Abstract
Here we report the genetic engineering and chemical modification of potato virus X (PVX) for the presentation of various peptides, proteins, and fluorescent dyes, or other chemical modifiers. Three different ways of genetic engineering are described and by these means, peptides are successfully expressed not only when the foot and mouth disease virus (FMDV) 2A sequence or a flexible Glycine–Serine linker is included, but also when the peptide is fused directly to the PVX coat protein. When larger proteins or unfavorable peptide sequences are presented, a partial fusion via the FMDV 2A sequence is preferable. When these PVX chimeras retain the ability to assemble into viral particles and are thus able to infect plants systemically, they can be utilized to inoculate susceptible plants for isolation of sufficient amounts of virus particles for subsequent chemical modification. Chemical modification is required for the display of nonbiological ligands such as fluorophores, polymers, and small drug compounds. We present three methods of chemical bioconjugation. For direct conjugation of small chemical modifiers to solvent exposed lysines, N-hydroxysuccinimide chemistry can be applied. Bioorthogonal reactions such as copper-catalyzed azide–alkyne cycloaddition or hydrazone ligation are alternatives to achieve more efficient conjugation (e.g., when working with high molecular weight or insoluble ligands). Furthermore, hydrazone ligation offers an attractive route for the introduction of pH-cleavable cargos (e.g., therapeutic molecules).
Keywords: Potato virus X (PVX), Genetic engineering, Chemical modification, Chemical bioconjugation, Bio-orthogonal reactions, Plant viral nanoparticles (VNPs)
1 Introduction
The nanomaterials formed by (plant) viruses have become popular tools in chemistry, materials science, nanotechnology, and biomedicine. For example, plant viral nanoparticles (VNPs) have been engineered to display foreign epitopes for application as vaccination platforms [1]. More recently, researchers have turned toward the development of VNP-based carrier system for delivery of therapeutic molecules (e.g., cytostatic drugs, siRNAs) or contrast agents for application in drug delivery or tissue-specific imaging [2]. Be it vaccine or drug delivery, the VNP functions as a scaffold for the desired ligands. A wide variety of molecules, including small chemical modifiers such as dyes or therapeutics, peptides or proteins, as well as polymers have been displayed on a variety of VNP scaffolds [3]. Two basic principles are applied to generate such hybrid VNP-based materials: genetic engineering or chemical conjugation. In this work, we will discuss the methods applied to modify the plant virus potato virus X (PVX) for applications in biomedicine.
PVX, the type member of the Potexvirus group, is a monopartite (+)-strand RNA plant virus [4]. Its particles are filamentous and flexible rods measuring 515 nm in length and 13 nm in diameter, comprising 1,270 identical 25-kDa coat protein (CP) subunits. Each CP contains at least one solvent-exposed lysine residue available for chemical modification [5]. The X-ray structure of PVX has not been reported yet; therefore the location of the chemically addressable lysine side chain within the CP remains elusive. It is thought that the N-terminal part of the CP is surface-exposed; therefore, most of the CP fusions reported to date are N-terminally situated [6, 7].
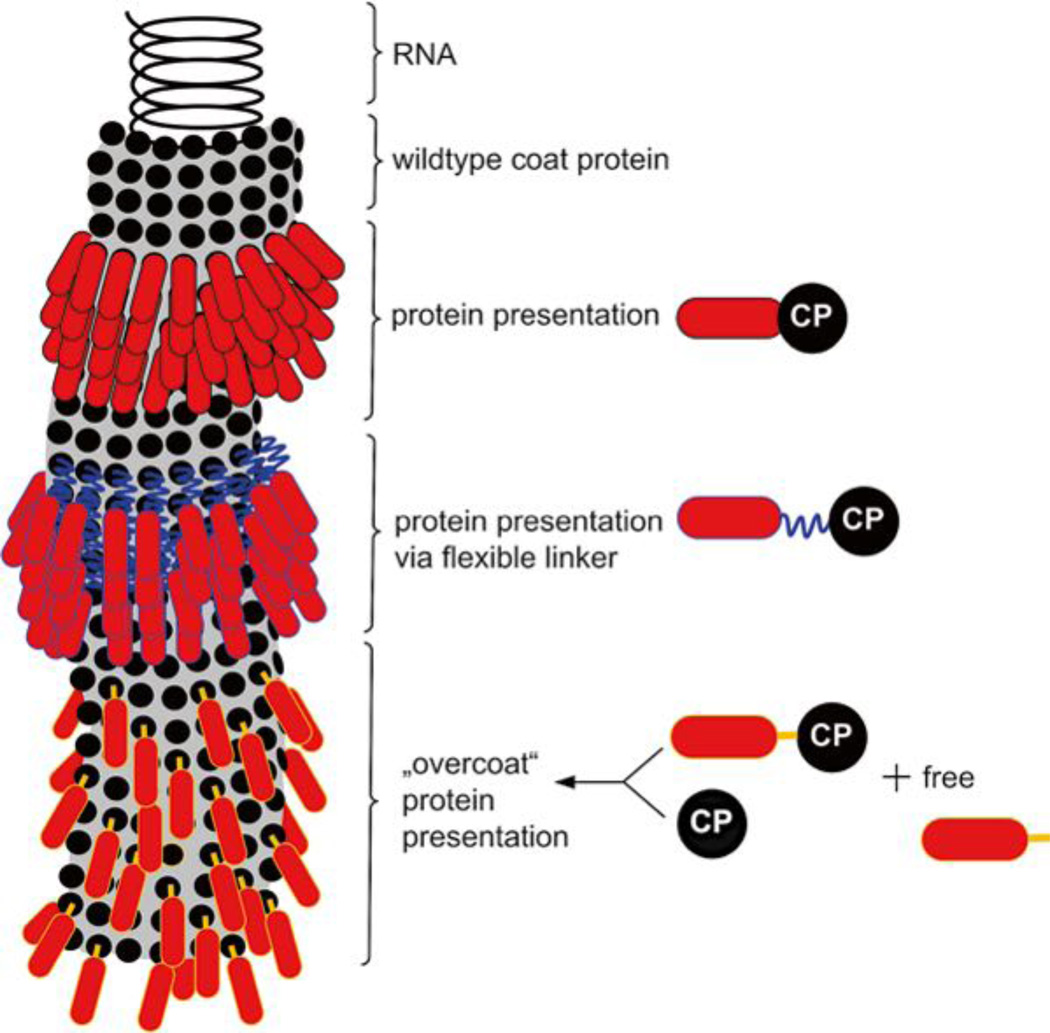
Several PVX-based vaccine formulations have been developed and tested, e.g., PVX-gp41 displaying HIV-1 epitopes [8], PVX-R9 displaying hepatitis C virus (HCV) epitopes [9], PVX-Staphylococcus aureus D2 FnBP [10], and PVX-16E7 formulations displaying human papillomavirus (HPV) epitopes [11]. Some investigations showed that, after fusion with the CPs, polypeptides of different size were presented on the surface of PVX particles only when wild type coat protein was co-expressed with the chimeric coat protein. This so-called “overcoat principle” is achieved when the 2A sequence of the foot and mouth disease virus (FMDV) is inserted between the foreign and the coat protein sequence as a translational fusion (Fig. 1) [12]. The 2A sequence induces a ribosomal skip leading to expression and free and fusion CP, the ratio is tunable by sequence variation [13, 14]. This expression strategy is typically applied when full length proteins are displayed, e.g., 238-amino-acid-long green fluorescent protein (GFP) [12].
