Abstract
Lysyl hydroxylase 2 (LH2) catalyzes the hydroxylation of lysine residues in the telopeptides of fibrillar collagens, which leads to the formation of stable collagen cross-links. Recently we reported that LH2 enhances the metastatic propensity of lung cancer by increasing the amount of stable hydroxylysine aldehyde-derived collagen cross-links (HLCCs), which generate a stiffer tumor stroma (Chen, Y., et al. (2015) J. Clin. Invest. 125, 125, 1147–1162). It is generally accepted that LH2 modifies procollagen α chains on the endoplasmic reticulum before the formation of triple helical procollagen molecules. Herein, we report that LH2 is also secreted and modifies collagen in the extracellular space. Analyses of lung cancer cell lines demonstrated that LH2 is present in the cell lysates and the conditioned media in a dimeric, active form in both compartments. LH2 co-localized with collagen fibrils in the extracellular space in human lung cancer specimens and in orthotopic lung tumors generated by injection of a LH2-expressing human lung cancer cell line into nude mice. LH2 depletion in MC3T3 osteoblastic cells impaired the formation of HLCCs, resulting in an increase in the unmodified lysine aldehyde-derived collagen cross-link (LCC), and the addition of recombinant LH2 to the media of LH2-deficient MC3T3 cells was sufficient to rescue HLCC formation in the extracellular matrix. The finding that LH2 modifies collagen in the extracellular space challenges the current view that LH2 functions solely on the endoplasmic reticulum and could also have important implications for cancer biology.
Keywords: collagen, extracellular matrix, hydroxylysine (Hyl), post-translational modification (PTM), secretion, lysine, lysyl hydroxylase 2
Introduction
Collagens are the main component of the extracellular matrix and the most abundant protein in mammals (1). Of the 29 types of collagen identified, fibrillar type I collagen is the most abundant and important in providing tissues and organs with form and mechanical strength. Type I collagen biosynthesis involves a series of post-translational modifications of nascent procollagen α chains on the endoplasmic reticulum; for example, specific lysine (Lys) and proline (Pro) residues undergo hydroxylation, and hydroxylysine (Hyl) residues undergo mono- and di-glycosylation. In the extracellular space, telopeptidyl Lys and Hyl residues on collagen molecules are oxidatively deaminated by lysyl oxidases (LOXs),4 producing the reactive aldehydes Lysald and Hylald, respectively, which initiates a series of condensation reactions to form various covalent intermolecular cross-links involving juxtaposed Lys, Hyl, and histidine (His) residues on the neighboring molecules, resulting in the formation of Hylald-derived collagen cross-links (HLCCs) (2).
HLCCs are generated through a series of intermediate steps involving multiple enzymatic reactions. For example, HLCCs are produced through the divalent iminium cross-links dehydro-hydroxylysinonorleucine (deH-HLNL) when paired with a juxtaposed Lys residue (i.e. HylaldXLys) on a neighboring molecule, and deH-dihydroxylysinonorlecine (deH-DHLNL) with a Hyl residue (i.e. HylaldXHyl). These are then spontaneously rearranged to form the stable ketoamines by Amadori rearrangement then mature into the formation of the trivalent pyridinium cross-links, pyridinoline (Pyr) (i.e. HylaldXHylaldXHyl) and deoxypyridinoline (d-Pyr) (i.e. HylaldXHylaldXLys). This pathway is predominant in skeletal tissues such as bone and cartilage. The Lysald-derived cross-linking pathway, on the other hand, is predominant in soft connective tissues. Telopeptidyl Lysald can condense with another Lysald residue within the same molecule to form an intramolecular aldol, which then eventually leads to a tetravalent intermolecular cross-link, deH-histidinohydroxymerodesmosine (deH-HHMD) (i.e. LysaldXLysaldXHisXHyl) (3). In skin and cornea, the Lysald-derived cross-linking pathway can also lead to a non-reducible, trivalent cross-link, histidinohydroxylysinonorleucine by involving the divalent, iminium cross-link, deH-HLNL (LysaldXHyl), and a His residue (i.e. LysaldXHylXHis) (4).
Lys hydroxylation is catalyzed by lysyl hydroxylases 1–3 (LH1–3; EC 1.14.11.4) in -X-Lys-Gly- sequences in a reaction that requires Fe2+, 2-oxoglutarate, O2, and ascorbate (5). In addition to the -X-Lys-Gly- sequence, -X-Lys-Ala-and -X-Lys-Ser- sequences present in the telopeptides (both N and C termini) of fibrillar collagens can be hydroxylated. It has been reported that LH2 catalyzes Lys hydroxylation in the telopeptides (6–8) and thereby drives the Hylald-derived collagen cross-linking pathway (9). Altered LH2 expression has a profound impact on the collagen matrix (10). Although all LH family members (LH1–3) appear to be capable of hydroxylating helical Lys residues, only LH2 modifies the telopeptidyl Lys residues (8, 11). Inherited skeletal disorders caused by inactivating mutations in the gene that encodes LH2 and a putative LH2 foldase, FKBP10 (12–14) demonstrate the importance of telopeptidyl Lys hydroxylation in normal collagen biosynthesis and function (15, 16).
It has been reported that LH3, a multifunctional enzyme possessing both LH and glycosyltransferase activities, can be secreted and modifies Lys residues on native proteins and possibly “microfolded” mature collagen molecules in the extracellular space (17). Further studies showed that the glucosyltransferase activity site is required for LH3 to be secreted into the extracellular space (18, 19). These findings are the basis of the current paradigm that LH3 is the only LH isoform with an extracellular function. However, whether other LH members are secreted and capable of modifying Lys residues in the extracellular space has not been thoroughly explored.
In breast cancer, lung cancer, sarcoma, and other tumor types, high LH2 expression increases tumor stiffness, promotes tumor cell invasion and metastasis, and is a predictor of poor clinical outcome (20–22). In lung cancer, high LH2 expression increases the amount of HLCCs in tumor stroma (21). Moreover, LH2 has been detected in the secretome of breast cancer cells (23), which raises the possibility that LH2 has an extracellular function. In this study we demonstrate that LH2 is associated with extracellular collagen fibrils in lung cancer tissue specimens and is secreted into the extracellular space in an active dimeric form. Importantly, our data strongly indicate that secreted LH2 is capable of hydroxylating telopeptidyl Lys residues of collagen in the extracellular space, thereby producing stable HLCCs. This implies that extracellular LH2 modifies telopeptidyl Lys residues before their LOX-catalyzed conversion to aldehyde, which challenges the current paradigm that LH2 modifies only intracellular nascent procollagen α chains.
