Abstract
Background
We investigated the relationship of the polymorphisms of SET and MYND domain-containing protein 3 (SMYD3) with risk and prognosis of ovarian cancer.
Material/Methods
The polymerase chain reaction (PCR) amplification method was applied to detect the polymorphisms of variable number of tandem repeats (VNTR) in the SMYD3 gene promoter region for 156 patients with ovarian cancer (case group) and 174 healthy people (control group). Quantitative reverse transcription polymerase chain reaction and Western blot were applied to detect SMYD3 mRNA and protein expressions.
Results
The frequencies of VNTR genotype 3/3 and allele genotype 3 in the case group were significantly higher than those in the control group, while the frequency of genotype 2/2 in the control group was significantly higher than that in case group (all P<0.05). The proportion of poorly differentiated patients carrying VNTR genotype 3/3 was significantly higher than the proportion of poorly differentiated patients carrying VNTR genotype 2/2+2/3, while the proportion of patients carrying genotype 3/3 with International Federation of Gynecology and Obstetrics (FIGO) stage III–IV disease was significantly higher than the proportion of patients carrying genotype 2/2 +2/3 with FIGO stage III–IV disease (all P<0.05). SMYD3 mRNA and protein expressions were higher in the patients carrying genotype 3/3 than they were in the patients with the 2/2+2/3 genotype (all P<0.05). The 5-year survival rate for patients carrying VNTR genotype 3/3 was significantly lower than that of patients carrying genotype 2/2+2/3, and Cox regression analysis showed that VNTR genotype 3/3 was an independent risk factor for ovarian cancer prognosis (all P<0.05).
Conclusions
VNTR genotype 3/3 of the SMYD3 gene was associated with the risk of ovarian cancer. The polymorphism of VNTR genotype could be recognized as an indicator for the poor prognosis of patients with ovarian cancer.
MeSH Keywords: Disease Susceptibility; Ovarian Neoplasms; Polymorphism, Genetic; Risk Assessment
Background
Ovarian cancer is the sixth most common cancer worldwide and also one of the reproductive system cancers with the highest incidence of mortality for women, accounting for 4.2% of the female cancer fatalities [1]. As the etiology of ovarian cancer remains controversial and the majority of patients are initially diagnosed with advanced ovarian cancer often accompanied by tumor metastasis, traditional surgeries and platinum-based chemotherapies fail to completely eradicate the tumor [2,3]. To the best of our knowledge, cancer antigen 125 (CA-125) exhibited a sensitivity of 80% along with a specificity of 97% in stage III or IV, while its sensitivity is just 30% in stage I; thus, CA-125 fails to be as a perfect biomarker for the diagnosis of ovarian cancer in this context [4,5]. The prognosis of ovarian cancer is unfavorable when the cancer cells have metastasized into other organs. For patients diagnosed at the early stage, the 5-year overall survival rate is approximately 90%, while patients in the advanced stage have a 5-year overall survival rate of less than 20% [6]. Therefore, investigation of the pathogenesis of ovarian cancer plays an important role in the treatment and prevention of the cancer [7].
Epigenetic alteration has a certain relationship with the occurrence of tumors; for instance, the dysfunction caused by histone modifications can regulate the function of chromatin and the gene expression [8]. SET and MYND domain-containing protein 3 (SMYD3), a histone methyltransferase, is involved in the development and progression of tumors [9,10]. SMYD3 has two functional domains: SET and MYD. The core SET domain consists of 130 amino acids with the same function as the methyl transferase enzyme, which can promote the chromosomal histone H3K4 to form dimethylation or trimethylation, so as to facilitate a loosened state of the chromosome’s spatial structure [11]. The zinc finger domain of myeloid translocation protein 8, Nervy, and DEAF1 (MYND) can bind to the promoter region of specific gene of 5′-CCCTCC-3 ‘or 5′-GGAGGG-3′, so as to promote the methylation function of the SET domain, which can affect gene transcription [12]. It was reported that the variable number of tandem repeats (VNTR) [(CCGCC) n] in the promoter region of SMYD3 can increase the risk of esophageal squamous cell carcinoma [13] and chronic lymphocytic leukemia [14]. However, no research is reported on the association between ovarian cancer and the polymorphism of the SMYD3 gene. In the current study, polymerase chain reaction (PCR) technology was applied to quantitatively analyze the VNTR polymorphism of the SMYD3 gene in ovarian cancer cases to explore the relevance of SMYD3 polymorphisms to the risk of ovarian cancer and its prognosis.
Material and Methods
Ethical statement
This study was performed with the approval of ethnics committee in The Affiliated Huaian Hospital of Xuzhou Medical University. Written informed consent was collected from all patients in this research.
