Abstract
Flagellated bacteria modulate their swimming behavior in response to environmental cues through the CheA/CheY signaling pathway. In addition to responding to external chemicals, bacteria also monitor internal conditions that reflect the availability of oxygen, light, and reducing equivalents, in a process termed “energy taxis.” In Escherichia coli, the transmembrane receptor Aer is the primary energy sensor for motility. Genetic and physiological data suggest that Aer monitors the electron transport chain through the redox state of its FAD cofactor. However, direct biochemical data correlating FAD redox chemistry with CheA kinase activity have been lacking. Here, we test this hypothesis via functional reconstitution of Aer into nanodiscs. As purified, Aer contains fully oxidized FAD, which can be chemically reduced to the anionic semiquinone (ASQ). Oxidized Aer activates CheA, whereas ASQ Aer reversibly inhibits CheA. Under these conditions, Aer cannot be further reduced to the hydroquinone, in contrast to the proposed Aer signaling model. Pulse ESR spectroscopy of the ASQ corroborates a potential mechanism for signaling in that the resulting distance between the two flavin-binding PAS (Per-Arnt-Sim) domains implies that they tightly sandwich the signal-transducing HAMP domain in the kinase-off state. Aer appears to follow oligomerization patterns observed for related chemoreceptors, as higher loading of Aer dimers into nanodiscs increases kinase activity. These results provide a new methodological platform to study Aer function along with new mechanistic details into its signal transduction process.
Keywords: chemotaxis, electron transport system (ETS), flavin adenine dinucleotide (FAD), oxidation-reduction (redox), redox signaling, bacterial energy taxis, Nanodiscs, bacterial chemotaxis
Introduction
Bacteria search out conditions that provide sufficient energy to sustain homeostasis and fuel growth. In Escherichia coli, so-called “energy taxis” employs a sensory transmembrane protein, Aer, to monitor the cellular energy state and convey that information to the chemotaxis system (1–5). Aer is related to bacterial chemoreceptors, also known as MCPs,2 that sense chemicals and ligand-binding proteins in the periplasmic space (5–8). However, unlike MCPs, the sensor domain of Aer resides on the cytoplasmic side of the membrane and Aer lacks the methylation-based adaptation mechanism that provides negative feedback in chemosensing (9, 10). Homodimeric Aer is composed of two N-terminal cytoplasmic PAS (Per-Arnt-Sim) domains, a membrane anchor region composed of two transmembrane helices per subunit (TM1 and TM2) (11, 12), a cytoplasmic HAMP domain (helices AS1 and AS2), and a kinase control module that binds and regulates the histidine kinase CheA. MCPs, CheA, and the coupling protein CheW form extended hexagonal arrays that generate signal gain and cooperative responses in chemotaxis (7, 13). Within these arrays, a core complex composed of two receptor trimer-of-dimers, a dimeric CheA, and two CheW proteins comprise a minimal unit for signaling (14). The Aer PAS domains bind FAD (3, 9, 15) and are believed to receive redox signals from components of the electron transport chain (ETC) in the inner membrane (15).
Genetic and cellular experiments have determined that Aer mediates aerotaxis (2, 3, 9); however, this behavior does not involve direct reaction of Aer with O2. Rather, Aer responds to the redox status of the ETC, which in turn depends on aerobic respiration (4, 15). The prevailing model for Aer signaling has been derived largely from physiological data and involves the three redox states of FAD cofactor. In the absence of substrates to provide reducing equivalents, the FAD remains oxidized and Aer activates CheA autophosphorylation. CheA then transfers phosphate to the response regulator, CheY. Phosphorylated CheY binds to the flagellar motor, reverses the direction of flagella rotation, and causes cell tumbling. When substrates and terminal electron acceptors are present, Aer inhibits CheA, causing smooth swimming (5, 15). Under these conditions, Aer is presumed to be reduced to the semiquinone state by the ETC (5, 15). With substrates present, but no terminal acceptors for the ETC, cells tumble again, which has been hypothesized to occur because of further FAD reduction to the hydroquinone (HQ) state (5, 15) (see Fig. 1A).