Results and Discussion
LH2 Is Secreted
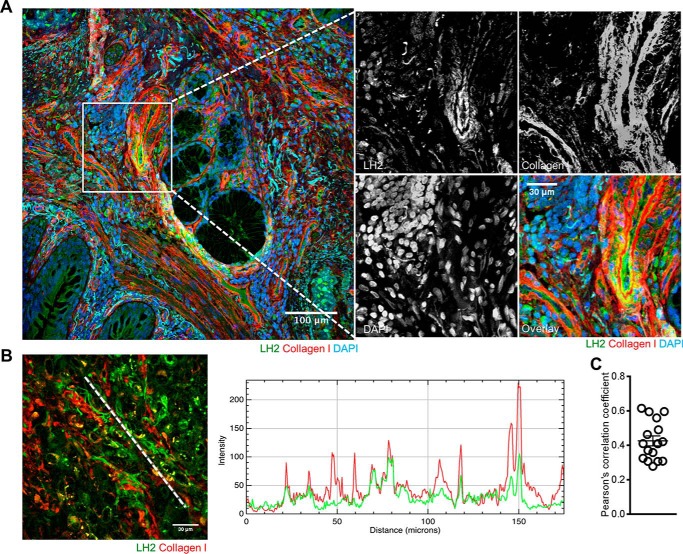
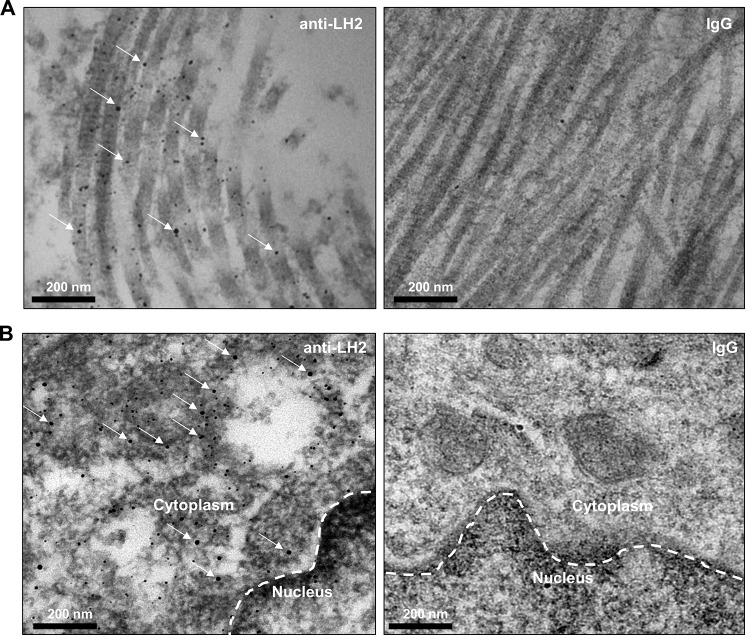
We initially performed stable isotope labeling of amino acids in cell culture on a panel of lung cancer cell lines (n = 52). By mass spectrometric sequencing of proteins, 27 cell lines had detectable LH2 in both cell lysates and conditioned media (Table 1). In the remaining 25 cell lines, LH2 was detected in only the cell lysates (n = 4) or conditioned media (n = 4) or neither fraction (n = 17) (Table 1). In contrast, LOX was detected in 21 cell lines and was almost exclusively in the conditioned media fraction (Table 1). Across the cell line panel, LH2 levels in the cell lysate and conditioned media were significantly correlated (Spearman correlation coefficient = 0.77, p = 1.31 × 10−11). Stable transfection of H1299 lung cancer cells, which have low endogenous LH2 levels (Table 1), with a vector that expresses FLAG-tagged full-length LH2 or an N-terminally truncated mutant LH2 that lacks the signal peptide showed that the full-length ectopic LH2 was present in both the cell lysate and conditioned media, whereas the signal peptide-deficient LH2 was detected only in the cell lysate (Fig. 1A). On the basis of the lower molecular weight of the signal peptide-deficient LH2 (Fig. 1A, left panel), we speculated that aberrant trafficking of the mutant LH2 led to a reduced level of glycosylation, as there are nine potential glycosylation sites in human LH2 (24). Supporting this possibility, treatment of the total cell lysates with peptide N-glycosidase F decreased the wild-type LH2 protein to a size similar to that of the mutant LH2 protein but had no effect on the size of the mutant LH2 protein (Fig. 1B). We then examined human lung cancers to determine if LH2 is localized in the extracellular space by confocal microscopic analysis. In a region of the tumor stroma containing dense collagen fibers (Fig. 2A), co-staining with antibodies against LH2 and type I collagen showed that LH2 and collagen indeed co-localized in the extracellular space. In a different region of the same tumor specimen, spatial overlapping between collagen and LH2 was also confirmed by plotting intensity of LH2 and collagen staining along a linear region of the tumor stroma (Fig. 2B). Quantification of LH2 and collagen I staining within multiple fields of tumor stroma showed that a fraction of LH2 co-localized with collagen I (Fig. 2C). Furthermore, staining of tumors derived from DFC1024 human lung cancer cells, which secreted LH2 in culture (Table 1), with immunogold-labeled anti-LH2 antibodies demonstrated that most, if not all, of extracellular LH2 was closely associated with type I collagen fibrils in the extracellular space (Fig. 3A), and LH2 was also detectable in the cytoplasm of tumor cells (Fig. 3B). Although we cannot determine if LH2 binds to collagen directly or indirectly through collagen-binding proteins such as small leucine-rich proteoglycans (25), these findings raised the possibility that LH2 directly binds to collagen and modifies telopeptidyl Lys residues in the extracellular space.
TABLE 1.
LH2 and LOX levels in total cell lysate (Cell lysates) and conditioned media (Media) collected from the indicated cell lines subjected to SILAC and mass spectrometry
Each value represents the total number of spectral counts within each cellular compartment from one biological sample in a single experiment.
| Cell line | LH2 |
LOX |
||
|---|---|---|---|---|
| Cell lysates | Media | Cell lysates | Media | |
| DFC1024 | 56.82 | 259.34 | 0 | 0 |
| H2030 | 28.97 | 130.94 | 0 | 0 |
| H1355 | 16.79 | 124.78 | 0 | 0 |
| H1568 | 10.77 | 112.29 | 0 | 0 |
| H1993 | 2.31 | 94.24 | 0 | 0 |
| HCC827 | 35.96 | 87.03 | 0 | 0 |
| H1651 | 27.05 | 70.13 | 0 | 0.91 |
| DFC1032 | 34.27 | 61.69 | 0 | 13.11 |
| H1437 | 5.28 | 46.34 | 0 | 0 |
| H1563 | 28.15 | 38.02 | 0 | 0 |
| H820 | 36.10 | 36.90 | 0 | 0 |
| H2342 | 0 | 30.39 | 0 | 0 |
| PC9 | 1.28 | 18.22 | 0 | 12.53 |
| H2228 | 5.88 | 17.57 | 1.60 | 35.15 |
| HCC2279 | 9.48 | 17.28 | 1.90 | 35.52 |
| H1395 | 0 | 14.70 | 1.32 | 54.38 |
| H1373 | 34.60 | 12.53 | 0 | 13.43 |
| H1792 | 16.63 | 11.49 | 2.02 | 42.13 |
| H2291 | 7.15 | 10.78 | 0 | 3.59 |
| HCC4017 | 4.47 | 7.73 | 0 | 5.16 |
| H1944 | 30.25 | 7.46 | 0 | 0 |
| H969 | 1.21 | 7.23 | 0 | 0 |
| H2122 | 0 | 6.56 | 0 | 0 |
| H650 | 12.40 | 6.13 | 0.77 | 18.39 |
| H1650 | 8.71 | 5.75 | 0 | 7.67 |
| H647 | 1.99 | 5.15 | 0 | 0 |
| H1573 | 2.53 | 3.64 | 0 | 0 |
| H69AR | 8.51 | 1.95 | 0 | 9.76 |
| H69 | 4.51 | 1.82 | 0 | 1.82 |
| HCC4006 | 4.59 | 1.31 | 0 | 7.86 |
| HCC1833 | 0 | 1.03 | 0 | 0 |
| DMS79 | 0 | 0 | 0 | 1.35 |
| H1299 | 1.28 | 0 | 0 | 0 |
| H1385 | 0 | 0 | 0 | 0 |
| H1435 | 0 | 0 | 0 | 0 |
| H1693 | 0 | 0 | 0 | 0 |
| H1703 | 0.48 | 0 | 0 | 0 |
| H1755 | 0 | 0 | 0 | 0 |
| H1793 | 0 | 0 | 0 | 9.03 |
| H1838 | 0 | 0 | 0 | 0 |
| H1975 | 0 | 0 | 0 | 0 |
| H2009 | 0 | 0 | 0 | 0 |
| H23 | 0 | 0 | 0 | 0 |
| H2405 | 0 | 0 | 0 | 0 |
| H3255 | 0 | 0 | 0 | 2.79 |
| H522 | 0 | 0 | 0 | 0 |
| H82 | 0 | 0 | 0 | 0 |
| H838 | 1.95 | 0 | 0 | 5.74 |
| H920 | 0 | 0 | 0 | 0.66 |
| HCC2935 | 0 | 0 | 0 | 0 |
| HCC4011 | 4.16 | 0 | 1.19 | 9.10 |
| HCC4019 | 0 | 0 | 0 | 0 |
FIGURE 1.