Study subjects
A total of 156 patients with ovarian cancer confirmed by histopathological and radiological diagnosis were selected as the case group from March 2007 to March 2010. The patients in the case group were from 22 to 78 years old, with a mean age of 50.5±12.2 years; age at first menstruation ranged from 11 to 19 years (mean: 14.5±2.3 years); age at first childbearing ranged from 16 to 33 years (mean: 22.6±4.3 years); and age at last childbearing ranged from 26 to 43 years (mean: 34.4±6.1 years). There were 106 premenopausal patients and 50 postmenopausal patients, and 53 of them had a history of abortion. According to the classification criteria of the International Federation of Gynecology and Obstetrics (FIGO) [15], these 156 patients can be classified into the following types: 96 cases of highly differentiated serous adenocarcinoma, 17 cases of endometrial adenocarcinoma, 18 cases of clear cell carcinoma, 19 cases of mucinous adenocarcinoma, and 6 cases of poorly differentiated serous adenocarcinoma. The patients were enrolled if a diagnosis of primary ovarian epithelial cancer was pathologically confirmed at the Affiliated Huaian Hospital of Xuzhou Medical University and the level of serum CA-125 was determined. Patients with other gynecological malignancies or systemic malignant tumors were excluded from the current study. All specimens obtained by surgical excision from the case group were pathologically confirmed and then stored at −80° for the detection of SMYD mRNA and protein expressions.
A total of 174 healthy women undergoing normal medical examinations in The Affiliated Huaian Hospital of Xuzhou Medical University were included as the control group. The patients in the control group were from 21 to 81 years old, with a mean age of 49.5±13.3 years; age at first menstruation ranged from 10 to 18 years (mean: 14.1±1.9 years); age at first childbearing ranged from 17 to 29 years (mean: 24.5±2.7 years); and age at last childbearing ranged from 23 to 46 years (mean: 34.0±5.3 years). There were 155 premenopausal women and 19 postmenopausal women, 62 of whom had a history of abortion. The inclusion criteria were as follows: acceptance of a gynecological physical examination, cervical cytology and gynecological ultrasound examination that showed no abnormality, and determination of serum CA-125 level. Women who had a history of ovarian cancer or gynecologic malignancies and systemic malignant tumor were removed from the control group. The ages of people in the control group were matched with the ages of patients in the case group. All the selected ones were Han people. Minorities and foreign patients were excluded.
Collection of peripheral venous blood specimen and extraction of genomic DNA
Peripheral venous blood specimens from patients in the case group and the control group were extracted and cryopreserved with the addition of 2% ethylene diamine tetraacetic acid (EDTA) anticoagulant. After the collection of samples, unified genomic DNA extractions were implemented with the blood genomic DNA extraction kit (Tiangen Biotech [Beijing] Co., Ltd.) according to the kit instructions. PCR analysis was applied after DNA extraction, and long-term preservation of the DNA samples in a refrigerator at −40°C was implemented afterwards.
Detection of SMYD3 polymorphisms through PCR
The primer sequences were as follows: F: 5′-GGCGTCT CACGGGCTGCCGGG-3; and R: 5′-CGGAGCCTTACGACCACCTTC-3′. There were 50 μL of PCR reaction system (purchased from Beijing Biomed Co. Ltd.) where there were 6 μL of human peripheral blood genomic DNA, 0.4 μL of Taq enzyme, 5 μL of 10× PCR buffer, and 2.5 μL of 5 moL/L deoxynucleotide mixture. Both the forward primer and the reserve primer were 80 pmol/L. The PCR reaction condition was initial denaturation at 94°C for 5 min, followed by 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s. The total circulation was 35 cycles with 7 min of extension at a temperature of 72°C. Electrophoresis was applied to the products of PCR amplification for 40 min at the voltage of 120 V with 3% agarose gel to analyze the enzyme-digested products, which were then observed and photographed under ultraviolet light at 300 nm. After gel electrophoresis testing, sequence detection was performed on the amplified PCR products using the 3730XL automatic genetic analyzer produced by US Apllid Biosystems Company.
Real-time quantitative polymerase chain reaction
Total RNA extraction was performed using the TRIzol one-step method, and then cDNA by reverse transcription was subject to real-time quantitative polymerase chain reaction (RT-qPCR). The primers were synthetized by Thermo Fisher Scientific (USA). They were as follows: SMYD3: forward, 5′-TGAATGTGACTGTTTCCGTTGC-3′; reverse, 5′-ATTGCTGC TTATGATCGCCTGG-3′; 172 bp. β-Actin (internal reference): forward, 5′-GAACGGTGAAGGTGACAG-3′; reverse, 5′-TAGAGAGA AGTGGGGTGG-3′. The reaction parameters were as follows: 95° for 2 min; 94° for 30 s; 57° for 30 s; 72° for 30 s, 45 cycles; 72° for 5 min. Obtained data was used for evaluation of sample difference after adjustment by 2−ΔΔCt.
