Abstract
Viral replication and shedding are key components of transmission and fitness, the kinetics of which are heavily dependent on virus, host, and environmental factors. To date, no studies have quantified the shedding kinetics of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss), or how they are associated with replication, making it difficult to ascertain the transmission dynamics of this pathogen of high agricultural and conservation importance. Here, the replication and shedding kinetics of two M genogroup IHNV genotypes were examined in their naturally co-evolved rainbow trout host. Within host virus replication began rapidly, approaching maximum values by day 3 post-infection, after which viral load was maintained or gradually dropped through day 7. Host innate immune response measured as stimulation of Mx-1 gene expression generally followed within host viral loads. Shedding also began very quickly and peaked within 2 days, defining a generally uniform early peak period of shedding from 1 to 4 days after exposure to virus. This was followed by a post-peak period where shedding declined, such that the majority offish were no longer shedding by day 12 post-infection. Despite similar kinetics, the average shedding rate over the course of infection was significantly lower in mixed compared to single genotype infections, suggesting a competition effect, however, this did not significantly impact the total amount of virus shed. The data also indicated that the duration of shedding, rather than peak amount of virus shed, was correlated with fish mortality. Generally, the majority of virus produced during infection appeared to be shed into the environment rather than maintained in the host, although there was more retention of within host virus during the post-peak period. Viral virulence was correlated with shedding, such that the more virulent of the two genotypes shed more total virus. This fundamental understanding of IHNV shedding kinetics and variation at the individual fish level could assist with management decisions about how to respond to disease outbreaks when they occur.
Keywords: Virus, Replication, Shedding, Competition, Fitness
1. Introduction
Replication and shedding kinetics are known to be important components of viral fitness (Wargo and Kurath, 2012). In general, the replication of a virus within a host is believed to govern the amount of virus shed into the environment, although this has not been proven quantitatively in most cases. Viral shedding is in turn thought to dictate the transmission of the virus, such that viruses which shed more infectious particles are predicted to have higher transmission rates (Wargo and Kurath, 2012). Likewise, viruses that shed from their host for longer are believed to have more opportunities for transmission (Alizon et al, 2009). As such, those viruses which can maintain highest rates of shedding from their host for the longest duration, will likely infect the greatest number of new hosts, which is the working definition of fitness for infectious agents (Kermack and McKendrick, 1927; Anderson and May, 1979).
Due to its importance for estimating transmission, shedding has been measured in several host-pathogen systems, including those involving fish hosts (Rose et al., 1989; Kocan et al., 1997; Bebak, 1998; McKibben and Pascho, 1999; Madetoja et al., 2000; Van der Goot et al., 2003; Hershberger et al, 2009; Munster et al, 2009; Kim and Faisal, 2012; Baigent et al., 2013; Garver et al., 2013; Purcell et al., 2016). However, the majority of these studies quantify shedding at one or two time points. Inferring overall viral fitness from such studies is difficult because in order to fully estimate the transmission success of a pathogen, it is necessary to quantify the shedding kinetics over the entire course of infection. Measuring shedding at one time point may estimate the instantaneous transmission rate, but cannot provide the total amount and duration of transmission (Diekmann et al., 1990). For example, it is possible a virus with a very high instantaneous shedding rate may have a short duration of shedding, and therefore ultimately have lower overall fitness than a virus which sheds at a lower rate but for a longer duration (Fenner and Ratcliffe, 1965; Anderson and May, 1982).
The goal of this study was to quantify the relationship between within host replication and shedding in vivo, over a large portion of the infectious period, and determine how this relationship is impacted by competition during coinfection. To do so, the replication and shedding kinetics of infectious hematopoietic necrosis virus (IHNV) was quantified in live rainbow trout hosts, in single and mixed genotype infections, over a 12-day course of infection. IHNV is a single stranded, negative sense, RNA virus in the family Rhabdoviridae (Bootland and Leong, 2011). The virus is known to cause an acute disease with clinical signs and mortality in experimental infections typically beginning about 5 days post-exposure and ending after approximately 21 days (Wolf, 1988; Garver et al., 2006). IHNV is believed to be primarily transmitted horizontally through virus shed from the host in urine and feces into surrounding water, which then enters and infects susceptible hosts through the gills, digestive tract, or skin, particularly at the fin bases (Mulcahy et al., 1983; Yamamoto et al., 1990; Traxler et al., 1993; Drolet et al., 1994; Bootland and Leong, 1999; Harmache et al., 2006). Phylogenetic analysis has shown the virus in North America can be separated into 3 major genogroups, M, U, and L (Kurath et al., 2003), with the M genogroup hypothesized to have emerged and evolved host specificity to rainbow trout in the 1970s (Troyer and Kurath, 2003).
This study focuses on two strains of M genogroup IHNV in rainbow trout. Previous studies of this system have shown that one of the strains has higher fitness than the other, as measured by within host viral loads in single and mixed infections at day 3 postexposure (Wargo et al, 2010). Viral shedding and within host viral load are also positively correlated at this time point, however, the study was stopped at day 3 and other time points were not measured (Wargo and Kurath, 2011). Other studies have examined the within host replication kinetics of IHNV through time, but they focused on comparisons between only one of the M genogroup strains used here and a U genogroup strain, tested in both rainbow trout and sockeye salmon (O. nerka) (Peñaranda et al., 2009; Purcell et al., 2009). These previous studies revealed significant within host replication differences between the M and U strains, and between the M strain in the different hosts, but they did not quantify shedding. Another study examined the shedding kinetics of a U genogroup virus strain in Atlantic Salmon, revealing a late onset and persistent duration of viral shedding (Garver et al., 2013). It is unknown if IHNV shedding kinetics observed in Atlantic Salmon will be similar in other host species. However, given the differences in virus replication kinetics between fish host species and viral genotypes (Peñaranda et al., 2009; Purcell et al., 2009), it is also possible that shedding kinetics will differ, particularly because Atlantic Salmon are an exotic host for IHNV. Furthermore, studies indicate that numerous factors, such as temperature, challenge dose, and exposure route, can influence the shedding and transmission of fish pathogens (Madetoja et al., 2000; Hershberger et al., 2009; Kim and Faisal, 2012; Purcell et al., 2016). This suggests that the shedding kinetics of one system under certain conditions, cannot be assumed to be predictive of the kinetics found in a different system, and thus patterns of transmission must be evaluated on a case-by-case basis. Such case studies provide a deeper and more thorough understanding of pathogen fitness and epidemiology and can be used to determine if there are generalities that can guide management decisions (Hershberger et al., 2009; Garver et al, 2013). For example, a management practice that targets the host when clinical signs appear is likely to be ineffective if the transmission of the pathogen is largely finished before hosts become symptomatic (Kennedy et al., 2016).
To date, no studies have been published examining the shedding kinetics of M genogroup IHNV in rainbow trout over multiple time points, or relating this to within host replication kinetics, as done here. Understanding the relationship between replication and shedding is critical because the majority of studies of pathogen fitness assume that transmission can be inferred from within host loads, although the association has often not been established (Wargo and Kurath, 2012). In addition, previous studies have generally measured pathogen shedding from fish held in batch, while this study quantified shedding from individual fish, providing insight into both population-based means and fish-to-fish variation in shedding patterns. As such, this study provides a unique evaluation of how viral transmission and fitness vary throughout the infection.
2. Methods
2.1. Virus and fish
Two isolates of IHNV were used in this study: HV and LV, which have elsewhere been referred to as 220:90 and WRAC, respectively, and previously described in detail (Wargo et al., 2010). These two isolates are herein referred to as genotypes, due to discrete nucleotide difference in the viral glycoprotein genes (Garver et al., 2006) that were used to differentiate the strains by genotype-specific RT-qPCR (described below). Previous studies showed that genotype HV has higher virulence and in vivo fitness than genotype LV (Garver et al., 2006; Wargo et al., 2010; Wargo and Kurath, 2011). The viruses were maintained and propagated in Epithelioma Papulosum Cyprini (EPC) cells (Fijan et al., 1983) and stored at-80 °C, as previously described (Wargo et al., 2010). Fish used in the experiments were research grade rainbow trout Oncorhynchus mykiss, provided by Clear Springs Foods Inc. A different lot of fish was used for each of the three experiments described below. All fish were 1–3g in size and maintained on sand filtered, UV irradiated fresh water at 15 °C prior to the experiments. All fish were taken off feed 24 h prior to the start of each experiment, remained off feed for the duration of the within host replication experiments, and were fed three times per week for the shedding experiment. All experiments were conducted at the USGS Western Fisheries Research Center, in Seattle, Washington.
