Abstract
Increasing evidence suggests that miR-194 is down-regulated in esophageal squamous cell carcinoma tumor tissue. However, the role and underlying mechanism of miR-194 in esophageal squamous cell carcinoma have not been well defined. We used DIANA, TargetScan and miRanda to perform target prediction analysis and found KDM5B is a potential target of miR-194. Based on these findings, we speculated that miR-194 might play a role in esophageal squamous cell carcinoma development and progression by regulation the expression of KDM5B. We detected the expression of miR-194 and KDM5B by quantitative real-time reverse transcription PCR (qRT-PCR) and Western blot assays, respectively, and found down-regulation of miR-194 and up-regulation of KDM5B existed in esophageal squamous cell carcinoma cell lines. By detecting proliferation, invasion and apoptosis of TE6 and TE14 cells transfected with miR-194 mimics or mimic control, miR-194 was found to inhibit proliferation and invasion and promote apoptosis of esophageal squamous cell carcinoma cells. miR-194 was further verified to regulate proliferation, apoptosis and invasion of esophageal squamous cell carcinoma cells by directly targeting KDM5B. Furthermore, animal studies were performed and showed that overexpression of miR-194 inhibited the growth of esophageal squamous cell carcinoma tumors in vivo. These results confirmed our speculation that miR-194 targets KDM5B to inhibit esophageal squamous cell carcinoma development and progression. These findings offer new clues for esophageal squamous cell carcinoma development and progression and novel potential therapeutic targets for esophageal squamous cell carcinoma.
Keywords: Esophageal squamous cell carcinoma, miR-194, histone demethylase lysine demethylase 5b, proliferation, apoptosis
Introduction
Esophageal cancer (EC) ranks eighth in order of occurrence and is the sixth most common cause of cancer deaths in the world, affecting males more than females.1 According to the etiologic and pathologic features, EC could be divided into two main forms, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). Although EAC is the most rapidly increasing cancer in Western countries, ESCC is still dominant in East Asia.2 The prognoses of ESCC patients are poor with a five-year survival rate of 10% despite the rapid development of therapeutic options such as surgery, radiotherapy and chemotherapy.3 At present, much effort has been spent on the research of the biological behavior of ESCC cells, which is aimed to develop effective treatment strategies for ESCC. Many oncogenes and tumor suppressor genes have been reported to be involved in the development of ESCC.4 However, few specific molecules modulating the initiation and progression of ESCC have been identified.
MicroRNAs (miRNAs) are a group of single-stranded and non-coding RNA with a length of 17–25 ribonucleotides, which regulate gene expression through targeting the 3′-untranslated region (UTR) of mRNAs for degradation, translational repression or both.5 miRNAs play important roles in various biological processes including cellular proliferation, apoptosis, differentiation and development.6 Mounting data have shown that aberrant expression of many miRNAs is associated with human diseases, including cancer. In fact, it has been widely reported that dysregulated expression of miRNAs occurs in various types of malignant tumors, some of which act as oncogenes or tumor suppressors.7,8 Furthermore, many recent studies have shown that many microRNAs are involved in essential tumor cell biological processes, such as proliferation, apoptosis and invasion.7,9 Owing to the critical role of miRNAs, miRNAs have been considered as putative targets for diagnosis and treatment of cancer. Recently, miR-194 is widely studied and its role in tumor development is complicated. For example, some studies showed that it is down-regulated and functions as a tumor suppressor in some malignant tumors such as non-small cell lung cancer and endometrial cancer.10,11 But some studies demonstrated that it is up-regulated and acts as an oncogene in EAC, prostate cancer, etc.12,13 miR-194 is down-regulated in ESCC tumor tissue compared to the matched normal tissue from a genome-wide microRNA expression profile assay.14 However, for miR-194, its role, the related mRNA targets and molecular mechanism in ESCC remain unclear. We used DIANA, TargetScan and miRanda to perform target prediction analysis and found that the histone demethylase lysine demethylase 5b (KDM5B) is a potential target of miR-194. Recent studies reported that KDM5B knockdown inhibits EC cell growth, sphere formation and invasion capacity.15 Therefore, we hypothesized that miR-194 might play a role in ESCC development and progression by regulation the expression of KDM5B.
In this study, the expression levels of miR-194 and KDM5B in ESCC cell lines were determined by qRT-PCR and Western blot, respectively, and the relationship between miR-194 expression and proliferation, apoptosis and invasion of ESCC cells was investigated. Luciferase reporter assay was performed to identify whether the 3′UTR of KDM5B mRNA is a binding target of miR-194. The effect of expression level of KDM5B on ESCC cell proliferation, apoptosis and invasion was also investigated. Moreover, animal studies were performed to explore the anti-tumor effect of miR-194 in vivo. These findings will provide new clues for ESCC development and progression and novel potential therapeutic targets for ESCC.
