Abstract
The Fas-activated serine/threonine kinase (FASTK) family of proteins has recently emerged as a central regulator of mitochondrial gene expression through the function of an unusual RNA-binding domain named RAP (for RNA-binding domain abundant in Apicomplexans), shared by all six members of the family. Here we describe the role of one of the less characterized members, FASTKD3, in mitochondrial RNA metabolism. First, we show that, in contrast to FASTK, FASTKD2, and FASTKD5, FASTKD3 does not localize in mitochondrial RNA granules, which are sites of processing and maturation of mtRNAs and ribosome biogenesis. Second, we generated FASTKD3 homozygous knock-out cell lines by homologous recombination and observed that the absence of FASTKD3 resulted in increased steady-state levels and half-lives of a subset of mature mitochondrial mRNAs: ND2, ND3, CYTB, COX2, and ATP8/6. No aberrant processing of RNA precursors was observed. Rescue experiments demonstrated that RAP domain is required for FASTKD3 function in mRNA stability. Besides, we describe that FASTKD3 is required for efficient COX1 mRNA translation without altering mRNA levels, which results in a decrease in the steady-state levels of COX1 protein. This finding is associated with reduced mitochondrial complex IV assembly and activity. Our observations suggest that the function of this family of proteins goes beyond RNA processing and ribosome assembly and includes RNA stability and translation regulation within mitochondria.
Keywords: gene expression, mitochondria, post-transcriptional regulation, RNA, translation control, FASTKD3, mitochondria, RNA metabolism, translation
Introduction
Mitochondria are thought to be descendants of endosymbiotic bacteria. During its evolution into the current “powerhouse” organelles of the eukaryotic cell, the endosymbiont transferred many of its essential genes to the nuclear chromosomes. In humans, mtDNA is a circular 16.6 kb that encodes 2 rRNAs, 22 tRNAs, and 13 protein-coding genes. The genome is transcribed into two long polycistronic heavy-strand and light-strand transcripts reminiscent of bacterial operons. The two rRNAs and most of the mRNAs are flanked by tRNAs. The polycistronic nature of the transcripts and the flanking tRNAs are the basis of the “tRNA punctuation” model. In this model, RNase P and Z recognize the secondary structure of the tRNA precursors and cleave the RNA, leading to the release of the tRNAs and the mRNAs from the precursor RNA (1). The non-canonical mRNAs do not have tRNAs flanking both ends. For these mRNAs, which include ATP8/6, Cox3, Cox1, CYTB, ND5, and ND6, the tRNA punctuation model is insufficient. Little is known about the mechanisms by which the 5′ and 3′ ends of these mRNAs are generated. A member of the pentatricopeptide repeat (PPR)5 protein family, PTCD2, has been reported to be involved in the processing of the pre-processed ND5-CYTB transcript (2).
The identification and characterization of a novel family of mitochondrial proteins named FASTK (Fas-activated serine/threonine kinase) have shed new light on the mechanisms controlling mitochondrial post-transcriptional RNA processing and translation. This family is composed of six members: FASTK, the founding member, and its homologs FASTKD1–5 (3). All members have been found only in vertebrates (3) and were identified as RNA-binding proteins on the basis of mRNA-bound proteome studies (4–6) and share three domains called FAST_1, FAST_2, and RAP (3). According to homology predictions, the RAP (an acronym for RNA-binding domain abundant in Apicomplexans) domain is a putative RNA-binding domain (7), whereas the functions of FAST_1 and FAST_2 domains remain unknown.
FASTK and FASTKD2 were recently found to accumulate into distinct foci that colocalize with newly synthesized mitochondrial RNA in mitochondrial RNA granules (MRGs) first observed over a decade ago (8), and whose function has been revealed recently (9). Different proteins have been found to localize in MRGs, such as GRSF1, RNase P, and DDX28, among other proteins (9–12). The biological properties of the proteins present in MRGs suggest that these foci are sites of processing and maturation of newly synthesized mtRNAs and ribosome assembly. Interestingly, recent studies have shown that both FASTK and FASTKD2 are required for the expression of the ND6 mRNA, which is the only mRNA encoded on the light strand, and has no tRNA at the 3′ end (13, 14). Besides that, the absence of FASTKD2 leads to aberrant processing and expression of 16S rRNA, which results in impaired mitochondrial translation and oxidative phosphorylation assembly defects (14, 15). It is important to note that Ghezzi et al. (16) found homozygosity for a nonsense mutation in FASTKD2 in two siblings with familial infantile mitochondrial encephalopathy, further underlying the importance of FASTKD2 in mitochondrial function. A recent study has shown that endogenous FASTKD5 partially colocalizes with MRGs (15), although our previous observations with a tagged version of FASTKD5 do not support this finding (3, 13). Despite conflicting data regarding the location of FASTKD5 in MRGs, it has been unequivocally demonstrated that FASTKD5 is essential for processing the three noncanonical transcripts encoded on the heavy chain (15). As a result of this property, FASTKD5 depletion renders COX1 mRNA almost undetectable, which severely reduces the synthesis of COX1 protein, resulting in a complex IV defect (15).
FASTKD1, FASTKD3, and FASTKD4 (TBRG4) do not localize in MRGs. FASTKD4 was found to modulate the half-lives of a subset of mitochondrial mRNAs and to associate with mtRNAs in vivo (17). Until now, little is known about the two members FASTKD1 and FASTKD3. We have previously reported that FASTKD3 is required for mitochondrial respiration and interacts with components of the RNA metabolism and translation machineries (3). In this study, we explore the role of FASTKD3 in mitochondrial RNA metabolism and translation.
Results
Generation of FASTKD3-deficient Cell Lines
All six FASTK family members have been annotated as RNA-binding proteins (RBPs) in independent mRNA-bound proteome studies (4–6). More recently, the three family members, namely FASTK, FASTKD2, and FASTKD5, have been reported to localize to mitochondrial RNA granules, which are considered centers for post-transcriptional RNA processing and ribosome biogenesis (13, 15). However, no colocalization with BrU-labeled RNA granules was observed for FASTKD1, FASTKD3, or FASTKD4 (13). Here we confirm that FASTKD3 does not concentrate in endogenous mitochondrial RNA granules stained with the anti-FASTKD2 antibody (supplemental Fig. 1), suggesting that its putative role in RNA metabolism may extend beyond these new recently described foci.