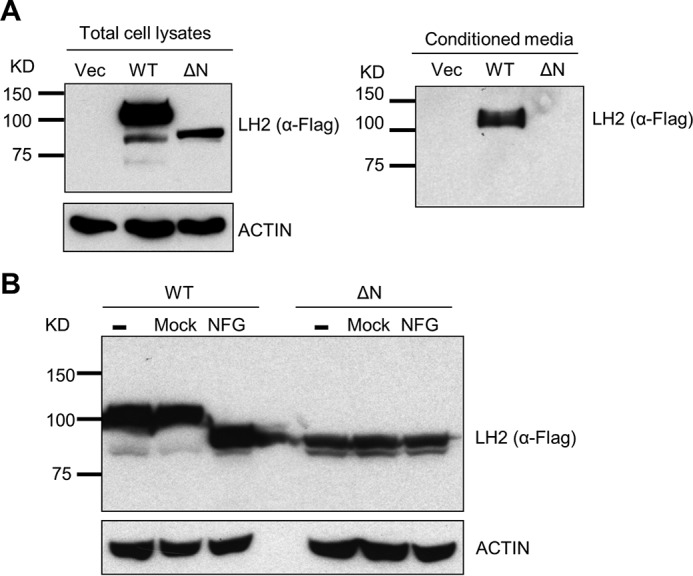
LH2 underwent glycosylation and was secreted. A, anti-FLAG Western blotting of total cell lysates or conditioned media isolated from H1299 cells that had been stably transfected with empty vector (Vec) or FLAG-tagged wild-type (WT) or mutant LH2 lacking the N-terminal signal peptide (ΔN). Actin was used as a loading control for the total cell lysates. B, anti-FLAG Western blotting of lysates from H1299 cells that had been stably transfected with FLAG-tagged wild-type (WT) or mutant LH2 lacking the N-terminal signal peptide (ΔN) after a 5-h incubation of the lysates with (NFG) or without (Mock) N-glycosidase F. As an additional control, Western blotting was performed on lysates that had not been subjected to a 5-h incubation (−). Actin was used as a loading control. Each result is representative of findings from two independent experiments.
FIGURE 2.
LH2 co-localized with extracellular collagen in a human lung adenocarcinoma. A, fluorescence micrograph of a human lung adenocarcinoma tissue specimen stained with antibodies against LH2 and collagen I. Nuclei were counterstained with DAPI. Scale bar, 100 μm. The inset shows higher magnification of a region in which LH2 co-localizes with collagen I. LH2 and collagen I signals are pseudocolored in the overlay images. Scale bar, 30 μm. Two human lung adenocarcinomas were stained, and the results shown are representative of findings from both tumor specimens. B, to illustrate overlap of LH2 and collagen I in a confocal micrograph of a human lung adenocarcinoma tissue specimen (left), LH2 (green) and collagen I (red) staining intensities were plotted against distance along a white line in the image (left). C, the fraction of LH2 that co-localizes with collagen I within single fields (circles) of two human lung adenocarcinomas as determined by Pearson's correlation coefficient.
FIGURE 3.
LH2 co-localized with extracellular collagen in tumors generated by subcutaneous injection of DFC1024 lung cancer cells into nude mice. Representative electron microscopic images of a DFC1024 tumor tissue section stained with gold particle-labeled IgG or anti-LH2 antibodies. Gold particles that localize on extracellular collagen fibers (A) and in the cytoplasm of tumor cells (B) are indicated (arrows). Dashes indicate location of nuclear membrane (B). Scale bar, 200 nm. The results shown are representative of a single tumor generated in 1 mouse.
Extracellular LH2 Is Functional
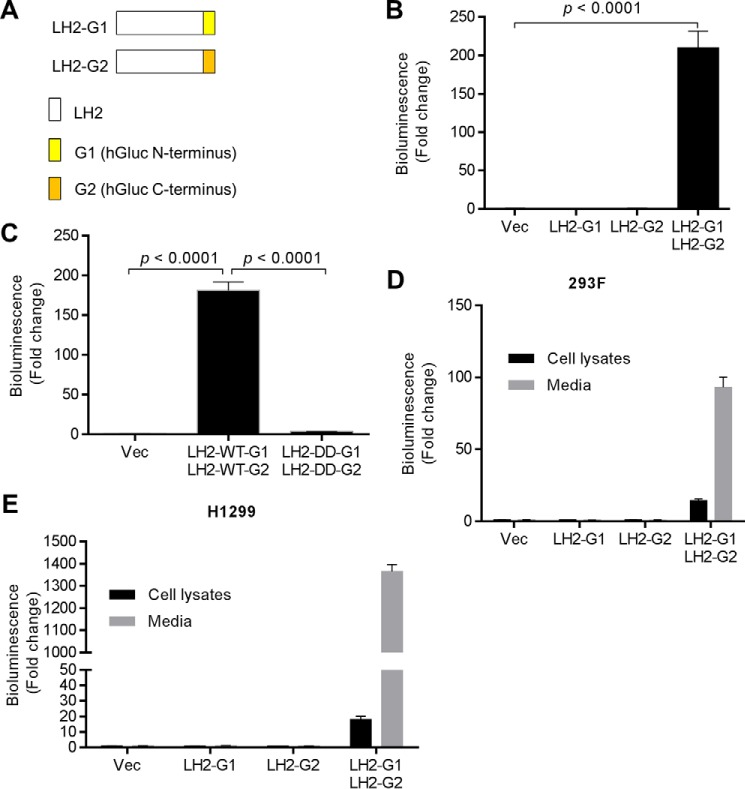
LH family members form homodimers and homodimerization is critical for LH enzymatic activity (26). To ascertain the presence of LH2 homodimeric complexes in the extracellular space, we utilized a protein-fragment complementation assay (27) in which the full-length LH2 protein is fused to the N- or C-terminal fragments of Gaussia luciferase (LH2-G1 and LH2-G2, respectively, Fig. 4A). As an initial validation step, we transfected the LH2-G1 and LH2-G2 reporters singly or in combination into 293F cells and measured luciferase activity in cell lysates. Compared with that achieved with transfection of either construct alone, co-transfection of the LH2-G1 and LH2-G2 constructs increased luciferase activity by ∼200-fold (Fig. 4B), and reconstitution of luciferase activity was abrogated by removal of LH2's putative dimerization motif (amino acids 562–568) (Fig. 4C). Using this assay, we measured luciferase activity in conditioned media samples and cell lysates, which showed the presence of dimeric, active LH2 in both compartments (Fig. 4, D and E).
