Abstract
Many bacterial pathogens secrete virulence (effector) proteins that interfere with immune signaling in their host. SpvD is a Salmonella enterica effector protein that we previously demonstrated to negatively regulate the NF-κB signaling pathway and promote virulence of S. enterica serovar Typhimurium in mice. To shed light on the mechanistic basis for these observations, we determined the crystal structure of SpvD and show that it adopts a papain-like fold with a characteristic cysteine-histidine-aspartate catalytic triad comprising Cys-73, His-162, and Asp-182. SpvD possessed an in vitro deconjugative activity on aminoluciferin-linked peptide and protein substrates in vitro. A C73A mutation abolished SpvD activity, demonstrating that an intact catalytic triad is required for its function. Taken together, these results strongly suggest that SpvD is a cysteine protease. The amino acid sequence of SpvD is highly conserved across different S. enterica serovars, but residue 161, located close to the catalytic triad, is variable, with serovar Typhimurium SpvD having an arginine and serovar Enteritidis a glycine at this position. This variation affected hydrolytic activity of the enzyme on artificial substrates and can be explained by substrate accessibility to the active site. Interestingly, the SpvDG161 variant more potently inhibited NF-κB-mediated immune responses in cells in vitro and increased virulence of serovar Typhimurium in mice. In summary, our results explain the biochemical basis for the effect of virulence protein SpvD and demonstrate that a single amino acid polymorphism can affect the overall virulence of a bacterial pathogen in its host.
Keywords: bacterial pathogenesis, crystal structure, cysteine protease, Salmonella enterica, structure-function, effector
Introduction
Salmonella enterica serovars Typhimurium and Enteritidis are broad host range intracellular bacterial pathogens causing gastrointestinal disease in humans and a typhoid-like systemic infection in certain mouse strains. Their virulence depends on the activity of two type III secretion systems (T3SSs),5 encoded by the pathogenicity islands Salmonella pathogenicity island 1 (SPI-1) and SPI-2 that enable translocation of effector proteins into host cells. The majority of effector proteins are encoded on the bacterial chromosome, but some are present on the pSLT virulence plasmid (1). An important virulence determinant of the pSLT plasmid is the Salmonella plasmid virulence locus (spv) (2–5), which comprises the spvABCD operon and its upstream regulator spvR (6). SpvB is an ADP ribosyl transferase that depolymerizes F-actin (7), and SpvC is a phosphothreonine lyase that inhibits mitogen-activated protein kinase signaling (8, 9). Mass spectrometry analysis of proteins secreted by Salmonella identified SpvD as an effector of both SPI-1 and SPI-2 T3SSs (10). Previously, we confirmed that SpvD contributes to virulence during systemic infection of mice (10, 11) and showed it inhibits the NF-κB signaling pathway and secretion of proinflammatory cytokines from infected macrophages (11). In this work we determined the structure of SpvD and showed that it is a hydrolase with a papain-like fold and a catalytic triad composed of Cys-73, His-162, and Asp-182. Although SpvD is highly conserved among different serovars of S. enterica, serovars Typhimurium and Enteritidis differ in containing an arginine or glycine, respectively, at residue 161 near the active site. We show that this polymorphism affects hydrolytic activity, degree of inhibition of the NF-κB pathway, and remarkably, bacterial virulence in mice.
Results
SpvD Adopts a Papain-like Fold
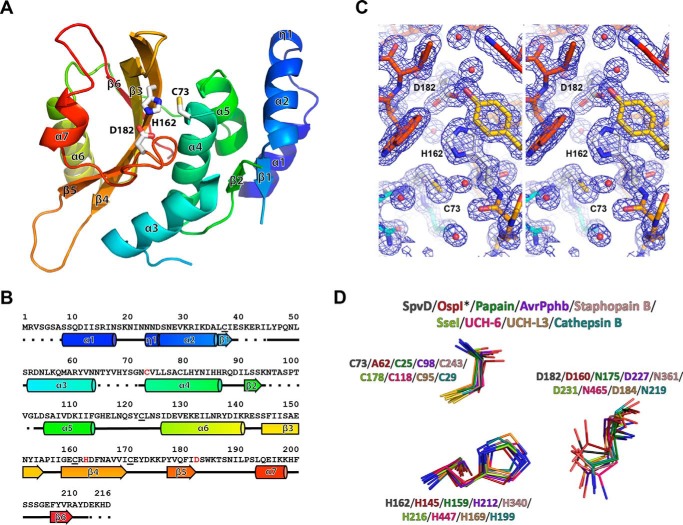
To gain insight into the function of SpvD, we attempted to determine its crystal structure. After unsuccessful crystallization of wild-type SpvD, we obtained crystals of SpvD in which its predicted surface cysteines were mutated (SpvDC37S/C122S/C160S/C170S, hereafter referred to as SpvD4CS), and these diffracted to 1.5 Å resolution (Table 1). Subsequent structure determination revealed that SpvD adopts an α+β fold with a total of 7 α-helices, 6 β-strands, and a short stretch of 310 helix (Fig. 1, A and B). Its central core is composed of a β-sheet formed by four antiparallel β-strands (β3–6) surrounded by five α-helices (α3–7). Additionally, there is a smaller β-sheet (β1–2) surrounded by α-helices 1–5 (Fig. 1, A and B). Close inspection of a shallow groove on the protein surface between helices α4/α5, strand β4, and the β5-α7 loop revealed a potential hydrolase active site including a catalytic triad of cysteine (Cys-73), histidine (His-162), and aspartic acid (Asp-182) (Fig. 1, A and C). A structure similarity search performed with the DALI software (12) revealed similarities to four other secreted effector proteins: OspI (a T3SS deamidase of the Gram-negative bacterial pathogen Shigella flexneri) (13, 14), SseI (a S. typhimurium SPI-2 T3SS effector protein of unknown enzymatic activity) (15), AvrPphB (a T3SS cysteine protease of a bacterial plant pathogen Pseudomonas syringae) (16), and SspB (a cysteine protease secreted by Staphylococcus aureus) (17) as well as other papain and papain-like cysteine proteases. These all have a characteristic central antiparallel β-sheet and catalytic cysteine-containing α-helix (Fig. 2). Superimposition of these structures revealed that the orientation of the catalytic triads in papain, OspI, AvrPphB, staphopain B, SseI, UCH-L3, UCH-6, and cathepsin B proteins are almost identical to that of SpvD (Fig. 1D) (14–20).
TABLE 1.
Data collection and refinement statistics
r.m.s.d., root mean square deviation; SIRAS, single isomorphous replacement with anomalous scattering (data set collected from crystals soaked in KAuCl4).
| SpvD4CS/A154/R161 | SpvD4CS/A154/G161/C73A | SpvD (Au SIRAS) | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.97625 | 0.97949 | 0.92000 |
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 43.7, 51.81, 47.27 | 44, 51.97, 47.22 | 43.84, 52.11, 47.38 |
| α, β, γ (°) | 90, 107.41, 90 | 90, 107.7, 90 | 90, 107.84, 90 |
| Resolution range (Å) | 51.81–1.48 (1.56–1.48)a | 51.97–1.60 (1.69–1.60) | 41.73–2.25 (2.31–2.25) |
| Reflections, total | 195,684 (21,204) | 156,479 (23,249) | |
| Reflections, unique | 33,404 (4614) | 26,737 (3,893) | |
| I/σI | 12.1 (2.1) | 10.4 (1.9) | 18.1 (3.8) |
| Rmerge | 0.076 (0.869) | 0.104 (1.018) | 0.108 (0.531) |
| Completeness (%) | 99.1 (94.1) | 99.4 (99.2) | 100 (60.8)b |
| Multiplicity | 5.9 (4.6) | 5.9 (6.0) | 6.4 (4.2)b |
| Refinement | |||
| Rfactor/Rfree (%) | 16.96/20.15 | 16.32/19.73 | |
| No. of atoms | |||
| Protein | 1,556 | 1,713 | |
| Ligand/ion | 1 | 18 | |
| Water | 182 | 205 | |
| Bfactors | |||
| Protein | 22.07 | 19.96 | |
| Ligand/ion | 17.18 | 28.19 | |
| Water | 30.76 | 30.61 | |
| r.m.s.d. bond length (Å) | 0.0151 | 0.0164 | |
| r.m.s.d. bond angle (°) | 1.573 | 1.410 | |
| Ramachandran favored/outliers (%) | 98.9/0 | 98.1/0.5 | |
a Values in parentheses correspond to the highest resolution shell.
b Completeness and multiplicity values given for anomalous data.