2.2. Within host virus replication and host response experiments
Two independent within host replication experiments were conducted. For both experiments, fish were exposed in batch to either virus genotype HV or LV alone at a concentration of 1 × 104 pfu virus/ml H2O, or mock exposed to MEM+ 10% fetal bovine serum (Gibco), in 5L buckets containing 1L of water for 12h. The virus exposure groups were separated into two buckets per treatment to reduce crowding stress. After exposure, water flow was resumed for 1 h to flush exposure virus, and then the fish were individually separated into 1L beakers with 400 ml of water. Isolation offish in individual beakers prevents cross-infection between fish. Experiment 1 contained 60 fish per virus genotype and 16 mock fish and experiment 2 contained 50 fish per virus genotype and 16 mock fish. Immediately after distribution into beakers (time zero) and then every day for 7 days, 5 fish exposed to each virus genotype and 2 mock fish (1 mock taken days 6 and 7) were euthanized by overdose of 0.27 mg/l tricaine methanesulfonate buffered with 0.09 mg/l sodium bicarbonate. The fish were collected aseptically and placed in Whirl-Pak (Nasco) bags and stored at −80 °C until further processing for fish viral load, immunity, and ARP housekeeping gene expression quantification. Because viral quantification of whole fish required euthanasia, it was not possible to track within host viral load of an individual fish through time, and as such different fish were harvested at each time point. Fish were held in static water conditions at 15 ± 2 °C, and 300 ml of water was exchanged in each beaker on day 3 post-exposure. Of the 176 fish sampled across the two experiments, 4 died before sampling.
2.3. Shedding experiment
To examine shedding kinetics, an independent study from the two within-host experiments was conducted. Groups of 10 fish were exposed either to genotypes HV or LV alone, a mixed infection of the two genotypes at a 1:1 ratio, or a mock exposure of MEM+ 10% fetal bovine serum (Gibco). A dosage of 1 × 104 pfu virus/ml H2O of each virus genotype was used, such that the mixed infections had a total of 2 × 104 pfu virus/ml H2O. This made it possible to compare the performance of a genotype alone to that in co-infection, while controlling for exposure dosage. Fish were exposed to the virus or media in batch by treatment in 1L of water for 12 h under static conditions at 15 °C. Water flow at 300 ml/min and 15 °C was then resumed for 1 h to remove exposure virus, after which the fish were separated into 1.5 l tanks in a tower rack system (Aquatic Ecosystems), with one fish per tank. Water flow to the individual tanks was set to approximately 200 ml/min and a 2 ml sample of water was collected from each tank immediately after the distribution of fish. The water flow was then turned off and the tanks were held static for 23 h to allow for accumulation of shed virus. Water was then sampled again from each tank after which flow was returned for 1 h, which was found to be sufficient time to wash out all detectable virus in the tank (data not shown). This process was repeated daily for 12 days. Water samples were stored at −80 °C until further processing. This sampling regime allowed us to quantify the total amount of virus shed by each fish per day. During the 23-h period where water was held static in the tanks, the water adjusted to ambient room temperature. This ranged from 7.8–15.7°C, with a median and mean temperature of 11.92 and 11.55°C respectively, over the 12-day study. Fish that died during the experiment were left in the tanks and water continued to be sampled daily until the end of the experiment. At day 12 post-exposure, all fish were euthanized with an overdose of tricaine methanesulfonate as above, harvested, and stored at −80 °C in Whirl-pak bags until further processing.
2.4. Sample processing
Fish were processed by homogenization in a guanadinium thiocynate based denaturing solution with a stomacher (Seward) followed by phenol-chloroform treatment to extract total RNA, as previously described (Wargo et al., 2010). For water samples, RNA was extracted from a 550ul volume using the QIAamp MinE-lute Virus Spin Kit (Qiagen) as previously described (Wargo and Kurath, 2011). All extracted RNA was stored at −80°C until further processing. The RNA samples were converted to cDNA using oligo(dT), random hexamers, and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega), with 11 µl of RNA in a 20 µl reaction volume, as previously described (Wargo et al, 2010). For quantification of viral RNA, cDNA was diluted 1:5, then underwent HV or LV genotype specific qPCR using MGB taqman probes (Applied Biosystems) and primers, no UNG Master Mix (Applied Biosystems), and genotype specific RNA transcript standards, as described previously (Wargo et al, 2010; Wargo and Kurath, 2011). Mixed infection samples underwent qPCR twice, once with each genotype specific assay. The qPCR quantifies RNA copy number of the viral glycoprotein (G) gene in either fish or water samples, presented as log copies per gram offish, or per ml of water, respectively. This is used as an indicator of within host virus replication and virus shedding and referred to in this way hereafter. The detection limit of the G gene qPCR assay for virus in water samples was estimated based on two relevant factors. Using dilutions of a G gene transcript RNA standard, the detection was between 1 (negative) and 10 (positive) viral RNA copies per reaction, which was equivalent to 66–660 copies per ml of water. Among the water samples tested in this study, the minimum viral load detected as positive for virus was 312 copies per ml of water. Thus the actual detection limit lies between 66 and 312 copies per ml of water, which equates to log 1.8–2.5. In experiment II, the cDNA generated for quantification of within host viral loads was also assayed for expression of the interferon immunity associated myxovirus resistance protein 1 (Mx-1) gene and acidic ribosomal protein (ARP) housekeeping, as previously described (Purcell et al., 2004).
Of the 51 water samples taken from mock exposed fish over the 12-day experiment, 3 were positive for genotype HV and 8 were positive for genotype LV. Given that in all but one case, these samples were not from the same fish on consecutive days, it is likely this was a result of sample processing contamination rather than accidental infection of mock fish. To account for the potential impact of this contamination on the results, all analyses of shedding data were rerun after subtracting the average viral load found in mock-exposed fish water samples (694 viral RNA copies/ml H2O) from the amount of virus found in water samples from virus-exposed fish. This did not affect the statistical results, likely because, viral RNA quantity in mock samples was generally an order of magnitude or more below the values of samples from virus exposed fish. The data and figures without this subtraction are presented.
2.5. Statistics
Statistical analyses were carried out in R, version 3.2.2 (R Core Team, 2015). For all analyses, the simplest model with the highest explanatory power was chosen under the assumption of parsimony. Minimal models were reached by comparing model fits using a likelihood ratio test and AIC value comparison, with p < 0.05 or differences in AIC value greater than 5 considered significant. Conformity to the assumptions of homogeneity of variance and normality were evaluated with residual plots, Levene’s tests, and Shapiro-Wilk tests. For all viral load comparisons, fish or water samples without detectable virus were dropped from the statistical analyses, unless otherwise noted.
2.5.1. Within host virus replication kinetics and host innate immune response
Within host viral replication data was analyzed with general linear models (GLM) using the lm function in R, with dependent factor log transformed viral load and explanatory factors day, genotype, and experiment. To examine differences in the number of fish positive for within host virus at each time point, Fisher exact tests were performed. To analyze Mx-1 data, the ΔΔCT transformation method was used to standardize expression against ARP levels and Mx-1 levels in mock fish (ΔCT Value = 2−[(CTMX treated fish–CT ARP treated fish)–(CT MX mock fish–CT ARP mock fish)]). Previous studies have shown ARP levels are stable across fish so this calculation controls for fish-to-fish variation in RNA yield while quantifying Mx-1 expression relative to uninfected fish (Purcell et al., 2004). A general linear model was then used with response variable transformed Mx-1 level and explanatory factors day and genotype. For ΔCT calculation of mock exposed fish, where multiple fish were used, the mean was calculated. For both the within host viral load and MX-1 expression analyses, differences between factors levels were compared using Tukey’s post-hoc tests, when factors were significant.
2.5.2. Viral shedding kinetics of individual fish
The day and quantity of peak shedding rate (determined by water sampling time point showing the highest viral load for each fish), as well as the number of days shedding, were analyzed using mixed effects models with the function lme from the nlme package in R (Pinheiro et al., 2015), with the maximum likelihood (ML) method of parameter estimation. The model included response variables log transformed virus load, day, or number of days; explanatory fixed factors genotype, competition, and their interaction; and random factor fish. Mixed models were used because quantifying HV and LV in mixed infections required a repeated measure from the same fish. F-tests were used to determine significance of fixed effects. Standard deviation in day and mean of peak shedding, as well as the number of days shedding, were analyzed using Levene’s test and a bootstrap method with a bootstrap value of 1000 (Amiri and Zwanzig, 2010,2011; Wargo et al., 2012).
2.5.3. Correlates of mortality
Correlates of mortality were investigated using survivorship analysis with a Cox proportional hazards regression. This was carried out with the coxph function from the package survival in R (Therneau, 2013). The response variable was day of death, with censoring of live fish on the last day of the experiment. Explanatory factors included log transformed peak shed viral load, percentage of days shedding before death, and infection type (HV, LV, or mix). The proportional hazards assumption of the model was validated using the cox.zph function, ensuring that the parameter rho was not significantly different from zero. A general liner model was also conducting to determine if the proportion of days shedding or peak daily virus shed differed between fish that died or survived.
2.5.4. Mean viral shedding kinetics
The number of fish shedding through time was analyzed using Fisher exact tests on a contingency table of the number of fish shedding detectable virus versus the number of fish tested, for each treatment (HV alone, LV alone, HV mix, LV mix). This was performed separately for each day (0–12), and resulted in 13 tests. To avoid inflated type I error, a Bonferroni corrected alpha value of 0.0039 (0.05/13) was used. The mean daily virus shed from virus positive fish through time (shedding rate) was analyzed using a mixed effects model, with the function lme (Pinheiro et al., 2015) and the maximum likelihood (ML) method of parameter estimation, as described above. This analysis was conducted from day 1 forward because genotype HV did not shed detectable virus until day 1. Including day 0 did not change the overall patterns (data not shown). Because shedding data was taken from the same fish through time, day was included as a covariate and day nested within fish was included as a random factor in the model. Examination of autocorrelation through time using the ACF function indicated no significant autocorrelation was present. The analysis showed that the variances between groups were not homogenous, so the varPower function was used as a weighting term, significantly improving the model fit (AIC = 583 vs 588, L Ratio = 7.32, P = 0.007). The minimal model was log (shed viral load) = genotype + competition + day + genotype*day + competition *day. The total amount of virus shed over the 12 days and amount of virus in fish on day 12 (at harvest), were also analyzed using mixed effects models with the lme function and response variable log transformed total virus shed, explanatory fixed factors genotype and competition, and random factor day. The total amount of virus shed from mixed infections compared to single infections was analyzed using a GLM with response variable log transformed viral load and explanatory factor infection (HV alone, LV alone, Mixed). For this analysis the quantity of HV and LV were summed in mixed infections.