Western blot
The total protein was extracted from 0.5 mg of ovarian cancer tissue, and the Bradford kit (Beyotime Biotechnology, Shanghai, China) was also used to detect the total protein content. Quantitative sample protein (20 μg in each well) was placed into sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which then was transferred into bromophenol blue at 200 V electrophoresis and underwent trarsmembrane for 70 min at 300 mA until 0.5 cm distant from the bottom of the separation gel. Well-prepared nitrocellulose membrane was shaken and then sealed in dried skimmed milk for 2 hours. With the addition of rabbit anti-human SMYD3 (1:400, Santa Cruz Company, USA) as the first antibody, the membrane was incubated at 4° overnight and then washed three times (each for 10 min) in Tris-buffered saline (TBS) buffer. The membrane with the addition of horseradish peroxidase-labeled second antibody (1:1000) was shaken for 1 hour, and then was washed three times (each for 10 min) in TBS buffer. After chemiluminescence and film exposure, the results were imaged using an ultraviolet polarimeter scanner.
Follow-up
Follow-up was done with all 156 patients for 5 years by telephone and questionnaire. At the end of the study, 53 patients had relapsed, 65 patients had died, 11 patients had lost contact, and 27 patients were found in a healthy condition. A total of 151 patients received routine oophorectomy and adjuvant chemotherapy, and only 5 cases of patients received conservative treatment.
Statistical analysis
The SPSS 19.0 statistical software package (IBM, USA) was used for data processing. The comparisons of tumor classification, differentiation, and FIGO staging were detected by the Wilcoxon rank-sum test, and the comparisons of other counting data were detected by the chi-square test. The measurement data were represented as mean±standard deviation. The t-test was applied for comparisons between the two groups; the chi-square test was applied to check whether the genotype was in accordance with the Hardy-Weinberg equilibrium. Multivariate logistic regression analysis was applied to determine ovarian cancer risk. The Kaplan-Meier method was applied to detect the survival curve, and Cox regression analysis was used for risk assessment. P<0.05 was considered to be statistically significant.
Results
General characteristics and clinical-pathological features
We summarized and analyzed the general characteristics of the study subjects (Table 1). First childbearing age of patients in the case group was significantly younger than that in the control group, and the proportion of menopausal patients in the case group was significantly increased compared with the proportion in the control group (all P<0.05). The other indicators, including age, age at first menstruation, last childbearing age, and abortion history, were not significantly different between the case group and control group (all P>0.05).
Table 1.
General characteristics of study subjects.
| Parameter | The case group (n=156) | The control group (n=174) | χ2 | P |
|---|---|---|---|---|
| Mean age (SD) | 50.5±12.2 | 49.5±13.3 | 0.709 | 0.478 |
| First menstruation age (SD) | 14.5±2.3 | 14.1±1.9 | 1.729 | 0.085 |
| First childbearing age (SD) | 22.6±4.3 | 24.5±2.7 | 4.858 | < 0.001 |
| Last childbearing age (SD) | 34.4±6.1 | 34.0±5.3 | 0.637 | 0.524 |
| Menstrual condition | ||||
| Premenopause | 106 (68.0%) | 155 (89.1%) | 22.21 | <0.001 |
| Postmenopause | 50 (32.0%) | 19 (10.9%) | ||
| Abortion history | ||||
| Yes | 53 (34.0%) | 62 (35.6%) | 0.01 | 0.752 |
| No | 103 (66.0%) | 112 (64.4%) | ||
| Tumor classification | ||||
| HGSC | 96 (61.5%) | |||
| EC | 17 (10.9%) | |||
| CCC | 18 (11.5%) | |||
| MC | 19 (12.2%) | |||
| LGSC | 6 (12.2%) | |||
| FIGO staging | ||||
| I–II | 100 (64.1%) | |||
| III–IV | 56 (35.9%) | |||
| Tumor differentiation | ||||
| Middle and high differentiation | 108 (69.2%) | |||
| Low differentiation | 48 (30.8%) | |||
Age was represented as mean ± standard deviation; EC – endometrial carcinoma; CCC – clear cell carcinoma; MC – mucin-like cell carcinoma; HGSC – high-grade serous adenocarcinoma; LGSC – low-grade serous adenocarcinoma; FIGO – International Federation of Gynecology and Obstetrics.
SMYD3 genotype determination and sequences
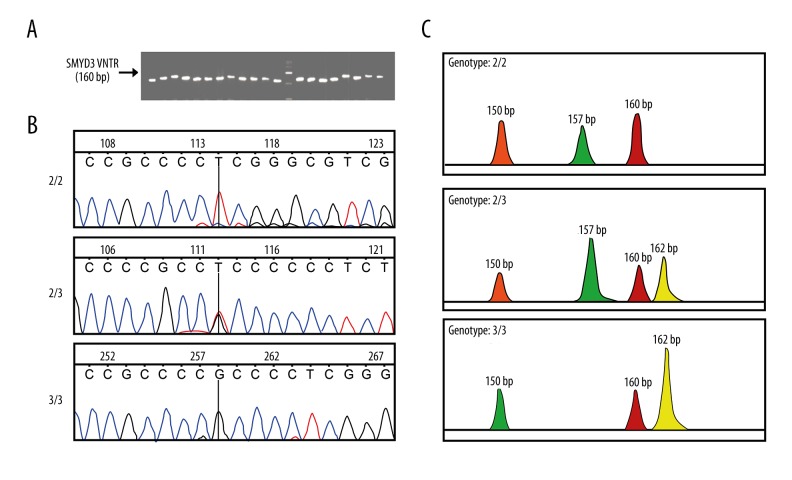
After the completion of the PCR reaction, electrophoresis was applied; comparison of the amplified product bands and the DL2000 DNA Marker could determine whether or not they were the expected PCR products (Figure 1A). The sequence detection of the products indicated that there were three genotypes: 2/2, 2/3, and 3/3; these are shown in Figure 1B and 1C.