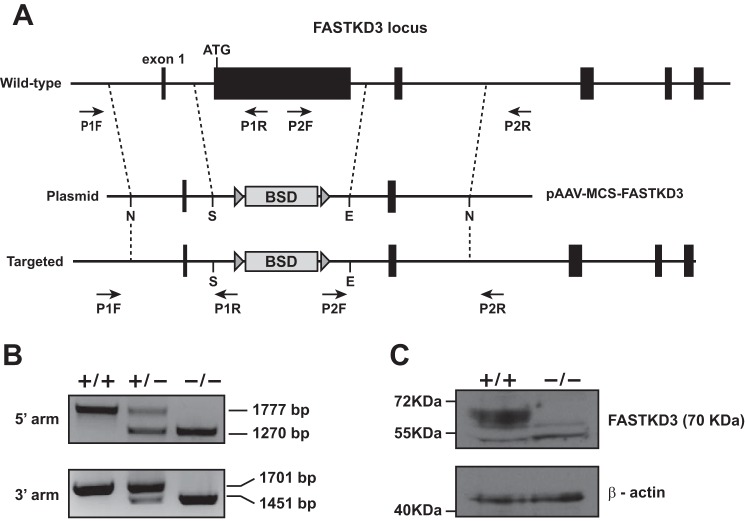
We generated FASTKD3 knock-out U2OS cells to investigate the importance of FASTKD3 in mitochondrial RNA metabolism. The genomic FASTKD3 locus contains seven exons. Exon 2 contains the first ATG and represents the majority of the coding region (78%). We constructed a targeting vector designed to remove the entire exon 2, replacing it with loxP-flanked blasticidin resistance cassette (Fig. 1A). We also engineered CRISPR/Cas9n against 20 nucleotides located at the 5′ end of intron 2 to facilitate homologous recombination. Puromycin-resistant colonies were checked for recombination by PCR (Fig. 1B). The single targeted cells obtained were treated with recombinant cell-permeant TAT-NLS-Cre recombinase to delete loxP-flanked blasticidin resistance cassette. Resultant cells were then subjected to a second round of gene targeting, and biallelic knock-out cells were identified by PCR (Fig. 1B). The absence of FASTKD3 in knock-out cells was confirmed by Western blotting (Fig. 1C).
FIGURE 1.
Generation of FASTKD3−/− cells. A, schematic illustration of FASTKD3 gene targeting with pAAV-MCS-FASTKD3 plasmid. N, NotI; S, SalI; E, EcoRI. Exons are indicated by black boxes. LoxP sites are depicted with triangles, with the orientation indicated by the direction of the triangle. BSD, blasticidin-S-deaminase resistance marker. B, representative PCR analysis for site-specific integration. Positions of the primers used for screening are designated by arrows in A, and expected size differences for PCR products are indicated. C, Western blotting analysis of whole-cell extracts from wild type and FASTKD3−/− with the indicated antibodies.
The Absence of FASTKD3 Leads to an Increase of the Steady-state Levels and Half-lives of a Subset of mtRNAs
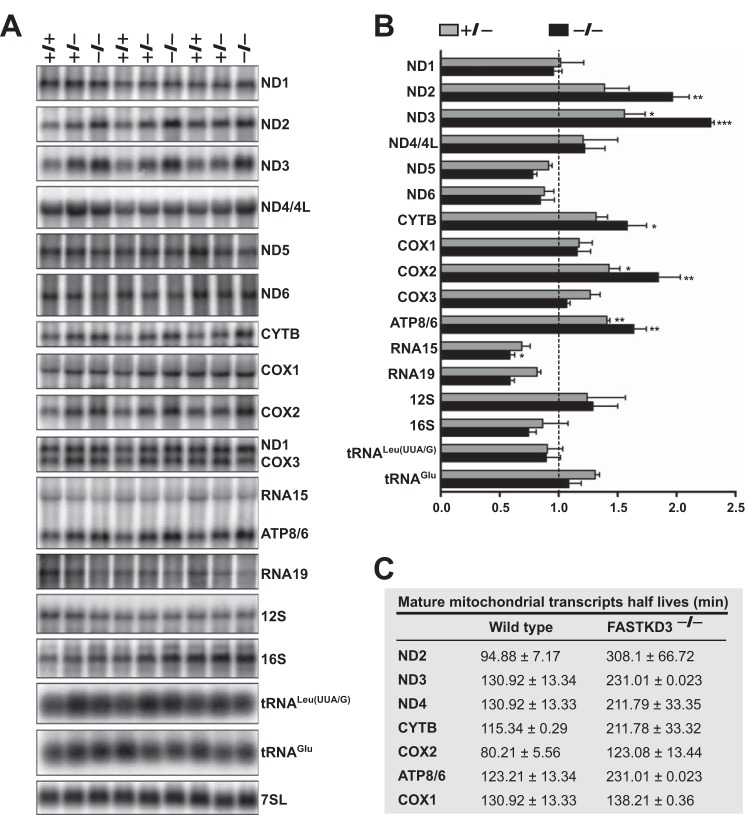
To explore the role of FASTKD3 in mitochondrial RNA metabolism, we first performed Northern blotting analysis to compare the expression of mitochondrial RNAs in wild type and FASTKD3−/− cells. FASTKD3−/− cells showed increased steady-state levels of mature mRNAs for ND2 (1.97 ± 0.14-fold increase), ND3 (2.29 ± 0.03-fold increase), CYTB (1.58 ± 0.16-fold increase), COX2 (1.85 ± 0.19-fold increase), and ATP8/6 (1.64 ± 0.11-fold increase) as compared with wild type cells (Fig. 2, A and B). The steady-state levels of all other mtDNA-encoded RNAs, including the other seven ORFs and the ribosomal RNAs 12S and 16S, were similar between wild type and FASTKD3−/− cells. As expected, FASTKD3+/− cells showed an intermediate phenotype. The increase in the steady-state levels of ND2, ND3, CYTB, COX2, and ATP8/6 transcripts in FASTKD3−/− cells was confirmed by qRT-PCR, and the results are shown in supplemental Table 1. No alterations in the precursor mRNA levels were observed in FASTKD3−/− cells, except for a 40% decrease in RNA15 precursor (encompassing ATPase8/6-COX3 mRNA) and a nonsignificant decrease RNA19 precursor (encompassing 16S rRNA-tRNALeu(UUA/G)-ND1 mRNA).
FIGURE 2.
Mitochondrial RNA analysis in FASTKD3−/− cells. A, RNA isolated from the indicated cell lines was analyzed by Northern blotting hybridization with probes specific for the mitochondrial mRNAs and rRNAs, and as a loading control, with probes for 7SL (signal recognition particle RNA). A representative experiment done in triplicates is shown. B, intensities of radioactive bands on Northern blots shown in A were quantified by densitometric analysis using ImageJ software. Data were normalized to 7SL RNA levels and presented relative to the wild type control (+/+, set as 1). Values represent the means ± S.E. (n = 3). *, p < 0.05, **, p < 0.01, ***, p < 0.001. C, analysis of mtRNA half-lives and mtDNA content in FASTKD3−/− cells. Total RNA was isolated at different times up to 6 h after treatment with ethidium bromide. Steady-state levels of mitochondrial mRNAs were quantified by qRT-PCR as described under “Experimental Procedures.” The half-life of each mitochondrial mRNA was calculated using the formula t½ = ln2/λ, where λ is the slope of mRNA decay. Data are expressed as means ± S.D. (n = 5).