FIGURE 1.
SpvD adopted a papain-like fold with Cys-73, His-162, and Asp-182 catalytic triad. A, crystal structure of S. typhimurium SpvD4CS colored starting from N terminus (blue) and ending at C terminus (red). Secondary structure elements and catalytic triad (sticks) are labeled in black. Cys-73/His-162/Asp-182 catalytic triad is shown in sticks. The image was generated using PyMOL software. B, secondary structure elements mapped according to their respective amino acid sequence and colored as in A. In the sequence, the catalytic triad residues are colored in red. Surface cysteines mutated to serines are underlined. C, walleye stereo view of the 2Fo − Fc electron density map of the active site region of the SpvD structure. D, superimposition of catalytic triad components of SpvD, OspIC62A (PDB code 3W31), papain (PDB code 1PPN), AvrPphB (PDB code 1UKF), staphopain B (PDB code 1Y4H), SseI (PDB code 4G29), UCH-6 (PDB code 1VJV), UCH-L3 (PDB code 1XD3), and cathepsin B (PDB code 3AI8).
FIGURE 2.
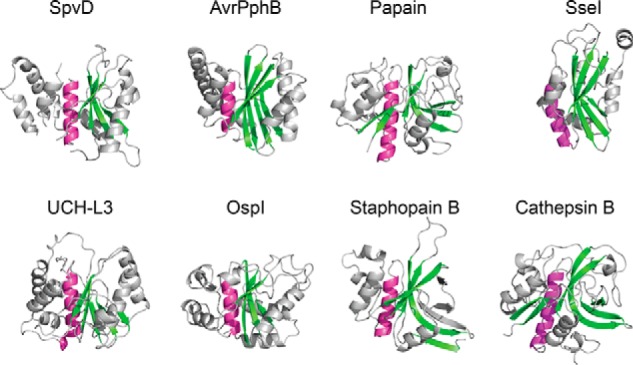
SpvD structure shared papain-like fold features. Schematic representation of SpvDA154/R161, OspIC62A (PDB code 3W31), papain (PDB code 1PPN), AvrPphB (PDB code 1UKF), staphopain B (PDB code 1Y4H), SseI (PDB code 4G29), UCH-L3 (PDB code 1XD3), and cathepsin B (PDB code 3AI8) is shown. The central α-helix containing the catalytic cysteine is colored in purple, and the main anti-parallel β-sheet is colored in green. Structures were oriented according to their superimposed catalytic triads.
A Naturally Occurring SpvD Variant with Enhanced Enzymatic Activity
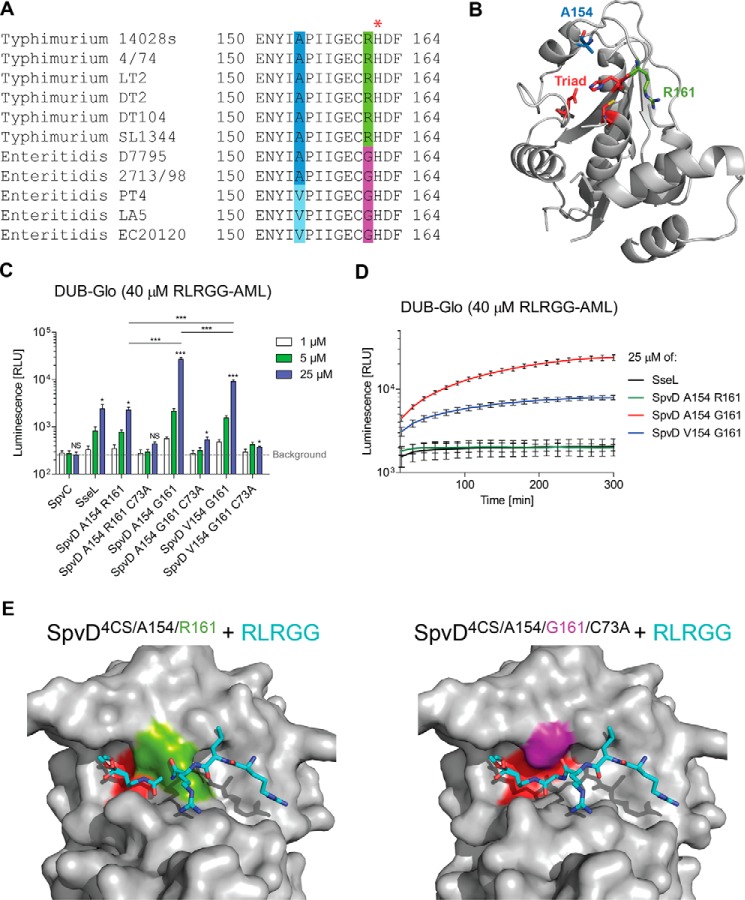
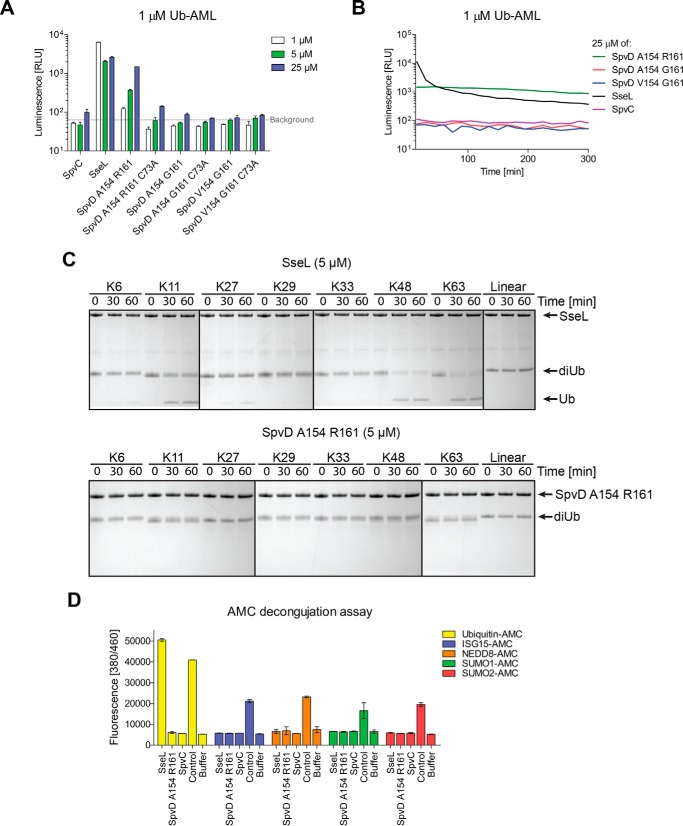
The amino acid sequence of SpvD varies among serovars of S. enterica (21). We carried out a protein BLAST search using the amino acid sequence of S. typhimurium 14028 SpvD. This confirmed previously described polymorphisms and found sequences homologous to SpvD in other bacterial species such as Pseudomonas psychrophila and Providencia burhodogranariea (supplemental Fig. S1). Within the broad host range serovars of S. enterica, Enteritidis and Typhimurium, there are only two positions that consistently vary in amino acid composition (154 and 161) (Fig. 3A and supplemental Fig. S1). Within Typhimurium serovars, an alanine is encoded at position 154, and an arginine is encoded at position 161. In contrast, all Enteritidis serovars sequenced to date have either alanine or valine at position 154 and glycine at position 161 (Fig. 3A). Both residues are in close proximity to the catalytic triad, and their side chains contribute to the structure of the active site groove (Fig. 3B, blue and green sticks). Positions 154 and 161 of SpvD homologues in species other than Salmonella are mainly occupied by small hydrophobic residues (leucine and isoleucine at 154 and glycine and alanine at 161; supplemental Fig. S1). Within Salmonella, the Gly-161 polymorphism appears to be unique to Enteritidis, Abortus-equi, and Bovismorbificans serovars as all others (Typhimurium, Dublin, Paratyphi C, Gallinarum, Pullorum, and Choleraesuis) contain arginine at this position (supplemental Fig. S1). We used a variety of assays to determine the potential enzymatic activity of SpvD. Structural similarity to ubiquitin hydrolases suggested that SpvD could be a deubiquitinase. In a screen that utilizes a C-terminal peptide of ubiquitin (RLRGG) fused to aminoluciferin for detection of ubiquitin and ubiquitin-like cysteine proteases (DUB-GloTM protease assay; Promega), we found that SpvD, like the known Salmonella deubiquitinase (DUB) SseL (22, 23), produced a stable and specific luminescence signal indicative of substrate hydrolysis. Consistent with the crystal structure, SpvD hydrolytic activity was dependent on cysteine 73 of its putative catalytic triad (Fig. 3C). Interestingly, conversion of the SpvD sequence to that of the Enteritidis strains increased the activity observed in the DUB-GloTM protease assay (Fig. 3, C and D). At the highest concentration tested, the most active variant, SpvDA154/G161, produced over 10 times more luminescence signal than SpvDA154/R161, 5 h after the start of the assay (Fig. 3, C and D). In this assay, protease and luciferase activities normally reach a steady state so that a maximal signal is reached after 30 min and maintained for several hours. In contrast to SseL and SpvDA154/R161, the luminescence signal produced by the SpvDA154/G161 and SpvDV154/G161variants did not stabilize after 30 min but continued to