Abstract
The activation of NF-κB downstream of T cell receptor (TCR) engagement is a key signaling step required for normal lymphocyte function during the adaptive immune response. During TCR signaling, the adaptor protein Bcl10 is inducibly recruited to the CARD11 scaffold protein as part of a multicomponent complex that induces IκB kinase (IKK) activity and NF-κB activation. Here, we show that a consequence of this recruitment is the TCR-induced conjugation of Bcl10 with linear-linked polyubiquitin chains to generate the signaling intermediate Lin(Ub)n-Bcl10, which is required for the association of Bcl10 with the NEMO subunit of the IKK complex. The TCR-induced generation of Lin(Ub)n-Bcl10 requires Bcl10 lysines 17, 31, and 63, CARD11, MALT1, and the HOIP subunit of the linear ubiquitin chain assembly complex (LUBAC) but not the HOIP accessory protein SHARPIN. CARD11 promotes signal-induced Lin(Ub)n-Bcl10 generation by co-recruiting Bcl10 with HOIP, thereby bringing substrate to enzyme. The CARD11-HOIP interaction is rendered TCR-inducible by the four autoinhibitory repressive elements in the CARD11 inhibitory domain and involves the CARD11 coiled-coil domain and two independent regions of HOIP. Interestingly, oncogenic CARD11 variants associated with diffuse large B cell lymphoma spontaneously induce Lin(Ub)n-Bcl10 production to extents that correlate with their abilities to activate NF-κB and with their enhanced abilities to bind HOIP and Bcl10. Our results define molecular determinants that control the production of Lin(Ub)n-Bcl10, an important signaling intermediate in TCR and oncogenic CARD11 signaling.
Keywords: gene regulation, NF-κB (NF-κB), oncogene, scaffold protein, signal transduction, Bcl10, CARD11, diffuse large B cell Lymphoma, LUBAC, T cell receptor
Introduction
The clearance of pathogens by the adaptive immune system relies upon signaling pathways that activate the NF-κB transcription factor following the recognition of foreign antigens by the T cell receptor (TCR)4 and B cell receptor (BCR) complexes on lymphocytes (1, 2). NF-κB controls the induction of programs of gene expression required for lymphocyte activation, proliferation, and survival. The failure to signal to NF-κB downstream of antigen receptor engagement can result in immunodeficiency and the increased susceptibility to infectious disease (3, 4).
TCR and BCR signaling to NF-κB proceeds through the multidomain scaffold protein CARD11, which inducibly recruits several signaling proteins into a complex for the activation of IKK complex kinase activity (5–14). The IKK complex then phosphorylates IκB proteins, leading to their ubiquitinylation and proteasomal degradation and the stable nuclear translocation of NF-κB. CARD11 scaffold activity is signal-inducible, due to the presence of an inhibitory domain (ID) in CARD11 that regulates the transition of CARD11 from an inactive to an active scaffold in response to TCR or BCR triggering. Prior to receptor signaling, the ID maintains CARD11 in a closed inactive state through intramolecular interactions that require the CARD and coiled-coil domains of CARD11 and that prevent the recruitment of signaling cofactors (15). The ID achieves its autoinhibition through four repressive elements (REs) that function cooperatively with redundancy (16, 17). Upon antigen receptor engagement, the inhibitory activity of the REs is neutralized, in part through the phosphorylation of specific serines within the ID, allowing CARD11 to assemble a multiprotein signaling complex (15, 18, 19). Factors that are recruited to CARD11 in a signal-dependent manner during normal antigen receptor signaling and that are required for activation of IKK kinase activity include Bcl10, MALT1, TRAF6, NEMO (also known as IKKγ), caspase-8, and CK1α (15, 20).
The antigen receptor signaling pathway is often dysregulated in leukemias and lymphomas, leading to the constitutive activation of NF-κB, which can contribute to the growth of transformed lymphocytes and potentially to the resistance to chemotherapeutic agents (21). Diffuse large B cell lymphoma (DLBCL), the most common form of non-Hodgkin lymphoma, has been divided into subtypes based on patterns of gene expression. The activated B cell-like (ABC) subtype is characterized by the constitutive activation of NF-κB, which is required for the proliferation and survival of ABC DLBCL cells in culture (22). The aberrant signaling to NF-κB in ABC DLBCL requires CARD11 (23). In addition CARD11 is mutated with gain-of-function mutations in ∼10% of human ABC DLBCL cases, which can account for their observed receptor-independent signaling to NF-κB (24). These oncogenic gain-of-function CARD11 mutations are located in the CARD, LATCH, and coiled-coil domains of CARD11 (24–29), and previous studies have demonstrated that they hyperactivate CARD11 activity by disrupting autoinhibition by the CARD11 ID (16, 17, 30, 31). Interestingly, the most active oncogenic CARD11 mutations selectively enhance the ability of the protein to associate with Bcl10 but not to other signaling cofactors so far examined (30, 31).
Several NF-κB-inducing signaling pathways have been shown to depend on the modification of one or more signaling proteins with linear ubiquitin chains, chains in which the N and C termini of ubiquitin are covalently linked (32). For example, the linear ubiquitinylation of RIP1 and NEMO is required for NF-κB activation by TNFα (33–35). The modification with linear ubiquitin chains can promote the binding of the modified substrate with other proteins that contain domains that specifically recognize linear chains. NEMO itself has been shown to bind linear ubiquitin chains with high affinity (36, 37). The E3 ligase responsible for linear ubiquitinylation is the linear ubiquitin chain assembly complex (LUBAC), which is commonly composed of three subunits, HOIP, HOIL-1L, and SHARPIN (33–35). Although a role for LUBAC in several pathways has been definitively demonstrated, it remains poorly understood how LUBAC targets its substrates for modification during signaling in a regulated, inducible manner.
Recently, several groups have linked LUBAC function to both normal and dysregulated antigen-receptor signaling, with seemingly disparate results. Yang et al. (38) discovered the enrichment of HOIP germline polymorphisms in DLBCL that increase LUBAC enzymatic activity and concluded that LUBAC-mediated modification of NEMO contributes to the constitutive BCR signaling to NF-κB that is a signature feature of ABC DLBCL. In contrast, Dubois et al. (39) concluded that LUBAC plays a non-catalytic role in antigen receptor signaling and in DLBCL based on the observation that a catalytically compromised mutant of HOIP was able to rescue HOIP-deficient Jurkat T cells in assays that measured TCR-induced NF-κB activation. Consistent with the latter, Sasaki et al. (40) reported that the inducible deletion of the RBR domain of HOIP, which is required for catalytic activity, in murine B cells had little effect on BCR signaling to NF-κB. However, in an unbiased survey of proteins ubiquitinylated during BCR signaling, Satpathy et al. (41) discovered that Bcl10 is conjugated with linear ubiquitin chains in response to BCR engagement. Furthermore, Yang et al. (42) also observed Bcl10 linear ubiquitinylation during the dysregulated chronic BCR signaling associated with ABC DLBCL, in a step proposed to lie downstream of the cIAP-mediated modification of Bcl10 with Lys-63-linked polyubiquitin chains. Therefore, it has remained unclear whether there is a different requirement for HOIP E3 ligase activity in TCR signaling, as opposed to BCR signaling, and whether linear-ubiquitinylated Bcl10 is an important intermediate in TCR signaling to NF-κB. Furthermore, because cIAP-inhibitory agents are ineffective for ABC DLBCL samples that harbor an oncogenic CARD11 allele (42), it has not been established to what extent oncogenic CARD11 variants that occur in ∼10% of ABC DLBCL cases also depend upon HOIP and the linear ubiquitinylation of Bcl10 for their dysregulated NF-κB activation, and if so, how these hyperactive variants promote Bcl10 ubiquitinylation.
In this report, we examine whether Bcl10 is modified with linear ubiquitin chains during TCR signaling and probe the mechanism and consequence of this modification. We find that CARD11 scaffold activity during TCR signaling inducibly recruits HOIP to Bcl10, leading to the linear ubiquitinylation of Bcl10, which is required for the association of Bcl10 with NEMO. Furthermore, we find that DLBCL-associated oncogenic mutations in CARD11 increase the ability of CARD11 to associate with the HOIP subunit of LUBAC and stimulate the linear ubiquitinylation of Bcl10 in a constitutive dysregulated manner even in the absence of antigen receptor engagement. We quantitatively assess the extent to which normal and oncogenic CARD11 signaling depends on HOIP activity and the polyubiquitinylation of Bcl10.
Results
Bcl10 Is Ubiquitinylated with Linear Chains during TCR Signaling
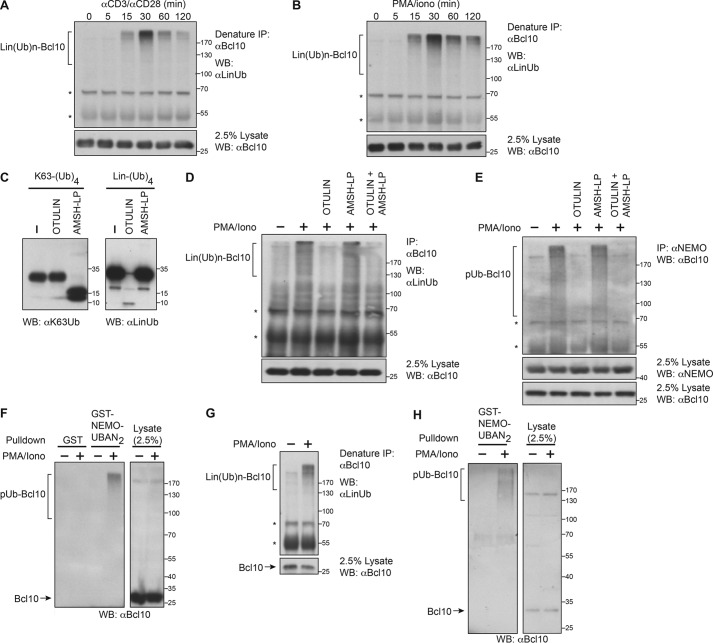
To determine whether Bcl10 is linearly ubiquitinylated as a consequence of TCR engagement, we stimulated Jurkat T cells with a time course of anti-CD3/anti-CD28 antibodies, immunoprecipitated Bcl10 under denaturing conditions to prevent isolation of co-associating proteins, and probed the precipitates with an antibody specific for linear ubiquitin chains. We observed signal-dependent linear ubiquitinylation of Bcl10, which peaked after 30 min of TCR cross-linking (Fig. 1A). We observed a similar result after treating Jurkat T cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin (PMA/iono) (Fig. 1B), indicating that TCR-proximal signaling events that are bypassed by PMA/iono treatment are not required for Bcl10 linear ubiquitinylation. To verify that the ubiquitin chains on Bcl10 were of a linear linkage, we treated Bcl10 precipitates with recombinant linkage-specific deubiquitylases. OTULIN is a linear ubiquitin-specific deubiquitylase that hydrolyzes linear polyubiquitin chains specifically (43, 44), whereas AMSH-LP cleaves Lys-63-pUb chains specifically (Fig. 1C) (37, 45). As expected, OTULIN but not AMSH-LP hydrolyzed nearly all of the linear polyubiquitin chains detected in the anti-Bcl10 precipitate (Fig. 1D). These data indicated that TCR signaling results in the generation of linear ubiquitinylated Bcl10, Lin(Ub)n-Bcl10.