Certain physiological circumstances, such as high energy needs, stimulate mitochondrial biogenesis, which leads to an increase in mitochondrial transcripts associated with increased mtDNA replication and transcription (18). This mechanism seemed unlikely to contribute to the mRNA phenotype found in FASTKD3−/− cells because that would lead to a global increase of steady-state levels of mtRNAs. Moreover, we have previously reported that siRNA-mediated FASTKD3 silencing does not affect mtDNA content (3). Thus, our most plausible hypothesis was that the increase in the steady-state levels of ND2, ND3, CYTB, COX2, and ATP8/6 transcripts in FASTKD3−/− cells was due to an increase in their half-lives. To measure the half-lives of the mitochondrial mRNAs, we blocked mitochondrial transcription with ethidium bromide. Cells were harvested at different times after the addition of the inhibitor, and mitochondrial mRNA steady-state levels were measured by qRT-PCR at each time point. The half-life of each mRNA was calculated as described previously (19). As shown in Fig. 2C, ND2, ND3, CYTB, COX2, and ATP8/6 transcripts in FASTKD3−/− cells showed longer half-lives as compared with those in control cells. As expected, the COX1 mRNA half-life was unaltered in the absence of FASTKD3.
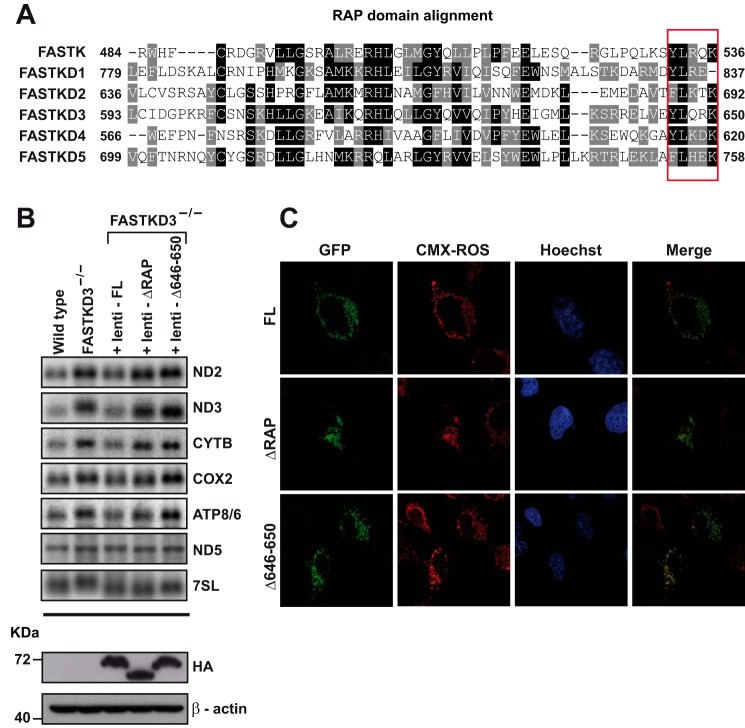
We next performed rescue experiments by stably expressing full-length FASTKD3-FLAG-HA, a FASTKD3 mutant that lacks the RAP domain (FASTKD3ΔRAP-FLAG-HA), or a FASTKD3 mutant that lacks the last five amino acid residues of the RAP domain (FASTKD3Δ646–650) in FASTKD3−/− cells. FASTKD3Δ646–650 mutant was generated on the basis of a previous report showing that point mutations Y616A, L617A, K618A, and K620A (NP_004740) at the C terminus end of FASTKD4 RAP domain lead to loss of function of FASTKD4 (17). In Fig. 3A, we show the sequence alignment of the RAP domain of all FASTK members and highlight the five partially conserved amino acids at the C terminus.
FIGURE 3.
Analysis of the domains required for the decay acceleration activity of FASTKD3. A, RAP domain protein sequence homology analysis among the different FASTK members was performed using ClustalW program. White letters on a black background highlight identical amino acids. White letters on a gray background highlight different but conserved amino acids. The red box indicates the five partially conserved amino acids at the C terminus end of the RAP domain of FASTK family members. Amino acid positions of the RAP domain boundaries are indicated. NCBI reference sequence identifiers for the aligned sequences are: FASTK (NP_006703), FASTKD1 (NP_001308975), FASTKD2 (NP_001129665), FASTKD3 (NP_076996), FASTKD4 (NP_004740), and FASTKD5 (NP_068598). B, RNA isolated from wild type, FASTKD3−/− cells, and FASTKD3−/− cells reconstituted with full-length (FL) FASTKD3-FLAG-HA, FASTKD3ΔRAP-FLAG-HA, or FASTKD3Δ646–650-FLAG-HA (by lentiviral vector-mediated transduction) was analyzed by Northern blotting with the indicated probes. The lower two panels show Western blotting analysis of whole-cell lysates using antibodies against HA or β-actin (loading control). C, confocal microscopy analysis of U2OS cells transiently overexpressing full-length FASTKD3-GFP, FASTKD3ΔRAP-GFP, or FASTKD3Δ646–650-GFP. Mitochondria were stained with MitoTracker (CMX-ROS), and Hoechst 33258 was used for nuclei staining.
As expected, overexpression of full-length FASTKD3-FLAG-HA was able to rescue the FASTKD3−/− phenotype (Fig. 3B), reaching steady-state levels of ND2 and ND3 transcripts similar to those seen in control wild type cells. Similarly, the levels of the transcripts CYTB, COX2, and ATP8/6 were rescued to levels comparable with those in control cells. As expected from our previous results, the levels of expression of ND5 mRNA were similar in all the cell lines. In agreement with previous work suggesting that the RAP domain is required for function of FASTK proteins (13, 17), expression of FASTKD3ΔRAP-FLAG-HA did not rescue the FASTKD3−/−phenotype. Finally, expression of FASTKD3Δ646–650-FLAG-HA was also unable to rescue the mRNA levels for the altered transcripts in FASTKD3−/− cells. The three FLAG-HA recombinant proteins were expressed at similar levels as determined by Western blotting (Fig. 3B). We also transiently expressed GFP-tagged full-length FASTKD3, FASTKD3ΔRAP, or FASTKD3Δ646–650 in U2OS cells and confirmed by confocal microscopy that they are all located to mitochondria and were expressed at similar levels (Fig. 3C). These results ruled out the possibility that the inability of mutants FASTKD3ΔRAP or FASTKD3Δ646–650 to rescue the FASTKD3−/− phenotype was simply due to altered expression or location within the cell.