increase up to 5 h (Fig. 3D). SpvDV154/G161 had an intermediate activity in the DUB-GloTM assay, in between that of SpvDA154/R161 and SpvDA154/G161 (Fig. 3C and D). Due to the more pronounced effect of the variation at position 161, we focused further studies on this polymorphism, maintaining alanine at position 154. To explain why the Gly-161 polymorphism leads to greater activity, we crystallized an inactive (C73A) form of the variant in the presence of RLRGG-AML substrate. Although density for the RLRGG-AML substrate was not observed, we were able to build additional loops (two of them near the active site) that were disordered in the previously obtained SpvD4CS/R161 structure (supplemental Fig. S2). To map a potential ligand binding site on SpvD, we superimposed the catalytic triad with that of the UCH-L3 ubiquitin hydrolase crystallized in complex with ubiquitin (Fig. 3E). The position of the superimposed five C-terminal amino acids of ubiquitin (RLRGG) is unobstructed in the SpvD4CS/G161/C73A variant (Fig. 3E, right), whereas in SpvD4CS/R161, binding would be impeded by the side chain of Arg-161, which restricts access to the active site (Fig. 3E, left). Therefore, the Arg-161 side chain could have an inhibitory steric or regulatory role in the activity of SpvD. These results demonstrate that SpvD is a hydrolytic enzyme with a catalytic triad that is characteristic of papain-like proteases. As the DUB-GloTM protease assay detects ubiquitin and ubiquitin-like proteases, we then assayed SpvD variants using full-length ubiquitin-AML as a substrate. As expected, the positive control SseL was active at all concentrations tested (1 μm, 5 μm, and 25 μm; Fig. 4A). Surprisingly, the only SpvD variant that produced a specific, Cys-73-dependent signal was SpvDA154/R161 (Fig. 4, A and B). The luminescent signal produced by SseL decreased over time from an initial level of 100-fold above background, indicating substrate depletion (Fig. 4B). In contrast SpvDA154/R161produced a level of luminescence that was ∼10-fold above background and remained relatively constant over the course of the assay. Interestingly, in this assay both SpvDV154/G161 and SpvDA154/G161 displayed virtually no activity (Fig. 4, A and B). To test if SpvDA154/R161 has linkage-specific deubiquitinating activity, diubiquitin molecules linked by seven different lysine residues or methionine M1 were tested for cleavage. SseL cleaved isopeptide molecules linked by Lys-11, Lys-48, and Lys-63 (Fig. 4C, top), but SpvDA154/R161 had no detectable activity on any of the substrates (Fig. 4C, bottom). To test if SpvDA154/R161 has activity against other ubiquitin-like proteins, we used substrates comprising full-length ubiquitin or four ubiquitin-like proteins (SUMO1, SUMO2, NEDD8, and ISG15) C-terminally linked to 7-amino-4-methylcoumarin (AMC). SpvDA154/R161 had no detectable activity on any of these substrates, whereas SseL produced a specific fluorescent signal with ubiquitin-AMC as a substrate (Fig. 4D).
FIGURE 3.
SpvD had a deconjugative activity that varied between serovars. A, zoom on SpvD amino acid alignment highlighting the two main (154 and 161) polymorphisms occurring in Enteritidis and Typhimurium serovars. The red asterisk indicates the catalytic histidine His-162. B, both Ala-154 (blue stick) and Arg-161 (green stick) positions are situated in the vicinity of the CHD catalytic triad (red sticks). C, DUB-GloTM assay with three (1, 5, and 25 μm) concentrations of tested proteins. Results are expressed as mean ± S.E. of three independent experiments. Asterisks above the blue bars represent significant differences when compared with the buffer control (background) as measured by the Student's t test. Comparison between SpvD variants at 25 μm concentration was made using one-way ANOVA statistical analysis with Bonferroni's multiple comparison test. *, p < 0.05; ***, p < 0.005; NS, not significant. RLU, relative light units. D, DUB-GloTM signal over time for the highest (25 μm) concentration tested for SseL, SpvDA154/R161, SpvDA154/G161, and SpvDV154/G161. Signal intensities were measured every 15 min for which ± S.E. of three independent experiments are shown. E, modeling of the RLRGG peptide onto the surface of SpvD4CS/A154/R161 (left) or SpvD4CS/A154/G161/C73A (right) using the C-terminal ubiquitin tail (RLRGG; cyan sticks) from the UCH-L3 ubiquitin hydrolase complexed with ubiquitin (PDB code 1XD3). The catalytic triad region of SpvD is colored in red, Arg-161 is in green, and Gly-161 is in purple.
FIGURE 4.
