 0 | 2 | |||
| AAQ56280.1 | NA | 0 | 1 | |||
| AAP94878.1 | NA | 0 | 1 | |||
| Q6WFW1.1 | NA | 0 | 1 | |||
| Q6X1C0.1 | NA | 0 | 1 | |||
| CCG85331.1 | NA | 0 | 1 | |||
| AAP94878.1 | NA | 0 | 1 | |||
| CAC95132.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 3 | |
| CAC95131.1 | http://amigo1.geneontology.org/cgi-bin/amigo/gp-details.cgi?gp=UniProtKB:Q8LIY8&session_id=9334amigo1418585215 | carotene catabolic process | GO:0016121 | 1 | 3 | |
If an association or linkage found through tools like Amigo2 and Genevestigator and if annotations are found for the query sequences, a score of 3 is given
Table 2c.
Identification of interologs, sub-cellular location, interactants and potential candidates among the query sequences
|
Classification 4
|
Classification 5
|
Classification 6
|
4+5+6
|
Total Reliability
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Back to back Orthology
|
Sorting signals
|
From known databases and visualizers
|
|
|
|||||||||||||||||||||
| Accession | Identity | E-Value | Score | TargetP | Psort | Score | Gene | Major approaches | Identity% | Score | |||||||||||||||
| AIF76151.1 | 54% | 0 | 0 | other | Peroxisomes | 0 | OsI_08991 | Os02g0755900 | Neighbouhood Databases, text mining | 54 | 0 | 0 | 1 | ||||||||||||
| AIF76152.1 | 54% | 0 | 0 | other | Peroxisomes | 0 | OsI_08991 | Os02g0755900 | Neighbourhood, Databases, text mining | 54 | 0 | 0 | 1 | ||||||||||||
| Q84KG5.1 | 83% | 0 | 1 | other | Peroxisomes | 0 | OsI_39285 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 83 | 1 | 2 | 4 | ||||||||||||
| AIF27228.1 | 57% | 0 | 0 | mitochondria | Mitochoandria | 1 | OsI_16897 | Os04g0550600 | Neighbourhood, Databases, text mining | 62 | 1 | 2 | 3 | ||||||||||||
| AIF27229.1 | 76% | 0 | 1 | other | Nucleus | 0 | OsI_03714 | Os01g0746400 | Neighbourhood, Databases, text mining | null | 0 | 1 | 3 | ||||||||||||
| AIF27230.1 | 76% | 0 | 1 | other | Cytoplasm | 0 | OsI_03714 | Os01g0746400 | Neighbourhood, Databases, text mining | 76 | 1 | 2 | 4 | ||||||||||||
| AIG94929.1 | 69% | 0 | 1 | other | Endoplasmic reticulum | 0 | OsI_39285 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 72 | 1 | 2 | 3 | ||||||||||||
| CAI60776.1 | 84% | 3.00E-83 | 1 | mitochondria | Mitochoandria | 1 | OsI_39199 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 84 | 1 | 3 | 5 | ||||||||||||
| CAI60777.1 | 79% | 6.00E-72 | 1 | other | Plasma membrane | 0 | OsI_06183 | Os02g0190600 | Neighbourhood, Databases, text mining | 79 | 1 | 2 | 5 | ||||||||||||
| CAC95133.1 | 60% | 7.00E-84 | 1 | chloroplast | Chloroplast | 1 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 59 | 0 | 2 | 3 | ||||||||||||
| ACD62475.1 | 67% | 0 | 1 | other | Endoplasmic reticulum | 0 | OsI_39285 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 71 | 1 | 2 | 4 | ||||||||||||
| ACD62477.1 | 60% | 0 | 1 | chloroplast | Plasma membrane | 0 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 59 | 0 | 1 | 2 | ||||||||||||
| ACD62477.1 | 60% | 0 | 1 | other | Plasma membrane | 0 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 60 | 1 | 2 | 3 | ||||||||||||
| ACM66950.1 | 46% | 7.00E-151 | 0 | other | Endoplasmic reticulum | 0 | OsI_06294 | Os02g0203300 | Neighbourhood, Databases, text mining | 46 | 0 | 0 | 1 | ||||||||||||
| CAD33262.1 | NA | 0 | other | Cytoplasm | 0 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 61 | 1 | 1 | 3 | |||||||||||||
