Abstract
Autotaxin (ATX) is a key enzyme that converts lysophosphatidylcholine (LPC) into lysophosphatidic acid (LPA), a lysophospholipid mediator that regulates cellular activities through its specific G protein-coupled receptors. The ATX-LPA axis plays an important role in various physiological and pathological processes, especially in inflammation and cancer development. Although the transcriptional regulation of ATX has been widely studied, the post-transcriptional regulation of ATX is largely unknown. In this study, we identified conserved adenylate-uridylate (AU)-rich elements in the ATX mRNA 3′-untranslated region (3′UTR). The RNA-binding proteins HuR and AUF1 directly bound to the ATX mRNA 3′UTR and had antagonistic functions in ATX expression. HuR enhanced ATX expression by increasing ATX mRNA stability, whereas AUF1 suppressed ATX expression by promoting ATX mRNA decay. HuR and AUF1 were involved in ATX regulation in Colo320 human colon cancer cells and the LPS-stimulated human monocytic THP-1 cells. HuR knockdown suppressed ATX expression in B16 mouse melanoma cells, leading to inhibition of cell migration. This effect was reversed by AUF1 knockdown to recover ATX expression or by the addition of LPA. These results suggest that the post-transcriptional regulation of ATX expression by HuR and AUF1 modulates cancer cell migration. In summary, we identified HuR and AUF1 as novel post-transcriptional regulators of ATX expression, thereby elucidating a novel mechanism regulating the ATX-LPA axis.
Keywords: AU-rich element RNA-binding protein 1 (AUF1), ELAV-like protein 1 (HuR (human antigen R)), lysophospholipid, post-transcriptional regulation, RNA-binding protein
Introduction
Autotaxin (ATX)4 is a secreted glycoprotein that was initially isolated from the culture medium of A2058 melanoma cells and was regarded as an “autocrine motility factor” because of its promotion of cell metastasis (1). ATX has lysophospholipase D (lysoPLD) activity and converts lysophosphatidylcholine (LPC) into lysophosphatidic acid (LPA) (2). LPA is a bioactive lysophospholipid that regulates multiple cellular activities (cell survival, migration, proliferation, and differentiation) via its specific G protein-coupled receptors (LPAR1 to LPAR6) (3). LPA functions are mediated by the various LPA receptors differentially coupled to distinct G proteins (Gq, Gi, and G12/13) and their downstream signaling molecules, including phospholipase C, PI3K, Ras-MAPK, Rac, and Rho (4). Complete knock-out of ATX in mice leads to embryonic lethality with impaired neurogenesis and vasculogenesis. Compared with the wild-type mice, heterozygous ATX knock-out mice develop normally but display a 50% reduction in plasma LPA levels (5). Therefore, ATX is a key enzyme in producing LPA in the blood and potentially other biological fluids.
Many, if not all, of the biological functions of ATX appear to be mediated by LPA signaling. There is growing evidence indicating that the ATX-LPA axis plays an important role in many physiological and pathological processes, including inflammatory diseases (6–8), obesity (9, 10), and tumorigenesis (11, 12). ATX is among the top 40 most up-regulated genes in metastatic cancer and is regarded as a potential cancer therapy target (13). Due to the biological significance of the ATX-LPA axis, the regulation of ATX expression has been widely studied. In neuroblastoma cells, the transcription factor Sp3 increases ATX expression by interacting with an upstream regulatory element, the GA motif (14). Studies in breast cancer cells have shown that NFAT1, a nuclear factor of activated T-cells, can up-regulate ATX expression by binding to the promoter region of the ATX gene (15). The WNT/β-catenin and PI3K pathways may contribute to the expression of ATX (16). In addition, many cytokines, including TNF-α, IL-1, and IL-4, have been reported to regulate ATX (17). We recently reported that ATX expression is up-regulated in immune cells in response to TLR activation through a type-I IFN-mediated pathway (18). These previous studies predominantly focused on the transcriptional regulation of ATX expression, but ATX regulation at the post-transcriptional level has not been elucidated.
RNA-binding proteins (RBPs) regulate gene expression by controlling mRNA stability and/or translation by binding to the 3′UTR of target mRNAs. Post-transcriptional regulation through the mRNA 3′UTR is a critical gene regulatory mechanism found in nearly all biological processes. Human antigen R (HuR) is a ubiquitously expressed RBP belonging to the Hu/Elav family. There are three conserved RNA recognition motifs in HuR. HuR can specifically bind to the uridylate (U)-rich or AU-rich elements (AREs) located in the 3′UTR to increase the mRNA stability and translation efficiency of target genes (19, 20). HuR is predominantly localized in the nucleus but can be translocated to the cytoplasm under specific cellular stimulations. The nucleus-cytoplasm translocation of HuR is critical to its function (21). HuR targets include many cell cycle regulators, proliferation-associated proteins, tumor-related factors, and cytokines, which are closely involved in cell division cycle, senescence, inflammation, and stress response. However, several other RBPs have been shown to promote ARE-mRNA decay. ARE/poly(U)-binding/degradation factor 1 (AUF1) is a RBP that primarily promotes ARE-mRNA degradation. AUF1 has been reported to antagonize HuR function under certain conditions. For example, AUF1 could antagonize HuR in regulating the mRNAs of cyclin D1 and p21 (22, 23).
In the present study, HuR and AUF1 were identified as the novel regulators of ATX expression. Our results showed that HuR and AUF1 directly interacted with the ATX mRNA 3′UTR, which contains AU-rich elements conserved among species. HuR enhanced ATX expression by increasing ATX mRNA stability, whereas AUF1 suppressed ATX expression by destabilizing ATX mRNA. HuR and AUF1 were involved in the regulation of ATX in the human colon cancer cell line Colo320 and the lipopolysaccharide (LPS)-stimulated human monocytic THP-1 cells, as well as B16 mouse melanoma cells. The post-transcriptional regulation by HuR and AUF1 affected the ATX-mediated B16 cell migration. Altogether, this study demonstrated that HuR and AUF1 can regulate ATX expression at the post-transcriptional level to modulate the biological function of the ATX-LPA axis.
Results
HuR Binds to the 3′UTR of ATX mRNA
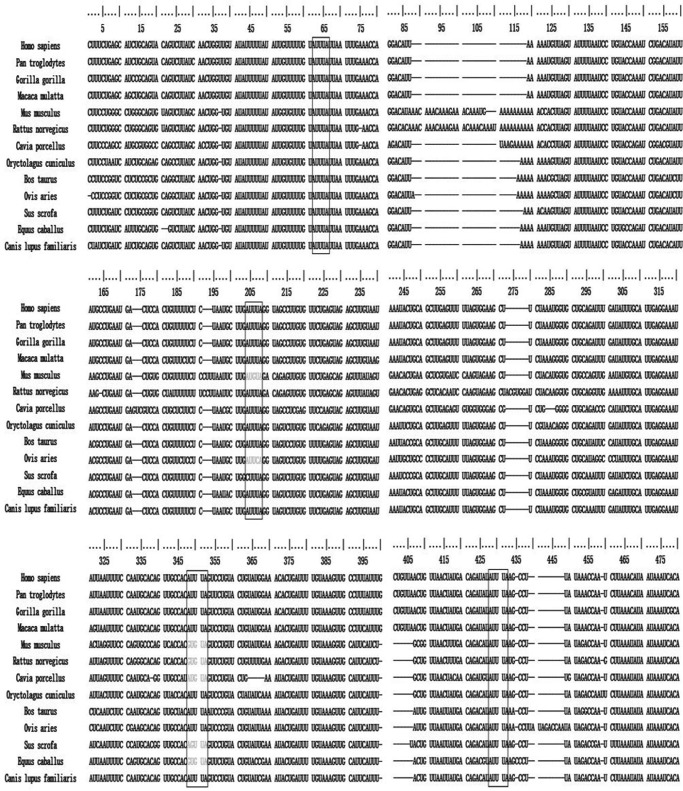
AREs have been found in the 3′UTR of many mammalian mRNAs and play an important role in regulating the stability and translational efficiency of transcripts by interacting with RNA-binding proteins. Analysis of the human ATX mRNA 3′UTR revealed four classic AUUUA motifs. Alignments of the ATX 3′UTR sequences indicated that the four AUUUA motifs in the ATX mRNA 3′UTR are conserved among different species, especially the first one proximal to the coding region and the fourth one close to the end of the ATX 3′UTR (Fig. 1), suggesting that the trans-acting RNA-binding proteins may interact with the ATX mRNA 3′UTR to regulate ATX expression at the post-transcriptional level.
