Abstract
Over the past three decades, the pathological classification of lymphoma has substantially improved. The early Rappaport classification included a handful of subtypes that did not reflect the cell of origin and, not surprisingly, resulted in diagnostic inaccuracies. The WHO currently classifies lymphoma into 30 major distinctive types. While this classification improved the accuracy and consistency of the histological diagnosis of lymphoma, it had little impact on advancing drug development or improving the cure rate of this disease. One reason for this lack of improvement is that recent developments in cancer genomics show these histopathological subtypes to be heterogeneous. Basing treatment decisions on histopathological subtypes is inefficient as it groups different underlying molecular characteristics into one category. Such a strategy exposes many patients to potentially toxic drugs without providing benefits. The recent approval of two new cancer drugs with companion diagnostics to allow selection and treatment of patients with melanoma and non-small-cell lung cancer has raised hope that a similar approach may also expedite successful drug development in lymphoma. We review the current status of biomarker development in lymphoma, and discuss novel biomarker-directed clinical trial designs for lymphoma.
Introduction
The treatment of patients with non-Hodgkin lymphoma has essentially remained the same for more than three decades, with the exception of the inclusion of monoclonal anti-CD20 agents in combination strategies.1 Front-line regimens continue to be predominantly based on cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), or on variations of this regimen.2 Salvage regimens are predominantly platinum-based regimens, such as DHAP (dexamethasone, high-dose cytarabine, and cisplatin) or ICE (ifosfamide, carboplatin, and etoposide).3 Similarly, the treatment of Hodgkin lymphoma continues to be based on ABVD (doxorubicin, bleomycin, vinblastine and dacarbazine), a regimen that was introduced in the 1970s.4 Over the past 30 years, however, there has been tremendous advances in the pathology, biology, and molecular characterization of lymphomas. For example, the histological classification of different types of lymphomas is now more precise using advanced diagnostic tools that incorporate morphology, immunophenotyping, and genetics analysis.5,6
The first widely used lymphoma classification was published by Rappaport in 1966, which broadly grouped lymphoma into nodular and diffuse entities. Today, the WHO classification of lymphoma includes morphological, genetic, and phenotypical features to describe more than 30 unique entities.5,7 Furthermore, a literature search in PubMed shows that thousands of papers were recently published on lymphoma genetics, gene-expression profiling and oncogenes associated with the disease. However, these recent discoveries have not yet translated into significant changes in treating lymphoma.
The discovery of a variety of tumour-suppressor genes and oncogenes led to the identification of numerous potential therapeutic targets, and to the development of more than 800 compounds that are being examined in clinical trials or in preclinical experiments for the treatment of cancer, including lymphoma.8 However, based on past experience, the development of the vast majority of these compounds is unlikely to succeed owing to the lack of anticancer benefit, excessive toxicity, poor understanding of the optimal dose and schedule, limited understanding of the patient subsets that benefit, or a combination of these reasons.9 Furthermore, although the number of studies enrolling lymphoma patients has increased, many of them lack focus, do not advance the field, and compete for a relatively small pool of eligible patients.1
A common outcome of clinical trials that test novel agents in unselected populations is a modest clinical activity with a reasonable safety profile.10–13 Such outcomes are not sufficient for securing approval by regulatory agencies, such as the FDA, or for affecting clinical practice. Moreover, an increasing number of costly phase III studies fail to meet their end points because these trials continue to use traditional randomization designs, and frequently compare empirical combination regimens with standard regimens in unselected patient populations.14 It is not surprising that only a handful of drugs have been approved in recent years by the FDA for the treatment of lymphoma; moreover, none of these newly approved drugs was developed based on a modern understanding of lymphoma biology.
These failures underline the importance of developing novel strategies that can translate the recent molecular and genetic discoveries into successful treatment regimens. With the recent success of new targeted agents for biomarker-selected patients with melanoma and non-small-cell lung cancer, the search for predictive biomarkers that may guide therapy for other cancers, including lymphoma, has become a focus in drug development. In this Review, we discuss how to incorporate biomarkers that may predict response to these agents, and how to use these biomarkers to develop rationally designed combination strategies that will help to produce higher response rates and durable remissions in patients with lymphoma.
Histology versus molecular pathway
Developments in cancer genetics and gene-expression profiling demonstrate that lymphoma histological subtypes are not as homogeneous as initially believed. For example, diffuse large B-cell lymphoma (DLBCL) comprises at least three distinctive subtypes: germinal center B-cell type (GCB), activated B-cell type (ABC) and primary mediastinal B-cell lymphoma (PMCL).15 Basing treatment decisions on histological classification can result in grouping tumours with different underlying molecular characteristics into one category. Because only a subset of patients is destined to benefit in clinical trials with eligibility based on histology, such trials fail unless they have very large sample sizes. Even if these trials show a statistically significant benefit for the experimental arm, the overall benefit is usually of only moderate clinical significance because the effect is diluted by the non-responders.16 This approach is not only inefficient for drug development, but it exposes many patients to potentially toxic drugs without providing them with any benefit.
Targeted agents that preferentially kill tumour cells while sparing normal cells have the potential of increasing therapeutic efficacy while minimizing treatment toxicity. Two broad strategies for developing targeted agents in lymphoma have been initiated. The first is based on targeting cell-surface antigens and receptors using monoclonal antibodies, and the second is based on using small molecules to target intracellular proteins that contribute to the oncogenic process by promoting cancer cell growth and survival.1
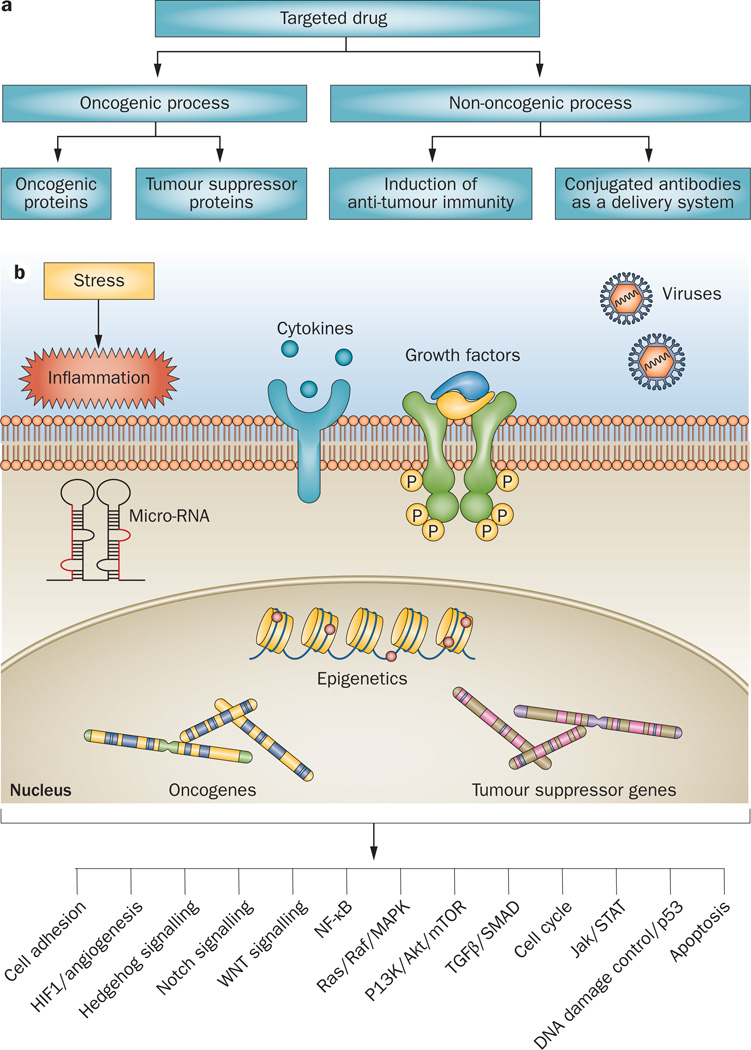
Although clinical responses have been observed with a variety of monoclonal antibodies that target non-oncogenic surface proteins, higher response rates will likely be achieved by targeting the underlying oncogenic process (Figure 1a). In general, tumorigenesis results from gain of function of oncogenic proteins, or loss of function of tumour-suppressor proteins.17 The recent success of developing cancer drugs that target single driver oncogenic proteins (such as Bcr–Abl, BRAF, and EL4–ALK) have raised hopes that such a strategy can be successfully applied to a variety of other cancers.
