Abstract
Antigen-stimulated T cells require elevated importation of essential and non-essential amino acids to generate large numbers of daughter cells necessary for effective immunity to pathogens. When amino acids are limiting, T cells arrest in the G1 phase of the cell cycle, suggesting that they have specific sensing mechanisms to ensure sufficient amino acids are available for multiple rounds of daughter generation. We found that activation of mTORC1, which is regulated by amino acid amounts, was uncoupled from limiting amino acids in the G1 phase of the cell cycle. Instead, we found that Rictor/mTORC2 has an essential role in T cell amino acid sensing. In the absence of Rictor, CD4+ T cells proliferate normally in limiting arginine or leucine. Our data suggest that Rictor/mTORC2 controls an amino acid-sensitive checkpoint that allows T cells to determine whether the microenvironment contains sufficient resources for daughter cell generation.
Keywords: amino acid, immunology, immunosuppression, mTOR complex (mTORC), T helper cells
Introduction
T cells, like all immune cells, are auxotrophs for the nine essential amino acids (1). Proliferating T cells also require external supplies of non-essential amino acids including arginine, cysteine, asparagine, and glutamine. Following T cell activation, amino acid transport increases, primarily by up-regulation of specific T cell receptor-regulated amino acid transporters (2, 3). Other immune cells exert regulatory control over T cell proliferation and function by exploiting amino acid auxotrophy of T cells. For example, regulated degradation of arginine and tryptophan by myeloid cells depletes these amino acids and blocks T cell proliferation (1).
Despite the importance of amino acids for lymphocyte proliferation, little is known about how these cells estimate what quantities are required for daughter cell generation (4). Once activated by antigen, the first CD4+ T division is preceded by a long G1 phase taking up ∼30–35 h in vitro. Once the irreversible commitment to cell division is made, 5–8 divisions occur over the subsequent 24–36 h. In vivo tracking of CD8+ division has confirmed that a similar situation occurs, with some fractions of the CD8+ T cell population dividing slowly, and others exhibiting rapid division kinetics observed in vitro (5, 6). Presumably, T cells need to determine whether sufficient resources are available for the generation of many daughter cells or whether they will not commit to cell cycle entry. How T cells measure the environmental concentrations of essential amino acids is unclear. The recognition and utilization of essential amino acids in T cell proliferation are also unresolved at the molecular level. For example, T cells have a specific way of determining the amount of glutamine in media; below 500 μm, T cells will not enter the cell cycle (7).
Recently, specific amino acid “sensors” have been described to integrate information about amino acid availability to the mTORC1 complex. mTORC1 is vital for T cell division, presumably because it signals the production of biopolymers and molecular machines needed for daughter generation (8). For example, sestrin2 was described to bind leucine, whereas Gatsl3 (also called Castor1) binds arginine, leading to inactivation of the GATOR-1 complex and eventual activation of mTORC1 (9–11). However, the precise roles of Gatsl3 and sestrin2 have yet to be evaluated in primary cells or in vivo settings where amino acid sensing is required (12). Other proteins have been implicated in amino acid detection including leucyl-tRNA synthase, SLC38A9, the TSC (tuberous sclerosis complex) complex, and various other modes of amino acid communication to DEPTOR (DEP domain-containing mTOR-interacting protein) and Rag GTPases (13–23). How these proteins assess information about amino acid amounts remains unclear. Another amino acid-sensing pathway is mediated by the stress kinase GCN2. However, in T cells, GCN2 is not required for integrating information about environmental amino acid amounts and cell cycle decision, and is instead essential for the efficiency and fidelity of cytotoxic, but not helper T cell proliferation (24).
Here we used quantitative cellular biochemistry and genetics to evaluate how activated CD4+ T cells use essential amino acids, focusing on arginine and leucine. This experimental platform exclusively employs primary cells. Our results indicate that although T cells require mTORC1 for completing the cell cycle, mTORC1 activation is uncoupled from the amino acid-sensing event(s) that license cell cycle progression in G1. We found that T cells use a threshold amino acid-sensing mechanism that has veto power over cell cycle entry; this mechanism has an obligatory requirement for Rictor, the defining subunit of the mTORC2 complex. Helper T cells lacking Rictor engage in proliferation at sub-threshold essential amino acid amounts.