FIGURE 1.
Sequence alignments of the ATX mRNA 3′UTRs. The computational analyses of AU-rich elements of RNA were conducted according to the methods described by Gruber et al. (26). The conserved AUUUA motifs are shown in boxes, and gaps (indicated by dashes) were introduced to optimize the alignment. The sequences are from Homo sapiens (NM_001040092.2), Pan troglodytes (XM_009455833), Gorilla gorilla (XM_004047471.1), Macaca mulatta (XM_015145919.1), Rattus norvegicus (NM_057104.2), Cavia porcellus (XM_013143751.1), Oryctolagus cuniculus (XM_008255849.1), Bos taurus (XM_015474534.1), Ovis aries (XM_012184197.2), Sus scrofa (XM_013996524.1), Equus caballus (XM_014728071.1), and Canis lupus familiaris (XM_014118597.1).
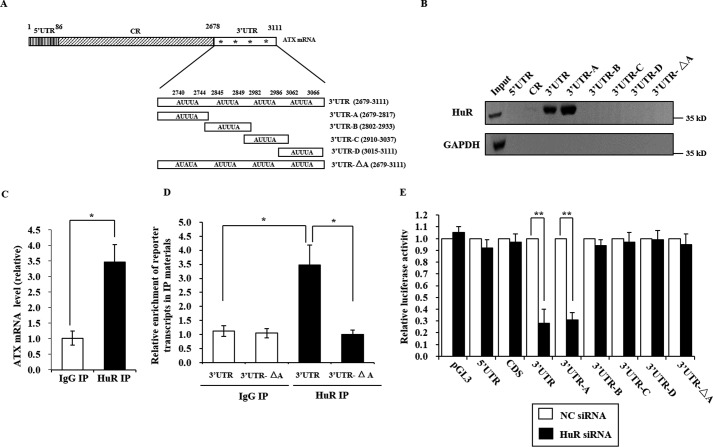
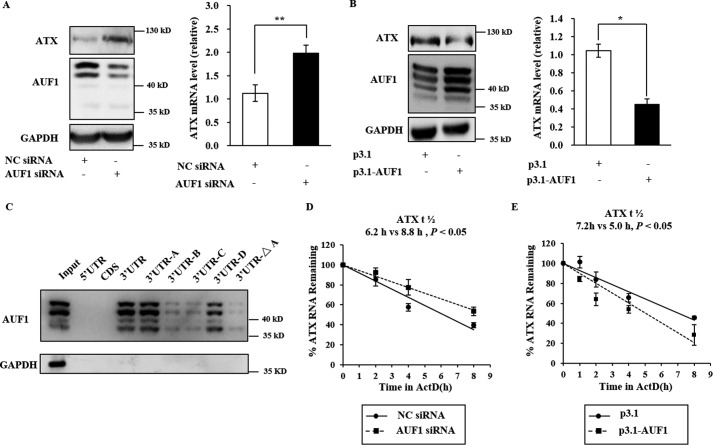
HuR is a ubiquitously expressed RNA-binding protein that interacts with U- or AU-rich elements in the 3′UTR. To determine whether HuR can bind to human ATX mRNA, we prepared biotinylated fragments of ATX mRNA, including the 5′UTR, the coding region, the full-length 3′UTR, and the 3′UTR-A, -B, -C, and -D fragments as indicated in Fig. 2A. The biotinylated ATX mRNA fragments were incubated with the whole cell extracts of Colo320 cells. The RNA protein pulldown analysis was performed as described under “Experimental Procedures” with GAPDH mRNA as a negative control. HuR was able to interact with the 3′UTR and the 3′UTR-A fragment but not with the 5′UTR, coding region, and the 3′UTR-B, -C, and -D fragments (Fig. 2B). In addition, when the first AUUUA motif in the ATX 3′UTR was mutated to AUAUA (named the 3′UTR-ΔA fragment), the HuR interaction with the 3′UTR was abolished (Fig. 2, A and B). These results indicated that HuR could bind to the ATX 3′UTR in vitro, and the binding site is the ARE in ATX mRNA 3′UTR-A fragment.
FIGURE 2.
HuR interacts with the 3′UTR of ATX. A, schematic representation of the ATX mRNA with the four AREs and the ATX mRNA fragments used for biotin pulldown assays. The AREs are indicated by a star (*). B, biotin pulldown assays were performed to detect the interaction between HuR and ATX mRNA. The Colo320 cell lysates were incubated with each of the biotinylated ATX mRNA fragments as indicated. HuR bound to the ATX mRNA fragments was detected by Western blotting. C, RNP-IP analysis was performed using nonspecific IgG or anti-HuR antibody to detect the interaction between endogenous HuR and ATX mRNA in Colo320 cells, in which ATX is endogenously expressed at a relatively high level. D, the pGL3 luciferase reporter vector fused to the ATX fragment 3′UTR or 3′UTR-ΔA was transformed into HEK293T cells, in which the endogenous ATX expression is undetectable. RNP-IP analysis was performed to detect the interaction of HuR with ATX 3′UTR and 3′UTR-ΔA. E, the pGL3 luciferase reporter vector fused to the indicated ATX mRNA fragment was transfected into HEK293T cells. The cells were treated with the NC siRNA or with HuR siRNA. At 48 h after siRNA transfection, the luciferase activity in each cell lysate was detected. Relative luciferase activity to the corresponding control sample was presented. Data are shown as the mean ± S.D. of three independent experiments and significance was analyzed using Student's t test. *, p < 0.05 and **, p < 0.01.
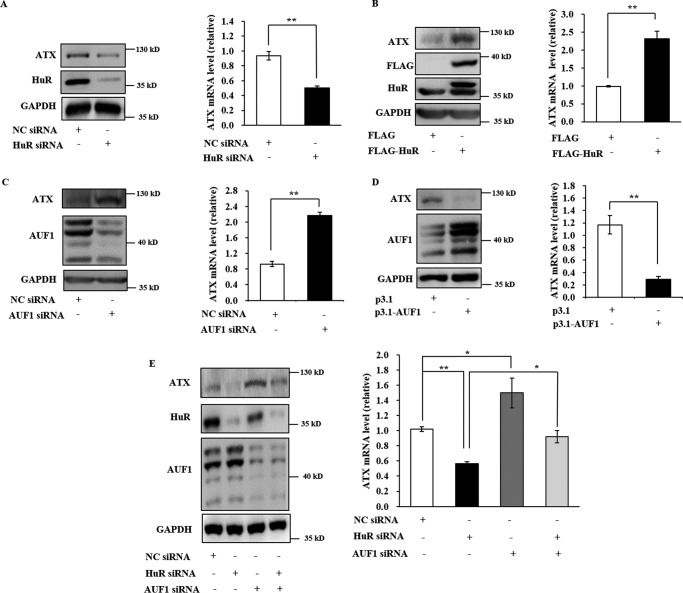
To assess the direct in vivo interaction between endogenous HuR and ATX mRNA, UV cross-linking RNP-IP analyses were performed using a specific antibody against HuR. As shown in Fig. 2C, ATX mRNA was significantly enriched in the IP sample using the HuR antibody, compared with that using a control antibody (IgG). Furthermore, RNP-IP analysis of the HEK293T cells harboring the pGL3 luciferase reporter vector fused to the ATX 3′UTR or 3′UTR-ΔA fragment was performed. Compared with the ATX 3′UTR fragment, the ATX 3′UTR-ΔA fragment was not enriched in the IP samples using a HuR antibody (Fig. 2D). To check the effects of the HuR and ATX mRNA interaction, a pGL3 luciferase reporter vector fused to different ATX fragments was transformed into the HEK293T cells, respectively. The cells were treated with HuR-specific siRNA or nonspecific (NC) siRNA, and then the luciferase activity in each cell lysate was detected. As shown in Fig. 2E, silence of HuR markedly reduced the luciferase activity in cells transfected with the pGL3 luciferase reporter vector fused to the 3′UTR or the 3′UTR-A fragment of ATX. The luciferase activity in the cells transfected with the pGL3 luciferase reporter vector fused to the 5′UTR, CDS, 3′UTR-B, -C, -D, or -ΔA fragment was not affected by the suppression of HuR. These results suggest that HuR positively regulate ATX expression at the post-transcriptional level by interacting with the first ARE motif in the ATX 3′UTR.