FIGURE 1.
Bcl10 is ubiquitinylated with linear chains and recognized by NEMO during TCR signaling. A and B, Jurkat T cells (1 × 108/sample) were stimulated with anti-CD3/anti-CD28 antibodies or PMA/iono for the indicated times, and lysates were immunoprecipitated (IP) under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted (WB) with the indicated antibodies. The migration of molecular mass markers is indicated at the right of the blot in kDa. The asterisks indicate nonspecific bands in the Western blotting. C, Lys-63-linked tetra-ubiquitin or linear tetra-ubiquitin recombinant proteins were incubated in the absence or presence of OTULIN or AMSH-LP deubiquitylases, resolved on SDS-PAGE, and immunoblotted with antibodies that recognize either linear (LinUb) or Lys-63-linked (K63Ub) ubiquitin chains. D, cell lysates from Jurkat T cells (1 × 108/sample) treated with or without PMA/iono for 30 min were immunoprecipitated with anti-Bcl10 antibody. Immunoprecipitates were incubated in the absence or presence of OTULIN, AMSH-LP, or both deubiquitylases, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. E, cell lysates from Jurkat T cells (1 × 108/sample) treated with or without PMA/iono for 30 min were immunoprecipitated with anti-NEMO antibody. Immunoprecipitates were incubated in the absence or presence of OTULIN, AMSH-LP, or both deubiquitylases, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. F, cell lysates from Jurkat T cells (1 × 108/sample) treated with or without PMA/iono for 30 min were subjected to pulldown with recombinant GST or GST-NEMO-UBAN2 protein as indicated. Precipitates were resolved on SDS-PAGE and immunoblotted with anti-Bcl10 antibody. G, cell lysates from purified primary murine CD4+ T cells (7.5 × 107/sample) treated with or without PMA/iono for 30 min were subjected to IP under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted (WB) with the indicated antibodies. H, cell lysates from purified primary murine CD4+ T cells (7 × 107/sample) treated with or without PMA/iono for 30 min were subjected to pulldown with recombinant GST-NEMO-UBAN2 protein. Precipitates were resolved on SDS-PAGE and immunoblotted with anti-Bcl10 antibody.
NEMO Recognizes Linear Ubiquitinylated Bcl10 in Stimulated T Cells
NEMO has previously been reported to recognize Bcl10 conjugated with Lys-63-linked ubiquitin chains during antigen receptor signaling (46). However, more recent studies have demonstrated that NEMO exhibits a much higher affinity for linear ubiquitin multimers than Lys-63-linked multimers through its UBAN domain (36, 37). We asked whether NEMO would bind the Lin(Ub)n-Bcl10 generated by TCR signaling. First, we immunoprecipitated NEMO from Jurkat T cells and probed the precipitate for high molecular weight species of Bcl10, which we did observe in cells stimulated with PMA/iono but not in unstimulated cells (Fig. 1E). These high molecular weight Bcl10 species were specifically susceptible to OTULIN treatment, but not to AMSH-LP, verifying that they represented Lin(Ub)n-Bcl10. The fact that OTULIN cleaved most, if not all, NEMO-associated polyubiquitinylated Bcl10 species indicated that the vast majority of polyubiquitinylated Bcl10 recognized by NEMO during TCR signaling is Lin(Ub)n-Bcl10 and not Lys-63-linked polyubiquitinylated Bcl10. Furthermore, the data suggest that Bcl10 is not conjugated appreciably with mixed linear and Lys-63-linked chains and that Lin(Ub)n-Bcl10 and Lys-63-linked polyubiquitinylated Bcl10 are distinct species. Next, we generated a recombinant GST fusion protein containing two copies of the NEMO UBAN domain, GST-NEMO-UBAN2. High molecular weight Bcl10 species associated with GST-NEMO-UBAN2, but not with the GST control, and only in lysates from stimulated Jurkat T cells (Fig. 1F). In addition, unmodified Bcl10 did not bind GST-NEMO-UBAN2 (Fig. 1F). These data suggested that in stimulated Jurkat T cells, NEMO binds Lin(Ub)n-Bcl10 through its UBAN domain. To verify that Lin(Ub)n-Bcl10 is generated during TCR signaling in primary cells, murine CD4+ T cells were purified from spleen and stimulated with PMA/ionomycin. We readily detected Lin(Ub)n-Bcl10 in lysates from stimulated primary CD4+ T cells after immunoprecipitating Bcl10 under denaturing conditions, followed by probing the precipitate with antibody specific for linear ubiquitin chains (Fig. 1G). Furthermore, Lin(Ub)n-Bcl10 was found in association with the GST-NEMO-UBAN2 fusion only after signaling (Fig. 1H), confirming the generation of this intermediate in primary T lymphocytes.
HOIP Is Required for Linear Ubiquitinylation of Bcl10 and TCR-induced NF-κB Activation
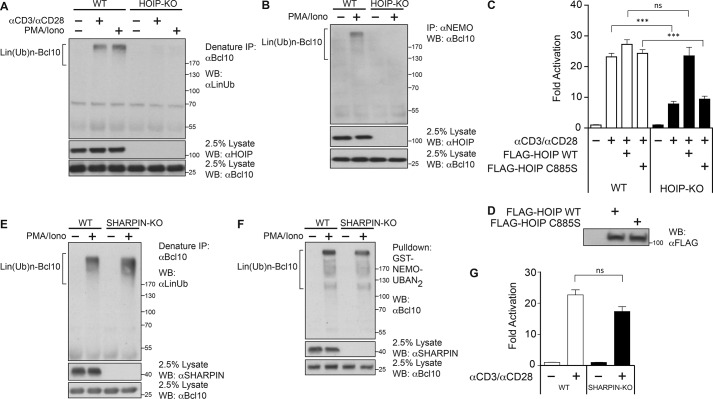
Because LUBAC is the only identified E3 ubiquitin ligase that is responsible for catalyzing the formation of linear polyubiquitin chains (33–35), we expected that LUBAC would be responsible for TCR-induced linear polyubiquitinylation of Bcl10. We used CRISPR/Cas9 targeting to generate a Jurkat T cell clone deficient in HOIP, the catalytic subunit of LUBAC. HOIP-deficient Jurkat T cells failed to generate Lin(Ub)n-Bcl10 after anti-CD3/anti-CD28 cross-linking or PMA/iono treatment (Fig. 2A). As expected, we failed to detect the binding of Lin(Ub)n-Bcl10 to NEMO in HOIP-deficient Jurkat T cells (Fig. 2B), consistent with the requirement of HOIP catalytic activity for the generation of Lin(Ub)n-Bcl10. Furthermore, the TCR-induced activation of an NF-κB-specific luciferase reporter, Igκ2-IFN-LUC (7), was significantly compromised in HOIP-deficient T cells (Fig. 2C). Importantly, this defect in TCR signaling to NF-κB activation could be rescued by the transient reintroduction of wild-type HOIP, but not by the catalytically dead C885S HOIP mutant (Fig. 2C), when expressed at equivalent levels (Fig. 2D). These results indicated that HOIP catalytic activity is required for TCR-mediated NF-κB activation.
FIGURE 2.
HOIP, but not SHARPIN, is required for linear ubiquitinylation of Bcl10 and TCR-induced NF-κB activation. A, WT or HOIP-deficient (HOIP-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without anti-CD3/anti-CD28 antibodies or PMA/iono for 30 min, and lysates were immunoprecipitated (IP) under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted (WB) with the indicated antibodies. B, WT or HOIP-deficient (HOIP-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were immunoprecipitated with anti-NEMO antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. C, WT or HOIP-deficient (HOIP-KO) Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC in the absence and presence of an expression vector for FLAG-HOIP WT or the FLAG-HOIP C885S mutant. About 40 h later, the cells were stimulated for 5 h with anti-CD3/anti-CD28 and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. *** indicates p < 0.001; ns (not significant) indicates p > 0.05. D, HEK293T cells were transfected with the expression vectors for FLAG-HOIP WT and FLAG-HOIP C885S used in C and lysates were analyzed by Western blotting with anti-FLAG antibody to indicate relative expression levels. E, WT or SHARPIN-deficient (SHARPIN-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were immunoprecipitated under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. F, WT or SHARPIN-deficient (SHARPIN-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were subjected to pulldown with recombinant GST-NEMO-UBAN2 protein. Precipitates were resolved on SDS-PAGE and immunoblotted with anti-Bcl10 antibody. G, WT or SHARPIN-deficient (SHARPIN-KO) Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC, and about 40 h later the cells were stimulated for 5 h with anti-CD3/anti-CD28, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. ns (not significant) indicates p > 0.05.
We also tested whether the SHARPIN subunit of LUBAC is also required for signal-induced linear ubiquitinylation of Bcl10 in T cells and for TCR signaling to NF-κB. We generated a Jurkat T cell clone deficient in SHARPIN using CRISPR/Cas9 and found, surprisingly, that Lin(Ub)n-Bcl10 was robustly generated in SHARPIN-deficient cells following PMA/ionomycin treatment (Fig. 2E). The absence of SHARPIN did not significantly diminish the signal-induced association of Lin(Ub)n-Bcl10 with the UBAN domain of NEMO (Fig. 2F) or the activation of NF-κB in response to TCR cross-linking (Fig. 2G). The data revealed that SHARPIN is not required for HOIP-dependent signaling to NF-κB downstream of TCR engagement.
CARD11 and MALT1 Are Required for Linear Ubiquitinylation of Bcl10
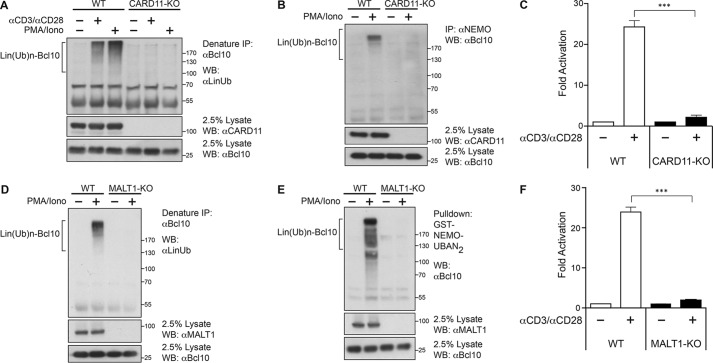
Bcl10 is inducibly recruited to CARD11 as a consequence of TCR signaling (5, 15, 47). To test whether CARD11 is required for the linear ubiquitinylation of Bcl10, we generated CARD11-deficient Jurkat T cells using CRISPR/Cas9 targeting. CARD11-deficient Jurkat T cells displayed a lack of Lin(Ub)n-Bcl10 generation (Fig. 3A), association of polyubiquitinylated Bcl10 with NEMO (Fig. 3B), and NF-κB reporter activation (Fig. 3C) following TCR cross-linking or PMA/iono treatment. Similarly, MALT1-deficient Jurkat T cells generated by CRISPR/Cas9 targeting also failed to generate Lin(Ub)n-Bcl10 upon PMA/ionomycin treatment (Fig. 3D) or exhibit the association of polyubiquitinylated Bcl10 with NEMO (Fig. 3E) or TCR-induced NF-κB activation (Fig. 3F). The data suggested that CARD11 and MALT1 are both required for the signal-induced action of LUBAC on Bcl10 during TCR signaling.