Figure 1.
Targeted therapy of lymphoma. a | Although targeting the underlying driver oncogenic process is logical, recent progress in lymphoma therapy has resulted from targeting non-oncogenic proteins, such as CD20 and CD30. b | Hundreds of genetic alterations in tumour cells have been identified in tumour-suppressor genes or oncogenes. Rather than designing unique drugs for each genetic alteration, several genetic defects can be grouped within well-defined oncogenic pathways. Such an approach may simplify drug development as only a dozen or so oncogenic pathways have been described. Within each pathway, multiple proteins can be targeted, regardless of their mutation status.
Unfortunately, only a minority of cancers is driven by a single genetic defect and, in most of these cases, the genetic alteration exists in only a fraction of the patients. For example, approximately 50% of melanomas contain driver BRAF mutations and 5% of non-small-cell lung cancers contain an activated ALK kinase, which are the defects that are targeted by successful modern agents.18 Despite this limitation, selecting patients based on these underlying genetic changes results in improved response rates of approximately 50% to agents targeting these driver genetic defects.19,20 Importantly, such selection can be achieved using simple diagnostic tests; PCR for BRAF, and fluorescence in situ hybridization (FISH) for ALK.18 It is more challenging to develop therapies when tumorigenesis results from a loss of a tumour-suppressor protein than from a gain of function mutation. In these cases, restoring tumour suppressor function may require re-introducing the protein through genetic transfer, or inducing its previously silenced expression by epigenetic modulating agents.
In the search for driver genetic abnormalities in lymphoma, several groups have reported genome sequencing results in a variety of lymphoma subtypes, including follicular lymphoma, DLBCL, peripheral T-cell lymphoma (PTCL), and mantle cell lymphoma (MCL).21–25 These studies revealed that the majority of recurrent genetic mutations occur at a low frequency. The roles of most of these genetic mutations, such as MLL2, in the oncogenic process of lymphoma remain unknown.22 Furthermore, the feasibility of selectively targeting the protein products of these genetic mutations in lymphoma remains to be demonstrated. In any case, it will be impractical to develop drugs that selectively target each mutant protein, especially in relatively uncommon lymphoma subtypes. A different and perhaps more practical approach is to group several oncogenic defects into well-defined oncogenic pathways. By doing so, it is conceivable that future treatment decisions can be based on the presence of specific deregulated oncogenic signalling pathways rather than basing treatment decisions on histological features. In such a new paradigm, it is possible to group most genetic defects into fewer than 20 oncogenic signalling pathways (Figure 1b). This approach may also allow the targeting of several key proteins in a given pathway, including some that are not mutated. An example is the successful use of agents that target mTOR, a non-mutated but hyper-activated protein in the PI3K pathway, for the treatment of MCL, renal-cell carcinoma, and neuroendocrine tumours.16,26,27
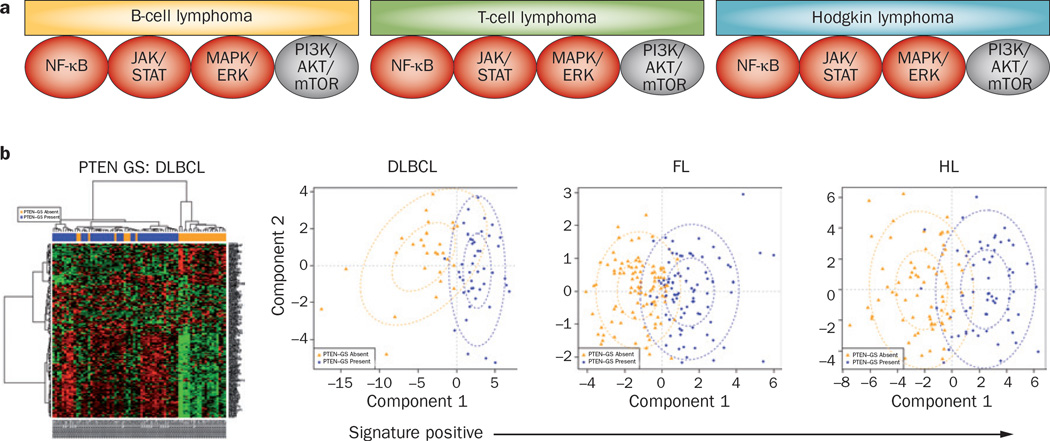
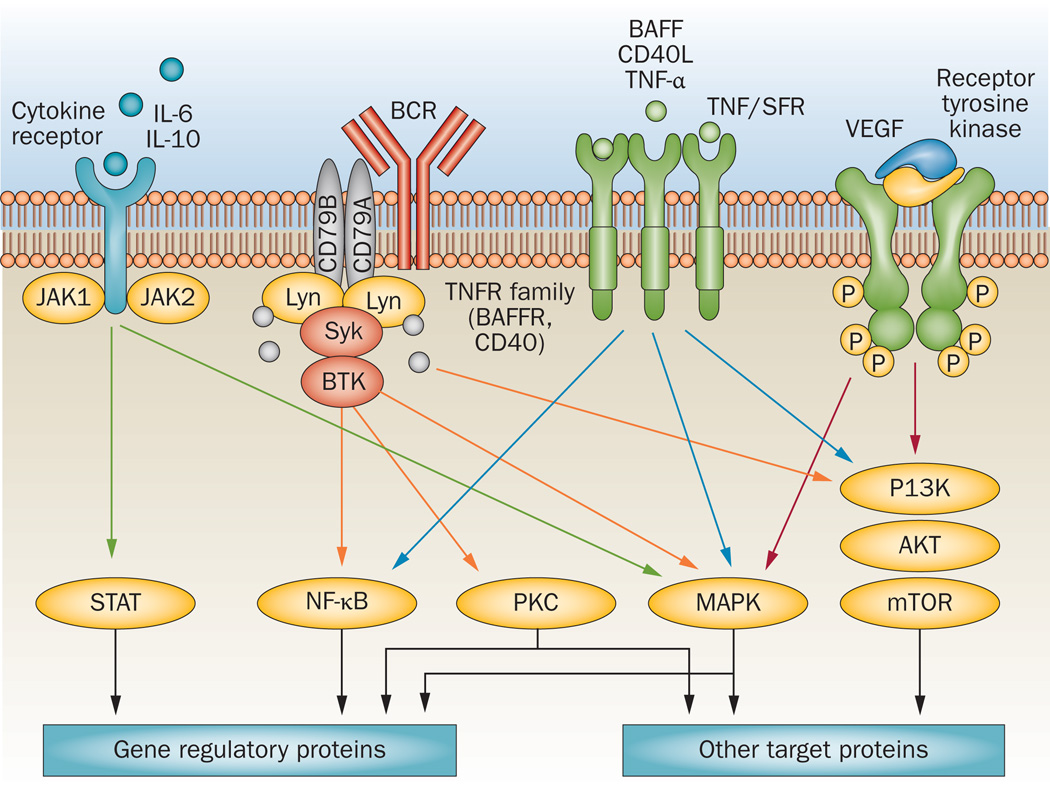
Among the most widely studied oncogenic pathways in lymphoma are PI3K/AKT/mTOR, JAK/STAT, B cell receptor (BCR) signalling, and NF-κB (Figure 2a and Table 1). In addition to harbouring possible oncogenic activating mutations, these pathways may also be activated by chromosomal translocations leading to overexpression of non-mutant proteins—such as Bcl-2—and by deletion of tumour-suppressor genes and the proteins they encode. Additionally, physiological pro-survival signalling pathways can be aberrantly activated by a variety of inflammatory cytokines and growth factors that are produced in the microenvironment. This concept is supported by emerging data demonstrating that targeting oncogenic pathways can produce clinical responses across different lymphoma histologies (Figure 2a and Table 1).13,16,28–33 For example, several agents that target the PI3K/AKT/mTOR pathway, such as CAL-101/GS-1101 (Gilead Sciences, Inc. Foster City, CA), everolimus, and temsirolimus, have produced clinical remissions in patients with follicular lymphoma, DLBCL, MCL, PTCL, and Hodgkin lymphoma.34 These data suggest that the growth and survival of a fraction of these lymphoma types is driven by an activated PI3K/AKT/mTOR signalling pathway. Similarly, agents that target the BCR pathway also demonstrated clinical activity across different B-cell lymphoma subtypes.13,33,35 Reliable and reproducible biomarkers for measuring pathway activation in clinical biospecimens will be required to select patients for pathway-targeted drugs, irrespective of the lymphoma histology.