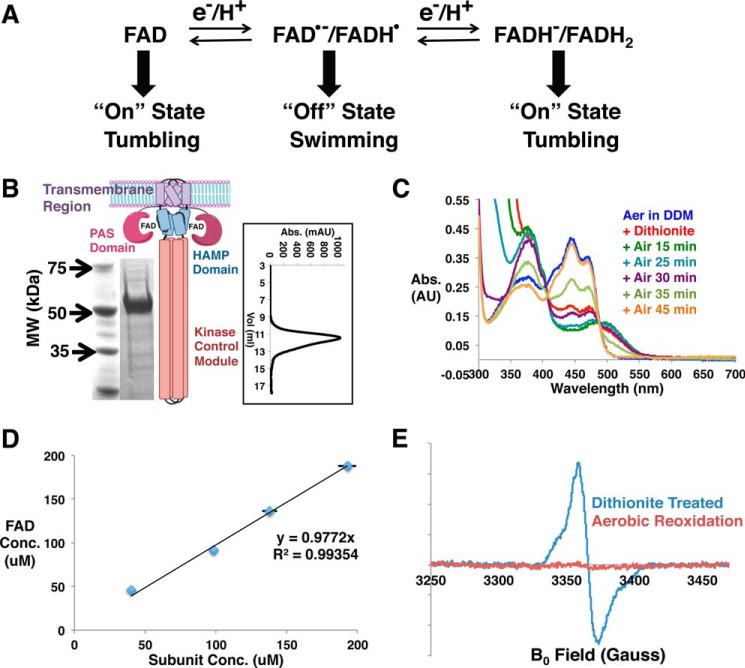
FIGURE 1.
Purification of Aer. A, schematic representation for the prevailing model of Aer-mediated energy taxis. B, the schematic of Aer dimer depicts the organization of the functional domains, and the SDS-PAGE gel shows the migration of detergent-purified Aer. Lane 1, molecular weight (MW) marker; lane 2, purified Aer. The inset displays the size exclusion profile of E. coli Aer in detergent in Superdex 200 10/300 column. Abs., absorbance; mAU, milliabsorbance units. C, UV-visible spectra of FAD in detergent-solubilized Aer after reduction with 10 mm sodium dithionite for the indicated times. AU, arbitrary units. D, Aer-bound FAD and the Aer subunit concentrations are highly correlated (black line: best fit). The FAD concentration (FAD Conc.) was calculated from UV-visible spectra of Aer with an extinction coefficient of 11.3 mm−1 cm−1 at 445 nm, and the Aer subunit concentration (Subunit Conc.) was measured with the Bradford assay. Error bars that are not evident are smaller than the data symbols. E, cw-ESR spectra of dithionite-reduced and aerobically reoxidized Aer-bound FAD.
There is currently no direct biochemical data correlating the redox state of the Aer PAS domain with kinase activity. Furthermore, although Aer clearly regulates CheA, it has not been determined whether Aer forms similar core complexes with CheA and CheW as do other MCPs (15, 16). Here, we report the purification of E. coli Aer and its incorporation into functional nanodiscs. We demonstrate that the full-length protein binds flavin as predicted, and that the cofactor undergoes chemical reduction to the anionic semiquinone (ASQ). Aer assembles into higher-order complexes with CheA and CheW, where it regulates CheA activity based on the redox state of FAD. No evidence is found for a stable hydroquinone form of the protein.