HuR Enhances ATX Expression by Increasing ATX mRNA Stability
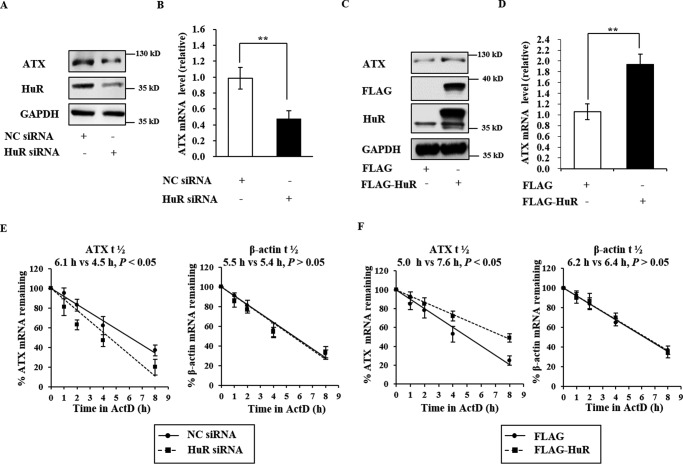
Having confirmed the binding of HuR to the ATX 3′UTR in vivo, we next examined the role of HuR in ATX expression regulation. As shown in Fig. 3, A and B, knockdown of HuR with HuR-specific siRNA markedly reduced both ATX mRNA and protein levels in Colo320 cells (Fig. 3, A and B). Conversely, overexpression of HuR in Colo320 cells with a vector expressing HuR fused to a FLAG tag (FLAG-HuR) significantly increased the ATX mRNA and protein levels (Fig. 3, C and D). It is well known that HuR is a key regulator of mRNA stability and/or translation efficiency of target genes. In further studies, we found that the half-life of the ATX mRNA was decreased from 6.1 h in Colo320 cells transfected with NC siRNA to 4.5 h in those transfected with HuR siRNA (Fig. 3E, left). However, overexpression of FLAG-HuR significantly increased the half-life of ATX mRNA in Colo320 cells from 5.0 to 7.6 h (Fig. 3F, left). As a negative control, the β-actin mRNA half-life was not changed by the alteration of HuR levels (Fig. 3, E, right, and F, right). These data indicate the HuR enhances ATX expression by increasing ATX mRNA stability.
FIGURE 3.
HuR promotes ATX expression by increasing ATX mRNA stability. A and B, the effects of HuR knockdown on ATX expression. Colo320 cells were transfected with HuR siRNA and the NC siRNA. At 48 h after transfection, the protein levels of HuR in the cell lysate and ATX in culture medium were detected by Western blotting analysis, and RNA isolated from the cells was subjected to RT-qPCR to assess the ATX mRNA levels. C and D, the effects of HuR overexpression on ATX expression. Colo320 cells were transfected with the plasmid expressing FLAG-HuR or the empty vector. The protein and mRNA levels of ATX were detected by Western blotting and RT-qPCR, respectively. E and F, the effects of HuR knockdown and overexpression on ATX mRNA stability. Colo320 cells were transfected with HuR siRNA (E) or with the plasmid expressing FLAG-HuR (F), respectively. At 48 h after the transfection, the cells were exposed to actinomycin D (1 μg/ml). The cellular RNA was isolated at the indicated time points after actinomycin D addition, and real-time RT-qPCR analyses were performed to assess the half-lives of ATX and β-actin mRNA. **, p < 0.01.
HuR Stabilizes ATX mRNA by Interacting with the First ARE Motif in the ATX 3′UTR
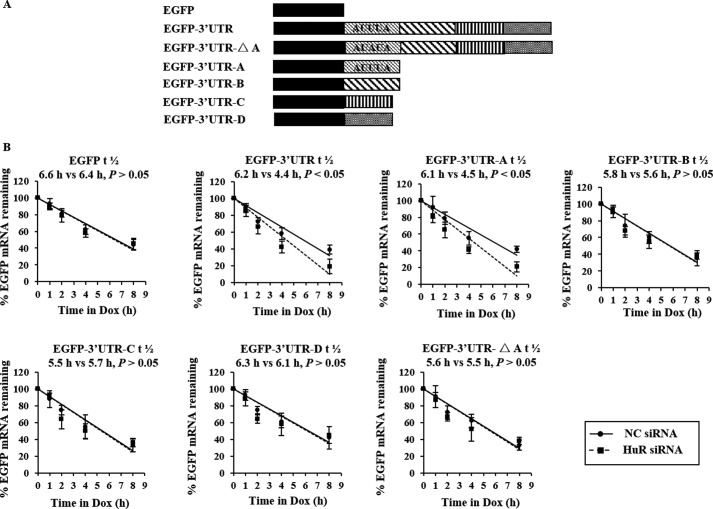
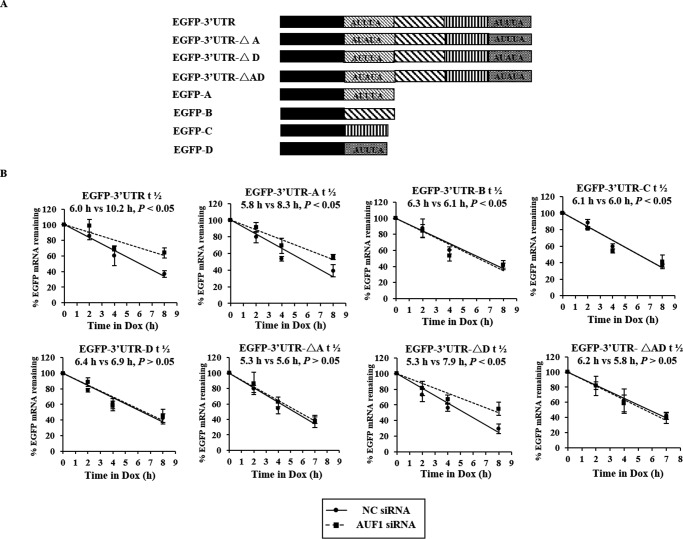
To determine the response element in ATX 3′UTR that interacts with HuR to affect the turnover of ATX mRNA, a series of EGFP-derived reporter gene plasmids bearing different ATX fragments (3′UTR, 3′UTR-A, -B, -C, -D, or -ΔA fragments) were constructed as indicated in Fig. 4A. HeLa cells stably transfected with the pTet-Off plasmid were individually transfected with each of the EGFP-ATX-3′UTR fragments vectors and then treated with HuR siRNA or NC siRNA. After siRNA treatment for 48 h, expression of the encoded chimeric RNA was blocked by the addition of Dox. Then, the total RNA was prepared at the indicated time points after Dox addition, and the levels of each reporter transcript were assessed to determine its half-life by RT-qPCR. As shown in Fig. 4B, knockdown of HuR significantly shortened the half-lives of EGFP-3′UTR (6.2 versus 4.4 h) and EGFP-3′UTR-A (6.1 versus 4.5 h) but did not influence the half-lives of the EGFP, EGFP-3′UTR-B, EGFP-3′UTR-C, and EGFP-3′UTR-D chimeric transcripts. Moreover, the half-life of EGFP-3′UTR-ΔA, which contains an AUUUA to AUAUA mutation in the ATX 3′UTR fragment, was not influenced by knockdown of HuR (Fig. 4B). These results indicate that HuR is capable of stabilizing the ATX 3′UTR chimeric transcripts and that the first ARE in the ATX 3′UTR is the response element for HuR.
FIGURE 4.
The effects of HuR on the stabilization of EGFP-ATX chimeric transcripts. A, schematic representation of the EGFP-ATX reporters studied. B, HeLa cells stably transfected with the pTet-Off plasmid were co-transfected with each of the EGFP-derived reporters bearing the ATX 3′UTR and 3′UTR fragments as indicated together with HuR siRNA or the nonspecific control siRNA (NC). At 48 h after transfection, cells were exposed to Dox (1 μg/ml) to inhibit the expression of the EGFP-ATX-3′UTR fragments chimeric transcripts, and cellular RNA was prepared at the indicated time points after Dox addition. Real-time qPCR was performed to assess the half-lives of the chimeric transcripts.