FIGURE 3.
CARD11 and MALT1 are required for TCR-induced linear ubiquitinylation of Bcl10. A, WT or CARD11-deficient (CARD11-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without anti-CD3/anti-CD28 antibodies or PMA/iono for 30 min, and lysates were immunoprecipitated (IP) under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted (WB) with the indicated antibodies. B, WT or CARD11-deficient (CARD11-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were immunoprecipitated with anti-NEMO antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. C, WT or CARD11-deficient (CARD11-KO) Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC. About 40 h later, the cells were stimulated with anti-CD3/anti-CD28 for 5 h, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. *** indicates p < 0.001. D, WT or MALT1-deficient (MALT1-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were immunoprecipitated under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. E, WT or MALT1-deficient (MALT1-KO) Jurkat T cells (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were subjected to pulldown with recombinant GST-NEMO-UBAN2 protein. Precipitates were resolved on SDS-PAGE and immunoblotted with anti-Bcl10 antibody. F, WT or MALT1-deficient (MALT1-KO) Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC, and about 40 h later the cells were stimulated for 5 h with anti-CD3/anti-CD28, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. ***, p < 0.001.
CARD11 Associates with HOIP and Promotes the Targeting of Bcl10 by HOIP
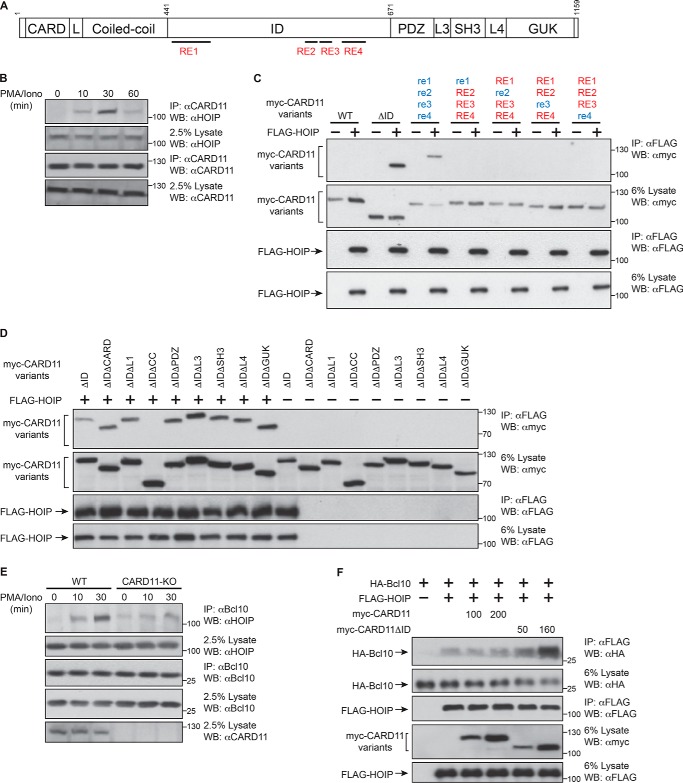
Next, we probed whether CARD11 (Fig. 4A) associates with HOIP in T cells. We treated Jurkat T cells with PMA/iono and found in immunoprecipitation experiments that CARD11 and HOIP associate in a signal-dependent manner to an extent that peaks 30 min after stimulation (Fig. 4B). CARD11 undergoes a transition during antigen receptor signaling from a closed inactive state to an open active scaffold. This transition is controlled by an ID that participates in intramolecular interactions involving the CARD, LATCH, and coiled-coil domains (15, 31). Antigen receptor ligation results in the neutralization of the inhibitory action of the ID, mediated in part by the phosphorylation of the ID on specific serine residues (18, 19). This neutralization of the ID causes it to disengage from the CARD, LATCH, and coiled-coil, which then allows these domains to recruit other signaling cofactors to CARD11, including Bcl10, TRAF6, NEMO, and caspase-8 (15). Recently, we reported that the inhibitory activity of the ID is accomplished by four REs, which function cooperatively with redundancy (16, 17). The fact that HOIP and CARD11 associated in a signal-dependent manner suggested that the CARD11 ID and the REs within it might control CARD11 binding to HOIP. Consistent with this prediction, we found that the ID-deleted CARD11 variant CARD11ΔID robustly associated with HOIP after co-expression in HEK293T cells, although wild-type CARD11 did not (Fig. 4C). Furthermore, mutation of all four REs allowed HOIP to associate with CARD11 (Fig. 4C), although single RE mutations did not (Fig. 4C), consistent with the conclusion that the REs cooperatively prevent HOIP from binding CARD11 in the absence of TCR engagement. We then used a panel of double-deletion CARD11 variants of CARD11ΔID to probe which CARD11 domains were required for the HOIP-CARD11 association. We found that the CARD11 coiled-coil was required for this association, although all other regions of CARD11 were dispensable (Fig. 4D).
FIGURE 4.
CARD11 inducibly associates with HOIP and promotes the targeting of Bcl10 by HOIP. A, CARD11 domain structure. The locations of the REs in the ID are indicated by the black bars. B, Jurkat T cells (6 × 107/sample) were stimulated with PMA/iono for the indicated times, lysed, and immunoprecipitated (IP) with anti-CARD11 antibody, resolved on SDS-PAGE, and analyzed by immunoblot with indicated antibodies. C, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of FLAG-HOIP expression vector and the following amounts of each CARD11 variant: WT (160 ng), ΔID (100 ng), re1 re2 re3 re4 (800 ng), re1 RE2 RE3 RE4 (600 ng), RE1 re2 RE3 RE4 (600 ng), RE1 RE2 re3 RE4 (600 ng), RE1 RE2 RE3 re4 (600 ng). The cell lysates were immunoprecipitated and analyzed by Western blotting (WB) with the indicated antibodies. Mutant repressive elements are indicated by blue lowercase letters, and wild-type repressive elements are indicated by red uppercase letters. D, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of FLAG-HOIP expression vector and the following amounts of each CARD11 variant: ΔID (100 ng), ΔIDΔCARD (480 ng), ΔIDΔL1 (120 ng), ΔIDΔCC (55 ng), ΔIDΔPDZ (50 ng), ΔIDΔL3 (200 ng), ΔIDΔSH3 (150 ng), ΔIDΔL4 (50 ng), and ΔIDΔGUK (90 ng). The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies. E, WT or CARD11-deficient (CARD11-KO) Jurkat T cells (6 × 107/sample) were stimulated with PMA/iono for the indicated times, and lysates were immunoprecipitated with anti-Bcl10 antibody, resolved on SDS-PAGE, and analyzed by immunoblot with indicated antibodies. F, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of FLAG-HOIP expression vector, 200 ng of HA-Bcl10, and the indicated amounts (ng) of either wild-type CARD11 or CARD11ΔID. The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies.
Because CARD11 inducibly binds both Bcl10 and HOIP during TCR signaling, we suspected that CARD11 might function to recruit HOIP to Bcl10 in response to TCR engagement. Consistent with this hypothesis, we found that endogenous HOIP and Bcl10 associated in a stimulation-dependent manner in Jurkat T cells (Fig. 4E) and that this induced HOIP-Bcl10 interaction was severely diminished in CARD11-deficient T cells (Fig. 4E). Furthermore, after overexpression in HEK293T cells, we showed that the association of HA-Bcl10 with FLAG-HOIP could be stimulated by co-expression of the constitutively open scaffold CARD11ΔID but not by wild-type CARD11, which is constitutively closed and inactive at these levels of expression in HEK293T cells (Fig. 4F). The results supported the notion that the scaffold activity of CARD11, induced by TCR triggering, promotes the targeting of Bcl10 by HOIP via co-recruitment to CARD11.
Two Independent HOIP Domains Associate with CARD11
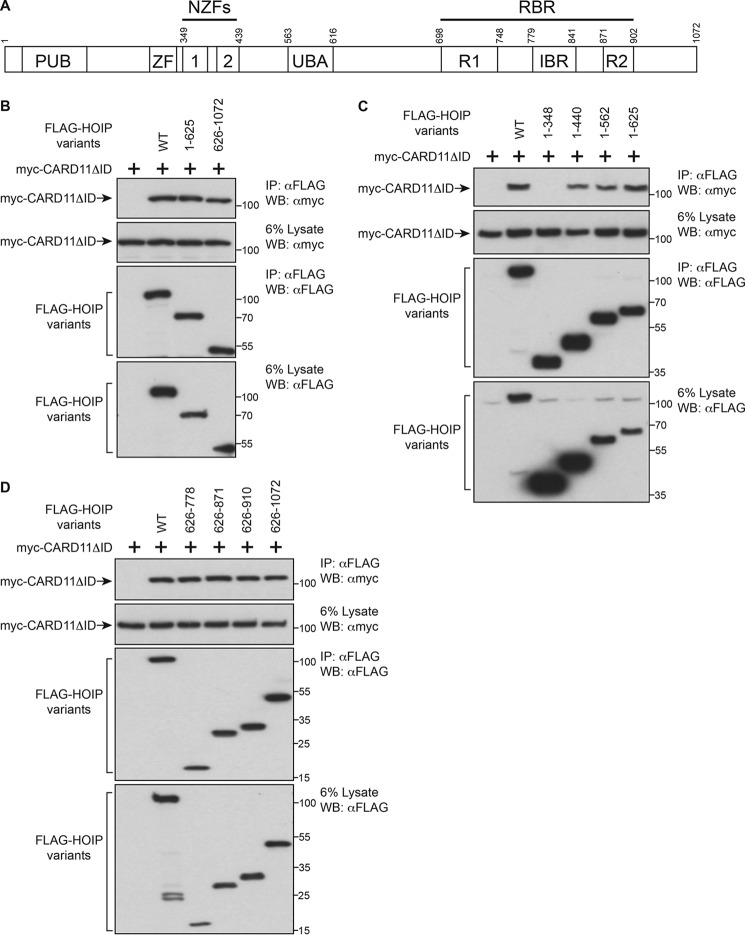
We next mapped the regions of HOIP (Fig. 5A) involved in the association with CARD11. After expression in HEK293T cells, we found that the 1–625 N-terminal portion of HOIP containing the PUB, ZF, and UBA domains (48) could readily associate with CARD11ΔID, and interestingly, the 626–1072 C-terminal portion containing the core RBR catalytic domain could independently co-immunoprecipitate with CARD11ΔID (Fig. 5B). A deletion series of the 1–625 fragment suggested that a region containing the two Npl4-type zinc fingers between residues 348 and 440 mediated CARD11 association (Fig. 5C). A similar approach with the 626–1072 fragment revealed that the region between 626 and 778, which contains Ring Finger 1 in the RBR domain was also sufficient for CARD11 association (Fig. 5D). Thus, two separate regions of the HOIP protein can associate with CARD11.