Results
Detergent-solubilized Aer Undergoes Reversible Chemical Reduction
N-terminally His6-tagged E. coli Aer was expressed in BL21(DE3) cells and purified in 0.1% n-dodecyl-β-d-maltopyranoside (DDM) using Co2+ affinity chromatography (Fig. 1B). The purified Aer shows signature UV-visible spectra of protein-bound FAD in the fully oxidized state with three absorption peaks at 416, 441, and 465 nm and a relatively broad shoulder centered at 365 nm (Fig. 1C). Aer binds approximately one FAD cofactor per subunit (Fig. 1D). Upon treatment with 10 mm dithionite in anaerobic conditions, the UV-visible spectrum of the Aer FAD converts to that of the single-electron reduced ASQ state as reflected by the concomitant increase at 372 and 500 nm, and disappearance of the 441 nm peak. A shoulder at 395 nm, which is a defining characteristic of the FAD ASQ state, could also be detected. The ASQ reverts back to the fully oxidized cofactor on exposure to air (Fig. 1C) (17). The continuous wave electron spin resonance (cw-ESR) spectrum confirms the radical nature of the dithionite-reduced FAD, which becomes ESR-silent upon reoxidation (Fig. 1E). The magnetic dipolar coupling between the flavin radicals can be used to measure their distance of separation by pulsed-dipolar ESR spectroscopy (PDS). PDS of the FAD ASQ state reveals one distance peak centered at 41.3 Å, suggesting purely dimeric association of Aer in DDM (Figs. 1B and 2A). These data also define a structural constraint on the disposition of the PAS domain in Aer (Fig. 2B). Interestingly, prolonged anaerobic incubation of Aer with 16 mm dithionite (DT) does not yield the two-electron reduced HQ state. DDM is detrimental to CheA activity, and thus nanodisc incorporation was applied to stabilize Aer in the absence of detergent (18–20). Nanodisc incorporation is extremely effective at reconstituting transmembrane MCPs in active forms (14, 21, 22), and the same holds true for Aer.
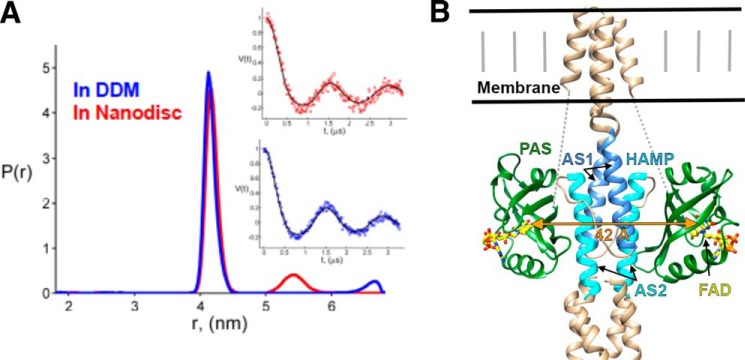
FIGURE 2.
PAS domain separations in Aer. A, PDS-derived distance distributions between ASQ states of FAD in Aer solubilized in DDM and incorporated into nanodiscs (Aer:MSP1E3D1 = 1.43). Both P(r)s are plotted normalized to the area of major peaks. The insets show the fits to the time domain DEER data for both cases. DQC data for detergent are very similar to respective DEER data (not shown). B, model of Aer based on data in Ref. 28. The 42 Å flavin separation agrees well with a tight interaction between the PAS β-sheet and HAMP helix AS2. Dotted lines represent an ∼40-residue unmodeled linker between PAS and the first transmembrane helix. An F1 linker helix between PAS and HAMP is also not shown.
Aer Reversibly Regulates CheA Activity Based on FAD Redox State
Aer was incorporated into nanodiscs assembled with E. coli polar lipids and the membrane scaffold protein (MSP), MSP1E3D1 (Fig. 3A, inset). As in detergent, Aer in nanodiscs can be reversibly reduced to the ASQ state upon dithionite treatment as revealed by UV-visible (Fig. 3A) and cw-ESR (Fig. 3B) spectroscopies. Nanodisc-incorporated Aer with fully oxidized FAD activates CheA to produce phosphorylated CheY, whereas upon chemical reduction to the ASQ, CheA activity is inhibited (Fig. 3C). Aer reoxidation by ambient oxygen recovers a majority of the activity. The reason for incomplete recovery is unknown, although it is not uncommon for DT to adversely affect protein activity. DT has little effect on CheA activity alone. Thus, Aer with oxidized FAD activates CheA and would therefore cause cell tumbling. Reduction of Aer to the ASQ reversibly inhibits kinase activity and would cause smooth swimming.