Materials and methods
Cell culture and transfection
Human ESCC cell lines Eca109 and TE1 were provided by the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China), and TE6, TE8, TE12 and TE14 were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Human esophageal epithelial cells (HEEpiC) were obtained from ScienCell (Carlsbad, CA, USA). All cells were grown in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 2 mmol/L glutamine (Invitrogen), 100 µg of streptomycin/mL (Sigma Chemical Company, St Louis, MO, USA) and 100 units of penicillin/mL (Sigma) and incubated in a humidified chamber containing 5% CO2 at 37℃.
miR-194 mimics (5′-UGU AAC AGC AAC UCC AUG UGG A-3′, 5′-CAC AUG GAG UUG CUG UUA CAU U-3′), inhibitors (5′-GAC AGU CCA CAU GGA GUU GCU GUU ACA CUU GA), miRNA control (5′-UUC UCC GAA CGU GUC ACG UTT-3′, 5′-ACG UGA CAC GUU CGG AGA ATT-3′), KDM5B siRNAs (siKDM5B) and control were obtained from GenePharma Company (Shanghai, China). Cell transfection was performed by using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) in accordance with manufacturer’s suggestions.
Quantitative real-time reverse transcription PCR
Total RNA was obtained from cultured cells using the mirVana microRNA isolation kit (Ambion, Austin, TX, USA), following manufacturer’s instructions. The expression level of miR-194 was determined by TaqMan qRT-PCR using the TaqMan microRNA assay (Applied Biosystems, Foster City, CA, USA) and normalized to U6 (Applied Biosystems) expression level.
Western blot assay
The proteins were obtained from the cultured cells by using a 1 × SDS lysis buffer. Protein concentration was determined by using BCA Protein Assay reagent kit (Thermo Fisher Scientific, Waltham, MA, USA), in accordance with manufacturer’s instructions. Then equal amounts of protein were separated by SDS-PAGE in 10% (w/v) polyacrylamide gels and transferred to nitrocellulose membranes (Immobilon Millipore Corporation, Bedford, MA, USA) with primary antibodies, against KDM5B (Chemicon, Temecula, CA, USA), Ki67 (Abcam, Cambridge, UK), Bax (Abcam), Bcl-2 (Abcam), MMP-2 (Sigma), MMP-9 (Sigma) and β-Actin (Chemicon). Secondary antibody was purchased from Genmed Scientifics (Arlington, MA, USA). The signal intensity of the band was quantified by Quantity One Software (Bio-Rad, Hercules, CA, USA).
Cell proliferation assay
Cell suspensions with 5 × 103 cells/well were seeded in 96-well plates, incubated in a humidified chamber containing 5% CO2 at 37℃ and examined at 0, 24, 48, 72 and 96 h. At each time-point, CCK-8 reagent (10 µL) was added into each well and further incubated for 2 h. The absorbance at 450 nm was measured by a multi-mode microplate reader (BioTek, Winooski, VT, USA), and the absorbance at 630 nm was used as reference.
Cell apoptosis assay
Cell apoptosis was analyzed with Annexin-V-FITC Apoptosis kit (Biovision Research Products, Mountain View, CA, USA) on a FACScalibur™ flow cytometer (Becton–Dickinson, San Jose, CA, USA), according to manufacturer’s instructions.
Cell invasion assay
Cell invasion was examined with transwell chambers. Cell suspensions with 2 × 104 cells in 100 µL of serum-free DMEM (Invitrogen) were added into the upper chamber precoated with Matrigel (24-well insert; 8 µm pore size; Becton–Dickinson). Medium supplemented with 10% FBS in the lower chamber served as the chemoattractant. After 22 h incubation, the non-invading cells were carefully wiped out by cotton-tipped swabs. The invasive cells on the lower surface of the membrane were fixed, stained and then analyzed under an inverted microscope (Olympus, Tokyo, Japan) by counting the cells in six random fields per insert.
Luciferase reporter assay
The wild-type 3′UTR (3′UTR-WT) of KDM5B containing the miR-194 binding sites was amplified by PCR. The mutant KDM5B 3′UTR (3′UTR-MUT) occurred in the binding sites for miR-194 was generated by using overlapping extension PCR. Normal or mutant KDM5B 3′UTR was inserted into XhoI-NotI digested psiCHECK-2 vector (Promega, Madison, WI, USA) which included both renilla and firefly luciferase reporter genes. Then the psiCHECK-2 vectors including 3′UTR-WT or 3′UTR-MUT of KDM5B were transfected into miR-194-overexpressing TE6 cells and control cells, respectively. The firefly and renilla luciferase activities were determined by using a dual luciferase assay kit (Promega), according to manufacturer’s instructions.