FIGURE 5.
Two HOIP regions can associate with CARD11. A, domain structure of HOIP adapted from Iwai et al. (32). B, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of myc-CARD11ΔID expression vector, and the following amounts of each FLAG-HOIP variant: WT (100 ng), 1–625 (100 ng), and 626–1072 (500 ng). The cell lysates were immunoprecipitated (IP) and analyzed by Western blotting (WB) with the indicated antibodies. C, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of myc-CARD11ΔID expression vector, and the following amounts of each FLAG-HOIP variant: WT (100 ng), 1–348 (50 ng), 1–440 (50 ng), 1–562 (70 ng), and 1–625 (100 ng). The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies. D, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of myc-CARD11ΔID expression vector and the following amounts of each FLAG-HOIP variant: WT (100 ng), 626–778 (150 ng), 626–871 (200 ng), 626–910 (300 ng),and 626–1072 (500 ng). The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies.
Linear Ubiquitinylation of Bcl10 Requires Lysines 17, 31, and 63
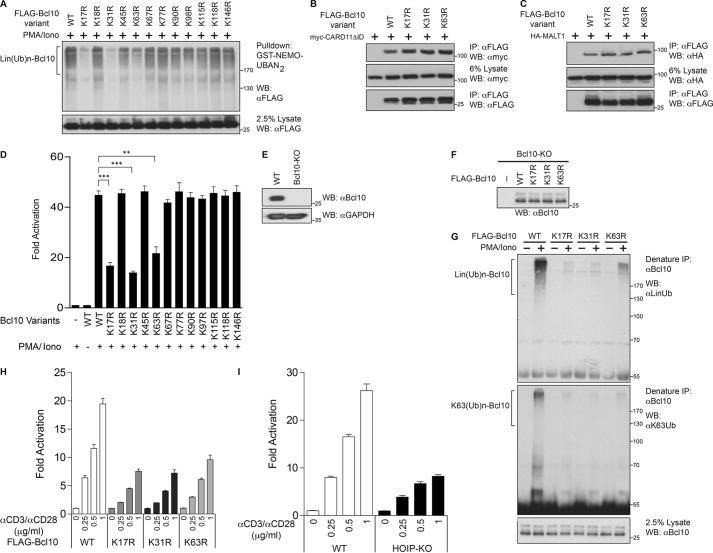
Our next goal was to map residues within Bcl10 that were required for its signal-induced linear ubiquitinylation. Because linear ubiquitinylation by LUBAC is thought to extend chains from an initial ubiquitin that is conjugated to a lysine residue on the substrate protein (48, 49), we individually mutated 12 lysines in Bcl10 to arginine in the context of FLAG-Bcl10 and assayed the mutants for linear ubiquitinylation after retrovirus-mediated stable expression in Jurkat T cells. We found that the mutation of Lys-17, Lys-31, or Lys-63 reduced the extent of Lin(Ub)n-Bcl10 generation following PMA/iono treatment, although the mutation of other lysines had no significant effect (Fig. 6A). Importantly, the K17R, K31R, and K63R Bcl10 mutants were fully competent to bind to both CARD11ΔID (Fig. 6B) and MALT1 (Fig. 6C) after co-expression in HEK293T cells, indicating that these lysine-to-arginine mutations did not affect Bcl10 folding or the interaction with its other known signaling partners.
FIGURE 6.
NF-κB activation by TCR signaling requires the linear ubiquitinylation of Bcl10. A, Jurkat T cells (1 × 108/sample) expressing the indicated FLAG-Bcl10 variants were stimulated with PMA/iono for 30 min, and cell lysates were subjected to pulldown with recombinant GST-NEMO-UBAN2 protein, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. B, HEK293T cells (1 × 106/sample) were co-transfected with myc-CARD11 ΔID (200 ng) and the indicated FLAG-Bcl10 variants (100 ng). The cell lysates were immunoprecipitated (IP) and analyzed by Western blotting (WB) with the indicated antibodies. C, HEK293T cells (1 × 106/sample) were co-transfected with HA-MALT1 (500 ng) and the indicated FLAG-Bcl10 variants (100 ng). The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies. D, Bcl10-deficient (Bcl10-KO) Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC and the indicated FLAG-Bcl10 variants (100 ng each). After about 40 h, the cells were stimulated with or without PMA/iono for 5 h, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. ***, p < 0.001; **, p < 0.01. E, lysates from WT or the Bcl10-KO Jurkat T cells used in D were resolved by SDS-PAGE and analyzed by Western blotting with the indicated antibodies. F, lysates from Bcl10-deficient Jurkat T cells (Bcl10-KO) and derivative lines stably expressing the indicated FLAG-Bcl10 variants were resolved by SDS-PAGE and analyzed by Western blotting with anti-Bcl10 antibody. G, Bcl10-KO Jurkat T cells stably reconstituted with the indicated FLAG-Bcl10 variants (1 × 108/sample) were stimulated with or without PMA/iono for 30 min, and lysates were immunoprecipitated under denaturing conditions with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. H, Bcl10-KO Jurkat T cells stably reconstituted with the indicated FLAG-Bcl10 variants were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC and stimulated with or without the indicated dose of anti-CD3/anti-CD28 for 5 h, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. I, WT or HOIP-KO Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC and stimulated with or without the indicated dose of anti-CD3/anti-CD28 for 5 h, and then luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. The Bcl10-deficient cells used in D and E were generated using TALEN technology, and the Bcl10-deficient cells used in F–H were generated using CRISPR/Cas9 technology.
We next tested whether these mutants could reconstitute TCR signaling to NF-κB in Jurkat T cells that had been made Bcl10-deficient by TALEN-mediated gene editing. PMA/iono treatment of these Bcl10-deficient Jurkat T cells failed to activate the Igκ2-IFN-LUC reporter, but signaling could be rescued upon transient expression of wild-type Bcl10 (Fig. 6, D and E). Transient expression of the K17R, K31R, or K63R Bcl10 mutants resulted in impaired Igκ2-IFN-LUC reporter activation by PMA/iono treatment (Fig. 6D), in contrast to the other mutants tested, suggesting a correlation between the linear ubiquitinylation of Bcl10 and the activation of NF-κB downstream of PMA/ionomycin treatment. However, Bcl10 residues Lys-31 and Lys-63 have also been reported to be required for the conjugation of Bcl10 with Lys-63-linked ubiquitin chains (46), although the contribution of Lys-17 to this process has not been previously addressed. To address whether Lys-17 was specifically required for linear and not Lys-63-linked ubiquitinylation of Bcl10, we reconstituted Bcl10-deficient Jurkat T cells with stable expression of Bcl10 K17R (Fig. 6F) and assayed its ability to be inducibly modified with polyubiquitin chains in comparison with wild-type Bcl10 and the K31R and K63R mutants. As shown in Fig. 6G, the K17R mutation significantly impaired the generation of both Lin(Ub)n-Bcl10 and Lys-63(Ub)n-Bcl10, similar to the effects of the K31R and K63R substitutions. We next used these Bcl10-reconstituted Jurkat T cells to assess the quantitative contribution of these Bcl10 modifications to NF-κB activation following a dose response of TCR cross-linking. The K17R mutation led to a 61–68% drop in TCR signaling to NF-κB; the K31R mutation reduced signaling by 63–69%, and the K63R mutation reduced signaling by 48–53% under these conditions (Fig. 6H). The deletion of the HOIP subunit of LUBAC assayed after the same dose response resulted in a 51–68% drop in TCR signaling to NF-κB (Fig. 6I), an effect similar in extent to that observed with the Lys-17, Lys-31, and Lys-63 mutations of Bcl10. The fact that the lysine mutations, which affect both Lin(Ub)n-Bcl10 and Lys-63(Ub)n-Bcl10, had effects comparable with the deletion of HOIP, which is a linear ubiquitin-specific E3 ligase, argues in favor of Lin(Ub)n-Bcl10 being the more critical signaling intermediate for TCR-mediated NF-κB activation. This notion is consistent with our observation (Fig. 1E) that most, if not all, of the polyubiquitinylated Bcl10 that binds to NEMO during signaling is Lin(Ub)n-Bcl10 and not Lys-63(Ub)n-Bcl10.
Oncogenic CARD11 Mutants Spontaneously Induce Linear Ubiquitinylation of Bcl10 in a Manner That Correlates with Signaling Output
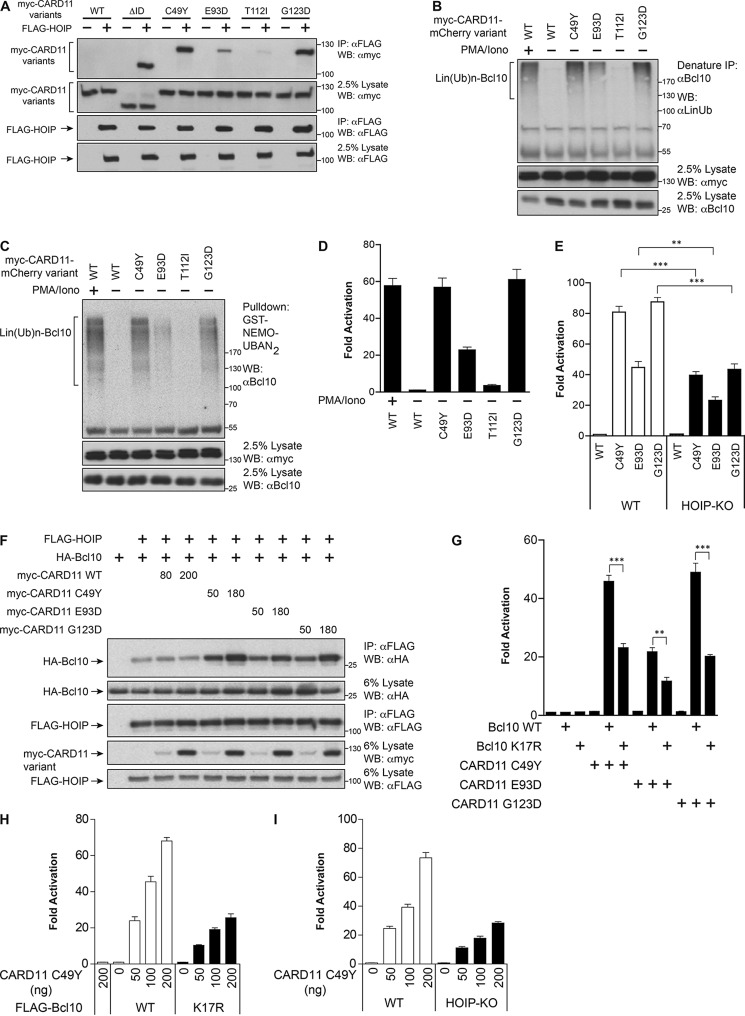
We have previously shown that many oncogenic CARD11 variants, associated with ABC DLBCL, have an enhanced ability to bind Bcl10 due to the fact that their gain-of-function mutations interfere with autoinhibition mediated by the ID (30, 31). We tested whether oncogenic mutations also confer enhanced binding to HOIP. We used four hyperactive alleles that display a range of signaling potency, C49Y, E93D, T112I, and G123D (31). After co-expression in HEK293T cells, each of these variants associated with HOIP at concentrations that the wild-type CARD11 did not (Fig. 7A). Their constitutive ability to bind both Bcl10 (31) and HOIP suggested that they might spontaneously promote the action of HOIP on Bcl10 and the generation of Lin(Ub)n-Bcl10, even in the absence of receptor signaling. To test this, we stably expressed these mutants as fusions to mCherry in Jurkat T cells using retroviral transduction. As shown in Fig. 7B, the oncogenic CARD11 variants did indeed elicit the spontaneous generation of Lin(Ub)n-Bcl10. In addition, the Lin(Ub)n-Bcl10 generated by each of these hyperactive variants was recognized by the UBAN domain of NEMO, as indicated by their association with the GST-NEMO-UBAN2 fusion protein (Fig. 7C). Furthermore, the steady state level of Lin(Ub)n-Bcl10 observed in the presence of each mutant correlated well with the degree of NF-κB activation elicited by each mutant, as measured by induction of the Igκ2-IFN-LUC reporter (Fig. 7D). To confirm a functional role for HOIP in oncogenic CARD11 signaling, we transiently expressed these mutants in HOIP-deficient and control Jurkat T cells. As predicted, the activity of each CARD11 variant was significantly reduced in the absence of HOIP (Fig. 7E).