The Absence of FASTKD3 Leads to a Selective Decrease in COX1 Translation
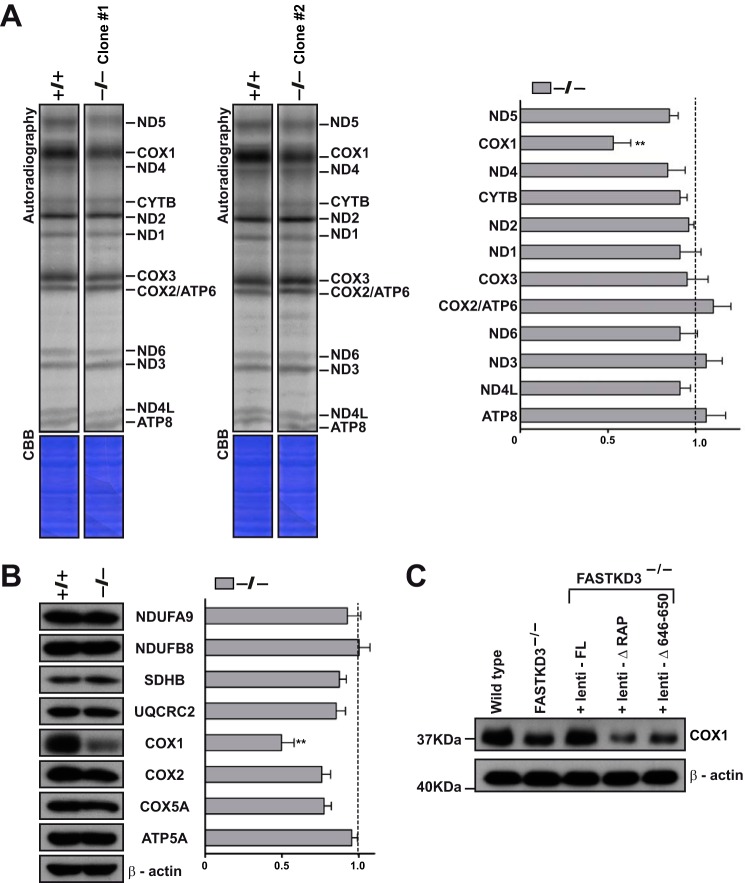
We next explored the role of FASTKD3 in mitochondrial translation. De novo protein synthesis was measured by metabolic labeling in the presence of emetine, which inhibits cytosolic translation. As shown in Fig. 4A, increase in the steady-state levels of ND2, ND3, CYTB, COX2, and ATP8/6 transcripts did not lead to over-synthesis of the corresponding proteins. Surprisingly, the absence of FASTKD3 caused a selective >40% reduction in the synthesis of COX1 (Fig. 4A). No significant alterations were detected in the de novo synthesis of other mitochondrial proteins in the FASTKD3−/− cells. We show representative experiments performed in two independent FASTKD3−/− clones.
FIGURE 4.
Mitochondrial protein synthesis in FASTKD3−/− cells. A, de novo mitochondrial protein synthesis by metabolic labeling in FASTKD3−/− cells (left panel). Two independent FASTKD3−/− cell lines (clone #1 and clone #2) were incubated in the presence of 200 μCi of [35S]cysteine and [35S]methionine for 1 h after the addition of emetine (100 μg/ml). Proteins were separated by SDS-PAGE in a 12–20% linear gradient gel. The gel was stained with Coomassie brilliant blue (CBB), and radioactivity was detected using a phosphorimaging device. Complete lanes from the same phosphorimaging device exposure were rearranged so that each clone and its corresponding wild type control were juxtaposed. This is indicated by leaving a space between the lanes. In the right panel, band intensities in FASTKD3−/− cells were determined by densitometric analysis, and data are expressed relative to those of wild type cells (set as 1). Values represent the means ± S.D. (n = 3). **, p < 0.01. B, representative Western blot showing expression of COX1 and other MRC subunits on whole cell lysates from wild type and FASTKD3−/− cells (left panel). The right panel shows quantification of Western blotting signals in FASTKD3−/− cells. Values were normalized to β-actin (loading control) and presented relative to wild type (set as 1). Values represent the means ± S.E. (n = 7).**, p < 0.01. C, representative Western blot of four independent experiments showing expression of COX1 in cell lysates from wild type, FASTKD3−/−, and FASTKD3−/− cells reconstituted with full-length (FL) FASTKD3-FLAG-HA, FASTKD3ΔRAP-FLAG-HA, or FASTKD3Δ646–650-FLAG-HA (by lentiviral vector-mediated transduction).
We next explored the steady-state levels of COX1 in the FASTKD3−/− cells. As expected, FASTKD3−/− showed decreased protein levels of COX1 (∼2-fold reduction) as compared with wild type cells (Fig. 4B). The steady-state levels of other mtDNA-encoded (COX2) and nuclear encoded (NDUFB8, NDUFA9, SDHB, UQCRC2, COX5A, and ATP5A) subunits of respiratory complexes were similar between FASTKD3−/− and wild type cells (Fig. 4B).
We next looked at the ability of full-length FASTKD3 and FASTKD3 mutants ΔRAP and Δ646–650 to restore the steady-state levels of COX1 protein. As described above, we used FASTKD3−/− cells stably expressing FLAG-HA-tagged full-length and truncated versions of FASTKD3. Only the full-length construct was able to rescue the steady-state levels of COX1 protein in FASTKD3−/− cells (Fig. 4C). Altogether, our observations demonstrate that FASTKD3 has a dual function affecting both mitochondrial mRNA stability and translation and that both activities seem to require the RAP domain.
The Absence of FASTKD3 Results in Defective Complex IV Assembly and Activity
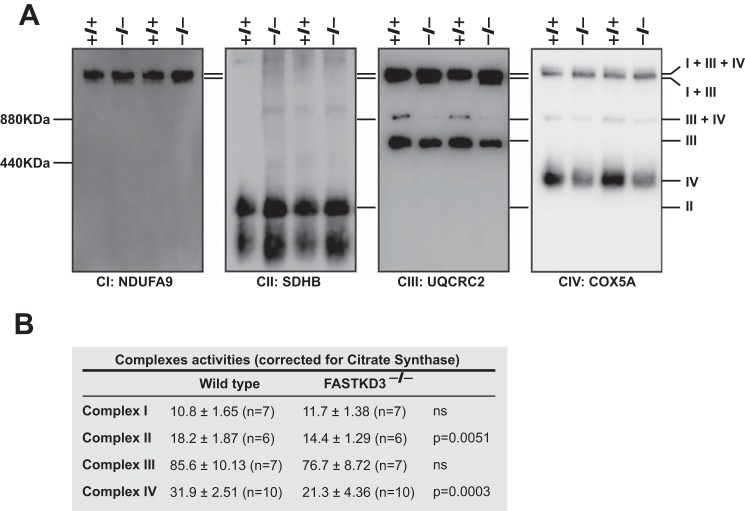
The decrease in COX1 steady-state levels by deleting FASTKD3 prompted us to investigate the integrity of the respiratory supercomplexes (SC) by 1D Blue Native-PAGE (BN-PAGE) using antibodies against NDUFA9 (complex I, CI), core 2 (complex III, CIII), and COX5a (complex IV, CIV) subunits that are reported to be incorporated in late steps of complex assembly (20). Our results showed that free CIV is reduced in FASTKD3−/− cells. The CIII included in the SC III+IV was strongly decreased in FASTKD3−/− cells, which in turn showed an accumulation of CIII in the SC I+III (Fig. 5A). However, there were no apparent changes in the SC I+III+IV (the so-called respirasome).