FIGURE 4.
LH2 formed homodimers in the extracellular space. A, schema of reporters containing full-length LH2 fused to the N-terminal (G1) or C-terminal (G2) domains of Gaussia luciferase (hGluc). LH2, G1, and G2 are color-coded (the legend). B, luciferase activities were measured in total cell lysates of 293F cells transiently transfected with the indicated reporters or empty vector (Vec). Data are the means ± S.D.) of triplicate samples. C, luciferase activities were measured in total cell lysates of 293F cells transfected with the indicated reporters or empty vector (Vec). Full-length LH2 (WT). Mutant LH2 lacking the putative dimerization motif (DD). Data are the means ± S.D. of triplicate samples. D, luciferase activities were measured in total cell lysates (Cell lysates) or conditioned media (Media) isolated from 293F cells transiently transfected with the indicated reporters or empty vector (Vec). Data are the means ± S.D. of triplicate samples. E, luciferase activities were measured in total cell lysates (Cell lysates) or conditioned media (Media) isolated from H1299 cells transiently transfected with the indicated reporters or empty vector (Vec). Data are the means ± S.D. of triplicate samples. The results shown are representative of two independent experiments.
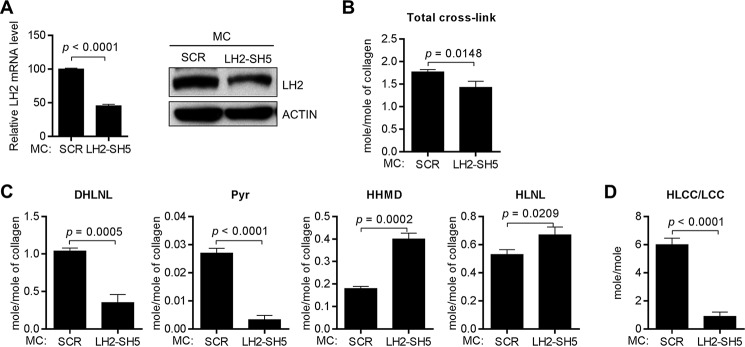
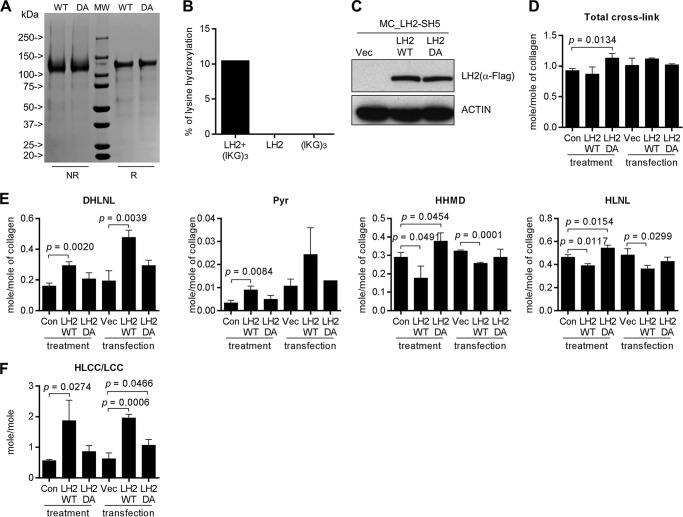
To determine whether LH2 can modify collagen in the extracellular space, we performed a rescue experiment on MC3T3 (MC) cells, which are murine calvaria-derived osteoblasts that have high LH2 expression and produce a matrix that is enriched in HLCCs and low in the LCC HHMD (9). After stable transfection of LH2-specific shRNAs to deplete LH2 in MC cells (Fig. 5A), LH2-deficient and scrambled control-transfected MC cells were cultured, and cell/matrix layers were collected and subjected to amino acid and cross-link analyses 2 weeks after the cells had achieved confluence. In all samples analyzed, d-Pyr levels were at the lower limits of detection and were, therefore, excluded from the analysis. In contrast to the total collagen cross-link concentrations, which were only minimally decreased by LH2 depletion (Fig. 5B), the LH2-deficient samples had sharply reduced DHLNL and Pyr (both HLCCs) and increased HHMD (LCC) (Fig. 5C), with a corresponding reduction in the HLCC-to-LCC ratio (5.998 ± 0.4661 versus 0.5094 ± 0.3050) (Fig. 5D). Because HLNL can be derived from either Lysald or Hylald by pairing with Hyl or Lys, respectively, HLNL was excluded from the calculation of the HLCC-to-LCC ratio. However, in the context of the changes in other cross-links, the moderate increase in HLNL concentrations observed in LH2-depleted samples was likely due to the decrease in the telopeptidyl Hylald levels. To rescue LH2 activity in the extracellular space of LH2-deficient MC cells, we added 45 nm (final concentration) recombinant LH2 proteins (wild-type or DA inactive mutant; Fig. 6A) to the media when the cells reached confluence. The wild-type recombinant LH2 hydroxylated a synthetic collagenous substrate, IKGIKGIKG ((IKG)3) (Fig. 6B), indicating that the protein has enzymatic activity. As a comparison to the recombinant LH2 protein treatments, LH2-deficient MC cells were rescued by stable transfection with a vector that expresses LH2 (wild-type or DA mutant) (Fig. 6C). The matrices were collected and subjected to collagen cross-link analysis 2 weeks after confluence. Reconstitution with wild-type LH2 by either approach had no effect on total collagen cross-link concentrations (Fig. 6D) but significantly increased the concentration of HLCCs (DHLNL and Pyr) and decreased LCCs (HHMD) (Fig. 6E), resulting in an increase in the HLCC-to-LCC ratio (Fig. 6F), whereas reconstitution with the mutant LH2 did not have this effect (Fig. 6, D–F). Treatment with recombinant LH2 also reversed the reduction in HLNL levels induced by LH2 depletion (Fig. 6E). These findings show that reconstitution of extracellular LH2 rescued the LH2-induced collagen cross-link switch, suggesting that LH2 is capable of hydroxylating collagen in the extracellular matrix.
FIGURE 5.
LH2 depletion decreased HLCCs and increased LCCs. A, the bar graph illustrates the results of Q-PCR analysis of LH2 mRNA levels in MC cells stably transfected with vectors expressing LH2 shRNA (SH5) or scrambled control shRNA (SCR). The results are expressed as the means ± S.D. of triplicate samples. The mRNA levels of the indicated transfectants are relative to those of the control transfectants, which were set at 100%. The blots on the right show the results of Western blot analysis of the MC transfectants. Actin was used as the loading control. The results shown are representative of two independent experiments. B–D, quantification of total collagen cross-links (B), HLCCs, LCCs, and HLNL (C) and the ratio of HLCCs-to-LCCs (D) in the MC transfectants. Total collagen cross-links was the sum of HLCCs (DHLNL+Pyr) + LCCs (HHMD) + HLNL. The HLCC-to-LCC ratio was calculated as (DHLNL+Pyr)/HHMD. Data are the means ± S.D. of triplicate biological samples in a single experiment. p values, 2-tailed Student's t test.
FIGURE 6.