FIGURE 7.
Oncogenic CARD11 mutants spontaneously induce the linear ubiquitinylation of Bcl10 in a manner that correlates with signaling output. A, HEK293T cells (1 × 106/sample) were co-transfected with FLAG-HOIP (100 ng) and the indicated myc-CARD11 variants (150 ng). The cell lysates were immunoprecipitated (IP) and analyzed by Western blotting (WB) with the indicated antibodies. B, Jurkat T cells (1 × 108/sample) stably expressing the indicated myc-CARD11-mCherry variants were stimulated with or without PMA/iono for 30 min, and cell lysates were immunoprecipitated (IP) with anti-Bcl10 antibody, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. C, Jurkat T cells (1 × 108/sample) stably expressing the indicated myc-CARD11-mCherry variants were stimulated with or without PMA/iono for 30 min, and cell lysates were subjected to pulldown with recombinant GST-NEMO-UBAN2 protein, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. D, Jurkat T cells stably expressing the indicated myc-CARD11-mCherry variants were transiently transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC. After about 40 h, the cells were stimulated with or without PMA/iono for 5 h, and then luciferase reporter assays were performed. E, WT or HOIP-KO Jurkat T cells were transfected with 200 ng of CSK-LacZ, 1500 ng of Igκ2-IFN-LUC, and 200 ng of the indicated CARD11 variants. After about 45 h, the cells were lysed, and luciferase assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. ***, p < 0.001; **, p < 0.01. F, HEK293T cells (1 × 106/sample) were co-transfected with 100 ng of FLAG-HOIP expression vector, 200 ng of HA-Bcl10, and the indicated amounts of the indicated CARD11 variants. The cell lysates were immunoprecipitated and analyzed by Western blotting with the indicated antibodies. G, Bcl10-KO Jurkat T cells were transfected with 200 ng of CSK-LacZ, 1500 ng of Igκ2-IFN-LUC, 100 ng of the indicated FLAG-Bcl10 variants (100 ng each), and 200 ng of the indicated myc-CARD11 variants. After about 45 h, the cells were lysed, and luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. ***, p < 0.001; **, p < 0.01. H, Bcl10-KO Jurkat T cells stably reconstituted with the indicated FLAG-Bcl10 variants were transfected with 200 ng of CSK-LacZ, 1500 ng of Igκ2-IFN-LUC, and the indicated amounts of CARD11 C49Y expression vector, and after 45 h, luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate. I, WT or HOIP-KO Jurkat T cells were transfected with 200 ng of CSK-LacZ and 1500 ng of Igκ2-IFN-LUC and the indicated amounts of CARD11 C49Y expression vector, and after 45 h, luciferase reporter assays were performed. Each graph represents the mean ± S.D. of three independent experiments done in triplicate.
We next tested whether the C49Y, E93D, and G123D oncogenic CARD11 variants could promote the physical association between HOIP and Bcl10. After co-expression in HEK293T cells, each of these hyperactive CARD11 variants could stimulate the amount of HA-Bcl10 that co-immunoprecipitated with FLAG-HOIP, although wild-type CARD11 could not (Fig. 7F).
We then used Bcl10-deficient Jurkat T cells to test whether Bcl10 Lys-17, which is required for both Lin(Ub)n-Bcl10 and Lys-63(Ub)n-Bcl10 generation, was also required for NF-κB activation by oncogenic CARD11 variants. As shown in Fig. 7G, each CARD11 variant tested displayed greater signaling to NF-κB in the presence of wild-type Bcl10 than in the presence of Bcl10 K17R. We then used a dose response of CARD11 C49Y expression to gauge the quantitative contribution of Bcl10 polyubiquitinylation and HOIP activity to oncogenic CARD11 signaling. As shown in Fig. 7H, the reconstitution of Bcl10-deficient cells with the K17R mutant resulted in a drop in oncogenic C49Y signaling of 57–62%. Similarly, the absence of HOIP led to a 56–61% drop in C49Y-mediated activation of NF-κB (Fig. 7I). The results support the conclusion that oncogenic CARD11 signaling proceeds maximally through HOIP-dependent polyubiquitinylation of Bcl10.
Discussion
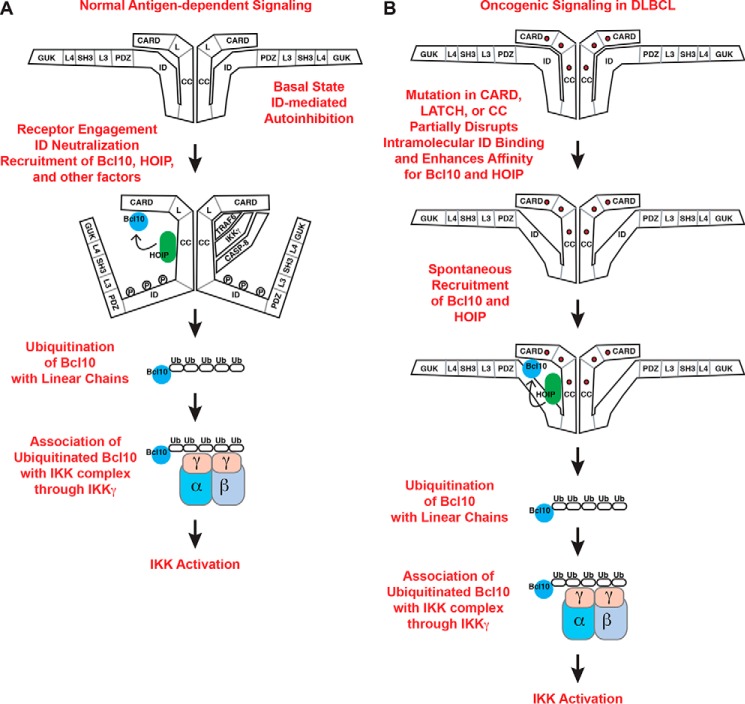
Our findings highlight Lin(Ub)n-Bcl10 as an important intermediate in TCR signaling to NF-κB and reveal several molecular determinants that allow CARD11 to promote HOIP action on Bcl10 in a signal-regulated manner (Fig. 8A). Prior to TCR engagement, the REs in the CARD11 ID prevent the association of Bcl10 (17) and HOIP to CARD11. Upon receptor triggering, the autoinhibitory roles of the REs are neutralized, allowing HOIP recruitment to CARD11 in an interaction that involves the CARD11 coiled-coil and two separate regions of HOIP, one containing Ring Finger 1 in the RBR domain, and another containing the two Ndl4-like zinc fingers. As previous studies have shown (5, 15, 47), Bcl10 is concomitantly recruited to CARD11 in a manner that requires both the CARD and coiled-coil domains of CARD11 and the CARD domain of Bcl10. The co-recruitment of HOIP and Bcl10 to CARD11 allows action of the enzyme on its substrate, leading to the generation of Lin(Ub)n-Bcl10, which subsequently associates with the NEMO subunit of the IKK kinase complex through its UBAN domain.
FIGURE 8.
Models depicting the CARD11-promoted linear ubiquitinylation of Bcl10 during normal (A) and oncogenic (B) signaling.
During TCR signaling, Bcl10 is conjugated with both Lys-63-linked ubiquitin chains to produce Lys-63(Ub)n-Bcl10 (46) and with linear ubiquitin chains to produce Lin(Ub)n-Bcl10 (this study). Our data suggest that these are separate species and that Bcl10 is not appreciably conjugated with mixed Lys-63-linked and Met-1-linked chains, because OTULIN, and not AMSH-LP, cleaves the Lin(Ub)n-Bcl10 species that we detect. It has been previously shown that the induced generation of Lys-63(Ub)n-Bcl10 requires CARD11, MALT1, and Bcl10 lysines Lys-31 and Lys-63 (46), and our results indicate that these same determinants, in addition to Bcl10 lysine 17, are also required for TCR induced Lin(Ub)n-Bcl10 generation. However, our analyses with linkage-specific deubiquitinases clearly indicate that it is predominantly Lin(Ub)n-Bcl10, not Lys-63(Ub)n-Bcl10, that associates with NEMO. The fact that mutation of Lys-17, Lys-31, and Lys-63 on Bcl10 has effects on NF-κB activation that are quantitatively comparable with that achieved by the knock-out of HOIP, the linear linkage-specific E3 ligase, supports the notion that it is Lin(Ub)n-Bcl10 that is important in signaling, although other interpretations of the data are possible. Several previous studies that have used the effect of Lys-31 and Lys-63 Bcl10 mutations to conclude an important functional role for Lys-63(Ub)n-Bcl10 in signaling (42, 46, 50, 51), including some from our own group (30), must be reevaluated in the light of our finding that these residues are also required for Lin(Ub)n-Bcl10. The functional effects observed in these studies may have been due to Lin(Ub)n-Bcl10 and not necessarily solely to Lys-63(Ub)n-Bcl10.