AUF1 Destabilizes ATX mRNA
AUF1 is an RNA-binding protein that primarily promotes ARE-mRNA decay. AUF1 has been shown to antagonize HuR function under certain conditions (22, 23). We further assessed whether AUF1 could regulate the decay of ATX mRNA to antagonize the effects of HuR on ATX mRNA turnover. It was found that ATX mRNA and protein levels were significantly increased by AUF1 knockdown with the specific siRNA and were decreased by exogenous overexpression of AUF1 in Colo320 cells (Fig. 5, A and B). RNA-protein pulldown analysis showed that AUF1 could bind to the ATX mRNA 3′UTR, with high affinity to the ATX mRNA 3′UTR-A and -D fragments. The AUF1 interaction with full-length ATX 3′UTR was disrupted when the first AUUUA motif in the ATX 3′UTR was mutated to AUAUA (named the 3′UTR-ΔA) (Fig. 5C). Furthermore, the half-life of ATX mRNA was markedly increased in the AUF1-silenced cells and decreased in the AUF1 overexpressing cells (Fig. 5, D and E), indicating that AUF1 may destabilize ATX mRNA.
FIGURE 5.
AUF1 interacts with ATX 3′UTR and destabilizes ATX mRNA. A and B, the effects of AUF1 knockdown and overexpression on ATX expression. Colo320 cells were transfected with the AUF1 siRNA (A) or with the plasmid expressing AUF1 (pcDNA3.1-AUF1) (B), respectively. At 48 h after the transfection, the protein levels of AUF1 in cell lysate and ATX in culture medium by Western blotting analysis, and RNA isolated from the cells were subjected to RT-qPCR to assess the ATX mRNA levels. C, the Colo320 cell lysates were incubated with each of the biotinylated ATX mRNA fragments as indicated. After biotin pulldown, AUF1 bound to the ATX mRNA fragments was detected by Western blotting. D and E, the effects of AUF1 knockdown and overexpression on ATX mRNA stability. Colo320 cells were transfected with the AUF1-specific siRNA (D) or with the plasmid expressing AUF1 (pcDNA3.1-AUF1) (E), respectively. At 48 h after the transfection, the cells were exposed to actinomycin D (1 μg/ml). The cellular RNA was isolated at the time points after actinomycin D addition as indicated, and real-time RT-qPCR analyses were performed to assess the half-lives of ATX mRNA. *, p < 0.05, **, p < 0.01.
AUF1 was able to interact with the 3′UTR-A and -D fragments of ATX mRNA in vitro. To determine whether the AREs in ATX 3′UTR-A and -D regions are the response elements for AUF1 in vivo, a series of EGFP-derived reporter gene plasmids bearing different ATX fragments (3′UTR, 3′UTR-A, 3′UTR-B, 3′UTR-C, 3′UTR-D, 3′UTR-ΔA, 3′UTR-ΔD, and 3′UTR-ΔAD fragment) were constructed as shown in Fig. 6A. The first ARE in ATX 3′UTR was mutated in 3′UTR-ΔA, the fourth ARE was mutated in 3′UTR-ΔD, and both the first and fourth ARE were mutated in 3′UTR-ΔAD. Under the treatment of AUF1 siRNA or NC siRNA, the half-lives of the EGFP reporter gene mRNA fused with the indicated ATX mRNA fragment were detected in HEK293T cells. It was found that the half-lives of EGFP-3′UTR and EGFP-3′UTR-A and -ΔD were significantly extended in AUF1-silenced cells. However, the half-lives of the EGFP-3′UTR-B, EGFP-3′UTR-C, EGFP-3′UTR-D, EGFP-3′UTR-ΔA, and EGFP-3′UTR-ΔAD chimeric transcripts were not prolonged by AUF1 knockdown. These results suggest that the first ARE in the ATX mRNA 3′UTR-A fragment is the response element for AUF1 but not the fourth ARE (Fig. 6B). Taken together, the RNA-binding proteins HuR and AUF1 exert opposing effects in the post-transcriptional regulation of ATX expression, and the first ARE in the ATX mRNA 3′UTR is the common regulatory site for HuR and AUF1.
FIGURE 6.
The effects of AUF1 on the stabilization of EGFP-ATX chimeric transcripts. A, schematic representation of the EGFP-ATX-3′UTR fragments reporters studied. B, HeLa cells stably transfected with the pTet-Off plasmid were co-transfected with each of the EGFP-derived reporters bearing the ATX 3′UTR and 3′UTR fragments as shown in A together with AUF1 siRNA or the NC siRNA. The half-lives of chimeric transcripts were assessed as described in the legend to Fig. 4B.
HuR and AUF1 Are Involved in ATX Regulation in LPS-stimulated Monocytic THP-1 Cells
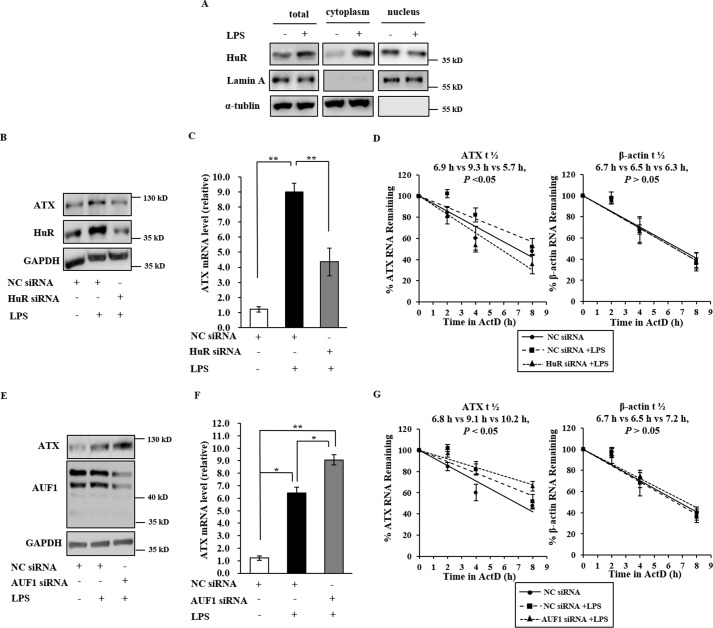
In our previous study, we reported that ATX expression is up-regulated in human monocytic THP-1 cells during LPS stimulation (18, 24, 25). HuR is known to play an important role in gene regulation at the post-transcriptional level during inflammation. Here, we assessed whether ATX expression is regulated at the post-transcriptional level by HuR during LPS stimulation. Exposure of THP-1 cells to LPS not only up-regulated ATX expression but also increased the protein levels of total HuR and promoted the nucleus-cytoplasm translocation of HuR, resulting in significant accumulation of HuR in the cytoplasm (Fig. 7A). When HuR was silenced in THP-1 cells by HuR-specific siRNA, ATX induction by LPS was obviously suppressed at both the mRNA and protein levels (Fig. 7, B and C). To further confirm the association between HuR and ATX mRNA turnover, the half-lives of ATX mRNA were tested in the LPS-stimulated THP-1 cells with or without HuR knockdown. The half-life of ATX mRNA increased after LPS stimulation in THP-1 cells transfected with NC siRNA, and the increase in ATX mRNA stability was inhibited by the knockdown of HuR (Fig. 7D). However, when AUF1 was silenced in THP-1 cells by its specific siRNA, the LPS-mediated ATX induction was enhanced (Fig. 7, E and F), and the half-life of ATX mRNA was further prolonged after LPS stimulation (Fig. 7G). These results suggest that HuR and AUF1 are involved in ATX regulation at the post-transcriptional level during the inflammatory response.
FIGURE 7.
The influence of HuR and AUF1 on ATX expression in the LPS-stimulated THP-1 cells. A, the effects of LPS treatment on HuR levels and cytoplasmic translocation of HuR in THP-1 cells. THP-1 cells were treated with LPS (1 μg/ml) for 16 h. Total HuR in the whole cell lysates and the HuR in the nuclear or cytosolic fractions were detected by Western blotting analysis. B and C, the effects of HuR knockdown on the ATX expression in LPS-stimulated THP-1 cells. THP-1 cells were transfected with HuR siRNA or NC siRNA. After transfection for 36 h the cells were starved for 8 h, then treated with LPS (1 μg/ml) for 16 h. The protein levels of HuR in the cell lysate and ATX in culture medium were detected by Western blotting. RNA isolated from the cells was subjected to RT-qPCR to assess the ATX mRNA levels. D, the effects of HuR knockdown on the ATX mRNA stability in LPS-stimulated THP-1 cells. THP-1 cells were transfected with HuR-specific siRNA or NC siRNA. After transfection for 36 h and cells starved for 8 h, the cells were then treated with LPS (1 μg/ml) for 16 h. Then, the cells were exposed to actinomycin D (1 μg/ml). The cellular RNA was isolated at the indicated time points after actinomycin D addition, and real-time RT-qPCR analyses were performed to assess the half-lives of ATX and β-actin mRNA. E and F, the effects of AUF1 knockdown on ATX expression in LPS-stimulated THP-1 cells. THP-1 cells were transfected with the AUF1 siRNA or the NC siRNA, and then treated as described in B and C. The protein levels of AUF1 in the cell lysate and ATX in culture medium were detected by Western blotting analysis. RNA isolated from the cells was subjected to RT-qPCR to assess the ATX mRNA levels. G, the effects of AUF1 knockdown on the ATX mRNA stability in LPS-stimulated THP-1 cells. THP-1 cells were transfected with AUF1 siRNA or the NC siRNA. The half-lives of ATX and β-actin mRNA were detected as described in D. **, p < 0.01.