Fig. 1.
Schematic presentation of a PVX particle. Black circle: wild type coat protein (CP), red shape: foreign protein, blue jagged line: flexible linker sequence, e.g., a glycine–serine linker (G4S)3, orange bar: 2A sequence from FMDV for expression of free and fusion CP
Shorter peptides can be presented on the PVX surface as direct CP fusion without intervening 2A sequence. In this case, every expressed CP subunit is chimeric. The inclusion of a short flexible linker or spacer, such as a 15 aa glycine–serine linker (G4S)3, however, has been found to be advantageous [15]. Here, we present a generalized procedure for genetic engineering of PVX with and without linker sequences.
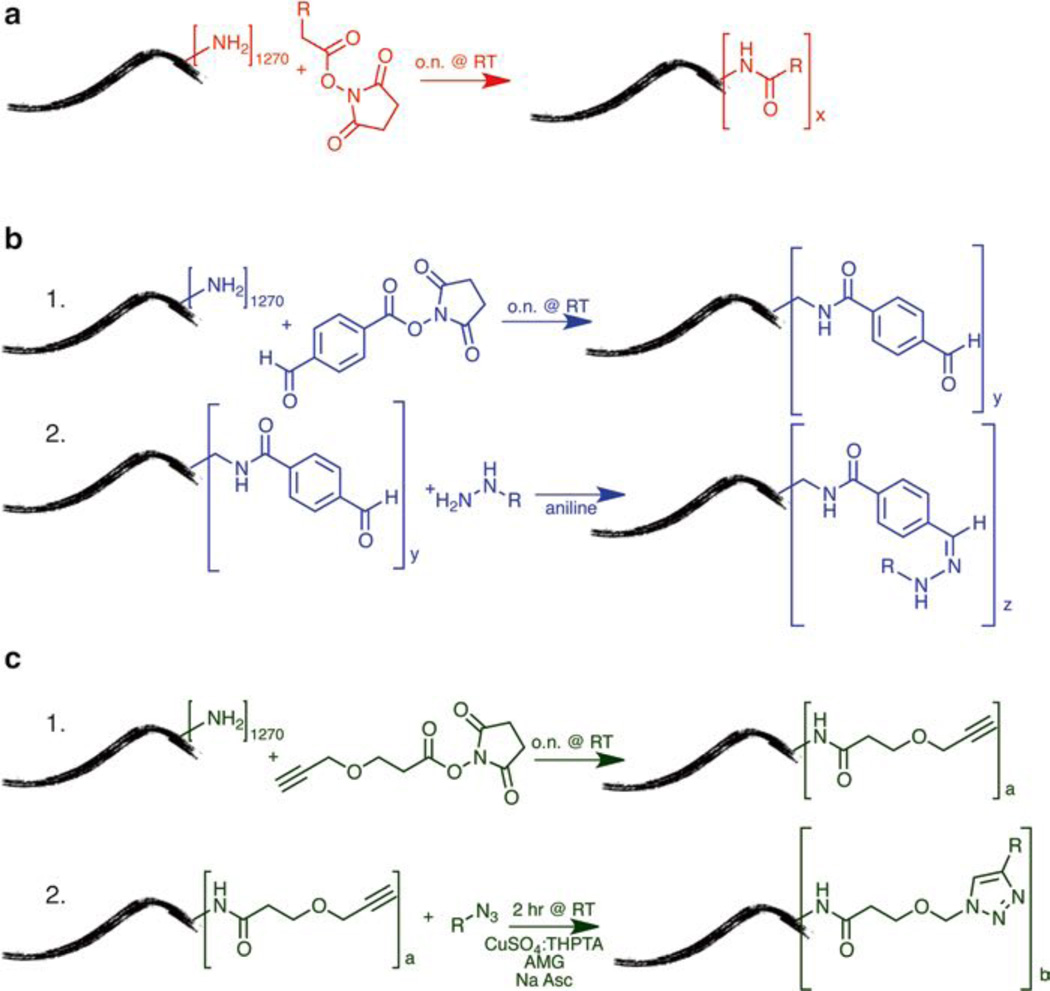
If genetic engineering is not an option or challenging, for example, for display of nonbiological materials such as contrast agents or fluorescent dyes, polymeric materials, or small molecule drugs, large protein complexes (e.g., antibodies), etc., chemical modification provides a reliable alternative. We have recently reported the chemical modification of PVX with near infrared dyes and polyethylene glycol (PEG) polymers. The dyes enabled tracking of the PVX platform in preclinical tumor models (mouse and avian), and the PEG chains are generally applied to enhance pharmacokinetics while reducing the immunogenic properties of the filaments [16]. In this work, we describe three methods for chemical modification of PVX: (1) N-hydroxysuccinimide (NHS) chemistry for direct conjugation to primary amines from lysine side chains [5], (2) hydrazone ligation to attach pH-cleavable cargos [17–19], and (3) copper-catalyzed azide–alkyne cycloaddition (CuCAAC, aka click chemistry) (Fig. 2) [20]. Standard coupling procedures using NHS-activated esters are simple and many reagents are commercially available, thus not requiring extensive knowledge in chemistry. However, NHS-lysine coupling reactions have slow reaction kinetics and large excesses of reagents have to be used to facilitate efficient labeling. Bio-orthogonal reactions such as hydrazone and click chemistry are more efficient and thus require low concentration of the reagent or ligand of interest. This is helpful when reagents are scarce, if solubility in aqueous conditions is a problem, or when conjugating high molecular weight ligands. Another advantage is lower cost, as less material is required.