| CAC79592.1 | NA | 0 | other | Chloroplast | 0 | OsI_39285 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 83 | 1 | 1 | 3 | |||||||||||||
| CAD33258.1 | 80% | 5.00E-52 | 1 | other | Peroxisomes | 0 | BGIOSIBCE015484 | annotation not avaliable | Neighbourhood, Databases, text mining | 78 | 1 | 2 | 4 | ||||||||||||
| CAD70567.1 | 79% | 0 | 1 | other | Cytoplasm | 0 | OsI_02651 | Os01g0591300 | Neighbourhood, Databases, text mining | 78 | 1 | 2 | 3 | ||||||||||||
| CAC95134.1 | 86% | 6.00E-91 | 1 | other | Cytoplasm | 0 | HMGR | Os09g0492700 | Neighbourhood, Databases, text mining | 87 | 1 | 2 | 4 | ||||||||||||
| CAC95130.2 | 87% | 3.00E-140 | 1 | chloroplast | Chloroplast | 1 | BGIOSIBCE009468 | Os03g0125100 | Neighbourhood, Databases, text mining | 74 | 1 | 3 | 4 | ||||||||||||
| CAI79433.1 | 90% | 2.00E-52 | 1 | mitochondria | Plasma membrane/ Mitochondria |
1 | BGIOSIBCE009468 | Os03g0125100 | Neighbourhood, Databases, text mining | 90 | 1 | 3 | 4 | ||||||||||||
| CAI79451.1 | NA | 0 | mitochondria | Plasma membrane/ Mitochondria |
1 | 0 | 0 | Neighbourhood, Databases, text mining | null | 0 | 1 | 3 | |||||||||||||
| ADA82242.1 | NA | 0 | other | Cytoplasm | 0 | 0 | 0 | Neighbourhood, Databases, text mining | null | 0 | 0 | 0 | |||||||||||||
| ACD44928.1 | 72% | 0 | 1 | chloroplast | Chloroplast | 1 | NCED3 | Os03g0645900 | Neighbourhood, Databases, text mining | 72 | 1 | 3 | 4 | ||||||||||||
| ADA82242.1 | 65% | 0 | 1 | other | Nucleus | 0 | OsI_06183 | Os02g0190600 | Neighbourhood, Databases, text mining | 69 | 1 | 2 | 3 | ||||||||||||
| Q84K96.1 | 61% | 9.00E-160 | 1 | other | Chloroplast | 0 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 61 | 1 | 2 | 4 | ||||||||||||
| AEO50759.1 | 73% | 0 | 1 | chloroplast | Chloroplast | 1 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 74 | 1 | 3 | 4 | ||||||||||||
| AAT84408.1 | 65% | 5.00E-108 | 1 | chloroplast | Chloroplast | 1 | BGIOSIBCE009468 | Os03g0125100 | Neighbourhood, Databases, text mining | 58 | 0 | 2 | 4 | ||||||||||||
| AAQ56280.1 | NA | 0 | other | Peroxisomes | 0 | OsI_16558 | Os04g0506000 | Neighbourhood, Databases, text mining | 38 | 0 | 0 | 1 | |||||||||||||
| AAP94878.1 | NA | 0 | mitochondria | Chloroplast | 0 | OsI_32059 | Os09g0518200 | Neighbourhood, Databases, text mining | 54 | 0 | 0 | 1 | |||||||||||||
| Q6WFW1.1 | 40% | 4.00E-96 | 0 | other | Peroxisomes | 0 | OsI_16558 | Os04g0506000 | Neighbourhood, Databases, text mining | 38 | 0 | 0 | 1 | ||||||||||||
| Q6X1C0.1 | 48% | 1.00E-144 | 0 | mitochondria | Mitochoandria/Chloroplast | 1 | OsI_32059 | Os09g0518200 | Neighbourhood, Databases, text mining | 49 | 0 | 1 | 2 | ||||||||||||
| CCG85331.1 | 52% | 3.00E-173 | 0 | other | Golgi body | 0 | BGIOSIBSE038738 | Os07g0503300 | Neighbourhood, Databases, text mining | 52 | 0 | 0 | 1 | ||||||||||||
| AAP94878.1 | NA | 0 | mitochondria | Chloroplast/ Mitochondria |
1 | OsI_32059 | Os09g0518200 | Neighbourhood, Databases, text mining | 49 | 0 | 1 | 2 | |||||||||||||
| CAC95131.1 | 66% | 7.00E-105 | 1 | mitochondria | Mitochondria /ER |
1 | OsI_39285 | Putative uncharacterized protein | Neighbourhood, Databases, text mining | 74 | 1 | 3 | 6 | ||||||||||||
| CAC95132.1 | 82% | 1.00E-99 | 1 | Signal peptide | Chloroplast stroma | 1 | OsI_08611 | Os02g0704000 | Neighbourhood, Databases, text mining | 65 | 1 | 3 | 6 | ||||||||||||