Figure 1.
SMYD3 gene classification and sequence. (A) PCR electrophoresis showing amplification band and DL2000 DNA Marker; the PCR product is 160 bp. (B) Sequence of PCR products including three genotypes: 2/2, 2/3, and 3/3. (C) Schematic view of three genotypes and allele 2 results in a PCR fragment of 157 bp, whereas allele 3 results in a PCR fragment of 162 bp. SMYD3 indicates SET and MYND domain-containing protein 3; PCR, polymerase chain reaction.
Distribution of SMYD3 VNTR genotype and allele frequency
The frequency distribution of the SMYD3 VNTR genotype and allele is shown in Table 2. The proportion of allele 3 in patients in the case group was 86.5%, significantly higher than the proportion of 74.1% in the control group, while the proportion of allele 2 in patients in the control group was 25.9%, significantly higher than the proportion of 13.5% in the case group (both P<0.05). The proportion of genotype 3/3 in case group patients was 78.6%, significantly higher than the proportion of 67.8% in the control group (P<0.05). Allele type 3 (odds ratio [OR]: 2.243, 95% confidence interval [CI]: 1.497~3.359; P<0.001) and genotype 3/3 (OR: 1.769, 95% CI: 1.074~2.913; P=0.024) may be risk factors for ovarian cancer (both P<0.05).
Table 2.
Frequency distribution of SYMD3 VNTR genotype and allele.
| Genotype | Case group n (%) | Control group n (%) | P | OR value (95%CI) |
|---|---|---|---|---|
| 2 | 42 (13.5%) | 90 (25.9%) | 1 (reference) | |
| 3 | 270 (86.5%) | 258 (74.1%) | <0.001 | 2.243 (1.497–3.359) |
| 2/2+2/3 | 33 (21.4%) | 56 (32.2%) | 1 (reference) | |
| 3/3 | 123 (78.6%) | 118 (67.8%) | 0.024 | 1.769 (1.074–2.913) |
P value was detected by chi-square test; OR value – odd ratio; CI – confidence interval.
Relationship between SMYD3 VNTR gene polymorphism and clinical pathological features
The results showed that the proportion of poorly differentiated patients carrying VNTR genotype 3/3 was significantly higher than the proportion of poorly differentiated patients carrying VNTR genotype 2/2+2/3, while the proportion of patients carrying genotype 3/3 in FIGO stage III–IV was significantly higher than that of patients carrying genotype 2/2+2/3 in FIGO stage III–IV (both P<0.05). However, the proportions of patients with different tumor differentiation who carried genotype 3/3 and genotype 2/2+2/3 were not significantly different (P>0.05). VNTR genotype was not correlated with the age, first menstruation age, first childbearing age, last childbearing age, menstruation status, or abortion history (all P>0.05) (Table 3).
Table 3.
Correlation of SMYD3 VNTR polymorphism with clinic-pathologic characteristics of patients.
