Abstract
Alterations to the epigenetic landscape of Diffuse Large B Cell Lymphoma (DLBCL) play a fundamental role in deregulating genes involved in normal lymphocyte differentiation. To determine whether targeted epigenetic therapy could reverse these pathogenic chromatin changes and suppress the expression of a lymphoma oncogene, we focused on BCL6, a transcriptional repressor whose aberrant expression is tightly linked to DLBCL proliferation and survival. We fused zinc-finger domains (ZF) specific for regulatory regions in the BCL6 locus to a repressive epigenetic modifier, the Kruppel-associated box repressor (KRAB). Distinct ZF-KRAB fusions repressed the local chromatin landscape, suppressed BCL6 expression, significantly impaired DLBCL growth and caused widespread cell death in a BCL6-dependent manner. Importantly, expression of ectopic BCL6 protein rescued ZF-KRAB-induced cell death, demonstrating the modifiers’ specificity. We show that sequence-specific epigenetic modifiers can alter oncogene expression and induce apoptosis in cancer cells, underscoring their potential for future development as targeted epigenetic protein therapies.
Keywords: Lymphoma and Hodgkin disease, Transcription factor changes, Chemotherapeutic approaches
INTRODUCTION
A hallmark of most cancers is the derangement of normal epigenetic patterns of chromatin and DNA modification, which are crucial for appropriate regulation of gene expression 1–3. We have recently profiled the chromatin landscape of normal germinal center B lymphocytes and Non-Hodgkin B cell lymphomas (NHL) 4. Our integrative analysis of epigenome data from primary NHL B cells identified regulatory elements with altered epigenetic states, demonstrating hyper- or hypo-activity relative to their normal germinal center B cell counterparts. These altered elements regulate the expression of a cohort of genes that play critical roles in lymphocyte activation, proliferation, and differentiation, as well as cellular transformation. The findings from our study and others indicate that the widespread gene deregulation observed in cancer is mediated, in part, by epigenetic-based mechanisms that alter the activation state of key cis-acting regulatory elements (i.e., promoters and enhancers).
Identification of pathogenic regulatory elements has enormous potential for developing new NHL therapeutics because, unlike genetic mutations, epigenetic modifications are reversible. For example, it may be possible to target cancer-specific regulatory elements with proteins that modify their pathogenic chromatin state, thereby reversing the expression pattern of their linked genes. Oncogenes, especially transcription factors, are attractive, yet challenging targets for new cancer therapies. Sustained down-regulation of specific genes via approaches such as RNAi is difficult, particularly for highly expressed oncogenes. Non-specific perturbation of gene expression may be achieved with broad spectrum epigenetic therapies such as histone deacetylase inhibitors; however, their efficacy in treating B cell lymphoma has been limited, perhaps due to non-specific effects and toxicities 5. An approach that specifically targets the expression of a single aberrantly expressed transcription factor would avoid such off-target effects, while simultaneously disrupting downstream components of the pathogenic transcriptional program. To evaluate the potential of targeted epigenetic therapies, we have focused on a subtype of NHL, Diffuse Large B Cell Lymphoma (DLBCL), which has an incidence of approximately ~20,000 cases in the US each year (www.cancer.org). Although >50% of DLBCL patients are cured by conventional therapies, up to one-third of cases are refractory to current regimens or relapse after treatment, highlighting the need for new therapeutics 6,7.
The transcription factor, BCL6, has been implicated in the pathogenesis of DLBCL by its aberrant expression, mutation, or translocation in a majority of tumors 8,9. BCL6 is a member of the BTB/POZ family of transcription factors and a key regulator of B cell proliferation and activation in germinal centers (GCs). Indeed, BCL6-null mice fail to form GCs or produce high-affinity antibodies 10,11. BCL6 functions as a transcriptional repressor at target promoters by directly recruiting histone deacetylases (HDACs) or co-repressor complexes 8,9. Expression of BCL6 is tightly controlled during mature B cell differentiation. Importantly, down-regulation of BCL6 activity is necessary for differentiation of GC B cells into plasma and memory B cells 8.
Although essential for B cell differentiation in GCs, the functions of BCL6 in promoting survival, proliferation, and tolerance of DNA damage can be oncogenic, leading to the development and maintenance of lymphoma. A direct role for deregulated BCL6 expression in lymphomagenesis was initially demonstrated by the development of lymphoma in a mouse model carrying a rearranged BCL6 locus in B cells 12. More recently, transient expression of BCL6 in hematopoietic stem/progenitor cells was sufficient to cause aggressive B-cell tumors comparable to human ABC-like (Activated B Cell) DLBCL13. Importantly, persistent BCL6 activity is required for survival of some lymphoma subtypes, both in cell lines and DLBCL xenotransplant models 14. In human B cell lymphomas, deregulation in the form of persistent BCL6 expression is common 9 and many potential mechanisms have been identified, including chromosomal translocation 15 or amplification of chromosome 3 13,16, somatic hypermutation that disrupts autoregulatory repression 17,18,15,19, and altered epigenetic landscapes 20,21. Thus, while different mechanisms are causative in individual tumors, the common outcome is the persistent expression of BCL6 in DLBCL, which contributes to proliferation and survival of lymphoma cells.
As an oncogene whose aberrant expression plays a fundamental role in pathogenesis, BCL6 is an attractive therapeutic target. We observed that the chromatin landscape surrounding regulatory regions within the BCL6 locus are highly enriched in activating histone marks. We reasoned that sustained BCL6 repression could be achieved by selective targeting of repressive epigenetic modifiers to these loci. Therefore, we designed targeted epigenetic modifiers composed of zinc-finger (ZF) DNA binding domains specific for BCL6 cis-regulatory elements fused to a potent transcriptional repressor. Using these sequence-specific epigenetic tools, we have selectively repressed the chromatin landscape of BCL6, reversing its pathogenic expression and causing widespread death of lymphoma cells.
METHODS
(See Supplemental Information for complete Methods)
ZF-KRAB proteins
BCL6-targeted zinc finger nuclease (ZFN) constructs were synthesized by Sigma (St. Louis, MO). Two ZF domains contain six zinc-fingers that each recognize a unique 18 bp sequence (ZFp: a region ~50 bp upstream of BCL6’s TATA boxes, ZFe1: first exon of BCL6 (Table S1).
Lentiviral constructs
3xHA-NLS-ZF-KRAB was cloned into a tet-inducible lentiviral vector (a gift from Barry Sleckman) that co-expresses reverse transcriptional transactivator rtTA2S-M2-IRES-Thy1.1 driven by ubiquitin C promoter.
Chromatin immunoprecipitation (ChIP)-Seq and -qPCR
ChIP was performed essentially as described previously 4.
Statistical analysis
Comparisons between doxycycline treated and untreated cells or rescue clones and parental clone were performed using T-test (GraphPad Prism software, La Jolla, CA).