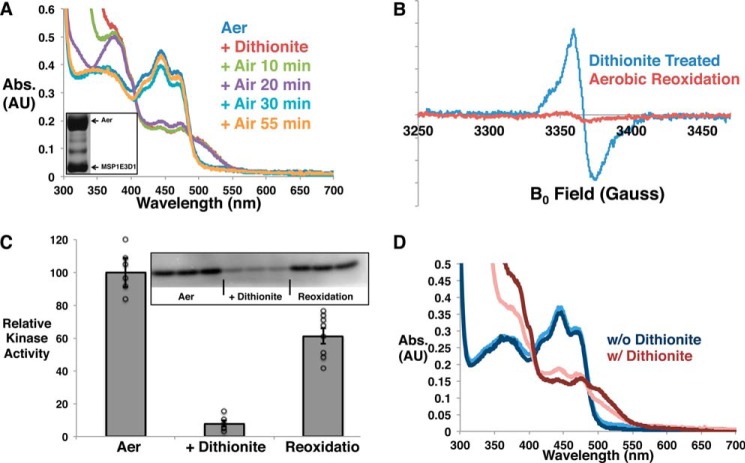
FIGURE 3.
Aer activity in nanodiscs depends on redox state. A and B, UV-visible spectra (A) and cw-ESR spectra (B) at different times during dithionite reduction and aerobic reoxidation of FAD in Aer incorporated in nanodiscs. Abs., absorbance; AU, absorbance units. The inset of A displays the bands for Aer and MSP1E3D1 on an SDS-PAGE gel after nanodisc reconstitution and purification. C, reversible regulation of CheA phospho-transfer activity based on the FAD redox state. Phospho-transfer to CheY is shown relative to that for fully oxidized FAD (designated as Aer), which has been normalized to 100. The error bars display the S.E. with n ≥ 6. The inset displays a representative autoradiograph, and the individual data points are shown as open black circles. D, the effect of additional free FAD on the chemical reduction of Aer. The UV-visible absorbance spectrum of Aer in nanodiscs with additional free FAD (0.1 mg/ml) is shown after subtracting the spectrum for free FAD alone. Likewise, the spectrum for dithionite-treated Aer in nanodiscs with additional FAD is shown after subtracting the spectrum for the reduced, free FAD. The light-colored spectra are for Aer without additional free FAD added, whereas the dark-colored spectra are for the Aer contribution in the presence of oxidized (blue) or fully reduced (red) free FAD.
In nanodiscs, we still could not generate a stable HQ state of Aer FAD by prolonged incubation with dithionite in anaerobic conditions even in the presence of redox mediators, which facilitate outer sphere electron and proton transfer reactions to FAD. Another reductant, Cr-EDTA, also failed to reduce the cofactor to the HQ state. As free FAD is reduced to the HQ state by dithionite, chemical reduction of Aer was tested in the presence of additional FAD with the expectation that the exchange of free FAD with that bound to Aer would lead to overall reduction to the HQ state. However, UV-visible spectroscopy could only detect the semiquinone state for the Aer-bound FAD (Fig. 3D), thereby indicating that exchange of the Aer FAD with free FAD HQ is not favored.
