Abstract
Structural polysaccharides like cellulose and chitin are abundant and their enzymatic degradation to soluble sugars is an important route in green chemistry. Processive glycoside hydrolases (GHs), like cellobiohydrolase Cel7A of Trichoderma reesei (TrCel7A) are key components of efficient enzyme systems. TrCel7A consists of a catalytic domain (CD) and a smaller carbohydrate-binding module (CBM) connected through the glycosylated linker peptide. A tunnel-shaped active site rests in the CD and contains 10 glucose unit binding sites. The active site of TrCel7A is lined with four Trp residues with two of them, Trp-40 and Trp-38, in the substrate binding sites near the tunnel entrance. Although addressed in numerous studies the elucidation of the role of CBM and active site aromatics has been obscured by a complex multistep mechanism of processive GHs. Here we studied the role of the CBM-linker and Trp-38 of TrCel7A with respect to binding affinity, on- and off-rates, processivity, and synergism with endoglucanase. The CBM-linker increased the on-rate and substrate affinity of the enzyme. The Trp-38 to Ala substitution resulted in increased off-rates and decreased processivity. The effect of the Trp-38 to Ala substitution on on-rates was strongly dependent on the presence of the CBM-linker. This compensation between CBM-linker and Trp-38 indicates synergism between CBM-linker and CD in feeding the cellulose chain into the active site. The inter-domain synergism was pre-requisite for the efficient degradation of cellulose in the presence of endoglucanase.
Keywords: carbohydrate-binding protein, cellobiohydrolase, cellulase, cellulose, processivity, Trichoderma reesei, binding, off-rate, on-rate, synergism
Introduction
Structural polysaccharides represent a vast resource available for clean energy and chemistry (1, 2). A lot of effort has been put into developing the industrial approaches for optimal deconstruction and valorization of structural polysaccharides including cellulose (the homopolymer of β-1,4-linked glucose units). The highly recalcitrant and insoluble crystalline matrix of cellulose is challenging for enzymatic degradation (2). In nature the most efficient degraders of cellulose are fungi, which secrete a myriad of cellulases for efficient decomposition of cellulose. For biotechnological applications the best known fungal isolate is the filamentous fungus Trichoderma reesei (teleomorph Hypocrea jecorina) (3). The key component of the enzyme system of T. reesei is glycoside hydrolase (GH)2 (4) family 7 enzyme TrCel7A, a processive cellobiohydrolase (CBH) acting from the reducing end of the cellulose chain (5–7). Most T. reesei cellulases including TrCel7A have a two-domain structure consisting of a non-catalytic cellulose binding module (CBM) connected by a flexible glycosylated linker peptide to the catalytic domain (CD) that is responsible for catalysis of the glycosidic bond hydrolysis (3, 8, 9) (Fig. 1A). The CBM has been shown to have a role in targeting (10–12), but also in disrupting the crystalline cellulose aggregates (13). A role of the linker peptide in activity and binding has also been recognized (9, 14–16). The CBM of TrCel7A belongs to CBM family 1 (8), a family of CBMs that primarily targets crystalline cellulose (17, 18). In line with this, the removal of the CBM has a more negative effect on the hydrolysis of crystalline cellulose compared with amorphous substrates (19). The CBM is beneficial for activity at low substrate concentrations, whereas at high substrate loads it is less important or even a disadvantage (20–22). The catalytic domain of TrCel7A contains a 5-nm long tunnel-shaped active site (23, 24). The tunnel is roofed by four surface loops and accommodates 10 glucose unit binding sites (−7 to +3), 7 substrate (denoted with − sign) and 3 product (denoted with + sign) binding sites (23, 24) (Fig. 1B). Characteristic for GHs, the active site of TrCel7A contains several aromatic residues that stack with sugar rings through π-interactions. Two of these, Trp-40 and Trp-38, are located at the entrance (−7) and in the middle (−4) of the tunnel, respectively. Another two, Trp-367 and Trp-376, are located near the scissile bond in binding sites −2 and +1, respectively (Fig. 1B). The negative effects of single aromatic substitutions on hydrolysis of crystalline substrates have been demonstrated for TrCel7A (7, 22, 25) as well as for bacterial cellulases (26–28) and chitinases (29–31). On the other hand the deficiency of Trp to Ala variants in hydrolysis of crystalline substrates has often accompanied the increased activity on amorphous and soluble substrates (29, 31). The hydrolysis of recalcitrant polysaccharides by processive enzymes is a complex process consisting of at least three steps (i) adsorption, recognition, and initial sliding of cellulose chain into the active site (3, 32), in this work we will collectively refer to this as feeding; (ii) processive sliding with concomitant hydrolysis of polymer chain; and (iii) dissociation (Fig. 1C). Computational studies have suggested that Trp-40 and Trp-38 of TrCel7A participate in substrate binding, whereas Trp-367 and Trp-376 are directly involved in catalysis of glycosidic bond hydrolysis by stabilizing the transition state (33, 34). Substitution of Trp-40 to Ala at the tunnel entrance (W40A) reduces the hydrolysis rates of highly crystalline cellulose by TrCel7A apparently through impairing the cellulose chain acquisition (7, 25). Substitution of Trp-38 to Ala (W38A) in TrCel7A has been shown to decrease the affinity of the variant to crystalline cellulose, Avicel, indicating the importance of Trp-38 in binding (22). In this study it was concluded that the decreased affinity was primarily governed by the increased off-rate constant (koff) rather than decreased on-rates (22). Increased koff of the W38A variant compared with the wild type (WT) enzyme was also proposed to be responsible for its higher steady-state hydrolysis rates at saturating substrate concentrations (22). This conclusion stems from the obstacle model of cellulose hydrolysis whereby the steady-state rate of hydrolysis by CBH acting in isolation is governed by the koff and apparent processivity (Papp) of CBH (19, 22, 35–39). Although the importance of koff and Papp is well recognized the experimental measurements of the values of these parameters are scarce. Furthermore, the reported koff and Papp values not only depend on the cellulose substrate but also the method used. The processivity values of TrCel7A measured on different cellulose substrates using different methods fall between 10 and 70 (6, 19, 35, 40–43). The differences in koff values measured for TrCel7A using different methods are even more prominent ranging from 10−6 to 10−1 s−1 (22, 35, 39, 43–45). Although several studies have supported the obstacle model, slow complexation (46) or a combination of slow complexation and slow dissociation (47) has been proposed as rate-limiting events. Furthermore, the inner processive cycle has been proposed to be rate-limiting when acting in synergy with endoglucanases (EGs) (37). These different proposals do not exclude one another as the rate-limiting step can change dependent on the experimental conditions. It stresses, however, the importance of the experimental conditions. Therefore, parallel assessment of binding, complexation, processivity, dissociation, and synergism is needed to elucidate the role of the CBM or aromatic residues in active site.
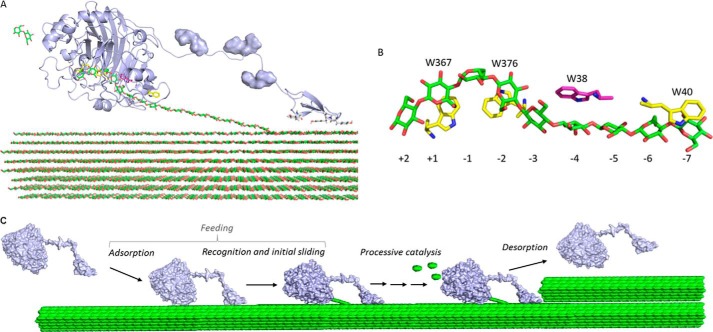
FIGURE 1.
Structure and mechanism of TrCel7A. A, schematic representation (light blue) of catalytic domain, linker, and CBM for TrCel7A in complex with a cellulose strand. The O-glycosylation of the linker is shown as a surface representation and the cellulose strand is represented as green sticks. The image was made using the crystal structure of the catalytic domain (Protein Data Bank code 8CEL) and CBM (Protein Data Bank code 1CBH). B, positions of the Trp residues in the active site tunnel of TrCel7A in complex with a cellodextrin chain (green). Numbers refer to the different binding subsites in the tunnel with −7 at the entrance and −1/+1 being the position of the scissile bond. Trp-38 investigated in this study is highlighted in magenta. C, molecular steps in the hydrolysis of cellulose for a processive enzyme. Steps leading from the free enzyme in the solution to the enzyme with the reducing end of the cellulose chain in the −1 binding site are collectively referred to as feeding. Processive catalysis includes the formation of a Michaelis complex (by sliding the chain end from binding site −1 to +2), hydrolysis of glycosidic bond, and expulsion of cellobiose (green ellipses). Processive catalysis is repeated until the enzyme meets an obstacle (depicted here as upper cellulose fibril) or happens to dissociate.