SpvDR161 was active on a ubiquitin-AML substrate. A, a modification of the DUB-GloTM assay with RLRGG substrate replaced by a full-length ubiquitin conjugated to aminoluciferin (Ub-AML). A representative assay result is shown. B, Ub-AML deconjugation assay over time measured for the highest concentration (25 μm) of the tested proteins. Signals were taken every 15 min. C, in vitro deubiquitination assay done with 5 μm of either SseL (top) or SpvDA154/R161 (bottom). Diubiquitin linkages were incubated with tested proteins for 0, 30, and 60 min followed by SDS-PAGE separation and Coomassie staining. Detectable cleavage product in the form of a single ubiquitin band is indicated with an arrow. D, AMC deconjugation assay using ubiquitin-AMC (yellow), ISG15-AMC (blue), NEDD8-AMC (orange), SUMO1-AMC (green), and SUMO2-AMC (red). Results are shown as mean fluorescence values with S.D. of three technical repeats obtained 1 h after start of the experiment. Isopeptidase T was used as a positive control for ubiquitin-AMC and ISG15-AMC. SENP2 was used as a control protease for SUMO1-AMC and SUMO2-AMC substrates, whereas NEDP1 was used in case of NEDD8-AMC.
Inhibition of the NF-κB Pathway by SpvD Is Dependent on Cys-73 and Is Affected by Variation at Residue 161
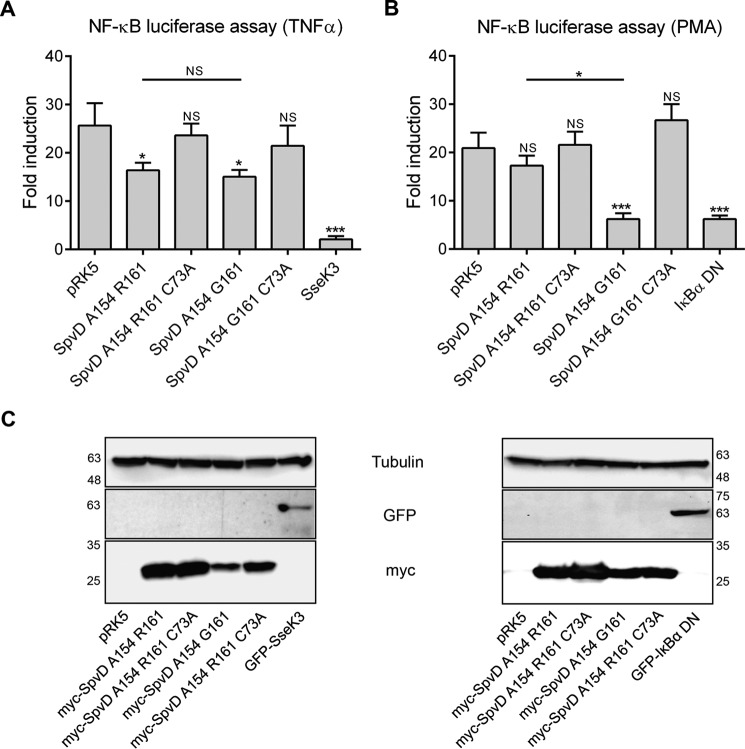
We showed previously that SpvD inhibits the NF-κB signaling pathway (11). To investigate the influence of the amino acid side chain at position 161 and the requirement for the putative catalytic cysteine (Cys-73) on the anti-inflammatory activity of SpvD, HEK 293 cells were co-transfected with a reporter plasmid encoding luciferase under the control of an NF-κB promoter and a plasmid encoding either myc alone (pRK5 vector), myc-SpvD variants with alanine at position 154, or GFP-tagged positive control inhibitory proteins: Salmonella SseK3 (24) and NF-κB inhibitor α (IκBα) S32A/S36A mutant protein (25). Luciferase activity was measured 16 h after stimulation with TNFα or phorbol 12-myristate 13-acetate (PMA) and normalized to that of non-stimulated cells. TNFα and PMA caused ∼25- and 20-fold increases in NF-κB activation, respectively (Fig. 5, A and B). Expression of tagged proteins was confirmed by Western blotting (Fig. 5C). SseK3 is an effector that is known to inhibit the NF-κB pathway upon TNFα stimulation (14). In this assay it inhibited the reporter activity by 92% (Fig. 5A). After TNFα stimulation, SpvDA154/R161 and SpvDA154/G161 reduced NF-κB fold activation by 36 and 41%, respectively, and these effects were dependent on Cys-73 (Fig. 5A). There was no significant difference in the level of NF-κB inhibition between the Arg-161 and Gly-161 variants (Fig. 5A). However, after stimulation with PMA, both the positive control protein IκBαS32A/S36A and SpvDA154/G161 inhibited the NF-κB pathway to a similar level (Fig. 5B), whereas SpvDA154/R161 had a much smaller effect. Therefore, the increased hydrolase activity of SpvDA154/G161 over SpvDA154/R161 in the DUB-GloTM protease assay (Fig. 3C) correlates with an enhanced ability of SpvDG161 to inhibit the NF-κB inflammatory pathway in cells stimulated with PMA.
FIGURE 5.
SpvD variants had differential NF-κB inhibitory activities in PMA- but not in TNFα-stimulated cells. NF-κB luciferase assay from transfected HEK293 cell line after 16 h stimulation with TNFα (A) or PMA (B). Results are shown as -fold activation in relation to unstimulated cells. Values are expressed as the mean ± S.E. of at least four independent experiments. Statistical significances were calculated using one-way ANOVA followed by Bonferroni's multiple comparison test against pRK5- or SpvDA154/R161-transfected cells (*, p < 0.05; ***, p < 0.005; NS, not significant). C, protein expression levels measured by SDS-PAGE and immunoblotting with anti-tubulin (top), anti-GFP (mid), and anti-myc (bottom) antibodies. Numbers on the sides correspond to molecular weight protein ladder in kDa.