Results
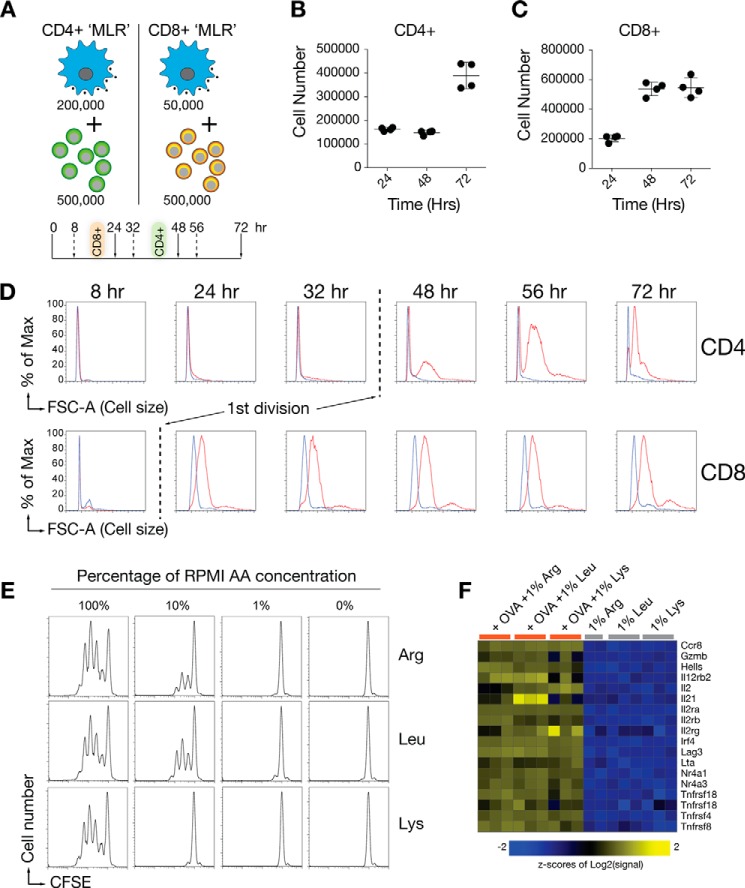
We developed a primary cell-based biochemistry platform to quantify the effects of environmental amino acids on pathways linked to cell cycle entry. The design used an antigen-specific equivalent of a “mixed lymphocyte reaction” (MLR)2 where CD3-depleted splenocytes and non-mesenteric lymph node cells were mixed in defined ratios with ovalbumin (OVA)-specific purified CD4+ DO11.10 T cells with the presence or absence of the specific OVA peptide recognized by DO11.10 T cells in the context of H-2Kd using Balb/c T cell-depleted splenocytes (Fig. 1A). CD4+ T cells first divided between 32 and 48 h (Fig. 1, B–D). In contrast to CD4+ cells, antigen-specific CD8+ cells first divided at ∼24 h (Fig. 1, C and D). The differences in time to the first division between CD4+ and CD8+ T cells suggested that each population uses distinct modes of decision-making to enter the cell cycle and for subsequent daughter generation. For this reason, the remainder of the experiments herein focuses on CD4+ T cell detection of amino acids.
FIGURE 1.
Kinetic responses of T cells to amino acid limitation. A, design of the MLR co-culture platforms. The optimal APC:T cell ratio differs, as defined by CFSE labeling experiments designed to induce maximal T cell proliferation in a 72-h period. B and C, viable cell numbers of CD4+ or CD8+ T cells recovered at 24, 48, or 72 h using the optimized MLR ratios in A. D, cell size differences between CD4+ and CD8+ T cells following exposure to antigen in the MLR co-culture systems. The time point of the first division, defined by parallel CFSE dye dilution assays, is shown by the dotted line. % of Max, percentage of maximum. E, T cell proliferation is dependent on threshold amino acid concentrations. DO11.10 CD4+ T cells were used in an MLR assay where the medium was adjusted to contain 10, 1, or 0% of the normal concentration of arginine, leucine, or lysine in RPMI medium. FSC-A, forward scatter area. F, microarray analysis of canonical mRNAs induced by TCR and co-stimulation stimulation (48-h time point) in purified CD4+ T cells. Shown are T cells in medium containing 1% of the normal concentration of amino acids, in both the presence and absence of antigen. Data in B–E are representative of 3–5 experiments. All MLR experiments also contain a separate CFSE control experiment to measure proliferation. Data in F represent individual 3 experiments (each column). One of the samples cultured in 1% Arg was excluded for quality control reasons.