RESULTS
BCL6 repression by targeted ZF-KRAB proteins
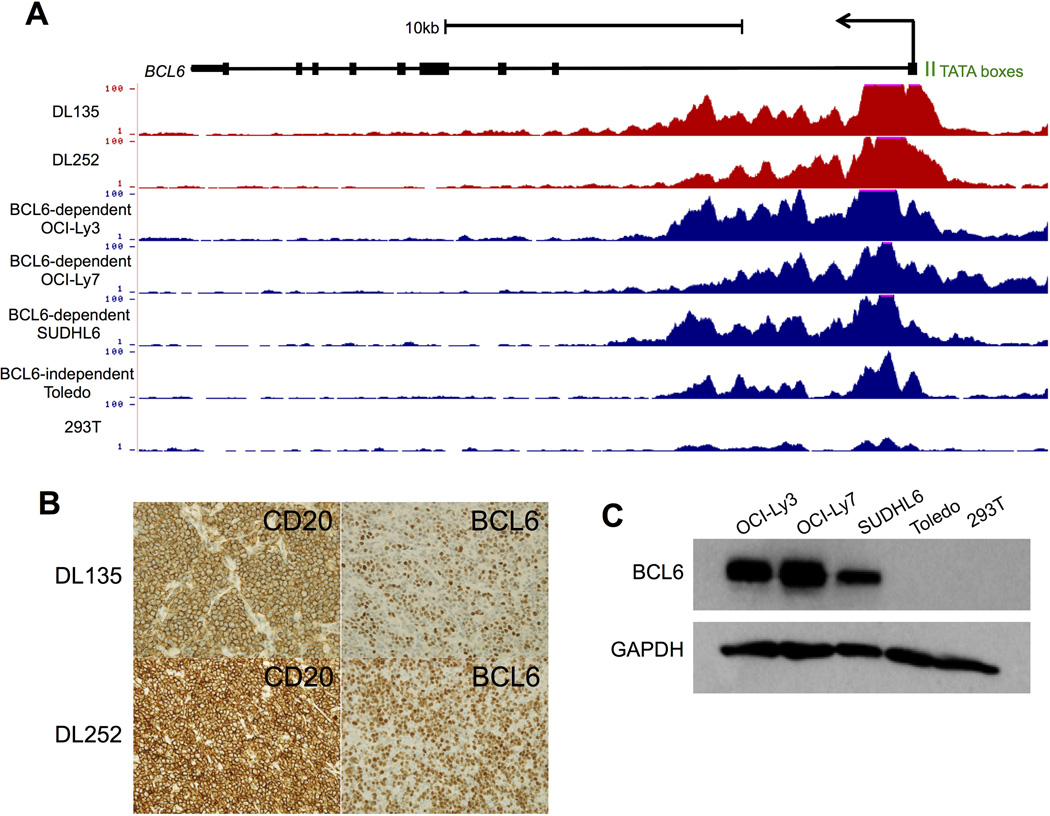
BCL6 is aberrantly expressed in a majority of DLBCL tumors 9, suggesting that the chromatin landscape surrounding the gene’s regulatory elements may also be altered. Active chromatin marks, including acetylation of histone H3 (H3ac), at promoters and gene bodies are required for ongoing transcription 22. Therefore, we performed chromatin immunoprecipitation for H3K9ac followed by high-throughput sequencing (ChIP-seq) 4 in primary DLBCL samples, as well as DLBCL and other cell lines. A region extending from the promoter TATA boxes to the first intron of BCL6 is highly enriched in H3ac in primary DLBCL samples and BCL6-dependent cell lines (OCI-Ly3, OCI-Ly7, SUDLH6) (Fig. 1A). Acetylation levels are considerably lower in BCL6-independent Toledo cells and in 293T cells, a non-lymphoid kidney epithelial line. Consistent with these epigenetic findings, BCL6 protein is highly expressed in primary DLBCL samples and BCL6-dependent lymphoma cell lines (Fig. 1B-C).
Figure 1. High levels of H3 acetylation at the BCL6 gene locus are associated with high levels of protein expression.
A) UCSC Genome Browser views of H3ac ChIP-seq data from DLBCL primary samples, DLBCL cell lines, and HEK293T cells illustrating the levels of acetylation located at the promoter and first exon/intron of the BCL6 locus. Data are presented as the number of reads per million mapped reads. B) Representative images for DL cases 135 and 252 (shown in A) with membranous CD20 staining (left, x400) and nuclear BCL6 staining (right, x400). C) Western blots for BCL6 or GAPDH (loading control) in cell lysates from cell lines (shown in A).
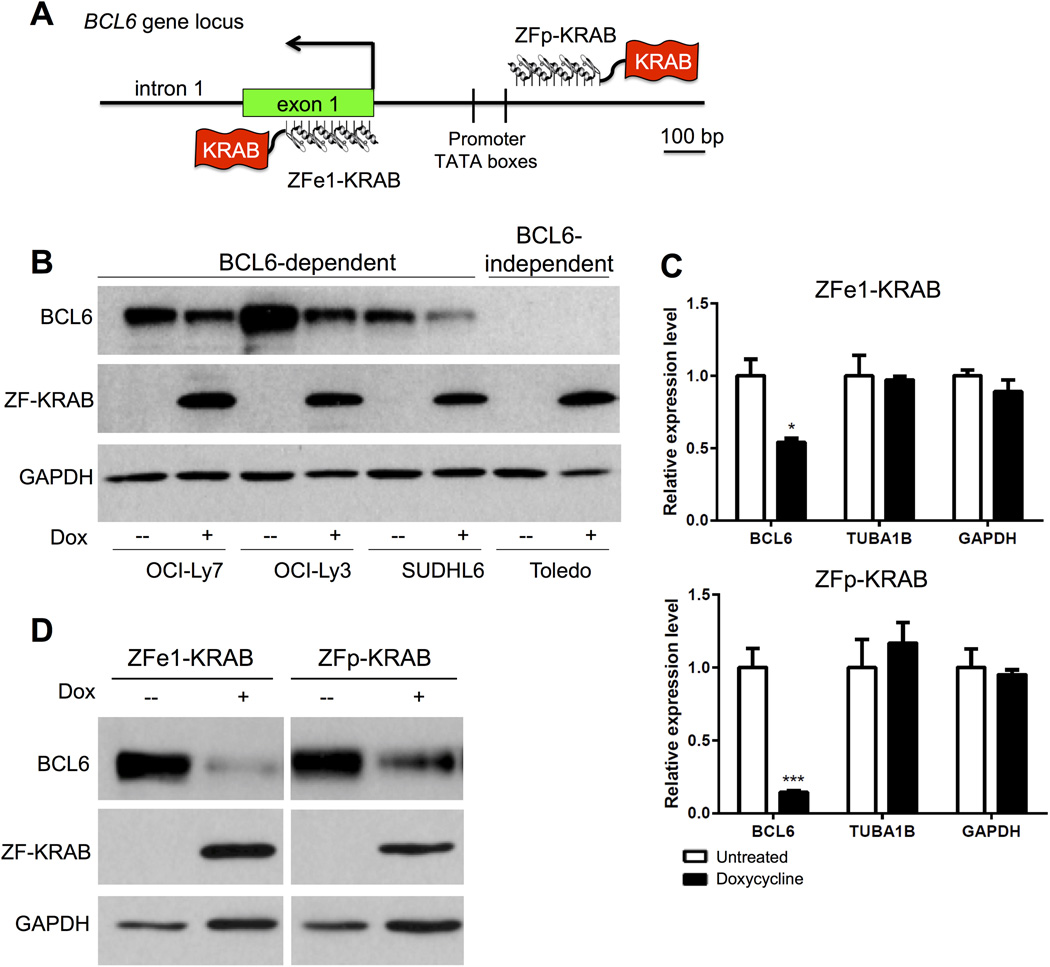
Based on these observations, we predicted that targeting of a repressive epigenetic modifier to a region near the promoter of BCL6 would result in substantial down-regulation of BCL6 transcripts 23,24. We selected a 1kb region in the BCL6 locus with high levels of H3ac, spanning the promoter TATA boxes and first untranslated exon, for the design of sequence-specific ZFs. Several ZF nucleases (ZFN) were commercially constructed (Sigma-Aldrich), each of which contained 4–6 zinc fingers fused to one of two variant FokI nuclease domains that require heterodimerization for DNA cleavage 25,26. Two of these ZFNs targeted adjacent sites in exon 1 and therefore were predicted to comprise an active heterodimer pair (Fig. S1A). Indeed, when transfected together, the ZFN pair demonstrated on-target DNA cleavage at exon 1, while neither ZFN cleaved DNA when transfected alone (Fig. S1B). We predicted that transcriptional repressors targeted to these locations would be effective, given recent reports that promoter-proximal targeting of artificial TFs provides maximum transcriptional activation 27. Unlike the obligate heterodimer design of the ZFNs, our approach required only one ZF repressor protein for activity. Therefore, we chose two ZFNs, each with six fingers, from pairs with robust cutting and with the fewest predicted off-target binding sites as determined by the Sangamo algorithm 26. One of the selected ZFs targets sequences ~50bp upstream of the TATA boxes in the BCL6 promoter (ZFp) and the other targets the 3’ portion of exon 1 (ZFe1) (Fig. 2A).