In vivo tumor formation assay
Animal studies were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University. Five-week-old male athymic nude mice (BALB/c nu/nu) were purchased from the National Laboratory Animal Center. Tumor formation in nude mice was performed with TE6 cells transfected with miR-194 mimics or miRNA control. We used six mice in this study, and each animal received single injection of 2 × 106 living cells suspended in 200 µL of PBS. TE6 cells transfected with miRNA control were injected into the left dorsal flank, and TE6 cells transfected with miR-194 mimics were injected into the right dorsal flank. The nude mice were examined for tumor formation over a period of one month. The tumor size was measured every five days with a slide caliper, and tumor volume was calculated as follows: tumor volume = length × width2/2. After 30 days, mice were sacrificed, and tumors were weighed. Moreover, the expression levels of miR-194, KDM5B, Ki67, Bax, Bcl-2, MMP-2 and MMP-9 protein in xenografts were examined by qRT-PCR and Western blot, respectively.
Statistical analysis
A statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA, USA). Values are expressed as the mean ± standard deviation (SD). Differences between groups were calculated with Student’s t test, and P < 0.05 was defined as being significant.
Results
miR-194 is down-regulated and KDM5B is up-regulated in ESCC cell lines
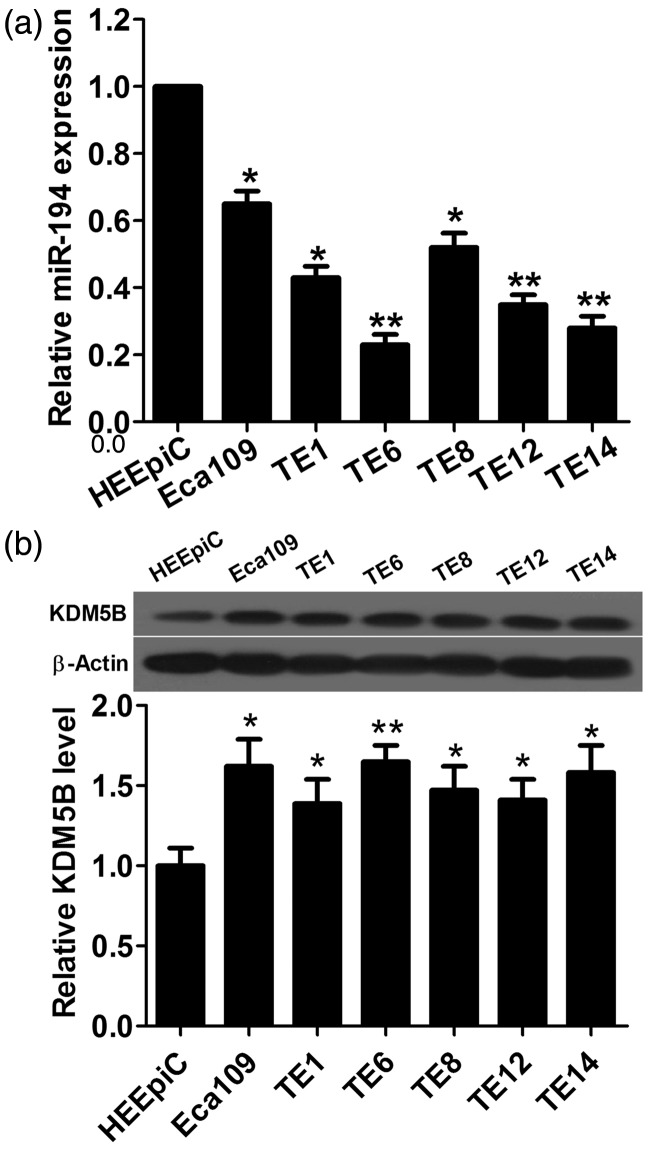
It has been reported that miR-194 was down-regulated14 and KDM5B was overexpressed in ESCC.16 To confirm the expression status of miR-194 and KDM5B in ESCC, the expression levels of miR-194 and KDM5B in six ESCC cell lines and HEEpiC were determined by qRT-PCR and Western blot, respectively. The results showed that the expression level of miR-194 in six ESCC cell lines was lower than that in HEEpiC, and the expression level of KDM5B was significantly increased in ESCC cell lines compared with that in HEEpiC (Figure 1(a) and (b)). These results indicated that miR-194 is down-regulated and KDM5B is up-regulated in ESCC cell lines.