3. Results
3.1. Within host virus replication kinetics and host innate immune response
In the two independent experiments to examine within host replication kinetics, within host viral loads were detected in all fish within 24 h after immersion exposure to virus. For the fish that had detectable virus, in both experiments a general trend of rapidly increasing mean viral load was seen for each genotype from day 0–2, after which the kinetics differed between the genotypes, and between the two experiments (Fig. 1). As typically observed in studies of this system (Wargo et al., 2012), between fish variation in within host viral load was high. As such daily trends were suggestive only, and when analyzing only the fish that were positive for virus in the two experiments neither genotype showed a significant difference in viral load between days (Day effect, Tukey post-hoc test, P > 0.05), except that days 1 through 7 all had significantly greater within host viral load than day 0 (F7,130 =8.57, P< 0.001). Furthermore, there was not a significant difference in within host viral load between genotypes HV and LV for any day (F1,129 = 2.67, P = 0.11), despite suggestive trends of higher viral load for genotype HV at some time points, particularly days 5 and 6 in experiment 2. A significant difference in within host viral load between experiments was observed, with more virus being produced in experiment 2 compared to experiment 1 (F1,130 = 6.48, P = 0.012). When examining the number of fish positive for virus over the course of infection, no significant differences were observed between genotypes or experiments (Fisher exact test, P > 0.05).
Fig. 1.
Cumulative within host viral load kinetics. Points show mean (±1 standard error) of log transformed within host viral RNA copies/g offish, of genotype HV (black circles) and LV (grey squares), in experiment 1 (left) and 2 (right), on each day of the 7-day experiment. Means were calculated from virus-positive fish only, and numbers along the bottom of the plot show number of fish positive for virus/number of fish sampled on each day, for genotype HV (black) and LV(grey). Different fish were sampled at each time point.
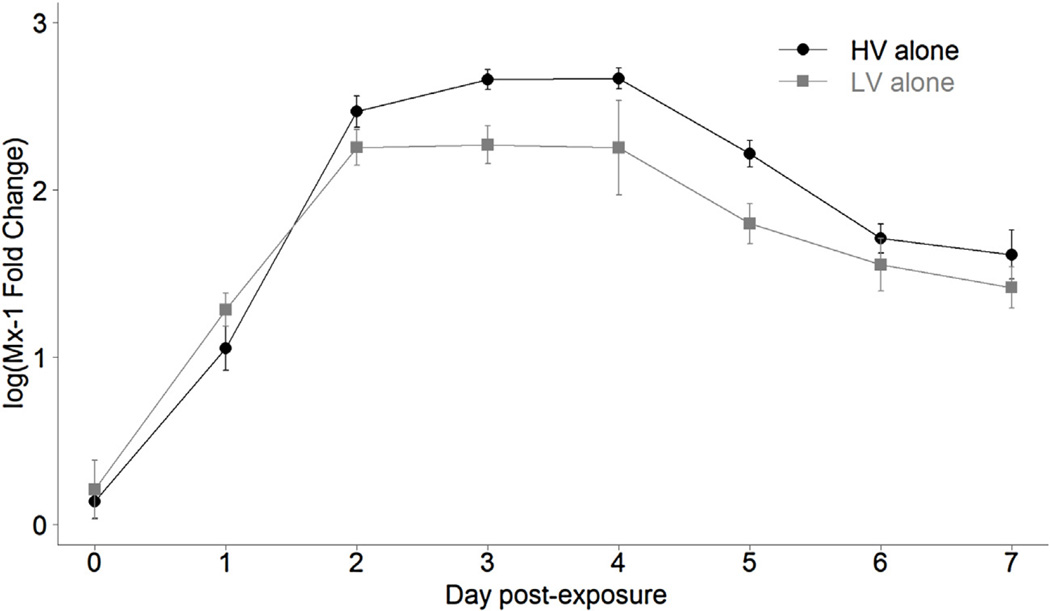
In experiment 2, expression of the host interferon stimulated gene Mx-1 was also quantified for each fish to assess kinetics of the host innate immune response. For both the LV and HV treated fish there was a significant increase in Mx-1 expression relative to mock fish from days 0–2, followed by a plateau in expression till day 4, and then a significant decrease on days 5–7 (Fig. 2, F7,62 = 60.31, P < 0.001, Tukey’s test P < 0.05). Thus, Mx-1 expression generally followed the same pattern of within fish viral load, as observed in previous studies (Purcell et al., 2004; Kell et al., 2013). Peak levels of Mx-1 expression were 476-fold and 262-fold above mock infected control fish for viral genotypes HV and LV, respectively. Overall, there was significantly greater relative Mx-1 induction in fish exposed to genotype HV compared to LV(F1,62 = 4.57, P = 0.04). Mx-1 expression was not quantified in experiment 1.
Fig. 2.
Innate immune response measured as interferon-induced Mx-1 gene expression. Points represent mean (±1 standard error) Log10transformed normalized Mx-1 fold changes relative to mock control group, in single infections of genotype HV (black circles) and LV (grey squares), over the 7-day course of the experiment. Value was calculated using the 2−ΔΔCT method where ΔΔCT equals mean (CTMx-1–CTARP individual infected fish)- mean (CTMx-1–CTARP individual mock fish). Standard error was calculated by propagating standard deviation through equation as described elsewhere (Schmittgen and Livak, 2008). ARP is a rainbow trout housekeeping gene (Purcell et al., 2004). Mx-1 expression was quantified for the same fish providing within host viral load in experiment 2 (Fig. 1).
3.2. Viral shedding kinetics of individual fish
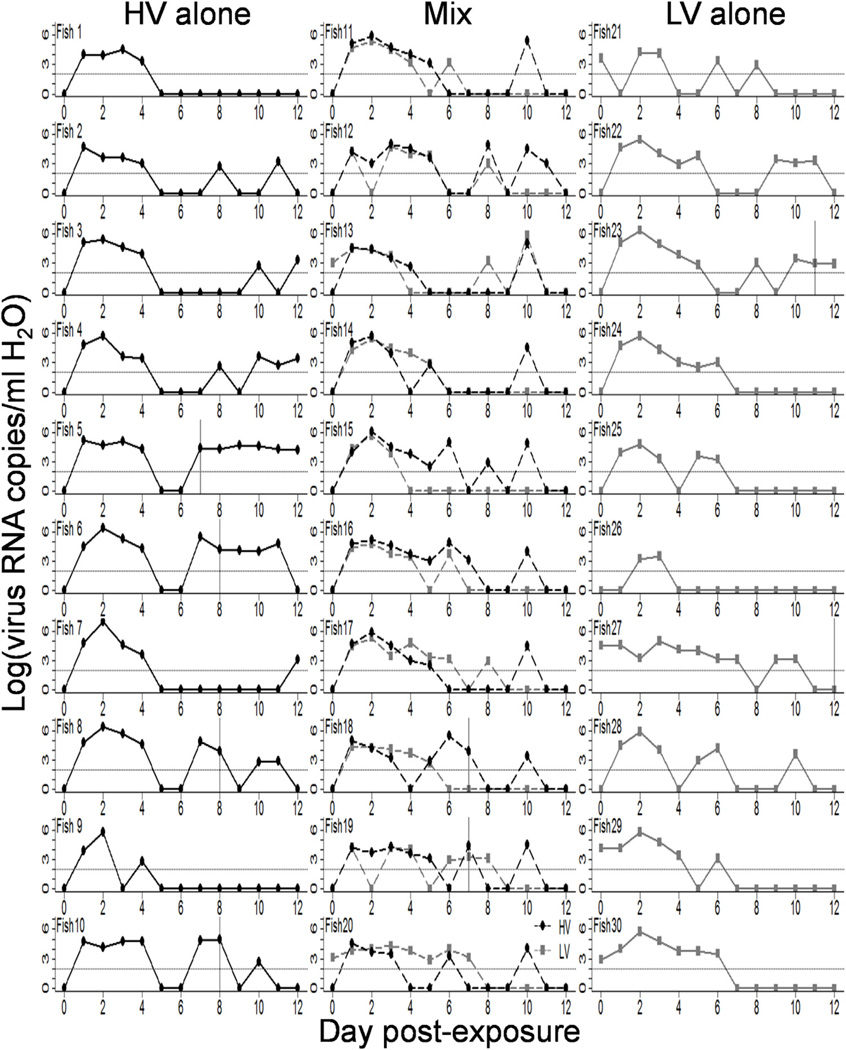
In the independent shedding experiment, detectable shedding began for 93% of the virus-exposed fish within 24 h and for all fish within two days post challenge regardless of treatment (Fig. 3). Shedding increased rapidly, with the peak amount of shed virus occurring between days 1–3 in 87.5% of fish (12.5% day 1, 60% day 2,15% day 3). There was no significant difference in the mean (2.88 days, mixed model, P > 0.05) or variance (5.75 days, bootstrap and Levene’s tests, P > 0.05) of the day of peak shedding between genotypes or single compared to mixed infections (Fig. 4A). Including fish in this analysis as a random factor accounted for 32% of the residual error in the model. When examining the peak amount of virus shed from individual fish in a 23-h period, irrespective of day (Fig. 4B), there was no difference in the mean or variance between single or mixed infections (mixed model, P > 0.05). However, the amount of peak virus shed was significantly higher for genotype HV compared to LV, when combining single and mixed infections (HV: 9.37 × 105, LV: 3.34 × 105; F1,9 =6.2, P = 0.035), although the variance in peak shedding was the same for the two genotypes (HV: 2 × 106, LV: 4.6 × 105; bootstrap and Levene’s tests, P > 0.05). Including fish as a random factor in this analysis accounted for 80% of the residual error in the model.