Figure 2. BCL6 expression is repressed by ZF-KRAB.
A) Cartoon representing the binding of ZF-KRAB proteins to targets within the promoter of BCL6 (ZFp-KRAB) or the first exon (ZFe1-KRAB). Relative positions of the TATA boxes within the promoter are shown. B) Western blots for BCL6, HA-tagged ZF-KRAB proteins, or GAPDH (loading control) in BCL6-dependent or -independent cells after culture for 3 days with (+) or without doxycycline (-) to induce expression of ZF-KRAB fusion proteins targeted to BCL6 exon 1. Data are representative of three independent experiments. C) Relative mRNA levels of BCL6, TUBA1B and GAPDH measured by qRT-PCR in lysates from OCI-Ly7 cells harboring Tet-inducible ZF constructs cultured for three days with or without doxycycline to induce expression of ZF-KRAB fusion proteins targeted to BCL6 exon 1 (ZFe1) or promoter (ZFp). Results represent the mean ± SD of three independent experiments. * p < 0.05; *** p < 0.001 D) Western blots demonstrate expression of BCL6, FLAG-tagged ZF-KRAB proteins, and GAPDH (loading control) in OCI-Ly7 cells harboring Tet-inducible ZF constructs cultured for three days with (+) or without doxycycline (-). Data are representative of five independent experiments.
For the creation of ZF chromatin modifier fusion proteins, we replaced the nuclease domain with a transcriptional repressor, the Kruppel-associated box (KRAB) domain, which recruits several repressive histone modifiers, including histone deacetylases, which remove acetyl groups on histone H3 24,28. The FLAG-tagged ZF domain fused to KRAB was then cloned into a Tet-inducible lentiviral vector and transduced into several BCL6-dependent or –independent DLBCL cell lines, OCI-Ly7, OCI-Ly3, SUDHL6 (dependent), and Toledo (independent). BCL6 dependence or independence for several B cell lymphoma cell lines has been previously determined based on cell survival and proliferation when BCL6 has been depleted or inhibited 14,29–32 After enrichment by flow sorting for co-expressed Thy1.1, ZF-KRAB expression was induced with doxycycline. Each of the BCL6-dependent cell lines demonstrated substantially decreased BCL6 protein expression upon induction of the ZF-KRAB targeted modifier (Fig. 2B and S1C). To establish a cell line with consistent and stable levels of ZF-KRAB expression in all cells, the Tet-inducible ZF-KRAB construct was introduced stably into the OCI-Ly7 lymphoma cell line. Isolated subclones were screened for ZF-KRAB expression following induction with doxycycline. Significant suppression by ZF-KRAB proteins targeting either the promoter (ZFp-KRAB) or first exon (ZFe1-KRAB) of BCL6 was observed at the level of BCL6 mRNA, but expression of non-targeted genes TUBA1B and GAPDH was unaffected (Fig. 2C). BCL6 protein expression was also substantially repressed by both ZF-KRABs (Figs. 2D and S1C). In contrast to the impact of ZF-KRAB fusions, BCL6 levels were unaffected by expression of the ZF domain alone (ZFe1 and ZFp, Fig. S1D). Together, our results demonstrate that ZF-KRAB proteins targeting the BCL6 promoter or first untranslated exon directly inhibit expression of this oncoprotein.