Our data also establish that several hyperactive oncogenic CARD11 variants found in ABC DLBCL spontaneously induce the generation of Lin(Ub)n-Bcl10 even in the absence of upstream signaling (Fig. 8B). These mutations enhance the association of CARD11 with both HOIP and Bcl10 (31), allowing the association between enzyme and substrate to occur without antigen receptor-induced neutralization of RE inhibitory function. Importantly, for the oncogenic variants we examined, the extent to which they spontaneously bind HOIP and Bcl10 (31) correlates with the steady-state levels of Lin(Ub)n-Bcl10 that they produce, the amount of conjugated Bcl10 that associates with NEMO, and with the signaling potential of each variant to activate NF-κB. Because the potential to activate NF-κB can determine the growth and survival of ABC DLBCL lymphoma cells (24, 31), it is highly likely that the quantitative ability of an oncogenic variant to generate Lin(Ub)n-Bcl10 could reflect its relative growth-promoting potential in lymphoma.
It is important to point out that our analyses in HOIP-KO cells and in Bcl10-KO cells reconstituted with lysine mutants indicate that HOIP activity and Bcl10 polyubiquitin modification only account for up to 69% of the normal TCR signaling pathway to NF-κB and only up to 62% of the signaling output of oncogenic CARD11. The deletion of Bcl10, MALT1, and CARD11 each results in a larger diminution of signaling, making it clear that other steps in signaling that are Bcl10-, CARD11-, and MALT1-dependent are critical for determining signaling output. The residual signaling that does not depend on HOIP may explain why some reports (39, 40) have not seen a dependence on HOIP catalytic activity on signaling, if their assays of activation were not performed under subsaturating conditions or measured outputs that are produced at equivalent levels beyond a certain threshold of pathway induction.
It is interesting that TCR signaling to NF-κB depends on the HOIP subunit of LUBAC but not on SHARPIN. The LUBAC complex involved in this TCR pathway must therefore function differently in the context of TNFα, IL-1, and CD40L signaling, which have been shown to be SHARPIN-dependent (33–35). Although many cell types contain a LUBAC complex containing HOIP, HOIL-1L, and SHARPIN, several studies have shown that HOIP·HOIL-1L and HOIP·SHARPIN complexes are sufficient to display LUBAC activity in vitro (33–35, 48) and that the binding of either HOIL-1L or SHARPIN to HOIP can relieve autoinhibition conferred by domains of HOIP that lie N-terminal to the RBR catalytic core (52–54). Because CARD11 can associate with the region in HOIP that contains the Npl4-type zinc fingers, as well as with a region that contains Ring Finger 1 in the RBR, it is possible that CARD11 binding relieves HOIP autoinhibition and obviates the need for SHARPIN in the TCR pathway. Further experiments will be required to explore this possibility. We were unable to isolate Jurkat T cells with complete knock-out of HOIL-1L and therefore could not evaluate whether HOIL-1L is required during TCR signaling. However, recent studies have shown that a key target of MALT1 proteolytic activity during antigen receptor signaling is the HOIL-1L protein and that MALT1-mediated cleavage of HOIL-1L attenuates antigen receptor induction of LUBAC activity (55, 56), which clearly implicates a role for HOIL-1L in supporting HOIP catalytic activity during antigen receptor signaling to NF-κB.
Lin(Ub)n-Bcl10 emerges from our studies as a signaling intermediate whose levels reflect the extent of TCR and oncogenic CARD11 signaling to NF-κB. Yang et al. (42) recently reported that inhibitors of cIAPs can prevent the generation of Lin(Ub)n-Bcl10 and NF-κB activation in ABC DLBCL samples that exhibit chronic BCR signaling. However, ABC DLBCL cells containing an oncogenic CARD11 allele were not affected by cIAP inhibitors (42), indicating that oncogenic CARD11 mutations that promote HOIP action on Bcl10, and spontaneously generate Lin(Ub)n-Bcl10 without upstream BCR signaling, bypass the need for cIAP activity in signaling to NF-κB. Therapeutic agents that directly target the HOIP-mediated generation of Lin(Ub)n-Bcl10 may be especially useful for reducing the levels of dysregulated constitutive NF-κB activity associated with cases of ABC DLBCL that harbor oncogenic CARD11 mutations.
Experimental Procedures
Cell Lines
HEK293T cells and Jurkat T cells were obtained from American Tissue Culture Collection and cultured as described previously (57).
Expression Constructs
CARD11 constructs, including CARD11ΔID and double deletion constructs (15), and constructs encoding oncogenic CARD11 mutants (31) and RE mutants (16, 17) have been previously described. pGEX4T-1-NEMO-UBAN (amino acids 242–350) was kindly provided by Jonathan D. Ashwell (NCI, National Institutes of Health, Bethesda), and this vector was modified by standard cloning techniques to generate pGEX4T-1-NEMO-UBAN2. FLAG-HOIP WT was a gift from Jae U. Jung (Keck School of Medicine, University of Southern California, Los Angeles), and the deletion mutants of FLAG-HOIP were cloned by standard molecular biology techniques. Mammalian expression plasmids for HA-tagged Bcl10 and MALT1 were constructed using standard techniques in the pRK7 vector. The Bcl10 KR mutants and HOIP-C885S mutant were created using QuikChange site-directed mutagenesis (Stratagene) of pMSCVpuro-FLAG-Bcl10 (Addgene plasmid 18718) and pCMV3FLAG8HOIP (Addgene plasmid 50015) vectors, respectively.
Antibodies
Antibodies used were purchased as follows: mouse monoclonal antibody against Bcl10 (331.3, sc-5273), rabbit polyclonal antibody against Bcl10 (H-197, sc-5611), rabbit polyclonal antibody against NEMO/IKKγ (F-419, sc-8330), mouse monoclonal antibody against c-Myc (9E10, sc-40), and mouse monoclonal antibody against HA (F-7, sc-7392) were purchased from Santa Cruz Biotechnology. Mouse monoclonal antibody against linear ubiquitin (clone LUB9, MABS451) was obtained from EMD Millipore. Mouse monoclonal antibody against Lys-63-linked ubiquitin (clone HWA4C4, 14-6077) was purchased from eBioscience. Rabbit polyclonal antibody against CARD11/CARMA1 was from ProSci Inc. Rabbit monoclonal antibody against MALT1 was from Abcam (ab33921). Rabbit polyclonal antibody against HOIP/RNF31 (A303-560) was purchased from Bethyl Laboratories Inc. Mouse monoclonal antibody against HOIP/RNF31 (MAB8039) was obtained from R&D Systems. Rabbit polyclonal antibody against SHARPIN (14626-1-AP) was purchased from Proteintech Group. Rabbit antibody against FLAG epitopes (Sigma F7425) and mouse antibodies against FLAG epitopes (M2, Sigma F1804) were from Sigma.
Recombinant Proteins
The linear tetra-ubiquitin (UC-710), the Lys-63-linked tetra-ubiquitin (UC-310), and the OTULIN recombinant protein (E-558) were obtained from BostonBiochem (Cambridge, MA). The AMSH-LP recombinant protein (pro-1354) was purchased from Protein Specialists Ltd. GST and GST-NEMO-UBAN2 proteins were expressed in Escherichia coli and purified as described previously (58).
Generation of HOIP-KO, CARD11-KO, Bcl10-KO, MALT1-KO, and SHARPIN-KO Jurkat T Cell Lines by CRISPR/Cas9-mediated Genome Editing
The lentiCRISPRv1 (Addgene plasmid 49535) vector was engineered to express guide RNAs targeting HOIP (5′-GAGAGCTGGCTAGTAGCGGC-3′), CARD11 (5′-GCGTCAGTGTAAGGTCATTG-3′), Bcl10 (5′-TCGGTGAGGGACGGTGCGGT-3′), MALT1 (5′-GCTGTTGGGGGACCCGCTAC-3′), or SHARPIN (5′-TGGCTGTGCACGCCGCGGTG-3′) by standard techniques. To package the lentiviruses, HEK293T cells were plated at 1 × 105 cells per well in a 24-well plate and transfected 24 h later using the calcium phosphate method with 116 ng of psPAX2 and 78 ng of pCMV-VSVg, and 156 ng of lentiCRISPR-based vector. The medium was replaced with 500 μl of RPMI 1640 medium 24 h post-transfection. Supernatants containing viral particles were harvested 48 h post-transfection after cell debris was cleared by centrifugation at 5000 rpm for 5 min. Viral supernatant (400 μl) was added to 5 × 106 Jurkat T cells in a final volume of 2.4 ml in a 6-well plate. Approximately 24 h postinfection, cells were resuspended in fresh RPMI 1640 medium containing 0.5 μg/ml puromycin and selected for 10 days. Single clones were isolated by diluting cells in 96-well plates to 0.5 cells per well in 100 μl. Gene knock-out was assessed by Western blotting analysis using anti-HOIP, anti-CARD11, anti-Bcl10, anti-MALT1 or anti-SHARPIN antibody.
Reconstitution of Bcl10-KO Jurkat T Cells with Bcl10 Variants
Bcl10 variants were expressed in the context of the pCLIB3B retroviral vector, a Moloney murine leukemia virus-based vector carrying blasticidin resistance. To package the retroviruses, HEK293T cells were plated at 1 × 105 cells per well in a 24-well plate and transfected 24 h later using the calcium phosphate method with 90 ng of pCL-SIN-Ampho, 90 ng of pCMV-VSVg, and 230 ng of retroviral vector. The medium was replaced with 500 μl of RPMI 1640 medium 24 h post-transfection. Supernatants containing viral particles were harvested 48 h post-transfection after cell debris was cleared by centrifugation at 5000 rpm for 5 min. Viral supernatant (400 μl) was added to 5 × 106 Bcl10-KO Jurkat T cells in a final volume of 2.4 ml in a 6-well plate. Approximately 24 h postinfection, cells were resuspended in fresh RPMI 1640 medium containing 0.5 μg/ml puromycin and 3 μg/ml blasticidin and selected for 2 weeks. Puromycin- and blasticidin-resistant Jurkat T cell lines were maintained in media containing 0.5 μg/ml puromycin and 3 μg/ml blasticidin.
Generation of Bcl10-KO Jurkat T Cell Lines by TALEN-mediated Genome Editing
A pair of TALENs was designed to target a region in the first coding exon of the human Bcl10 gene (ensemble ID, ENSG00000142867). The TALEN binding sequences were 5′-GGAGCCCACCGCACC-3′ and 5′-CACTTCAGTGAGGTCC-3′ and were positioned with a 15-bp spacer, 5′-GTCCCTCACCGAGGA-3′, in-between. The TALENs were cloned into the pC-GoldyTALEN vector (Addgene plasmid 38143) and then transfected into Jurkat T cells using Lipofectamine 2000. 10 days later, single clones were isolated by diluting cells in 96-well plates to 0.5 cells per well. Bcl10 knock-out was assessed by Western blotting analysis using anti-Bcl10 antibody.