As expected, CD4+ T cells were sensitive to the amounts of arginine, leucine, and lysine in the medium (Fig. 1F). No cell division occurred at 1% of the normal RPMI concentration of arginine (1148 μm), leucine (381 μm), or lysine (218 μm). As a first step to understand how T cells determine the amount of amino acids they need for division, we performed transcriptomic studies on CD4+ T cells from co-cultures with antigen-presenting cells after 48 h of exposure to limiting arginine, leucine, or lysine and in the presence or absence of antigen stimulation (Fig. 1F). Two unexpected findings emerged from analysis of the transcriptomes of each population. First, there were no specific transcriptional differences when cells starved for each amino acid were compared in the presence or absence antigen. Therefore, we concluded that CD4+ T cells did not induce specific transcriptional responses in response to individual limiting amino acids.3 In other words, the transcriptional response to leucine, arginine, or lysine was indistinguishable between the three amino acids. Second, antigen signaling, which induces the coordinate regulation of thousands of genes (Fig. 1E, example of canonical T cell activation mRNAs), was unaffected by the limiting amino acids. Therefore, amino acid deprivation did not blunt gene activation downstream of the TCR and costimulatory pathways within the time frame of the assay. Therefore, amino acid amounts blocked cell cycle progression, but did not affect TCR and co-stimulatory signaling to the mRNAs required for CD4+ T cell activation.
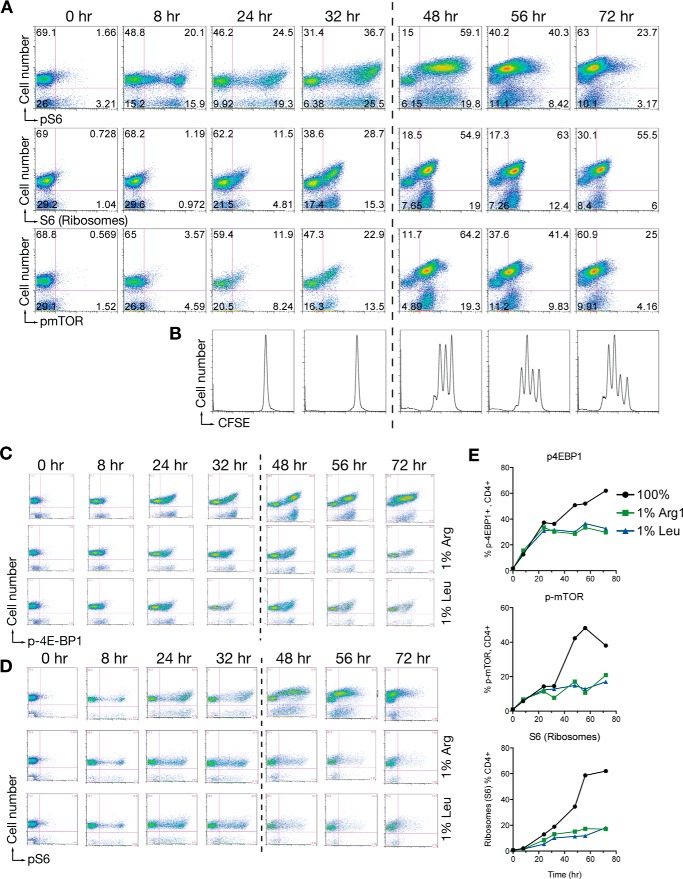
mTORC1 is a key signaling hub necessary for T cell division, and is involved in amino acid sensing at multiple levels (9, 11, 19, 21). To investigate links between cell cycle entry, amino acid amounts, and mTORC1 activity, we adapted the MLR cultures to quantify different mTORC1-dependent events across time, and in the presence of limiting amino acids in the culture system. When we measured phosphorylated S6 (p-S6, a key mTORC1 target downstream of S6 kinase), total S6 (as a surrogate for ribosome number and biogenesis), and phospho-mTOR (p-mTOR) across time after antigen stimulation, we observed a biphasic response where p-S6 and p-mTOR were initially activated by exposure to antigen in the G1 phase of the cell cycle. After the first division, p-S6, S6, and p-mTOR increased substantially (Fig. 2, A and B). These data suggest that mTORC1 activity in CD4+ T cells is differentially activated at different points in the cell cycle.