FIGURE 5.
Assembly and activity of MRC complexes in FASTKD3−/− cells. A, BN-PAGE and Western blotting analyses of digitonin-solubilized mitochondrial extracts from wild type and FASTKD3−/− cells. Membranes were incubated with the indicated oxidative phosphorylation subunits. I+III+IV, SC containing CI, CIII, and CIV; I+III, SC containing CI and CIII; and III+IV, SC containing CIII and CIV. Data are representative of two independent experiments out of four. B, the specific activities of complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (cytochrome c reductase), and complex IV (cytochrome c oxidase) were measured in cell lysates, and then normalized to citrate synthase activity and expressed as: (nmol/min/mg of protein)/(specific activity of citrate synthase) × 100. Data are presented as means ± S.E. The number of independent experiments and the p value using Mann Whitney U test are indicated. ns, not significant.
Next, we measured the activity of the complexes of the respiratory chain in crude mitochondria isolated from FASTKD3 or the control wild type cell lines (Fig. 5B). All activities were normalized to citrate synthase activity measured in the respective mitochondrial preparations. Consistent with a reduced level of assembled CIV, COX activity in FASTKD3−/− cells showed a 33% decrease as compared with that of wild type cells. Unexpectedly, we also observed that FASTKD3−/− cells had a mild but significant decrease in complex II activity. Our data demonstrate that FASTKD3 is required for a fully assembled and active CIV.
Discussion
Here we report the role of FASTKD3 in the regulation of mitochondrial gene expression. We show that targeted disruption of FASTKD3 gene leads to an increase in the half-lives and steady-state levels of mature mitochondrial transcripts ND2, ND3, CYTB, COX2, and ATP8/6, and that this activity requires the RAP domain of FASTKD3. In addition, we show that FASTKD3 is necessary for the translation of COX1, but not for the stability of its mRNA.
Previous work on other members of the FASTK family has shed light on the function of this emerging family of proteins in the regulation of mitochondrial RNAs. This includes the regulation of RNA processing as well as the maturation and assembly of the mitochondrial ribosomes (13–15, 17). Interestingly, this is the first report that demonstrates that a FASTK family member negatively regulates the stability of a subset of mitochondrial transcripts and selectively enhances translation. These findings raise interesting questions about the mechanism of action of FASTK proteins. FASTK has been reported to bind to ND6 mRNA and prevent its 3′ end degradation by the mitochondrial degradosome, thus allowing correct ND6 processing (13). This “barrier mechanism” is not expected to be responsible for the phenotype found in FASTKD3−/− cells because the deletion of FASTKD3 leads to the accumulation of mitochondrial mRNA rather than their depletion. However, FASTKD3 might bind to target RNAs in a way similar to how FASTK binds to ND6 and then degrade the target RNAs either directly or indirectly through the recruitment of the degradation machinery. Likewise, the apparent precursor processing defect reported for FASTKD5-depleted cells could be explained by binding to the atypical ATP8/6-COX3 junction and subsequent cleavage by FASTKD5 or by proteins recruited by FASTKD5 with nuclease activity. Therefore, it is likely that FASTK proteins have different (even opposite) molecular functions based on their ability to interact with RNA and would involve trans-acting factors such as the RNA degradation machinery (as in the case of FASTK), among others still to be defined. In this context, it is interesting to note that FASTKD2 has been recently identified in a 16S regulatory complex together with three pseudouridine synthases that are essential for 16S stability (21).
FASTK proteins share FAST_1, FAST_2, and RAP domains. The RAP domain is particularly abundant in a phylum of parasitic protists known as apicomplexans, and it is thought to be an RNA-binding domain (7). The exact biological importance of the RAP domain still needs to be clarified. For example, it seems to be required for Raa3-mediated chloroplast group II intron trans-splicing activity in Chlamydomonas reinhardtii (22). Concerning its importance in the functions of FASTK proteins, we have previously reported that the RAP domain is required for the alternative splicing activity of nuclear FASTK (23). More recently, Wolf et al. (17) have reported that four characteristic RAP domain residues located at the end of the domain are required for the ability of FASTKD4 to stabilize a subset of mitochondrial transcripts. Also, Jourdain et al. (13) have reported that the RAP domain is essential for the interaction of FASTK with ND6 mRNA. Here we report that the RAP domain and also the final five residues of the domain are required for FASTKD3 function in mRNA stability. Interestingly, homology modeling of the RAP domain of FASTKD2 revealed an endonuclease-like fold that generates an interface rich in basic and aromatic residues that might be involved in RNA binding (5). The similarity between the RAP domain and endonucleases was further highlighted by structural modeling by Boulouis et al. (24). The conserved residues at the end of the RAP domain, which are critical for the role of FASTKD2 and FASTKD5 in mitochondrial mRNA stability, have not been identified as important for catalysis in the endonucleases based on the alignments of these domains. Further biochemical and structural studies will be required to determine whether the RAP domain has endonuclease activity and the role of this domain in each member of the FASTK family. As for FAST_1 and FAST_2 domains, their function remains unknown. Interestingly, Eberhard et al. (25) suggested that FAST_1 domain is structurally related to octotricopeptide repeat (OPR) domains, which have been proposed to structurally and functionally resemble PPR domains. Both OPR and PPR domains are predicted to fold into a pair of antiparallel α-helices. Most OPR and PPR proteins are predicted to be targeted to organelles where several have been shown to control the post-transcriptional steps of gene expression such as RNA maturation, stability, and translation (24–28). It will therefore be important to study the structure of FAST_1 and FAST_2 domains and their contribution to the function of FASTK proteins.
Interestingly, the translation rate of ND2, ND3, CYTB, COX2, and ATP8/6 was not faster in FASTKD3−/− cells despite the increase in their mRNA levels. It has been previously demonstrated that mammalian mitochondria have a great excess of transcripts under basal physiological conditions (29). We speculate that the mitochondrial translation machinery is saturated under basal physiological circumstances, and therefore increases in transcript steady-state levels by 1.5–2-fold are not accompanied by faster translation rates.
Importantly, we also describe that FASTKD3 is required for efficient COX1 mRNA translation. Our data support a role for FASTKD3 as a translational activator of mitochondrially encoded COX1 because its absence causes a selective defect in the translation of COX1 mRNA and decreased steady-state protein levels without altering its mRNA levels. TACO1 has been reported to selectively promote COX1 mRNA translation without altering mRNA levels in mammalian cells (30). Mutations in TACO1 cause cytochrome c oxidase deficiency and late-onset Leigh syndrome (30). The depletion of another FASTK protein, FASTKD5, also leads to a selective COX1 translation defect; however, and in contrast to FASTKD3−/− cells, depletion of FASTKD5 is accompanied by almost undetectable levels of COX1 mature mRNA (15). The leucine-rich pentatricopeptide repeat-containing (LRPPRC) protein has been reported in in vitro studies to be necessary for the stability and translation of the COX1 and COX3 transcripts in the mitochondria, and an amino acid substitution of this protein causes the French-Canadian type of Leigh syndrome (LSFC) (31). We have previously reported that LRPPRC takes part in a protein complex with FASTKD3 (3), and we speculate that LRPPRC and FASTKD3 could act cooperatively in the regulation of COX1 translation. It will be important to unravel the molecular mechanisms underlying the ability of FASTKD3 to promote COX1 mRNA translation and to decipher the precise role that the RAP domain plays in this function.