Here we studied the role of CBM-linker and Trp-38 in TrCel7A by measuring their effect on total and active site mediated binding, off-rates, on-rates, processivity, and synergism with EG. A novel method relying on measuring the exchange rate of the enzyme between non-labeled and 14C-labeled cellulose was introduced for measuring the dissociation of TrCel7A. We found that the CBM-linker had more influence in determining on-rates than off-rates. The Trp-38 to Ala substitution resulted in decreased processivity and ∼2–3-fold increase in off-rate constant on both crystalline and amorphous cellulose. The effect of Trp-38 to Ala substitution on on-rates was strongly dependent on the presence of CBM-linker indicating compensation between CBM-linker and Trp-38. The presence of both CBM-linker and Trp-38 was required for the efficient degradation of cellulose in the presence of EG.
Results
Measuring the Off-rates of TrCel7A with Substrate Exchange Experiments
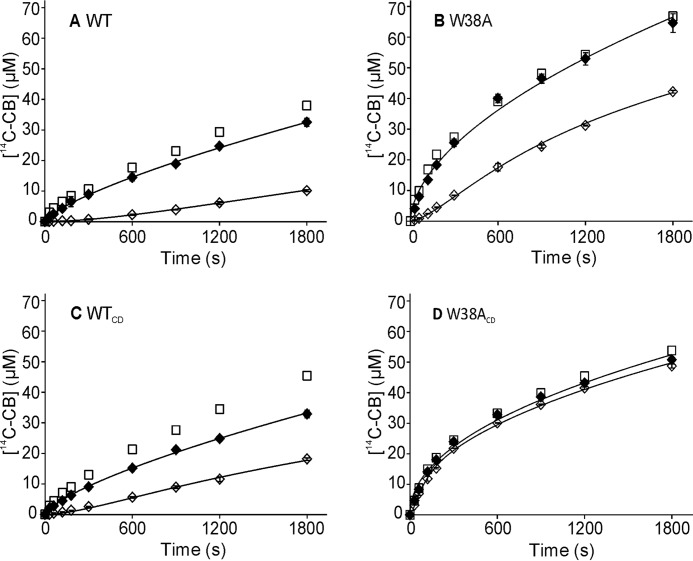
Recently a new method, referred to as a substrate exchange experiment (SEE), for measuring koff for dissociation of chitinase from chitin was developed (31). The method is based on measuring the exchange rate of the enzyme between a non-labeled (substrate of interest) and a 14C-labeled polymeric substrate (reference substrate). The method does not rely on the assumptions about the rate-limiting step, although the experimental conditions must be adjusted so that both substrates are at saturating concentrations for the enzyme. Here we adapted the SEE for measuring the koff values of cellulase TrCel7A. Besides intact WT enzyme its catalytic domain lacking the CBM and linker peptide (WTCD) was studied. To reveal the contribution of Trp-38 at binding site −4 the W38A mutants of both variants, W38A and W38ACD, respectively, were also included. Because of its high binding capacity 14C-labeled amorphous cellulose ([14C]AC) was used as a reference substrate. The koff values were measured for the dissociation from Avicel, the wood-derived model cellulose often used in cellulase studies. Comparison of the reference time curves (Avicel and [14C]AC were mixed together before the addition of the enzyme) with the time curves of the hydrolysis of [14C]AC present alone reveals that [14C]AC at 2 mg ml−1 effectively outcompetes Avicel at 50 mg ml−1 (Fig. 2). In SEE, Avicel at 100 mg ml−1 was preincubated with 0.4 μm TrCel7A for 1 h before an equal volume of [14C]AC was added. With W38ACD as an exception a lag phase in the formation of soluble 14C-product (expressed in [14C]cellobiose ([14C]CB) equivalents) characteristic to SEE was observed. The koff values were found by comparison of SEE with the reference time curves (see ”Experimental Procedures“) and are listed in Table 1. The absence of a clear lag-phase with W38ACD is apparently a mixed effect of fast dissociation and non-complete saturation with Avicel before the addition of [14C]AC (the Km for W38ACD for Avicel is 24.3 mg ml−1 (22)). Working with Avicel at concentrations significantly higher than 100 mg ml−1 was not possible for technical reasons. Unfortunately the SEE attempts, using bacterial cellulose (BC) or amorphous cellulose (AC) as a substrate, were not successful. The absence of lag-phase with BC can be caused by the difficulties in obtaining saturating conditions with BC and/or multiple binding modes of TrCel7A on this substrate (48, 49). Using AC, we found that [14C]AC was not efficient enough to compete with AC at saturating concentration for the enzymes.
FIGURE 2.
SEE with Avicel. In SEE, Avicel (100 mg ml−1) was preincubated with 400 nm TrCel7A for 1 h, after which an equal volume of [14C]AC (final concentration 2 mg ml−1) was added, and the release of radioactivity (expressed in [14C]CB equivalents) in time was followed (♢). In reference time curves (♦) the same conditions were used as in SEE but Avicel was mixed with [14C]AC before the addition of TrCel7A. Control experiments (□) of hydrolysis of [14C]AC (2 mg ml−1) in the absence of Avicel are also shown (made in one parallel). Error bars show S.D. and are from two independent experiments. Solid lines represent the best fit from non-linear regression analysis (see “Experimental Procedures”). The TrCel7A variants used were: A, WT; B, W38A; C, WTCD; D, W38ACD.
TABLE 1.
Kinetic parameters of TrCel7A and its variants measured with crystalline celluloses
| Enzyme | Avicel |
rBC |
|||
|---|---|---|---|---|---|
| koffa | Kmb | konc | kIRGd | Pappe | |
| 10−3 s−1 | g l−1 | 10−3 l g−1s−1 | 10−3 s−1 | ||
| WT | 0.7 ± 0.1 | 1.54 ± 0.23 | 0.46 ± 0.07 | 0.65 ± 0.03 | 70 ± 10 |
| W38A | 2.3 ± 0.1 | 5.95 ± 0.34 | 0.39 ± 0.02 | 32 ± 1 | |
| WTCD | 1.7 ± 0.2 | 12.6 ± 1.82 | 0.14 ± 0.02 | 1.17 ± 0.25 | 58 ± 7 |
a The koff values measured using SEE (Fig. 2).
b Data from Kari et al. (22).
c kon values are calculated according to kon = koff/Km using listed koff and Km values.
d kIRG represents the off-rate constant measured from the generation of IRGs on rBC (Fig. 3B).
e Papp is found as a slope of the linear regression line of the data in coordinates [RG]tot versus [IRG] (Fig. 3C).
Apparent Processivity and Rate Constant of the Initiation of Processive Runs
Because the koff is directly related to the enzymes intrinsic processivity (50) the enzyme variants with increased off-rates are expected to have also lower processivity. Therefore we also measured the processivity of TrCel7A and its variants. Here we used the method that relies on the hydrolysis of reduced cellulose under single-hit conditions (35, 51). The apparent processivity is given by the ratio of the number of processive cuts (Ncatal) and the number of initiations of processive runs (Ninit). It has been shown that for the enzymes employing reducing-end exo and/or endo-mode initiation the Ninit is given by the number of insoluble reducing ends (IRG) generated on reduced cellulose. The Ncatal is represented by the total amount of enzyme generated reducing groups (RGtot). Therefore, Papp = [RGtot]/[IRG]. The concentration of IRGs was measured by fluorescence labeling of enzyme-generated reducing ends with anthranilic acid (AA).
First the Papp was measured using reduced bacterial cellulose (rBC) as a substrate. Wild type enzymes, WT and WTCD, produced more soluble reducing groups (SRGs) compared with their W38A variants (Fig. 3A). At shorter hydrolysis times the highest amount of IRGs was produced by W38A. However, this high initial activity was followed by a rapid slowdown. The slowdown in IRG production was most evident for W38ACD where the increase in IRGs after the first 10 min remained below the limit of detection. The Papp was found as the slope of the linear regression line to the data plotted in coordinates [RGtot] versus [IRG] (Fig. 3C). The highest Papp of 70 ± 10 was measured for WT. This figure is in a good agreement with the Papp of 61 ± 14 measured for TrCel7A with rBC substrate using diaminopyridine labeling of IRGs (35). The W38A had 2.3-fold lower Papp compared with the WT. The Papp of WTCD remained between the corresponding figures of WT and W38A (Table 1). Because of the very low amount of IRGs we could not measure the Papp of W38ACD.
FIGURE 3.