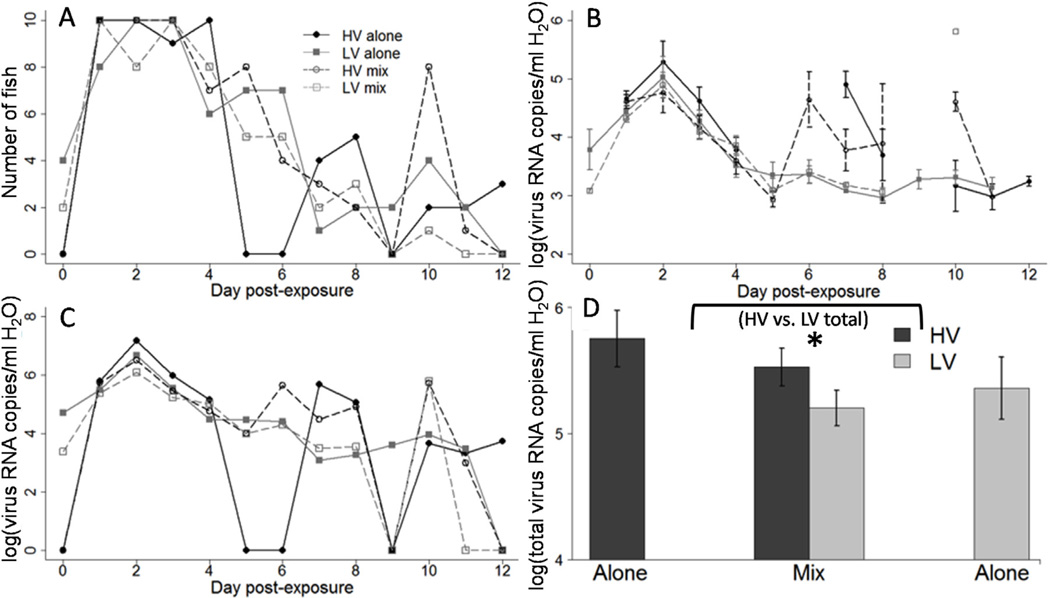
Fig. 3.
Daily quantity of virus shed from individual fish. Each panel shows the log [(virus RNA copies +1)/ml of H2O] for individual fish, per 23 h, over the 12-day course of the experiment. The left column shows 10 fish exposed to IHNV genotype HV alone, the right column shows 10 fish exposed to IHNV genotype LV alone, and the middle column shows 10 fish exposed to a mixed infection of genotypes HV and LV at a 1:1 ratio with separate lines for HV (black) and LV (grey). The vertical lines on some panels show the day a fish was found dead in the tank. Note some fish shed detectable virus RNA by qPCR after death, but this was not tested for infectivity to cells. The horizontal dotted lines on each panel show the approximate minimum detection threshold of the qPCR assay, which was estimated as being between 66 and 312 viral RNA copies/ml H2O (see Methods). Data points below the limit of detection were set to a value of 1, to indicate that they fell somewhere between zero and the limit of detection.
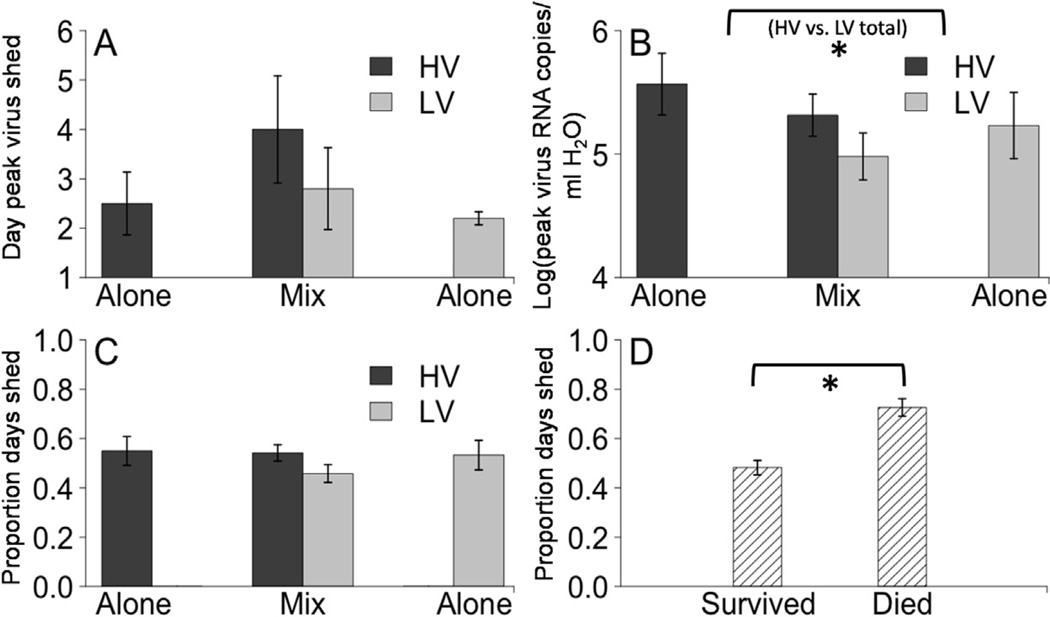
Fig. 4.
Means for shedding patterns of individual fish. (A) Mean day (±1 standard error) of peak shedding of each genotype in each fish. (B) Mean of log transformed (±1 standard error) peak quantity of virus shed for each genotype in each fish, irrespective of day. Bracket with asterisk below indicates an overall significant difference between HV and LV when single and mixed infections were combined (F1,9 = 6.2, P = 0.035) (C) Mean proportion (±1 standard error) of days shedding out of the 12-day observation period, calculated for each genotype in each fish. In panels A-C, genotype HV is shown in black and genotype LV shown in grey. Single infections are shown on far left and right, mixed infections are shown in middle. The values for the two genotypes in mixed infections come from the same fish. (D) Mean proportion (±1 standard error) of days that fish shed detectable virus for fish that died (left, n = 8) or survived (right, n = 22), during the period the fish was alive over the 12-day experiment. Bracket with asterisk below indicates a significant difference in the proportion of days shedding between fish than died and those that survived (F1,28 = 21, P < 0.001).
After the initial peak of shedding, individual fish varied in shedding pattern during a post-peak period where all fish dropped below the level of detectable shedding at some points and several fish showed secondary peaks of smaller magnitude and/or duration (Fig. 3). The number of days a fish shed was fairly dispersed, with only 37.5% of fish shedding for the median value of 6 out of 12 days (Fig. 4C). However there was no significant difference in the mean (6.25 out of 12 days, mixed model, P > 0.05) or variance (3.32 days, bootstrap and Levene’s tests, P > 0.05) of the days fish shed detectable virus between genotypes or single versus mixed infections. Fish accounted for less than 0.1% of the random error in this model.
3.3. Correlates of mortality and shedding from fish that died
Among the 30 fish in the shedding experiment 4 died in the HV group and 2 died in the mixed infection group on days 7–8 post-exposure, and 2 died in the LV group on days 11–12. No mock exposed fish died during the experiment. In general, fish that died shed virus for a higher percentage of the days prior to their death (73% ± 3.5% of days) than the percentage of total days fish that survived shed (48% ± 3% of days) (Fig. 4D; F1,28 = 21, P < 0.001). Infection type and the percentage of days shedding virus were found to be correlates of mortality (Wald Test = 9, degrees of freedom =3, P = 0.03), such that for each percent of increase in the days a fish shed there was a 28% increase in the rate of mortality (95% CI = 8.7%-52%, Z = 2.9, P = 0.003). Likewise, the rate of mortality (number of fish that died per day) was significantly lower for genotypes LV alone (95% CI = 28%-99.8% lower, Z =−2.2, P=0.03) and mixed infections (95% CI = 17%-99.9% lower, Z= −2.1, P=0.04) compared to single infections of genotype HV. Peak viral load was not found to be a correlate of mortality (P > 0.05).
All fish that died shed detectable virus by qPCR after they died, except fish 27 which died on the last day of the study. One fish (Fig. 3, fish 5) shed detectable viral RNA by qPCR for a full 5 days after it died. The infectivity of virus shed from dead fish was not quantified and all dead fish were removed from all shedding statistical analyses from the day of death onward, unless noted otherwise. All fish died after their shed viral loads had peaked and begun to decline.