Higher in Vitro Deconjugative and Anti-inflammatory Activities of Naturally Occurring SpvD Variant Correlate with Salmonella Virulence in Vivo
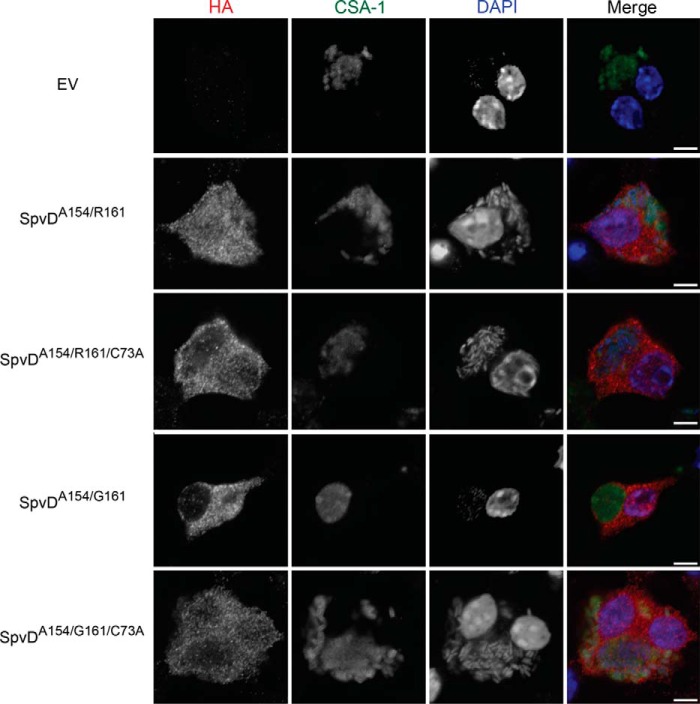
Salmonella strains lacking SpvD were shown to be attenuated for growth in mice when compared with the wild-type strain (10, 11). Having detected different in vitro deconjugation and anti-inflammatory activities of SpvD variants, we tested if the side chain at position 161 within the SpvD active site influences Salmonella growth in vivo. First, we confirmed bacterial translocation of C-terminal double hemagglutinin (HA)-tagged SpvDA154/G161/C73A and SpvDA154/R161/C73A into iBMDMs, showing that catalytic triad mutations did not affect effector delivery (Fig. 6). Mice were then inoculated by intraperitoneal injection with a mixture of equivalent cfu of ΔspvD mutant strain and chloramphenicol-resistant ΔspvD mutant strains harboring various pACYC184-based complementation plasmids (Fig. 7). After 72 h of infection, bacteria were quantified after plating spleen homogenates on LB agar medium and LB agar medium containing chloramphenicol to distinguish the strains. The competitive index (CI) of ΔspvD mutant and ΔspvD mutant harboring an empty pACYC184 plasmid (EV) was 0.34 ± 0.04, showing that the presence of pACYC184 incurred a fitness cost (Fig. 7). When mixed with ΔspvD strains carrying pACYC184 encoding either SpvDA154/R161 or SpvDA154/G161, the CI of the ΔspvD mutant strain dropped from 0.34 to −0.12 and −0.39, respectively (Fig. 7). Interestingly, the ΔspvD mutant was more efficiently outcompeted by the SpvDA154/G161 strain compared with the SpvDA154/R161 strain, demonstrating that the more active SpvDA154/G161 variant is more potent in complementing the ΔspvD mutation than SpvDA154/R161 (Fig. 7). When mixed with ΔspvD strains carrying plasmids encoding the C73A mutant versions of SpvD, the ΔspvD strain had CIs of 0.15 ± 0.14 and 0.32 ± 0.08 for SpvDA154/R161/C73A and SpvDA154/G161/C73A, respectively) similar to the CI of the ΔspvD strain mixed with ΔspvD strain harboring empty pACYC184 plasmid (Fig. 7). Therefore, the ability of both SpvDA154/R161 and SpvDA154/G161 to complement the virulence defect was dependent on a functional catalytic triad.
FIGURE 6.
Catalytically dead variants of SpvD were translocated from bacteria in iBMDMs. Cells were infected for 18 h with S. typhimurium spvD strain carrying either an empty pWSK29 plasmid (EV) or a pWSK29 plasmid encoding C-terminally 2×HA-tagged variants of SpvD under SPI-2 promoter. In the last 2 h of infection, cells were treated with 10 μm MG-132 proteasomal inhibitor. DNA, Salmonella, and HA-tagged SpvD were stained using DAPI dye, CSA-1, and HA11 antibodies, respectively. The white scale bar represents 5 μm.
FIGURE 7.
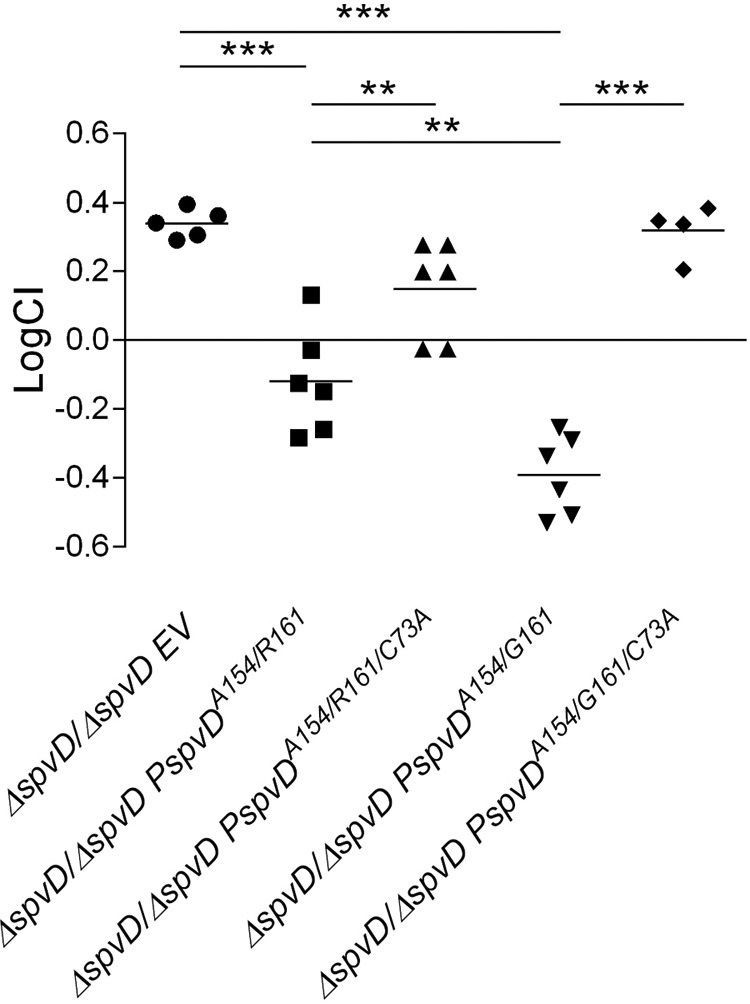
SpvDA154/G161 conferred increased virulence over SpvDA154/R161. C57BL/6 mice were inoculated by intraperitoneal injection with equal numbers (5 × 102 cfu of each of the two strains) of the indicated bacteria. Bacteria were recovered from infected spleens 3 days post-inoculation. CI values were calculated as described under “Experimental Procedures.” The scatter plot displays values obtained for individual mice and the mean are indicated (line). Statistical significances were calculated using one-way ANOVA followed by Bonferroni's multiple comparison test. **, p < 0.01; ***, p < 0.005.