Figure 2.
Clinical rationale for targeting oncogeneic pathways. a | Patients with different lymphoma histologies show response to pathway-targeted therapy (Table 1), indicating that a fraction of these histologies is driven by specific pathway activation. b | Applying gene-expression signature of PI3K pathway activation to primary lymphoma biopsy specimens demonstrated that the signature can dichotomize the patients into signature-positive and signature-negative cases (Table 3). Abbreviations: DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; GS, gene signature; HL, Hodgkin lymphoma.
Table 1.
Response rates according to histology
| Pathway | Drug | Target | Response rate in different histologies (%) | |||||
|---|---|---|---|---|---|---|---|---|
| DLBCL | FL | MCL | SLL/CLL | T-cell | HL | |||
| PI3K/ AKT/m TOR |
Everolimus Temsirolimus CAL-101 |
mTOR mTOR PI3K |
30 36 0 |
50 56 55 |
32 38 67 |
18 10 30 |
63 – – |
53 – – |
| B-cell receptor |
Fostamatinib PC132765 |
Syk Btk |
22 17 |
10 23 |
11 69 |
55 67 |
0 – |
– – |
Abbreviations: CLL, chronic lymphocytic leukaemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; HL, Hodgkin lymphoma; MCL, mantle-cell lymphoma; SLL, small lymphocytic lymphoma.
Biomarkers for patient selection
As our knowledge of cancer biology has improved, countless publications have advocated the use of biomarkers for prognostic classification and for guiding therapy.36–39 Yet, only a handful of biomarkers have been approved by regulatory agencies or shown to be useful in the clinic.40,41 The high failure rate in clinical biomarker development reflects the high false-positive rate associated with biomarker studies and the multiplicity of biomarkers and their combinations. It also reflects the prevalence of biomarker analyses from single institutions that use small biospecimen sample sizes, and without proper quality control of biospecimen collection and processing.42,43 For these reasons, and owing to an increased demand for identifying reliable and clinical biomarkers that can aid in selecting patients for specific therapy, the National Cancer Institute and the European Organisation for Research and Treatment of Cancer published guidelines for reporting recommendations for tumour marker prognostic studies (REMARK).44
As there is no single best method for clinical biomarker development, diverse platforms are currently used in clinical diagnostic assays for guiding cancer therapy (Table 2). Some of these assays were pre-existing and became widely used for diagnostic purposes, including assessment of CD20 and CD30 expression. Other assays were specifically developed as companion diagnostics that gained approval by the FDA, including BRAF mutation analysis and EML4–ALK fusion status.45–47
Table 2.
Diagnostic platforms used to guide treatment decisions
| Drug | Cancer | Target/biomarker | Diagnostic method |
|---|---|---|---|
| Rituximab | B-cell NHL | CD20 | IHC, flow cytometry |
| Brentuximab vedotin |
Hodgkin lymphoma and ALCL |
CD30 | IHC, flow cytometry |
| Tamoxifen | Breast | Oestrogen receptor | IHC |
| Imatinib | CML | ABL | PCR |
| Trastuzumab | Breast | HER2 HER2 |
IHC FISH |
| Crizotinib | NSCLC | EML4/ALK | FISH |
| Vemurafinib | Melanoma | BRAF V600 | RT-PCR |
| Adjuvant therapy for breast carcinoma |
Breast | Oestrogen receptor, progesterone receptor, HER2 |
RT-PCR |
| Adjuvant therapy for breast carcinoma |
Breast | 70 genes (MammaPrint) |
GEP |
| Adjuvant therapy for colon carcinoma |
Colon | 12 genes | RT-PCR |
| Gefitinib or erlotinib | Lung | EGFR mutation | PCR |
| Gefitinib or erlotinib | Colon, lung | KRAS mutation | PCR |
Abbreviations: ALCL, anaplastic large-cell lymphoma; CML, chronic myeloid leukaemia; FISH, fluorescence in situ hybridization; GEP, gene-expression profiling; IHC, immunohistochemistry; NHL, non-Hodgkin lymphoma; NSCLC, non-small-cell lung cancer; RT-PCR, reverse transcription-PCR.
From the beginning of developing therapeutic monoclonal antibodies, patients were selected for these trials based on biomarker expression. For example, all patients who were enrolled on trials assessing the monoclonal antibody rituximab were required to have CD20-expressing lymphoma.48 CD20 expression could easily be examined in any clinical diagnostic laboratory by either immunohistochemistry (IHC) or flow cytometry methods. Similarly, the recently approved antibody drug-conjugate (ADC) brentuximab vedotin was evaluated in patients with CD30-expressing lymphomas.49,50 This simple selection method, while intuitive, is generally not sufficient to predict response or resistance to monoclonal antibody therapy. Therefore, additional biomarkers that are independent of the target may be required to better predict treatment response to these agents. For example, response to EGFR-targeted therapy in head and neck cancer is influenced by mutations in the EGFR gene encoding the extracellular domain of the protein, which may predict resistance to the EGFR antibody cetuximab,51 whereas activation mutations in the EGFR tyrosine kinase domain in patients with lung cancer can predict sensitivity to the small-molecule inhibitor gefitinib.52,53 On the other hand, KRAS mutations are associated with resistance to cetuximab in colorectal carcinoma.54,55 Thus, EGFR expression status alone is not sufficient to guide therapy in patients with these cancers.56
Furthermore, the predictive value of these biomarkers is dependent on the targeted drugs. For example, mutations in the binding domain of the EGFR may predict resistance to one monoclonal antibody, but have no effect on another.56 Similarly, in patients with relapsed follicular lymphoma, CD22 expression predicts approximately 25% response rate to the naked anti-CD22 antibody epratuzumab, but predicts an 80% response rate to another drug that targets CD22, the ADC inotuzumab ozogamicin.57,60 More recently, targeting CD30 with the naked anti-CD30 antibody SGN-30 (Seattle Genetics, Bothell, WA) produced no meaningful clinical responses in patients with relapsed Hodgkin lymphoma and anaplastic large-cell lymphoma,61,62 whereas targeting the same protein using the ADC brentuximab vedotin produced response rates ranging from 74% to 85%.50,63,64 Nevertheless, compared with small-molecule inhibitors, the development of predictive biomarkers for monoclonal antibodies is relatively simple. Unlike many small molecules, monoclonal antibodies are infrequently associated with off-target effects that may complicate the predictive significance of the biomarker.
Biomarkers for oncogenic pathways
For a successful pathway-based therapy, it is imperative to develop clinical assays that accurately measure activated oncogenic pathways. As shown in Table 2, several diagnostic platforms have been successfully used to select patients for specific therapies. Many of these platforms are also being explored to measure pathway activation based on evaluating tumour DNA, RNA, and protein expression status.
Genetic mutations
Modern sequencing platforms have improved the efficiency of testing for genetic aberrations and substantially decreased its cost. In lymphoma, whole-genome, exome, and transcriptome sequencing methods have been used to study recurrent mutations.21–25 In the near future, whole-genome sequencing results will be achieved in a matter of days, and possibly in a few hours.65 A limiting factor is the ability to perform data analysis in a timely manner, and to provide simple and clear results that can be used by investigators and treating physicians to make therapeutic decisions.