3.4. Mean viral shedding kinetics
The number of fish shedding virus increased rapidly through time such that all fish in every treatment group shed virus early in infection (days 1–3). The numbers then tapered from day 3 forward, such that by day 12, only 3 out of the 22 fish that remained alive were still shedding virus (Fig. 5A). This pattern of the number of fish shedding was similar between virus exposure treatment groups, except for genotype HV alone, which completely dropped below the detection threshold of shedding in all fish on days 5–6, then resumed shedding at detectable levels on days 7 and 8. All treatments also showed an increase in the number of fish shedding on day 10, with the increase being qualitatively largest for HV in mixed infection. Statistical analysis of the number of fish shedding through time indicated that the only days there were significant differences in the number offish shedding between virus exposure treatment groups were days 5, 6, and 10 (Fisher exact test, P < 0.0038). This was likely due to the drop in the number of fish shedding in the HV alone treatments on days 5 and 6, and the increase in the number offish shedding HV in the mixed infection treatments, compared to the other virus treatment groups.
Fig. 5.
Mean shedding kinetics. (A) Mean number of fish shedding detectable virus per day for the 12-day course of infection, out of 10 total fish in each treatment group. (B) Mean (±1 standard error) log transformed virus shed per ml of H2O for each treatment group per day, for the 12-day course of infection. Only fish with detectable shed virus were included in the mean amount of virus shed. Number of fish for each data point is given by Fig. 5A. (C) Total virus shed per treatment group per day, for the 12-day course of infection. (D) Mean total virus shed from all fish per treatment group over the entire 12-day course of infection. Bracket with asterisk below indicates an overall significant difference between HV and LV when single and mixed infections were combined (F1,9 = 8.21, P = 0.019). In all panels HV is shown with black lines and shapes, LV is shown with grey lines and shapes. Dotted lines of respective colors represent each genotype in mixed infections. Figure legend for panels A-C is found in panel A. Dead fish are excluded from all figures from the day after they were found dead onward. Inclusion of dead fish did not change overall trends.
The mean daily shedding rate for live fish that were shedding detectable virus showed a peak in all treatments on day 2 and then uniform tapering until reaching a plateau by day 5 post-exposure, at approximately 2 logs below peak shedding values (Fig. 5B). There also appeared to be a secondary peak in viral shedding rate for genotype HV between days 6–8 post-exposure, in both single and mixed infections. Analysis of the daily amount of virus shed from virus positive fish from day 1 forward (day 0 was dropped because only 6 fish had started shedding), with day as a covariate, revealed that there was a significant interaction between day and genotype (F1,210 = 5.5, P = 0.02). The analysis indicated that there was a negative slope for the change in viral load through time, such that viral load significantly decreased over the course of infection for both genotypes, in single and mixed infections. The slope was significantly steeper for genotype LV than HV, indicating that viral load decreased more rapidly. However, the mean shedding rate did not differ (P > 0.05) between the two genotypes at the beginning (day 2) or the end of the study (day 11). Furthermore, single infections overall had a significantly higher shedding rate through time compared to mixed infections (Fig. 5B; F1,28 =4.3, P = 0.048). The random factor day nested within fish accounted for 64% of the residual variance in the model.
To examine the overall transmission potential on a population basis, the total amount of each virus genotype shed from all fish per treatment, per day was calculated, which can be approximated by the number of fish shedding multiplied by the mean rate of shedding for each day. From day 1 through 4 of infection, all virus exposure treatments showed a similar peak pattern of total shedding, with genotype HV alone appearing to have a slight advantage (Fig. 5C). During the post-peak period after day 4, the virus treatment with the shedding advantage depended on the day. If examining across both single and mixed infections, there were more time points where HV had the advantage compared to LV.
When analyzing the total virus shed summed across the 12-day course of infection (Fig. 5D), the HV treatment group shed more virus than the LV group (F1,9 = 8.21, P = 0.019). There was no significant difference in the total amount of virus shed between single and mixed infections for either genotype (F1,28 =0.62, P = 0.44), although both genotypes showed a qualitative trend of less total virus shed in mixed infections. This analysis included fish as a random factor, which accounted for 79% of the residual error in the model. The lack of a competition effect would suggest that more total virus should be shed in mixed infections when summing across the two viral genotypes. However, the total virus shed did not differ between the combined quantity of HV and LV in a mixed infection (7.64 × 105 ±1.81 × 105 virus copies/ml) and the most prolific single infection, HV alone (1.71 × 106 ±0.92 × 106 virus copies/ml) (F2,27 = 1.15, P = 0.332), indicating a cap in viral shedding.
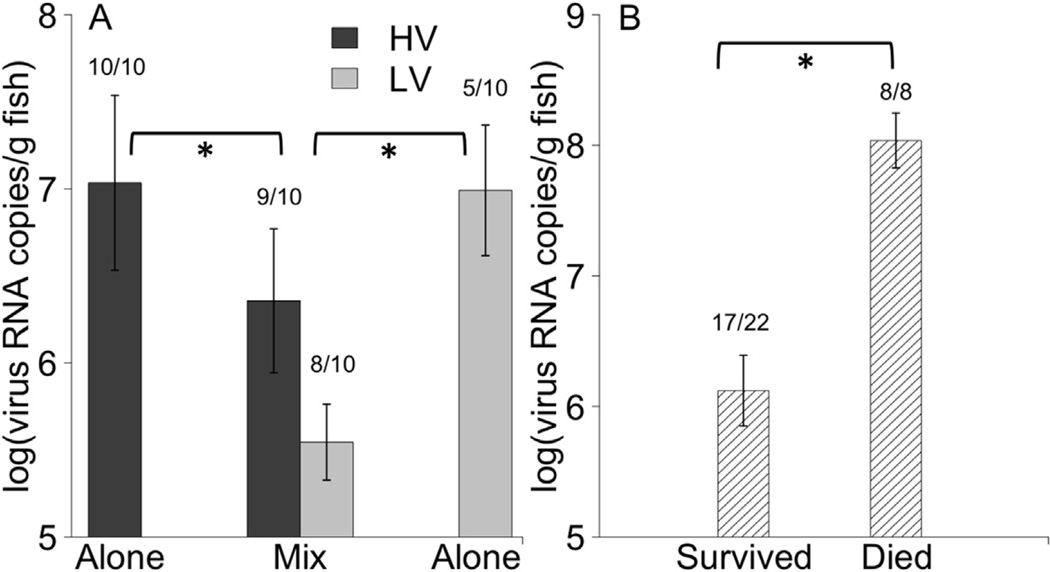
On day 12 post-infection, all fish used for the shedding kinetics study were harvested for viral load quantification. Among virus-positive fish, more virus was found within fish in single compared to mixed infections for both genotypes (Fig. 6A, F1,29 = 9.61, P = 0.004). Fish that died prior to day 12 also had significantly higher viral loads, by approximately 100-fold, than those that were still alive on day 12 (Fig. 6B, F1,29 = 18.74, P < 0.001). There was no difference in viral loads within fish on day 12 between genotypes HV or LV in single or mixed infections (P > 0.05). The random factor fish was found to account for less than 0.0001% of the model variance, so the term was dropped from the analysis. When examining the numbers offish positive for within host virus on day 12, all fish in the HV and mixed infection groups were positive, but only half of the fish in the LV group remained positive. Thus significantly more fish had detectable virus genotype HV compared to LV (Fisher exact test, p < 0.05), suggesting more rapid clearance of genotype LV There was no significant difference in the number offish positive for virus between dead and alive fish at the time of harvest on day 12 (Fisher exact test, p > 0.05).
Fig. 6.
Mean within host virus RNA copies (i.e. viral loads) in fish at the end of the 12-day shedding experiment. (A) Mean(±1 standard error) log transformed viral loads for fish exposed to genotype HV (black) and LV (grey), in single and mixed infections. Values for genotypes in mixed infections came from the same fish. Brackets with asterisk below indicate significantly more virus found in fish with single compared to mixed infections for both genotypes (F1,29 = 9.61, P = 0.004). (B) Mean (±1 standard error) log transformed viral load within fish at the end of the 12-day shedding experiment, for fish that survived (left) or died (right). Viral load in each fish for B is combined quantity of genotype HV and LV. Bracket with asterisk below indicates significantly more virus found in fish that died compare to those that survived (F1,29 = 18.74, P < 0.001). For both panels A and B mean viral load included values from fish that died during the experiment, and was only calculated for fish with detectable virus. Numbers above bars indicate number of positive fish/number of fish sampled per category.