ZF-KRAB proteins bind target sites in BCL6 and revise its epigenetic landscape
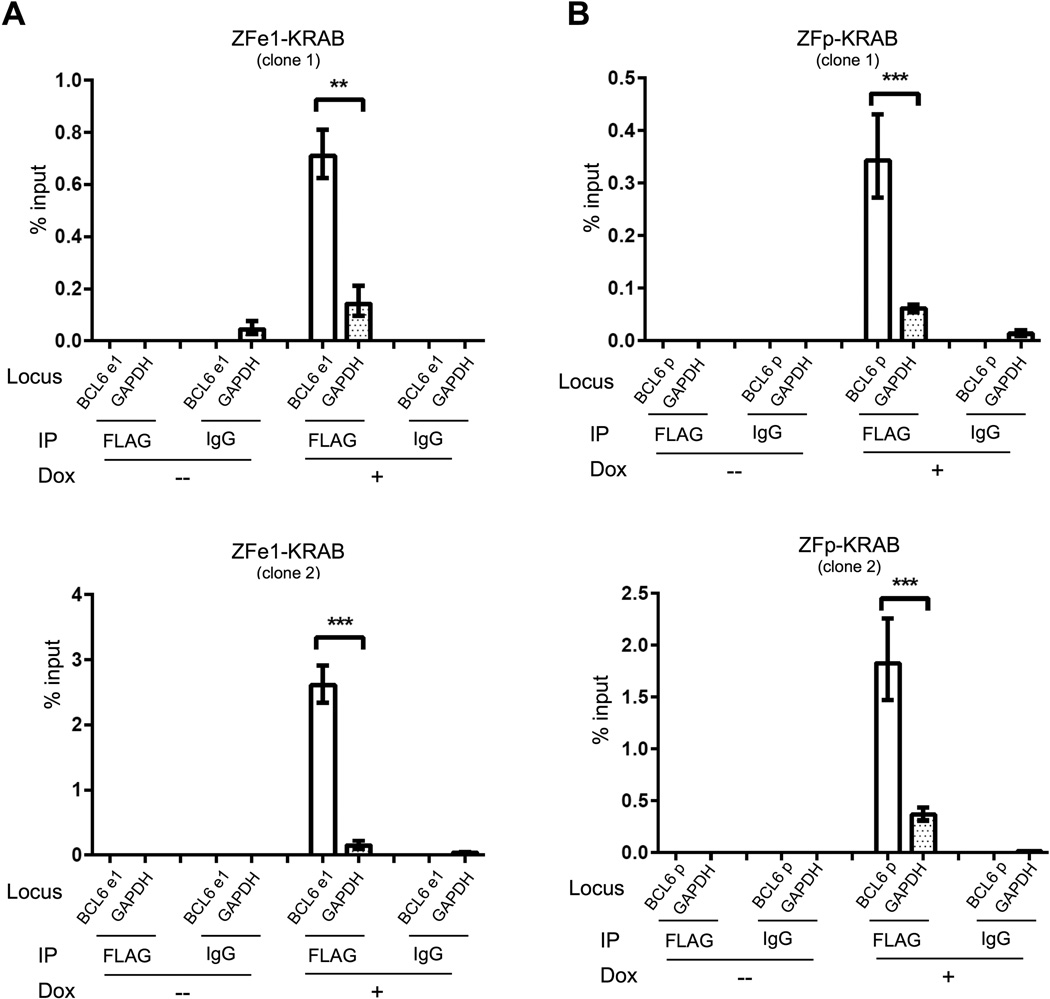
We next sought to verify that BCL6 suppression was due to direct binding of ZF-KRAB proteins and subsequent repressive histone modification of the target BCL6 locus. To determine whether the ZF-KRAB proteins bind specifically to their target regions in the BCL6 locus, we performed chromatin immunoprecipitation (ChIP) using an anti-FLAG antibody on cross-linked chromatin from OCI-Ly7 cells after induction with doxycycline. Both ZFe1- and ZFp-KRAB fusions demonstrate substantially enriched binding to their target regions compared to control regions (GAPDH and TUBA1B, Figs. 3 and S2).
Figure 3. ZF-KRAB proteins bind preferentially to target sites in the BCL6 gene locus.
A) Binding of FLAG-tagged ZFe1-fusion protein to exon 1 of BCL6 or to a control locus, GAPDH, measured by anti-FLAG ChIP-qPCR in OCI-Ly7 cells harboring the Tet-inducible ZFe1-KRAB construct. Cells were cultured for three days with (+) or without (−) doxycycline. IgG ChIP represents the negative control. Results are representative of three independent experiments. B) Binding of FLAG-tagged ZFp-fusion protein to the promoter region of BCL6 or to a control locus, GAPDH, measured by anti-FLAG ChIP-qPCR in OCI-Ly7 cells harboring the Tet-inducible ZFp-KRAB construct. Cells were cultured and treated as in A. IgG ChIP represents the negative control. Results are representative of three independent experiments.
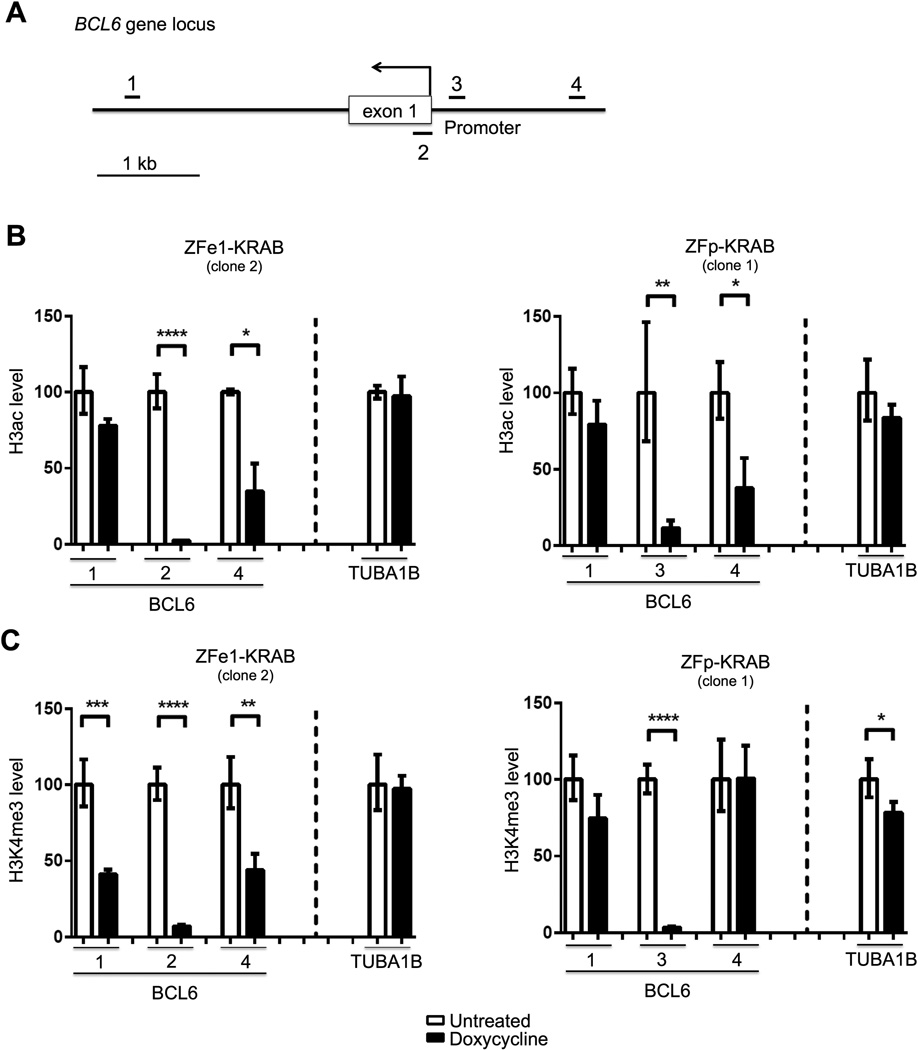
KRAB domains recruit histone modifiers, including deacetylases (HDACs), which cause chromatin repression by removing activating histone marks (e.g. H3ac) 23,28. We therefore examined H3ac levels at ZF-KRAB-targeted regions of BCL6 using ChIP assays, which were normalized for total histone H3 to adjust for nucleosome density. To determine the extent of ZF-KRAB-mediated alteration to the chromatin landscape, we evaluated the ZF target loci as well as regions 3 kb downstream and 1.5 kb upstream of the BCL6 transcription start site (TSS) (Fig. 4A). Indeed, H3ac is nearly depleted at each BCL6 target site (numbered 2 and 3) after induction of either ZF-KRAB protein, while levels at a non-targeted locus are relatively preserved in comparison (Figs. 4B and S3B). The repressive effect of ZF-KRAB binding is also detectable at regions a short distance up- and downstream of the BCL6 targets (sites 1 and 4) as demonstrated by H3ac levels that are decreased, though not to the same degree as at the ZF binding sites. The activating histone mark H3K4me3 is enriched at active promoters and has been shown to be depleted at loci bound by KRAB fusion proteins 33. Therefore, we performed ChIP for H3K4me3 and normalized to total H3 as above. We observe a similar diminution of H3K4me3 levels at BCL6 target loci in the presence of ZF-KRAB proteins, whereas levels at the control locus are relatively maintained in comparison (Figs. 4C and S3C). Similar to H3ac, H3K4me3 levels in regions up- and down-stream of the ZF-KRAB binding sites are somewhat less attenuated than the ZF targets. Collectively, these results demonstrate that ZF-KRAB proteins bind specifically to their target sites in BCL6, causing repressive changes to the epigenetic landscape by reducing the levels of active histone marks. As expected, these chromatin changes extended a limited distance beyond the target ZF binding sites, demonstrating the ability of KRAB-recruited histone modifiers to cause localized epigenetic repression 34. Importantly, these repressive chromatin effects correlated with diminished expression of BCL6.