A more-practical approach would be to sequence a panel of genes that are known to be recurrently mutated in lymphoma. Such a panel may consist of one to two hundred genes. As the technologies improve they can be performed on existing formalin-fixed paraffin-embedded tissue sections, avoiding the need for fresh tissue biopsies. This strategy is currently being explored by several clinical diagnostic laboratories, such as Foundation Medicine.66 The advantage of using mutation status to assign patients to specific therapy is the robustness of the assay. This approach was successful in obtaining the FDA approval of vemurafenib for the treatment of melanoma patients carrying BRAF V600E mutations.19 A similar approach can be applied to evaluate specific inhibitors in patients with lymphoma carrying frequent recurrent mutations, such as EZH2, CD79B, and MYD88.25,67,68 However, although mutation analysis assays can be easily developed, the majority of mutated genes in lymphoma have low prevalence. For example, PIK3CA mutations are observed in 27% of breast carcinoma cases, but in less than 5% of lymphoma cases.22,69 Therefore, it is important to develop additional biomarkers that may be relevant for lymphoma and that can be assessed in clinical laboratory settings.
Gene-expression profiling
Gene-expression profiling provides a detailed analysis of the composition of disease subtypes. Even in histologically uniform lymphoma subtypes, gene-expression profiling reveals heterogeneity at the molecular level. Such broad molecular subsets carry different prognostic features, and in some cases may be useful in stratifying patients for treatment on clinical trials. A good example is the molecular classification of DLBCL into the subtypes GCB, ABC and PMCL.15 Although this classification is widely accepted, it is infrequently used in clinical practice, mainly because of the requirement for fresh tissue samples and the need for special equipment and expertise. Attempts to translate gene-expression profiling classification into more practical IHC-based classification produced concordant results in only 80% of cases.70 More recently, the procedure has been simplified by the ability to perform gene-expression profiling analysis on archived formalin-fixed paraffin-embedded tissue sections.71 However, such a broad classification results in combining several pathways into a single category. Because gene-expression-profiling signatures are different among histological subtypes,72–74 there is no one unifying signature that can be applied across different histologies.
An alternative approach is to examine a gene signature that is associated with a specific oncogenic pathway activation. These signatures are generated by genetic transfer of driver oncogenes into tumours cells, including MYC, RAS, SRC, and CTNNB1. Other pathway signatures have been characterized by examining gene-expression profiling in cells that carry mutant genes, such as PIK3CA. In other models, deletion of a tumour-suppressor gene, such as PTEN, has been used to develop an activated pathway gene signature.75 Some of these signatures have been shown to have prognostic significance across different tumour types, including breast, bladder, and prostate carcinomas.75
To investigate whether these signatures are relevant for lymphoma, we applied two activated PI3K pathway gene signatures to different primary lymphoma histological subtypes using publicly available gene-expression profiling data (Y. Yuan, D. Berry & A. Younes, unpublished work). Using Gene Expression Omnibus, we selected uniformly processed primary tissues representing DLBCL, follicular lymphoma, MCL, PTCL and Hodgkin lymphoma. An activated PI3K pathway gene signature that was developed from PTEN-null tumour models was successfully applied to these histological subsets (Figure 2b and Table 3). The proportion of cases with an activated PI3K pathway gene signature was within the range of response rates reported with agents targeting this oncogenic pathway (Figure 2 and Tables 1 and 3). Further studies should address whether only the patients who benefit from these agents are those that have this gene signature.
Table 3.
Predicting response rates for therapies based on pathway signatures
| Histology | n | Incidence of activated PI3K pathway gene signature (%) |
Predicted response rate to PI3K and mTOR pathway inhibitors (%) |
|
|---|---|---|---|---|
| Absent | Present | |||
| DLBCL | 73 | 38 | 62 | 30–36 |
| FL | 184 | 54 | 46 | 50–56 |
| HL | 130 | 52 | 48 | 53 |
| MCL | 15 | 73 | 27 | 32–67 |
| PTCL | 50 | 44 | 56 | 63 |
Abbreviations: DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; HL, Hodgkin lymphoma; MCL, mantle-cell lymphoma; PTCL, peripheral T-cell lymphoma.
In an ongoing phase II clinical trial, we are treating patients with relapsed lymphoma with an oral AKT inhibitor (MK-2206, Merck, Whitehouse Station, NJ). Pretreatment tissue biopsies are obtained from all patients to assess whether they have the activated PI3K gene signature and to correlate this signature with clinical responses.76 This approach can also be used to link gene signatures of other oncogenic pathways and agents that target components of these pathways. Showing a therapeutic benefit for patients with an activated gene signature is possible by focusing exclusively on tumours having the signature, but it leaves open the question of whether the signature can predict which patients derive a benefit. To address that question requires randomly assigning patients who do not have the signature to the experimental and control agent. Ethical concerns can be mitigated by taking an adaptive approach and dropping patients with signature-negative tumours from the trial, possibly by gradually lowering their probability of being assigned to the drug in question, should interim results indicate this to be appropriate.48,77 However, when dealing with a large data set, simple errors can result in major problems leading to the wrong conclusions. Because of the potential harms of incorrectly applying large genomic data in clinical practice, the Institute of Medicine recently published recommendations for developing ‘omics’-based tests in the clinical setting.78–80
Immunohistochemistry
Although IHC is widely used for establishing the diagnosis of most cancers, this diagnostic method is rarely used to guide the use of targeted therapy (Table 2). Although IHC is more practical to use and is widely available, the method frequently produces inconsistent results among different laboratories, mainly because of the lack of standardization of reagents and methods, scoring criteria, and interobserver variations.38 The Lunenburg Lymphoma Biomarker Consortium recently reported how the prognostic significance of commonly assessed IHC-based biomarkers can change with newer therapies.81 The development of a standardized clinical diagnostic test using IHC that accurately measures the expression level of key targets on oncogenic pathways is challenged by technical difficulties. For example, the expression of the active phosphorylated form of AKT (p-AKT) is clearly associated with an activated PI3K pathway status, yet no simple or inexpensive test has been established on formalin-fixed, paraffin-embedded tissue specimens. Instead, other phospho-proteins are being explored as surrogate biomarkers, including p-PRAS40 and p-S6.82 Many proteins that can be therapeutically targeted can be examined by IHC in diagnostic specimens, including Bcl-2 and Myc.83,84 However, the lack of standardized technical and scoring methods complicate their potential use for patient selection.81 Even simple tests to measure proliferation using the Ki-67 antigen have failed to give reproducible results.85 Continued collaborative efforts among investigators—similar to the Lunenburg Lymphoma Biomarker Consortium—to standardize these clinical diagnostic methods are needed to help develop reliable diagnostic tests that can guide future therapeutic decisions.30,54
Designs of biomarker-driven trials
In a recent analysis, the success rate of cancer drug development that entered clinical trials was estimated to be around 20%.56 Approximately two-thirds of failures occur in phase III trials, which is the most expensive part of drug development. This high failure rate can be attributed to several factors, including excessive toxicity and lack of significant efficacy in unselected patients. This problem is compounded by the fact that although the current traditional design of phase II studies may accurately identify ineffective agents, it fails to predict the success of follow-up randomized phase III trials.56,57 Furthermore, the end point in phase II studies (for example, tumour response) may bear little relationship with clinical end points such as progression-free survival or overall survival that are standard in phase III studies. Another culprit is the erstwhile convention of using single-arm, non-randomized designs in phase II trials. Comparisons with historical control data usually overestimate but sometimes underestimate the efficacy of a drug, leading to unexpected results in randomized phase III trials. Importantly, drugs are typically effective in only a subset of patients. To see a benefit in unselected patient populations requires very large randomized trials.
Drawing conclusions about responding patient subsets is fraught with inferential traps. Foremost among them are false positives. Consider investigating the effect of a drug as it relates to a moderate number of biomarkers, say 20. Consider the simplest case in which the markers are dichotomous and consider the 40 single-marker subsets (one each for each marker being positive and negative). Under the conventional assumption of a 5% false-positive rate, two of the 40 subsets are expected to show a statistically significant drug effect when the drug has no effect at all. Hence, the conventional practice of adjusting for multiple comparisons is to claim statistical significance only when the P value of drug effect in a subset is less than 0.05/40 or 0.00125. Paradoxically, considering more biomarkers can make discovering predictive biomarkers more difficult. Combinations of biomarkers are more interesting scientifically, but they are even more problematic statistically. For 20 markers there are over a million possible ‘drug signatures,’ and subsets of patients who benefit from a drug (more precisely, there are 220 − 1 = 1,048,576 subsets). Suppose a drug is effective for exactly one of these subsets. Even if one is lucky in picking the right 20 biomarkers for defining this subset, it is very difficult—one chance in a million if it’s a guess—to correctly identify a drug’s signature.