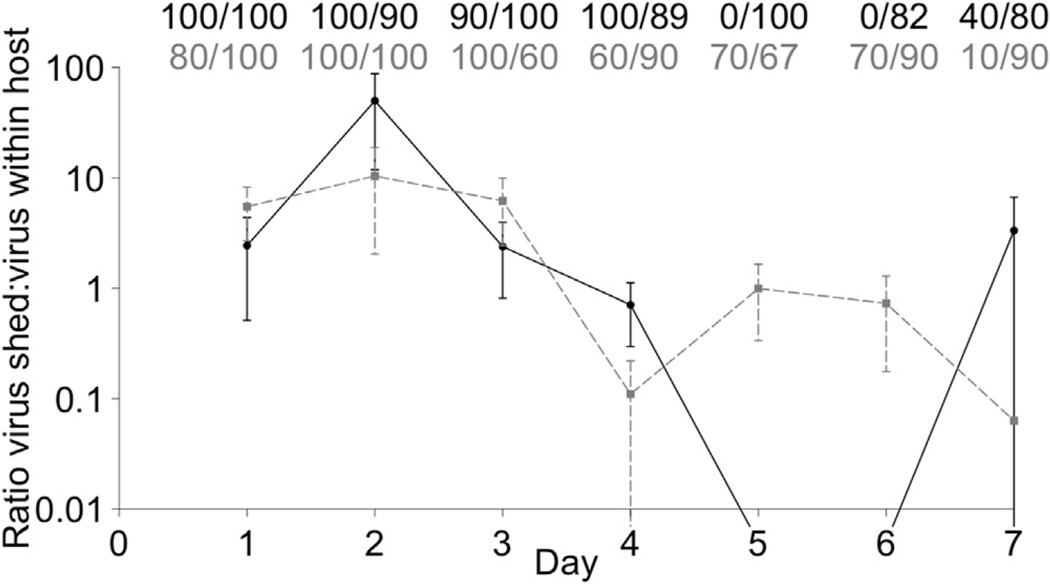
3.5. Ratio of shed virus to within host virus
Within host viral loads from the two replication experiments was compared with data from the independent shedding experiment to address the question of what proportion of the virus produced during infection is shed into the environment? It was found that the ratio of shed to within host viral RNA followed a peak and post-peak pattern over the course of infection (Fig. 7), similar to that observed for mean virus shed (Fig. 5B). In the peak period (days 1–3) the ratio of shed to within host virus was typically greater than 1, reaching a maximum value of approximately 50 for genotype HV and 10 for genotype LV on day 2. In the post-peak period (days 4–8), the ratio was typically at or below 1 for both genotypes, and fell to 0 on days 5 and 6 for genotype HV because no shed virus was detected on those days. There was a large degree of variability in the ratios of shed to within host virus, such that on most days the standard error overlapped for genotypes HV and LV, indicating no significant difference between the genotypes. The exception to this was day 5 and 6 where genotype HV appeared to have a lower ratio of within host to shed virus compared to genotype LV, but a statistical comparison could not be made because genotype HV shedding fell below the threshold of detection. When comparing the percentage offish positive for shed viral RNA to the percentage positive for within host viral RNA, there was no significant difference for genotype HV on days 1–4 and genotype LV on days 1–6 (P > 0.05). This pattern then changed such that on days 5 and 6 for genotype HV and day 7 for genotype LV, there were significantly more fish positive for within host than shed viral RNA (Fisher exact test, P < 0.001). These patterns collectively indicate that in the peak period more virus was shed than harbored in the host, and in the post-peak period an equal amount or more virus is harbored in the host than is shed. In some cases, there was a higher percentage of fish shedding virus than positive for within host virus, although this was never statistically significant. This trend was possible because the shedding data was not taken from the same fish as the within host data. These comparisons were only made for single infections because within host virus was not quantified in mixed infections.
Fig. 7.
Ratio of shed to within host virus. Lines show mean total viral RNA shed within tanks (virus RNA copies/ml *1460ml tank) divided by mean total viral RNA within fish (virus RNA copies/g * weight of fish), for each genotype on each day. Samples negative for virus RNA were not included in the calculation. Genotypes HV and LV are shown in black and dotted grey respectively. Error bars show ±1 standard error, which was calculated by the formula , where X = shed viral load, and Y = within host viral load. The Y-axis is on the log scale. Fractions above figure show percent offish shedding detectable virus/percent offish positive for within host virus, for genotype HV (black, top row) and genotype LV (grey, bottom row). Within host and shed viral copies are from different experiments. Data from the two within host replication experiments was pooled to generate the data in the figure. On days 5 and 6 no detectable virus was shed by genotype HV, so the ratio of shed to within host virus dropped to approximately 0. This was indicated by setting the values to below the y-axis minimum, because an exact ratio could not be calculated. Since mixed infections were not conducted for within host experiments, the ratio of shed to within host virus in mixed infections could not be calculated.
4. Discussion and conclusions
The replication and shedding kinetics of a pathogen can provide valuable insights into its overall prevalence and transmission in the field, which is vital for informing disease epidemiology and management (LaPatra et al., 2001; Bjornstad et al, 2002; Ogbunugafor et al, 2010; Garver et al, 2013). Studies of these traits can also be used to elucidate how other pathogen traits, such as virulence, might be linked to fitness and thus evolve (Alizon et al., 2009; Wargo and Kurath, 2012). Here, two genotypes of the pathogen IHNV, previously shown to differ in virulence, were examined for their replication and shedding kinetics in rainbow trout. In general, both within host and shed viral loads approached peak values very rapidly, by day 2 post-exposure to virus. Within host cumulative viral loads remained around maximum levels and did not significantly increase or decrease for 3–7 days, despite qualitative trends suggesting otherwise (Fig. 1). In contrast, daily viral shedding tapered off quickly, such that it dropped by 2–3 orders of magnitude from day 2 to 5 post-exposure. These kinetics created a peak (day 1–4) and post-peak (days 5 onward) period in shedding, akin to what is seen in other viruses such as HIV (Piatak et al., 1993). This was observed in both the amount of virus shed per fish and the number of fish shedding. The findings here indicate that for IHNV the majority of shedding occurs well before host mortality begins. Whether this is an evolved strategy of the host or the virus in unknown.
The shedding dynamics observed here were different from those found in the only other study of IHNV shedding kinetics, where shedding by infected Atlantic salmon did not become detectable until day 4 and peaked between day 7–10 post-exposure to virus (Garver et al, 2013). There were several differences between our study and this previous one, such as host species, holding temperatures, and use of IHNV strains from different phylogenetic genogroups, all of which are known to influence the replication and virulence of the virus (Garver et al., 2006) and thus potentially shedding. The rapid replication kinetics and early peak viral loads presented here were consistent with a previous study of within host replication of IHNV genotype HV in the same host species O. mykiss (Peñaranda et al., 2009). Collectively, these studies highlight the importance of environmental, host, and virus factors in driving pathogen transmission.
To our knowledge, this is the first published study of IHNV to link replication kinetics in the host with shedding kinetics through time. The data indicated that in general, most of the virus produced is shed into the environment rather than harbored in the host. This suggests that the virus maximizes its fitness by investing heavily in transmission rapidly after infection. However, the ratio of shed to within host virus dropped substantially in the post-peak period of infection, indicating that much less of the virus produced in the host was shed. Why this occurred is unknown. A potential factor could be the destruction of viral RNA within the host by the immune response just prior to packaging and release of virions, perhaps in specific tissues from which shedding occurs. This is supported by the finding that host innate immunity typically peaks and remains at high levels around 48 h post-infection (Fig. 2), when shedding begins to decline (Purcell et al., 2010). Shedding may also be limited by host tissue damage, such as necrosis of the kidneys, which inhibits fish urination (Amend and Smith, 1975) and is the main cause of mortality in IHNV infected fish (Bootland and Leong, 2011). This hypothesis is supported by the finding that the post-peak shedding rate reached minimal levels about the time morbidity and mortality began, around day 5 post-infection (Garver et al, 2006). Furthermore, during the post-peak period for shedding, it is unknown if the maintenance of cumulative within host viral load levels at or below maximum levels represents a lack of new viral replication, or if there is continued replication that is counterbalanced by degradation. A decline in active viral RNA replication could contribute to a drop in shedding if only actively replicating or recently synthesized viral genomic RNA is packaged into virions. These potential mechanisms driving the decline in shedding warrant further investigation.
When comparing the two IHNV genotypes studied here, it was found that within host replication and shedding kinetics were similar for the first two days of infection. After this time point, the amount of virus shed dropped significantly faster for the low virulence genotype (LV) compared to the high virulence genotype (HV). Furthermore, within host viral load and the number of fish shedding did not significantly differ between the genotypes except at day 12, at the end of the shedding experiment, when the number of fish positive for within host virus was significantly lower for genotype LV than HV. Thus, fish appear to be reducing the shedding of LV more rapidly than genotype HV, through potentially a combination of clearance and shedding blockage of within host virus. Exceptions to this pattern were on day 5, 6, and 9 post-exposure, where none of the fish exposed to HV alone were shedding detectable virus. It is unclear why the shedding of HV in single infections dropped so low on these days and then rebounded to detectable levels of virus afterwards. Interestingly, all fish, regardless of treatment, dropped below the detectable level of shedding at some point and then the majority resumed detectable shedding within the 12-day observation period. Furthermore, on days 6 and 7 genotype HV was shed at detectable levels from mixed infections. It has been reported for a related fish rhabdovirus, viral hemorrhagic septicemia virus, that fish that have stopped detectable shedding for an extended period of time may resume shedding upon stress (Kim and Faisal, 2012). It is possible that the temperature fluctuations in our study caused stress on the fish and thus impacted shedding. Regardless, even if genotype HV shed virus on days 5–6, the overall pattern would have remained the same: genotype HV shed more total virus than genotype LV over the entire course of infection. Furthermore, if day was excluded from the analysis, genotype HV shed higher peak quantities of virus than genotype LV. Thus genotype HV appeared to have the overall fitness advantage. This is consistent with previous studies of this system, although the fitness difference in those studies was larger than that observed here (Wargo et al, 2010; Wargo and Kurath, 2011). It may be that the fitness differences were less here because average temperatures were lower (Garver et al., 2006), but this remains to be further investigated.