Fig. 2.
Chemical modification of PVX (a) NHS reaction, (b) Hydrazone Ligation, (c) Click Chemistry, where R = fluorescent label, PEG, or peptide
We discuss genetic and chemical modification methods and provide a description of multiple techniques for the characterization of modified PVX particles: a combination of UV–Visible spectros-copy, SDS-PAGE, size exclusion fast protein liquid chromatography (FPLC), and transmission electron microscopy (TEM) is typically used to confirm expression of chimeric CPs or covalent modification of cargos, as well as to determine the number of labels per PVX, and to verify the stability of PVX hybrids [3, 16].
2 Materials
2.1 Genetic Engineering of PVX
TE buffer: 10 mM Tris–HCl, pH 7.5, 1 mM EDTA.
Taq polymerase: GoTaq® DNA Polymerase (Promega).
Nucleotides (dNTPs, Promega).
T4 ligase (Promega).
Internal PVX backward primers: universe, CX1, CX4 (see Table 1 for sequences).
Electrophoresis: 1.2 % (w/v) agarose gel in 1× TAE buffer.
Gel purification kit (e.g., QIAquick Gel Extraction Kit Qiagen).
Plasmids: CXI [12], PVXG4SI (Uhde-Holzem et al. unpublished).
pCR2.1-TOPO: TOPO® TA Cloning® Kit for subcloning.
E. coli DH5α cells, electro or heat shock competent.
Table 1.
Primer sequences
| Sequence | CXI binding site | |
|---|---|---|
| Universe | GTTGTAAAACGACGGCCAGT | Backward primer binding ~200 nt downstream of the CP stop codon |
| Reverse | ACACAGGAAACAGCTATGAC | Forward primer binding ~460 nt upstream of the RdRp start codon |
| CXI | TTGAAGAAGTCGAATGCAGC | Backward primer binding ~465 nt downstream of the first CP codon |
| CX2 | CTAGATGCAGAAACCATAAG | Forward primer binding ~100 nt upstream of the GFP start codon |
| CX3 | ATAGCAGTCATTAGCACTTC | Forward primer binding ~180 nt upstream of the GFP start codon |
| CX4 | CGGGCTGTACTAAAGAAATC | Backward primer binding ~100 nt downstream of the first CP codon |
2.2 PVX Purification
Phosphate buffers: Prepare 0.2 M Na2HPO4 and 0.2 M NaH2PO4 and use Table 2 for production of the various phosphate buffers.
Homogenization buffer: 0.1 M phosphate buffer pH 8.0 with 0.2 % (v/v) of 2-mercaptoethanol and 10 % (v/v) ethanol (see Note 1).
Waring (USA) blender, preferably with a glass container.
Gauze: Miracloth (Calbiochem, Cat 475855).
1 M NaCl/20 % (w/v) PEG: weigh 58.44 g NaCl, 100 g polyethylene glycol 6000, and 100 g polyethylene glycol 8000, and make up to 1 l with DI H2O while continuously stirring; autoclave for better solubility and sterilization.
Sucrose gradient: prepare a 10 % (w/v) and a 45 % (w/v) sucrose solution (see Note 2) in 0.01 M phosphate buffer pH 7.2 with 0.01 M EDTA (mix 12.25 ml 0.2 M Na2HPO4, 17.75 ml 0.2 M NaH2PO4, 5 ml 0.5 M EDTA (see Note 3), and 220 ml DI H2O to obtain 250 ml), use 14 ml 10 % (w/v) and 14 ml 45 % (w/v) sucrose solution to prepare a continuous 28 ml sucrose gradient with a gradient mixer.
Sucrose gradient centrifugation: Beckman Ultra-Clear Tubes (25 × 89 mm), Rotor: Beckman SW32Ti (Beckman coulter Optima L-100 XP Ultracentrifuge).
Centrifugation tubes for virus sedimentation: Beckman Ultra-Clear Tubes (14 × 89 mm), Rotor SW41Ti (Beckman coulter Optima L-100 XP Ultracentrifuge).
PBS: phosphate buffered saline, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4.
Table 2.
Na2HPO4–NaH2PO4 buffer solutions, pH 6.8–8.0 at 25 °C [21]
| pH, 25 °C | xml 0.2 M Na2HPO4 | yml 0.2 M NaH2PO4 |
|---|---|---|
| 7.0 | 30.5 | 19.5 |
| 7.2 | 36.0 | 14.0 |
| 7.4 | 40.5 | 9.5 |
| 7.6 | 43.5 | 6.5 |
| 7.8 | 45.75 | 4.25 |
| 8.0 | 47.35 | 2.65 |
xml 0.2 M Na2HPO4, yml 0.2 M NaH2PO4; diluted to 100 ml with H2O → 0.1 M solution
x/2 ml 0.2 M Na2HPO4, y/2 ml 0.2 M NaH2PO4; diluted to 100 ml with H2O → 0.05 M solution
xml 0.2 M Na2HPO4, yml 0.2 M NaH2PO4; diluted to 1,000 ml with H2O → 0.01 M solution
…
2.3 Chemical Modification of PVX
Potassium phosphate (KP) buffer (0.1 M): 10.7 g K2HPO4, 5.23 g KH2PO4 in 1 l water, pH 7.0.
Propargyl-dPEG®1-NHS ester (Quanta Biodesign, Ltd.) (see Note 4).
50 mM Copper(II) sulfate pentahydrate (Sigma).
50 mM Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) (see Note 5).
100 mM l-ascorbic acid sodium salt (Na Asc., Sigma).
100 mM Aminoguanidine Hydrochloride (AMG, Sigma).
Dimethyl sulfoxide (DMSO, Sigma).
Oregon Green 488 succinimidyl ester *6-isomer* (Invitrogen) (see Note 6).
Oregon Green 488 Azide, 6-isomer (Invitrogen) (see Note 6).
Sulfo-succinimidyl 4-formylbenzoate (Sulfo-S-4FB) (Solulink).
Fluorescein-hydrazide (see Note 6).
Aniline (Fisher).
2.4 Characterization of PVX Particles
SDS-PAGE gel: 4–12 % Bis–Tris NuPAGE SDS Gel (Invitrogen).
Pre-stained Protein Standard: SeeBlue Plus2 Pre-stained standard (Invitrogen).
Sample Buffer: NuPAGE LDS Sample Buffer (4×) (Invitrogen).
Running buffer: 50 ml 20× MOPS buffer (Invitrogen), bring final volume to 1 l with deionized water.