FIGURE 2.
mTORC1 regulation in T cells in low amino acid environments. A and B, time course of pS6, S6, or p-mTOR in gated CD4+ cells from an MLR assay. The dotted line indicates the time zone of the first division defined by the independently performed CFSE dye dilution assay (B). C, time course of p-4E-BP1 in gated CD4+ T cells as in A where cultures were in normal RPMI or RPMI with 1% arginine or 1% leucine. D, time course of pS6 in gated CD4+ T cells as in C. E, quantification of flow data from the experiment in panels C and D. Experiments were done 3 independent times for arginine starvation. Panel B is the internal CFSE control experiment for data in panel A. Data in panel E is a summary figure from a single representative experiment.
Because limiting amino acids block cell cycle entry in G1 (25, 26), connections between mTORC1 activity and amino acids are likely to be first integrated in the G1 phase of the cell cycle. To test whether mTORC1 activity was blocked in G1 by limiting amino acids, we used the MLR system in control normal RPMI, or in RPMI containing 1% arginine or 1% leucine of the normal RPMI concentration. We measured p-S6, p-4E-BP1, and p-mTOR activity, which are collectively used as markers of mTORC1 activity. Following antigen stimulation, phosphorylation of S6, 4E-BP1, and mTOR was similar to RPMI controls regardless of the amino acid amounts, up to the time window of the first division (Fig. 2, C–E). These data indicate that overall mTORC1 activity measured by activation of key substrates was uncoupled from the decision-making process that connects cell cycle entry to available amino acids. Once cell division began (between 32 and 48 h), the secondary phase of mTORC1 activity (Fig. 2A, 48–72 h) was not observed in non-dividing, amino acid-restricted, CD4+ T cells. When considered with the kinetic studies in Fig. 2A, these data suggest that mTORC1 may play a more complex role in T cell amino acid sensing than previously appreciated.
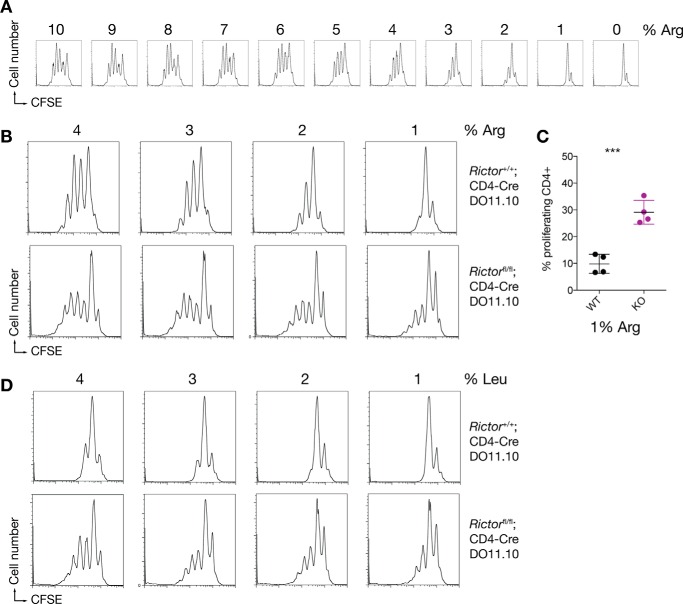
A key question about the relationship between T cell division and amino acid availability concerns the threshold amount required to enter the cell cycle after activation. To estimate this concentration, we titrated the amount of arginine in the MLR system in single percentage increments from 10% to 1% of the normal RPMI concentration (Fig. 3A). The results of these experiments indicated that the minimum amount of arginine necessary for one division is ∼2% (23 μm). To identify mechanisms linking amino acids to cell cycle entry, we took a candidate approach to focus on links between antigen and mitogen (IL-2) signaling and cell cycle entry. Our goal was to identify a mutant(s) that permitted antigen-stimulated cell cycle entry in low amino acid environments. For example, we used T cells from transgenic mice engineered to express a constitutive version of STAT5 (27), which is a key IL-2-activated transcription factor required for T cell division. However, T cells constitutively expressing active STAT5 failed to proliferate to the same extent as controls in any in vitro conditions we tested.4 Similarly, T cells from phosphatase and tensin homolog (PTEN)-deficient mice, which regulates a key negative step in IL-2 receptor signaling (28) did not show differences in amino acid starvation responses.4 Another protein complex that could be linked to antigen or mitogen signaling regulation is Rictor/mTORC2, as this complex phosphorylates AKT, a key kinase in T cell proliferation (29). We therefore generated Rictorflox/flox; CD4-Cre; DO11.10 mice (CD4+). Unlike all other mutants tested, we observed that T cells from these mice proliferated in 1% arginine (Fig. 3, B and C). Rictor-deficient CD4+ T cells proliferated at all concentrations of arginine tested. Therefore, Rictor is involved in conveying information to antigen-stimulated T cells about how much arginine is available. To determine whether the effects of Rictor deficiency were specific to arginine or more general in nature, we used the MLR culture system to test growth in 1% leucine. Again, loss of Rictor was permissive for T cell growth in limiting amino acids (Fig. 3D). Therefore, loss of Rictor/mTORC2 bypasses the amino acid-sensing mechanisms used by CD4+ T cells to block cell cycle entry. These data also indicate that 1% arginine or leucine is not limiting for T cell proliferation so long as mTORC2 signaling is disabled.