Discussion
In this work we have determined the structure of SpvD, a S. typhimurium SPI-2 T3SS effector protein that inhibits the NF-κB signaling pathway (11). Structurally, SpvD can be classified as belonging to the CA clan of cysteine proteases that contains papain and related proteases (MEROPS peptidase database). A hallmark of this clan is a CHD (or CHN) catalytic triad, which in the case of SpvD is composed of Cys-73, His-162, and Asp-182. Using a DUB-GloTM assay, we showed that SpvD is an enzyme with deconjugative activity. Taken together, the results of our experiments strongly suggest that SpvD acts as a protease. Due to the chemical nature of the DUB-GloTM substrate (aminoluciferin fused to the C terminus of an RLRGG peptide), the DUB-GloTM assay would potentially not only detect deubiquitinases or ubiquitin-like proteases but any protease that recognizes and cleaves after a sequence similar to GG. In all the assays tested (DUB-GloTM, NF-κB inhibition, virulence in mouse) the activity of SpvD was abolished by a C73A mutation, demonstrating the necessity of an intact catalytic triad for its function. Previously, we reported that SpvD specifically interacts with exportin-2 to inhibit nuclear transport of p65 (11). Although the results presented here provide additional evidence for a function of SpvD in inhibiting the NF-κB pathway, we have not been able to detect cleavage of endogenous exportin-2 in HEK293 cells transiently expressing SpvDA154/R161 or SpvDA154/G161 (data not shown). This suggests that there might be another as yet unidentified substrate that is modified by SpvD. Analysis of naturally occurring SpvD polymorphisms revealed that position 161 is critical for the deconjugative activity of SpvD in the DUB-GloTM assay. Modeling of the RLRGG DUB-GloTM substrate onto the Arg-161 or Gly-161 structures provides a rationale for this observation, clearly showing that the side chain at position 161 forms part of the active site groove and that an arginine side chain here restricts access to the catalytic triad. Despite being less active in the DUB-GloTM assay, SpvDA154/R161 was the most active SpvD variant in the Ub-AML deconjugation assay. This suggested that SpvDA154/R161 might be a DUB or ubiquitin-like protease. However, we were unable to obtain evidence for this in subsequent AMC deconjugation and in vitro deubiquitination assays. When compared with the Typhimurium SpvDA154/R161 variant, SpvD with a single polymorphism (Gly-161) was more potent in (i) the DUB-GloTM protease assay, (ii) inhibition of the NF-κB luciferase assay in PMA-stimulated cells, and (iii) virulence in mice. The observation that SpvDA154/G161 is more potent at inhibiting NF-κB signaling than SpvDA154/R161 in PMA- but not TNFα-stimulated HEK293 cells might help guide the identification of its substrate. PMA mimics diacylglycerol in stimulating NF-κB through activation of protein kinase C (26). Therefore, our results suggest that the more active SpvD variant targets a diacylglycerol-stimulated branch of NF-κB pathway. There are very few descriptions of single amino acid polymorphisms influencing the potency of bacterial effectors. A naturally occurring polymorphism within S. typhimurium SseI at position 103 was shown to affect binding to the cell migration adaptor protein TRIP6 in a yeast two-hybrid assay and bacterial dissemination in mice (27). However, the position of the SseI polymorphism in relation to the catalytic site is unknown as only partial SseI (amino acids 145–313) structure is currently available (15). The SpvD polymorphism that we have investigated here is in very close vicinity of the catalytic triad. Depending on the substrate used in vitro, the Arg-161 or Gly-161 variant displayed greater potency. However, the Gly-161 variant conferred stronger inhibition of a PMA-stimulated NF-κB reporter and greater virulence of Salmonella in mice. Regardless of the identity of the physiological substrate for hydrolytic activity of SpvD, it is remarkable that a single amino acid substitution that alters hydrolytic potency can influence the complex process of virulence in a host. The fact that the SpvDR161 variant is present in all Salmonella serovars except Enteritidis, Bovismorbificans, and Abortus-equi is intriguing. It is possible that SpvDR161 has gained a different substrate specificity that is not related to NF-κB signaling. Collectively, our results indicate SpvD is a papain-like hydrolase that functions through a cysteine-mediated hydrolysis reaction to inhibit gene expression from NF-κB promoters. Furthermore, we demonstrate that a natural SpvD polymorphism significantly affects virulence and NF-κB inhibition in PMA-stimulated cells, leading us to hypothesize that the substrate hydrolyzed by SpvDG161 might be part of a signaling pathway involving protein kinase C.
Experimental Procedures
Bacterial Strains and Plasmids
Bacterial strains and plasmids used in this study are listed in supplemental Table S1. The Salmonella strains were grown in Luria Bertani (LB) medium at 37 °C with shaking and supplemented with ampicillin (100 μg/ml) or chloramphenicol (34 μg/ml) as appropriate. Complementation plasmids (pACYC184 or pWSK29) containing non-tagged or double HA-tagged spvD were obtained using restriction enzyme cloning. Site-directed mutagenesis of SpvD was performed by inverse PCR using pRK5-myc-SpvDA154/R161 vector as DNA template.
Protein Production
For crystallography, SpvDC73S/C122S/C160S/C170S and SpvDC73S/C122S/C160S/C170S/C73A/G161 were expressed with a precision protease-cleavable N-terminal His6 tag from pHiSH, a pET15b-derived vector (28) in PC2 cells (29). Cultures were grown in Terrific Broth or LB to an A600 of 0.8 at 37 °C and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 18 °C. Cells were lysed by sonication in 50 mm Tris/Cl, pH 7.5, 500 mm NaCl, 20 mm imidazole, 0.5 mm phenylmethylsulfonyl fluoride. After clarification by centrifugation, the lysate was mixed with nickel-nitrilotriacetic acid resin for 1 h at 4 °C. The resin was extensively washed with 50 mm Tris/Cl, pH 7.5, 500 mm NaCl, and 20 mm imidazole, and SpvD was eluted with 50 mm Tris/Cl, pH 7.5, 500 mm NaCl, and 500 mm imidazole. The His tag was removed by the addition of 10 mm dithiothreitol and HRV 3C protease at a ratio of 1 mg protease to 30 mg SpvD and overnight incubation at 4 °C. The SpvD solution was loaded onto a Superdex75 gel filtration column (GE Healthcare) pre-equilibrated in 50 mm Tris/Cl, pH7.5, 150 mm NaCl, and 10 mm β-mercaptoethanol. For biochemical analyses, GST-SseL, SpvC-His6, and SpvDA154/R161-His6 proteins were purified from previously generated pGEX and pET22b vectors (9, 11, 22). Variants of SpvD were made using site-directed mutagenesis, and their protein-coding DNA sequences were ligated into pET22b using identical restriction sites. Protein was expressed in Escherichia coli BL21(DE3) cells that were grown to an A600 of 0.8–1.0 at 37 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 16–20 h at 20 °C. Depending on the protein fusion tag, proteins were purified to apparent homogeneity using either nickel- or glutathione-based affinity chromatography as described previously (9, 22).
DUB-GloTM and Ub-AML Assays
Purified SpvC, SseL, and SpvD proteins were each used in the DUB-GloTM assay at 1, 5, and 25 μm concentrations. The DUB-GloTM assay was performed using the manufacturer's instructions (Promega). Briefly, 50 μl of 2, 10, and 50 μm of tested proteins diluted in reaction buffer (50 mm HEPES, pH 7.4, 50 mm NaCl, 0.5 mm EDTA, 10 mm DTT) was mixed with 50 μl of RLRGG-AML substrate containing luciferase reporter enzyme. Luciferase activity was measured at 22 °C every 15 min up to 5 h with an Infinite M200Pro plate reader (Tecan). In a modification of the assay, 40 μm RLRGG-AML was replaced with 1 μm Ub-AML, whereas the remaining buffer composition and luciferase enzyme remained unchanged.
AMC Deconjugation Assay
SseL, SpvC, SpvDA154/R161, and relevant control proteins were incubated with 1 μm concentrations of ubiquitin-, SUMO1-, SUMO2-, ISG15-, and NEDD8-AMC (Boston Biochem) in 50 mm HEPES, pH 8.0, and 1 mm DTT. Liberation of AMC at 22 °C was measured after 1 h of incubation using Infinite M200Pro plate reader (Tecan; excitation/emission wavelengths 380/460 nm).
In Vitro Deubiquitination Assay
A panel of M1 linear Lys-6-, Lys-11-, Lys-27-, Lys-29-, Lys-33-, Lys-48-, and Lys-63-linked diubiquitin chains was acquired from UbiQ. The reaction mixture contained 50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 5 mm DTT, and 5 μm diubiquitin chains. Diubiquitin hydrolysis reactions were performed at 37 °C by adding 5 μm concentrations of either SseL (positive control) or SpvD. The assay was started by mixing the tested protein and diubiquitin solutions, and 10-μl aliquots were taken at the indicated time points. The reaction was resolved on 4–12% SDS-PAGE gradient gels run in MES buffer (Invitrogen) and stained with Coomassie Brilliant Blue R-250 (Bio-Rad).