Our data also demonstrate that FASTKD3 is required for a fully assembled and active CIV. Our 1D BN-PAGE results showed that in FASTKD3−/− cells, the free CIV content is decreased, which could be explained by the diminished COX1 protein steady-state levels. We also observed that the amount of CIII taking part in the SC III+IV was strongly decreased in FASTKD3−/− cells in which CIII seems to be incorporated into the SC I+III. We can speculate that the low steady-state levels of COX1 protein lead to both a drop in free complex IV and a reorganization of SC III+IV and SC I+III, with no apparent changes in the respirasome (SC I+III+IV). An intense debate exists concerning different models of supercomplexes biogenesis and assembly (32–35). However, CIV seems to be functional both in free form and also when incorporated into supercomplexes (36), so the reduced levels of free CIV and SC III+IV could account for the decreased complex IV enzyme activity seen in the FASTKD3−/− cells. In vitro COX activity in FASTKD3−/− cells showed a 33% decrease as compared with that of wild type cells. This finding correlates well with our earlier observation that FASTKD3 depletion causes a decrease in oxygen consumption (3). We have also observed that FASTKD3−/− cells have a mild but significant decrease in complex II activity. This result was unexpected because complex II is exclusively nuclear encoded; however, native complex II content was similar to that of wild type cells, when an antibody against subunit SDHC, which anchors other subunits of the complex II to the internal mitochondrial membrane, was used in BN-PAGE experiments. Further studies are required to explain the differences between content and enzyme activity of complex II.
In conclusion, we have established the consequences of a genomic deletion of FASTKD3 in mitochondrial RNA metabolism, and our results demonstrate that FASTKD3 has dual functions: 1) as a modulator of the stability of a subset of mature mitochondrial mRNAs ND2, ND3, CYTB, COX2, and ATP8/6; and 2) promoting COX1 mRNA translation. Our observations suggest that the function of this emerging family of RNA-binding proteins goes beyond RNA processing and ribosome assembly, and more detailed studies on FASTKD3 and the other members will help us understand the interplay between them and proteins relevant to post-transcriptional regulation of mitochondrial gene expression.
Experimental Procedures
Cell Lines and Antibodies
The human osteosarcoma cell line U2OS was obtained from Dr. Paul Anderson (Harvard University). Rabbit polyclonal antibodies raised against the 350 C-terminal amino acids of human FASTKD3 and FASTKD2 were purchased from Proteintech (catalog numbers 18392-1-AP and 17464-1-AP, respectively). Rabbit polyclonal antibody against actin was purchased from Sigma (catalog number AC-40). Mouse monoclonal antibodies used were as follows: anti-NDUFB8 (clone 20E9DH10C12), anti-SDHB (clone 21A11AE7), anti-UQCRC2 (clone 13G12AF12BB11), anti-COX1 (clone 1D6E1A8), and anti-ATP5A (clone 15H4C4) from Abcam; anti-NDUFA9 (clone 20C11B11B11), anti-COX2 (clone 12C4F12), and anti-COX5A (clone 6E9B12D5) from MitoSciences; anti-FLAG (clone M2) from Sigma; and anti-HA (clone 16B12) from Covance. All secondary fluorescent antibodies were purchased from Molecular Probes.
Fluorescence Microscopy
Briefly, FASTKD3-FLAG transfected U2OS cells were fixed in 4% paraformaldehyde, and immunostaining with anti-FLAG and anti-FASTKD2 antibodies was performed in PBS containing 0.1% Triton X-100 and 3% w/v BSA (Sigma-Aldrich). When indicated, EGFP-tagged full-length FASTKD3 and FASTKD3 mutants ΔRAP and Δ646–650 plasmids were transfected into cells. EGFP plasmids were generated by subcloning the respective cDNAs into the pEGFP-N1 vector (Clontech). Mitochondria were stained with 100 nm MitoTracker Red CMXRos (Thermo Fisher Scientific), and nuclei were stained with Hoechst 33258 (Sigma). Digital images were captured using a Zeiss Axiophot microscope.
Generation of FASTKD3-deficient Cell Lines
The targeting vector pAAV-MCS-FASTKD3 was designed to delete exon 2 of human FASTKD3, which encodes the initiator ATG, replacing it with loxP-flanked blasticidin resistance cassette. pAAV-MCS-FASTKD3 plasmid was obtained through the following steps. First, the 5′ homology arm (982-bp fragment) was amplified by PCR from U2OS genomic DNA using the primers 1F (5′-ATTGCGGCCGCCTGGAAAGCGCCTAGAAC-3′, containing a NotI site) and 1R (5′-TAAGTCGACGCTCTATGCATCTGAAAATCAGCGAGGTTAGAGCAAGGCAAG-3′, containing a SalI site, and a FASTKD3 exon 2 sequence artificially introduced as a tool for screening). Second, a SalI/EcoRI fragment (1,582 bp) containing a blasticidin resistance cassette flanked by loxP sequences was obtained from a pBluescript II-based plasmid previously generated in our laboratory. Third, the 3′ homology arm (1,380-bp fragment) was amplified by PCR from U2OS genomic DNA using the primers 2F (5′-ATAGAATTCTAGAAAGCTGGAAAACGTGCCCTGAAGTTAACAGATGCTGG-3′, containing an EcoRI site, and a FASTKD3 exon 2 sequence artificially introduced as a tool for screening) and 2R (5′-TATGCGGCCGCATTTGGGCGTAGAAACTGA-3′, containing a NotI site). The NotI/SalI-digested 5′ homology arm, the SalI/EcoRI fragment containing a blasticidin resistance cassette, and the EcoRI/NotI-digested 3′ homology arm were cloned into the NotI site of pAAV-MCS (Addgene). We also engineered two CRISPR/Cas9n nickases, targeting 20-nucleotide genomic sequences 5′-GACCTGAAGTTAACAGATGC-3′ and 5′-TAGTGATCCGAAGATGAGAC-3′, respectively, to facilitate homologous recombination. Annealed oligonucleotides were cloned into the BbsI site of pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) containing humanized Streptococcus pyogenes Cas9 (D10A) nickase (a gift from Feng Zhang, Addgene plasmid number 42335) (37), and the resulting plasmids were named as pX335-FASTKD3-sgRNA#1 and pX335-FASTKD3-sgRNA#2, respectively. U2OS cells were transfected with the targeting vector pAAV-MCS-FASTKD3, pX335-FASTKD3-sgRNA#1, and pX335-FASTKD3-sgRNA#2 using TransIT reagent (Mirus Bio). Transfected cells were selected with 5 μg/ml blasticidin, and single colonies were checked for homologous recombination. Homologous recombination at the 5′ arm was verified by PCR using primers P1F (5′-CCCATGAAACACACATCCTG-3′) and P1R (5′-GCTCTATGCATCTGAAAATCAG-3′), and expected fragment sizes were 1,777 bp for the wild type allele and 1,261 bp for the targeted allele. Homologous recombination at the 3′ arm was verified by PCR using primers P2F (5′-TAGAAAGCTGGAAAACGTGC-3′) and P2R (5′-TTTGATTAGTGAGTCTCATTCC-3′), and expected fragment sizes were 1,750 bp for the wild type allele and 1,450 bp for the targeted allele. The single targeted cells obtained were treated with recombinant cell-permeant TAT-NLS-Cre enzyme to delete loxP-flanked blasticidin resistance cassette. Resultant cells were then subjected to a second round of gene targeting with pAAV-MCS-FASTKD3 vector as described above, to obtain null mutant cells for FASTKD3 (FASTKD3−/−).