The Post-transcriptional Regulation of ATX Expression by HuR and AUF1 in Melanoma Cells and Its Role in Cell Migration
ATX was originally isolated from the culture medium of melanoma cells and termed an autocrine motility factor due to its ability to promote cancer cell migration. Autotaxin is overexpressed in various cancer cells and is believed to be a target for cancer therapy. In this study, the role of HuR and AUF1 in ATX expression was investigated in B16 mouse melanoma cells. In B16 cells, ATX mRNA and protein levels were decreased by HuR knockdown and increased by HuR overexpression (Fig. 8, A and B). In contrast, ATX expression was enhanced by AUF1 knockdown and suppressed by AUF1 overexpression (Fig. 8, C and D). When B16 cells were transfected with HuR or AUF1 siRNA alone or co-transfected with HUR and AUF1 siRNAs together, the reduction in ATX protein and mRNA levels resulting from HuR knockdown could be significantly reversed by the knockdown of AUF1 (Fig. 8E).
FIGURE 8.
The effect of HuR and AUF1 on ATX expression in B16 melanoma cells. A and B, the effects of HuR knockdown and HuR overexpression on ATX expression in B16 cells. B16 cells were transfected with HuR siRNA (A) or the plasmid expressing FLAG-HuR (B). At 48 h after the transfection, ATX protein and mRNA expression levels were assessed by Western blotting and RT-qPCR, respectively. C and D, the effects of AUF1 knockdown or AUF1 overexpression on ATX expression in B16 cells. B16 cells were transfected with AUF1 siRNA (C) or the plasmid pcDNA3.1-AUF1 (D). At 48 h after the transfection, ATX protein and mRNA expression levels were assessed by Western blotting and RT-qPCR, respectively. E, the functional interaction between HuR and AUF1 in ATX expression regulation. B16 cells were transfected with HuR siRNA and/or AUF1 siRNA as indicated. At 48 h after the transfection, ATX protein and mRNA levels were assessed by Western blotting and RT-qPCR. *, p < 0.05, **, p < 0.01.
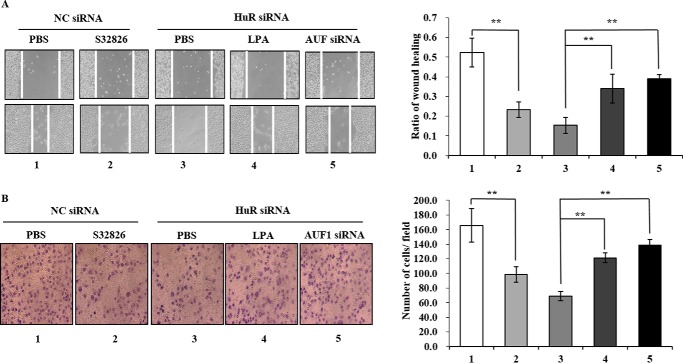
The effects of ATX post-transcriptional regulation on B16 cell migration were detected by wound-healing and transwell assays. In the wound-healing assay, the migration of B16 cells was inhibited following addition of the ATX inhibitor S32826 to the culture medium, indicating that the ATX-LPA axis plays a role in B16 cell migration. Knockdown of HuR by siRNA inhibited B16 cell migration, whereas the inhibition of B16 cell migration by HuR knockdown was suppressed by either addition of LPA or AUF1 knockdown (Fig. 9A). Similar results were observed in the transwell assay (Fig. 9B). These data suggested that the post-transcriptional regulation of ATX expression by HuR and AUF1 is involved in the modulation of B16 cell migration.
FIGURE 9.
The effects of HuR/AUF1-mediated ATX post-transcriptional regulation on B16 cell migration. A, scratch wound-healing assay. B16 cells treated with the indicated siRNA and/or the chemical compound were used for the scratch wound-healing assay. 1, NC siRNA; 2, NC siRNA and ATX inhibitor S32826 (2 μm); 3, HuR siRNA; 4, HuR siRNA and LPA (2 μm); 5, HuR siRNA and AUF1 siRNA. Images of the cells migrating into the wound were captured at 0 and 12 h by an inverted microscope (×10). The relative migration rate was calculated by dividing the change in the gap distance between the scratch edges by the initial distance. B, transwell assay. B16 cells were treated with the indicated siRNA and/or the chemical compound as described in A and subjected to the transwell assay. Images of cells on the lower surface of the upper chamber were taken 24 h after the cells were seeded into the upper chamber. The relative migration was calculated by counting the cell number in the lower surface of the upper chamber, and data were obtained from three randomly chosen fields. Representative data were from three independent experiments. The p value derived from Student's t test is *, p < 0.05 and **, p < 0.001.
Discussion
ATX is a secreted glycoprotein with lysoPLD activity. It is a key enzyme that converts LPC into LPA, a lysophospholipid mediator regulating cell proliferation, differentiation, apoptosis, and migration through its specific receptor on cellular membranes. The ATX-LPA axis is involved in the regulation of various physiological and pathological processes, especially inflammation and cancer development. Therefore, the mechanism of ATX regulation is an important issue that should be studied. Although transcriptional regulation of ATX has been extensively investigated, the post-transcriptional regulation of ATX expression is largely unknown. Here, we demonstrated that the RNA-binding proteins HuR and AUF1 acted as post-transcriptional regulators of ATX expression. Both HuR and AUF1 interacted directly with the ATX mRNA 3′UTR and had a common response element in the ATX mRNA 3′UTR. HuR promoted ATX expression by increasing ATX mRNA stability, whereas AUF1 suppressed ATX expression by decreasing ATX mRNA stability, which implied that HuR and AUF1 acted as antagonistic functions in ATX regulation. To our knowledge, this is the first report on the effects of RNA-binding proteins on ATX expression.
Post-transcriptional regulation of gene expression is a powerful adaptive mechanism in eukaryotic cells. One of the major post-transcriptional regulatory mechanisms is the binding of RNA-binding proteins to the ARE in the mRNA 3′UTR to alter mRNA processing, stability, and/or translation. It has been reported that as many as 5–8% of human transcripts belong to ARE-mRNAs (26). There are four AREs in the human ATX mRNA 3′UTR. These AREs in ATX mRNA 3′UTR are conserved among different species, especially the first one proximal to the coding region termed as ARE-A, and the fourth one close to the end of ATX mRNA 3′UTR termed as ARE-D. Notably, the ARE-A is responsible for HuR binding, whereas both ARE-A and ARE-D are major AUF1 binding sites. However, AUF1 could not bind to ATX 3′UTR-ΔA containing an AUUUA to AUAUA mutation in ARE-A along with the wild-type ARE-D, suggesting that ARE-D itself cannot mediate the interaction of AUF1 to full-length ATX 3′UTR, which may be due to the second structure of ATX mRNA 3′UTR. Our data further proved that only the first ARE, ARE-A, was the regulatory site for HuR and AUF1 together. The regulation of ATX expression by HuR and AUF1 was observed in both human (Colo320 and THP-1) and mouse (B16) cells, suggesting that it may be a conserved mechanism for ATX post-transcriptional regulation.