Figure 4. ZF-KRAB proteins cause repressive chromatin changes at the BCL6 locus.
A) Schematic of the BCL6 locus, including ChIP-qPCR regions (labeled 1 – 4) and binding sites for ZFe1- (exon 1, #2) and ZFp-KRAB (promoter, #3) proteins. B) H3ac ChIP assays in OCI-Ly7 cells cultured for three days with or without doxycycline to induce expression of the ZFe1- or ZFp-KRAB fusion protein. Associated DNA was analyzed via qPCR using primers spanning the following regions: ZFe1-KRAB binding site in exon 1 of BCL6 (2), ZFp-KRAB binding site in the promoter of BCL6 (3), sites 3kb downstream (1) or 1.5kb upstream (4) of the BCL6 TSS, and a control site at the TUBA1B gene. H3ac was normalized to total histone H3. Representative of three independent experiments. C) H3K4me3 ChIP assays in OCI-Ly7 cells cultured and treated as in B. Associated DNA was analyzed via qPCR and normalized to total histone H3 as in B. Representative of three independent experiments.
ZF-KRAB proteins induce apoptosis in BCL6-dependent lymphoma cells
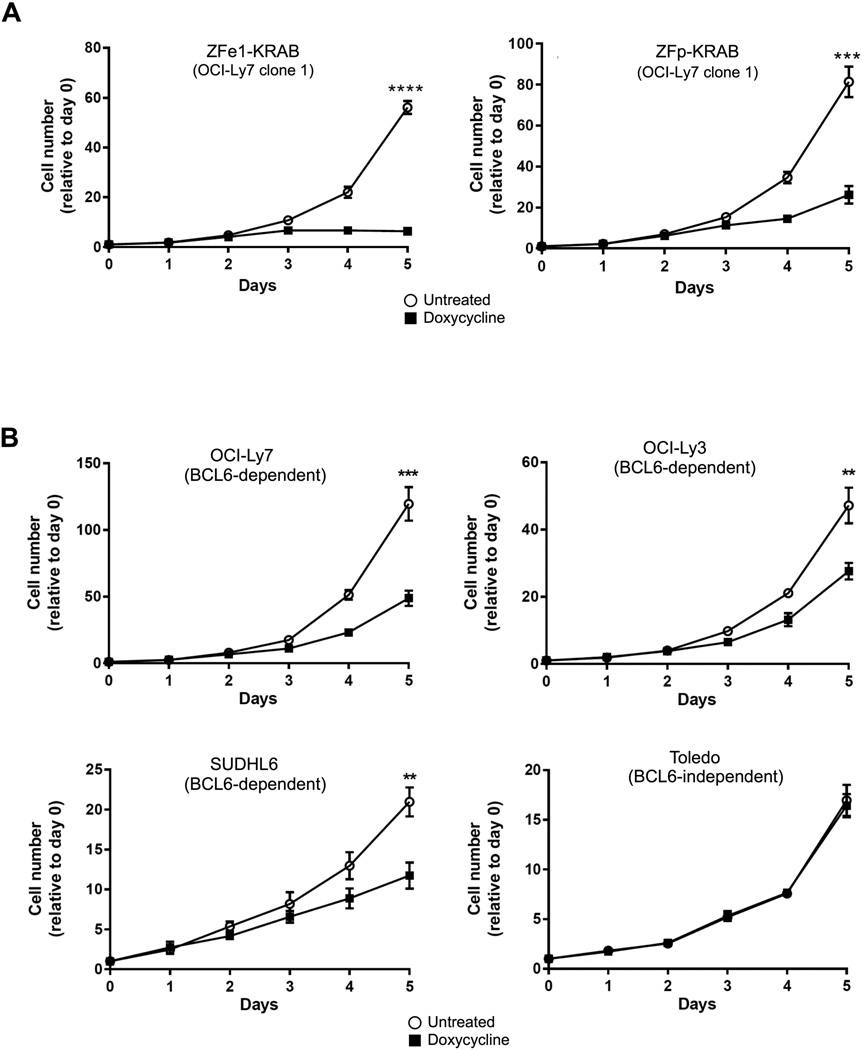
Persistently elevated BCL6 expression is a common feature of DLBCL, and has been linked to survival and proliferation of malignant cells. Moreover, BCL6 activity in a subset of DLBCL cell lines, including OCI-Ly7, OCI-Ly3, and SUDHL6, is essential for survival; knockdown or inhibition suppresses proliferation and causes cell death. These cells are therefore termed BCL6-dependent. Other DLBCL cell lines, such as Toledo, do not require BCL6 for survival, are resistant to knockdown and inhibitor treatment, and are therefore referred to as BCL6-independent14. Thus, we assessed the growth and survival of BCL6-dependent and -independent cells after induction of ZF-KRAB proteins over a five-day time course. Expression of either ZFp- or ZFe1-KRAB proteins in stably expressing clones or lentivirally transduced BCL6-dependent DLBCL cells resulted in significantly diminished cell numbers compared to uninduced cells, while cell growth was unaffected in BCL6-independent cells (Figs. 5A-B, S4A). Stable expression of the ZFe1-KRAB fusion protein in Raji, a Burkitt’s lymphoma cell line that expresses BCL6 and has been shown to be BCL6-dependent 29, demonstrated a substantial suppression of BCL6 protein and a negative effect on cell growth (Figs. S4B and C). In contrast, cell growth was unaffected by ZF-KRAB expression in a non-lymphoma cancer cell line, the osteosarcoma line U2OS (Fig. S4D and E). Collectively, these results suggest that ZF-KRAB-mediated suppression of BCL6 significantly decreases growth in cells that are dependent upon BCL6 for proliferation and survival. In contrast, cells that are not reliant on BCL6 demonstrated no change in cell growth with ZF-KRAB expression, suggesting that ZF-KRAB has a targeted anti-proliferative effect on cells that require BCL6 for growth and survival.