Antibodies
Immunofluorescence analysis was done using goat anti-Salmonella (CSA-1, Kirkegaard and Perry Laboratories) and mouse anti-HA (HA11, Covance) antibodies. Immunoblotting of tagged proteins was performed using 9e10 (Roche Applied Science) and GFP (Life Technologies) antibodies. Tubulin was probed with E7 antibody (Developmental Studies Hybridoma Bank).
Cell Culture
HEK293 (human embryonic kidney) cells used in this study were obtained from the European Collection of Animal and Cell Cultures (Salisbury, UK) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal calf serum (FCS) (PAA Laboratories or Sigma) at 37 °C in 5% CO2. Bone marrow-derived macrophages (BMDM) were infected with the v-myc/v-raf expressing J2 retrovirus (30) and differentiated in 20% LCM (L929 cell (ATCC) conditioned medium). Cells were then maintained in DMEM (Sigma), 10% FCS, 20% LCM, and 1 mm sodium pyruvate at 37 °C, 5% CO2.
Crystallization and Structure Determination
SpvD4CS and SpvD4CS/C73A/G161 proteins at 9 mg/ml in 50 mm Tris/Cl, pH 7.5, 50 mm NaCl, and 10 mm β-mercaptoethanol were crystallized by vapor diffusion in hanging drops against a reservoir of 100 mm Tris/Cl, pH 7.5, 200 mm NaCl, 22% PEG3350 at 4 °C. Two different crystal morphologies grew in the drops, and seeding was used to propagate the three-dimensional crystal form, which diffracted consistently to high resolution. SpvD4CS crystals were transferred to a cryoprotectant solution of 100 mm Tris/Cl, pH 7.5, 200 mm NaCl, 22% PEG3350, and 20% glycerol and flash-frozen in liquid nitrogen, and diffraction data were collected to 1.48 Å on beamline I03 at Diamond Light Source (Oxfordshire, UK) (Table 1). Anomalous data were collected on I04-1 at Diamond Light Source from a crystal soaked for 1 h in 100 mm Tris/Cl, pH 7.5, 200 mm NaCl, 22% PEG3350, 20% glycerol, 1 mm KAuCl4. SpvD4CS/C73A/G161 crystals were grown as above and transferred stepwise with increasing concentrations of dimethyl sulfoxide (DMSO) and Z-RLRGG-aminoluciferin (Promega) to a final solution of 100 mm Tris/Cl, pH 7.5, 200 mm NaCl, 22% PEG3350, 25% DMSO, 1 mm Z-RLRGG-aminoluciferin before flash-freezing and collecting data at Diamond Light Source beamline I02. Native data were integrated in Mosflm (31) and merged in Scala (32) of the CCP4 suite (33), and anomalous data were processed with the xia2 pipeline using XDS and XSCALE (34–37). The structure was solved by single isomorphous replacement with anomalous scattering in Phenix using the AutoSol wizard (38, 39) with one site and an initial figure of merit of 0.35. The initial structure was built by Phenix AutoBuild wizard (40) and refined using Refmac and Phenix (38, 41) iterated with manual model building in Coot (42). Structures were refined to final resolutions of 1.48 Å (Arg-161) and 1.60 Å (Gly-161) with good geometry (Table 1). The coordinates and structure factors of the structures of SpvD4CS/R161 and SpvD4CS/C73A/G161 have been deposited in the protein data bank with PDB codes 5LQ6 and 5LQ7, respectively.
DNA Transfection
HEK293 cells were seeded in 24-well plates at a concentration of 2 × 104 cells/well 24 h before transfection with Lipofectamine 2000 (Life Technologies) following the manufacturer's protocol. Cells were used 24 h after transfection.
Fixation, Permeabilization, Fluorescence Labeling, and Microscopy
All samples were fixed in 3% paraformaldehyde. Permeabilization was done in 0.2% Triton X-100 together with incubation with primary antibodies. All antibodies were diluted to the appropriate concentrations in PBS containing 10% horse serum and 0.2% Triton-X100. The coverslips were washed once in PBS, incubated with primary antibodies for 2 h, washed 3 times in PBS, and incubated with secondary antibodies and nucleic acid dye DAPI (Thermo Fisher) for 1 h. Coverslips were washed and mounted onto glass slides using Mowiol mounting medium. Cells were analyzed using a confocal laser-scanning microscope (LSM710; Zeiss GmbH).
Luciferase Reporter Assay
HEK293 cells were seeded at a density of 5 × 104 cells per well in a 24-well plate 24 h before transfection. Cells were transfected for 24 h with 50 ng of luciferase reporter plasmid (NF-κB dependent luciferase reporter plasmid), 30 ng of pTK-Renilla luciferase, and 500 ng of expression vectors (SpvD, SseK3, IκBαS32A/S36A or pRK5 vector alone). Cells were then incubated either with 10 ng/ml TNFα or 100 nm PMA for 16 h and harvested in 100 μl of passive lysis buffer (Promega). Luciferase activity was measured using the Dual Luciferase reporter assay system (Promega) and Infinite M200Pro plate reader (Tecan) and normalized according to Renilla luciferase intensity. The data presented are from at least four independent experiments.
Mice Infections
To prepare the inocula, bacteria were first grown overnight in LB broth and then subcultured at a dilution of 1:100 for a further 2 h. Cultures were diluted to a concentration of 5 × 103 cfu/ml in physiological saline. For CI measurements, bacterial cultures were mixed for intraperitoneal inoculation (0.2 ml per mouse). Viable bacteria in inocula were quantified by dilution and plating onto LB agar plates with appropriate antibiotics to distinguish between strains. Female C57BL/6 mice (Charles River, 6–8 weeks) were sacrificed at 3 days post inoculation. The spleens were removed aseptically and homogenized in distilled water by mechanical disruption. Serial dilutions were plated on LB agar for cfu enumeration. Strains were distinguished by differential counting or replica plating on antibiotic-supplemented plates. For each mouse, the CI was calculated by dividing the output ratio (i.e. strain a versus strain b) divided by the input ratio. The log CI values were used to calculate means and for statistical analyses.
Ethics Statement
Animals were used in accordance with UK Home Office regulations. The Imperial College Animal Welfare and Ethical Review Body (AWERB) committee approved the project license for animal research (70/7768). The following people formed the panel: Applicant Scientist, CBS site manager/NACWO, NVS, peer scientist, and a lay person.
Statistical Analysis
All results are reported as the mean ± S.E. Statistical analyses were done using one-tailed paired Student's t test or ANOVA followed by Bonferroni's multiple comparison test. Differences denoted in the text as significant represent p values lower than 0.05.
Author Contributions
G. J. G., S. A. H., and D. W. H. designed the project. N. R. and Y. Y. designed and purified SpvD4CS. Y. Z., M. P., and S. A. H. performed crystal structures determination. J. N. P. and D. K. provided the amino acid sequence and structural insights. G. J. G. performed SpvD activity assays, immunofluorescence, and mouse infections. G. J. G., S. A. H., and D. W. H. wrote the paper with contributions from the other authors.
Supplementary Material
Acknowledgments
We thank colleagues from Centre for Molecular Bacteriology and Infection for critical reading of the manuscript and Steve Matthews for helpful discussions throughout the project. We are grateful to Izabela Glegola-Madejska for technical support with mouse infections. We thank Felix Randow and Regina Günster for the luciferase reporter, SseK3, and IκBαS32A/S36A plasmids. We acknowledge the help of staff from Diamond Light Source beamlines I03 and I02 with data collection.