Staining buffer: 0.25 % (w/v) Coomassie Brilliant Blue R-250, 30 % (v/v) methanol, 10 % (v/v) acetic acid in deionized water.
Destaining buffer: 20 % (v/v) methanol, 10 % (v/v) acetic acid in deionized water.
2 % (w/v) uranyl acetate (Fisher).
Carbon type-B TEM grid (Ted Pella).
Transmission electron microscope (Zeiss, Libra 200FE).
Protein liquid chromatography (FPLC) using a Superose 6 size-exclusion column and the ÄKTA Explorer (GE Healthcare).
Cutoff Spin Filter: Amicon Ultra-0.5 ml Centrifugal Filters (10 kDa cutoff) (Millipore) (see Note 7).
NuPage LDS Sample Buffer (4×) (Invitrogen).
3 Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1 Genetic Engineering of PVX
Choose epitope/peptide or protein of interest.
Design appropriate nucleotides for cloning by adapting the codon usage to that of the PVX coat protein (e.g., see NCBI Gene Bank) and add appropriate restriction sites at the ends (see step 3).
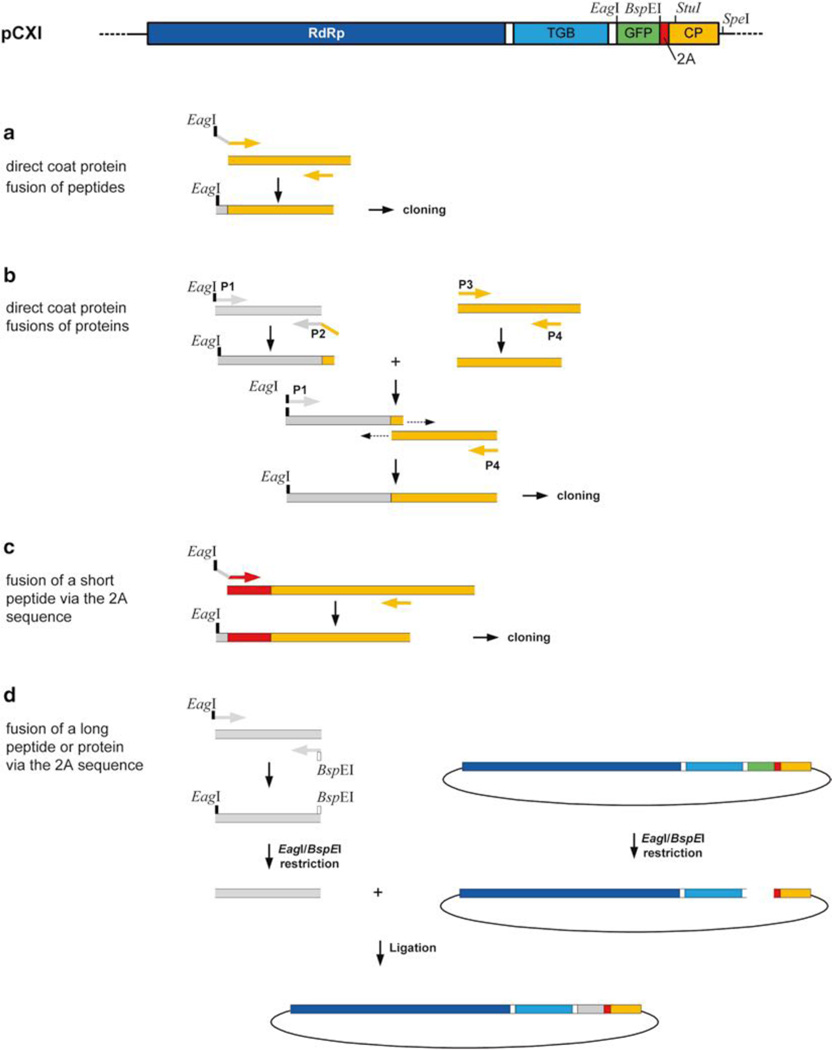
- Generate CP fusions (Fig. 3)
- For direct coat protein fusion of peptides (see Note 8), design one nucleotide (see Note 9), starting with the EagI restriction site (see Note 10), followed by the sequence of the peptide of interest, and terminated by the first 19 nucleotides of the PVX coat protein (see Note 11). Use this oligonucleotide in combination with an appropriate primer binding internally or downstream of the coat protein (CX4, CX1, or universe, see Table 1), to amplify the peptide of interest fused to the 5′-end of the coat protein coding sequence.
-
For direct coat protein fusions of proteins, do a SOE-PCR, using four primers:
- Primer 1. Forward primer binding to the 5′-end of the protein of interest, including the EagI restriction site at the 5′-end (see Note 12).
- Primer 2. Backward primer binding to the 3′-end of the protein of interest which contains as well the first 19 nt of the 5′-end of the coat protein (see Note 13).
- Primer 3. Forward primer binding to the 5′-end of the coat protein, this sequence is complementary to the 19 nucleotides used for the second primer (see Note 14).
- Primer 4. Backward primer binding internally to or downstream of the coat protein coding sequence (CX4, CX1, or universe, see Table 1).
First perform two separate PCRs using primers sets 1 + 2 and 3 + 4, respectively, and appropriate templates (nucleotide sequence of protein of interest, CXI plasmid). Purify PCR products using gel electrophoresis and gel extraction (e.g., Qiagen Gel Extraction Kit). Perform a second PCR reaction, using these two PCR fragments in combination with the outer primers (1 + 4) to amplify the whole sequence (Fig. 3b). - For fusion of a short peptide (≤ 15 aa) via the 2A sequence to the coat protein, design one primer, which should contain the EagI restriction at the 5′-end, followed by the foreign sequence and the sequence complementary to the 5′-end of the 2A sequence (see Note 15).
- For fusion of a long peptide (≥ 15AA) or protein via the 2A sequence or the glycine–serine (G4S)3 linker sequence to the coat protein, design two primers. The 5′-primer should contain the EagI restriction site and the 3′-primer the BspEI restriction site.
Insert the final PCR products into the TOPO vector (Invitrogen) for DNA amplification. Follow the manufacturer’s instructions to obtain positive TOPO clones (see Note 16) and do a plasmid preparation for the subsequent cloning procedure.
Cut the insert sequence from the TOPO vector with appropriate restriction enzymes (EagI/BspEI or EagI/SpeI), at the same time cut the PVX vector containing plasmid DNA using the same restriction enzymes. Purify insert and vector backbone using gel electrophoresis and gel extraction (e.g., Qiagen Gel Extraction Kit).