Figure 1.
miR-194 expression is obviously low and KDM5B protein expression is significantly high in ESCC cell lines compared with those in human esophageal epithelial cells (HEEpiC). (a) qRT-PCR shows that miR-194 is down-regulated in ESCC cell lines compared with that in HEEpiC. (b) The expression level of KDM5B protein is significantly increased in ESCC cell lines compared with that in HEEpiC. *P < 0.05 and **P < 0.01
miR-194 inhibits proliferation and invasion and promotes apoptosis of ESCC cells
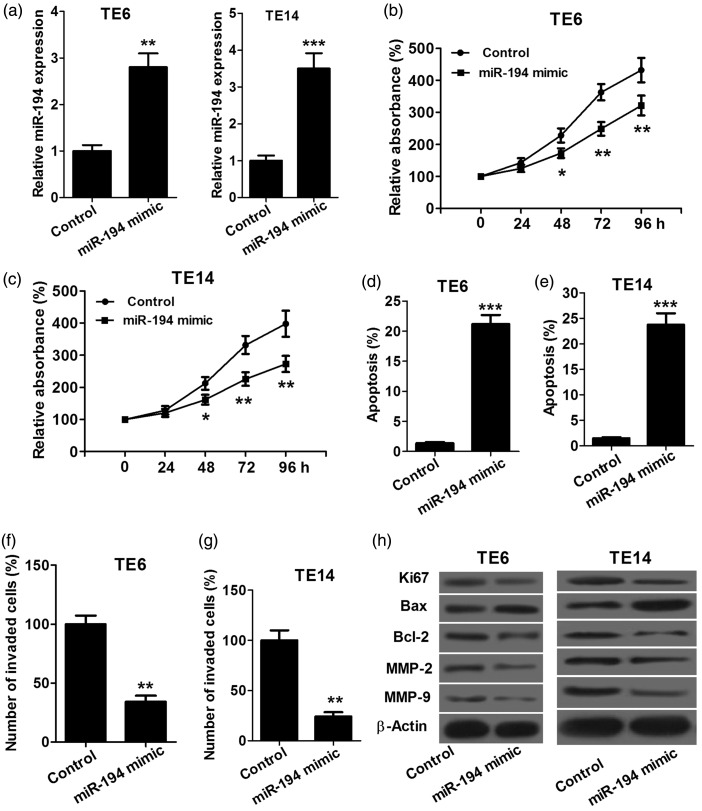
To investigate the effect of miR-194 on proliferation, apoptosis and invasion of ESCC cells, we transfected TE6 and TE14 cells with miR-194 mimics or miRNA control, respectively, and then performed proliferation, apoptosis and invasion assays. The miR-194 level was higher in TE6 and TE14 cells transfected with miR-194 mimics than in control cells, which confirmed that transfection is successful (Figure 2(a)). Proliferation assay showed that TE6 and TE14 cells transfected with miR-194 mimics had a significantly lower proliferation rate than controls (Figure 2(b) and (c)). Apoptosis assay showed that TE6 and TE14 cells transfected with miR-194 mimics had a significantly higher apoptosis rate than controls (Figure 2(d) and (e)). Invasion assay showed that the invasion capacity of TE6 and TE14 cells transfected with miR-194 mimics was significantly reduced compared with the control (Figure 2(f) and (g)). In addition, we performed Western blot assay to detect the expression levels of proliferation-related protein Ki67, apoptosis-related proteins Bax and Bcl-2 and markers of tumor invasion MMP-2 and MMP-9 and found that TE6 and TE14 cells transfected with miR-194 mimics had obvious decreases in Ki67, Bcl-2, MMP-2 and MMP-9 protein expression, and significant increases in Bax protein expression (Figure 2(h)). These results indicated that miR-194 inhibits proliferation and invasion and promotes apoptosis of ESCC cells.
Figure 2.