Rescue of FASTKD3−/− Cells and Overexpression of FASTKD3 Mutants
A full-length FASTKD3 human cDNA (NM_024091) with C-terminal FLAG-HA tag was cloned into the lentiviral vector pSin-EF2-Sox2-Pur digested with EcoRI and SpeI. pSin-EF2-Sox2-Pur was a gift from James Thomson (Addgene plasmid number 16577) (38). FASTKD3 mutants ΔRAP (deletion of amino acids 593–650) and Δ646–650 (deletion of amino acids 646–650) were created via QuikChange Site-Directed mutagenesis (Agilent). Amino acid positions refer to the National Center for Biotechnology Information (NCBI) protein database accession number NP_076996. Lentiviral stocks were produced by transient cotransfection into the human 293FT cell line (Thermo Fisher Scientific) with the appropriate lentiviral expression plasmid and lentiviral helper plasmids (psPAX2 packaging vector and pMD2.G envelope-encoding vector) using Lipofectamine 2000. psPAX2 (Addgene plasmid number 12260) and pMD2.G (Addgene plasmid number 12259) were gifts from Didier Trono. FASTKD3−/− cells were then transduced and selected with 2 μg/ml puromycin for at least 2 weeks before assaying.
RNA Isolation, qRT-PCR, and Northern Blotting
Total RNA was extracted using TRIzol (Invitrogen). DNA contamination from RNA samples was removed by treatment with DNase I (Ambion). RNA (1 μg) was reverse-transcribed using iScript (Bio-Rad Laboratories) to generate cDNA that was quantified by real-time PCR analysis (LightCycler 480 System; Roche Life Science) using SYBR Green PCR Master Mix (Applied Biosystems). The primers used for amplification were described in detail by Nagao et al. (19).
Northern blotting was performed as in Jourdain et al. (11), in which total RNA was extracted with TRI Reagent (Sigma-Aldrich) and 5–15 μg of RNA were separated on a denaturing formaldehyde agarose gel and transferred via electrophoresis to a nylon membrane (GE Healthcare). Strand-specific [32P]UTP-labeled riboprobes were transcribed using T7 polymerase (Bio-Rad), and hybridization was performed at 60 °C in 50% formamide, 7% SDS, 0.2 m NaCl, 80 mm sodium phosphate (pH 7.4), and 100 μg/ml salmon sperm DNA. Imaging was done with a phosphorimaging device (Bio-Rad). The RNA bands were quantified by densitometric analysis using National Institutes of Health ImageJ software.
Measurement of Mitochondrial mRNAs Half-lives
Mitochondrial transcription was disrupted with ethidium bromide (0.5 μg/ml). Cells were harvested at different time points after the addition of the inhibitor (0, 30, 60, 90, 120, 150, 180, 240, 300, and 360 min), and mitochondrial mRNA steady-state levels were measured by qRT-PCR as described above. The half-life of each mitochondrial mRNA was calculated using the formula t½ = ln2/λ, where λ is the slope of mRNA decay (19).
Mitochondrial Translation Assay
Cells were grown to 70% confluency in DMEM with 10% FBS and then washed two times with PBS. They were then incubated for 30 min with DMEM without methionine and cysteine, supplemented with 10% dialyzed FBS, GlutaMAX (Gibco), and 1 mm sodium pyruvate. Cytoplasmic translation was inhibited by the addition of 100 μg/ml emetine, and cells were incubated for 5 min. Finally, translation products were labeled by the addition of 200 μCi of [35S]cysteine and [35S]methionine (Easy Tag protein labeling mix, PerkinElmer) for 1 h. Cells were washed three times with PBS, harvested, and lysed. Proteins extracts were separated by SDS-PAGE in a 12–20% linear gradient gel. The gel was stained with Coomassie brilliant blue and dried, and radioactive bands were visualized using a phosphorimaging device.
Respiratory Chain Activity
Respiratory activity of complexes I, II, III, and IV was determined in a Shimadzu UV-1800 Spectrophotometer as described previously (39) with slight modifications.
BN-PAGE
Mitochondrial pellets were isolated from wild type and FASTKD3−/− cells, and native mitochondrial proteins were prepared as described previously (33, 34). Native PAGE Novex 3–12% Bis-Tris Protein Gels (Invitrogen) were loaded with 30 μg of mitochondrial protein. After electrophoresis, proteins were transferred to nitrocellulose or PVDF membranes at 1.3 A (constant) for 10 min and probed with specific antibodies.
Statistics
All analyses were performed using Prism software (GraphPad). Data are expressed as means ± S.D. or means ± S.E. and were analyzed using one-way analysis of variance with Bonferroni correction, the unpaired Student t test, or Mann-Whitney U test, as appropriate. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Author Contributions
E. B. and M. Z. conducted most of the experiments, analyzed the results, made the figures, and contributed to the writing of the paper. A. A. J. conducted some of the NB experiments and edited the manuscript. I. G. C. conducted BN-PAGE analyses. R. T. M. obtained one of the FASTKD3−/− cell lines and provided excellent technical assistance in making constructs and qRT-PCR studies. A. D. M. and M. A. M. conducted mtDNA quantification and complexes activities measurements. J. C. M. and A. O. provided knowledge and infrastructure support. M. S. and M. A. D. l. F. conceived and coordinated the study and wrote the paper with E. B. and M. Z. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Roberto Cantalapiedra and Raquel Carretero for technical support.
This work was supported by Consejería de Sanidad JCyL Grants BIO/VA20/15 (to M. S.) and BIO/VA21/15 (to M. A. D. l. F.), Roche Diagnostics S.L. (to M. S.), the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Förschung Grant 310030B_160257/1 (to J. C. M.), iGE3, and the State of Geneva. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. 1 and supplemental Table 1.