| Clinical charateristics | 2/2+2/3 | 3/3 | OR value (95%CI) | P |
|---|---|---|---|---|
| Age (age) | 0.309 | |||
| <45 | 13 (39.4%) | 37 (30.1%) | 1 (reference) | |
| ≥45 | 20 (60.6%) | 86 (69.9%) | 0.662 (0.298–1.470) | |
| First menstruation (age) | 0.699 | |||
| <13 | 16 (48.5%) | 55 (44.7%) | 1 (reference) | |
| ≥13 | 17 (51.5%) | 68 (55.3%) | 0.859 (0.398–1.856) | |
| First childbearing age | 0.399 | |||
| <20 | 15 (45.5%) | 46 (37.4%) | 1 (reference) | |
| ≥20 | 18 (54.5%) | 77 (62.6%) | 0.717 (0.329–1.559) | |
| Last childbearing age | 0.988 | |||
| <40 | 19 (57.6%) | 71 (57.7%) | 1 (reference) | |
| ≥40 | 14 (42.4%) | 52 (42.3%) | 1.006 (0.462–2.190) | |
| Mesntruation codition | 0.508 | |||
| Premenopause | 24 (72.7%) | 82 (66.7%) | 1 (reference) | |
| Postmenopause | 9 (27.3%) | 41 (33.3%) | 0.750 (0.319–1.760) | |
| Abortion history | 0.333 | |||
| No | 23 (69.7%) | 80 (65.0%) | 1 (reference) | |
| Yes | 10 (30.3%) | 43 (35.0%) | 1.545 (0.637–3.748) | |
| Tumor classification | ||||
| HGSC | 17 (51.5%) | 79 (64.2%) | 1 (reference) | |
| EC | 4 (12.1%) | 13 (10.6%) | 1.105 (0.327–3.729) | 0.872 |
| CCC | 5 (15.2%) | 13 (10.6%) | 1.381 (0.444–4.296) | 0.576 |
| MC | 5 (15.2%) | 14 (11.4%) | 1.282 (0.416–3.952) | 0.664 |
| LGSC | 2 (6.0%) | 4 (3.2%) | 1.795 (0.308–10.46) | 0.509 |
| Tumor differentiation | 0.017 | |||
| Middle/high differentiation | 29 (87.9%) | 89 (72.3%) | 1 (reference) | |
| Low differentiation | 4 (12.1%) | 44 (35.7%) | 3.584(1.186–10.840) | |
| FIGO staging | 0.045 | |||
| I–II | 27 (81.8%) | 78 (63.4%) | 1 (reference) | |
| III–IV | 6 (18.2%) | 45 (36.6%) | 0.385 (0.148–1.004) |
P value was detected by Wilcoxon rank-sum test; OR – odd ratio; CI – confidence interval; EC – endometrial carcinoma; CCC – clear cell carcinoma; MC – mucin-like cell carcinoma; HGSC – high-grade serous adenocarcinoma; LGSC – low-grade serous adenocarcinoma; FIGO – International Federation of Gynecology and Obstetrics.
SMYD3 mRNA and protein expression and SMYD3 genotype
As shown in Figure 2, RT-qPCR results demonstrated that genotype 3/3 exhibited higher SMYD3 mRNA expression than genotype 2/2+2/3 (P<0.05), and Western blot detection revealed that genotype 3/3 showed higher SMYD3 protein expression than genotype 2/2+2/3 (P<0.05).
Figure 2.
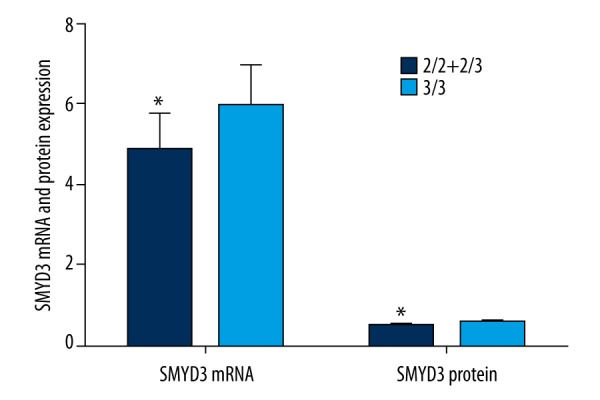
The comparisons of SMYD3 mRNA and protein expressions between 2/2+2/3 and 3/3. SMYD3 mRNA and protein expressions were significantly higher in genotype 3/3 than in genotype 2/2+2/3. * P<0.05 compared with genotype 2/2+2/3. SMYD3 indicates SET and MYND domain-containing protein 3.
Logistic regression analysis of SMYD3 gene polymorphism with risk of ovarian cancer
Logistic regression analysis was applied to analyze the general information on the case group and the control group, including age, first menstruation age, first childbearing age, last childbearing age, menstruation status, abortion history, and VNTR polymorphism of SMYD3. The statistical results suggested that low first childbearing age (<20 years old), menopause, abortion, and VNTR genotype 3/3 may be independent risk factors for ovarian cancer prevalence (all P<0.05). The remaining factors were not significantly correlated with risk of ovarian cancer (all P>0.05) (Table 4).
Table 4.
Logistic regression analysis on risk factors for ovarian cancer.
| Parameter | B | S.E. | Wald | Sig. | Exp (B) | 95%CI for Exp (B) | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| Age | 0.007 | 0.01 | 0.566 | 0.452 | 1.007 | 0.988 | 1.027 |
| First menstruation | 0.107 | 0.061 | 3.092 | 0.079 | 1.113 | 0.988 | 1.253 |
| First childbearing age | −0.146 | 0.034 | 18.177 | <0.001* | 0.864 | 0.808 | 0.924 |
| Last childbearing age | 0.003 | 0.022 | 0.023 | 0.878 | 1.003 | 0.961 | 1.048 |
| Menstruation condition | 2.511 | 0.456 | 30.339 | <0.001* | 12.314 | 5.04 | 30.088 |
| Abortion history | 1.599 | 0.405 | 15.558 | <0.001* | 4.949 | 2.236 | 10.957 |
| SMYD3 VNTR | 0.648 | 0.297 | 4.78 | 0.029* | 1.913 | 1.069 | 3.42 |
CI – confidence interval;
represents that difference of P<0.05 is statistically significant.