LPA plays an important role in inflammation by regulating the motility of T cells, B cells, and macrophages, enhancing the survival of lymphocytes, and inducing cytokine expression through the Gi/Rho signal pathway (27). Recently, we demonstrated that ATX was induced by LPS in human monocytic THP-1 cells and that the LPS-mediated ATX induction is dependent on Type I interferon production and the IFNAR-mediated activation of the JAK-STAT pathway (18, 25). The RNA-binding protein HuR is known to regulate inflammatory responses. The results presented in this study demonstrate that the cytoplasmic HuR level in THP-1 cells is increased after LPS treatment and that HuR contributes to the high ATX expression in LPS-treated THP-1 cells. Thus, TLR4 activation by LPS promotes both transcriptional and post-transcriptional increases in ATX expression in a coordinated manner. The post-transcriptional regulation by HuR may contribute to maintain ATX expression at high levels during inflammatory responses and enhance the LPA level in the cellular microenvironment to modulate the immune process.
ATX-LPA signaling is positively correlated with invasion and the metastatic potential of several cancers, including melanoma, breast cancer, ovarian cancer, and neuroblastoma (3). HuR has also been correlated with many cancers. Yong et al. (28) found lower levels of HuR in normal colon tissues but higher HuR expression in colon adenomas and adenocarcinomas. Cytoplasmic HuR localization is increased in cancer tissues, and HuR is considered a cancer prognostic factor (29, 30). Our results in this study revealed that HuR promotes ATX expression in Colo320 and B16 cells. Knockdown of HuR by its specific siRNA decreased ATX expression and inhibited the migration of B16 cells, which was reversed by addition of LPA or AUF1 knockdown. These results indicate that the post-transcriptional regulation of ATX expression by HuR and AUF1 are involved in cancer metastasis.
AUF1 is an ARE-binding protein that promotes transcript destabilization. It has been reported that AUF1 appeared to antagonize HuR function under certain conditions, as HuR and AUF1 bind to common AU-rich target mRNAs and exert opposing effects on target mRNA stability. In this study, ATX mRNA was shown to share the joint ARE response element for HuR and AUF1 in the ATX mRNA 3′UTR. In contrast to HuR, AUF1 promotes ATX mRNA decay as a negative regulatory factor. ATX expression is decreased by AUF1 overexpression and increased by AUF1 knockdown. The ATX down-regulation and inhibition of cell migration resulting from HuR knockdown were significantly recovered by AUF1 knockdown. All these results suggested that, in accordance with the past study, there may be an antagonism between HuR and AUF1 that existed in the regulation of ATX mRNA stability. The balance between HuR and AUF1 levels may play a role in post-transcriptional regulation of ATX expression. The HuR and AUF1 levels can be regulated under certain conditions. For example, HuR is regulated by its post-translational modification, especially phosphorylation (31), and AUF1 is modulated by signal-induced phosphorylation and ubiquitination (32, 33). Further studies are warranted to clarify the role of the signals affecting HuR and AUF1 subcellular location on ATX expression.
In conclusion, we demonstrated that ATX expression is regulated by HuR and AUF1 at the post-transcriptional level. HuR and AUF1 are able to bind to the ATX mRNA 3′UTR and share a common ARE response element. HuR increased ATX expression by stabilizing the ATX mRNA, whereas AUF1 decreased ATX expression by promoting ATX mRNA decay. This post-transcriptional regulation was involved in ATX expression during inflammatory response (LPS stimulation) and the ATX expression cancer cells, which contributes to cancer cell migration. This study establishes the relationship between ATX, the key enzyme in LPA production, and RNA-binding proteins HuR and AUF1, elucidating a novel mechanism in the regulation of bioactive lipid metabolism.
Experimental Procedures
Reagents and Antibodies
LPS (from Escherichia coli serotype 055:B5), actinomycin D, and doxycycline (Dox) were all purchased from Sigma. The ATX primary antibody was generated as described previously (24). The HuR antibody, AUF1 antibody, GAPDH antibody, α-tublin antibody, Lamin-A antibody, and FLAG tag antibody were all obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture and Transfection
Colo320, THP-1, and B16 cells were cultured in RPMI 1640 medium (Macgnen). HeLa cells were maintained in Dulbecco's modified Eagle's medium (Macgnen). The media were supplemented with 10% fetal bovine serum (Gibco), 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Life Technologies). In experiments to detect secreted ATX, cells were cultured in serum-free medium for 24 h. All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. The plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Unless otherwise indicated, cells were collected for analysis 48 h after transfection.
RNA Interference
For transient silencing of HuR or AUF1, cells were transfected with a siRNA targeting HuR or AUF1 using Lipofectamine 2000 (Invitrogen), according to the protocol supplied by the manufacturer. All siRNAs were synthesized by GenePharma (Shanghai, China). In each siRNA transfection experiment, a nonspecific siRNA (NC) was used as a control. For the THP-1 cells, the siRNA-transfected cells were treated with 1 μg/ml of LPS and then collected for analysis 24 h later. The siRNA sequences are as follows: human HuR siRNA sequence, AATCTTAAGTTTCGTAAGTTA; mouse HuR siRNA sequence, AAGAGGCAATTACCAGTTTCA; human AUF1 siRNA sequence, GATCCTATCACAGGGCGAT; and mouse AUF1 siRNA sequence, GATCCTATCACAGGGCGAT.
Construction of Vectors Expressing FLAG-HuR and AUF1
For construction of the vector expressing FLAG-HuR, the full-length coding region of HuR was amplified by PCR using primers CCGGAATTCAATGTCTAATGGTTATGAAGA and CTAGTCTGAGTTATTTGTGGGACTTGTTGG and then inserted between the EcoRI and XbaI sites of the 3× FLAG-CMV10 vector (Clontech). For construction of the vector expressing AUF1 (four splice variants), the full-length coding region of four AUF1 isoforms was amplified by PCR using the primers CCGGAATTCATGTCGGAGGAGCAGTTCGG and CCGCTCGAGTTAGTATGGTTTGTAGCTATT and then inserted between the EcoRI and XhoI sites of the pcDNA3.1 vector (Clontech).
RNA Isolation and Quantitative Real-time PCR
Total cellular RNA was extracted from cells with TRIzol regent (Sigma). RNA (1 μg) from cells was reverse-transcribed using an anchored oligo(dT) primer and a reverse transcription system (Promega). The levels of ATX mRNA, GAPDH mRNA, β-actin mRNA, and transcripts derived from pEGFP plasmids (chimeric EGFP transcripts) were detected using the following primer pairs: TATGCTTCGGAAAGAAATGGAG and ATGTTCAATGTCACGCACCCT-3′ for ATX mRNA; TTAGCACCCCTGTCCAAGG and CCTACTCCTTGGAGGCCATG for GAPDH mRNA; GGGCATGGGTCAGAAGGATT and TCGATGGGGTACTTCAGGGT for β-actin mRNA; and TACAACTACAACAGCCACAACG and ATCCTGCTCCTCCACCTCC for pEGFP-derived reporter transcripts (EGFP RNA). Each RT-PCR experiment was repeated at least three times with 3 replicate samples. Real-time RT-PCR was performed using SYBR Green Supermix (Thermo) with an iCycler iQ real-time RT-PCR detection system (Bio-Rad). Relative expression of each target gene was determined by normalization to the expression of GAPDH.
Western Blotting Analysis
Whole cell extracts were prepared in RIPA buffer for 30–60 min. After centrifugation at 4 °C, the supernatants were quantified by bicinchoninic acid assays (Micro BCA; Pierce Biotechnology, Rockford, IL). For ATX detection in the culture medium, cell culture medium was concentrated (∼30-fold) using an Amicon Ultra 30000 (Millipore). Protein quantification was conducted, and equal protein amounts were loaded for each sample. Protein samples were subjected to SDS-PAGE and analyzed by polyclonal ATX antibody, polyclonal HuR antibody, polyclonal AUF1 antibody, monoclonal α-tublin antibody, polyclonal Lamin-A antibody, or monoclonal GAPDH antibody.
Nuclear and Cytosolic Extract Preparation
Cells were washed with cold phosphate-buffered saline, and cellular fractions were isolated using a Nuclear and Cytoplasmic Extraction Kit (CWbio). Nuclear and cytoplasm fractions were analyzed for HuR protein levels by Western blotting. Lamin A and α-tubulin were used as the nuclear and cytoplasm markers, respectively.