The FDA was correct with their Critical Path Initiative: in a 2006 update they announced that their “outreach efforts uncovered a consensus that the two most important areas for improving medical product development are biomarker development (Topic 1) and streamlining clinical trials (Topic 2)”.58 In a subsequent guidance document they addressed adaptive clinical trials.59 Two successful combination ‘biomarker signatures’ were developed for oestrogen-receptor-positive, tamoxifen-treated adjuvant breast cancer: Oncotype DX® and MammaPrint.86–89 The developers took the elegant but simple approach of combining a variety of biomarkers (numbering 21 and 70, respectively) into a single index. This approach makes confirmation easier, but it is difficult to change the indices by adding or subtracting biomarkers or otherwise modifying their contributions to the index. Also, such an index does not help in learning which patients benefit from which therapies (although both indices have been shown to predict the benefits of chemotherapy in the general sense).
Novel clinical trial designs will be needed to expedite drug development and to save on the high cost of potentially futile large-scale phase III trials.77,90–92 Indeed, the need for phase III randomized studies in the era of highly effective biomarker-driven targeted agents has been recently questioned.91 Furthermore, because many small-molecule inhibitors are not highly selective, they may have favourable off-target effects that can influence biomarker prediction. For this reason, evaluating treatment outcome in both biomarker-positive and biomarker-negative patient subpopulations may provide valuable information that could assist in generating new hypothesis and developing new biomarkers that can improve the accuracy of predicting clinical responses. Positive and negative interactions between various biomarkers may need to be examined to improve prediction of treatment outcome. For this reason, there is no uniquely optimal design for biomarker-driven clinical trials. Instead, there are several trial designs that can be used based on biomarker prevalence, causality of the target in lymphoma pathogenesis, availability of patients, and the robustness of the clinical biomarker assays.67 Furthermore, the selection of trial design is influenced by the availability of rapid turnaround time for obtaining and using the results of a biomarker status in a multicentre setting.
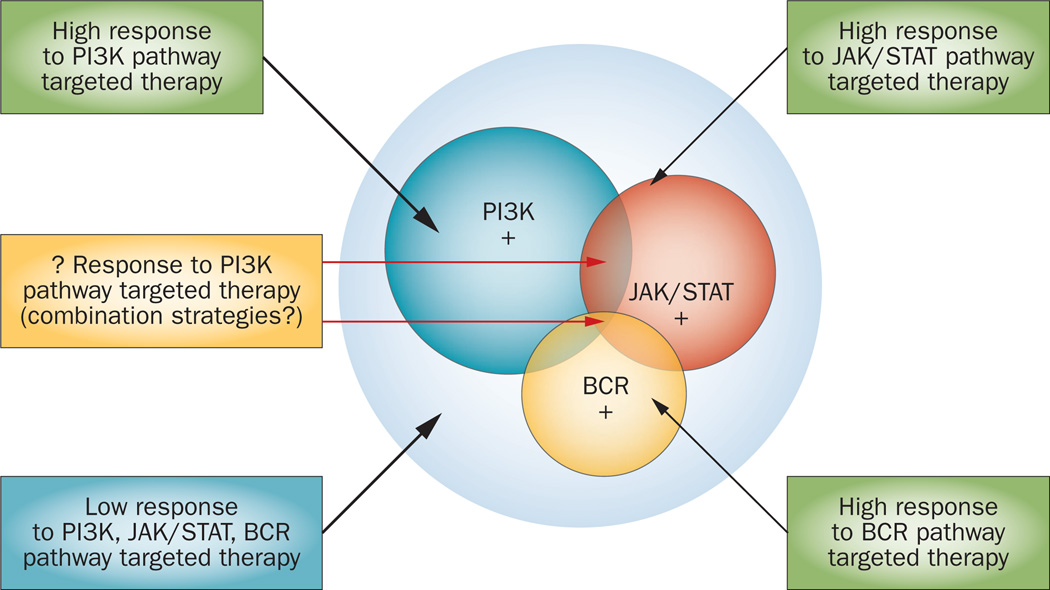
The dependence of tumour cells on a single driver oncogenic defect can lead to the identification of one predictive biomarker that can assist in selecting patients for a specific therapy (such as BRAF mutation in patients with melanoma). However, lymphoma cells frequently use several parallel survival mechanisms that may require identification of a combination of predictive biomarkers (Figure 3). This situation may also require the use of rationally designed drug combinations to achieve the best treatment outcome. In the breast cancer field, oncologists are used to evaluating three biomarkers to assist in the selection of the most-appropriate therapy (according to the biomarkers oestrogen receptor, progesterone receptor, and HER2). In patients with lymphoma, using three biomarkers that are associated with distinctive oncogenic pathways will result in eight different biomarker-defined subgroups that can be linked to therapeutic outcome (Figure 4). Such an approach can be adopted to uniformly screen pathway-targeted agents to define the subsets of patients that are likely to have the most benefit.
Figure 3.
Rationale for combination therapy in lymphoma. Lymphoma cells frequently use several activated oncogenic pathways to promote their growth and survival. In some cases, activation of one receptor or several receptors is involved in the oncogenic process. Furthermore, within one pathway, such as the PI3K/AKT/mTOR pathway, several proteins can be therapeutically targeted.
Figure 4.
Defining lymphoma subsets based on biomarkers of activated pathways. This approach to select patients for pathway-directed therapy is expected to result in higher response rates in biomarker-enriched populations. In this example, the inclusion of three biomarkers will result in eight unique subsets of biomarker sets that may have different response to therapy.
Biomarker-selected and biomarker-enriched trials
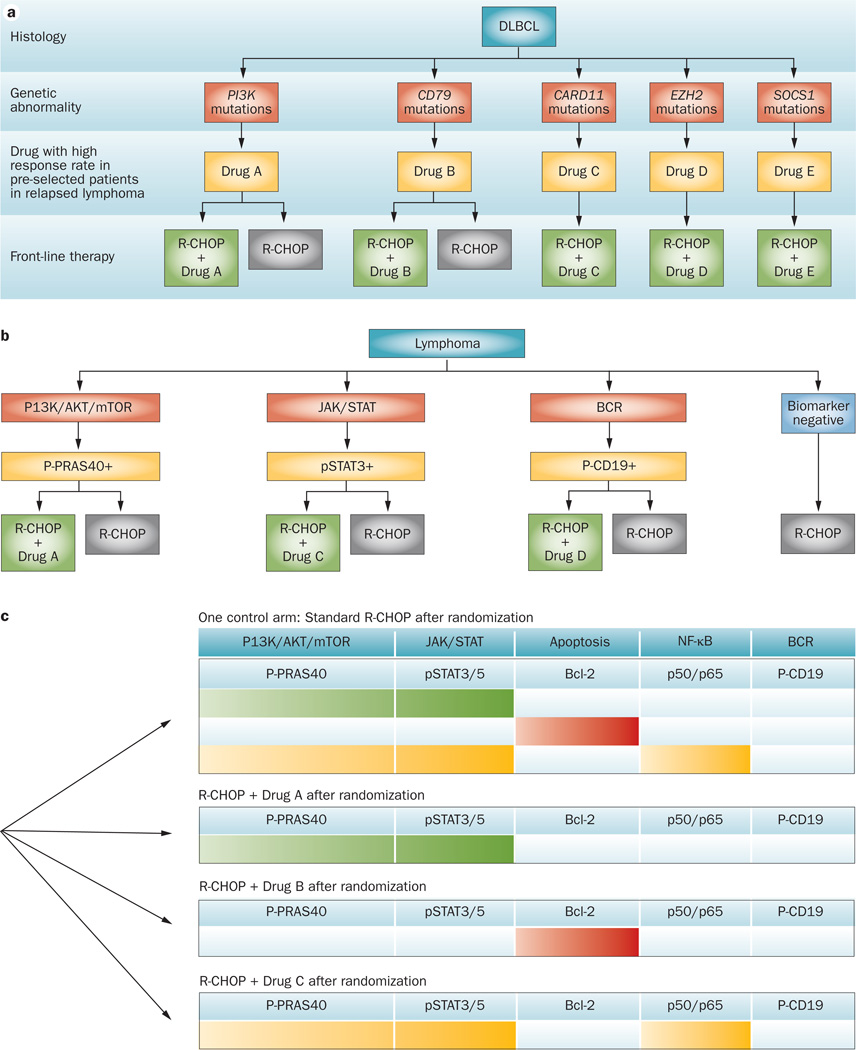
The simplest clinical trial addressing performance of a drug targeted to a biomarker is to restrict patients whose tumours have the biomarker target in question (Figure 5a).68 This may be appropriate in some circum-stances, but it prevents defining whether and how well the drug is actually hitting its target (as measured in the trial). A simple but still useful adaptive biomarker-driven trial is one involving a single dichotomous biomarker and two treatment arms; an experimental arm compared with control (Figure 5b). Treatment effect is evaluated periodically or even continuously during the course of the trial, separately within the biomarker-positive and biomarker-negative subpopulations. If the experimental arm shows little or no improvement over control in either subset then that subset is dropped from consideration in the trial. If the experimental arm performs sufficiently poorly in both subsets then the trial is stopped. Rather better from the perspective of efficiency and, arguably, ethics as well, is to employ response-adaptive randomization throughout the trial within both subsets.77,93 The poorer performing arm in a subset is assigned to a smaller proportion of patients within that subset as the trial proceeds, until the randomization ceases within that subset. This trial design addresses at least three questions: first, is the experimental arm effective within the biomarker-positive subset? Second, is the experimental arm effective within the biomarker-negative subset? Third, is the experimental arm effective in the entire patient population?