Figure 5. ZF-KRAB proteins reduce the growth of BCL6-dependent lymphoma cells.
A) Cell growth of Tet-inducible ZFe1-KRAB or ZFp-KRAB expressing OCI-Ly7 cells (BCL6-dependent) cultured with or without doxycycline was measured by Trypan blue exclusion. Results represent the mean ± SD of three independent experiments. **** p < 0.0001 B) Cell growth of Tet-inducible ZFe1-KRAB expressing BCL6-dependent or -independent cells measured as in A.
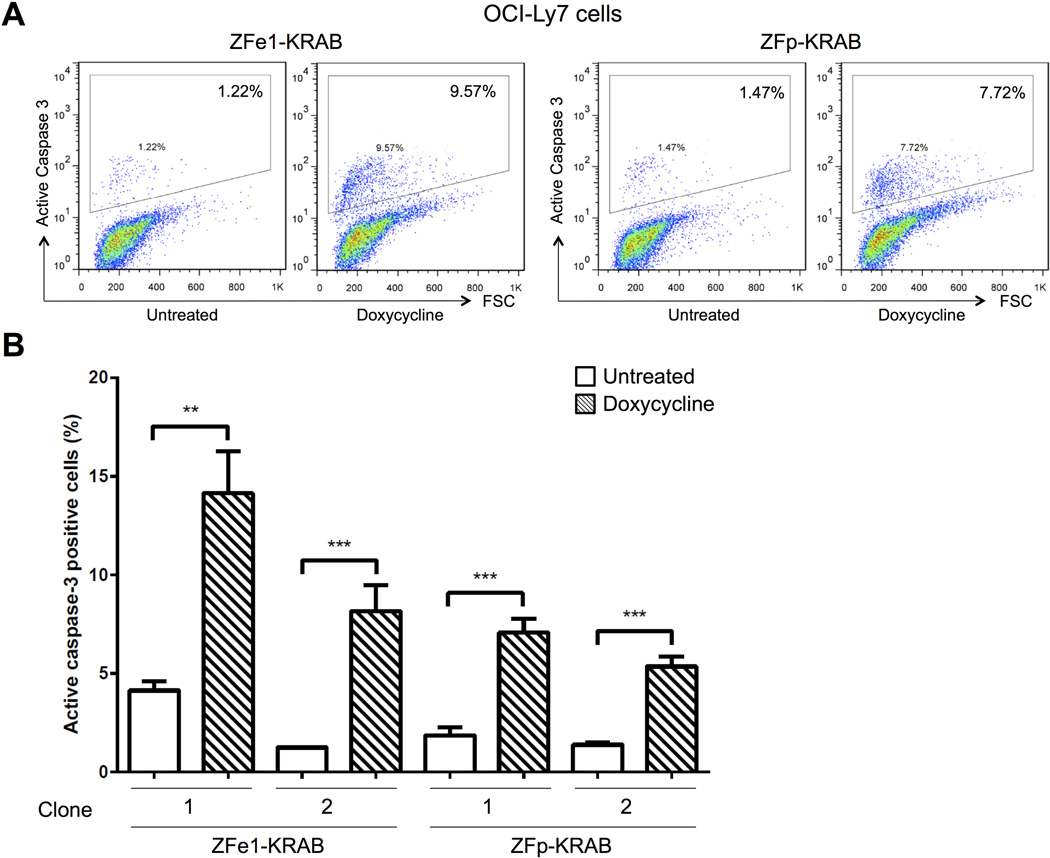
Decreased growth in the ZF-KRAB expressing cells could be due to reduced proliferation or to enhanced cell death, or a combination of both. To differentiate between these two mechanisms, we assessed apoptosis by measuring the active form of caspase-3 in BCL6-dependent OCI-Ly7 cells. The proportion of active caspase-3 positive cells is three to five-fold higher in cells expressing either ZFe1- or ZFp-KRAB, compared to uninduced cells (Fig. 6A-B). Together, these data indicate that the ZF-KRAB modifiers suppress expression of the pro-proliferative BCL6 protein, reducing the growth of BCL6-dependent lymphoma cells via reduced proliferation and apoptosis. These results suggest a specific, targeted activity for the ZF chromatin modifier fusions in B cell lymphomas.
Figure 6. ZF-KRAB proteins induce apoptosis in BCL6-dependent lymphoma cells.