Third classifier (2c) starts with searching for orthologs in saffron using Oryza as query with the help of BLAST. This is followed by assigning a sub-cellular location to the saffron query proteins, double-checked by two localization tools followed by searching for interactants using tools such as AtPID and IntAct. A consensus is reached combining all the three classifiers (Tables 2a, 2b and 2c) based on a final score >3
Conclusions
With wide interest in increasing numbers of therapeutics, there remains a challenge in studying several non-curative compounds that could be significantly obtained from saffron. This therapeutic potential of the compounds in the form of chemotherapy or radiotherapy can allow us to find novel insights to study effects on diseases. With saffron as a chemical modulator derived from wide number of plant nutrients, employing omics technologies is the need of the hour so as to enhance the potential for drugs through possible anti-disease agents like colorants, stigmas, etc. We imagine these technologies, if integrated together can not only attribute to a better understanding of drug targets but also allow us to consider new case studies for saying ‘ome’ for medicinal plants.
References
- Abdullaev FI. Antitumor effect of saffron (Crocus sativus L): overview and perspectives. Acta Hort. 2004:491–500. [Google Scholar]
- Abrishami MH. Mashhad. Iran: AstanGhods Razavi Publication; 1997. Iranian saffron. Historic, cultural and agronomic prospects; pp. 1–10. [Google Scholar]
- Agayev YMO, Fernandez JA, Zarifi E. Clonal selection of saffron (Crocus sativus L): the first optimistic experimental results. Euphytica. 2009;169:81–99. [Google Scholar]
- Álvarez-Ortí M, Gómez LG, Rubio A, Escribano J, Pardo J, Jiménez F, Fernández JA. Development and gene expression in saffron corms. Acta Hortic. (ISHS) 2004;650:141–153. [Google Scholar]
- Amin A, Hamza AA, Bajbouj K, Ashraf SS, Daoud S. Saffron: a potential candidate for a novel anticancer drug against hepatocellular carcinoma. Hepatol. 2011;54:857–867. doi: 10.1002/hep.24433. [DOI] [PubMed] [Google Scholar]
- TorelliAssimiadis MK, Tarantilis PA, Polissiou MG. UV-Vis, FT-Raman and H-NMR spectroscopies of cis-trans carotenoids from saffron (Crocus sativus L) Appl Spect. 1998;52:519–522. [Google Scholar]
- Bathaie SZ, Farajzade A, Hoshyar R. A review of the chemistry and uses of crocins and crocetin, the carotenoid natural dyes in saffron, with particular emphasis on applications as colorants including their use as biological stains. Biotechnic Histochem. 2014;89:401–411. doi: 10.3109/10520295.2014.890741. [DOI] [PubMed] [Google Scholar]
- Beale MH, Sussman , MR Metabolomics of Arabidopsis thaliana. Ann Plant Rev. 2011;43:157–180. [Google Scholar]
- Bhattacharjee B, Vijayasarathy S, Karunakar P, Chatterjee J. Comparative reverse screening approach to identify potential anti-neoplastic targets of saffron functional components and binding mode. Asian Pac J Cancer Prev. 2012;13:5605–11. doi: 10.7314/apjcp.2012.13.11.5605. [DOI] [PubMed] [Google Scholar]
- Broadhead GK, Chang A, Grigg J, McCluskey P. Efficacy and Safety of Saffron Supplementation: Current Clinical Findings. Crit Rev Food Sci Nutr. 2015;15:0. doi: 10.1080/10408398.2013.879467. [DOI] [PubMed] [Google Scholar]