Ligate insert and vector using T4 Ligase, follow the manufacturer’s instructions.
Transform E. coli DH5α with the ligation product.
Produce sufficient DNA amounts using appropriate Midi/ Maxi or Mega DNA preparation kits (see Note 17).
Inoculate three leaves of a Nicotiana benthamiana plant (approximately 4 weeks after seeding, ~8 leave stage) each with 5–10 µg DNA (in 50 µl PBS) by rubbing the recombinant PVX vector containing plasmid DNA onto carborundum or Celite (503 Roth) dusted leaves (see Note 18).
After 10 min rinse the leaves with tap water to remove abrasive and excess DNA.
14 days post infection (dpi) systemically infected leaves are harvested and first used for expression analysis via appropriate methods such as SDS-PAGE, western blot, ELISA, and electron microscopy (see Note 19).
If the recombinant particles are tested positive, start a virus purification (see Subheading 3.2).
Fig. 3.
Cloning strategies to generate PVX CP fusions. RdRp RNA-dependent RNA polymerase gene, TGB triple gene block, GFP green fluorescent protein coding sequence. 2A FMDV 2A sequence, CP coat protein gene, grey sequences foreign peptide or protein coding sequence
3.2 Purification of Chimeric PVX Particles
Inoculate at least ten Nicotiana benthamiana plants with recombinant PVX vector containing plasmid DNA (see above) (see Note 20).
Harvest systemically infected leaves 14 dpi and freeze them immediately at −80 °C.
Homogenize frozen plant material (100 g) with 2 volumes (w/v) ice-cold homogenization buffer (see Note 1).
Filter through three layers of gauze and clarify by centrifugation at 7,800 × g for 20 min at 4 °C (see Note 21).
Process the supernatant by adding 1 % (v/v) Triton X-100 (see Note 22) and stir for 1 h at 4 °C.
Clarify by centrifugation at 5,500 × g for 20 min at 4 °C.
Process the supernatant by adding 1/5 vol. 1 M NaCl and 20 % (w/v) PEG and stir first for 1 h at 4 °C and then incubate for 1 h at room temperature.
Precipitate the viral particles by centrifugation at 7,800 × g for 10 min at 4 °C (see Note 23).
Resuspend the pellet in 4 ml 0.05 M phosphate buffer pH 8.0 with 1 % (v/v) Triton X-100, rinse the tubes immediately with 2 ml of the same buffer and combine the samples.
Clarify by centrifugation at 7,800 × g for 10 min at 4 °C.
Load the supernatant onto a sucrose gradient (10–45 % (w/v) in 0.01 M phosphate buffer pH 7.2 with 0.01 M EDTA produced using a gradient mixer) and centrifuge in a swinging bucket rotor at 104,000 × g for 75 min at 4 °C (SW32Ti, 23,700 rpm).
Collect 1.5 ml gradient fractions from bottom to top using a syringe needle connected to a flexible tube (see Note 24).
Analyze all fractions by SDS PAGE and if necessary by western blotting.
Select and combine fractions with highest concentration of PVX coat protein.
Dilute combined fractions at least in the same or ideally in two volumes of 0.01 M phosphate buffer pH 7.2.
Sediment virus particles by at least 3 h of ultracentrifugation at 248,000 × g (SW41Ti, 38,000 rpm, 4 °C).
Resuspend the pellet in 0.2 ml 0.01 M phosphate buffer pH 7.2 each and stir overnight at 4 °C (see Note 25).
Combine virus fractions and clarify the solution by centrifugation at 5,000 × g, 4 °C, for 10 min.
- Read absorbance 260 nm in a spectrophotometer and calculate the concentration using the PVX extinction coefficient 2.97:
- Concentration (mg/ml) = A260 value × dilution factor/extinction coefficient.
Analyze PVX particles by SDS PAGE, western blotting, and electron microscopy.
3.3 Chemical Modification of PVX
Carry out all reactions using 1 mg of PVX at a protein concentration of 2 mg/ml, unless otherwise noted.
3.3.1 N-Hydroxysuccinimide Chemistry
Prepare 50 mg/ml solution of O488-NHS in DMSO (see Note 26).
Add 2,000 molar excess of O488-NHS (see Note 27) per PVX particle and adjust the final DMSO concentration to 10 % (v/v) (see Note 28).
Shield solution from light and react overnight at room temperature with agitation (see Note 29).
Purify PVX particles from excess dye using sucrose density gradient ultracentrifugation (as described above, Subheading 3.2) and/or using dialysis using 10-kDa cutoff spin filters. Wash filter with 0.1 M potassium phosphate (KP) buffer until the dye is no longer detectable in the flow through (typically after 5–10 washes) (see Note 30).
Resuspend PVX particles in KP buffer, store at 4 °C until further processing or characterization (see below).
3.3.2 Hydrazone Ligation
This protocol discusses the addition of benzaldehydes to PVX surface lysines followed by attachment of O488-hydrazide; this protocol could also be extended to reaction with aminooxyacetyl-functionalized compounds [18].
Modify PVX lysines with benzaldehydes using a 2,000 molar excess of NHS-4-formylbenzamide (e.g., Solulink) and react overnight with agitation followed by purification from excess benzaldehydes (see Subheading 3.3.1).
Determine the efficiency of 4FB decoration into PVX using the aromatic 2-hydrazinopyridine-dihydrochloride hydrazine, which reacts specifically with 4FB-modified groups to form a UV-traceable and quantifiable hydrazone bond. Follow the manufacturer’s procedure, see Solulink.
Prepare a 35 nM solution of fluorescein-hydrazide in DMSO and a 50 mM solution of aniline in KP buffer.
To benzaldehyde-modified PVX, add a 10,000 molar excess fluorescein-hydrazide per PVX (assuming 1,000 benzaldehydes available) and aniline to a final concentration of 10 mM (see Note 31).
Protect solution from light and react for 2 h at room temperature with agitation (see Note 31).
Purify the modified PVX particles from excess reagents using density gradient ultracentrifugation and/or 10-kDa cutoff spin filters as described above (Subheading 3.3.1).
Characterize particles (see protocol below, Subheading 3.4).
3.3.3 Copper-Catalyzed Azide–Alkyne Cycloaddition (CuCAAC, aka Click Chemistry)
This protocol discusses the addition of alkynes to PVX surface lysines followed by attachment of O488-azide. However, the reaction can also be performed by adding azides to the lysines of PVX followed by an O488-alkyne (or any other biomedically relevant molecule).