Figure 5.
Examples of biomarker-driven clinical trials for lymphoma. a | Patients can be preselected based on single driver genetic mutations. In each biomarker-defined subset, patients can be randomized to standard R-CHOP regimens or experimental R-CHOP + drug x. Depending on the incidence of the biomarker, this approach may not be practical because the patient population may be too small to perform a timely randomized study. This approach was successful in developing therapy for patients with BRAF mutant melanoma, and may be more practical for screening single agents in the relapsed lymphoma setting. b | Several mutations can be grouped in well-defined signalling pathways. This strategy may be more practical as the number of patients can be higher than those with a single genetic defect. However, a standardized approach to accurately measure pathway activation, using surrogate biomarkers, will be required. Examples of biomarkers are shown. c | A multi-arm randomized trial design comparing several experimental arms with one standard control arm. Initially patients are enrolled in an unselected manner. After treating a certain number of patients, clinical responses are evaluated within each of the biomarker-defined subgroups and responses are compared between the experimental and control groups. Biomarker-defined subsets that have low probability of improved outcome are subsequently closed to patient enrollment, and patients are enrolled prospectively in a biomarker-selected manner. Coloured cells indicate where the highest clinical responses are observed in a theoretical example.
Combined dynamic multi-arm biomarker trials
A biomarker-driven trial that addresses many questions, and consequently is very complicated, is I-SPY 2.77,94 This study is more a process than a trial. The setting is neoadjuvant breast cancer and the primary end point is pathological complete response (pCR). The focus is on 10 predefined tumour biomarker signatures depending on combinations of hormone-receptor status, HER2 status, and MammaPrint.77 Experimental agents are added to the trial as they become available and are compared with the control using response-adaptive randomization within biomarker-defined subsets. Experimental agents that are successful in the trial are matched with one or more of the signatures and are ‘graduated’ into a small confirmatory trial. Experimental agents that show insufficient improvement in pCR in all 10 signatures are dropped from the trial.77
A critical aspect of both simple and complicated adaptive trials as described above is confirmation that preliminary observations in the trial are real. Asking many questions is possible in a single trial without much cost in terms of sample size, but it does have a cost. Biomarker-driven trials have to be somewhat larger than traditional one-question clinical trials to enable addressing positive conclusions (for example, that a drug is effective for a particular signature) to decide whether they are true positives or false positives. To be specific, the total number of patients assigned to any experimental agent in I-SPY 2 ranges from 20 to 120, with a total of at least 60 patients required for any agent for a conclusion that it has a matching biomarker signature.77 Applying the I-SPY 2 model to lymphoma is feasible, but requires modifications. Having a common control arm in a process of evaluating many experimental therapies (including combinations of experimental drugs) is a simple device that makes for greater efficiency in any setting. This study includes experimental drugs from different companies in a single trial, which is not a trivial matter, but I-SPY 2 shows that it is possible. Combinations of lymphoma histologies are possible signatures, and biomarkers can be incorporated within those signatures, possibly applying across histologies. The neoadjuvant approach does not apply to lymphoma end points, such as tumour response and times to event (progression-free survival). As some of the new agents may modulate glucose uptake and metabolism, the use of interim FDG-PET scan results should be used with caution to guide therapy.71 On the other hand, the adaptive randomization that improves efficiency in I-SPY 2 relies on information available before the primary end point becomes available: MRI volume measured at various times while the patient is receiving systemic therapy. The patient’s tumour burden is modelled over time to help inform the primary end point of pCR at surgery. More generally, including in lymphoma, auxillary information, such as imaging or other biomarkers, can be incorporated in a longitudinal model that is updated based on data from the trial to help predict the course of disease for individual patients. These are not necessarily ‘surrogate’ markers, and might turn out to be uncorrelated with the primary end point. If they are correlated then the early information about each patient improves the efficiency of the design. Should the accumulating data show that the early end points are not predictive of the primary end point then the model will be updated accordingly and the information from the early end points will not be used in making adaptations or other decisions in the trial. This is an example of the range of questions that can be addressed and at least partially answered in an adaptive clinical trial. So, trial end points in lymphoma will be different and will be subject to practical and ethical standards. A multi-arm trial that considers combinations of targeted agents with standard R-CHOP (that is, R-CHOP + drug X) in comparison with R-CHOP alone will need to be tailored based on the natural history of the disease and treatment outcome (Figure 5c).
Conclusions
In summary, as more drugs and molecular targets are identified, proper inclusion of clinical biomarkers in the development plans of novel agents is becoming important for successful outcome. Incorporating such biomarkers in novel clinical trial designs may expedite drug development and reduce their cost. Such an approach has been moderately successful in a variety of solid tumours, and it will be important to follow a similar path to develop new agents for the treatment of lymphoma. We also have the advantage of learning from their mistakes. Finally, adopting collaborative multi-arm randomized studies with one control arm is likely to improve the time for clinical trial completion. Careful preclinical assessment and clinical biomarker evaluation in pre-treatment and on-treatment specimens will be required to better understand mechanisms of resistance to targeted agents, and to develop rationally designed novel combination regimens that may produce higher response rates, longer duration of response, and may overcome resistance due to tumour heterogeneity.72,73
Key points.
-
■
Recent studies demonstrated that histological subtypes of lymphoma consist of diverse molecular entities creating new challenges for basing treatment decisions on histological classification
-
■
Most single targeted agents produce low response rates in unselected lymphoma patients
-
■
Preselecting patients based on predictive biomarkers is needed to improve treatment outcome
-
■
Different clinical diagnostic platforms that are currently being evaluated for patient selection include genetic sequencing, immunohistochemistry, and gene-expression profiling tests
-
■
Multi-arm randomized biomarker-driven clinical trials using one control arm provide a rapid and efficient strategy to screen novel agents before conducting phase II studies
Review criteria.
Information for this Review was compiled by searching the PubMed, Highwire Press, and ClinicalTrials.gov databases for articles published before June 2012, including abstracts. Search terms included “lymphoma”, “Hodgkin”, “biomarker”, “sequencing” and “mutations”. Only articles published in English were considered and references were chosen based on the best clinical evidence.
Acknowledgments
This work is partially supported by NCI lymphoma SPORE grant 1P50CA136411-01A1, Robert and Vicki Smith Fund, Prince Fund for lymphoma research, and the Young Texans Against Cancer Fund.