- Busconi M, Colli L, Sánchez RA, Santaella M, De-Los-Mozos PM, Santana O, Roldán M, Fernández JA. AFLP and MS-AFLP analysis of the variation within saffron crocus (Crocus sativus L) germplasm. PLoS One. 2015;10:e0123434. doi: 10.1371/journal.pone.0123434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani LR, Culeddu N, Chessa M, Consonni R. NMR investigations for quality assessment of Italian PDO saffron (Crocus sativus L) Food Contrl. 2015;50:342–348. [Google Scholar]
- Calsteren MR, Bissonnette MC, Cormier F, Dufrense C, Ichi T, Le BlancCY, et al. Spectroscopic characterization of crocetin derivatives from Crocus sativus and Gardenia jasminoides. J Agri Food Chem. 1997;45:1055–1061. [Google Scholar]
- Carmona M, Zalacain A, Salinas MR, Alonso GL. A new approach to saffron aroma. Crit Rev Food Sci. Nutr. 2007;47:145–159. doi: 10.1080/10408390600626511. [DOI] [PubMed] [Google Scholar]
- Castillo R, Fernández JA, Gómez-Gómez L. Implications of Carotenoid Biosynthetic Genes in Apocarotenoid Formation during the Stigma Development of Crocus sativus and Its Closer Relatives. Plant Physiol. 2005;139:674–689. doi: 10.1104/pp.105.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahine N, Nader M, Duca L, Martiny L, Chahine R. Saffron extracts alleviate cardiomyocytes injury induced by doxorubicin and ischemia-reperfusion in vitro. Drug Chem Toxicol. 2015;17:1–10. doi: 10.3109/01480545.2015.1036281. [DOI] [PubMed] [Google Scholar]
- D'Agostino N, Pizzichini D, Chiusano ML, Giuliano G. An EST database from saffron stigmas. BMC Plant Biol. 2007;7:53–61. doi: 10.1186/1471-2229-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I, Das S, Saha T. Saffron suppresses oxidative stress in DMBA-induced skin carcinoma: a histopathological study. Acta Histo Chem. 2010;112:317–327. doi: 10.1016/j.acthis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Fernández JA. Biology, biotechnology and biomedicine of saffron. In: Pandalai, S. G., editor. Recent Res Dev Plant Sci. Vol. 2. 2004. pp. 127–159. [Google Scholar]
- Festuccia C, Mancini A, Gravina GL, Scarsella L, Llorens S, Alonso , G L, Carmona M. Antitumor effects of saffron-derived carotenoids in prostate cancer cell models. Bio Med Res Intl. 2014:135048. doi: 10.1155/2014/135048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas CA. Computational methods in protein structure prediction. Biotechnol Bioeng. 2007;97:207–13. doi: 10.1002/bit.21411. [DOI] [PubMed] [Google Scholar]
- Formisano C, Rigano D, Senatore F, Celik S, Bruno M, Rosselli S. Volatile constituents of aerial parts of three endemic Centaurea species from Turkey: Centaurea amanicola Hub-Mor, Centaurea consanguinea DC And Centaurea ptosimopappa Hayek and their antibacterial activities. Nat Prod Res. 2008;22:833–839. doi: 10.1080/14786410701218259. [DOI] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Giuliano G. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. PNAS. 2014;111:12246–12251. doi: 10.1073/pnas.1404629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadrdoost B, Vafaei AA, Rashidy-Pour A, Hajisoltani R, Bandegi AR, Motamedi F, Pahlvan S. Protective effects of saffron extract and its active constituent crocin against oxidative stress and spatial learning and memory deficits induced by chronic stress in rats. Euro J Pharmacol. 2011;667:222–229. doi: 10.1016/j.ejphar.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Moraga-Rubio A, Ahrazem O. Understanding carotenoid metabolism in saffron stigmas: unravelling aroma and color formation. Func Plant Sci Biotech. 2010;4:56–63. [Google Scholar]