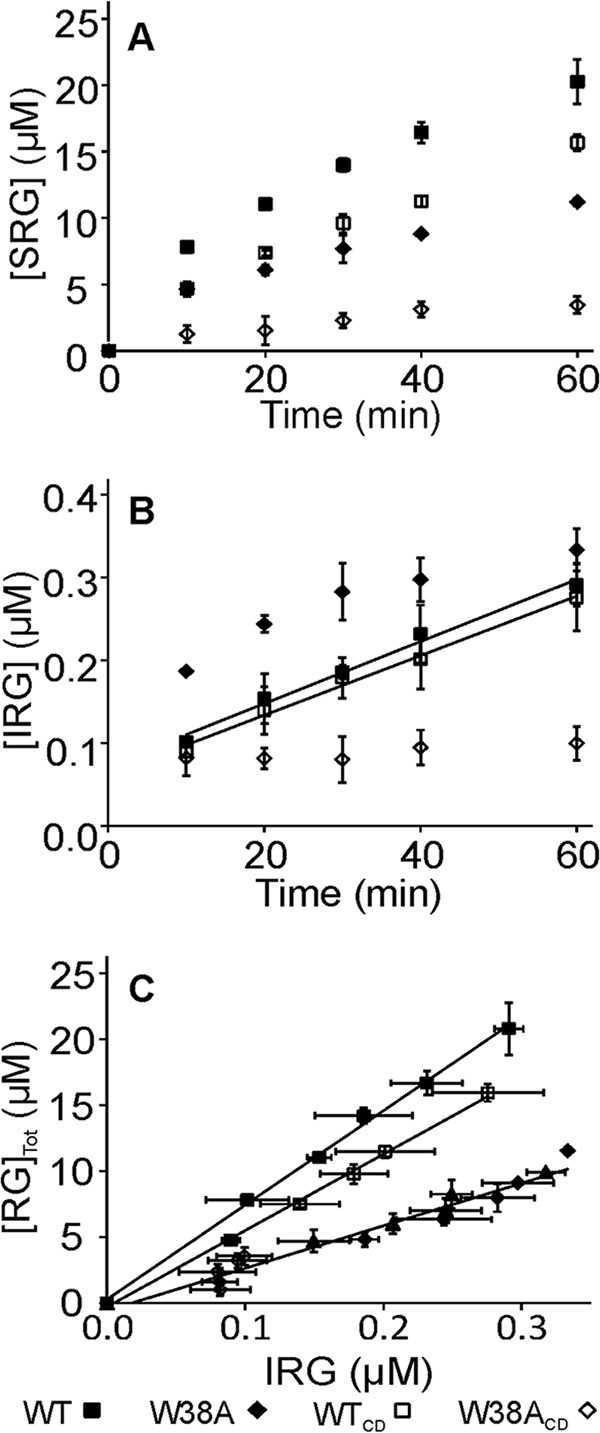
Formation of soluble and insoluble reducing groups in hydrolysis of rBC. rBC (1 mg ml−1) was incubated with 100 nm TrCel7A and the formation of SRGs (A) and IRGs (B) in time was followed. Solid lines in B for the series with WT and WTCD are from linear regression and the slopes were used to calculate the values of kIRG. In panel C, the data are plotted in coordinates [RG]tot versus [IRG]. Solid lines are from linear regression and the slope equals the Papp value. Error bars show S.D. and are from three independent experiments. With W38A, experiments were also made using 62 nm enzyme concentrations (▴).
The Papp values were also measured using reduced amorphous cellulose (rAC) as a substrate. Contrary to rBC the W38A variants outperformed the WT and WTCD in both, the production of SRGs and IRGs (Fig. 4, A and B). However, the Papp of W38A variants was lower than that of their wild type counterparts (Fig. 4 C, Table 2) indicating that Papp is not the sole determinant of activity. With all enzymes the Papp measured with rAC was significantly lower than that found with rBC. The Papp of 17 ± 2 found for WT on rAC was within the error limits with Papp of 20.8 ± 1.2 measured for the same system using diaminopyridine labeling of reducing ends before (35).
FIGURE 4.
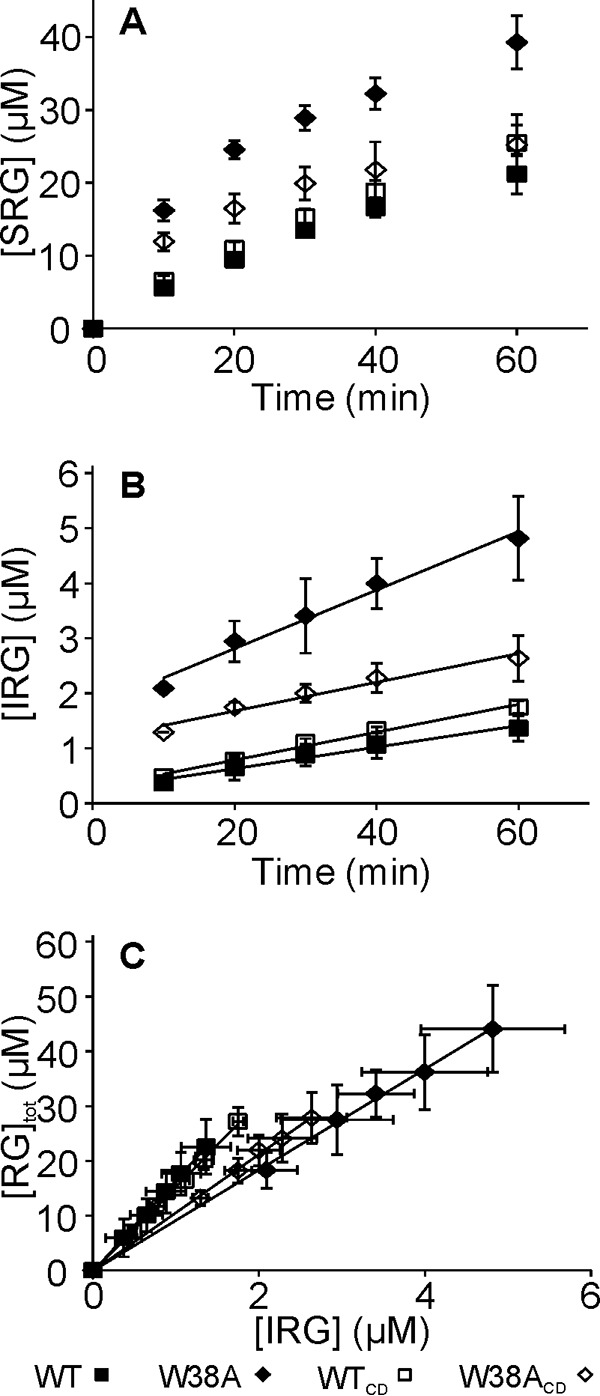
Formation of soluble and insoluble reducing groups in hydrolysis of rAC. rAC (1 mg ml−1) was incubated with 100 nm TrCel7A and the formation of SRGs (A) and IRGs (B) in time was followed. Solid lines in B are from liner regression and the slopes were used to calculate the values of kIRG. In panel C, the data are plotted in coordinates [RG]tot versus [IRG]. Solid lines are from linear regression and the slope equals the Papp value. Error bars show S.D. and are from three independent experiments.
TABLE 2.
Kinetic parameters of TrCel7A and its variants measured with amorphous cellulose
| Enzyme | kIRGa | Pappb | [S]0.5c | kond |
|---|---|---|---|---|
| 10−3 s−1 | 10−3 g l−1 | l g−1s−1 | ||
| WT | 3.4 ± 0.5 | 17 ± 2 | 1.7 ± 0.1 | 2.0 ± 0.3 |
| W38A | 8.9 ± 1.5 | 9 ± 2 | 4.5 ± 0.5 | 1.9 ± 0.3 |
| WTCD | 4.4 ± 0.2 | 15 ± 1 | 10.9 ± 0.5 | 0.40 ± 0.02 |
| W38ACD | 10.2 ± 2.9 | 11 ± 0 | 520 ± 60 | 0.020 ± 0.006 |
a kIRG represents the off-rate constant measured from the generation of IRGs on rAC (Fig. 4B).
b Papp is found as a slope of the linear regression line of the data in coordinates [RG]tot versus [IRG] (Fig. 4C).
c [S]0.5 was found by the non-linear regression analysis of the data in Fig. 6A according to Equation 2.
d kon values are calculated according to kon = kIRG/[S]0.5 using listed kIRG and [S]0.5 values.