Extracellular LH2 was sufficient to rescue the collagen cross-link switch. A, SDS-PAGE of immobilized metal affinity chromatography/gel filtration-purified wild-type LH2 (WT) or enzymatically-inactive D689A mutant LH2 (DA) under reducing (R) and non-reducing (NR) conditions. The positions of the molecular weight markers (MW) are indicated. The results shown are representative of two independent experiments. B, Hyl levels by amino acid analysis of (IKG)3 reacted with wild-type LH2. Control reactions were performed in the absence of (IKG)3 (LH2) or LH2 ((IKG)3). Each result is from one sample in a single experiment. C, Western blot analysis of MC_LH2-SH5 cells transfected with control vector (Vec) or vectors expressing WT LH2 or inactive LH2 (DA) to reconstitute intracellular and extracellular LH2. Actin was used as the loading control. The results shown are representative of two independent experiments. D–F, quantification of total collagen cross-links (D), HLCCs, LCCs, and HLNL (E), and the ratio of HLCCs-to-LCCs (F). Total collagen cross-links was the sum of HLCCs (DHLNL+Pyr) + LCC (HHMD) + HLNL. For each assay, the bars on the left are from cells treated with or without (Con) recombinant LH2 protein (treatment), and the bars on the right are from cells stably transfected with empty (Vec) or LH2 expression vectors (transfection). The HLCC-to-LCC ratio was calculated as (DHLNL+Pyr)/HHMD. The data are the means ± S.D. from triplicate biological samples in a single experiment. p values, 2-tailed Student's t test.
The results shown here demonstrate for the first time that LH2 is secreted and modifies collagen in the extracellular space. These findings raise many important questions, such as the way in which LH2 secretion is regulated, the sites on which LH2 binds type I collagen, and whether LH2 has extracellular substrates other than collagen telopeptidyl Lys residues. These findings also have important implications. First, they challenge the current understanding that LH2 modifies only the nascent pro-α chains before they form a triple helical procollagen molecule (2, 28) and imply that telopeptidyl Lys residues may remain accessible to LH2 after procollagen triple helix formation. Second, because extracellular LH2-catalyzed Lys hydroxylation must precede LOX-catalyzed conversion of telopeptidyl Lys to aldehyde, which occurs on the fibril form of collagen but not on denatured collagen or a collagen monomer (29), our findings suggest that LH2-induced collagen modifications may occur in the extracellular space before or at an early stage of fibrillogenesis, i.e. during procollagen processing and/or initial fibrillogenesis. Third, because collagen telopeptidyl Lys hydroxylation impacts the biomechanical properties of collagen cross-links, LH2 secretion may be an important regulatory step in collagen biosynthesis. Finally, disease states related to LH2 overproduction such as cancer (20–22, 30) and fibrosis (31, 32) may be effectively treated with anti-LH2 neutralizing antibodies that can inhibit extracellular LH2.
Experimental Procedures
Plasmids
To create the pLVX-Puro2 plasmid, we modified the pLVX-Puro plasmid (Clontech, Mountain View, CA) by adding the SbfI and NotI cutting sites after the XbaI site. The C-terminal 3× FLAG-tagged WT LH2 cDNAs were cloned into the XbaI and NotI sites on pLVX-Puro2 to generate the LH2–3FLAG-WT expression plasmid. The LH2–3FLAG cDNA without the signal peptide was amplified by PCR and cloned into the XbaI and NotI sites on pLVX-Puro2 to generate the LH2–3FLAG-ΔN expression plasmid. The N terminus (G1, amino acids 1∼93) or the C terminus (G2, amino acids 94∼169) of Gaussia luciferase were fused to the WT LH2 C terminus with the polypeptide linker (Gly-Gly-Gly-Gly-Ser)2 and cloned into the XbaI and NotI sites on pLVX-Puro2 to generate the LH2-G1 and LH2-G2 expression plasmids. The dimerization motif (amino acids 562∼568) on LH2 was deleted by overlapping PCR to generate the LH2-DD-G1 and LH2-DD-G2 expression plasmids. The pLKO.1_scramble and pLKO.1_LH2-SH5 plasmids were constructed as described previously (21). To create the pEF-bsr plasmid for the rescue experiment, we modified the pLVX-Puro plasmid by replacing the CMV promoter with the EF1α promoter, replacing the puromycin-resistant gene with the blasticidin-resistant gene, and adding the SbfI and NotI cutting sites after the XbaI site. The WT LH2 cDNA was released from the pLVX-Puro2-LH2–3FLAG and subcloned into the XbaI and NotI sites on the pEF-bsr plasmid. The D689A mutant was generated by site-directed mutagenesis using overlapping PCR.
Cell Culture
All cells were obtained from ATCC and grown in a humidified atmosphere with 5% CO2 at 37 °C. LH2-deficient MC3T3-E1 (MC) cells were cultured in minimum essential medium α supplemented with 10% FBS. 293T and 293F were cultured in a 1:1 mixture of DMEM and Ham's F-12 medium supplemented with 10% FBS. Human lung cancer cells were grown in RPMI 1640 with 10% FBS and 1% penicillin/streptomycin. Stable isotope labeling by amino acids in cell culture (SILAC) was done as described previously (33). Briefly, cells were grown for seven passages in RPMI 1640 supplemented with [13C]lysine and 10% dialyzed FBS according to the standard SILAC protocol (34).
Proteomic Analysis of Human Lung Cancer Cells
For protein fractionation, we used a Shimadzu HPLC system equipped with 2 LC-20AD pumps, 1 dual-wavelength SPD-20A UV detector (220 nm and 280 nm), 1 FRC-10A fraction collector, 1 CTO-20A column oven, and 1 SCL-10 controller. The whole system was controlled with an EZ Start work station (version 7.4 SP3). Protein samples from the total cell extract or the fraction secreted in media were reduced and alkylated with acrylamide, loaded onto an RPGS reversed-phase column (4.6 mm (inner diameter) × 150 mm, 15 μm, 1000 Å, Column Technology Inc., Fremont, CA), and subjected to desalting with 95% mobile-phase A (0.1% TFA in 95% H2O) at a flow rate of 3 ml/min for 5 min. Proteins were eluted from the column at a flow rate of 2.1 ml/min with gradient elution that included an increase from 5% to 70% mobile phase B (0.1% TFA in 95% acetonitrile) over 25 min, 70% to 95% mobile phase B for 3 min, a wash step to hold at 95% mobile phase B for 2 min, and a re-equilibration step at 95% mobile phase A for 5 min. Fractions were collected in 1.1-ml microtubes (Dot Scientific Inc., Burton, MI) at 20-s intervals and stored at −80 °C until use.
The lyophilized protein samples were dissolved in 100 mm NH4HCO3 (pH 8.5) followed by overnight in-solution digestion with trypsin at 37 °C. before LC-MS/MS analysis, the digestion was quenched by adding 5 μl of 1.0% formic acid solution. Briefly, peptides were separated by reversed-phase chromatography using a nano-HPLC system (Eksigent, Dublin, CA) coupled online with an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA). Mass spectrometer parameters were as follows: spray voltage, 2.5 kV; capillary temperature, 200 °C; FT resolution, 100,000; FT target value, 8 × 105; LTQ target value, 1 × 104; 1 FT microscan with 850-ms injection time; 1 LTQ microscan with 100-ms injection time. Mass spectra were acquired in a data-dependent mode with the m/z range of 400–2000. The full mass spectrum (MS scan) was acquired by the FT, and the tandem mass spectrum (MS/MS scan) was acquired by the LTQ with 35% normalized collision energy. The acquisition of each full mass spectrum was followed by the acquisition of MS/MS spectra for the 5 most intense +2 or +3 ions within a 1-s duty cycle. The minimum signal threshold (counts) for triggering an MS/MS scan for a precursor occurring during an MS scan was set at 1000.