miR-194 inhibits proliferation and invasion and promotes apoptosis in ESCC cells. (a) qRT-PCR confirmed successful transfection TE6 and TE14 cells with miR-194 mimics. (b and c) CCK-8 assay shows that TE6 and TE14 cells transfected with miR-194 mimics both have a significantly lower proliferation rate than controls. (d and e) Flow cytometer assay shows that TE6 and TE14 cells transfected with miR-194 mimics both have an obviously high apoptosis compared with controls. (f and g) Transwell chamber shows that the invasion capacity of TE6 and TE14 cells transfected with miR-194 mimics is significantly reduced compared with the control. (h) TE6 and TE14 cells transfected with miR-194 mimics both have obvious decreases in Ki67, Bcl-2, MMP-2 and MMP-9 protein expression, and significant increases in Bax protein expression compared with controls. *P < 0.05, **P < 0.01 and ***P < 0.001
miR-194 directly targets KDM5B
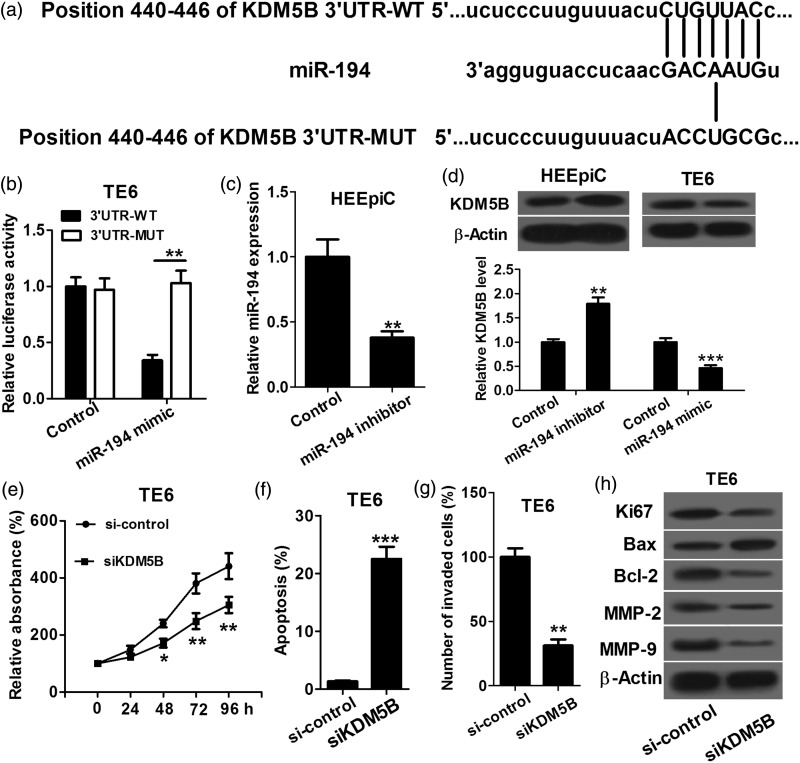
We used DIANA, TargetScan and miRanda to perform target prediction analysis, and found KDM5B is a potential target of miR-194 (Figure 3(a)). Then dual luciferase reporter assay was performed on TE6 cells to identify whether the 3′UTR of KDM5B mRNA is a binding target of miR-194. As shown in Figure 3(b), in the miR-194 mimic group, the luciferase activity driven by KDM5B 3′UTR was significantly lower than that in the control, while miR-194-mediated decrease of luciferase activity was abolished by mutation in the presumptive binding site, which suggested that KDM5B is indeed a direct target of miR-194. To determine the regulatory effect of miR-194 on KDM5B, we transfected HEEpiC with miR-194 inhibitors or miRNA control, and TE6 cells with miR-194 mimics or miRNA control. The miR-194 level was lower in HEEpiC transfected with miR-194 inhibitors than in control cells, which indicated that transfection is successful (Figure 3(c)). Western blot assay was carried out to determine the expression level of KDM5B protein in response to the miR-194 expression alteration in HEEpiC and TE6 cells. As shown in Figure 3(d), down-regulation of miR-194 in HEEpiC cells increased KDM5B protein expression, and up-regulation of miR-194 in TE6 cells significantly decreased KDM5B protein expression, which indicated that miR-194 negatively regulates KDM5B. To further confirm if miR-194 regulates ESCC cell proliferation, apoptosis and invasion by targeting KDM5B, we transfected TE6 cells with pcDNA-KDM5B, siKDM5B or control and then conducted proliferation, apoptosis and invasion assays. As shown in Figure 3(e)–(g), the siKDM5B group had significantly lower cell proliferation rate and invasion capacity and higher apoptosis rate than si-control group. Furthermore, the siKDM5B group had significant decreases in Ki67, Bcl-2, MMP-2 and MMP-9 protein expression, and obvious increase in Bax protein expression compared with si-control group (Figure 3(h)). Taken together, these results suggested that miR-194 regulates ESCC cell proliferation, apoptosis and invasion by directly targeting KDM5B.
Figure 3.