Besides measuring Papp the production of IRGs on reduced cellulose can be used as a measure of koff (31, 35). As originally developed the rate constant of IRG production (kIRG) was taken equal to the koff using the following relationship: koff ≈ vIRG/[E]tot. Using the total concentration of enzyme ([E]tot) instead of the concentration of active site bound enzyme assumes saturating substrate concentration. Furthermore, it assumes that the dissociation is rate-limiting for the enzyme recruitment so that the rate of the initiation of processive runs is governed by koff. However, kIRG represents the dissociation rate constant of a polymer chain from the active site if [E]tot is replaced with the concentration of the enzyme with active site occupied by the polymer chain ([Enzyme]bound-OA),
| (Eq. 1) |
[Enzyme]bound-OA is measured in parallel with the rate of IRG formation (vIRG) by measuring the inhibition of the hydrolysis of the low molecular weight reporter molecule like 4-methylumbelliferyl-β-lactoside (MUL) by reduced cellulose. In this way we found that at 100 nm total enzyme concentration and rBC at 1 mg ml−1, the concentration of [WT]bound-OA, [WTCD]bound-OA, and [W38A]bound-OA were 94.9 ± 0.8, 52.9 ± 6.6, and 56.7 ± 8.9 nm, respectively. The level of active site bound enzyme was constant within the studied time intervals (10–60 min) (data not shown). In the case of W38ACD the concentration of active site bound enzyme was below the limit of detection suggesting that the low rate of IRG production was caused by inefficient binding of the cellulose chain in the active site. Provided with the concentration of enzyme with occupied active site and rates of IRG formation the kIRG values on rBC were calculated for WT and WTCD (Table 1). Because of the nonlinear time curve of IRG formation the kIRG for W38A was not found. Notably, the kIRG values of WT and WTCD on rBC were close to their koff values measured with Avicel using SEE. The consistency between koff and kIRG is expected when (i) exo-mode initiation from the non-reducing end is negligible, and (ii) the lateral diffusion on cellulose surface is not the prevalent mode of targeting the hydrolysis initiation sites. If the former assumption is violated, kIRG will underestimate koff. If the latter assumption does not hold kIRG will overestimate koff because the enzyme makes the number of initiations (revealed by kIRG) before the full dissociation from the surface takes place (revealed by SEE koff). The latter scenario was recently demonstrated for chitinase ChiA from bacterium Serratia marcescens (SmChiA) (31). However, lateral diffusion was suggested to be unimportant for TrCel7A (49).
With rAC (1 mg ml−1) the concentrations of active site bound enzymes was close to the total enzyme concentration (100 nm). An exception here was W38ACD with an active site bound enzyme concentration of 42.4 ± 3.7 nm. The kIRG values on rAC (Table 2) were significantly higher than corresponding figures measured with rBC (Table 1). Like in the case of koff values measured with Avicel using SEE (Table 1) the removal of the CBM-linker and/or W38A substitution resulted in increased kIRG values on rAC.
Binding to BC and AC
Different levels of binding to rBC at 1 mg ml−1 prompted us to study the binding in more detail. For that the binding to BC was studied at different cellulose concentrations. Furthermore, the binding was studied on the level of total bound enzyme ([Enzyme]bound-tot) as well as on the level of active site bound enzyme. The difference between [Enzyme]bound-tot and [Enzyme]bound-OA represents the population of bound enzyme with the active site free from cellulose chain ([Enzyme]bound-FA). The binding of core domains was too weak to be saturated at the highest practical possible BC concentration. Hence the binding studies to BC were conducted only with the intact enzymes. The binding of WT to BC was stronger than that of W38A. Another major difference between the WT enzyme and W38A variant is that the variant has a significant population of bound enzyme with free active sites. Still, the concentration of active site bound W38A seems to level off at the value of total enzyme concentration with increasing BC concentration (Fig. 5). Because the binding of TrCel7A involves different binding modes, which relative contribution depends on the enzyme to substrate ratio (48) a more thorough quantitative analysis of binding to BC was omitted.
FIGURE 5.
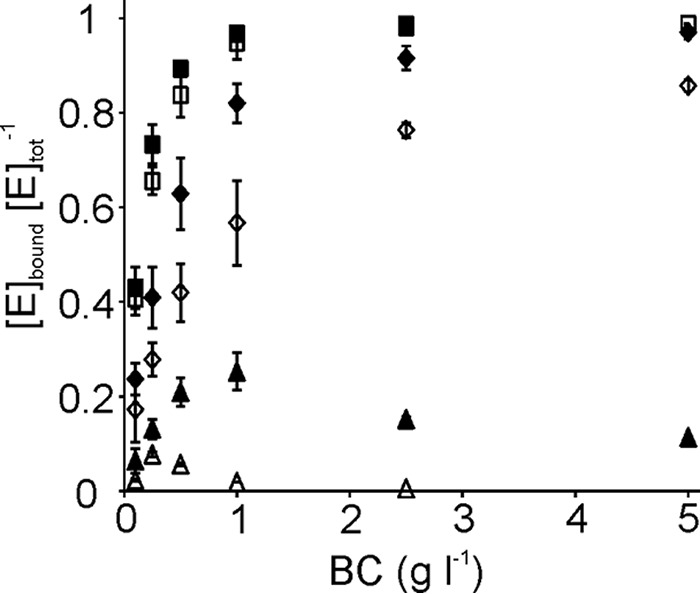
Binding of WT TrCel7A and its W38A variant to BC. Total bound, WT (■) and W38A (♦); active site bound WT (□) and W38A (♢); bound enzyme with free active site, WT (▵) and W38A (▴). The concentration of total bound enzyme ([TrCel7A]bound-tot) was found as a difference between the total enzyme concentration and the concentration of enzyme free in the solution. Concentration of active site bound enzyme ([TrCel7A]bound-OA) was found from the strength of inhibition of MUL hydrolysis by BC. Concentration of bound enzyme with the free active site ([Cel7A]bound-FA) was found as a difference between [TrCel7A]bound-tot and [TrCel7A]bound-OA. Error bars show S.D. and are from three independent experiments.
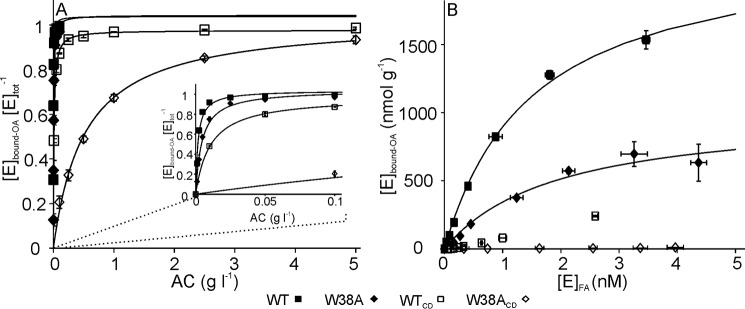
To get insight into the binding of core domains we also measured the binding to AC, a substrate with high binding capacity. Here, only the active site level bound enzyme was measured (Fig. 6A). The binding curves were analyzed using a simple hyperbolic relationship between [Enzyme]bound-OA and the substrate concentration ([S]),
| (Eq. 2) |
with all enzyme variants the maximum levels of active site bound enzyme ([Enzyme]bound-OAmax) approached the [E]tot. However, the values of half-saturating substrate concentration ([S]0.5) were largely different for the variants (Table 2). The most remarkable is the different effect of the CBM-linker to the bindings of WT and W38A enzymes. In the case of WT, removal of the CBM-linker resulted in about 2-fold increase in [S]0.5, whereas the corresponding effect to the W38A was an order of magnitude higher (Table 2). We also rearranged the data to show a conventional binding isotherm that implies the saturation of the substrate with the enzyme (Fig. 6B). The data were analyzed according to the one binding site Langmuir isotherm. Because of the weak binding the concentration of CDs was far from reaching saturating. For this reason only the binding of intact enzymes was analyzed. The WT had about 2-fold higher binding capacity compared with W38A, 2.2 ± 0.1 and 1.0 ± 0.1 μmol g−1, respectively. The half-saturating free enzyme concentrations of WT and W38A were 1.5 ± 0.1 and 1.9 ± 0.6 nm, respectively.
FIGURE 6.
The active site mediated binding of TrCel7A and its variants to AC. A, binding was measured with 5 nm total concentration of TrCel7A ([E]tot) by varying the concentration of AC between 0.001 and 5 g liter−1. Inset shows the enlargement of the region of low AC concentrations. Solid lines represent the best fit of non-linear regression according to Equation 2. B, binding data from panel A re-plotted in coordinates of concentration of the active site bound enzyme versus the concentration of enzyme with free active site. Solid lines represent the best fit of non-linear regression according to one binding site Langmuir isotherm. Error bars show S.D. and are from three independent experiments.