Activity Depends on Oligomerization of Aer Dimers
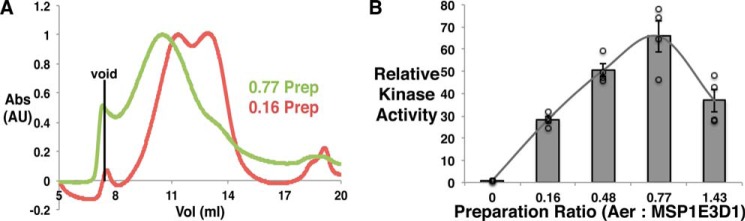
Ternary signaling complexes of the major E. coli MCPs Tar (aspartate receptor) and Tsr (serine receptor) with CheA and CheW require a trimer of receptor dimers to activate CheA (14, 23–26). To investigate whether association of dimers is important for Aer activity, we varied the molar ratio of Aer to MSP1E3D1 for the reconstitutions to alter the average number of Aer dimers in each nanodiscs, as was demonstrated previously for Tar (14). Notably, two MSP proteins are required for nanodisc formation, and thus the preparation with Aer to the MSP1E3D1 ratio of 0.16 will mainly produce single Aer dimers in the discs. Size exclusion chromatograms for two preparations (Aer:MSP1E3D1 = 0.16 and 0.77) reveal a shift of the nanodisc-incorporated Aer to a smaller elution volume in the 0.77 preparation, implying substantially larger particles with higher Aer:MSP subunit ratios (Fig. 4A). Increasing the molar ratio of the receptor in the preparations (14, 27) raises the probability of three dimers aligned on the same side of the nanodiscs. Interestingly, distance distribution measurement between FAD ASQs in the highest ratio preparation (1.43) gives two peaks centered at 41.5 and 52.4 Å, respectively (Fig. 2A). The peak at smaller distance arises from the intersubunit separation (which matches well to the distance in detergent). The narrow breadth of the shorter distance distribution indicates a relatively stable juxtaposition of the PAS domains, whereas the ∼42 Å separation is too far for a PAS-PAS dimer. However, a close association between the two PAS and HAMP domains (Fig. 2B), in accordance with identified PAS-HAMP interface residues (28), agrees well with the PDS data. Although the PDS peak at longer separation is rather weak and cannot be confirmed with full certainty, the longer distance (absent in detergent) may arise from different dimers on the same side of the nanodiscs. Accordingly, the ability of CheA to catalyze phospho-transfer to CheY is modulated based on the preparation ratio for the Aer incorporation into nanodiscs (Fig. 4B). The kinase activity shows a bell-shaped dependence on the molar ratio, which is very similar to what has been observed for Tar (14), suggesting that Aer requires the similar trimeric assembly of other MCPs to achieve maximum kinase activation. However, due to the low cellular abundance of Aer (∼2% of all MCPs (29, 30), it may associate as mixed trimers with the related high abundance chemoreceptors Tar and Tsr (31, 32).
FIGURE 4.
Aer oligomerization affects activity. A, higher molar ratios of Aer to MSP1E3D1 increases Aer loading into nanodiscs. The analytical size exclusion chromatograms for two preparations (Prep) of Aer into nanodiscs are shown after the areas of the peaks have been normalized. Larger particles with a higher average number of Aer dimers per nanodisc result from increased Aer:MSP1E3D1 ratios. The solid black line marks the position of the void peak. Abs., absorbance; AU, arbitrary units. B, CheA activities in the presence of various Aer preparations are compared. The activities are measured relative to the control sample (designated by 0), which contains only CheA, CheW, and CheY. The continuous line is drawn for the aid of inspection. Individual data points are shown as open black circles. The error bars are S.E. with n ≥ 4.
Discussion
Here, we show that Aer modulates CheA activity based on FAD redox states, and like other chemoreceptors, likely functions as a trimer-of-dimers. The prevailing model (5, 15) conjectured a third HQ state to explain the cellular tumbling in the presence of substrate and absence of terminal electron acceptors. However, in our in vitro experimental conditions, even the low-potential reductant DT was unable to produce a fully reduced HQ state. Notably, the related flavin-binding protein Azotobacter vinelandii NifL reduces to the HQ upon DT treatment (33, 34). Further reduction of Aer may be limited by its preference to form the anionic instead of the neutral semiquinone. Reduction to the hydroquinone requires proton transfer, which may be kinetically disfavored under these conditions. If oxygen were to react directly with Aer, it would not be expected to produce the observed smooth swimming response because this behavior requires reduced FAD, which O2 destabilizes. However, oxygen can favor FAD reduction by rather acting as an acceptor to the ETC.
Interface studies indicate that the HAMP AS2 helix and the PAS β-sheet closely associate in the kinase-off state (28). Such interactions are fully consistent with the well defined ∼42 Å separation between the reduced flavin cofactors that we observe. Thus, tight sandwiching of the HAMP domain by the reduced PAS domains appears to send an “off” signal through Aer to CheA.