KDM5B is a direct target of miR-194. (a) Bioinformatics-based target prediction analysis shows that KDM5B is a potential target of miR-194. (b) Luciferase reporter assay shows that in the miR-194 mimic group, the luciferase activity driven by 3′UTR of KDM5B is obviously decreased compared with that in the negative control and 3′UTR-MUT group. (c) qRT-PCR shows that miR-194 level is lower in HEEpiC transfected with miR-194 inhibitors than in control cells. (d) Down-regulation of miR-194 in HEEpiC cells increased KDM5B protein expression, and up-regulation of miR-194 in TE6 cells significantly decreased KDM5B protein expression. (e) CCK-8 assay shows that the siKDM5B group has a lower cell proliferation rate than si-control group. (f) Flow cytometer assay shows that the siKDM5B group has a significantly higher apoptosis rate than si-control group. (g) Transwell chamber shows that the siKDM5B group has a markedly weaker invasion capacity than si-control group. (h) Western blot assay shows that the siKDM5B group has obvious decreases in Ki67, Bcl-2, MMP-2 and MMP-9 protein expression, and significant increase in Bax protein expression compared with si-control group. *P < 0.05, **P < 0.01 and ***P < 0.001
Overexpression of miR-194 suppresses growth of ESCC tumors in vivo
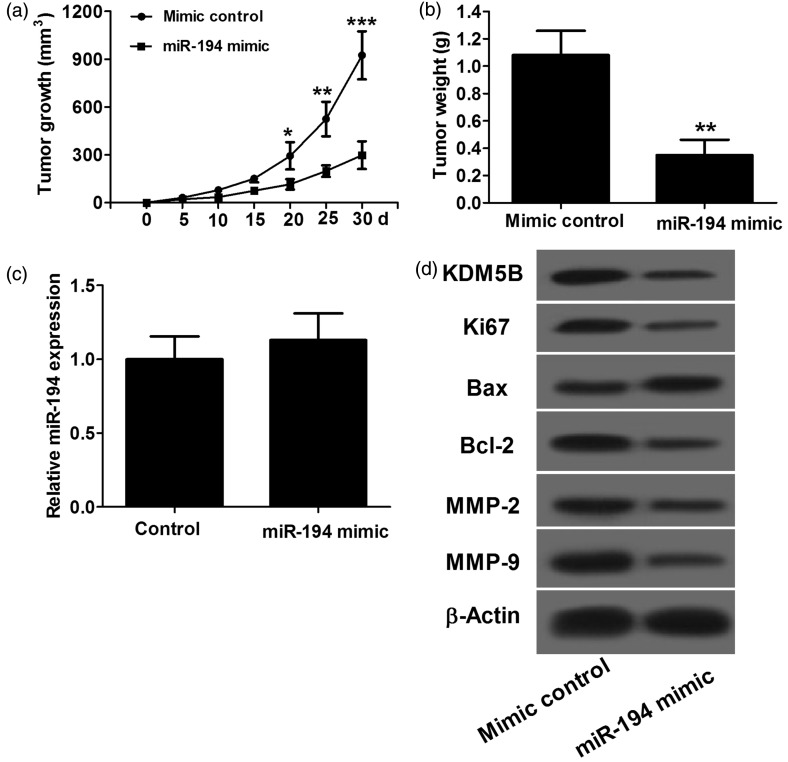
In vitro studies indicated that miR-194 suppresses ESCC cell proliferation and invasion and promotes apoptosis by targeting KDM5B. We further investigated the effect of miR-194 in mouse xenograft models. TE6 cells transfected with miR-194 mimics and miRNA control were subcutaneously injected into the right and left dorsal flanks of nude mice, respectively. We measured the tumor size weekly and calculated the tumor volume. As illustrated in Figure 4(a), the miR-194 mimic group had smaller tumor volume than control. After 30 days, mice were sacrificed, and tumors were isolated and weighed. Accordingly, the tumor weight in the miR-194 mimic group was significantly lower than that in the control (Figure 4(b)). Furthermore, qRT-PCR was performed to detect the expression level of miR-194. As shown in Figure 4(c), miR-194 level in the miR-194 mimic group was not significantly different from that in control group. Western blot assay was performed to determine the expression levels of KDM5B, Ki67, Bax, Bcl-2, MMP-2 and MMP-9 protein in xenografts. As shown in Figure 4(d), the miR-194 mimic group had obvious decreases in KDM5B, Ki67, Bcl-2, MMP-2 and MMP-9 expression and remarkable increase in Bax expression compared with control. Taken together, these results indicated that overexpression of miR-194 represses growth of ESCC tumors in vivo.
Figure 4.