Association between SMYD3 gene polymorphism and ovarian cancer prognosis
After five-year follow-up of the 156 patients with ovarian cancer, the Kaplan-Meier survival curves showed that the 5-year survival rate for patients carrying VNTR genotype 3/3 was significantly lower than that for patients carrying genotype 2/2+2/3 (P<0.05) (Figure 3). Meanwhile, Cox regression analysis was applied to the risk factors for ovarian cancer, including age, first menstruation age, first childbearing age, last childbearing age, menstruation status, abortion history, tumor differentiation, FIGO staging, and VNTR genotype polymorphism of SYMD3. The statistical results suggested that later last childbearing age (P=0.015), poor differentiation (P=0.024), and VNTR genotype 3/3 (P<0.001) may be independent risk factors for poor prognosis of ovarian cancer. The other indicators including age, first menstruation age, last childbearing age, and abortion history were not significantly associated with the prognosis of ovarian cancer (all P>0.05) (Table 5).
Figure 3.
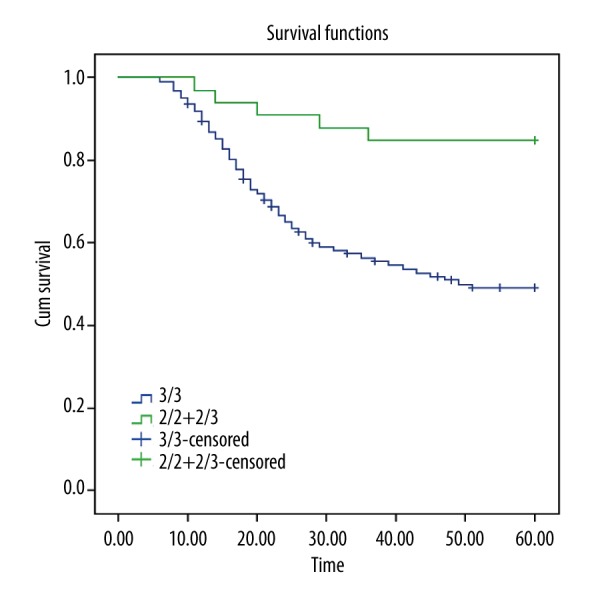
Application of the Kaplan-Meier survival curve to determine the effect of SYMD3 VNTR polymorphism on the 5-year survival of patients. The 5-year survival rate for the patients carrying VNTR genotype 3/3 was significantly lower than that of the patients carrying genotype 2/2+2/3. SMYD3 indicates SET and MYND domain-containing protein 3; VNTR, variable number of tandem repeats.
Table 5.
COX multivariate regression analysis on risk assessment of ovarian cancer.
| Parameter | B | S.E. | Wald | Sig. | Exp (B) | 95%CI for Exp (B) | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| First childbearing age (<20 vs. ≥20) | −0.449 | 0.733 | 0.376 | 0.54 | 0.638 | 0.152 | 2.682 |
| Last childbearing age (<40 vs. ≥40) | −1.358 | 0.559 | 5.909 | 0.015* | 0.257 | 0.086 | 0.769 |
| Menstruation (premenopause vs. postmenopause) | 0.157 | 0.803 | 0.038 | 0.845 | 1.17 | 0.243 | 5.646 |
| Abortion (N vs. Y) | 0.078 | 0.935 | 0.007 | 0.934 | 1.081 | 0.173 | 6.753 |
| Degree of differentiation (mid-high vs. low) | −0.951 | 0.422 | 5.088 | 0.024* | 0.386 | 0.169 | 0.883 |
| FIGO staging (I vs. II/III vs. IV) | −1.526 | 0.858 | 3.163 | 0.075 | 0.217 | 0.04 | 1.169 |
| SYMD3 VNTR (2/2+2/3 vs. 3) | 2.238 | 0.507 | 19.472 | <0.001* | 9.371 | 3.468 | 25.316 |
CI – confidence interval;
represents that P<0.05;
N – no; Y – yes.
Discussion
To the best of our knowledge, the development and progression of tumors comprise a complex process influenced by the interaction between genetics and epigenetics. Epigenetics was expressed by methylation, histone modification, and other means of gene regulation to regulate and adjust genes, which was reversible and was considered to be a hot topic of cancer research [16]. In this study, quantitative fluorescence was applied to detect whether the SMYD3 polymorphism was a risk factor for ovarian cancer, and the result showed that VNTR genotype 3/3 of SMYD3 increased the probability of ovarian cancer.
Most importantly, we found that the frequency distribution of the SMYD3 VNTR genotype and allele were significantly different between the case group and the control group. Genotype 3/3 exhibited higher SMYD3 mRNA and expressions than genotype 2/2+2/3. The possible reason is that VNTR of CCGCC in the promoter region of SMYD3 can combine E2F1, which is an important transcription factor in the cell cycle and can regulate the expression of the downstream gene and the synthesis, repair, proliferation, and apoptosis of DNA [17]. VNTR genotype 3/3 of the SMYD3 gene can effectively increase its affinity with E2F1, the trans-activation of which was enhanced afterward compared to genotype 2/2 [18]. It has been reported that overexpression of SMYD3 can promote the activity of histone methyltransferase and the growth of tumor cells [19]. The interference of siRNA with the expression of SMYD3 can effectively inhibit the growth of tumor [20]. Hamamoto et al. showed that high expression of SMYD3 VNTR genotype 3/3 could increase the risk of colon cancer and breast cancer [21].