In addition to a genotype difference, there was indication of a competition effect on viral shedding, such that, when examining just the fish shedding detectable virus, both genotypes shed at a significantly higher rate per day in single infections than they did in mixed infections (Fig. 4B). Interestingly, the effect of competition was the same for each genotype, such that one genotype did not have greater reductions in viral shedding rate compared to the other. However, this competition effect was not strong enough to impact the total amount of virus shed, which was not found to be significantly different between single and mixed infections (Fig. 4D). This is likely because although the overall rate of shedding averaged across the entire 12 days was higher for single compared to mixed infections, there was no difference in the peak of viral shedding, and on average 65 ± 5% of the total shed virus was shed on the peak day (Fig. 4B). This is in line with previous studies which showed a lack of competition effect at the peak of within host viral loads (Wargo et al, 2010; Wargo and Kurath, 2011). As such, this data suggests that competition only becomes apparent during the post-peak phase of the infection. This is further supported by the finding that the within host viral loads offish on day 12 of the shedding study were higher for single compared to mixed infections. The mechanisms driving this hypothesized delayed competition dynamic are unknown. One possibility is that the virus genotypes are competing for entry into cells, which only occurs when naive cells become scarce in the post-peak infection period. This is supported by the finding that the total amount of virus produced in a mixed infection (combined quantity of HV and LV) was equal to that produced by the most prolific single infection, genotype HV alone, suggesting there is an upper limit to the total amount of virus that can be produced in a host of this size. Previous studies have also indicated that there is superinfection exclusion in this system such that viral genotypes are restricted from initiating a secondary infection in a host that is already infected with a different viral genotype, when the exposure time between the genotypes is 12 h or greater (Kell et al., 2013). If this is also occurring at the cellular level, it may indicate why there is a delay in the observed competition effect in this study. Competition may also be mediated by the innate immune response, which can take 24–48 h to fully activate in fish (Purcell et al., 2012). The studies on superinfection indicated that the interferon response was likely not the primary factor responsible for competitive exclusion (Kell et al., 2013, 2014), but it may be that other constitutive immune mechanisms are at play (Purcell et al., 2010, 2011). Ultimately, the mechanisms shaping competition for IHNV require further study.
The mechanisms driving mortality in this system are also an area that warrants further investigation. Mortality in this study was most correlated with the length of time a fish was shedding virus, such that fish that shed detectable virus for a greater percentage of the days that they were alive, had a higher incidence of mortality. In contrast, the peak amount of virus shed by fish that died was no different from those that survived. However, it was also found that fish that died had higher within host viral loads than those that survived, at the end of the study on day 12. As such, there may be an interplay between peak or persistence of within host IHNV loads and shedding duration, which is correlated with fish immune-pathology (Purcell et al., 2012) and mortality. The cause and effect of these relationships remain to be elucidated. There was also found to be a lower incidence of mortality for fish infected with genotype LV alone and mixed infections of the two genotypes, compared to genotype HV alone. This result supports previous studies showing lower virulence for genotype LV compared to HV (Garver et al., 2006). It was surprising that mixed infections had a lower incidence of mortality than genotype HV, but it should be noted this difference was small (2 compared to 4 fish), and the experiment was completed before the bulk of mortality typically occurs (days 14–21). It is expected that mixed infections would show equal or greater mortality than single infections of genotype HV if the experiment extended for 30 days. Interestingly, all the fish that died shed detectable virus after death, in some cases for up to 5 days. The virus shed from dead fish was not included in the analyses because the infectivity of the virus could not be verified. This had no impact on the overall patterns observed, because very few fish died, and the bulk of shedding occurred before mortality began. If virus shed from dead fish is infectious this would reinforce our finding that host mortality has a minimal impact on reducing vial transmission and fitness in this system.
Overall, this study indicated that viral shedding for IHNV is a very acute process that begins with a rapid peak after exposure to virus and tapers off quickly to a post-peak period of lower shedding magnitude that can extend for at least 12 days. This comprises the first detailed characterization of the magnitude and kinetics of IHNV shedding from rainbow trout. It is also the first report to present aquatic pathogen shedding patterns from individual fish, providing insight into the variability of different shedding measures on a fish-to-fish basis. The high level of variability between fish in shedding kinetics illustrates the importance of considering individual hosts when trying to infer epidemiological patterns. For example, the total amount of virus shed ranged from 5.1 × 104 to 9.5 × 106 virus copies/ml H2O for genotype HV, and 5.0 × 103 to 2.2 × 106 virus copies/ml H2O for genotype LV. The duration of shedding was likewise highly variable, ranging from 3 to 9 of the 12 days, much of which could be attributed to variation in the post-peak period. The sources of fish-to-fish variation have not been defined, but previous studies have indicated that host genetic variation is not the only contributing factor (Wargo et al., 2012). Fish size is likely to be a factor associated with viral load variability (LaPatra et al., 1990), such that larger fish typically have higher viral loads than smaller fish, likely due to greater resource availability for the virus. This was controlled for here by using fish uniform in size, ranging from 1 to 3 g. If the fish in this study had been larger, within and shed viral loads might have also been higher, and the kinetics may have been different. This is an area that warrants further investigation.
These findings suggest that viral transmission may primarily occur within the first week of infection and management decisions must be swift. However, it should be noted that although mean shedding tapered consistently in the post-peak period, some fish were still shedding detectable virus on day 12, at the end of this study. It is important to consider how shedding may continue over longer periods of time and what epidemiological impact this could have. Even though such persistent shedders may be rare, they could provide a vital role in the transmission and movement of the virus over large temporal and spatial scales. Expanded studies with different IHNV genotypes and different hosts will be essential to assess if shedding kinetics is a variable viral phenotype. Analysis of variation in individual fish shedding patterns as well as mean shedding kinetics is important for understanding the ecological factors that may drive the virus to evolve to become an acute or persistent shedder, which would be a valuable goal.
Acknowledgments
We would like to thank Alison Kell, Maureen Purcell, Rachel Breyta, Douglas McKenney, and Jim Winton for valuable discussions. We are grateful to Scott LaPatra of Clear Springs Foods Inc. for providing research grade rainbow trout. We also thank an anonymous reviewer for insightful comments. This work was supported by the USGS Western Fisheries Research Center, the University of Washington, National Institute of Health R.L. Kirschstein NRSA University of Washington Institutional Service Award 5 T32 AI007509 and National Science Foundation Ecology of Infectious Diseases grant 0812603. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mention of trade names does not imply U.S. Government endorsement.