The antitumor effect of miR-194 in vivo. TE6 cells are used in this study. (a) In the miR-194 mimic group, the tumor volume is significantly lower than that in control. (b) After 30 days, in the miR-194 mimic group, the tumor weight is significantly lighter than that in control. (c) qRT-PCR shows that after 30 days, miR-194 level in the miR-194 mimic group was not significantly different from than in control. (d) Western blot assay shows that compared with control, the miR-194 mimic group has marked decreases in KDM5B, Ki67, Bcl-2, MMP-2 and MMP-9 protein expression, and significant increase in Bax protein expression. *P < 0.05, **P < 0.01 and ***P < 0.001, n = 6
Discussion
The incidence of ESCC is broadly varied by geographic locations. The highest incidence rates are found in China, South America, Iran and South Africa, and almost one-half of all ESCC cases in the world occur in China.17 Although a myriad of improvements in diagnostic and therapeutic techniques have been achieved over the last 30 years, ESCC continues to have a poor prognosis.3 Previous studies have identified many genetic alterations relating to induction of ESCC. However, the molecular mechanisms which are responsible for the cellular deregulation in ESCC have not been completely clear. It has been widely reported that many miRNAs are aberrantly regulated in various types of malignant tumors, some of which function as oncogenes or tumor suppressors and are involved in essential tumor cell biological processes, such as proliferation, apoptosis and invasion.7,8,18 Recent studies found that the expression of some miRNAs was aberrant in ESCC, which suggested that these miRNAs might play an important role in the development and progression of ESCC.19,20 A number of studies have demonstrated the roles of abnormally expressed miRNAs in the carcinogenesis of ESCC.21–24 miR-21 was reported to be up-regulated and act as an oncogene in ESCC by targeting PDCD4 and PTEN.21,22 miR-205 and miR-29 c were down-regulated in ESCC and inhibited ESCC cell proliferation and invasion.23,24 Recently, miR-194 was found to be down-regulated in ESCC tumor tissue, but its role in the carcinogenesis of ESCC remains unclear.14 In this study, we determined the expression level of miR-194 in ESCC cell lines and found that the expression level of miR-194 is higher in ESCC cell lines than that in HEEpiC. We also detected the effect of miR-194 on ESCC cell proliferation, apoptosis and invasion and found miR-194 suppresses proliferation and invasion and promotes apoptosis in TE6 and TE14 cells. Furthermore, we further explored the related mRNA targets and molecular mechanisms of miR-194 in ESCC cells.
KDM5B, the specific demethylase of histone H3 lysine 4 (H3K4), is predicted to be a potential target of miR-194 by miRNA target analysis tools, and its up-regulation can reduce H3K4 methylation level without affecting other histone lysine methylation status. Histone lysine methylation plays a crucial role in the epigenetic regulation of eukaryotic genes, and histone methylation disorders can cause cancer.25 Wong et al. reported that KDM5B collaborates with TFAP2C and Myc to suppress the cell cycle inhibitor p21cip.26 Enkhbaatar et al. reported that ectopic expression of KDM5B promotes epithelial-mesenchymal transition (EMT) of cancer cells.27 Li et al. reported that KDM5B is highly expressed in prostate cancer cells and promotes proliferation and inhibits apoptosis of prostate cancer cells.28 For the role of KDM5B in EC, Nishida et al. reported that KDM5B knockdown results in the inhibition of EC cell growth, sphere formation and invasion ability.15 The above findings are the reason why we selected KDM5B for further analysis. In this study, KDM5B protein expression level are found significantly higher in ESCC cell lines than that in HEEpiC, and KDM5B promotes proliferation and invasion and inhibits apoptosis of ESCC cells, which are completely consistent with the experimental result of Nishida et al.15 Luciferase reporter assay confirmed that miR-194 targets KDM5B directly. An inverse correlation is also found between miR-194 and KDM5B in HEEpiC and TE6 cells. These findings implied that miR-194 represses KDM5B protein expression by directly binding on the 3′UTR of KDM5B mRNA to inhibit proliferation and invasion and promote apoptosis of ESCC cells. We also explored the anti-tumor effect of miR-194 in vivo and reached the conclusion that overexpression of miR-194 suppresses growth of ESCC tumors in vivo.
All together, down-regulation of miR-194 and up-regulation of KDM5B protein exist in ESCC cell lines, and miR-194 represses proliferation and invasion and promotes apoptosis of ESCC cells. We identified and verified that KDM5B is a direct functional target of miR-194, and miR-194 regulates proliferation, apoptosis and invasion of ESCC cells by targeting KDM5B. Animal studies had confirmed that overexpression of miR-194 suppresses the growth of ESCC tumors in vivo. miR-194 which functions as a tumor suppressor provides a novel therapeutic target for ESCC treatment, and miR-194/KDM5B pathway might be exploited by a therapeutic strategy for ESCC treatment in future.
Acknowledgment
There is no funding for this research. Thank to all the members in our department for their help in data collection.