Analysis of clinical-pathological features in this study showed that the proportion of poorly differentiated patients carrying VNTR genotype 3/3 was significantly higher than that of poorly differentiated patients carrying VNTR genotype 2/2+2/3, which indicated that VNTR genotype 3/3 of SMYD3 can significantly promote the proliferation of ovarian cancer. Chen et al. also reported that SMYD3 can enhance cell proliferation and promote the transformation to malignancy [22]. In addition, the FIGO staging for patients carrying the VNTR genotype 3/3 was significantly higher than that for patients carrying genotype 2/2+2/3. The polymorphism of SMYD3 may be related to the FIGO staging and tumor differentiation in ovarian cancer, which indicates that high expression of SMYD3 can up-regulate the expression of DNA topoisomerase II J3 [11]. It was reported that histone methyltransferase SMYD3 was correlated with cell cycles, which could accelerate the process of S phase and promote the cell transition from S phase to G2 phase to facilitate the promotion of cell proliferation [23]. Kunizaki et al. suggested that SMYD3 methylation was capable of targeting vascular endothelial growth factor 1 (VEGFR1), which could accelerate the methylation of the 831st lysine [24]. The activity of VEGFR1 was enhanced afterward, and the tumor metastasis was promoted as well [24]. It was also reported that SMYD3 could be correlated with a number of signaling pathways. It could be methylated through MAP3K2 mediation, activated through the MAP kinase pathway, and amplified through Ras signaling cascade so as to promote the tumor formation [25]. This study found out that SMYD3 VNTR genotype 3/3 was a significant risk factor for the occurrence of ovarian cancer, which indicated that the methylation of DNA could not only regulate the sequence of tumor suppressor gene, but also inhibit the interaction of proteins through the expression of histone modifications. Cedar et al. reported that DNA methylation and histone modification pathways can be dependent on one another, and this crosstalk can be regulated through biochemical interactions between SET domain histone methyltransferases as well as DNA methyltransferases. Thus associations between DNA methylation and histone modification play an important role in understanding normal development and somatic cell reprogramming as well as tumorigenesis [26].
The progression of ovarian cancer was influenced by many factors, such as environment, genetics, and patients’ own physical features. After the statistical analysis of factors and prognosis of ovarian cancer in this study, we found that the polymorphism of SMYD3 gene and younger first childbearing age (<20 years), menopause, and abortion may be independent risk factors for ovarian cancer; the polymorphism of SMYD3, older last childbearing age (≥40 years), and FIGO stage of ovarian cancer may be independent risk factors for poor prognosis of ovarian cancer. It’s reported that non-ovariectomized artificial menopause and early age of menopause are protective factors for ovarian cancer, while late menopause may increase the probability of ovarian cancer [27]. All these results suggested that the menstrual cycle may be one of the factors that may influence the occurrence of ovarian cancer. In addition, a meta-analysis showed that first menstruation, first childbearing age, history of dysmenorrhea, infertility history and history of multiple pregnancy, history of prolific production, multiple abortions, history of birth control pills, and intrauterine history of birth control were protective factors for ovarian cancer [28]. This study showed that the SMYD3 polymorphism, childbearing age, and menstrual cycle are somehow correlated with the occurrence of ovarian cancer.