- Guan H, Kiss-Toth E. Advanced technologies for studies on protein interactomes. In: Werther M, Seitz H, editors. Protein–Protein Interaction. Springer Berlin Heidelberg; 2008. pp. 1–24. [DOI] [PubMed] [Google Scholar]
- Hamid B, Sam S, Islam T, Singh P, Sharma M. The free radical scavenging and the lipid peroxidation inhibition of Crocin isolated from Kashmiri saffron (Crocus sativus) occurring in northern part of India. Int J Pharm Tech Res. 2009;1:1317–1321. [Google Scholar]
- Harrold JM, Ramanathan M, Mager DE. Network-based approaches in drug discovery and early development. Clin Pharmacol Ther. 2013;94:651–8. doi: 10.1038/clpt.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HausenblasHA , Heekin K, Mutchie HL Anton S. A systematic review of randomized controlled trials examining the effectiveness of saffron (Crocus sativus L) on psychological and behavioral outcomes. J Integr Med. 2015;13:231–240. doi: 10.1016/S2095-4964(15)60176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinzadeh H, Noraei NB. Anxiolytic and hypnotic effect of Crocus sativus aqueous extract and its constituents, crocin and safranal, in mice. Phytother Res. 2009;23:768–774. doi: 10.1002/ptr.2597. [DOI] [PubMed] [Google Scholar]
- Hosseinzadeh H, Karimi G, Niapoor M. Antidepressant effect of Crocus sativus L stigma extracts and their constituents, crocin and safranal, in mice. Acta Hort. 2004;650:435–445. [Google Scholar]
- Hosseinzadeh H, Ziaee T, Sadeghi A. 2008. Phytomed. 15:491–495. doi: 10.1016/j.phymed.2007.09.020. https://en.wikipedia.org/wiki/Trade_and_use_of_saffron. [DOI] [PubMed] [Google Scholar]
- Husaini AM. Challenges of climate change: Omics-based biology of saffron plants and organic agricultural biotechnology for sustainable saffron production. GM crops food. 2014;5:97–105. doi: 10.4161/gmcr.29436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husaini AM, Wani SA, Sofi P, Rather AG, Parray GA, Shikari AB, Mir JI. Bioinformatics for saffron (Crocus sativus L) improvement. Comm Biomet Crop Sci. 2009;4:3–8. [Google Scholar]
- Jessie SW, Krishnakantha TP. Inhibition of human platelet aggregation and membrane lipid peroxidation by food spice, saffron. Mol Cell Biochem. 2005;278:59–63. doi: 10.1007/s11010-005-5155-9. [DOI] [PubMed] [Google Scholar]
- Jiang C, Cao L, Yuan Y, Chen M, Jin Y, Huang L. Barcoding Melting Curve Analysis for Rapid, Sensitive, and Discriminating Authentication of Saffron (Crocus sativus L) from Its Adulterants. Bio Med Res Int. 2014;2014:809037. doi: 10.1155/2014/809037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlem P, Newfeld SJ. Informatics approaches to understanding TGFβ pathway regulation. Dev (Cambridge, England) 2009;136:3729–3740. doi: 10.1242/dev.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakoudi A, Ordoudi AS, Roldán-Medina M, Tsimidou ZM. Saffron, a functional spice. Austin J Nutri Food Sci. 2015;3:1059. [Google Scholar]
- Leffingwell JC. Saffron. Leffingwell Rep. 2002;2:1–6. [Google Scholar]
- Mathews-Roth MM. Effect of crocetin on experimental skin tumors in hairless mice. Oncol. 1982;39:362–364. doi: 10.1159/000225672. [DOI] [PubMed] [Google Scholar]
- Mir JI, Ahmed N, Singh DB, Khan MH, Zffer S, Shafi W. Breeding and biotechnological opportunities in saffron crop improvement. Afri J Agri Res. 2015;10:1970–1974. [Google Scholar]