Transient Transfections and NF-κB Luciferase Reporter Assay of Jurkat T Cells
Jurkat T cells were plated in 6-well plates at 5 × 105 cells/ml and 2 ml/well. LT-1 (Mirus) was used with 3 μg of total DNA per manufacturer's instructions. For the NF-κB luciferase assay, Jurkat T cells were transfected with the indicated vectors plus 200 ng of pCSK-LacZ and 1500 ng of Igκ2-IFN-LUC. In each experiment, each sample was supplemented with empty parental expression vector to keep the total amount of expression vector constant. Approximately 40 h after transfection, cells were stimulated in 1 ml of media alone or with 1 μg/ml each of mouse anti-human CD3 (Pharmingen 555329), mouse anti-human CD28 (Pharmingen 555725), anti-mouse IgG1 (Pharmingen 02231D), or with 50 ng/ml PMA (Sigma) and 1 μm ionomycin (Sigma). Luciferase and β-galactosidase activities were determined, and fold activation was calculated as described previously (15).
Denatured Co-immunoprecipitation
Jurkat T cells were treated with or without 1 μg/ml each of mouse anti-human CD3 (Pharmingen 555329), mouse anti-human CD28 (Pharmingen 555725), anti-mouse IgG1 (Pharmingen 02231D), or with 50 ng/ml PMA (Sigma) and 1 μm ionomycin (Sigma) for the indicated times at 37 °C. Cells (1 × 108/sample) were lysed in Den-IP lysis buffer (DIPLB) (20 mm Tris·HCl, pH 7.5, 150 mm NaCl, 30 mm NaF, 2 mm Na4P2O7, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 5 mm N-ethylmaleimide, 1:1000 protease inhibitor mixture (Sigma P8340)). For analyzing Bcl10 polyubiquitination, SDS was added to the cell lysates to a concentration of 1%, and the lysates were heated at 95 °C for 5 min, diluted with DIPLB to a final concentration of 0.1% SDS, and incubated with anti-Bcl10 antibody for 12 h at 4 °C with rotation, followed by incubation with protein G-conjugated Sepharose for 2 h at 4 °C with rotation. The immunoprecipitates and cell lysate input samples were boiled in SDS loading buffer, resolved by SDS-PAGE, and transferred to PVDF membranes for Western blotting with the indicated antibodies. Mouse CD4+ T cells were purified from the spleen of B6 mice using a CD4+ T cell isolation kit (Miltenyi Biotec). Purified mouse CD4+ T cells (7.5 × 107/sample) were stimulated with or without 50 ng/ml PMA (Sigma) and 1 μm iono (Sigma) for 30 min at 37 °C, prior to lysis with DIPLB and analysis as described above.
Immunoprecipitations in HEK293T Cells
At 1 day prior to transfection, 5 × 105 HEK293T cells were plated in each well of a 6-well plate. A total of 2 μg of DNA per well was transfected using the calcium phosphate method. The medium was changed 24 h post-transfection, and the cells were harvested about 40 h post-transfection. Cells were lysed in 500 μl of immunoprecipitation lysis buffer (IPLB, 150 mm NaCl, 50 mm HEPES, pH 7.9, 1 mm EDTA, 10% glycerol, 1% Igepal) and incubated 20 min on ice, and debris was cleared by centrifugation at 13,000 rpm at 4 °C. Cell lysate (30 μl) was saved for Western blotting analysis, and 450 μl was incubated with 1 μg of anti-FLAG antibody (Sigma F7425) for 4 h at 4 °C with rotation. A 10-μl bed volume of protein G-Sepharose 4 Fast Flow (GE Healthcare) was added, and the mixture was incubated for 2 h at 4 °C with rotation. The resulting immunocomplex was washed with IPLB four times for 5 min at 4 °C with rotation, eluted twice with FLAG peptide (Sigma), pooled, and resolved for Western blotting analysis using the indicated antibodies.
Treatment with Deubiquitylase
The ubiquitinylated Bcl10 captured by GST-NEMO-UBAN2, anti-Bcl10, or anti-NEMO antibody was washed three times with 1 ml of DUB-lysis buffer (20 mm Tris·HCl, pH 7.5, 150 mm NaCl, 30 mm NaF, 2 mm Na4P2O7, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 5 mm N-ethylmaleimide, 1:1000 protease inhibitor mixture (Sigma P8340)) and once with 1 ml of wash buffer (50 mm Tris·HCl, pH 7.5, 50 mm NaCl, and 5 mm DTT). Following the last wash, the beads were resuspended in 30 μl of reaction buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT, 1 mm MnCl2, 0.01% (w/v) Brij-35) with or without OTULIN (1 μm) or AMSH-LP (5 μm). After incubation for 60 min at 30 °C, incubations were subjected to SDS-PAGE, and the protein gels were transferred to PVDF membranes and immunoblotted with the appropriate antibodies.
Stable Expression of myc-CARD11 Variants and FLAG-Bcl10 Variants in Jurkat T Cells
CARD11 and Bcl10 variants were expressed in the context of pCLIP3A-myc-CARD11-mCherry (31) and pMSCVpuro-FLAG-Bcl10 (46) retroviral vectors, respectively. To package the retroviruses, HEK293T cells were plated at 1 × 105 cells per well in a 24-well plate and transfected 24 h later using the calcium phosphate method with 90 ng of pCL-SIN-Ampho, 90 ng of pCMV-VSVg, and 230 ng of retroviral vector. The medium was replaced with 500 μl of RPMI 1640 medium 24 h post-transfection. Supernatants containing viral particles were harvested 48 h post-transfection after cell debris was cleared by centrifugation at 5000 rpm for 5 min. Viral supernatant (400 μl) was added to 5 × 106 Jurkat T cells in a final volume of 2.4 ml in a 6-well plate. Approximately 24 h postinfection, cells were resuspended in fresh RPMI 1640 medium containing 0.5 μg/ml puromycin and selected for 2 weeks. Puromycin-resistant Jurkat T cell lines were maintained in media containing 0.5 μg/ml puromycin.
Glutathione-Sepharose Pulldowns
Mouse CD4+ T cells were purified from the spleens of B6 mice using a CD4+ T cell isolation kit (Miltenyi Biotec). Purified mouse CD4+ T cells (7 × 107/sample) or Jurkat T cells (1 × 108/sample) were stimulated with or without 50 ng/ml PMA (Sigma) and 1 μm iono (Sigma) for 30 min at 37 °C. Cells were lysed in IPLB. Cell lysates were supplemented with 1 μg of purified GST protein or GST-NEMO-UBAN2 protein and then incubated for 12 h at 4 °C with rotation, followed by incubation with a 20-μl bed volume of glutathione-Sepharose 4B beads (Amersham Biosciences) for 2 h at 4 °C with rotation. The bead-bound complexes were washed with IPLB four times for 5 min at 4 °C with rotation, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies.
Endogenous Co-immunoprecipitations in Jurkat T Cells
6 × 107 Jurkat T cells/sample were stimulated for the indicated times and then centrifuged for 5 min at 4600 rpm. Cell pellets were resuspended in 1.2 ml of IP Lysis Buffer, incubated for 20 min on ice, and cell debris was cleared by centrifugation at 13,000 rpm 4 °C. Lysates were then precleared by incubating with 7-μl bed volume protein G-Sepharose (Amersham Biosciences) twice for 30 min at 4 °C with rotation. A 30-μl aliquot was removed for input analysis, and the remaining lysate was incubated with rotation overnight at 4 °C with 2 μg of the indicated antibodies. The next day, protein G-Sepharose (10-μl bed volume) preblocked with 1% human insulin was added to the samples and incubated for 1 h at 4 °C with rotation. The beads were then washed with rotation four times for 5 min at 4 °C with IP Lysis Buffer and then resolved by SDS-PAGE. Immunoprecipitates were analyzed by Western blotting using the indicated antibodies.
Statistical Analysis
All experiments presented have been performed a minimum of three times. A two-tailed unpaired Student's t test with unequal variance was used to analyze data, and p values of <0.05 were considered significant.
Author Contributions
Y. K. Y. and J. L. P. designed and interpreted the experiments. Y. K. Y., C. Y., W. C., Z. W., and K. E. D. performed the experiments. Y. K. Y. and J. L. P. wrote the manuscript with input from all authors.
Acknowledgments
We thank all the members of the Pomerantz laboratory and Mollie Meffert for helpful discussions and advice.
Note Added in Proof
In Fig. 4E, an incorrect immunoblot was inadvertently used for Bcl10, 2.5% lysate in the version of this manuscript that was published on October 24, 2016. This error has now been corrected.
This work was supported in part by Quest for Cures Grant 0793-14 from the Leukemia and Lymphoma Society, Grant RO1CA177600 from the National Institutes of Health, and funds from The Johns Hopkins University Institute for Cell Engineering. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCR
- T cell receptor
- BCR
- B cell receptor
- IKK
- IκB kinase
- ID
- inhibitory domain
- LUBAC
- linear ubiquitin chain assembly complex
- ABC
- activated B cell-like
- DLBCL
- diffuse large B cell lymphoma
- Ub
- ubiquitin
- PMA
- phorbol 12-myristate 13-acetate
- iono
- ionomycin
- RE
- repressive element
- cIAP
- cellular inhibitor of apoptosis.