FIGURE 3.
Loss of Rictor in T cells bypasses the amino acid depletion signal that blocks entry into cell cycle. A, quantification of the limiting amount of arginine needed for cell cycle entry. CD4+ T cells were stimulated in the MLR assay where arginine was titrated from 10 to 0% of the normal RPMI concentration. CD4+ proliferation was assessed at 72 h. B, CD4+ T cells from control or T cell-specific Rictor-deficient mice on a DO11.10 background were used in an MLR assay where arginine was 4, 3, 2, or 1% of the normal RPMI concentration. CD4+ T cell proliferation was assessed at 72 h. C, summary data of 4 independent experiments comparing proliferation in 1% arginine between control cells (WT) and Rictor-deficient CD4+ T cells (KO). ***, p = 0.0005 two-tailed t test. D, as in B, but using limiting leucine. Data are representative of two independent experiments.
Discussion
T cells programmed to engage in proliferation have obligate requirements for essential and non-essential amino acids. When environmental amino acids become limiting, T cells arrest at a point in approximately mid-G1 (26). We found that Rictor controls this process, because in the absence of Rictor, CD4+ T cells divide in limiting arginine or leucine. Therefore, we postulate that Rictor, and presumably mTORC2, is part of an amino acid-sensing mechanism that regulates the uptake of amino acids at their source: the extracellular milieu.
Different mechanisms have been proposed to mediate amino acid sensing. GCN2 plays a key role in detecting uncharged tRNAs and activates stress-dependent transcription in limiting amino acids. In T cells, GCN2 is required to activate transcription factors ATF3, ATF4, and CHOP (CCAAT-enhancer-binding homologous protein) in response to low amino acid environments. However, it is dispensable for coupling this information to cell division, as GCN2-deficient T cells are blocked from entering into cell division cycles in a manner identical to wild-type controls (24). Instead, GCN2 has a key role in modulating the efficiency of CD8+ but not CD4+ proliferation, and regulates CD8+ T cell trafficking in vivo (24). Other pathways have been specifically linked to mTORC1 in transformed cells including Rag GTPases, sestrin2, Gatsl3 (Castor1), leucyl tRNA synthase, and the lysosomal transporter SLC38A9 (10, 11, 13, 18, 21, 30, 31). However, definitive evidence that these pathways are required for amino acid sensing in normal physiology is lacking (12). One possibility is that the different pathways implicated in direct and indirect amino acid sensing are diversified across different cell types. For example, amino acid sensors upstream of mTORC1 may be active in T cells and may modulate mTORC1 activity, but have less important roles when rapid cell division is programmed by antigen stimulation; there Rictor has a key modulating role in CD4+ cells. One study has shown that mTORC2 is activated by exogenous amino acids (32). However, how these data correlate to our work is unclear at present as the authors' experimental approach is considerably different from our experiments in primary T cells.
Previous work on mTOR signaling in T cells has revealed a complex and obligate requirement for mTORC1 in emergence from quiescence following antigen stimulation, Treg (regulatory T cell) proliferation, and normal homeostasis of the T cell compartment (8, 33–35). However, mTOR can be eliminated from the entire T cell compartment, a genetic alteration that causes shifts in T cell phenotype (33). Through the measurement of mTORC1 substrates by flow cytometry, our experiments revealed additional aspects of mTORC1 activity in antigen-stimulated T cells. We observed that antigen stimulation provokes early and heterogeneous activation of p-S6 and p-4E-BP1, which would be expected for cells preparing to expand biomass in preparation for division. However, this early activation of mTORC1 is independent of extracellular amino acid amounts. Closer to the first division (32 h) and after the first division when CD4+ T cells engage in rapid division, mTORC1 activity was increased and more uniform within the dividing T cells. At this point, amino acid-starved T cells did not increase mTORC1 activity. We interpret these data in the context of the cell cycle, as the amino acid-starved T cells are G1-arrested at this point, and have no requirement for the “second” phase of mTORC1 activity. These data suggest that mTORC1 activity may have differential roles across the different phases of the cell cycle, which requires further exploration using proteomic and metabolomics approaches. We further speculate that the previously identified amino acid-sensitive mTORC1 modulators may have differential roles across the cell cycle.