Luciferase Reporters Construction and Luciferase Activity Assays
The cDNA fragments corresponding to the ATX 5′UTR, CDS, and 3′UTR, as well as the 3′UTR-A, -B, -C, and -D fragments, the following primer pairs were used: GCTCTAGAAATAGACTAAACCCAGAGCCTC and GCTCTAGAGTCGAGGATTCTTGGAAAGC for 5′UTR; GCTCTAGAATGGCAAGGAGGAGCTCGTTC and GCTCTAGATTAAATCTCGCTCTCATATGTATG for CDS; GCTCTAGACTTTCTGAGCATCTGCAGTAC and GCTCTAGAAACTGAATGTGTGATTTATTATG for 3′UTR; GCTCTAGACTTTCTGAGCATCTGCAGTAC and GCTCTAGAATTCAGGCATAATATGTCAGATTTG for A; GCTCTAGAATATTATGCCTGAATGACTCCAC and GCTCTAGAAATCTGCAGCACCATTTAGAAGC for B; GCTCTAGAGCTTCTAAATGGTGCTGCAG and GCTCTAGACAGCAAATAAAGGCAACTTTAC for C; GCTCTAGAGTAAAGTTGCCTTTATTTGCTG and GCTCTAGAAACTGAATGTGTGATTTATTATG for D, then inserted between the XbaI site of the pGL3-Control vector (Promega). After producing pGL3-ATX-3′UTR vector, we used the vector as a model, and constructed pGL3-ATX-3′UTR-ΔA with the following primers: TTATATTGTTTTTGTATATATTAATTTGAAACCAGGAC and CTGGTTTCAAATTAATATATACAAAAACAATATAAAAA. The luciferase activity was detected by a Dual-luciferase Reporter Assay System Kit (Promega).
RNA-Protein Interaction Assays and UV Cross-link RNP IP Assays
pGL3-ATX-3′UTR was used as a template for the PCR amplification of different fragments of ATX mRNA. All 5′ primers contained the T7 promoter sequence CCAAGCTTCTAATACGACTCACTATAGGGAGA-3 (T7). To prepare templates for the 5′UTR (positions 1 to 86), CDS (positions 87 to 2678), 3′UTR (positions 2679 to 3111), and 3′UTR-A (positions 2679 to 2817), -B (positions 2802 to 2933), -C (positions 2910 to 3037), -D (positions 3015 to 3111), the following primer pairs were used: (T7)AATAGACTAAACCCAGAGCCTC and GTCGAGGATTCTTGGAAAGC for 5′UTR; (T7)ATGGCAAGGAGGAGCTCGTTC and TTAAATCTCGCTCTCATATGTATG for CDS; (T7)CTTTCTGAGCATCTGCAGTAC and AACTGAATGTGTGATTTATTATG for 3′UTR; (T7)CTTTCTGAGCATCTGCAGTAC and ATTCAGGCATAATATGTCAGATTTG for A; (T7)ATATTATGCCTGAATGACTCCAC and AATCTGCAGCACCATTTAGAAGC for B; (T7)GCTTCTAAATGGTGCTGCAG and CAGCAAATAAAGGCAACTTTAC for C; (T7)GTAAAGTTGCCTTTATTTGCTG and AACTGAATGTGTGATTTATTATG for D. pGL3-ATX-3′UTR-ΔA was used as template for ΔA (the 3′UTR fragment with the first AUUUA mutated to AUAUA in ARE motif) fragments with the primers: (T7)CTTTCTGAGCATCTGCAGTAC and AACTGAATGTGTGATTTATTATG. For biotin pulldown assays, PCR-amplified DNAs were used for the in vitro transcription of the RNA probe in the presence of biotin-UTP (BIOTIUM) with an in vitro Transcript Aid T7 High Yield Transcription Kit purchased from Thermo Scientific (USA). One microgram of purified biotinylated transcript was incubated with 100 μg of whole cell lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo), and the pulldown samples were analyzed by Western blotting. For cross-link RNP IP assays, cells were exposed to UVC (400 mJ/cm2), and the cytoplasmic extracts were prepared for immunoprecipitation using a monoclonal anti-HuR antibody (Santa Cruz, CA). The protocol was previously described by Lopez de Silanes et al. (34). The primers used in the RNP-IP assays were the ATX-specific primers shown in Fig. 1C (TATGCTTCGGAAAGAAATGGAG and ATGTTCAATGTCACGCACCCT) and the luciferase-specific primers shown in Fig. 1D (GATTACCAGGGATTTCAGT and GACACCTTTAGGCAGACC).
mRNA Half-life Measurement
To determine the influence of the ATX 3′UTR and 3′UTR fragments on mRNA stability, a series of EGFP reporter vectors were constructed. The cDNA fragments corresponding to the ATX 3′UTR and the ATX 3′UTR-A, -B, -C, -D, and -ΔA fragments were obtained by RT-PCR using the primers without the T7 promoter sequence as described above. The PCR product was cloned downstream of the EGPP open reading frame of the pTRE-d2EGFP vector (Clontech). To assess the half-lives of the EGFP-ATX-3′UTR fragment chimeric transcripts, HeLa cells stably transfected with the pTet-Off plasmid were further transfected with each of the EGFP-ATX-3′UTR fragment constructs. At 24 h after transfection, the expression of the EGFP-ATX-3′UTR chimeric transcript was inhibited by addition of Dox (1 μg/ml), and total RNA was prepared at the indicated times. Then, the transcript half-lives were evaluated by real-time qPCR using EGFP-specific primers (TACAACTACAACAGCCACAACG and ATCCTGTCCTCCACCTCC). To measure the half-life of endogenous ATX mRNA in different cell lines, the expression of ATX mRNA was inhibited by adding actinomycin D (1 μg/ml) to the cell culture medium, and total RNA was prepared at the indicated times and subjected to RT-qPCR analysis using the ATX-specific primers (TATGCTTCGGAAAGAAATGGAG and ATGTTCAATGTCACGCACCCT for human cell, TCCTCCCTATCTGAGCTCTTC and GGTGATGATGCTGTAGTAGTGG for mouse cell).
Scratch Wound-healing Assay
B16 cells were transfected with nonspecific siRNA (NC), HuR siRNA, and/or AUF1 siRNA as indicated. For the scratch wound-healing assays, 48 h after siRNA transfection, B16 cells were trypsinized and seeded equally into six-well tissue culture plates and grown to almost total confluence at 24 h. The cells were starved for 8 to 12 h, and then artificial homogenous wounds were created on the monolayer with a sterile 200-μl tip. After scratching, the cells were incubated with serum-free medium in the absence or presence of the ATX inhibitor S32826 (2 μm) or 18:1 LPA (2 μm) as indicated. Images of the cells migrating into the wound were captured at 0 and 12 h by an inverted microscope (×10), and all experiments were repeated at least 3 times.
Transwell Assay
Cell migration was measured based on the ability of the cells to migrate across a transwell filter (8 μm pores, Costar, Cambridge, MA). After siRNA transfection for 48 h, B16 cells (5 × 105) were suspended in 200 μl of serum-free RPMI 1640 medium and added to the upper chamber. 500 μl of serum-free RPMI 1640 medium was added to the lower chamber. The ATX inhibitor S32826 (2 μm) or 18:1 LPA (2 μm) was added in the medium in the upper and lower chambers as indicated. After 24 h incubation, the non-migrated cells were scraped off from the filter using a cotton swab, and the cells that migrated to the lower side of the upper chamber were fixed with 4% paraformaldehyde and stained with hematoxylin. The cells per microscopic field (B16 cells, ×20) were imaged and counted in 3 randomly chosen fields.
Computational and Statistical Analysis
The computational analyses of AU-rich elements in RNA were conducted according to the methods described by Gruber et al. (26) using online resources. All data are represented as the mean ± S.D. from three independent experiments. The differences between two independent groups were analyzed using Student's t test. A p value <0.05 was considered statistically significant.
Author Contributions
J. Z. and X. Z. designed the experiments. J. Z., X. Z., and S. S. wrote the manuscript. X. Z. tested the half-lives of mRNA and performed luciferase activity assay. S. S. performed the Western blotting, real-time PCR, transwell, and wound-healing assays. X. L. completed the sequence alignments and analysis of the ATX mRNA 3′UTR. L. L. and S. Y. contributed in the plasmid construction.
This work was supported by Grants 31500619 and 31470765 from the National Natural Science Foundation of China, Grant 2014NT24 from the Young Teacher Fund of Beijing Normal University, and the Open Fund of the Key Laboratory of Cell Proliferation and Regulation Biology (Beijing Normal University), Ministry of Education. The authors declare that they have no conflicts of interest with the contents of this article.