Synergistic Hydrolysis of BC
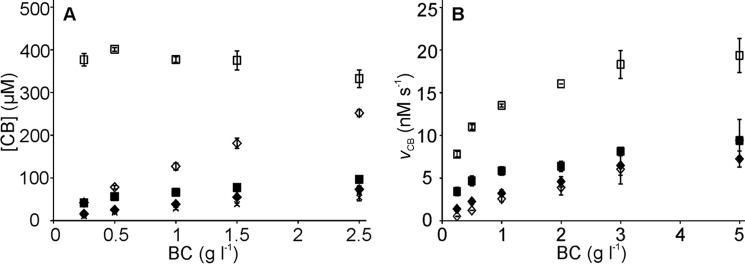
It is well known that efficient and complete degradation of recalcitrant polysaccharides can be achieved only by using synergistic enzyme mixtures instead of individual enzymes. Therefore we assessed the performance of W38A in synergistic hydrolysis of BC. It has been shown that, at the optimal substrate concentration BC is very efficiently degraded by a mixture of just three enzyme components, TrCel7A, EG, and β-glucosidase (BG) (37). The activity of the W38ACD variant was too low to be measured accurately and the performance of WTCD in synergistic hydrolysis of BC has been described before (37). Therefore, only intact enzymes were included in the studies of synergism here. First we followed the release of 14C-product in hydrolysis of [14C]BC by WT and W38A supplemented with EG TrCel5A and BG. [14C]BC was used at low concentration (0.25 mg ml−1), a condition where the highest synergism between TrCel7A and EG has been reported (37). Indeed, the activity of synergistic mixtures containing WT was very high with more than 50% of [14C]BC solubilized during 1 h of hydrolysis. In sharp contrast the mixture containing W38A showed only a very low activity that abruptly decayed after first few minutes of hydrolysis (data not shown). In tracking the causes we found that the low activity of W38A was caused by an inefficient binding through the active site, which remained below the limit of detection. Next, we assessed the synergistic effect at different BC concentrations. As seen in Fig. 7A the deficiency of W38A compared with WT decreased with the increasing BC concentration. This was also reflected in the degree of synergistic effect (DSE), a parameter showing the ratio of the activity of an enzymes mixture over the sum of the activities of the individual components. Whereas the DSE of WT decreased with increasing BC concentration the DSE of W38A showed little dependence (supplemental Table S1). We note that the performance of W38A in synergistic hydrolysis of BC is similar to that of the WTCD described before (37). Because the active site mediated binding of WTCD has been shown to be negatively affected by the presence of EG we also measured the concentration of W38A bound through the active site in the presence of EG. Binding was measured after 20 min of hydrolysis at different BC concentrations. The active site mediated binding of W38A was clearly depressed by the presence of EG (supplemental Fig. S1). This is in contrast to WT binding, which has been shown to be stimulated by EG (37). Finally we tested the effect of pre-treatment of BC with EG to the following hydrolysis by TrCel7A. For that the BC (1 mg ml−1) was incubated with 0.5 μm TrCel5A for 1 h. After removal of the EG by washing with alkali, the pre-treated BC was used as substrate for individual WT or W38A enzymes. As also shown in earlier reports (52, 53) the pre-treatment of BC with EG stimulated the following hydrolysis by TrCel7A (Fig. 7B). The stimulating effect of EG pre-treatment for WT was about 2-fold and was independent of the BC concentration. In contrast the EG pre-treatment of BC had a negative effect to the following hydrolysis by W38A at low BC concentrations and a slightly stimulating effect at highest BC concentration used (Fig. 7B).
FIGURE 7.
Synergistic hydrolysis of BC by TrCel7A or its W38A variant in the presence of EG TrCel5A. A, release of radioactivity (in [14C]CB equivalents) in hydrolysis of [14C]BC by 250 nm TrCel7A acting in isolation, WT (■) and W38A (♦), or in the presence of 25 nm EG, WT + EG (□), and W38A + EG (♢). The release of radioactivity by 25 nm EG is also shown (x). B, the hydrolysis of native (filled labels) and EG pre-treated (open labels) BC by 100 nm TrCel7A. TrCel7A variants were: WT (■ and □), and W38A (♦ and ♢). The rate of cellulose hydrolysis (vCB) was found from the formation of soluble reducing groups (in CB equivalents) after 1 h of hydrolysis. Error bars show S.D. and are from three independent experiments.
Discussion
Processive GHs utilize a complex multistep catalytic mechanism (Fig. 1C) and the identification of the rate-limiting step of these enzymes has been in a focus of intensive research. In this study we zoomed in on different parts of these molecular steps by varying the experimental conditions and the selected parts of the enzyme. More specifically the enzyme was changed by removing the CBM-linker and changing a Trp-38 in the initial part of the tunnel to an Ala. The combined effect of W38A and CBM-linker was also tested to see if an inter-domain synergy existed between the interaction of the CBM-linker and CD.
Dissociation (Off-rate)
The importance of koff in determining the hydrolysis rate of recalcitrant polysaccharides by processive GHs is well recognized (19, 31, 35, 36, 38, 54). The results of many studies support the obstacle model whereby the hydrolysis rate is directly related to the Papp and koff according to vCB = koff Papp [Enzyme]bound-OA (36). For TrCel7A this simple relationship is supported by measurements showing that dissociation is much slower than the processive run with concomitant release of the cellobiose product (37). Because the Papp has been shown to be limited by the length of the obstacle-free path on the substrate (35) most of the population of [Enzyme]bound-OA is expected to be non-productive, e.g. stalled behind obstacles (38, 55, 56) (see Fig. 1C). In line with this, the CBH variants with higher off-rates have been shown to have higher maximal rates (22). The positive correlation between activity and off-rates of GHs is most prominent on amorphous (31) and soluble polymeric substrates (29, 30). However, in most of the studies the higher off-rates of enzyme variants were suggested rather than measured directly. Here, we adapted the SEE for measuring the koff values of dissociation of TrCel7A from Avicel. The TrCel7A variants with higher expected off-rates, WTCD, W38A, and W38ACD were also included. The pVmax values measured with Avicel have been published before (22) and they increased in the order of WT < WTCD < W38A < W38ACD. Although we could not measure the koff for W38ACD, the same sequence seems to hold also for the koff values (Table 1). Higher activity of the variants with higher off-rates is also consistent with the hydrolysis of rAC. Even though their Papp was reduced (Table 2) the W38A variants clearly outperformed their WT counterparts in production of SRGs (Fig. 4A, note that in this experiment less than 50% of W38ACD was bound to rAC, whereas other enzymes were saturated). Unfortunately we were not able to measure the koff values using SEE with AC and BC. Therefore, with rAC and rBC as substrates the kIRG was used as the measure of koff. The good consistency between the koff and kIRG values measured on crystalline substrates Avicel and rBC (Table 1) suggests that for TrCel7A these two approaches provide equivalent rate constants. For separate domains of TrCel7A only the dissociation of its CBM from bacterial microcrystalline cellulose has been measured before with the resulting koff value of 0.029 s−1 (57). This figure is more than an order of magnitude higher than the koff value of WTCD measured here (Table 1), suggesting that dissociation of the WT should be governed by dissociation of the CD. This interpretation also parallels the conclusions in a recent, independent study (49). The 2.3-fold higher koff value of WTCD compared with that of the WT measured here may indicate the effect of physical linkage between two domains on the dissociation/association. Indeed, the synergistic effect of the physical linkage between two CBMs in the binding to different celluloses has been demonstrated (58). However, one must bear in mind that the koff of 0.029 s−1 has been measured for TrCel7A CBM having no glycosylated linker (57). The presence of the glycosylated linker has been shown to increase the affinity of the binding of TrCel7A CBM to BC by a factor of 10 (9). Unfortunately the relative contribution of on- and off-rate components in a binding affinity of the glycosylated linker is not known.
Feeding of Cellulose Chain (On-rate)
Dissecting the [S]0.5 values for binding to AC into its on- and off-rate components reveals that the CBM-linker had little effect on the dissociation from the amorphous cellulose (Table 2, Table 3). In contrast the CBM-linker had a prominent effect on the on-rates especially in the case of the W38A variant where the removal of the CBM-linker resulted in a 100-fold decrease in the kon value (Table 3). Comparison of the effects on the on-rates reveals that the absence of Trp-38 can be compensated in a large part by the CBM-linker and vice versa. Contrary to what was observed with on-rates the increase in off-rate (about 2.5-fold) caused by the Trp-38 to Ala substitution was independent on the presence of the CBM-linker (Table 3). With crystalline substrates, Avicel and BC, we do not have a full data set to analyze the effect of the CBM-linker and Trp-38 on both, on- and off-rates. A moderate effect (about 2–3-fold increase) of the CBM-linker (in WT enzyme) and the Trp-38 (in intact enzyme) on the off-rates (Table 1) in parallel with the drastically reduced binding affinity of the W38ACD (too low to be measured with BC) suggest that the CBM-linker/Trp-38 compensation in on-rates holds also with crystalline substrates. Compensation of the reduced binding affinity upon W40A substitution in the −7 binding site of TrCel7A by CBM has been reported before (25). Altogether these results suggest that there is a strong synergistic effect between the CBM-linker and the entrance region of the active site tunnel in feeding the cellulose chain into the active site. About 2-fold lower maximum adsorption capacity to AC of W38A compared with WT (Fig. 6B) suggests a role of Trp-38 in chain end recognition. A similar effect of Trp-38 was reported also with crystalline Avicel (22). Apart from this suggestion, the data presented here does not allow us to single out which elementary step in feeding is synergistically targeted.
TABLE 3.