Convert available amine groups to alkynes using a 10,000 molar excess of propargyl-dPEG®1-NHS ester (as described above, see Subheading 3.3.1) (see Note 32).
Purify PVX–alkyne and store at 4 °C or proceed to step 3.
- Prepare the following solutions fresh, in water.
- Cu (II) pentahydrate: 50 mM.
- THPTA: 50 mM.
- Sodium ascorbate: 100 mM.
- AMG: 100 mM.
- O488-azide: 50 mg/ml.
To 1 mg of PVX–alkyne at a concentration of 2 mg/ml in KP buffer, add in the following order: a 10,000 molar excess of O488-azide, AMG at 5 mM final concentration, CuSO4– THPTA mix at 0.25:1.25 mM final concentration, and sodium ascorbate at 5 mM final concentration (see Note 33).
Protect solution from light and react 2 h at room temperature with agitation.
Purify PVX from excess reagents via density gradient ultracentrifugation and/or 10-kDa cutoff spin filters as described above (see Subheading 3.3.1).
Characterize particles (see below, Subheading 3.4).
3.4 Characterization of Particles
3.4.1 UV–Visible Spectroscopy
Read absorbance at 260, 280, and 496 nm on a spectrophotometer (see Note 34).
Compare A260:280 ratio to determine if the sample is pure. A ratio of 1.2 ± 0.1 indicates pure PVX preparations.
Determine the concentration of PVX particles and dyes using the Beer–Lambert law (A = εcl), where A is the absorbance, ε is the extinction coefficient, c is the concentration, and l is the path length. The extinction coefficients of PVX and O488 are 2.97 cm−1 mg−1 ml (at 260 nm) and 70,000 cm−1 M−1 (at 496 nm), respectively (see Note 35).
Determine the number of dyes per particle using the ratio of the concentration of dye to that of PVX.
3.4.2 Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Add 3 µl of 4× LDS sample buffer to 10 µg of PVX particles in 9 µl of KP buffer (see Note 36).
Incubate in heat block for 5 min at 100 °C (for denaturation of the CPs) (see Note 37).
Load samples (modified vs. non-modified PVX) onto 4–12 % Bis–Tris NuPAGE SDS gel. In one additional lane, include 10 µl SeeBlue Plus2 Pre-stained protein standard.
Run samples at 200 V for 1 h in 1× NuPAGE MOPS SDS running buffer.
Document gel under UV light to observe fluorescent bands, indicating covalent decoration of the dye to the CPs (Fig. 4b).
Stain with 0.25 % (w/v) Coomassie blue for 1 h at room temperature (see Note 38).
Soak in destaining buffer overnight at room temperature (see Note 39).
Document gel under white light (Fig. 4b).
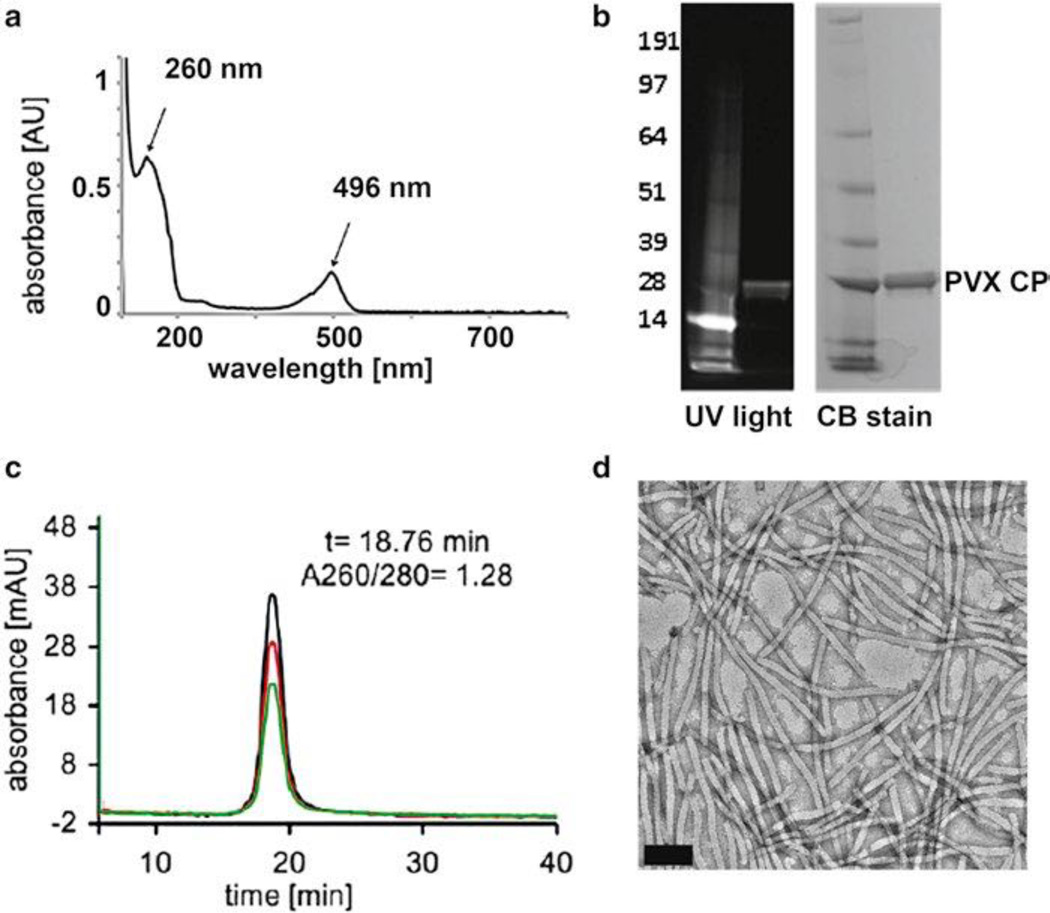
Fig. 4.
Characterization of PVX-O488 (a) UV–Visible spectrum indicating the PVX and O488 characteristic peaks at 260 nm (PVX nucleic acids) and 496 (O488), (b) SDS-PAGE gel before and after Coomassie blue stain; the fluorescence of the coat protein indicates covalent labeling with the O488 dye, (c) FPLC spectra, black line = 260 nm, red line = 280 nm, green line = 496 nm; nucleic acid, protein, and dye co-elute indicating successful labeling and intact PVX (reproduced from Steinmetz et al. 2010, Nano Letters), and (d) TEM of negatively stained PVXO488, scale bar is 100 nm
3.4.3 Fast Protein Liquid Chromatography (FPLC)
Analyze particle integrity by size exclusion chromatography (SEC) using the ÄKTAvExplorer and a Superose 6 size-exclusion column.