Experimental Procedures
E. coli Aer Expression and Purification
BL21(DE3) cells were transformed with pET28 vector containing N-terminally His6-tagged E. coli Aer, plated on a kanamycin-added LB-agar plate, and incubated at 37 °C overnight. A single colony was added to an autoclaved and kanamycin-mixed LB medium for overnight growth under 200 rpm of shaking at 37 °C. 1 liter of the autoclaved and antibiotic-added Terrific Broth medium was inoculated with 20 ml of the overnight culture. The cells were then grown at 37 °C until A600 reached ∼0.3. The temperature was turned down to 16 °C, whereas cell growth continued until A600 reached 0.6–0.8. At this point, the expression was induced by adding 0.5 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested after overnight expression at 16 °C and stored at −80 °C.
The cells were resuspended in KCl extraction buffer (50 mm Tris, pH 8.0; 500 mm KCl; 10% glycerol), sonicated for 5–6 min, and centrifuged at 10,000 rpm for 30 min to obtain the membrane fractions in the low-spin pellet, which was again resuspended in KCl extraction buffer added with 1 mm PMSF and 1% DDM. The resuspended low-spin pellet was first sonicated at low power with the 4.5 power setting on the instrument to ensure fine resuspension and then at high power (7.5 setting) for 2 min before rocking at 4 °C for 5 h and centrifuging at 22,000 rpm for 1 h. The supernatant was incubated with Co(II) resin overnight after 0.1 mg/ml FAD was added to it. The next morning, the resin was washed once with wash buffer I (50 mm Tris, pH 8.0; 300 mm NaCl; 10% glycerol) containing 0.1% DDM and 0.1 mg/ml FAD. The resin was then washed several times with wash buffer II (25 mm Tris, pH 8.0; 150 mm NaCl; 10% glycerol) containing 0.1% DDM and 0.1 mg/ml FAD, and then with the buffer without FAD, and finally, Aer protein was eluted from the resin with elution buffer (wash buffer II; 250 mm imidazole, pH 8.0). UV-visible spectrum was acquired to ensure that Aer contained FAD. Normally, 1 liter of Terrific Broth culture typically gave ∼12 mg of Aer (1 ml of 100 μm dimer).
Aer Incorporation into Nanodiscs
Detergent-purified Aer was incorporated into lipidic nanodiscs following the protocols described in various articles (35–37). Briefly, chloroform was evaporated from E. coli polar lipid stock by blowing argon and then by keeping under vacuum overnight. The lipid was then dissolved in sodium cholate by sonication to form vesicles and stored at −80 °C. Membrane scaffold protein His6-MSP1E3D1 was purified from BL21(DE3) as described elsewhere (36, 37). The lipid amount to be used in the nanodisc preparations with variable molar ratio of Aer to MSP1E3D1 was determined empirically. 25 mm sodium cholate, 4% glycerol, and 0.05 mg/ml FAD were maintained into the reaction mixture to ensure solubility and stability. The mixture was rocked at 4 °C for 60 min before pre-hydrated Bio-Beads (SM2) of two-thirds of the volume of the mixture were added and then incubated at 4 °C overnight. The mixture was then centrifuged at 13,000 rpm for 30 min, and the supernatant was further purified either through size exclusion chromatography or in a desalting column to separate the nanodiscs from free FAD and excess lipids. The size exclusion chromatography experiments were done with the Superdex 200 10/300 column from GE Healthcare.
Sodium Dithionite Reduction of Aer and Aerobic Reoxidation
10 mm DT was added to Aer from a stock of 100 mm prepared in 1 m Tris, pH 8.0, in an anaerobic glove box, and UV-visible spectra were recorded with a cuvette of 0.2-cm path length that was sealed with Parafilm. To follow aerobic reoxidation, the Parafilm seal was removed, the cuvette was stored in a 4 °C refrigerator, and at different times, the spectra were recorded.
cw-ESR Spectra Collection
cw-ESR spectra were collected at 4 °C on Bruker Elexsys E500 EPR instrument at 9.4 GHz with 100-kHz modulation frequency and 1.5- or 2-G modulation amplitude. DT-reduced spectra of Aer contained narrow features because of DT radicals, which were subtracted in MATLAB (MathWorks Inc.).