The acquired LC-MS/MS data were processed by the Computational Proteomics Analysis System (18). Briefly, LC-MS/MS data were first converted to mzXML format using ReAdW software (version 1.2) to generate the peak list for protein database searching. The X!Tandem search engine (version 2005.12.01) parameters included cysteine alkylated with acrylamide (71.0371@C) as a fixed modification and methionine oxidation (15.99491@M) as a variable modification. Data were searched against the International Protein Index human protein knowledgebase (version 3.57), which contains entries for 76,542 proteins. The minimum criterion for peptide matching was a Peptide Prophet Score of ≥0.2. Peptides meeting this criterion were grouped to protein sequences using the Protein Prophet algorithm at an error rate of ≤5%. The total peptide count in each fraction was used as a measure of protein concentration within that fraction.
The proteomic search results from lung cancer cell lines was obtained and combined in a MySQL database application. The total number of spectral counts for each protein group within each sample from the total cell extract and the secreted media compartment was normalized to total spectral count set at 50,000 using R (v3.0.1)/RStudio (v0.97.551; RStudio, Inc). The data were further used for differential protein analysis between cell lines.
Generation of Stable Cell Lines
Parental MC cells were infected with lentiviruses carrying LH2-SH5 shRNA or the scramble control that had been packaged in 293T cells using the pMD2.G and psPAX2 plasmids. After 48 h, the cells were selected with puromycin (10 μg/ml) for 2 weeks to generate LH2-deficient MC cells (MC_LH2-SH5 cells). To restore LH2 expression, the MC_LH2-SH5 cells were infected with lentiviruses carrying the cDNA of WT LH2 or inactive LH2 (D689A) and selected with blasticidin (10 μg/ml) for 2 weeks.
Protein Complementation Assay
Plasmids with the LH2 and Gaussia luciferase fusions were co-transfected at a 1:1 ratio into 293F or H1299 cells plated on 24-well plates using the FuGENE 6 transfection reagent (Promega, Madison, WI). After 48 h, the cells were washed with PBS, 400 μl of passive lysis buffer (Promega) was added to each well, and the plate was shaken for 15 min at room temperature. Then, 100 μl of the lysate was mixed with 100 μl of native coelenterazine (Nanolight Technology) stock solution. Native coelenterazine was used at a final concentration of 20 μm. The signal intensities of the reaction were read using a Synergy 2 microplate reader (BioTek, Winooski, VT).
Quantitative Real-time PCR
Quantitative real-time PCR (Q-PCR) was done as described previously (21). Briefly, total RNA was isolated from the cells using TRIzol Q-PCR assays and reverse-transcribed into cDNA with qScript cDNA SuperMix (Quanta BioSciences, Gaithersburg, MD). The mRNA level was analyzed using a SYBR Green-based system (Applied Biosystems, Waltham, MA). mRNA levels were normalized on the basis of mRNA for ribosomal protein L32 (Rpl32). The primers for Q-PCR of Lh2 and Rpl32 have been described previously (21).
Western Blotting
Western blotting was performed as described previously (21). Briefly, cells were washed with PBS and lysed with cell lysis buffer (Cell Signaling Technology, Danvers, MA) to extract total proteins. Cell lysates were separated by SDS-PAGE, transferred onto nitrocellulose transfer membrane (GE Healthcare), and then incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (GE Healthcare). Protein bands were visualized with Pierce ECL Western blotting substrate (Thermo Fisher Scientific).
Purification of Recombinant LH2 Protein
Human LH2b recombinant proteins (residues 33–758, WT and inactive D689A mutant) were produced from FreeStyle CHO-S cells (Invitrogen) via large-scale transient transfection with polyethyleneimine as described previously (35, 36). The recombinant LH2b proteins were then purified from CHO-S cell-conditioned media utilizing immobilized metal affinity chromatography and gel filtration chromatography.
LH2 Activity Assay
An LH2 activity assay was performed in reaction buffer (20 mm HEPES buffer (pH 7.4), 150 mm NaCl) at 37 °C for 1 h with 1 μm LH2 enzyme, 50 μm FeSO4, 100 μm α-ketoglutarate, 500 μm ascorbate, 1 mm peptide substrate ((IKG)3), and 1.5 μm catalase as described previously (37). Peptide substrate hydroxylation was measured directly by amino acid analysis (see below).
Recombinant LH2 Protein Treatment
MC_LH2-SH5 cells were cultured in 20-cm culture plates, and the treatment started when the cells reached confluence. Recombinant LH2-WT and LH2-DA protein were added every other day with fresh medium supplemented with 100 μg/ml ascorbic acid (Sigma). The final concentration for the recombinant protein was 45 nm. The cells were treated for 2 weeks, and then cell matrixes were collected for the collagen cross-linking analysis.
Amino Acid Analysis
The peptide substrates with or without LH2 treatment were hydrolyzed with 6 n HCl and subjected to amino acid analysis as described (38). The % of Lys hydroxylation was calculated by Hyl/(Lys+Hyl). Amino acid analysis of the reduced collagen from cell/matrix layer was also performed in the same manner to determine the amount of Hyp (22).
Immunofluorescent Staining and Colocalization Analysis
Immunofluorescent staining of human lung cancer tissue sections was performed as described elsewhere (39). The LH2 antibody (R&D systems, catalog #MAB4445) was used in our previous study and showed high specificity (21). Anti-type I collagen antibody (ab34710) (Abcam) that recognizes native type I collagen molecule was used to stain type I collagen fibrils in the extracellular space. For widefield fluorescence microscopy, images were acquired with a 20× (0.75 NA) objective on a Nikon Eclipse Ti inverted widefield microscope equipped with DS-Qi2 monochrome sCMOS camera (for fluorescence) and DS-Ri2 color sCMOS camera (for color brightfield) using NIS Elements software (Nikon). For quantitative colocalization analysis, confocal images were acquired with Nyquist sampling criteria using a 40× (0.95 NA) objective on a Nikon A1+ confocal microscope using NIS Elements software. Raw images were iteratively deconvolved with CMLE algorithm in Huygens Professional version 15.05 (Scientific Volume Imaging, The Netherlands). Colocalization was estimated with colocalization analyzer plugin in Huygens Pro. Intensity plots of signal intensity (y axis) against distance in microns (x axis) were generated from 8-bit thresholded images with RGB profile plot in ImageJ (National Institutes of Health, Bethesda, MD).
Immunogold Labeling and Transmission Electron Microscopy
Two million DFC1024 human lung cancer cells were injected subcutaneously into nude mice to generate tumors. Tumors were fixed for 2 h at room temperature in phosphate-buffered saline (PBS) containing 2% paraformaldehyde. Fixed tissues were washed in PBS and then blocked overnight in PBS containing 1% bovine serum albumin and 0.1% Triton X-100 at 4 °C. This was followed by overnight incubation in blocking buffer containing anti-LH2 antibody (10 μg/ml) or control non-immune isotyped matched IgG2b antibody (10 μg/ml). After incubation with the primary antibody, the samples were washed in PBS and incubated overnight in NanogoldTM-anti-mouse secondary antibody (Nanoprobes, catalog #2002, 1.4 nm Nanogold® particle) diluted 1:40 in blocking buffer. After a PBS wash, the samples were incubated in GoldEnhance EMTM toner (Nanoprobes, catalog #2113) for 7 min and then washed in deionized water before fixing in 0.1 m sodium cacodylate buffer containing 2.5% glutaraldehyde for 60 min. The samples were then post-fixed in 1% osmium, dehydrated through an acetone series, infiltrated, and embedded in Embed-812 (Electron Microscopy Sciences, Hatfield, PA), sectioned (80–100 nm), and stained with lead and uranyl acetate. Sections were viewed on an FEI Tecnai 12 transmission electron microscope operating at 80 keV.