Author contributions
GC and SZ conception and design of research; DL,WL,YHL, YGL and WS performed experiments; GC, DL and SZ analyzed data; GC, DL and WL interpreted results of experiments; GC and SZ drafted manuscript; all authors approved final version of manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003; 349: 2241–52. [DOI] [PubMed] [Google Scholar]
- 2.Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res 2009; 15: 1915–22. [DOI] [PubMed] [Google Scholar]
- 3.Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, Fadda P, Ozer HG, Huebner K, Farber JL, Croce CM. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis 2012; 33: 1736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Leung AC, Ko JM, Lo PH, Tang JC, Srivastava G, Oshimura M, Stanbridge EJ, Daigo Y, Nakamura Y. Tumor suppressive role of a 2.4 Mb 9q33–q34 critical region and DEC1 in esophageal squamous cell carcinoma. Oncogene 2005; 24: 697–705. [DOI] [PubMed] [Google Scholar]
- 5.Hobert O. Gene regulation by transcription factors and microRNAs. Science 2008; 319: 1785–6. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 2008; 26: 462–9. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–69. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302: 1–12. [DOI] [PubMed] [Google Scholar]
- 9.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83–6. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Liu T, Fang O, Leach L, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27kip1. Oncogene 2014; 33: 1506–14. [DOI] [PubMed] [Google Scholar]
- 11.Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J, Sakuragi N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer 2011; 10: 99–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijnhoven B, Hussey D, Watson D, Tsykin A, Smith C, Michael M. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg 2010; 97: 853–61. [DOI] [PubMed] [Google Scholar]
- 13.Tong A, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund A, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther 2009; 16: 206–16. [DOI] [PubMed] [Google Scholar]
- 14.Kong KL, Kwong DLW, Chan TH-M, Law SY-K, Chen L, Li Y, Qin Y-R, Guan X-Y. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2011; 61: 33–42. [DOI] [PubMed] [Google Scholar]
- 15.Nishida N, Kano Y, Koseki J, Konno M, Kawamoto K, Doki Y, Mori M, Ishii H. KDM5B plays a central role in esophageal cancer progression. Cancer Res 2015; 75: 102–2. [Google Scholar]
- 16.Sun L-L, Sun X-X, Xu X-E, Zhu M-X, Wu Z-Y, Shen J-H, Wu J-Y, Huang Q, Li E-M, Xu L-Y. Overexpression of Jumonji AT-rich interactive domain 1B and PHD finger protein 2 is involved in the progression of esophageal squamous cell carcinoma. Acta Histochem 2013; 115: 56–62. [DOI] [PubMed] [Google Scholar]
- 17.Parkin DM, Bray F, Devesa S. Cancer burden in the year 2000. The global picture. EurJ Cancer 2001; 37: 4–66. [DOI] [PubMed] [Google Scholar]
- 18.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell 2005; 122: 6–7. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res 2008; 68: 26–33. [DOI] [PubMed] [Google Scholar]
- 20.Wu B-L, Xu L-Y, Du Z-P, Liao L-D, Zhang H-F, Huang Q, Fang G-Q, Li E-M. MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J Gastroenterol 2011; 17: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori Y, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Ogawa R, Katada T, Harata K, Tanaka T, Shiozaki M. MicroRNA-21 induces cell proliferation and invasion in esophageal squamous cell carcinoma. Mol Med Rep 2009; 2: 235–9. [DOI] [PubMed] [Google Scholar]
- 22.Ma W-j, Lv G-d, Tuersun A, Liu Q, Liu H, Zheng S-T, Huang C-G, Feng J-G, Wang X, Lin R-Y. Role of microRNA-21 and effect on PTEN in Kazakh’s esophageal squamous cell carcinoma. Mol Biol Rep 2011; 38: 3253–60. [DOI] [PubMed] [Google Scholar]
- 23.Ding D-P, Chen Z-L, Zhao X-H, Wang J-W, Sun J, Wang Z, Tan F-W, Tan X-G, Li B-Z, Zhou F. miR-29 c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 2011; 32: 1025–32. [DOI] [PubMed] [Google Scholar]
- 24.Matsushima K, Isomoto H, Yamaguchi N, Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S, Nagayasu T. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med 2011; 9: 30–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varier RA, Timmers HM. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 2011; 1815: 75–89. [DOI] [PubMed] [Google Scholar]
- 26.Wong P-P, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21cip (CDKN1A). Mol Cell Biol 2012; 32: 1633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enkhbaatar Z, Terashima M, Oktyabri D, Tange S, Ishimura A, Yano S, Suzuki T. KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle 2013; 12: 2100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wan X, Qiang W, Li T, Huang W, Huang S, Wu D, Li Y. MiR-29 a suppresses prostate cell proliferation and induces apoptosis via KDM5B protein regulation. Int J Clin Exp Med 2015; 8: 5329–39. [PMC free article] [PubMed] [Google Scholar]