Footnotes
Competing interests
D. A. Berry declares associations with the following company: Berry Consultants LLC. See the article online for full details of the relationship. A. Younes declares no competing interests.
Author contributions
Both authors contributed to researching data for the article, and to writing, editing and reviewing the manuscript before submission and after the peer review and editing stages.
References
- 1.Younes A. Beyond chemotherapy: new agents for targeted treatment of lymphoma. Nat. Rev. Clin. Oncol. 2011;8:85–96. doi: 10.1038/nrclinonc.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg JW. New strategies in diffuse large B-cell lymphoma: translating findings from gene expression analyses into clinical practice. Clin. Cancer Res. 2011;17:6112–6117. doi: 10.1158/1078-0432.CCR-11-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagberg H, Gisselbrecht C. Randomised phase III study of R-ICE versus R-DHAP in relapsed patients with CD20 diffuse large B-cell lymphoma (DLBCL) followed by high-dose therapy and a second randomisation to maintenance treatment with rituximab or not: an update of the CORAL study. Ann. Oncol. 2006;7(Suppl. 4):iv31–iv32. doi: 10.1093/annonc/mdj996. [DOI] [PubMed] [Google Scholar]
- 4.Younes A. Novel treatment strategies for patients with relapsed classical Hodgkin lymphoma. Hematology Am. Soc. Hematol. Educ. Program. 2009;2009:507–519. doi: 10.1182/asheducation-2009.1.507. [DOI] [PubMed] [Google Scholar]
- 5.Chen WL, et al. The clinicopathological analysis of 303 cases with malignant lymphoma classified according to the World Health Organization classification system in a single institute of Taiwan. Ann. Hematol. 2010;89:553–562. doi: 10.1007/s00277-009-0870-z. [DOI] [PubMed] [Google Scholar]
- 6.Good DJ, Gascoyne RD. Classification of non-Hodgkin’s lymphoma. Hematol. Oncol. Clin. North Am. 2008;22:781–805. doi: 10.1016/j.hoc.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Tan LH. A practical approach to the understanding and diagnosis of lymphoma: an assessment of the WHO classification based on immunoarchitecture and immuno-ontogenic principles. Pathology. 2009;41:305–326. doi: 10.1080/00313020902884501. [DOI] [PubMed] [Google Scholar]
- 8.LoRusso PM, Boerner SA, Seymour L. An overview of the optimal planning, design, and conduct of phase I studies of new therapeutics. Clin. Cancer Res. 2010;16:1710–1718. doi: 10.1158/1078-0432.CCR-09-1993. [DOI] [PubMed] [Google Scholar]
- 9.Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- 10.Robertson MJ, et al. Phase II study of enzastaurin, a protein kinase C beta inhibitor, in patients with relapsed or refractory diffuse large B-cell lymphoma. J. Clin. Oncol. 2007;25:1741–1746. doi: 10.1200/JCO.2006.09.3146. [DOI] [PubMed] [Google Scholar]
- 11.Advani R, et al. Phase I study of the humanized anti-CD40 monoclonal antibody dacetuzumab in refractory or recurrent non-Hodgkin’s lymphoma. J. Clin. Oncol. 2009;27:4371–4377. doi: 10.1200/JCO.2008.21.3017. [DOI] [PubMed] [Google Scholar]
- 12.Younes A, et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: an open-label, single-arm, phase 2 trial. Lancet Oncol. 2011;12:1222–1228. doi: 10.1016/S1470-2045(11)70265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg JW, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115:2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrowsmith J. Trial watch: phase III and submission failures: 2007–2010. Nat. Rev. Drug Discov. 2011;10:87. doi: 10.1038/nrd3375. [DOI] [PubMed] [Google Scholar]
- 15.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 16.Hess G, et al. Phase III study to evaluate temsirolimus compared with investigator’s choice therapy for the treatment of relapsed or refractory mantle cell lymphoma. J. Clin. Oncol. 2009;27:3822–3829. doi: 10.1200/JCO.2008.20.7977. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat. Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 19.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cairns RA, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kridel R, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 25.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao JC, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 28.Johnston PB, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed Hodgkin lymphoma. Am. J. Hematol. 2010;85:320–324. doi: 10.1002/ajh.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witzig TE, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011;25:341–347. doi: 10.1038/leu.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witzig TE, et al. Phase II trial of single-agent temsirolimus (CCI-779) for relapsed mantle cell lymphoma. J. Clin. Oncol. 2005;23:5347–5356. doi: 10.1200/JCO.2005.13.466. [DOI] [PubMed] [Google Scholar]
- 31.Smith SM, et al. Activity of single agent temsirolimus (CCI-779) in non-mantle cell non-Hodgkin lymphoma subtypes [abstract] J. Clin. Oncol. 2008;26(Suppl.):a8514. [Google Scholar]
- 32.Kahl B, et al. Significant clinical activity of cal-101, an isoform-selective inhibitor of phosphatidylinositol 3 kinase p110d, in patients with relapsed or refractory indolent and mantle cell lymphoma [abstract] Ann. Oncol. 2011;22:199. [Google Scholar]
- 33.Fowler N, et al. A phase I trial of Btk inhibitor PCI-32765 in patients with relapsed non-Hodgkin’s lymphoma: evidence of antitumor activity [abstract] Haematologica. 2010;95:371. [Google Scholar]
- 34.Younes A, Samad N. Utility of mTOR inhibition in hematologic malignancies. Oncologist. 2011;16:730–741. doi: 10.1634/theoncologist.2010-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang LH, et al. The Bruton’s tyrosine kinase inhibitor PCI-32765 is highly active as single-agent therapy in previously-treated mantle cell lymphoma (MCL): preliminary results of a phase II trial [abstract] Blood. 2011;118:203–204. [Google Scholar]
- 36.Iqbal J, et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J. Clin. Oncol. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 37.Casasnovas RO, et al. Plasma cytokine and soluble receptor signature predicts outcome of patients with classical Hodgkin’s lymphoma: a study from the Groupe d’Etude des Lymphomes de l’Adulte. J. Clin. Oncol. 2007;25:1732–1740. doi: 10.1200/JCO.2006.08.1331. [DOI] [PubMed] [Google Scholar]
- 38.de Jong D, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications--a study from the Lunenburg Lymphoma Biomarker Consortium. J. Clin. Oncol. 2007;25:805–812. doi: 10.1200/JCO.2006.09.4490. [DOI] [PubMed] [Google Scholar]
- 39.Korenberg MJ, Farinha P, Gascoyne RD. Predicting survival in follicular lymphoma using tissue microarrays. Methods Mol. Biol. 2007;377:255–268. doi: 10.1007/978-1-59745-390-5_16. [DOI] [PubMed] [Google Scholar]
- 40.Halait H, et al. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF gene, used as the companion diagnostic test for the novel BRAF inhibitor vemurafenib in metastatic melanoma. Diagn. Mol. Pathol. 2012;21:1–8. doi: 10.1097/PDM.0b013e31823b216f. [DOI] [PubMed] [Google Scholar]
- 41.Shaw AT, Solomon B, Kenudson MM. Crizotinib and testing for ALK. J. Natl Compr. Canc. Netw. 2011;9:1335–1341. doi: 10.6004/jnccn.2011.0115. [DOI] [PubMed] [Google Scholar]
- 42.Liotta LA, Petricoin E. Cancer biomarkers: closer to delivering on their promise. Cancer Cell. 2011;20:279–280. doi: 10.1016/j.ccr.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner PD, Srivastava S. New paradigms in translational science research in cancer biomarkers. Transl. Res. 2012;159:343–353. doi: 10.1016/j.trsl.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McShane LM, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat. Clin. Pract. Oncol. 2005;2:416–422. [PubMed] [Google Scholar]
- 45.Kelley RK, et al. Predictive biomarkers in advance of a companion drug: ahead of their time? J. Natl Compr. Canc. Netw. 2012;10:303–309. doi: 10.6004/jnccn.2012.0031. [DOI] [PubMed] [Google Scholar]
- 46.Parkinson DR, Johnson BE, Sledge GW. Making personalized cancer medicine a reality: challenges and opportunities in the development of biomarkers and companion diagnostics. Clin. Cancer Res. 2012;18:619–624. doi: 10.1158/1078-0432.CCR-11-2017. [DOI] [PubMed] [Google Scholar]
- 47.Ross JS. Cancer biomarkers, companion diagnostics and personalized oncology. Biomark. Med. 2011;5:277–279. doi: 10.2217/bmm.11.29. [DOI] [PubMed] [Google Scholar]
- 48.Maloney DG, et al. Phase I clinical trial using escalating single-dose infusion of chimeric anti-CD20 monoclonal antibody (IDEC-C2B8) in patients with recurrent B-cell lymphoma. Blood. 1994;84:2457–2466. [PubMed] [Google Scholar]
- 49.Younes A, et al. Brentuximab vedotin (SGN-35) for relapsed CD30-positive lymphomas. N. Engl. J. Med. 2010;363:1812–1821. doi: 10.1056/NEJMoa1002965. [DOI] [PubMed] [Google Scholar]
- 50.Younes A, Yasothan U, Kirkpatrick P. Brentuximab vedotin. Nat. Rev. Drug Discov. 2011;11:19–20. doi: 10.1038/nrd3629. [DOI] [PubMed] [Google Scholar]
- 51.Sok JC, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin. Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 52.Mitsudomi T. Erlotinib, gefitinib, or chemotherapy for EGFR mutation-positive lung cancer? Lancet Oncol. 2011;12:710–711. doi: 10.1016/S1470-2045(11)70194-2. [DOI] [PubMed] [Google Scholar]
- 53.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 54.De Roock W, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 55.Lièvre A, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 56.Bardelli A, Jänne PA. The road to resistance: EGFR mutation and cetuximab. Nat. Med. 2012;18:199–200. doi: 10.1038/nm.2646. [DOI] [PubMed] [Google Scholar]
- 57.Leonard JP, et al. Epratuzumab, a humanized anti-CD22 antibody, in aggressive non-Hodgkin’s lymphoma: phase I/II clinical trial results. Clin. Cancer Res. 2004;10:5327–5334. doi: 10.1158/1078-0432.CCR-04-0294. [DOI] [PubMed] [Google Scholar]
- 58.FDA. Critical Path Opportunities Report [online] 2006 http://www.fda.gov/downloads/ScienceResearch/SpecialTopics/CriticalPathInitiative/CriticalPathOpportunitiesReports/UCM077254.pdf.