Conclusions
VNTR genotype 3/3 of SMYD3 is closely correlated with the occurrence of ovarian cancer. The VNTR polymorphism of SMYD3 might be a sensitive predictor for poor prognosis of patients with ovarian cancer, which provides a theoretical basis for treatment options and individualized diagnosis. Due to the insufficient study samples, this study may be limited regarding its results. Therefore, further studies are in need to better clarify the exact mechanism relating VNTR polymorphism of SMYD3 and ovarian cancer.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Fustino N, Rakheja D, Ateek CS, et al. Bone morphogenetic protein signalling activity distinguishes histological subsets of paediatric germ cell tumours. Int J Androl. 2011;34:e218–33. doi: 10.1111/j.1365-2605.2011.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Li JFeng L. Hedgehog signaling pathway as a therapeutic target for ovarian cancer. Cancer Epidemiol. 2015;40:152–57. doi: 10.1016/j.canep.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 4.El Ayed M, Bonnel D, Longuespee R, et al. MALDI imaging mass spectrometry in ovarian cancer for tracking, identifying, and validating biomarkers. Med Sci Monit. 2010;16(8):BR233–45. [PubMed] [Google Scholar]
- 5.Kumar P, Rehani MM, Kumar L, et al. Tumor marker CA-125 as an evaluator and response indicator in ovarian cancer: Its quantitative correlation with tumor volume. Med Sci Monit. 2005;11(2):CR84–89. [PubMed] [Google Scholar]
- 6.Li H, Xu Y, Qiu W, et al. Tissue miR-193b as a novel biomarker for patients with ovarian cancer. Med Sci Monit. 2015;21:3929–34. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pradjatmo H. Methylation status and expression of BRCA2 in epithelial ovarian cancers in Indonesia. Asian Pac J Cancer Prev. 2015;16:8599–604. doi: 10.7314/apjcp.2015.16.18.8599. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons RJ. Histone modifying and chromatin remodelling enzymes in cancer and dysplastic syndromes. Hum Mol Genet. 2005;14(Spec No 1):R85–92. doi: 10.1093/hmg/ddi106. [DOI] [PubMed] [Google Scholar]
- 9.Guo N, Chen R, Li Z, et al. Hepatitis C virus core upregulates the methylation status of the RASSF1A promoter through regulation of SMYD3 in hilar cholangiocarcinoma cells. Acta Biochim Biophys Sin (Shanghai) 2011;43:354–61. doi: 10.1093/abbs/gmr021. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Liu Y, Tan W, et al. Association of the variable number of tandem repeats polymorphism in the promoter region of the SMYD3 gene with risk of esophageal squamous cell carcinoma in relation to tobacco smoking. Cancer Sci. 2008;99:787–91. doi: 10.1111/j.1349-7006.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamamoto R, Furukawa Y, Morita M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Wu J, Sun B, et al. Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding. Nucleic Acids Res. 2011;39:4438–49. doi: 10.1093/nar/gkr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Liu Y, Tan W, et al. Association of the variable number of tandem repeats polymorphism in the promoter region of the SMYD3 gene with risk of esophageal squamous cell carcinoma in relation to tobacco smoking. Cancer Sci. 2008;99:787–91. doi: 10.1111/j.1349-7006.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira-Santos W, Rabello DA, Lucena-Araujo AR, et al. Residual expression of SMYD2 and SMYD3 is associated with the acquisition of complex karyotype in chronic lymphocytic leukemia. Tumour Biol. 2016 doi: 10.1007/s13277-016-4846-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Genotype analysis of the VNTR polymorphism in the SMYD3 histone methyltransferase gene: lack of correlation with the level of histone H3 methylation in NSCLC tissues or with the risk of NSCLC. Int J Cancer. 2008;122:1441–42. doi: 10.1002/ijc.23227. [DOI] [PubMed] [Google Scholar]
- 16.Foreman KW, Brown M, Park F, et al. Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS One. 2011;6:e22290. doi: 10.1371/journal.pone.0022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Z, Ren N, Jiang Y, et al. Adenovirus-mediated E2F-1 gene transfer augments gemcitabine-induced apoptosis in human colon cancer cells. Clin Lab. 2015;61:1435–44. doi: 10.7754/clin.lab.2015.150104. [DOI] [PubMed] [Google Scholar]
- 18.Tsuge M, Hamamoto R, Silva FP, et al. A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5′ flanking region of SMYD3 is a risk factor for human cancers. Nat Genet. 2005;37:1104–7. doi: 10.1038/ng1638. [DOI] [PubMed] [Google Scholar]
- 19.Peserico A, Germani A, Sanese P, et al. A SMYD3 small-molecule inhibitor impairing cancer cell growth. J Cell Physiol. 2015;230:2447–60. doi: 10.1002/jcp.24975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank B, Hemminki K, Wappenschmidt B, et al. Variable number of tandem repeats polymorphism in the SMYD3 promoter region and the risk of familial breast cancer. Int J Cancer. 2006;118:2917–18. doi: 10.1002/ijc.21696. [DOI] [PubMed] [Google Scholar]
- 21.Hamamoto R, Silva FP, Tsuge M, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–18. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LB, Xu JY, Yang Z, et al. Silencing SMYD3 in hepatoma demethylates RIZI promoter induces apoptosis and inhibits cell proliferation and migration. World J Gastroenterol. 2007;13:5718–24. doi: 10.3748/wjg.v13.i43.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Wang C, Wang K, et al. SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J Natl Cancer Inst. 2013;105:1719–28. doi: 10.1093/jnci/djt304. [DOI] [PubMed] [Google Scholar]
- 24.Kunizaki M, Hamamoto R, Silva FP, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–65. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 25.Mazur PK, Reynoird N, Khatri P, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–87. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 27.La Vecchia C. Ovarian cancer: Epidemiology and risk factors. Eur J Cancer Prev. 2016 doi: 10.1097/CEJ.0000000000000217. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Shim SH, Kim SN, Jung PS, et al. Impact of surgical staging on prognosis in patients with borderline ovarian tumours: A meta-analysis. Eur J Cancer. 2015;54:84–95. doi: 10.1016/j.ejca.2015.11.005. [DOI] [PubMed] [Google Scholar]