The effect of the Trp-38 to Ala substitution and CBM-linker to off- and on-rates on AC
| Parameter |
||
|---|---|---|
| kIRGa | konb | |
| Effect of W38A substitution | ||
| WTCD/W38ACD | 0.4 ± 0.1 | 20.6 ± 5.9 |
| WT/W38A | 0.4 ± 0.1 | 1.0 ± 0.2 |
Synergism
We started this discussion with demonstrating the consistency between the experimental data and the off-rate limited hydrolysis by individual CBHs. The situation may be different with optimal synergistic enzyme mixtures where the rate of cellulose hydrolysis may be limited by the rate of the glycosidic bond hydrolysis (37). Several studies have pointed to that the CBM has no effect on kcat of glycosidic bond hydrolysis (7, 37, 42). Likewise, analysis of the rates of Avicel hydrolysis measured in both, steady- and pre-steady-state regimes suggests that W38A substitution has no significant effect on kcat (supplemental Fig. S2). The poor performance of WTCD (37) and W38A (Fig. 7) in synergistic hydrolysis of BC under conditions that are optimal for WT apparently reflects deficiency in feeding of a cellulose chain. The deficiency of W38A in feeding of a cellulose chain is much more evident in synergistic hydrolysis than when the enzyme acts in isolation. This suggest that the synergistic interaction between CBM-linker and the tunnel entrance region is a pre-requisite in feeding EG-generated chain ends. A possible mechanistic interpretation is that EG generates so called blunt chain ends (59) that are more tightly attached to the bulk cellulose crystal and are more difficult to feed into the active site.
Based on literature data and observations made here, the role of the active site aromatic residues near the tunnel entrance and the CBM-linker of TrCel7A can be envisaged. The primary role of Trp at the tunnel entrance, Trp-40, is chain end recognition (7, 25, 60). A similar role of Trp at the tunnel entrance has been suggested for other GHs like TrCel6A (61) and TrCel7B (62), and chitinases (63). Our data point toward the importance of Trp-38 at subsite −4 for progression of the cellulose chain end in the active site tunnel during the feeding. Trp-38 is located in the active site tunnel just before the cellulose chain is twisted (Fig. 1B) (24, 33). In this position the medium affinity region of the active site tunnel borders with the low affinity binding region (64). Therefore one can speculate that Trp-38 is needed for holding the cellulose chain end in the position, long enough to proceed through the region of twist. Once the chain end has proceeded through the region of twist the further sliding and processive hydrolysis is not affected by W38A. This is supported by a limited effect of Trp-38 to Ala substitution on kcat of glycosidic bond hydrolysis. Because Papp is independent on the on-rate the reduced Papp of the W38A variants apparently reflects the increased koff values. Computational studies have revealed that Trp-38 to Ala substitution in TrCel7A results in the decrease in binding free energy by 3.8 kcal mol−1 (62). The similar conclusions about the role of Trp-167 (analogous to the Trp-38 in TrCel7A) in determining the binding free energy (63), Papp and koff values (31), can be made for chitinase, SmChiA. Thus, the Trp residue in binding site −4/−3 appears to be functionally equivalent in two enzymes with different active site architectures, i.e. tunnel shaped active site of TrCel7A versus deep cleft shaped active site of SmChiA. The role of the CBM in increasing the enzymes affinity for the substrate has been recognized for a long time (65) and we suggest that this is mainly due to increased on-rate.
Conclusion
Taken together, the primary role of aromatic residues near the active site entrance is in feeding the cellulose chain into the active site tunnel. They also contribute to processivity via a reduction of the off-rate. Decreasing the binding strength in the active site entrance region leads to increased off-rates. This may be reflected in an increased overall activity under conditions where the availability of the chain ends compensates for the deficient feeding, i.e. at saturating substrate concentrations. The inefficient feeding of the chain end by the Trp to Ala substitution near the tunnel entrance can be compensated in large part by the CBM-linker. It is therefore possible that, analogous to the Trp-38, the CBM-linker is required for efficient chain end feeding into the active site.
Experimental Procedures
Materials
Avicel was purchased from Fluka. Anthranilic acid, sodium borohydride, sodium cyanoborohydride, bovine serum albumin (BSA), and MUL were purchased from Sigma. The scintillation mixture was purchased from Merck. All chemicals were used as purchased.
Enzymes
Intact TrCel7A (WT) and its W38A variant were heterologously expressed in Aspergillus oryzae and purified as described previously (22). W38ACD was constructed as described in Kari et al. (22). For WTCD the intact TrCel7A was first purified from the culture filtrate of T. reesei QM 9414 (66) and WTCD was prepared by limited proteolysis of intact TrCel7A with papain (67). Aspergillus BG was purified from Novozyme® 188 (C6105, Sigma) as described before (68).
Celluloses
BC was prepared by laboratory fermentation of the Gluconobacterium xylinum strain ATCC 53582 as described elsewhere (69). [14C]BC was prepared as BC but the glucose carbon source was supplemented with [14C]glucose as described Jalak et al. (37). The specific radioactivity of [14C]BC preparation was 6.4 × 105 dpm mg−1. AC was prepared from Avicel as described before (69). [14C]AC was prepared from 14C-bacterial microcrystalline cellulose by dissolution in and regeneration from phosphoric acid (69) and had specific radioactivity of 6.4 × 105 dpm mg−1. rBC and rAC were prepared from BC and AC, respectively, using reduction with sodium borohydride (69).
Substrate Exchange Experiment (SEE)
In SEE, Avicel (100 mg ml−1) was preincubated with 0.4 μm TrCel7A and 50 nm BG in 50 mm sodium acetate buffer, pH 5 (containing BSA 0.2 mg ml−1), at 25 °C for 1 h with magnetic stirring. 1 ml of the above reaction mixture was added to an equal volume of [14C]AC and the release of radioactivity in time was followed. Final concentrations of Avicel, [14C]AC, TrCel7A, and BG were 50 mg ml−1, 2 mg ml−1, 0.2 μm, and 25 nm, respectively. After mixing with [14C]AC the stirring was omitted but the reaction mixture was gently suspended with pipette before each sampling. For time points, 0.2-ml aliquots were withdrawn and stopped by adding NaOH to 0.1 m. For zero time points, NaOH was added before enzymes. Cellulose was separated by centrifugation (5 min at 104 × g) and radioactivity in the supernatant was quantified using a liquid scintillation counter. The released radioactivity was expressed in [14C]CB equivalents. The reference curves were made using the same experimental conditions as in SEE (see above) but Avicel and [14C]AC were mixed together before the addition of the enzymes. Hydrolysis time courses of [14C]AC present as a sole substrate were also made. In this case the experimental set-up was identical to that used to generate reference curves but Avicel was replaced with an equal volume of buffer.
The koff values were found by comparing the release of radioactivity in reference curves and SEE as described before (31). In short, the reference curves were analyzed using non-linear regression according to [14C-CB] = A × tb, where t is time and A and b are empirical constants (70). The SEE time curves were analyzed using non-linear regression according to [14C-CB] = A ×(1-e−t×koff)tb. In the latter regression the value of the constant b was fixed to the value found from the analysis of reference curves so that only the values of koff and A were let free (31).
Measuring Apparent Processivity and Rate Constant of the Initiation of Processive Runs
rBC or rAC (1 mg ml−1) was incubated with 0.1 μm TrCel7A in 50 mm sodium acetate buffer, pH 5 (containing BSA 0.1 mg ml−1), at 25 °C. At selected times the reaction was stopped by the addition of NaOH to 0.2 m. For zero time points NaOH was added before the enzyme. Cellulose was separated by centrifugation (2 min at 104 × g) and the supernatant was used for measuring SRGs using 3-methyl-2-benzothiazolinone hydrazone hydrochloride (MBTH) method, as described in Horn and Eijsink (71). The amount of IRGs was measured using fluorescence labeling of cellulose with AA (69). The cellulose was washed 2 times with 1 ml of water, once with 1 ml of sodium acetate buffer, and once with 1 ml of water. All washings were performed through centrifugation (2 min at 104 ×g) and re-suspension steps. After washing, cellulose was suspended in 0.2 ml of water and AA labeling was carried in 80% buffered methanol at 80 °C, 2 h in the presence of 0.5 m NaCNBH3 and 50 mm AA (69). Cellulose concentration was 1.6 mg ml−1. After AA labeling the cellulose was washed 3 times with 1 ml of water, incubated 1 h at room temperature with 0.2 m sodium hydroxide to remove the unspecific labeling. After alkali treatment the AA-labeled cellulose was further washed 3 times with 50 mm NaAc buffer, pH 5. Before fluorescence measurement the AA-labeled cellulose was solubilized by incubating overnight with a crude mixture of T. reesei cellulases. The fluorescence of the hydrolysate was measured at excitation and emission wavelengths of 330 and 425 nm, respectively.