- ATX
- autotaxin
- lysoLPD
- lysophospholipase D
- LPC
- lysophosphatidylcholine
- LPA
- lysophosphatidic acid
- RBP
- RNA-binding protein
- ARE
- AU-rich element
- Dox
- doxycycline
- qPCR
- quantitative PCR
- IP
- immunoprecipitation
- EGFP
- enhanced green fluorescent protein.
References
- 1. Stracke M. L., Krutzsch H. C., Unsworth E. J., Arestad A., Cioce V., Schiffmann E., and Liotta L. A. (1992) Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 267, 2524–2529 [PubMed] [Google Scholar]
- 2. van Meeteren L. A., and Moolenaar W. H. (2007) Regulation and biological activities of the autotaxin-LPA axis. Prog. Lipid Res. 46, 145–160 [DOI] [PubMed] [Google Scholar]
- 3. Samadi N., Bekele R., Capatos D., Venkatraman G., Sariahmetoglu M., and Brindley D. N. (2011) Regulation of lysophosphatidate signaling by autotaxin and lipid phosphate phosphatases with respect to tumor progression, angiogenesis, metastasis and chemo-resistance. Biochimie 93, 61–70 [DOI] [PubMed] [Google Scholar]
- 4. Lin M. E., Herr D. R., and Chun J. (2010) Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 91, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moolenaar W. H., Houben A. J., Lee S. J., and van Meeteren L. A. (2013) Autotaxin in embryonic development. Biochim. Biophys. Acta 1831, 13–19 [DOI] [PubMed] [Google Scholar]
- 6. Wang L., Knudsen E., Jin Y., Gessani S., and Maghazachi A. A. (2004) Lysophospholipids and chemokines activate distinct signal transduction pathways in T helper 1 and T helper 2 cells. Cell Signal. 16, 991–1000 [DOI] [PubMed] [Google Scholar]
- 7. Rubenfeld J., Guo J., Sookrung N., Chen R., Chaicumpa W., Casolaro V., Zhao Y., Natarajan V., and Georas S. (2006) Lysophosphatidic acid enhances interleukin-13 gene expression and promoter activity in T cells. Am. J. Physiol. Lung Cell Mol. Physiol. 290, L66–L74 [DOI] [PubMed] [Google Scholar]
- 8. Park G. Y., Lee Y. G., Berdyshev E., Nyenhuis S., Du J., Fu P., Gorshkova I. A., Li Y., Chung S., Karpurapu M., Deng J., Ranjan R., Xiao L., Jaffe H. A., Corbridge S. J., et al. (2013) Autotaxin production of lysophosphatidic acid mediates allergic asthmatic inflammation. Am. J. Respir. Crit. Care Med. 188, 928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boucher J., Quilliot D., Pradères J. P., Simon M. F., Grès S., Guigné C., Prévot D., Ferry G., Boutin J. A., Carpéné C., Valet P., and Saulnier-Blache J. S. (2005) Potential involvement of adipocyte insulin resistance in obesity-associated up-regulation of adipocyte lysophospholipase D/autotaxin expression. Diabetologia 48, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferry G., Tellier E., Try A., Grés S., Naime I., Simon M. F., Rodriguez M., Boucher J., Tack I., Gesta S., Chomarat P., Dieu M., Raes M., Galizzi J. P., Valet P., et al. (2003) Autotaxin is released from adipocytes, catalyzes lysophosphatidic acid synthesis, and activates preadipocyte proliferation: up-regulated expression with adipocyte differentiation and obesity. J. Biol. Chem. 278, 18162–18169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu S., Umezu-Goto M., Murph M., Lu Y., Liu W., Zhang F., Yu S., Stephens L. C., Cui X., Murrow G., Coombes K., Muller W., Hung M. C., Perou C. M., Lee A. V., et al. (2009) Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell 15, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nam S. W., Clair T., Campo C. K., Lee H. Y., Liotta L. A., and Stracke M. L. (2000) Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene 19, 241–247 [DOI] [PubMed] [Google Scholar]
- 13. Euer N., Schwirzke M., Evtimova V., Burtscher H., Jarsch M., Tarin D., and Weidle U. H. (2002) Identification of genes associated with metastasis of mammary carcinoma in metastatic versus non-metastatic cell lines. Anticancer Res. 22, 733–740 [PubMed] [Google Scholar]
- 14. Farina A. R., Cappabianca L., Ruggeri P., Di Ianni N., Ragone M., Merolle S., Sano K., Stracke M. L., Horowitz J. M., Gulino A., and Mackay A. R. (2012) Constitutive autotaxin transcription by Nmyc-amplified and non-amplified neuroblastoma cells is regulated by anovel AP-1 and SP-mediated mechanism and abrogated by curcumin. FEBS Lett. 586, 3681–3691 [DOI] [PubMed] [Google Scholar]
- 15. Chen M., and O'Connor K. L. (2005) Integrin α6β4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene 24, 5125–5130 [DOI] [PubMed] [Google Scholar]
- 16. Black E. J., Clair T., Delrow J., Neiman P., and Gillespie D. A. (2004) Microarray analysis identifies Autotaxin, a tumour cell motility and angiogenic factor with lysophospholipase D activity, as a specific target of cell transformation by v-Jun. Oncogene 23, 2357–2366 [DOI] [PubMed] [Google Scholar]
- 17. Kehlen A., Englert N., Seifert A., Klonisch T., Dralle H., and Langner J., and Hoang-Vu C. (2004) Expression, regulation and function of autotaxinin thyroid carcinomas. Int. J. Cancer 109, 833–838 [DOI] [PubMed] [Google Scholar]
- 18. Song J., Guan M., Zhao Z., and Zhang J. (2015) Type I interferons function as autocrine and paracrine factors to induce autotaxin in response to TLR activation. PLoS ONE 10, e0136629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nabors L. B., Gillespie G. Y., Harkins L., and King P. H. (2001) HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 61, 2154–2161 [PubMed] [Google Scholar]
- 20. Hinman M. N., and Lou H. (2008) Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan X. C., and Steitz J. A. (1998) HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U.S.A. 95, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J. L., and Gorospe M. (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23, 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan Y. X., Chen H., and Kilberg M. S. (2005) Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J. Biol. Chem. 280, 34609–34616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li S., and Zhang J. (2009) Lipopolysaccharide induces autotaxin expression in human monocytic THP-1 cells. Biochem. Biophys. Res. Commun. 378, 264–268 [DOI] [PubMed] [Google Scholar]
- 25. Li S., Xiong C., and Zhang J. (2012) ATX and LPA receptor 3 are coordinately up-regulated in lipopolysaccharide stimulated THP-1 cells through PKR and SPK1-mediated pathways. FEBS Lett. 586, 792–797 [DOI] [PubMed] [Google Scholar]
- 26. Gruber A. R., Fallmann J., Kratochvill F., Kovarik P., and Hofacker I. L. (2011) AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 39, D66–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang C. L., Lin M. E., Hsu H. Y., Yao C. L., Hwang S. M., Pan C. Y., Hsu C. Y., and Lee H. (2008) Lysophosphatidic acid-induced interleukin-1β expression is mediated through Gi/Rho and the generation of reactive oxygen species in macrophages. J. Biomed. Sci. 15, 357–363 [DOI] [PubMed] [Google Scholar]
- 28. Young L. E., Sanduja S., Bemis-Standoli K., Pena E. A., Price R. L., and Dixon D. A. (2009) The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 136, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yi X., Zhou Y., Zheng W., and Chambers S. K. (2009) HuR expression in the nucleus correlates with high histological grade and poor disease-free survival in ovarian cancer. Aust. N. Z. J. Obstet. Gynaecol. 49, 93–98 [DOI] [PubMed] [Google Scholar]
- 30. Zhu Z., Wang B., Bi J., Zhang C., Guo Y., Chu H., Liang X., Zhong C., and Wang J. (2013) Cytoplasmic HuR expression correlates with P-gp, HER-2 positivity, and poor outcome in breast cancer. Tumour Biol. 34, 2299–2308 [DOI] [PubMed] [Google Scholar]
- 31. Grammatikakis I., and Abdelmohsen K., and Gorospe M. (2016) Posttranslational control of HuR function. Wiley Interdiscip. Rev. RNA 10.1002/wrna.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zucconi B., and Wilson G. (2013) Modulation of neoplastic gene regulatory pathways by the RNA binding factor AUF1. Front. Biosci. 16, 2307–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gratacós F. M., and Brewer G. (2010) The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 1, 457–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. López de Silanes I., Zhan M., Lal A., Yang X., and Gorospe M. (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]