Collagen Cross-link Analysis
For the collagen cross-link analysis, MC_LH2-SH5 cells treated or non-treated with recombinant LH2 or inactive LH2 (n = 3/group) were cultured for 2 weeks as described (9, 40). The cell/matrix layer was first washed with cold PBS, scraped, and collected by centrifugation at 10,000 rpm for 30 min. The pellets were pulverized in liquid nitrogen using a Spex Freezer Mill (Spex, Metuchen, NJ), washed with cold PBS and cold distilled water, lyophilized, and weighed. Aliquots were reduced with standardized NaB3H4 and hydrolyzed with 6 n HCl. The hydrolysates were then subjected to amino acid and cross-linking analyses as described previously (38). Upon reduction, the reducible cross-links, i.e. deH-DHLNL/ketoamine (Hylald X Hyl), deH-HLNL/ketoamine (Hylald X Lys or Lysald X Hyl), and deH-HHMD (Lysald X Lysald X His X Hyl) are reduced to DHLNL, HLNL, and HHMD, respectively. Hereafter, the terms DHLNL, HLNL, and HHMD will be used for both the unreduced and reduced forms. These reducible cross-links were identified and measured as their reduced forms. The mature trivalent cross-links, Pyr and d-Pyr, were simultaneously analyzed by their fluorescence. All cross-links were quantified as the mol/mol collagen based on the value of 300 residues of Hyp per collagen molecule.
Author Contributions
Y. C., H. G., and M. T. conducted the experiments, including generation of stable cell lines, purification of the recombinant proteins, and performance of in vitro hydroxylase activity assays, biochemical and mass spectrometric analyses, and analysis of and interpretation of the data. A. A. M., H. K., and S. M. H. conducted and interpreted the stable isotope labeling of amino acids in cell culture analyses. P. B. performed the immunofluorescent staining and colocalization analysis. A. R. B. conducted the immunogold labeling and transmission electron microscopy experiment. G. B. F., J. Y., and X. L. provided technical assistance. J. M. K., M. Y., and Y. C. conceived the project, interpreted all of the data, and wrote the paper with other co-authors. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R21AR060978 (NIAMS; to M. Y.) and R01CA105155 (NCI; to J. M. K. and M. Y.). This work was also supported by the Elza A. and Ina S. Freeman Professorship in Lung Cancer (to J. M. K.) and MD Anderson Cancer Center Support Grant CA016672. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- LOX
- lysyl oxidase
- LH2
- lysyl hydroxylase 2
- HLCCs
- hydroxylysine aldehyde-derived collagen cross-links
- deH-HLNL
- dehydro-hydroxylysinonorleucine
- deH-DHLNL
- deH-dihydroxylysinonorlecine
- Pyr
- pyridinoline
- d-Pyr
- deoxypyridinoline
- deH-HHMD
- deH-histidinohydroxymerodesmosine
- LCCs
- Lys aldehyde-derived cross-links
- MC
- MC3T3-E1
- SILAC
- stable isotope labeling by amino acids in cell culture
- Q-PCR
- quantitative real-time PCR
- FT
- Fourier transform
- (IKG)3
- IKGIKGIKG.
References
- 1. Di Lullo G. A., Sweeney S. M., Korkko J., Ala-Kokko L., and San Antonio J. D. (2002) Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 277, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 2. Yamauchi M., and Sricholpech M. (2012) Lysine post-translational modifications of collagen. Essays Biochem. 52, 113–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanzer M. L., Housley T., Berube L., Fairweather R., Franzblau C., and Gallop P. M. (1973) Structure of two histidine-containing crosslinks from collagen. J. Biol. Chem. 248, 393–402 [PubMed] [Google Scholar]
- 4. Yamauchi M., London R. E., Guenat C., Hashimoto F., and Mechanic G. L. (1987) Structure and formation of a stable histidine-based trifunctional cross-link in skin collagen. J. Biol. Chem. 262, 11428–11434 [PubMed] [Google Scholar]
- 5. Kivirikko K. I., and Pihlajaniemi T. (1998) Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv. Enzymol. Relat. Areas Mol. Biol. 72, 325–398 [DOI] [PubMed] [Google Scholar]
- 6. Uzawa K., Grzesik W. J., Nishiura T., Kuznetsov S. A., Robey P. G., Brenner D. A., and Yamauchi M. (1999) Differential expression of human lysyl hydroxylase genes, lysine hydroxylation, and cross-linking of type I collagen during osteoblastic differentiation in vitro. J. Bone Miner. Res. 14, 1272–1280 [DOI] [PubMed] [Google Scholar]
- 7. Mercer D. K., Nicol P. F., Kimbembe C., and Robins S. P. (2003) Identification, expression, and tissue distribution of the three rat lysyl hydroxylase isoforms. Biochem. Biophys. Res. Commun. 307, 803–809 [DOI] [PubMed] [Google Scholar]
- 8. van der Slot A. J., Zuurmond A. M., Bardoel A. F., Wijmenga C., Pruijs H. E., Sillence D. O., Brinckmann J., Abraham D. J., Black C. M., Verzijl N., DeGroot J., Hanemaaijer R., TeKoppele J. M., Huizinga T. W., and Bank R. A. (2003) Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 278, 40967–40972 [DOI] [PubMed] [Google Scholar]
- 9. Pornprasertsuk S., Duarte W. R., Mochida Y., and Yamauchi M. (2004) Lysyl hydroxylase-2b directs collagen cross-linking pathways in MC3T3-E1 cells. J. Bone Miner. Res. 19, 1349–1355 [DOI] [PubMed] [Google Scholar]
- 10. Pornprasertsuk S., Duarte W. R., Mochida Y., and Yamauchi M. (2005) Overexpression of lysyl hydroxylase-2b leads to defective collagen fibrillogenesis and matrix mineralization. J. Bone Miner. Res. 20, 81–87 [DOI] [PubMed] [Google Scholar]
- 11. Takaluoma K., Lantto J., and Myllyharju J. (2007) Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix Biol. 26, 396–403 [DOI] [PubMed] [Google Scholar]
- 12. Kelley B. P., Malfait F., Bonafe L., Baldridge D., Homan E., Symoens S., Willaert A., Elcioglu N., Van Maldergem L., Verellen-Dumoulin C., Gillerot Y., Napierala D., Krakow D., Beighton P., Superti-Furga A., De Paepe A., and Lee B. (2011) Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J. Bone Miner. Res. 26, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwarze U., Cundy T., Pyott S. M., Christiansen H. E., Hegde M. R., Bank R. A., Pals G., Ankala A., Conneely K., Seaver L., Yandow S. M., Raney E., Babovic-Vuksanovic D., Stoler J., Ben-Neriah Z., et al. (2013) Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum. Mol. Genet. 22, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaheen R., Al-Owain M., Faqeih E., Al-Hashmi N., Awaji A., Al-Zayed Z., and Alkuraya F. S. (2011) Mutations in FKBP10 cause both Bruck syndrome and isolated osteogenesis imperfecta in humans. Am. J. Med. Genet. A 155A, 1448–1452 [DOI] [PubMed] [Google Scholar]