Limitations of our work concern the role of mTORC2 activation and its subsequent substrate range. In our study, we could not detect the most widely used readout of mTORC2 kinase activity, Ser-473 phosphorylation of AKT (36), which meant that we could not track when mTORC2 was active in control MLR cultures. This may be a technical issue, or a consequence of the cell type used or the amount of p-AKT in T cells. A second limitation concerns the molecular connections between mTORC2 and the events leading to cell cycle entry. For example, recent proteomic data have shown that the IL-2 receptor activates a wide range of pathways involving hundreds of downstream effects (37). Thus, it is possible that a proteomic approach could connect mTORC2 and specific mediators downstream from the IL-2 receptor, the T cell receptor, or other costimulatory receptors that eventually trigger the events leading to the irreversible commitment to cell cycle entry. Nevertheless, our data indicate that Rictor/mTORC2 is an essential component of a pathway that determines external amino acid amounts.
In T cells with genetically disabled Rictor/mTORC2, the sensing system that determines whether sufficient amino acid amounts are available is bypassed, and CD4+ T cells divide in amino acid amounts normally limiting for proliferation. How Rictor/mTORC controls the uptake of amino acids is unclear, and may be through direct modulation of another sensing pathway, or an indirect mechanism that affects other pathways involved in amino acid metabolism. Munder and colleagues (38) have previously proposed that activated T cells might have a counting mechanism to ensure that enough material is available for both the present and future needs. However, how such a mechanism would work at the molecular level is unknown. Our data suggest that Rictor/mTORC2 comprises an essential component of the way T cells measure amino acid availability.
Experimental Procedures
Mice
Rictorflox/flox (Rictortm1.1Klg/SjmJ), CD4-Cre (B6.Cg-Tg(CD4-cre)1Cwi/BfluJ), OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J), DO11.10 (C.Cg-Tg(DO11.10)10Dlo/J), C57BL/6J, and Balb/c mice were purchased from The Jackson Laboratory. Rictorflox/flox mice were crossed with CD4-Cre and DO11.10 mice and genotyped according to Jackson Laboratory genotyping protocols. All animals were maintained and used according to the policies and procedures of the St. Jude Children's Research Hospital Institutional Animal Care and Use committee.
Formulations and Modifications of Media
For non-titration-related experiments, RPMI 1640 (Corning) was supplemented with 10% FBS (Thermo Fisher) and penicillin/streptomycin (Gibco). For l-arginine, l-leucine, and l-lysine titration experiments, stable isotope labeling with amino acids in cell culture (SILAC) RPMI 1640 medium (Sigma) was supplemented with 10% dialyzed FBS (Gibco) and penicillin/streptomycin solution. Cell culture grade amino acids were purchased from Sigma.
Cell Purification and Cell Culture Systems
Spleens and non-mesenteric lymph nodes were isolated, and single cell suspensions were obtained by grinding through a 70-μm nylon mesh. Red blood cells were removed with RBC Lysis buffer (BioLegend) prior to purification. Antigen-presenting cells (APCs) were obtained by using the CD3ϵ MicroBead Kit (Miltenyi) according to the manufacturer's instructions. Purified CD4+ or CD8+ populations were obtained by negative selection. Briefly, cells were incubated with biotinylated anti-CD45R (B220, clone RA3-6B2), anti-I-A/I-E (clone M5/114.15-2), anti-CD11b (clone M1/70), anti-CD49b (clone DX5), and either anti-CD4 (clone GK1.5) or anti-CD8α (clone 53-607) (BioLegend). Cells were washed and incubated with streptavidin microbeads (Miltenyi), and then washed and passed over a magnetic column (Miltenyi). Purified T cells were then labeled with 2 μm carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) (Invitrogen) and resuspended in the appropriate medium for the given assay. 5 × 104 (OT-I system) or 2 × 105 (DO11.10 system) APCs were mixed with 5 × 105 T cells in medium containing 0.1 μm OVA peptide. Proliferation was assessed at 72 h by flow cytometry.