References
- 1. Schulze-Luehrmann J., and Ghosh S. (2006) Antigen-receptor signaling to nuclear factor κB. Immunity 25, 701–715 [DOI] [PubMed] [Google Scholar]
- 2. Vallabhapurapu S., and Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 3. Turvey S. E., Durandy A., Fischer A., Fung S. Y., Geha R. S., Gewies A., Giese T., Greil J., Keller B., McKinnon M. L., Neven B., Rozmus J., Ruland J., Snow A. L., Stepensky P., and Warnatz K. (2014) The CARD11-BCL10-MALT1 (CBM) signalosome complex: stepping into the limelight of human primary immunodeficiency. J. Allergy Clin. Immunol. 134, 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pérez de Diego R., Sánchez-Ramón S., López-Collazo E., Martínez-Barricarte R., Cubillos-Zapata C., Ferreira Cerdán A., Casanova J. L., and Puel A. (2015) Genetic errors of the human caspase recruitment domain-B-cell lymphoma 10-mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (CBM) complex: molecular, immunologic, and clinical heterogeneity. J. Allergy Clin. Immunol. 136, 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaide O., Favier B., Legler D. F., Bonnet D., Brissoni B., Valitutti S., Bron C., Tschopp J., and Thome M. (2002) CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat. Immunol. 3, 836–843 [DOI] [PubMed] [Google Scholar]
- 6. Wang D., You Y., Case S. M., McAllister-Lucas L. M., Wang L., DiStefano P. S., Nuñez G., Bertin J., and Lin X. (2002) A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3, 830–835 [DOI] [PubMed] [Google Scholar]
- 7. Pomerantz J. L., Denny E. M., and Baltimore D. (2002) CARD11 mediates factor-specific activation of NF-κB by the T cell receptor complex. EMBO J. 21, 5184–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hara H., Wada T., Bakal C., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E. K., Krawczyk C., Bauer B., D'Acquisto F., Ghosh S., Yeh W. C., Baier G., Rottapel R., and Penninger J. M. (2003) The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18, 763–775 [DOI] [PubMed] [Google Scholar]
- 9. Jun J. E., Wilson L. E., Vinuesa C. G., Lesage S., Blery M., Miosge L. A., Cook M. C., Kucharska E. M., Hara H., Penninger J. M., Domashenz H., Hong N. A., Glynne R. J., Nelms K. A., and Goodnow C. C. (2003) Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18, 751–762 [DOI] [PubMed] [Google Scholar]
- 10. Egawa T., Albrecht B., Favier B., Sunshine M. J., Mirchandani K., O'Brien W., Thome M., and Littman D. R. (2003) Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 13, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 11. Newton K., and Dixit V. M. (2003) Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 13, 1247–1251 [DOI] [PubMed] [Google Scholar]
- 12. Stepensky P., Keller B., Buchta M., Kienzler A. K., Elpeleg O., Somech R., Cohen S., Shachar I., Miosge L. A., Schlesier M., Fuchs I., Enders A., Eibel H., Grimbacher B., and Warnatz K. (2013) Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J. Allergy Clin. Immunol. 131, 477–485 [DOI] [PubMed] [Google Scholar]
- 13. Greil J., Rausch T., Giese T., Bandapalli O. R., Daniel V., Bekeredjian-Ding I., Stütz A. M., Drees C., Roth S., Ruland J., Korbel J. O., and Kulozik A. E. (2013) Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J. Allergy Clin. Immunol. 131, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 14. Hayden M. S., and Ghosh S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCully R. R., and Pomerantz J. L. (2008) The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-κB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol. Cell. Biol. 28, 5668–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jattani R. P., Tritapoe J. M., and Pomerantz J. L. (2016) Cooperative control of caspase recruitment domain-containing protein 11 (CARD11) signaling by an unusual array of redundant repressive elements. J. Biol. Chem. 291, 8324–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jattani R. P., Tritapoe J. M., and Pomerantz J. L. (2016) Intramolecular interactions and regulation of cofactor binding by the four repressive elements in the caspase recruitment domain-containing protein 11 (CARD11) inhibitory domain. J. Biol. Chem. 291, 8338–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sommer K., Guo B., Pomerantz J. L., Bandaranayake A. D., Moreno-García M. E., Ovechkina Y. L., and Rawlings D. J. (2005) Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 23, 561–574 [DOI] [PubMed] [Google Scholar]
- 19. Matsumoto R., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Wang D., and Lin X. (2005) Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity 23, 575–585 [DOI] [PubMed] [Google Scholar]
- 20. Bidère N., Ngo V. N., Lee J., Collins C., Zheng L., Wan F., Davis R. E., Lenz G., Anderson D. E., Arnoult D., Vazquez A., Sakai K., Zhang J., Meng Z., Veenstra T. D., Staudt L. M., and Lenardo M. J. (2009) Casein kinase 1α governs antigen-receptor-induced NF-κB activation and human lymphoma cell survival. Nature 458, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim K. H., Yang Y., and Staudt L. M. (2012) Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev. 246, 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis R. E., Brown K. D., Siebenlist U., and Staudt L. M. (2001) Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194, 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ngo V. N., Davis R. E., Lamy L., Yu X., Zhao H., Lenz G., Lam L. T., Dave S., Yang L., Powell J., and Staudt L. M. (2006) A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 [DOI] [PubMed] [Google Scholar]
- 24. Lenz G., Davis R. E., Ngo V. N., Lam L., George T. C., Wright G. W., Dave S. S., Zhao H., Xu W., Rosenwald A., Ott G., Muller-Hermelink H. K., Gascoyne R. D., Connors J. M., Rimsza L. M., et al. (2008) Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 [DOI] [PubMed] [Google Scholar]
- 25. Compagno M., Lim W. K., Grunn A., Nandula S. V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., Bhagat G., Chadburn A., Dalla-Favera R., and Pasqualucci L. (2009) Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 459, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohr J. G., Stojanov P., Lawrence M. S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y. W., Slager S. L., Novak A. J., Dogan A., Ansell S. M., Link B. K., Zou L., et al. (2012) Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 3879–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Montesinos-Rongen M., Schmitz R., Brunn A., Gesk S., Richter J., Hong K., Wiestler O. D., Siebert R., Küppers R., and Deckert M. (2010) Mutations of CARD11 but not TNFAIP3 may activate the NF-κB pathway in primary CNS lymphoma. Acta Neuropathol. 120, 529–535 [DOI] [PubMed] [Google Scholar]
- 28. Bu R., Bavi P., Abubaker J., Jehan Z., Al-Haqawi W., Ajarim D., Al-Dayel F., Uddin S., and Al-Kuraya K. S. (2012) Role of NF-κB regulators-TNFAIP3 and CARD11 in Middle Eastern diffuse large B cell lymphoma. Leuk. Lymphoma. 53, 1971–1977 [DOI] [PubMed] [Google Scholar]
- 29. Dong G., Chanudet E., Zeng N., Appert A., Chen Y. W., Au W. Y., Hamoudi R. A., Watkins A. J., Ye H., Liu H., Gao Z., Chuang S. S., Srivastava G., and Du M. Q. (2011) A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin. Cancer Res. 17, 1440–1451 [DOI] [PubMed] [Google Scholar]
- 30. Lamason R. L., McCully R. R., Lew S. M., and Pomerantz J. L. (2010) Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry 49, 8240–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan W., Schaffer T. B., and Pomerantz J. L. (2013) A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival. Mol. Cell. Biol. 33, 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwai K., Fujita H., and Sasaki Y. (2014) Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 [DOI] [PubMed] [Google Scholar]
- 33. Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S. J., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., et al. (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., and Iwai K. (2011) SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 [DOI] [PubMed] [Google Scholar]
- 35. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., and Walczak H. (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 [DOI] [PubMed] [Google Scholar]
- 36. Hadian K., Griesbach R. A., Dornauer S., Wanger T. M., Nagel D., Metlitzky M., Beisker W., Schmidt-Supprian M., and Krappmann D. (2011) NF-κB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-κB activation. J. Biol. Chem. 286, 26107–26117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emmerich C. H., Ordureau A., Strickson S., Arthur J. S., Pedrioli P. G., Komander D., and Cohen P. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 110, 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Y., Schmitz R., Mitala J., Whiting A., Xiao W., Ceribelli M., Wright G. W., Zhao H., Yang Y., Xu W., Rosenwald A., Ott G., Gascoyne R. D., Connors J. M., Rimsza L. M., et al. (2014) Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov. 4, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dubois S. M., Alexia C., Wu Y., Leclair H. M., Leveau C., Schol E., Fest T., Tarte K., Chen Z. J., Gavard J., and Bidère N. (2014) A catalytic-independent role for the LUBAC in NF-κB activation upon antigen receptor engagement and in lymphoma cells. Blood 123, 2199–2203 [DOI] [PubMed] [Google Scholar]
- 40. Sasaki Y., Sano S., Nakahara M., Murata S., Kometani K., Aiba Y., Sakamoto S., Watanabe Y., Tanaka K., Kurosaki T., and Iwai K. (2013) Defective immune responses in mice lacking LUBAC-mediated linear ubiquitination in B cells. EMBO J. 32, 2463–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Satpathy S., Wagner S. A., Beli P., Gupta R., Kristiansen T. A., Malinova D., Francavilla C., Tolar P., Bishop G. A., Hostager B. S., and Choudhary C. (2015) Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 11, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y., Kelly P., Shaffer A. L. 3rd, Schmitz R., Yoo H. M., Liu X., Huang da W., Webster D., Young R. M., Nakagawa M., Ceribelli M., Wright G. W., Yang Y., Zhao H., Yu X., et al. (2016) Targeting non-proteolytic protein ubiquitination for the treatment of diffuse large B cell lymphoma. Cancer Cell 29, 494–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keusekotten K., Elliott P. R., Glockner L., Fiil B. K., Damgaard R. B., Kulathu Y., Wauer T., Hospenthal M. K., Gyrd-Hansen M., Krappmann D., Hofmann K., and Komander D. (2013) OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rivkin E., Almeida S. M., Ceccarelli D. F., Juang Y. C., MacLean T. A., Srikumar T., Huang H., Dunham W. H., Fukumura R., Xie G., Gondo Y., Raught B., Gingras A. C., Sicheri F., and Cordes S. P. (2013) The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Komander D., Reyes-Turcu F., Licchesi J. D., Odenwaelder P., Wilkinson K. D., and Barford D. (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu C. J., and Ashwell J. D. (2008) NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 105, 3023–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang C., and Lin X. (2012) Regulation of NF-κB by the CARD proteins. Immunol. Rev. 246, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., and Iwai K. (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., and Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 50. Paul S., Kashyap A. K., Jia W., He Y. W., and Schaefer B. C. (2012) Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity 36, 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alexia C., Poalas K., Carvalho G., Zemirli N., Dwyer J., Dubois S. M., Hatchi E. M., Cordeiro N., Smith S. S., Castanier C., Le Guelte A., Wan L., Kang Y., Vazquez A., Gavard J., Arnoult D., and Bidère N. (2013) The endoplasmic reticulum acts as a platform for ubiquitylated components of nuclear factor κB signaling. Sci. Signal. 6, ra79. [DOI] [PubMed] [Google Scholar]
- 52. Smit J. J., Monteferrario D., Noordermeer S. M., van Dijk W. J., van der Reijden B. A., and Sixma T. K. (2012) The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stieglitz B., Morris-Davies A. C., Koliopoulos M. G., Christodoulou E., and Rittinger K. (2012) LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yagi H., Ishimoto K., Hiromoto T., Fujita H., Mizushima T., Uekusa Y., Yagi-Utsumi M., Kurimoto E., Noda M., Uchiyama S., Tokunaga F., Iwai K., and Kato K. (2012) A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO Rep. 13, 462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klein T., Fung S. Y., Renner F., Blank M. A., Dufour A., Kang S., Bolger-Munro M., Scurll J. M., Priatel J. J., Schweigler P., Melkko S., Gold M. R., Viner R. I., Régnier C. H., Turvey S. E., and Overall C. M. (2015) The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat. Commun. 6, 8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elton L., Carpentier I., Staal J., Driege Y., Haegman M., and Beyaert R. (2016) MALT1 cleaves the E3 ubiquitin ligase HOIL-1 in activated T cells, generating a dominant negative inhibitor of LUBAC-induced NF-κB signaling. FEBS J. 283, 403–412 [DOI] [PubMed] [Google Scholar]
- 57. Lamason R. L., Kupfer A., and Pomerantz J. L. (2010) The dynamic distribution of CARD11 at the immunological synapse is regulated by the inhibitor kinesin GAKIN. Mol. Cell 40, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pedersen S. M., Chan W., Jattani R. P., Mackie dS., and Pomerantz J. L. (2015) Negative regulation of CARD11 signaling and lymphoma cell survival by the E3 ubiquitin ligase RNF181. Mol. Cell. Biol. 36, 794–808 [DOI] [PMC free article] [PubMed] [Google Scholar]