Load 100 µg of native or modified particles in 200 µl of KP buffer (see Note 40).
Analyze at a flow rate of 0.5 ml/min.
Set detectors to 260, 280, and 496 nm (see Note 41).
The elution profile indicates intactness of the preparation (see Fig. 4c).
3.4.4 Transmission Electron Microscopy (TEM)
Acknowledgments
This work was supported by Ohio Cancer Research Associates (to N.F.S.), Mt. Sinai Foundation (N.F.S.), NIH grant NCI R25 CA148052 Cancer Pharmacology training grant (K.L.L.).
Footnotes
Any work with 2-mercaptoethanol should be carried out using proper engineering controls (e.g., chemical hood). To avoid exposure to fumes, the homogenization buffer can be prepared without 2-mercaptoethanol; 2-mercaptoethanol is then added after the leaves extract filtration through gauze.
Weight 45 g sucrose into a heat stable beaker, fill up to ~80 ml with H2O, use vigorous stirring and moderate heat, after completely dissolved add H2O to reach 100 ml and let solution cool down.
For preparation of a 0.5 M EDTA stock solution: weight 93.06 g EDTA (ethylenediamine-tetraaceticacid 2Na·2H2O) under the hood wearing a mask (EDTA dust is irritant and inhalation or skin contact needs to be avoided), add to a small volume of H2O and start immediately to adjust the pH with NaOH. EDTA will not be soluble until the pH reaches 8.0. Use vigorous stirring, moderate heat (if desired) and time. After completely dissolved, add H2O to reach 500 ml.
For click chemistry modification, this protocol describes the addition of alkynes to the lysines of PVX. However, it is equally valid to attach azides to the lysines of PVX. For this modification, purchase an NHS-azide instead of Propargyl-dPEG®1-NHS ester (Quanta Biodesign, Ltd.).
The THPTA ligand was a gift from M.G. Finn (The Scripps Research Institute). For synthesis protocol see Hong et al. [20].
This protocol describes the attachment of Oregon Green 488 (or fluorescein) using various chemical modification protocols. However, these protocols can be translated too other cargos including (but not limited to): polyethylene glycol (PEG) chains of various molecular weights and other fluorescent dyes, peptide ligands, proteins, small drugs, e.g., doxorubicin. For the addition of these cargos, purchase the appropriate derivative of the cargo of interest for the chemical modification technique chosen (i.e., NHS ester, azide, alkyne, hydrazide).
Spin filters with different molecular weight cutoffs (3-kDa to 100-kDa) can be purchased for different reagents.
Check the length of the peptide/protein sequence to be added as well as the isoelectric point. If the peptide is longer than 15 aa and/or the isoelectric point is higher than that of the PVX wild type coat protein (6.73), a direct fusion most probably is not feasible. Instead, try a fusion via the FMDV 2A sequence.
Oligonucleotides (e.g., Eurofins MWG Operon) of up to 73 nt length have been used successfully for PCR amplification of the hybrid coat protein DNA.
If the peptide or protein does not start with a Serine, we highly recommend to add this amino acid to the N-terminus of your peptide or protein. This will help to stabilize the protein.
5′-AAACGGCCGATG(AGT)-peptide sequence-CCCGCGAGCACAACACAGC-3′ (underlined sequence: EagI restriction site, double underlined sequence: Serine codon). When designing primers, always use 1–3 A at the 5′-end, to enable better direct restriction of the PCR product. At least two nucleotides at the very 3′-end should be C or G for solid binding.
Forward primer P1: 5′-AAACGGCCGATG(AGT)-5′-protein sequence (underlined sequence: EagI restriction site, double underlined sequence: Serine codon).
Backward primer P2: 5′-GCTGTGTTGTGCTCGCGGG-3′-reverse-protein sequence.
P3: 5′-CCCGCGAGCACAACACAGC-3′.
5′-AAACGGCCGATG(AGT)-peptide sequence-TCCGGATCTAGAAATTTTG-3′ underlined sequence: EagI restriction site, double underlined sequence: Serine codon, bold underlined sequence: BspEI restriction site.
We recommend colony screening by PCR using primers universe and reverse (see Table 1) to select positive clones directly from the agar plate. Just pick a bit of the colony with a tooth pick and shortly dip it into the PCR Mix.
We recommend using the PureYield Plasmid Maxiprep System (Promega), which gives high plasmid DNA concentration in a minimum of time.
Rub the plasmid DNA very carefully into the leaves by using your thumb (wearing gloves).
Pestle leave material in two volumes of PBS and use this as a “stock solution” for further analysis.
14 dpi about 65–100 g of systemically infected leave material can be harvested from ten plants.
Place four layers of Miracloth in a big cone, wring out the plant sap holding three layers and leaving one in the cone in case the three tear.
Triton X-100: prepare a 20 % (v/v) working solution of Triton X-100, stir thoroughly and autoclave for better solubility, always shake the solution before usage.
After centrifugation save the pellet and as well the supernatant. Some recombinant virus particles are not precipitated with NaCl/PEG. If particles are not precipitated with PEG, they can be sedimented from the supernatant using ultracentrifugation: 4 h, 122,000 × g, 4 °C. Afterwards, dilute the virus pellets in ~5 ml 0.05 M phosphate buffer (pH 7.2) and continue with the sucrose gradient step (step 11, Subheading 3.2).
Place the hollow needle at the flank about 0.5 cm above the bottom of the ultracentrifuge bottle (not directly at the base). Collect fraction 0 (the bottom fraction) at the end and dissolve a possible pellet in it.
Place a small magnetic stir bar into the centrifugation tube, seal the tube with Parafilm (Roth) and stir on a magnetic stirrer overnight in the fridge or cold room.
Concentration can be varied; for example, if a large amount of reagent is required, it may be helpful to make a more concentrated stock solution. NHS-reactive compounds are best prepared fresh and in dry DMSO. The NHS ester is susceptible to aqueous hydrolysis rendering the compound nonreactive toward lysines.
Molar excess will vary depending on the cargo that is being attached. For example, higher molecular weight cargos (i.e., PEG5000) require a larger molar excess.
The final DMSO concentration should be kept at 10 % by volume to give good yields. PVX remains stable at DMSO concentrations up to 20 % by volume and overnight exposure (yields may be reduced).