- Moraga ÁR, Mozos AT, Ahrazem O, Gómez-Gómez L. Cloning and characterization of a glucosyltransferase from Crocus sativus stigmas involved in flavonoid glucosylation. BMC Plant Bio. 2009;9:109. doi: 10.1186/1471-2229-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SZ, Bathaie SZ. Historical uses of saffron: Identifying potential new avenues for modern research. Avicenna Journal of Phytomedicine. 2011;1:57–66. [Google Scholar]
- Naghshineh A, Dadras A, Ghalandari B, Riazi GH, Modaresi SMS, Afrasiabi A, AslaniM K. Safranal as a novel anti-tubulin binding agent with potential use in cancer therapy: An in vitro study. Chemico-biological interactions. 2015;238:151–160. doi: 10.1016/j.cbi.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Nemati Z, Mardi M, Majidian P, Zeinalabedini M, Pirseyedi SM, Bahadori M. Saffron (Crocus sativus L), a monomorphic or polymorphic species? Spanish J Agri Res. 2014;12:753–762. [Google Scholar]
- Nithya G, Sakthisekaran D. In Silico Docking Studies On The Anti-Cancer Effect Of Thymoquinone On Interaction With Pten- A Regulator Of Pi3k/ Akt Pathway. Asian J Pharma Clin Res. 2015;1:192–195. [Google Scholar]
- Omidi A, Riahinia N, Torbati MBM, Behdani MA. Hepatoprotective effect of Crocus sativus (saffron) petals extract against acetaminophen toxicity in male Wistar rats. Avicenna J Phytomed. 2014;4:330. [PMC free article] [PubMed] [Google Scholar]
- Ordoudi SA, Tsimidou MZ. Production practices and quality assessment of food crops. In: Dris R, Jain SM, editors. Saffron Quality: Effect of agricultural practices, processing and storage. Netherlands: Kluwer Academic Publ.Dordrecht; 2004. pp. 209–260. [Google Scholar]
- Ordoudi SA, Cagliani LR, Lalou S, Naziri E, Tsimidou MZ, Consonni R. H NMR-based metabolomics of saffron reveals markers for its quality deterioration. Food Res Int. 2015;70:1–6. [Google Scholar]
- Pentony MM, Winters P, Penfold-Brown D, Drew K, Narechania A, DeSalle R, Bonneau R, Purugganan MD. The Plant Proteome folding project: structure and positive selection in plant protein families. Genome BiolEvol. 2012;4:360–371. doi: 10.1093/gbe/evs015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F. 2006. MODBASE: a database of annotated comparative protein structure models and associated resources. Nuc Acid Res. Sali A;34:291–295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piqueras A, Han BH, Escribano’ J, Rubio’ C, Hellín E, Fernández JA. Development of cormogenic nodules and microcorms by tissue culture, a new tool for the multiplication and genetic improvement of saffron. Agron. 1999;19:603–610. [Google Scholar]
- Poma A, Fontecchio G, Carlucci G, Chichiricco G. Anti-inflammatory properties of drugs from saffron crocus. Anti-Inflamm Anti-Aller Agents in Med Chem. 2012;11:37–51. doi: 10.2174/187152312803476282. [DOI] [PubMed] [Google Scholar]
- Rezaee R, Hosseinzadeh H. Safranal: from an aromatic natural product to a rewarding pharmacological agent. Iran J Basic Med Sci. 2013;16:12. [PMC free article] [PubMed] [Google Scholar]
- Rubio-Moraga A, Ahrazem O, Pérez-Clemente RM, Gómez-Cadenas A, Yoneyama K, López-Ráez JA, Molina RV, Gómez-Gómez L. Apical dominance in saffron and the involvement of the branching enzymes CCD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 2014;14:171. doi: 10.1186/1471-2229-14-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffari B, Mohabatkar H Mohsenzadeh S. T and B-cell epitopes prediction of Iranian saffron (Crocus sativus) profiling by bioinformatics tools. Protein Pept Lett. 2008;15:280–5. doi: 10.2174/092986608783744270. [DOI] [PubMed] [Google Scholar]