- 59.FDA. Guidance for Industry. Adaptive Design Clinical Trials for Drugs and Biologics [online] 2010 http://www.fda.gov/downloads/DrugsGuidanceComplianceRegulatoryInformation/Guidances/UCM201790.pdf.
- 60.Advani A, et al. Safety, pharmacokinetics, and preliminary clinical activity of inotuzumab ozogamicin, a novel immunoconjugate for the treatment of B-cell non-Hodgkin’s lymphoma: results of a phase I study. J. Clin. Oncol. 2010;28:2085–2093. doi: 10.1200/JCO.2009.25.1900. [DOI] [PubMed] [Google Scholar]
- 61.Bartlett NL, et al. A phase 1 multidose study of SGN-30 immunotherapy in patients with refractory or recurrent CD30+ hematologic malignancies. Blood. 2008;111:1848–1854. doi: 10.1182/blood-2008-01-127118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forero-Torres A, et al. A phase II study of SGN-30 (anti-CD30 mAb) in Hodgkin lymphoma or systemic anaplastic large cell lymphoma. Br. J. Haematol. 2009;146:171–179. doi: 10.1111/j.1365-2141.2009.07740.x. [DOI] [PubMed] [Google Scholar]
- 63.Younes A, et al. Results of a pivotal phase II study of brentuximab vedotin for patients with relapsed or refractory Hodgkin’s lymphoma. J. Clin. Oncol. 2012;30:2183–2189. doi: 10.1200/JCO.2011.38.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pro B, et al. Brentuximab vedotin (SGN-35) in patients with relapsed or refractory systemic anaplastic large-cell lymphoma: results of a phase II study. J. Clin. Oncol. 2012;30:2190–2196. doi: 10.1200/JCO.2011.38.0402. [DOI] [PubMed] [Google Scholar]
- 65.Eisenstein M. Oxford Nanopore announcement sets sequencing sector abuzz. Nat. Biotechnol. 2012;30:295–296. doi: 10.1038/nbt0412-295. [DOI] [PubMed] [Google Scholar]
- 66.Foundation Medicine. Cancer diagnostics [online] 2012 http://www.foundationmedicine.com/diagnostics.php. [Google Scholar]
- 67.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc. Natl Acad. Sci. USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu P, et al. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 71.Linton K, et al. Microarray gene expression analysis of fixed archival tissue permits molecular classification and identification of potential therapeutic targets in diffuse large B-cell lymphoma. J. Mol. Diagn. 2012;14:223–232. doi: 10.1016/j.jmoldx.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 72.Staudt LM. Gene expression profiling of lymphoid malignancies. Ann. Rev. Med. 2002;53:303–318. doi: 10.1146/annurev.med.53.082901.103941. [DOI] [PubMed] [Google Scholar]
- 73.Kuppers R, et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J. Clin. Invest. 2003;111:529–537. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staudt LM. Molecular diagnosis of the hematologic cancers. N. Engl. J. Med. 2003;348:1777–1785. doi: 10.1056/NEJMra020067. [DOI] [PubMed] [Google Scholar]
- 75.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl Acad. Sci. USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.US National Library of Medicine. ClinicalTrials.gov [online] 2012 http://www.clinicaltrials.gov/ct2/show/NCT01258998.
- 77.Berry DA. Adaptive clinical trials in oncology. Nat. Rev. Clin. Oncol. 2011;9:199–207. doi: 10.1038/nrclinonc.2011.165. [DOI] [PubMed] [Google Scholar]
- 78.MacReady N. New report sets guidelines for omics research. Lancet Oncol. 2012;13:e191. [Google Scholar]
- 79.Tuma RS. Prepping omics for the clinic. J. Natl Cancer Inst. 2012;104:727–728. doi: 10.1093/jnci/djs248. [DOI] [PubMed] [Google Scholar]
- 80.Vucic EA, et al. Translating cancer ‘omics’ to improved outcomes. Genome Res. 2012;22:188–195. doi: 10.1101/gr.124354.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salles G, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: a study from the Lunenburg Lymphoma Biomarker Consortium. Blood. 2011;117:7070–7078. doi: 10.1182/blood-2011-04-345256. [DOI] [PubMed] [Google Scholar]
- 82.Andersen JN, et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci. Transl. Med. 2010;2:43ra55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 83.Kluk MJ, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS ONE. 2012;7:e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iqbal J, et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin. Cancer Res. 2011;17:7785–7795. doi: 10.1158/1078-0432.CCR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chabot-Richards DS, Martin DR, Myers OB, Czuchlewski DR, Hunt KE. Quantitative image analysis in the assessment of diffuse large B-cell lymphoma. Mod. Pathol. 2011;24:1598–1605. doi: 10.1038/modpathol.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 87.van ‘t veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 88.Albain KS, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat. Rev. Clin. Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- 90.Kirk R, Hutchinson L. Oncology trials—the elephant in the room. Nat. Rev. Clin. Oncol. 2012;9:185–186. doi: 10.1038/nrclinonc.2012.33. [DOI] [PubMed] [Google Scholar]
- 91.Sharma MR, Schilsky RL. Role of randomized phase III trials in an era of effective targeted therapies. Nat. Rev. Clin. Oncol. 2011;9:208–214. doi: 10.1038/nrclinonc.2011.190. [DOI] [PubMed] [Google Scholar]
- 92.Meurer WJ, Lewis RJ, Berry DA. Adaptive clinical trials: a partial remedy for the therapeutic misconception? JAMA. 2012;307:2377–2378. doi: 10.1001/jama.2012.4174. [DOI] [PubMed] [Google Scholar]
- 93.Berry DA. Adaptive clinical trials: the promise and the caution. J. Clin. Oncol. 2011;29:606–609. doi: 10.1200/JCO.2010.32.2685. [DOI] [PubMed] [Google Scholar]
- 94.Barker AD, et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]