To adjust number of dyes attached per particle, incubation time can be altered. The less time the solution is allowed to react, the fewer dyes will be attached. However, the number of dyes attached will eventually level off, and will not increase indefinitely. This is due to the hydrolysis of the NHS esters, as well as the decreasing availability of lysines.
Purification may take more than seven spins. Continue to discard waste and add sterile KP until flow through is clear. Alternative methods include use of density gradients or ultrapelleting.
If the final number of dyes/particle is not the desired amount, reaction conditions should be optimized: variables are molar excess used, reaction time, concentration of reagents and of the catalyst aniline.
At this point, a coumarin assay [22] can be used to determine the number of alkynes conjugated to PVX. This may be important for the next steps of the reaction.
If the final number of dyes/particle is not the desired number, reaction conditions can be altered and optimized: the variables are molar excess, concentration of copper–ligand, protein concentration, and reaction time. CuSO4–THPTA should be added as a mix to the reaction. Sodium ascorbate acts as a reducing agent generating catalytically active Cu(I); sodium ascorbate must be prepared fresh for each reactions. Aminoguanidine is a useful additive to prevent protein crosslinking due to ascorbate oxidation [20].
Absorbance at 260 and 280 nm detect nucleic acid and protein, respectively. Absorbance at 496 nm is unique for Oregon Green 488. For different fluorescent tags, absorbance should be read at absorbance maximum (available from the manufacturer).
The extinction coefficient of 70,000 cm−1 M−1 is unique to Oregon Green 488. For different fluorescent tags, refer to manufacturer manuals for extinction coefficients.
If 10 µg of sample is more than 9 µl, add more 4× LDS sample buffer to prepare larger sample volume. In all cases, dilute LDS sample dye to 1× and do not exceed 30 µl total volume.
If heat block is unavailable, this step may also be performed in a water bath. During this step, pressure within tubes may increase and cause tops to pop off. To avoid this, punch a hole in the top of each tube with a needle prior to heating and seal with Parafilm.
If the gel is not completely stained by the end of the hour, let it continuing staining for longer.
Adding a Kimwipe to the container during destaining will decrease the amount of time needed.
Between 50 and 200 µg of sample can be loaded. Volume of sample is determined by the available sample loop holder (ranges between 100 µl and 2 ml). For full protocol prior to loading sample (i.e., cleaning out the sample loop), see instruction manual of FPLC system.
Detectors are set at 260 and 280 nm for nucleic acid and protein, respectively. A third detector can be set to measure the dye-specific absorbance, e.g., 496 nm is unique for Oregon Green 488. For different fluorescent tags, detector should be set at the excitation wavelength (available from the manufacturer).
If particles aggregate, appear too dense or are sparse, the concentration can be altered; typically a 0.1 mg/ml solution is a good starting point.
TEM staining protocol can be altered to optimize the amount of sample on each grid. Different methods include (but are not limited to) increasing the amount of time the grid is placed on the sample and uranyl acetate, removing the wash steps, and using smaller volumes of sample and allowing it to completely dry on the grid.
References
- 1.Plummer EM, Manchester M. Viral nanoparticles and virus-like particles: platforms for contemporary vaccine design. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;3(2):174–196. doi: 10.1002/wnan.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinmetz NF. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine. 2010;6:634. doi: 10.1016/j.nano.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokorski JK, Steinmetz NF. The art of engineering viral nanoparticles. Mol Pharm. 2011;8:29. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koenig RJ, Lesemann DE. Potato virus X, potexvirus group. Assoc Appl Biol Warwick. 1989;354:1. [Google Scholar]
- 5.Steinmetz NF, Mertens ME, et al. Potato virus X as a novel platform for potential biomedical applications. Nano Lett. 2010;10:305. doi: 10.1021/nl9035753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhde K, Fischer R, Commandeur U. Expression of multiple foreign epitopes presented as synthetic antigens on the surface of Potato virus X particles. Arch Virol. 2005;150:327. doi: 10.1007/s00705-004-0402-z. [DOI] [PubMed] [Google Scholar]
- 7.Uhde-Holzem K, Fischer R, Commandeur U. Genetic stability of recombinant potato virus X virus vectors presenting foreign epitopes. Arch Virol. 2007;152:805. doi: 10.1007/s00705-006-0892-y. [DOI] [PubMed] [Google Scholar]
- 8.Marusic C, Rizza P, et al. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J Virol. 2001;75:8434. doi: 10.1128/JVI.75.18.8434-8439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhde-Holzem K, Schlösser V, Viazov S, Fischer R, Commandeur U. Immunogenic properties of chimeric potato virus X particles displaying the hepatitis C virus hypervariable region I peptide R9. J Virol Methods. 2010;166:12. doi: 10.1016/j.jviromet.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Brennan FR, Jones TD, et al. Immunogenicity of peptides derived from a fibronectin-binding protein of S. aureus expressed on two different plant viruses. Vaccine. 1999;17:1846. doi: 10.1016/s0264-410x(98)00485-x. [DOI] [PubMed] [Google Scholar]
- 11.Massa S, Simeone P, et al. Antitumor activity of DNA vaccines based on the human papillomavirus-16 E7 protein genetically fused to a plant virus coat protein. Hum Gene Ther. 2008;19:354. doi: 10.1089/hum.2007.122. [DOI] [PubMed] [Google Scholar]
- 12.Cruz SS, Chapman S, et al. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci U S A. 1996;93:6286. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly ML, Hughes LE, et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82:1027. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly ML, Luke G, et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 15.Werner S, Marillonnet S, Hause G, Klimyuk V, Gleba Y. Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc Natl Acad Sci U S A. 2006;103:17678. doi: 10.1073/pnas.0608869103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla S, Ablack AL, et al. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle potato virus X. Mol Pharm. 2012;10(1):33–42. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunel FM, Lewis JD, et al. Hydrazone ligation strategy to assemble multifunctional viral nanoparticles for cell imaging and tumor targeting. Nano Lett. 2010;10:1093. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirksen A, Dawson PE. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjug Chem. 2008;19:2543. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z, Chen K, et al. Development of viral nanoparticles for efficient intracellular delivery. Nanoscale. 2012;4:3567. doi: 10.1039/c2nr30366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48:9879. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomori G. Preparation of buffers for use in enzyme studies. In: Colowick SP, Kaplan NO, editors. Methods in enzymology. New York: Academic; 1955. pp. 138–143. [Google Scholar]
- 22.Sivakumar K, Xie F, et al. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett. 2004;6:4603. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]