Measuring the Total and Active Site Mediated Binding of TrCel7A with BC
For quantifying Cel7Abound-OA the initial rates of MUL hydrolysis by TrCel7A in the presence of BC were measured (48). For that the BC (0.1–5 mg ml−1) was incubated with 0.1 μm TrCel7A and 0.1 μm β-glucosidase in 50 mm sodium acetate buffer, pH 5 (containing BSA, 0.1 mg ml−1), at 25 °C. After 20 min MUL was added to a final concentration of 5 μm and after further incubation for 2 min the reaction was quenched by the addition of ammonium hydroxide to 0.1 m. After separation of cellulose (2 min at 104 × g) the concentration of released MU was found by measuring the fluorescence in the supernatant. The concentration of TrCel7A with the free active site was found from the rate of MUL hydrolysis, and the concentration of Cel7Abound-OA was found as a difference of total concentration of TrCel7A and TrCel7A with free active site (48).
For quantifying the concentration of Cel7Abound-tot, BC (0.1–5 mg ml−1) was incubated with 0.1 μm TrCel7A and 0.1 μm BG in 50 mm sodium acetate buffer, pH 5 (containing BSA 0.1 mg ml−1), at 25 °C. After 20 min the cellulose was separated by filtration through glass microfiber filter. Filtrate was further centrifuged (2 min at 104 × g) and the concentration of TrCel7A free from cellulose was measured by following the MUL (5 μm) hydrolyzing activity in the supernatant using appropriate calibration curves (48). The concentration of Cel7Abound-tot was found as the difference between the total concentration of TrCel7A and the concentration of TrCel7A free from cellulose.
Measuring the Active Site-mediated Binding of TrCel7A with AC
[Cel7A]bound-OA with AC was determined analogously to that with BC. The AC (0.001–5 mg ml−1) was incubated with 5 nm TrCel7A and 10 nm β-glucosidase in 50 mm sodium acetate buffer, pH 5 (containing BSA 0.1 mg ml−1), at 25 °C. After 20 min the MUL was added to a final concentration of 5 μm and after further incubation for 1 h the reaction was quenched by the addition of ammonium hydroxide to 0.1 m. AC was separated by centrifugation (2 min for 104 × g) and the concentration of Cel7Abound-OA was measured as described for BC (see above).
Synergism between EG Cel5A and TrCel7A
[14C]BC (0.25–2.5 mg ml−1) was incubated with 0.25 μm TrCel7A and 0.1 μm β-glucosidase in 50 mm sodium acetate buffer, pH 5 (containing BSA 0.1 mg ml−1), at 25 °C for 1 h. If present, the concentration of EG TrCel5A was 25 nm. The reaction was stopped by adding NaOH to 0.1 m. Cellulose was separated by centrifugation (5 min at 104 × g) and the concentration of hydrolysis products was measured from the radioactivity in the supernatant using liquid scintillation counter.
Pre-treatment of BC with EG and Further Hydrolysis with Cel7A
For pre-treatment BC (1 mg ml−1) was incubated with 0.5 μm EG Cel5A for 1 h in 50 mm sodium acetate buffer, pH 5, at 25 °C. The reaction was stopped by adding NaOH up to 0.1 m. To remove EG, pre-treated BC was thoroughly washed with 0.1 m NaOH, water, and 50 mm sodium acetate buffer, pH 5, through centrifugation (10 min at 4500 × g) and re-suspension steps. For the following hydrolysis with TrCel7A, pre-treated BC (0.25–2.5 mg ml−1) was incubated with 0.1 μm TrCel7A in 50 mm sodium acetate buffer, pH 5 (containing BSA, 0.1 mg ml−1), at 25 °C for 1 h. The reaction was stopped by the addition of NaOH to 0.2 m. After separation of cellulose by centrifugation (2 min at 104 × g) the concentration of reducing sugars in the supernatant was measured using the MBTH method (71). Experiments without EG pre-treatment were performed exactly as described above (including washing steps) but the EG was omitted.
Author Contributions
P. V., R. K., K. B., and P. W. conceived and coordinated the study and wrote the paper. R. K., J. K., and P. V. designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by the Danish Agency for Science, Technology, and Innovation, Programme Commission on Sustainable Energy and Environment Grant 2104-07-0028, Carlsberg Foundation Grant 2013-01-0208, and Estonian Science Agency Grant PUT1024. K. B. works at Novozymes, which is a major enzyme producing company.

This article contains supplemental Table S1 and Figs. S1 and S2.
- GH
- glycoside hydrolase
- AA
- anthranilic acid
- AC
- amorphous cellulose
- BC
- bacterial cellulose
- BG
- β-glucosidase
- CB
- cellobiose
- CBH
- cellobiohydrolase
- CBM
- carbohydrate binding module
- CD
- catalytic domain
- TrCel7A
- cellobiohydrolase Cel7A from Trichoderma reesei
- IRG
- insoluble reducing groups
- MBTH
- 3-methyl-2-benzothiazolinone hydrazone hydrochloride
- MU
- 4-methyl-umbelliferone
- MUL
- 4-methyl-umbelliferyl-β-d-lactoside
- rAC
- reduced amorphous cellulose
- rBC
- reduced bacterial cellulose
- RGtot
- the total number of reducing groups
- SEE
- substrate exchange experiment
- SRG
- soluble reducing groups
- DSE
- degree of synergistic effect
- EG
- endoglucanase.
References
- 1. Ragauskas A. J., Williams C. K., Davison B. H., Britovsek G., Cairney J., Eckert C. A., Frederick W. J. Jr., Hallett J. P., Leak D. J., Liotta C. L., Mielenz J. R., Murphy R., Templer R., and Tschaplinski T. (2006) The path forward for biofuels and biomaterials. Science 311, 484–489 [DOI] [PubMed] [Google Scholar]
- 2. Himmel M. E., Ding S.-Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., and Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 3. Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., and Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 [DOI] [PubMed] [Google Scholar]
- 4. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., and Henrissat B. (2009) The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynd L. R., Weimer P. J., van Zyl W. H., and Pretorius I. S. (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kipper K., Väljamäe P., and Johansson G. (2005) Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as “burst” kinetics on fuorescent polymeric model substrates. Biochem. J. 385, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., and Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Payne C. M., Resch M. G., Chen L., Crowley M. F., Himmel M. E., Taylor L. E. 2nd, Sandgren M., Ståhlberg J., Stals I., Tan Z., and Beckham G. T. (2013) Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc. Natl. Acad. Sci. U.S.A. 110, 14646–14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carrard G., Koivula A., Söderlund H., and Béguin P. (2000) Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. U.S.A. 97, 10342–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McLean B. W., Boraston A. B., Brouwer D., Sanaie N., Fyfe C. A., Warren R. A., Kilburn D. G., and Haynes C. A. (2002) Carbohydrate-binding modules recognize fine substructures of cellulose. J. Biol. Chem. 277, 50245–50254 [DOI] [PubMed] [Google Scholar]
- 12. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., and Knox J. P. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Din N., Damude H. G., Gilkes N. R., Miller R. C. Jr., Warren R. A., and Kilburn D. G. (1994) C1-Cx revisited: intramolecular synergism in a cellulase. Proc. Natl. Acad. Sci. U.S.A. 91, 11383–11387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srisodsuk M., Reinikainen T., Penttilä M., and Teeri T. T. (1993) Role of the interdomain linker peptide of Trichoderma reesei cellobiohydrolase I in its interaction with crystalline cellulose. J. Biol. Chem. 268, 20756–20761 [PubMed] [Google Scholar]
- 15. Sammond D. W., Payne C. M., Brunecky R., Himmel M. E., Crowley M. F., and Beckham G. T. (2012) Cellulase linkers are optimized based on domain type and function: insights from sequence analysis, biophysical measurements, and molecular simulation. Plos One 7, e48615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox J. M., Jess P., Jambusaria R. B., Moo G. M., Liphardt J., Clark D. S., and Blanch H. (2013) A single-molecule analysis reveals morphological targets for cellulase synergy. Nat. Chem. Biol. 9, 356–361 [DOI] [PubMed] [Google Scholar]
- 17. Sugimoto N., Igarashi K., Wada M., and Samejima M. (2012) Adsorption characteristics of fungal family 1 cellulose-binding domain from Trichoderma reesei cellobiohydrolase I on crystalline cellulose: negative cooperative adsorption via a steric exclusion effect. Langmuir 28, 14323–14329 [DOI] [PubMed] [Google Scholar]
- 18. Guo J., and Catchmark J. M. (2013) Binding specificity and thermodynamics of cellulose-binding modules from Trichoderma reesei Cel7A and Cel6A. Biomacromolecules 14, 1268–1277 [DOI] [PubMed] [Google Scholar]
- 19. Cruys-Bagger N., Tatsumi H., Ren G. R., Borch K., and Westh P. (2013) Transient kinetics and rate-limiting steps for the processive cellobiohydrolase Cel7A: effects of substrate structure and carbohydrate binding domain. Biochemistry 52, 8938–8948 [DOI] [PubMed] [Google Scholar]