References
- Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Amend DF, Smith L. Pathophysiology of infectious hematopoietic necrosis virus disease in rainbow trout: hematological and blood chemical changes in moribund fish. Infect. Immun. 1975;11:171–179. doi: 10.1128/iai.11.1.171-179.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri S, Zwanzig S. An Improvement of the nonparametric bootstrap test for the comparison of the coefficient of variations. Commun. Stat. Simul. 2010;39:1726–1734. [Google Scholar]
- Amiri S, Zwanzig S. Assessing the coefficient of variations of chemical data using bootstrap method. J. Chemom. 2011;25:295–300. [Google Scholar]
- Anderson RM, May RM. Population biology of infectious diseases: part I. Nature. 1979;280:361–367. doi: 10.1038/280361a0. [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM. Coevolution of host and pathogen. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Baigent SJ, Kgosana LB, Gamawa AA, Smith LP, Read AF, Nair VK. Relationship between levels of very virulent MDV in poultry dust and in feather tips from vaccinated chickens. Avian Dis. 2013;57:440–447. doi: 10.1637/10356-091012-Reg.1. [DOI] [PubMed] [Google Scholar]
- Bebak J. Infectious pancreatic necrosis virus: transmission from infectious to susceptible rainbow trout fry. J. Aquat. Anim. Health. 1998;10:287–293. [Google Scholar]
- Bjornstad ON, Finkenstadt BF, Grenfell BT. Dynamics of measles epidemics: estimating scaling of transmission rates using a Time series SIR model. Ecol. Monogr. 2002;72:169–184. [Google Scholar]
- Bootland LM, Leong JC. Infectious hematopoietic necrosis virus. In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders. Vol. 3. CAB International; 1999. pp. 57–112. [Google Scholar]
- Bootland LM, Leong JC. Infectious hematopoietic necrosis virus. In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders. Vol. 3. CAB International; 2011. pp. 66–110. [Google Scholar]
- Diekmann O, Heesterbeek JAP, Metz JAJ. On the definition and the computation of the basic reproduction ratio R0 in models for infectious-diseases in heterogeneous populations. J. Math. Biol. 1990;28:365–382. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- Drolet BS, Rohovec JS, Leong JC. The route of entry and progression of infectious hematopoietic necrosis virus in Oncoryhynchus-mykiss (Walbaum)—a sequential immunohistochemicalstudy. J. Fish Dis. 1994;17:337–347. [Google Scholar]
- Fenner F, Ratcliffe R. Myxomatosis. London: Cambridge University Press; 1965. [Google Scholar]
- Fijan N, Sulimanovic E, Bearzotti M, Muzinic D, Zwillenberg LO, Chilmonczyk S, Vautherot JF, de Kinkelin P. Some properties of the epithelioma papulosum cyprini (EPC) cell line from carp Cyprinus carpi . Ann. Inst. Pasteur Vir. 1983;134:207–220. [Google Scholar]
- Garver K, Batts W, Kurath G. Virulence comparisons of infectious hematopoietic necrosis virus U and M genogroups in sockeye salmon and rainbow trout. J. Aquat. Animal Health. 2006;18:232–243. doi: 10.1577/H05-038.1. [DOI] [PubMed] [Google Scholar]
- Garver KA, Mahony AAM, Stucchi D, Richard J, Van Woensel C, Foreman M. Estimation of parameters influencing waterborne transmission of infectious hematopoietic necrosis virus (IHNV) in Atlantic Salmon (Salmo salar) PLoS One. 2013;8:e82296. doi: 10.1371/journal.pone.0082296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmache A, LeBerre M, Droineau S, Giovannini M, Bremont M. Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus. J. Virol. 2006;80:3655–3659. doi: 10.1128/JVI.80.7.3655-3659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger P, Gregg J, Grady C, Collins R, Winton J. Kinetics of viral shedding provide insights into the epidemiology of viral hemorrhagic septicemia in Pacific herring. Mar. Ecol.-Prog. Ser. 2009;400:187. [Google Scholar]
- Kell AM, Wargo AR, Kurath G. The role of virulence in in vivo superinfection fitness of the vertebrate RNA virus infectious hematopoietic necrosis virus. J. Virol. 2013;87:8145–8157. doi: 10.1128/JVI.00089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell AM, Wargo AR, Kurath G. Viral fitness does not correlate with three genotype displacement events involving infectious hematopoietic necrosis virus. Virology. 2014;464:146–155. doi: 10.1016/j.virol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DA, Kurath G, Brito IL, Purcell MK, Read AF, Winton JR, Wargo AR. Potential drivers of virulence evolution in aquaculture. Evol. Appl. 2016;9:344–354. doi: 10.1111/eva.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermack WO, McKendrick AG. A contribution to the mathematical theory of epidemics. P. Roy. Soc. Lond. B Bio. 1927;115:700–721. [Google Scholar]
- Kim R, Faisal M. Shedding of viral hemorrhagic septicemia virus (Genotype IVb) by experimentally infected muskellunge (Esox masquinongy) J. Microbiol. 2012;50:278–284. doi: 10.1007/s12275-012-1145-2. [DOI] [PubMed] [Google Scholar]
- Kocan R, Bradley M, Elder N, Meyers T, Batts WN, Winton J. North American strain of viral hemorrhagic septicemia virus is highly pathogenic for laboratory-reared Pacific herring. J. Aquat. Anim. Health. 1997;9:279–290. [Google Scholar]
- Kurath G, Garver KA, Troyer RM, Emmenegger EJ, Einer-Jensen K, Anderson ED. Phylogeography of infectious hematopoietic necrosis virus in North America. J. Gen. Virol. 2003;84:803–814. doi: 10.1099/vir.0.18771-0. [DOI] [PubMed] [Google Scholar]
- LaPatra SE, Groberg WJ, Rohovec JS, Fryer JL. Size-related susceptibility of salmonids to two strains of infectious hematopoietic necrosis virus. Trans. Am. Fish. Soc. 1990;119:25–30. [Google Scholar]
- LaPatra SE, Batts WN, Overturf K, Jones GR, Shewmaker WD, Winton JR. Negligible risk associated with the movement of processed rainbow trout, Oncorhynchus mykiss (Walbaum), from an infectious haematopoietic necrosis virus (IHNV) endemic area. J. Fish Dis. 2001;24:399–408. [Google Scholar]
- Madetoja J, Nyman P, Wiklund T. Flavobacterium psychrophilum, invasion into and shedding by rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 2000;43:27–38. doi: 10.3354/dao043027. [DOI] [PubMed] [Google Scholar]
- McKibben CL, Pascho RJ. Shedding of Renibacterium salmoninarum by infected chinook salmon Oncorhynchus tschawytscha. Dis. Aquat. Organ. 1999;38:75–79. doi: 10.3354/dao038075. [DOI] [PubMed] [Google Scholar]
- Mulcahy D, Pascho RJ, Jenes CK. Detection of infectious hematopoietic necrosis virus in river water and demonstration of waterborne transmission. J. Fish Dis. 1983;6:321–330. [Google Scholar]
- Munster VJ, de Wit E, van den Brand JMA, Herfst S, Schrauwen EJA, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus A, Fouchier RAM. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbunugafor CB, Basu S, Morales NM, Turner PE. Combining mathematics and empirical data to predict emergence of RNA viruses that differ in reservoir use. Philos. Trans. R. Soc. B Biol. Sci. 2010;365:1919–1930. doi: 10.1098/rstb.2010.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñaranda MMD, Purcell MK, Kurath G. Differential virulence mechanisms of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J. Gen. Virol. 2009;90:2172–2182. doi: 10.1099/vir.0.012286-0. [DOI] [PubMed] [Google Scholar]
- Piatak M, Saag MS, Yang LC, Clark SJ, Kappes JC, Luk KC, Hahn BH, Shaw GM, Lifson JD. High-levels of HIV-1 plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, Core Team R. nlme: Linear and Nonlinear Mixed Effects Models. 2015 [Google Scholar]
- Purcell M, Kurath G, Garver K, Herwig R, Winton JR. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immun. 2004;17:447–462. doi: 10.1016/j.fsi.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Garver KA, Conway C, Elliott DG, Kurath G. Infectious haematopoietic necrosis virus genogroup-specific virulence mechanisms in sockeye salmon, Oncorhynchus nerka (Walbaum), from Redfish Lake, Idaho. J. Fish Dis. 2009;32:619–631. doi: 10.1111/j.1365-2761.2009.01045.x. [DOI] [PubMed] [Google Scholar]
- Purcell MK, LaPatra SE, Woodson JC, Kurath G, Winton JR. Early viral replication and induced or constitutive immunity in rainbow trout families with differential resistance to Infectious hematopoietic necrosis virus (IHNV) Fish Shellfish Immun. 2010;28:98–105. doi: 10.1016/j.fsi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Marjara IS, Batts W, Kurath G, Hansen JD. Transcriptome analysis of rainbow trout infected with high and low virulence strains of Infectious hematopoietic necrosis virus. Fish Shellfish Immun. 2011;30:84–93. doi: 10.1016/j.fsi.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Laing K, Winton JR. Immunity to fish rhabdoviruses. Viruses. 2012;4:140–166. doi: 10.3390/v4010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell MK, McKibben CL, Pearman-Gillman S, Elliott DG, Winton JR. Effects of temperature on Renibacterium salmoninarum infection and transmission potential in Chinook salmon, Oncorhynchus tshawytscha (Walbaum) J. Fish Dis. 2016;39:787–798. doi: 10.1111/jfd.12409. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Rose A, Ellis A, Munro A. The infectivity by different routes of exposure and shedding rates of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon Salmo salar L., held in sea water. J. Fish Dis. 1989;12:573–578. [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Therneau T. A package for survival analysis in S. 2013 [Google Scholar]
- Traxler GS, Roome JR, Kent ML. Transmission of infectious hematopoietic necrosis virus inseawater. Dis. Aquat. Org. 1993;16:111–114. [Google Scholar]
- Troyer RM, Kurath G. Molecular epidemiology of infectious hematopoietic necrosis virus reveals complex virus traffic and evolution within southern Idaho aquaculture. Dis. Aquat. Organ. 2003;55:175–185. doi: 10.3354/dao055175. [DOI] [PubMed] [Google Scholar]
- Van der Goot JA, De Jong MCM, Koch G, Van Boven M. Comparison of the transmission characteristics of low and high pathogenicity avian influenza A virus (H5N2) Epidemiol. Infect. 2003;131:1003–1013. doi: 10.1017/s0950268803001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo AR, Kurath G. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J. Virol. 2011;85:3959–3967. doi: 10.1128/JVI.01891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo AR, Kurath G. Viral fitness: definitions, measurement, and current insights. Curr. Opin. Virol. 2012;2:538–545. doi: 10.1016/j.coviro.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo AR, Garver KA, Kurath G. Virulence correlates with fitness in vivo fortwo M group genotypes of infectious hematopoietic necrosis virus (IHNV) Virology. 2010;404:51–58. doi: 10.1016/j.virol.2010.04.023. [DOI] [PubMed] [Google Scholar]
- Wargo AR, Kell AM, Scott RJ, Thorgaard GH, Kurath G. Analysis of host genetic diversity and viral entry as sources of between-host variation in viral load. Virus Res. 2012;165:71–80. doi: 10.1016/j.virusres.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. Fish Viruses and Fish Viral Diseases. Ithaca, NY: Cornell University Press; 1988. Infectious hematopoietic necrosis virus; pp. 83–114. [Google Scholar]
- Yamamoto T, Batts WN, Arakawa CK, Winton JR. Multiplication of infectious hematopoietic necrosis virus in rainbow trout following immersion infection: whole-body assay and immunohistochemistry. J. Aquat. Anim. Health. 1990;2:271–280. [Google Scholar]