Pulsed-dipolar ESR
Distance distributions between ASQ FADs in Aer were calculated from PDS data as described elsewhere (26, 38, 39). Briefly, ∼60 μm DT-treated Aer solutions containing 35% (v/v) glycerol were prepared in ESR tubes. The dipolar evolution was recorded for ∼20 h (DDM sample) and for ∼40 h (nanodisc sample) at 17.35 GHz on a home-built 2D Fourier transform-ESR instrument using four-pulse double electron-electron resonance (DEER) and double quantum coherence (DQC) ESR. Pump and detection π-pulses were 40 ns, and a 60-MHz frequency separation was used. Data averaging was significantly faster with DQC (4 h for DDM) and is further reduced by the use of a new wavelet-based data denoising method (40).3 The baseline of the time domain data was corrected with a log-linear polynomial function. The data were then converted to distance distributions between ASQ FAD centers with Tikhonov regularization (41) followed by maximum entropy refinement (42) where the regularization parameter λ of 0.05 was used to achieve acceptable fits of time-domain data (Fig. 2A).
CheY Phospho-transfer Assay
As described elsewhere (26) samples containing 2 μm CheA subunit, 3 μm CheW, 15 μm Aer subunit, and 50 μm CheY were produced and treated with no DT (for the fully oxidized state), or 16 mm DT (for the ASQ state). The 20-μl assay mixtures contained ∼10% glycerol (v/v) and 2 μl of buffer composed of 62.5 mm Tris (pH 7.5), 625 mm KCl, 6.25 mm EDTA, and 125 mm MgCl2. 16 mm DT was utilized to ensure that the Aer stayed reduced even after the addition of 5 μl of radioactive ATP solution (which was prepared aerobically) during the assays. For the reoxidation experiment, Aer alone was first reduced with 10 mm DT and aerobically reoxidized, and then this reoxidized Aer was mixed with the other proteins.
Author Contributions
D. S. and B. R. C. designed the research, D. S., J. W., and P. P. B. performed the experiments, D. S., P. P. B., and B. R. C. analyzed the data, and D. S. and B. R. C. wrote the manuscript with input from all authors.
Acknowledgments
We thank Boris Dzikovski for valuable suggestions and discussions, Madhur Srivastava for help with data analysis, and Zachary Maschmann for help with sample preparation.
This work was supported by National Institutes of Health Grants GM066775 (to B. R. C.) and P41GM103521 and R01EB003150 (to J. H. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
M. Srivastava, E. R. Georgieva, and J. H. Freed, manuscript in preparation.
- MCP
- methyl-accepting chemotaxis protein
- ETC
- electron transport chain
- MSP
- membrane scaffold protein
- ASQ
- anionic semiquinone
- HQ
- hydroquinone
- DT
- dithionite
- cw-ESR
- continuous wave electron spin resonance
- PDS
- pulsed-dipolar ESR spectroscopy
- DEER
- double electron-electron resonance
- DQC
- double quantum coherence
- DDM
- n-dodecyl-β-d-maltopyranoside.
References
- 1. Taylor B. L., Zhulin I. B., and Johnson M. S. (1999) Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53, 103–128 [DOI] [PubMed] [Google Scholar]
- 2. Rebbapragada A., Johnson M. S., Harding G. P., Zuccarelli A. J., Fletcher H. M., Zhulin I. B., and Taylor B. L. (1997) The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. 94, 10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bibikov S. I., Biran R., Rudd K. E., and Parkinson J. S. (1997) A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179, 4075–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor B. L., and Zhulin I. B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor B. L. (2007) Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65, 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wadhams G. H., and Armitage J. P. (2004) Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5, 1024–1037 [DOI] [PubMed] [Google Scholar]
- 7. Hazelbauer G. L., Falke J. J., and Parkinson J. S. (2008) Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parkinson J. S. (2010) Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64, 101–122 [DOI] [PubMed] [Google Scholar]
- 9. Bibikov S. I., Barnes L. A., Gitin Y., and Parkinson J. S. (2000) Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bibikov S. I., Miller A. C., Gosink K. K., and Parkinson J. S. (2004) Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186, 3730–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amin D. N., Taylor B. L., and Johnson M. S. (2006) Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 188, 894–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amin D. N., Taylor B. L., and Johnson M. S. (2007) Organization of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 189, 7206–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parkinson J. S., Hazelbauer G. L., and Falke J. J. (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boldog T., Grimme S., Li M., Sligar S. G., and Hazelbauer G. L. (2006) Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. 103, 11509–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Repik A., Rebbapragada A., Johnson M. S., Haznedar J. O., Zhulin I. B., and Taylor B. L. (2000) PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36, 806–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taylor B. L., Rebbapragada A., and Johnson M. S. (2001) The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3, 867–879 [DOI] [PubMed] [Google Scholar]