This work was supported by Medical Research Council Grants MR/K027077/1 (to D. H.) and G1100332 (to S. H.), Wellcome Trust Grant 095484/Z/11/Z (to D. H.), and Wellcome Trust Fellowship 093617/Z/10/Z (to G. G.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (codes 5LQ6 and 5LQ7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T3SS
- type III secretion system
- SPI-1
- Salmonella pathogenicity island 1
- spv
- plasmid virulence locus
- DUB
- deubiquitinase
- 4CS
- SpvDC37S/C122S/C160S/C170S
- AML
- aminoluciferin
- AMC
- 7-amino-4-methylcoumarin
- PMA
- 12-myristate 13-acetate
- CI
- competitive index
- (i)BMDM
- (immortalized) bone marrow-derived macrophage
- ANOVA
- analysis of variance
- Ub
- ubiquitin.
References
- 1. Figueira R., and Holden D. W. (2012) Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158, 1147–1161 [DOI] [PubMed] [Google Scholar]
- 2. Beninger P. R., Chikami G., Tanabe K., Roudier C., Fierer J., and Guiney D. G. (1988) Physical and genetic mapping of the Salmonella dublin virulence plasmid pSDL2: relationship to plasmids from other Salmonella strains. J. Clin. Invest. 81, 1341–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gulig P. A., and Curtiss R. 3rd (1988) Cloning and transposon insertion mutagenesis of virulence genes of the 100-kilobase plasmid of Salmonella typhimurium. Infect. Immun. 56, 3262–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones G. W., Rabert D. K., Svinarich D. M., and Whitfield H. J. (1982) Association of adhesive, invasive, and virulent phenotypes of Salmonella typhimurium with autonomous 60-megadalton plasmids. Infect. Immun. 38, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Libby S. J., Adams L. G., Ficht T. A., Allen C., Whitford H. A., Buchmeier N. A., Bossie S., and Guiney D. G. (1997) The spv genes on the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect. Immun. 65, 1786–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krause M., Fang F. C., and Guiney D. G. (1992) Regulation of plasmid virulence gene expression in Salmonella dublin involves an unusual operon structure. J. Bacteriol. 174, 4482–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lesnick M. L., Reiner N. E., Fierer J., and Guiney D. G. (2001) The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39, 1464–1470 [DOI] [PubMed] [Google Scholar]
- 8. Zhu Y., Li H., Long C., Hu L., Xu H., Liu L., Chen S., Wang D.-C., and Shao F. (2007) Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol. Cell. 28, 899–913 [DOI] [PubMed] [Google Scholar]
- 9. Mazurkiewicz P., Thomas J., Thompson J. A., Liu M., Arbibe L., Sansonetti P., and Holden D. W. (2008) SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol. Microbiol. 67, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niemann G. S., Brown R. N., Gustin J. K., Stufkens A., Shaikh-Kidwai A. S., Li J., McDermott J. E., Brewer H. M., Schepmoes A., Smith R. D., Adkins J. N., and Heffron F. (2011) Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rolhion N., Furniss R. C., Grabe G., Ryan A., Liu M., Matthews S. A., and Holden D. W. (2016) Inhibition of nuclear transport of NF-κB p65 by the Salmonella type III secretion system effector SpvD. PLOS Pathog. 12, e1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holm L., and Sander C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 13. Sanada T., Kim M., Mimuro H., Suzuki M., Ogawa M., Oyama A., Ashida H., Kobayashi T., Koyama T., Nagai S., Shibata Y., Gohda J., Inoue J., Mizushima T., and Sasakawa C. (2012) The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 [DOI] [PubMed] [Google Scholar]
- 14. Nishide A., Kim M., Takagi K., Himeno A., Sanada T., Sasakawa C., and Mizushima T. (2013) Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI. J. Mol. Biol. 425, 2623–2631 [DOI] [PubMed] [Google Scholar]
- 15. Bhaskaran S. S., and Stebbins C. E. (2012) Structure of the catalytic domain of the Salmonella virulence factor SseI. Acta Crystallogr. D. Biol. Crystallogr. 68, 1613–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu M., Shao F., Innes R. W., Dixon J. E., and Xu Z. (2004) The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate binding site. Proc. Natl. Acad. Sci. U.S.A. 101, 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Filipek R., Potempa J., and Bochtler M. (2005) A comparison of staphostatin B with standard mechanism serine protease inhibitors. J. Biol. Chem. 280, 14669–14674 [DOI] [PubMed] [Google Scholar]
- 18. Pickersgill R. W., Harris G. W., and Garman E. (1992) Structure of monoclinic papain at 1.60 Å resolution. Acta Crystallogr. Sect. B Struct. Sci. 10.1107/S0108768191006572 [DOI] [PubMed] [Google Scholar]
- 19. Misaghi S., Galardy P. J., Meester W. J., Ovaa H., Ploegh H. L., and Gaudet R. (2005) Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 280, 1512–1520 [DOI] [PubMed] [Google Scholar]
- 20. Mirković B., Renko M., Turk S., Sosič I., Jevnikar Z., Obermajer N., Turk D., Gobec S., and Kos J. (2011) Novel mechanism of cathepsin B inhibition by antibiotic nitroxoline and related compounds. ChemMedChem. 6, 1351–1356 [DOI] [PubMed] [Google Scholar]
- 21. Bauerfeind R., Barth S., Weiss R., and Baljer G. (2001) Sequence polymorphism of the Salmonella plasmid virulence factor D (SpvD) in Salmonella enterica isolates of animal origin. Berl. Munch. Tierarztl. Wochenschr. 114, 404–408 [PubMed] [Google Scholar]
- 22. Rytkönen A., Poh J., Garmendia J., Boyle C., Thompson A., Liu M., Freemont P., Hinton J. C., and Holden D. W. (2007) SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 3502–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pruneda J. N., Durkin C. H., Geurink P. P., Ovaa H., Santhanam B., Holden D. W., and Komander D. (2016) The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell. 63, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z., Soderholm A., Lung T. W., Giogha C., Hill M. M., Brown N. F., Hartland E., and Teasdale R. D. (2015) SseK3 Is a Salmonella Effector That Binds TRIM32 and Modulates the Host's NF-κB Signalling Activity. PLoS ONE. 10, e0138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., and Maniatis T. (1995) Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 26. Holden N. S., Squires P. E., Kaur M., Bland R., Jones C. E., and Newton R. (2008) Phorbol ester-stimulated NF-κB-dependent transcription: roles for isoforms of novel protein kinase C. Cell. Signal. 20, 1338–1348 [DOI] [PubMed] [Google Scholar]
- 27. Thornbrough J. M., and Worley M. J. (2012) A naturally occurring single nucleotide polymorphism in the Salmonella SPI-2 type III effector srfH/sseI controls early extraintestinal dissemination. PLoS ONE 7, e45245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong C. T., Xu Y., Gupta A., Garnett J. A., Matthews S. J., and Hare S. A. (2015) Structural analysis of haemoglobin binding by HpuA from the Neisseriaceae family. Nat. Commun. 6, 10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cherepanov P. (2007) LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blasi E., Mathieson B. J., Varesio L., Cleveland J. L., Borchert P. A., and Rapp U. R. (1985) Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318, 667–670 [DOI] [PubMed] [Google Scholar]
- 31. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winter G., Lobley C. M., and Prince S. M. (2013) Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69, 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sauter N. K., Grosse-Kunstleve R. W., and Adams P. D. (2004) Robust indexing for automatic data collection. J. Appl. Crystallogr. 37, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Z., Sauter N. K., van den Bedem H., Snell G., and Deacon A. M. (2006) Automated diffraction image analysis and spot searching for high-throughput crystal screening. J. Appl. Crystallogr. 39, 112–119 [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D. Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.