A) Number of OCI-Ly7 cells with active Caspase-3 staining measured by flow cytometry. OCI-Ly7 cells harboring Tet-inducible ZFe1- or ZFp-KRAB were cultured for four days with or without doxycycline. Representative of three independent experiments. B) Percentage of cells with active Caspase-3 staining measured by flow cytometry. OCI-Ly7 cells (BCL6-dependent) harboring Tet-inducible ZFe1-KRAB or ZFp-KRAB were cultured and treated as in (A). Results represent the mean ± SD of three independent experiments. ** p < 0.01 *** p < 0.001**** p < 0.0001
Ectopic BCL6 expression rescues ZF-KRAB-induced cell death
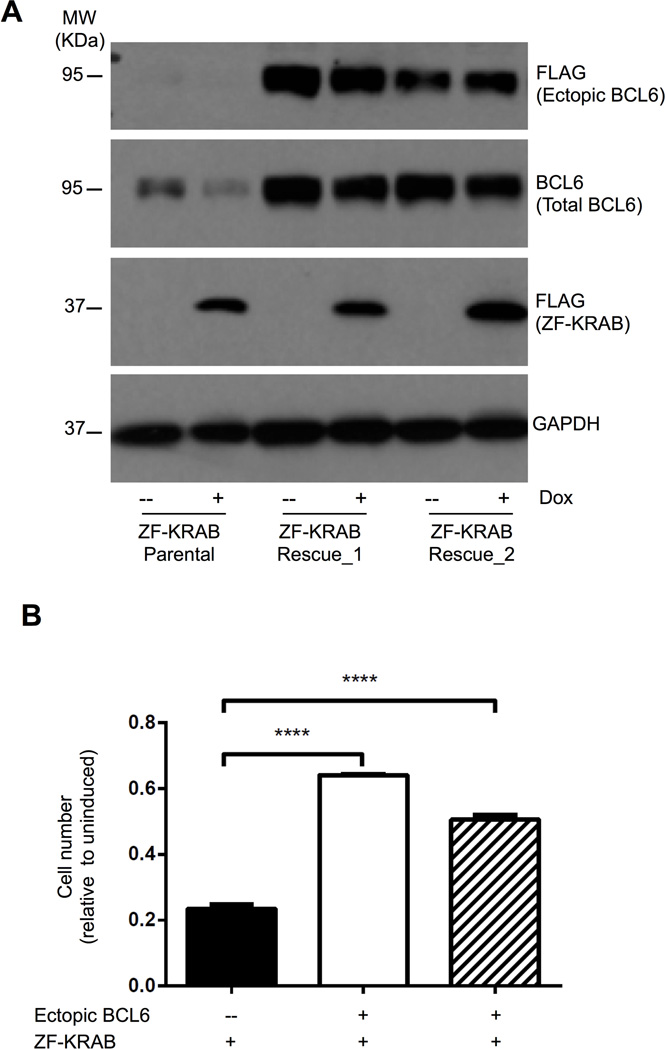
The potent cytotoxic effects of ZF-KRAB in BCL6-dependent cells likely are direct consequences of its specific targeting and repression of the BCL6 locus. However, the data presented thus far do not preclude off-target effects by this chromatin modifier, such as the attenuation of other genes via direct or indirect mechanisms. To distinguish these possibilities, we expressed BCL6 ectopically in ZF-KRAB-expressing OCI-Ly7 cells. We predicted that ectopic BCL6 would rescue cell viability if the dominant impact of ZF-KRAB were suppression of BCL6 in these dependent cells. Importantly, the BCL6 expression construct lacked binding sites for either ZFe1 or ZFp domains, which was possible because translation of full length BCL6 protein begins with exon 3. As shown in Figure 7A, two independent subclones harboring these vectors express ectopic flag-tagged BCL6 in the presence or absence of ZF-KRAB protein, while endogenous BCL6 is nearly abolished in the ZF-KRAB parental clone. Importantly, enforced expression of BCL6 was sufficient to rescue viability in cells expressing ZF-KRAB, which exhibit a two- to three-fold increase in cell growth compared to the parental cells (Fig. 7B). These results demonstrate that the negative impact of ZF-KRAB fusions on cell growth and survival is specific, occurring as a direct consequence of repression of the pro-proliferative, pro-survival BCL6 protein.
Figure 7. Ectopic BCL6 expression rescues cell death induced by ZF-KRAB proteins.
A) Western blots for FLAG-tagged BCL6 (ectopic), total BCL6, FLAG-tagged ZFe1-KRAB, or GAPDH (loading control) in OCI-Ly7 cells after three days in culture with (+) or without (-) doxycycline. The parental OCI-Ly7 cells contain Tet-inducible ZFe1-KRAB; two subclones also express ectopic FLAG-tagged BCL6 protein. Representative of three independent experiments. B) Cell growth of parental OCI-Ly7 cells and two subclones expressing FLAG-tagged ectopic BCL6 measured by Trypan blue exclusion. Cells were cultured for five days with doxycycline to induce expression of ZFe1-KRAB fusion protein. Cell number is presented relative to uninduced cells for each clone or subclone. Results represent the mean ± SD of three independent experiments. **** p < 0.0001
DISCUSSION
In the study presented here, we set out to demonstrate that a targeted epigenetic modifier can specifically reverse the aberrant chromatin features that drive pathogenic expression of a lymphoma oncogene. To accomplish this goal, we created sequence-specific chromatin modifiers that fused zinc-finger domains (ZF) specific for BCL6 regulatory regions with the repressive KRAB domain. We showed that these ZF-KRAB fusion proteins directly altered the epigenetic landscape of cis-regulatory elements in the BCL6 locus, causing repressive chromatin changes and suppressing expression of BCL6. In BCL6-dependent lymphoma cells, expression of the ZF-KRAB proteins decreased cell growth and caused apoptosis. Importantly, the specificity of the ZF-KRAB fusions was demonstrated by rescue of the apoptotic phenotype with ectopic BCL6 protein.
The epigenetic landscape of normal B lymphocytes is disrupted at key regulatory regions in NHL, leading to alterations in the binding of cognate transcription factors and deregulated expression of critical genes involved in lymphocyte differentiation and activation 4. The therapeutic potential of these findings is further underscored by our present study, in which the pathogenic activity of a regulatory region for a lymphoma oncogene was restored to its normal state by targeting of a sequence-specific epigenetic modifier. Although frequently involved in carcinogenesis, the dysregulation of transcription factors is difficult to target therapeutically, since there is no enzyme activity to inhibit and few options exist to control the expression of individual genes. Direct targeting of repressive chromatin modifications to a gene locus, as shown here for BCL6, circumvents these obstacles and provides sustained suppression of transcription.
In this regard, the first criterion for designing a targeted repressor is to ensure its specificity for the locus of interest, avoiding off-target effects in other genes. Three different classes of proteins have been employed as sequence-specific DNA binding domains/molecules for genome editing or for transcriptional control: zinc-fingers, TALEs, and CRISPR/Cas9 28,35–37. Each has a range of specificities, and thus off-target effects, which can be partially controlled by designing the DNA binding domain/molecule for optimal specificity (for ZF proteins ~ 6 zinc fingers) 36. Here we have demonstrated the specificity of two BCL6-targeted ZF-KRAB proteins, each of which contain 6-finger DNA binding domains, in several ways. First, the binding of ZF-KRAB fusions was substantially enriched at their target sites, the BCL6 promoter or its first exon, as compared to multiple control loci. In addition, we detected significant decreases in active histone marks, H3ac and H3K4Me3, at the targeted BCL6 loci compared to control loci. Most importantly, ZF-KRAB proteins significantly suppressed the levels of BCL6 transcript and protein, having no impact on the expression of control genes. Thus, we have demonstrated specific abrogation of target gene expression with sparing of control genes using this sequence-specific epigenetic modifier approach. The robust effects of the promoter-targeted ZF-KRAB highlight the potential of this approach for targeting the regulatory regions of other oncogenes or tumor suppressors 27.