- PPR
- pentatricopeptide repeat
- FASTKD3
- FAST kinase domains 3
- FASTK
- Fas-activated serine/threonine kinase
- LRPPRC
- leucine-rich pentatricopeptide repeat-containing
- MRG
- mitochondrial RNA granule
- OPR
- octotricopeptide repeat
- qRT-PCR
- quantitative RT-PCR
- BN-PAGE
- Blue Native-PAGE
- SC
- supercomplex(es)
- CI
- complex I
- CIII
- complex III
- CIV
- complex IV
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Ojala D., Montoya J., and Attardi G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 [DOI] [PubMed] [Google Scholar]
- 2. Xu F., Ackerley C., Maj M. C., Addis J. B., Levandovskiy V., Lee J., Mackay N., Cameron J. M., and Robinson B. H. (2008) Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 416, 15–26 [DOI] [PubMed] [Google Scholar]
- 3. Simarro M., Gimenez-Cassina A., Kedersha N., Lazaro J. B., Adelmant G. O., Marto J. A., Rhee K., Tisdale S., Danial N., Benarafa C., Orduña A., and Anderson P. (2010) Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 401, 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., Wyler E., Bonneau R., Selbach M., Dieterich C., and Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 5. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., and Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 6. Kwon S. C., Yi H., Eichelbaum K., Föhr S., Fischer B., You K. T., Castello A., Krijgsveld J., Hentze M. W., and Kim V. N. (2013) The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122–1130 [DOI] [PubMed] [Google Scholar]
- 7. Lee I., and Hong W. (2004) RAP: a putative RNA-binding domain. Trends Biochem. Sci. 29, 567–570 [DOI] [PubMed] [Google Scholar]
- 8. Iborra F. J., Kimura H., and Cook P. R. (2004) The functional organization of mitochondrial genomes in human cells. BMC Biol. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jourdain A. A., Boehm E., Maundrell K., and Martinou J. C. (2016) Mitochondrial RNA granules: compartmentalizing mitochondrial gene expression. J. Cell Biol. 212, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antonicka H., Sasarman F., Nishimura T., Paupe V., and Shoubridge E. A. (2013) The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 17, 386–398 [DOI] [PubMed] [Google Scholar]
- 11. Jourdain A. A., Koppen M., Wydro M., Rodley C. D., Lightowlers R. N., Chrzanowska-Lightowlers Z. M., and Martinou J. C. (2013) GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 17, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tu Y. T., and Barrientos A. (2015) The human mitochondrial DEAD-Box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 10, 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jourdain A. A., Koppen M., Rodley C. D., Maundrell K., Gueguen N., Reynier P., Guaras A. M., Enriquez J. A., Anderson P., Simarro M., and Martinou J. C. (2015) A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 10, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 14. Popow J., Alleaume A. M., Curk T., Schwarzl T., Sauer S., and Hentze M. W. (2015) FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 21, 1873–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Antonicka H., and Shoubridge E. A. (2015) Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 10, 920–932 [DOI] [PubMed] [Google Scholar]
- 16. Ghezzi D., Saada A., D'Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V., Elpeleg O., and Zeviani M. (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 83, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolf A. R., and Mootha V. K. (2014) Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 7, 918–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cámara Y., Asin-Cayuela J., Park C. B., Metodiev M. D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., Franz T., Erdjument-Bromage H., Tempst P., Hallberg B. M., Gustafsson C. M., and Larsson N. G. (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 13, 527–539 [DOI] [PubMed] [Google Scholar]
- 19. Nagao A., Hino-Shigi N., and Suzuki T. (2008) Measuring mRNA decay in human mitochondria. Methods Enzymol. 447, 489–499 [DOI] [PubMed] [Google Scholar]
- 20. Fernández-Vizarra E., Tiranti V., and Zeviani M. (2009) Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793, 200–211 [DOI] [PubMed] [Google Scholar]
- 21. Arroyo J. D., Jourdain A. A., Calvo S. E., Ballarano C. A., Doench J. G., Root D. E., and Mootha V. K. (2016) A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 10.1016/j.cmet.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rivier C., Goldschmidt-Clermont M., and Rochaix J. D. (2001) Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20, 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simarro M., Mauger D., Rhee K., Pujana M. A., Kedersha N. L., Yamasaki S., Cusick M. E., Vidal M., Garcia-Blanco M. A., and Anderson P. (2007) Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 104, 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N. J., Pascal K., Girard-Bascou J., Vallon O., Wollman F. A., and Choquet Y. (2015) Spontaneous dominant mutations in Chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell 27, 984–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., and Wollman F. A. (2011) Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 26. Barkan A., and Small I. (2014) Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 [DOI] [PubMed] [Google Scholar]
- 27. Kleinknecht L., Wang F., Stübe R., Philippar K., Nickelsen J., and Bohne A. V. (2014) RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lefebvre-Legendre L., Choquet Y., Kuras R., Loubéry S., Douchi D., and Goldschmidt-Clermont M. (2015) A nucleus-encoded chloroplast protein regulated by iron availability governs expression of the photosystem I subunit PsaA in Chlamydomonas reinhardtii. Plant Physiol. 167, 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lagouge M., Mourier A., Lee H. J., Spåhr H., Wai T., Kukat C., Silva Ramos E., Motori E., Busch J. D., Siira S., German Mouse Clinic Consortium, Kremmer E., Filipovska A., and Larsson N. G. (2015) SLIRP regulates the rate of mitochondrial protein synthesis and protects LRPPRC from degradation. PLoS Genet. 11, e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J. E., Lochmüller H., Chevrette M., Kaufman B. A., Horvath R., and Shoubridge E. A. (2009) Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 41, 833–837 [DOI] [PubMed] [Google Scholar]
- 31. Xu F., Morin C., Mitchell G., Ackerley C., and Robinson B. H. (2004) The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lapuente-Brun E., Moreno-Loshuertos R., Acín-Pérez R., Latorre-Pellicer A., Colás C., Balsa E., Perales-Clemente E., Quirós P. M., Calvo E., Rodríguez-Hernández M. A., Navas P., Cruz R., Carracedo Á., López-Otín C., Pérez-Martos A., et al. (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 [DOI] [PubMed] [Google Scholar]
- 33. Moreno-Lastres D., Fontanesi F., García-Consuegra I., Martín M. A., Arenas J., Barrientos A., and Ugalde C. (2012) Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 15, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mourier A., Matic S., Ruzzenente B., Larsson N. G., and Milenkovic D. (2014) The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab. 20, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pérez-Pérez R., Lobo-Jarne T., Milenkovic D., Mourier A., Bratic A., García-Bartolomé A., Fernández-Vizarra E., Cadenas S., Delmiro A., García-Consuegra I., Arenas J., Martín M. A., Larsson N. G., and Ugalde C. (2016) COX7A2L is a mitochondrial complex III binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep. 16, 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Enríquez J. A. (2016) Supramolecular organization of respiratory complexes. Annu. Rev. Physiol. 78, 533–561 [DOI] [PubMed] [Google Scholar]
- 37. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 39. Medja F., Allouche S., Frachon P., Jardel C., Malgat M., Mousson de Camaret B., Slama A., Lunardi J., Mazat J. P., and Lombès A. (2009) Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion 9, 331–339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.