- 15. Puig-Hervás M. T., Temtamy S., Aglan M., Valencia M., Martínez-Glez V., Ballesta-Martínez M. J., López-González V., Ashour A. M., Amr K., Pulido V., Guillén-Navarro E., Lapunzina P., Caparrós-Martín J. A., and Ruiz-Perez V. L. (2012) Mutations in PLOD2 cause autosomal-recessive connective tissue disorders within the Bruck syndrome: osteogenesis imperfecta phenotypic spectrum. Hum. Mutat. 33, 1444–1449 [DOI] [PubMed] [Google Scholar]
- 16. Ha-Vinh R., Alanay Y., Bank R. A., Campos-Xavier A. B., Zankl A., Superti-Furga A., and Bonafé L. (2004) Phenotypic and molecular characterization of Bruck syndrome (osteogenesis imperfecta with contractures of the large joints) caused by a recessive mutation in PLOD2. Am. J. Med. Genet. A 131, 115–120 [DOI] [PubMed] [Google Scholar]
- 17. Salo A. M., Wang C., Sipilä L., Sormunen R., Vapola M., Kervinen P., Ruotsalainen H., Heikkinen J., and Myllylä R. (2006) Lysyl hydroxylase 3 (LH3) modifies proteins in the extracellular space, a novel mechanism for matrix remodeling. J. Cell Physiol. 207, 644–653 [DOI] [PubMed] [Google Scholar]
- 18. Wang C., Ristiluoma M. M., Salo A. M., Eskelinen S., and Myllylä R. (2012) Lysyl hydroxylase 3 is secreted from cells by two pathways. J. Cell Physiol. 227, 668–675 [DOI] [PubMed] [Google Scholar]
- 19. Salo A. M., Sipilä L., Sormunen R., Ruotsalainen H., Vainio S., and Myllylä R. (2006) The lysyl hydroxylase isoforms are widely expressed during mouse embryogenesis, but obtain tissue- and cell-specific patterns in the adult. Matrix Biol. 25, 475–483 [DOI] [PubMed] [Google Scholar]
- 20. Gilkes D. M., Bajpai S., Wong C. C., Chaturvedi P., Hubbi M. E., Wirtz D., and Semenza G. L. (2013) Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol. Cancer Res. 11, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Chen Y., Terajima M., Yang Y., Sun L., Ahn Y. H., Pankova D., Puperi D. S., Watanabe T., Kim M. P., Blackmon S. H., Rodriguez J., Liu H., Behrens C., Wistuba I. I., Minelli R., et al. (2015) Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J. Clin. Invest. 125, 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eisinger-Mathason T. S., Zhang M., Qiu Q., Skuli N., Nakazawa M. S., Karakasheva T., Mucaj V., Shay J. E., Stangenberg L., Sadri N., Puré E., Yoon S. S., Kirsch D. G., and Simon M. C. (2013) Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 3, 1190–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blanco M. A., LeRoy G., Khan Z., Alečković M., Zee B. M., Garcia B. A., and Kang Y. (2012) Global secretome analysis identifies novel mediators of bone metastasis. Cell Res. 22, 1339–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valtavaara M., Papponen H., Pirttilä A. M., Hiltunen K., Helander H., and Myllylä R. (1997) Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle. J. Biol. Chem. 272, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 25. Kalamajski S., and Oldberg A. (2010) The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 29, 248–253 [DOI] [PubMed] [Google Scholar]
- 26. Heikkinen J., Risteli M., Lampela O., Alavesa P., Karppinen M., Juffer A. H., and Myllylä R. (2011) Dimerization of human lysyl hydroxylase 3 (LH3) is mediated by the amino acids 541–547. Matrix Biol. 30, 27–33 [DOI] [PubMed] [Google Scholar]
- 27. Remy I., and Michnick S. W. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 28. Suokas M., Myllyla R., and Kellokumpu S. (2000) A single C-terminal peptide segment mediates both membrane association and localization of lysyl hydroxylase in the endoplasmic reticulum. J. Biol. Chem. 275, 17863–17868 [DOI] [PubMed] [Google Scholar]
- 29. Siegel R. C. (1974) Biosynthesis of collagen crosslinks: increased activity of purified lysyl oxidase with reconstituted collagen fibrils. Proc. Natl. Acad. Sci. U.S.A. 71, 4826–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sada M., Ohuchida K., Horioka K., Okumura T., Moriyama T., Miyasaka Y., Ohtsuka T., Mizumoto K., Oda Y., and Nakamura M. (2016) Hypoxic stellate cells of pancreatic cancer stroma regulate extracellular matrix fiber organization and cancer cell motility. Cancer Lett. 372, 210–218 [DOI] [PubMed] [Google Scholar]
- 31. van der Slot A. J., van Dura E. A., de Wit E. C., De Groot J., Huizinga T. W., Bank R. A., and Zuurmond A. M. (2005) Elevated formation of pyridinoline cross-links by profibrotic cytokines is associated with enhanced lysyl hydroxylase 2b levels. Biochim. Biophys. Acta 1741, 95–102 [DOI] [PubMed] [Google Scholar]
- 32. Remst D. F., Blaney Davidson E. N., Vitters E. L., Blom A. B., Stoop R., Snabel J. M., Bank R. A., van den Berg W. B., and van der Kraan P. M. (2013) Osteoarthritis-related fibrosis is associated with both elevated pyridinoline cross-link formation and lysyl hydroxylase 2b expression. Osteoarthritis Cartilage 21, 157–164 [DOI] [PubMed] [Google Scholar]
- 33. Schliekelman M. J., Taguchi A., Zhu J., Dai X., Rodriguez J., Celiktas M., Zhang Q., Chin A., Wong C. H., Wang H., McFerrin L., Selamat S. A., Yang C., Kroh E. M., Garg K. S., Behrens C., et al. (2015) Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 75, 1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ong S. E., and Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 35. Cao Y., Hoeppner L. H., Bach S., Guanni E., Guo Y., Wang E., Wu J., Cowley M. J., Chang D. K., Waddell N., Grimmond S. M., Biankin A. V., Daly R. J., Zhang X., and Mukhopadhyay D. (2013) Neuropilin-2 promotes extravasation and metastasis by interacting with endothelial α5 integrin. Cancer Res. 73, 4579–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kivirikko K. I., and Myllylä R. (1982) Posttranslational enzymes in the biosynthesis of collagen: intracellular enzymes. Methods Enzymol. 82, 245–304 [DOI] [PubMed] [Google Scholar]
- 38. Yamauchi M., and Shiiba M. (2008) Lysine hydroxylation and cross-linking of collagen. Methods Mol. Biol. 446, 95–108 [DOI] [PubMed] [Google Scholar]
- 39. Robertson D., and Isacke C. M. (2011) Multiple immunofluorescence labeling of formalin-fixed paraffin-embedded tissue. Methods Mol. Biol. 724, 69–77 [DOI] [PubMed] [Google Scholar]
- 40. Sricholpech M., Perdivara I., Nagaoka H., Yokoyama M., Tomer K. B., and Yamauchi M. (2011) Lysyl hydroxylase 3 glucosylates galactosylhydroxylysine residues in type I collagen in osteoblast culture. J. Biol. Chem. 286, 8846–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]