Overexpression of BCL6 is critical for the proliferation and survival of malignant germinal center B cells, and thus presented an ideal target for focused epigenetic repression. In this regard, transcriptional repression by custom DNA binding domains fused to epigenetic modifiers has been demonstrated for other genes including ERBB2, SOX2 and CXCR4 23,28,37,38. Although these studies and others demonstrate transcriptional control with targeted modifiers, most fail to show a cellular phenotype, such as an effect on signaling, differentiation, or cell death. In contrast, our studies demonstrate that ZF-KRAB-mediated repression of BCL6 essentially halted cell growth in BCL6-dependent lymphoma cells. Importantly, this inhibition of cell growth was accompanied by a significant increase in apoptosis in BCL6-dependent DLBCL cells. Most importantly, our results are the first, to our knowledge, to demonstrate the functional specificity of targeted modifiers by incorporating a rescue experiment. Specifically, ectopic BCL6 protein reversed the growth defects and cell death caused by BCL6-targeted ZF-KRAB fusions. Our results solidify the specificity of these BCL6-targeted ZF-KRAB proteins, underscoring the potential of this approach as a novel cancer therapeutic.
Although effective immuno- and chemotherapies exist for B cell lymphomas, many are refractory to initial therapy or relapse within a short period, and these malignancies often prove fatal despite aggressive treatment. Therefore, novel treatment modalities that are targeted and tolerable are urgently needed. The ZF-KRAB fusions presented here, which target BCL6 regulatory regions and caused potent and specific abrogation of BCL6 expression and consequent lymphoma cell death, present a potential new treatment avenue. Although the particular ZF domain tested here would not target lymphomas with BCL6 translocations, ZF domains that would bind to those altered loci could be easily designed. Delivery of ZF fusions could not be achieved in the same manner in patients. However, recent data suggests that zinc finger nuclease fusions, delivered as intact proteins, can penetrate cell membranes, transit to the nucleus, and effectively and specifically cleave target genomic loci 39. Advances in in vivo targeting, including the tethering of tumor-specific ligands to nanocarrier systems, provide a path toward the effective deployment of sequence-specific epigenetic modifiers for lymphoma therapy. Collectively, our results support the potential of targeted epigenetic therapies as a precision medicine approach to reverse the pathogenic expression of transcription factors that mediate many cancers.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA156690 and CA188286 (J.E.P. and E.M.O.), UL1 TR000448 (WU-ICTS, from NCATS of the NIH), and CA91842 (Siteman Cancer Center). We thank B. Sleckman and O. Koues for helpful comments on the experiments and manuscript.
Footnotes
Authorship Contributions
H.L., E.M.O., and J.E.P. designed the research. H.L., J.A.S., and Y.L. performed the research. H.L. and J.E.P analyzed and interpreted the data. J.E.P. wrote the manuscript with help from E.M.O.
Disclosure of Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Stergachis AB, Neph S, Reynolds A, et al. Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell. 2013;154(4):888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawkins RD, Larjo A, Tripathi SK, et al. Global Chromatin State Analysis Reveals Lineage-Specific Enhancers during the Initiation of Human T helper 1 and T helper 2 Cell Polarization. Immunity. 2013;38(6):1271–1284. doi: 10.1016/j.immuni.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koues OI, Kowalewski RA, Chang L-W, et al. Enhancer Sequence Variants and Transcription Factor Deregulation Synergize to Construct Pathogenic Regulatory Circuits in B Cell Lymphoma. Immunity. 2015;42(1):186–198. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 6.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat. Rev. Clin. Oncol. 2014;11(1):12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2014;125(1):22–32. doi: 10.1182/blood-2014-05-577189. [DOI] [PubMed] [Google Scholar]
- 8.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 2012;247(1):172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 9.Hatzi K, Melnick A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014;20(6):343–352. doi: 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dent aL. Control of Inflammation, Cytokine Expression, and Germinal Center Formation by BCL-6. Science (80-. ) 1997;276(5312):589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 11.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997;16(2):161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 12.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Green MR, Vicente-Dueñas C, Romero-Camarero I, et al. Transient expression of Bcl6 is sufficient for oncogenic function and induction of mature B-cell lymphoma. Nat. Commun. 2014;5:3904. doi: 10.1038/ncomms4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerchietti LC, Ghetu AF, Zhu X, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17(4):400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye BH, Lista F, Lo Coco F, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262(5134):747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 16.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc. Natl. Acad. SciUS.A. 2002;99(23):15018–15023. doi: 10.1073/pnas.232581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasqualucci L, Migliazza A, Basso K, et al. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101(8):2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 19.Saito M, Gao J, Basso K, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12(3):280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Koues OI, Kowalewski RA, Chang L-W, et al. Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity. 2015;42(1):186–198. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai AY, Fatemi M, Dhasarathy A, et al. DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J. Exp. Med. 2010;207(9):1939–1950. doi: 10.1084/jem.20100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, Birney E, Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnenat L, Schwimmer LJ, Barbas CF. Drug-inducible and simultaneous regulation of endogenous genes by single-chain nuclear receptor-based zinc-finger transcription factor gene switches. Gene Ther. 2008;15(17):1223–1232. doi: 10.1038/gt.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osipovich O, Milley R, Meade A, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5(3):309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 25.Szczepek M, Brondani V, Büchel J, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25(7):786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 26.Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 27.Ji Q, Fischer AL, Brown CR, et al. Engineered zinc-finger transcription factors activate OCT4 (POU5F1), SOX2, KLF4, c-MYC (MYC) and miR302/367. Nucleic Acids Res. 2014;42(10):6158–6167. doi: 10.1093/nar/gku243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong L, Zhou R, Kuo Y-C, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo JM, Dell’Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 2004;10(12):1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 30.Polo JM, Juszczynski P, Monti S, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc. Natl. Acad. SciUS.A. 2007;104(9):3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerchietti LC, Yang SN, Shaknovich R, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113(15):3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerchietti LC, Lopes EC, Yang SN, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat. Med. 2009;15(12):1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupo A, Cesaro E, Montano G, et al. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genomics. 2013;14(4):268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meylan S, Groner AC, Ambrosini G, et al. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug Discov. 2003;2(5):361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 36.Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert LA, Larson MH, Morsut L, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falahi F, Huisman C, Kazemier HG, et al. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol. Cancer Res. 2013;11(9):1029–1039. doi: 10.1158/1541-7786.MCR-12-0567. [DOI] [PubMed] [Google Scholar]
- 39.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9(8):805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.