Flow Cytometry
Cultured cells were harvested and surface-stained with anti-CD4 antibody prior to fixation with 4% paraformaldehyde for 10 min at 37 °C. Cells were washed and resuspended in ice-cold methanol and incubated on ice for a minimum of 1 h. Cells were washed and resuspended in PBS, 1% FBS containing a 1:50 dilution of phospho-antibody and incubated at room temperature for 1 h. Data were acquired with a FACS Canto II (BD Biosciences) and analyzed using FlowJo software version 9 (TreeStar).
Additional Antibodies
Anti-CD4 (clone RM4-5) and anti-CD8α (clone 53-6.7) were purchased from BioLegend. The following intracellular antibodies were used: anti-S6 ribosomal protein (clone 54D2, Cell Signaling), anti-phospho-S6 ribosomal protein (Ser-235/236) (clone D57.2.2E, Cell Signaling), anti-phospho-4E-BP1 (Thr-37/46) (clone 236B4, Cell Signaling), and anti-phospho-mTOR (clone MRRBY, eBioscience).
Author Contributions
L. V. d. V. did the experiments. P. J. M and L. V. d. V. designed the experiments and wrote the manuscript.
Acknowledgments
We thank Mike Farrar for the gift of lymph nodes and spleens from STAT5 CA mice, Geoff Neale for bioinformatics analysis, and Hongbo Chi for lymph nodes from T cell-specific PTEN-deficient mice and for lymph nodes from T cell specific Rictor-deficient mice used in pilot studies.
This work was supported by National Institutes of Health Grant CA189990 and Cancer Center Core Grant P30 CA21765 (to P. J. M.) and the American Lebanese Syrian Associated Charities. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The microarray data set reported in this paper has been submitted to the GEO Database under accession number GSE68804.
Details and statistical analysis of this microarray data set will be reported in a separate manuscript (L.-A. Van de Velde et al., submitted for publication). Data can be retrieved in GEO (GSE68804).
L.-A. Van de Velde and P. J. Murray, unpublished observations.
- MLR
- mixed lymphocyte reaction
- mTOR
- mammalian target of rapamycin
- OVA
- ovalbumin
- TCR
- T cell receptor
- p
- phosphorylated
- APC
- antigen-presenting cell
- CFSE
- carboxyfluorescein succinimidyl ester.
References
- 1. Murray P. J. (2016) Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol. 17, 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sinclair L. V., Rolf J., Emslie E., Shi Y. B., Taylor P. M., and Cantrell D. A. (2013) Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakaya M., Xiao Y., Zhou X., Chang J. H., Chang M., Cheng X., Blonska M., Lin X., and Sun S. C. (2014) Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang R., and Green D. R. (2012) Metabolic checkpoints in activated T cells. Nat. Immunol. 13, 907–915 [DOI] [PubMed] [Google Scholar]
- 5. Hwang L. N., Yu Z., Palmer D. C., and Restifo N. P. (2006) The in vivo expansion rate of properly stimulated transferred CD8+ T cells exceeds that of an aggressively growing mouse tumor. Cancer Res. 66, 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kinjyo I., Qin J., Tan S. Y., Wellard C. J., Mrass P., Ritchie W., Doi A., Cavanagh L. L., Tomura M., Sakaue-Sawano A., Kanagawa O., Miyawaki A., Hodgkin P. D., and Weninger W. (2015) Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat. Commun. 6, 6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carr E. L., Kelman A., Wu G. S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A. M., and Frauwirth K. A. (2010) Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang K., Shrestha S., Zeng H., Karmaus P. W., Neale G., Vogel P., Guertin D. A., Lamb R. F., and Chi H. (2013) T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chantranupong L., Scaria S. M., Saxton R. A., Gygi M. P., Shen K., Wyant G. A., Wang T., Harper J. W., Gygi S. P., and Sabatini D. M. (2016) The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saxton R. A., Chantranupong L., Knockenhauer K. E., Schwartz T. U., and Sabatini D. M. (2016) Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536, 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saxton R. A., Knockenhauer K. E., Wolfson R. L., Chantranupong L., Pacold M. E., Wang T., Schwartz T. U., and Sabatini D. M. (2016) Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J. H., Cho U. S., and Karin M. (2016) Sestrin regulation of TORC1: Is Sestrin a leucine sensor? Sci. Signal. 9, re5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., and De Virgilio C. (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110 [DOI] [PubMed] [Google Scholar]