- Saxena RB. Botany, Taxonomy and Cytology of Crocus sativus series. Ayu. 2010;31 doi: 10.4103/0974-8520.77153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahihi M. In-Silico Study on the Interaction of Saffron Ligands and beta-Lactoglobulin by Molecular Dynamics and Docking Approach. J Macromol Sci Ph. 2015 doi: 10.1080/00222348.2015.1125066. [Google Scholar]
- Schmidt M, Betti G, Hensel A. Saffron in phytotherapy: pharmacology and clinical uses. Wien Med Wochenschr. 2007;157:315–319. doi: 10.1007/s10354-007-0428-4. [DOI] [PubMed] [Google Scholar]
- Sharifi G, Ebrahimzadeh H, Ghareyazie B, Gharechahi J, Vatankhah E. Identification of differentially accumulated proteins associated with embryogenic and non-embryogeniccalli in saffron (Crocus sativus L) Proteome Sci. 2012;10:3–18. doi: 10.1186/1477-5956-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Luo YM, Ding XY, Mao SG. Authentication of Crocus sativus L and its adulterants by rDNA ITS sequences and allele-specific PCR. J Nanjing Normal Univ. 2007;30:89–92. [Google Scholar]
- Singla RK, Bhat , VG Crocin: An Overview. Indo Global J Pharma Sci. 2011;1:281–286. [Google Scholar]
- Suravajhala P, Sundararajan VS. A classification scoring schema to validate protein interactors. Bioinformation. 2012;8:34–9. doi: 10.6026/97320630008034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, TsafouKP , Kuhn M, Bork P, Jensen LJ. von Mering C STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015 Jan;43(Database issue):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum N, Hamdani M. Plants used to treat skin diseases. Pharmacognosy Rev. 2014;8:52. doi: 10.4103/0973-7847.125531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli A, Marieschi M, Bruni R. Authentication of saffron (Crocus sativus L) in different processed, retail products by means of SCAR markers. FoodContrl. 2014;36:126–131. [Google Scholar]
- Tsaftaris A, Pasentsis K, Makris A, Darzentas N, Polidoros A, Kalivas A, Argiriou A. The study of the E-class SEPALLATA3-like MADS-box genes in wild-type and mutant flowers of cultivated saffron (Crocus sativus L) and its putative progenitors. J Plant Physiol. 2011;168:1675–1684. doi: 10.1016/j.jplph.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Tsatsaroni E, Liakopoulou-Kyriakides M. Effect of enzymatic treatment on the dyeing of cotton and wool fibers with natural dyes. Dyes and pig. 1995;29:203–209. [Google Scholar]
- Vandamme D, Minke BA, Fitzmaurice W, Kholodenko BN, Kolch W. Systems biology‐embedded target validation: improving efficacy in drug discovery. Wiley Interdiscip Rev SystBiol Med. 2014;6:1–11. doi: 10.1002/wsbm.1253. [DOI] [PubMed] [Google Scholar]
- Wetie AGN, Sokolowska I, Woods AG, Roy U, Deinhardt K, Darie CC. Protein–protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches. Cell Mol Life Sci. 2014;71:205–228. doi: 10.1007/s00018-013-1333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Nyberg NT, Molgaard P, Asili J, Jaroszewsk JW. HNMR metabolic fingerprinting of saffron extracts. Metab. 2010;6:511–517. [Google Scholar]
- Zargari A. Medicinal Plants. Tehran: Tehran University Press; 1990. pp. 574–578. [Google Scholar]
- Zeraatkar M, Khalili K, Foorginejad A. Studying and Generation of Saffron Flower's 3D Solid Model. Procedia Tech. 2015;19:62–69. [Google Scholar]