- 17. Liu B., Liu H., Zhong D., and Lin C. (2010) Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 13, 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bayburt T. H., Grinkova Y. V., and Sligar S. G. (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 [Google Scholar]
- 19. Denisov I. G., Grinkova Y. V., Lazarides A. A., and Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 20. Nath A., Atkins W. M., and Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 21. Amin D. N., and Hazelbauer G. L. (2010) The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J. Bacteriol. 192, 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amin D. N., and Hazelbauer G. L. (2012) Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J. Biol. Chem. 287, 41697–41705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim K. K., Yokota H., and Kim S.-H. (1999) Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 400, 787–792 [DOI] [PubMed] [Google Scholar]
- 24. Studdert C. A., and Parkinson J. S. (2004) Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U.S.A. 101, 2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Briegel A., Li X., Bilwes A. M., Hughes K. T., Jensen G. J., and Crane B. R. (2012) Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 3766–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Samanta D., Borbat P. P., Dzikovski B., Freed J. H., and Crane B. R. (2015) Bacterial chemoreceptor dynamics correlate with activity state and are coupled over long distances. Proc. Natl. Acad. Sci. U.S.A. 112, 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bayburt T. H., Grinkova Y. V., and Sligar S. G. (2006) Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 450, 215–222 [DOI] [PubMed] [Google Scholar]
- 28. Garcia D., Watts K. J., Johnson M. S., and Taylor B. L. (2016) Delineating PAS-HAMP interaction surfaces and signalling-associated changes in the aerotaxis receptor Aer. Mol. Microbiol. 100, 156–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li M., and Hazelbauer G. L. (2004) Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186, 3687–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gosink K. K., Burón-Barral M. C., and Parkinson J. S. (2006) Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 188, 3487–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng X., Baumgartner J. W., and Hazelbauer G. L. (1997) High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179, 6714–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barnakov A. N., Barnakova L. A., and Hazelbauer G. L. (1998) Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180, 6713–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hill S., Austin S., Eydmann T., Jones T., and Dixon R. (1996) Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl. Acad. Sci. U.S.A. 93, 2143–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macheroux P., Hill S., Austin S., Eydmann T., Jones T., Kim S. O., Poole R., and Dixon R. (1998) Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem. J. 332, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boldog T., Li M., and Hazelbauer G. L. (2007) Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 423, 317–335 [DOI] [PubMed] [Google Scholar]
- 36. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., and Sligar S. G. (2009) Chapter 11: Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karasawa A., Swier L. J. Y. M., Stuart M. C. A., Brouwers J., Helms B., and Poolman B. (2013) Physicochemical factors controlling the activity and energy coupling of an ionic strength-gated ATP-binding cassette (ABC) transporter. J. Biol. Chem. 288, 29862–29871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Airola M. V., Sukomon N., Samanta D., Borbat P. P., Freed J. H., Watts K. J., and Crane B. R. (2013) HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol. 11, e1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhatnagar J., Sircar R., Borbat P. P., Freed J. H., and Crane B. R. (2012) Self-association of the histidine kinase CheA as studied by pulsed dipolar ESR spectroscopy. Biophys. J. 102, 2192–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Srivastava M., Anderson C. L., and Freed J. H.. (2016) A new wavelet denoising method for selecting decomposition levels and noise thresholds. IEEE Access 4, 3862–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chiang Y.-W., Borbat P. P., and Freed J. H. (2005) The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 172, 279–295 [DOI] [PubMed] [Google Scholar]
- 42. Chiang Y.-W., Borbat P. P., and Freed J. H. (2005) Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J. Magn. Reson. 177, 184–196 [DOI] [PubMed] [Google Scholar]