- 14. Carroll B., Maetzel D., Maddocks O. D., Otten G., Ratcliff M., Smith G. R., Dunlop E. A., Passos J. F., Davies O. R., Jaenisch R., Tee A. R., Sarkar S., and Korolchuk V. I. (2016) Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. Elife 5, e11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dyachok J., Earnest S., Iturraran E. N., Cobb M. H., and Ross E. M. (2016) Amino acids regulate mTORC1 by an obligate two-step mechanism. J. Biol. Chem. 291, 22414–22426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han J. M., Jeong S. J., Park M. C., Kim G., Kwon N. H., Kim H. K., Ha S. H., Ryu S. H., and Kim S. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 [DOI] [PubMed] [Google Scholar]
- 17. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., and Sabatini D. M. (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rebsamen M., Pochini L., Stasyk T., de Araújo M. E., Galluccio M., Kandasamy R. K., Snijder B., Fauster A., Rudashevskaya E. L., Bruckner M., Scorzoni S., Filipek P. A., Huber K. V., Bigenzahn J. W., Heinz L. X., et al. (2015) SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., and Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith E. M., Finn S. G., Tee A. R., Browne G. J., and Proud C. G. (2005) The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 280, 18717–18727 [DOI] [PubMed] [Google Scholar]
- 21. Wang S., Tsun Z. Y., Wolfson R. L., Shen K., Wyant G. A., Plovanich M. E., Yuan E. D., Jones T. D., Chantranupong L., Comb W., Wang T., Bar-Peled L., Zoncu R., Straub C., Kim C., et al. (2015) Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X., Fonseca B. D., Tang H., Liu R., Elia A., Clemens M. J., Bommer U. A., and Proud C. G. (2008) Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 283, 30482–30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoon M. S., Son K., Arauz E., Han J. M., Kim S., and Chen J. (2016) Leucyl-tRNA synthetase activates Vps34 in amino acid-sensing mTORC1 signaling. Cell Rep. 16, 1510–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van de Velde L. A., Guo X. J., Barbaric L., Smith A. M., Oguin T. H., Thomas P. G., and Murray P. J. (2016) Stress kinase GCN2 controls the proliferative fitness and trafficking of cytotoxic T cells independent of environmental amino acid sensing. Cell Rep., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blagosklonny M. V., and Pardee A. B. (2002) The restriction point of the cell cycle. Cell Cycle 1, 103–110 [PubMed] [Google Scholar]
- 26. Rodriguez P. C., Quiceno D. G., and Ochoa A. C. (2007) l-Arginine availability regulates T-lymphocyte cell-cycle progression. Blood 109, 1568–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taylor D. K., Walsh P. T., LaRosa D. F., Zhang J., Burchill M. A., Farrar M. A., and Turka L. A. (2006) Constitutive activation of STAT5 supersedes the requirement for cytokine and TCR engagement of CD4+ T cells in steady-state homeostasis. J. Immunol. 177, 2216–2223 [DOI] [PubMed] [Google Scholar]
- 28. Brennan P., Babbage J. W., Burgering B. M., Groner B., Reif K., and Cantrell D. A. (1997) Phosphatidylinositol 3-kinase couples the interleukin-2 receptor to the cell cycle regulator E2F. Immunity 7, 679–689 [DOI] [PubMed] [Google Scholar]
- 29. MacIver N. J., Michalek R. D., and Rathmell J. C. (2013) Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wolfson R. L., Chantranupong L., Saxton R. A., Shen K., Scaria S. M., Cantor J. R., and Sabatini D. M. (2016) Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jewell J. L., Russell R. C., and Guan K. L. (2013) Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tato I., Bartrons R., Ventura F., and Rosa J. L. (2011) Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J. Biol. Chem. 286, 6128–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delgoffe G. M., Kole T. P., Zheng Y., Zarek P. E., Matthews K. L., Xiao B., Worley P. F., Kozma S. C., and Powell J. D. (2009) The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pollizzi K. N., and Powell J. D. (2015) Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 36, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Procaccini C., De Rosa V., Galgani M., Abanni L., Calì G., Porcellini A., Carbone F., Fontana S., Horvath T. L., La Cava A., and Matarese G. (2010) An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity 33, 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 37. Ross S. H., Rollings C., Anderson K. E., Hawkins P. T., Stephens L. R., and Cantrell D. A. (2016) Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8+ T cells. Immunity 45, 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Werner A., Amann E., Schnitzius V., Habermeier A., Luckner-Minden C., Leuchtner N., Rupp J., Closs E. I., and Munder M. (2016) Induced arginine transport via cationic amino acid transporter-1 is necessary for human T-cell proliferation. Eur. J. Immunol. 46, 92–103 [DOI] [PubMed] [Google Scholar]