- 20. Várnai A., Siika-Aho M., and Viikari L. (2013) Carbohydrate-binding modules (CBMs) revisited: reduced amount of water counterbalances the need for CBMs. Biotechnol. Biofuels 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sørensen T. H., Cruys-Bagger N., Windahl M. S., Badino S. F., Borch K., and Westh P. (2015) Temperature effects on kinetic parameters and substrate affinity of Cel7A cellobiohydrolases. J. Biol. Chem. 290, 22193–22202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kari J., Olsen J., Borch K., Cruys-Bagger N., Jensen K., and Westh P. (2014) Kinetics of cellobiohydrolase (Cel7A) variants with lowered substrate affinity. J. Biol. Chem. 289, 32459–32468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Divne C., Ståhlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., and Jones T. A. (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 [DOI] [PubMed] [Google Scholar]
- 24. Divne C., Ståhlberg J., Teeri T. T., and Jones T. A. (1998) High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275, 309–325 [DOI] [PubMed] [Google Scholar]
- 25. Nakamura A., Tsukada T., Auer S., Furuta T., Wada M., Koivula A., Igarashi K., and Samejima M. (2013) The tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is important for initiation of degradation of crystalline cellulose. J. Biol. Chem. 288, 13503–13510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kostylev M., Alahuhta M., Chen M., Brunecky R., Himmel M. E., Lunin V. V., Brady J., and Wilson D. B. (2014) Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues. Biotechnol. Bioeng. 111, 664–673 [DOI] [PubMed] [Google Scholar]
- 27. Zhang S., Irwin D. C., and Wilson D. B. (2000) Site-directed mutation of noncatalytic residues of Thermobifida fusca exocellulase Cel6B. Eur. J. Biochem. 267, 3101–3115 [DOI] [PubMed] [Google Scholar]
- 28. Li Y., Irwin D. C., and Wilson D. B. (2007) Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Cel9A. Appl. Environ. Microbiol. 73, 3165–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., and Eijsink V. G. (2006) Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., and Eijsink V. G. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurašin M., Kuusk S., Kuusk P., Sørlie M., and Väljamäe P. (2015) Slow off-rates and strong product binding are required for processivity and efficient degradation of recalcitrant chitin by family 18 chitinases. J. Biol. Chem. 290, 29074–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beckham G. T., Ståhlberg J., Knott B. C., Himmel M. E., Crowley M. F., Sandgren M., Sørlie M., and Payne C. M. (2014) Towards a molecular-level theory of carbohydrate processivity in glycoside hydrolases. Curr. Opin. Biotechnol. 27, 96–106 [DOI] [PubMed] [Google Scholar]
- 33. Knott B. C., Haddad Momeni M., Crowley M. F., Mackenzie L. F., Götz A. W., Sandgren M., Withers S. G., Ståhlberg J., and Beckham G. T. (2014) The mechanism of cellulose hydrolysis by a two-step, retaining cellobiohydrolase elucidated by structural and transition path sampling studies. J. Am. Chem. Soc. 136, 321–329 [DOI] [PubMed] [Google Scholar]
- 34. Knott B. C., Crowley M. F., Himmel M. E., Ståhlberg J., and Beckham G. T. (2014) Carbohydrate-protein interactions that drive processive polysaccharide translocation in enzymes revealed from a computational study of cellobiohydrolase processivity. J. Am. Chem. Soc. 136, 8810–8819 [DOI] [PubMed] [Google Scholar]
- 35. Kurasin M., and Väljamäe P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jalak J., and Väljamäe P. (2010) Mechanism of initial rapid rate retardation in cellobiohydrolase catalyzed cellulose hydrolysis. Biotechnol. Bioeng. 106, 871–883 [DOI] [PubMed] [Google Scholar]
- 37. Jalak J., Kurašin M., Teugjas H., and Väljamäe P. (2012) Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 287, 28802–28815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cruys-Bagger N., Elmerdahl J., Praestgaard E., Tatsumi H., Spodsberg N., Borch K., and Westh P. (2012) Pre-steady state kinetics for the hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J. Biol. Chem. 287, 18451–18458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruys-Bagger N., Elmerdahl J., Praestgaard E., Borch K., and Westh P. (2013) A steady-state theory for processive cellulases. FEBS J. 280, 3952–3961 [DOI] [PubMed] [Google Scholar]
- 40. von Ossowski I., Ståhlberg J., Koivula A., Piens K., Becker D., Boer H., Harle R., Harris M., Divne C., Mahdi S., Zhao Y., Driguez H., Claeyssens M., Sinnott M. L., and Teeri T. T. (2003) Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A: a comparison with Phanerochaete chrysosporium Cel7D. J. Mol. Biol. 333, 817–829 [DOI] [PubMed] [Google Scholar]
- 41. Medve J., Karlsson J., Lee D., and Tjerneld F. (1998) Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59, 621–634 [PubMed] [Google Scholar]
- 42. Brady S. K., Sreelatha S., Feng Y., Chundawat S. P., and Lang M. J. (2015) Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps. Nat. Commun. 6, 10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamura A., Watanabe H., Ishida T., Uchihashi T., Wada M., Ando T., Igarashi K., and Samejima M. (2014) Trade-off between processivity and hydrolytic velocity of cellobiohydrolases at the surface of crystalline cellulose. J. Am. Chem. Soc. 136, 4584–4592 [DOI] [PubMed] [Google Scholar]
- 44. Luterbacher J. S., Walker L. P., and Moran-Mirabal J. M. (2013) Observing and modeling BMCC degradation by commercial cellulase cocktails with fluorescently labeled Trichoderma reseii Cel7A through confocal microscopy. Biotechnol. Bioeng. 110, 108–117 [DOI] [PubMed] [Google Scholar]
- 45. Shibafuji Y., Nakamura A., Uchihashi T., Sugimoto N., Fukuda S., Watanabe H., Samejima M., Ando T., Noji H., Koivula A., Igarashi K., and Iino R. (2014) Single-molecule imaging analysis of elementary reaction steps of Trichoderma reesei cellobiohydrolase I (Cel7A) hydrolyzing crystalline cellulose Iα and IIII. J. Biol. Chem. 289, 14056–14065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fox J. M., Levine S. E., Clark D. S., and Blanch H. W. (2012) Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry 51, 442–452 [DOI] [PubMed] [Google Scholar]
- 47. Shang B. Z., Chang R., and Chu J.-W. (2013) Systems-level modeling with molecular resolution elucidates the rate-limiting mechanisms of cellulose decomposition by cellobiohydrolases. J. Biol. Chem. 288, 29081–29089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jalak J., and Väljamäe P. (2014) Multi-mode binding of cellobiohydrolase Cel7A from Trichoderma reesei to cellulose. Plos One 9, e108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cruys-Bagger N., Alasepp K., Andersen M., Ottesen J., Borch K., and Westh P. (2016) Rate of threading a cellulose chain into the binding tunnel of a cellulase. J. Phys. Chem. B 120, 5591–5600 [DOI] [PubMed] [Google Scholar]
- 50. Payne C. M., Jiang W., Shirts M. R., Himmel M. E., Crowley M. F., and Beckham G. T. (2013) Glycoside hydrolase processivity is directly related to oligosaccharide binding free energy. J. Am. Chem. Soc. 135, 18831–18839 [DOI] [PubMed] [Google Scholar]
- 51. Horn S. J., Sørlie M., Vårum K. M., Väljamäe P., and Eijsink V. G. (2012) Measuring processivity. Methods Enzymol. 510, 69–95 [DOI] [PubMed] [Google Scholar]
- 52. Väljamäe P., Sild V., Nutt A., Pettersson G., and Johansson G. (1999) Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 266, 327–334 [DOI] [PubMed] [Google Scholar]
- 53. Väljamäe P., Pettersson G., and Johansson G. (2001) Mechanism of substrate inhibition in cellulose synergistic degradation. Eur. J. Biochem. 268, 4520–4526 [DOI] [PubMed] [Google Scholar]
- 54. Praestgaard E., Elmerdahl J., Murphy L., Nymand S., McFarland K. C., Borch K., and Westh P. (2011) A kinetic model for the burst phase of processive cellulases. FEBS J. 278, 1547–1560 [DOI] [PubMed] [Google Scholar]
- 55. Kuusk S., Sørlie M., and Väljamäe P. (2015) The predominant molecular state of bound enzyme determines the strength and type of product inhibition in the hydrolysis of recalcitrant polysaccharides by processive enzymes. J. Biol. Chem. 290, 11678–11691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jung J., Sethi A., Gaiotto T., Han J. J., Jeoh T., Gnanakaran S., and Goodwin P. M. (2013) Binding and movement of individual Cel7A cellobiohydrolases on crystalline cellulose surfaces revealed by single-molecule fluorescence imaging. J. Biol. Chem. 288, 24164–24172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Linder M., and Teeri T. T. (1996) The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc. Natl. Acad. Sci. U.S.A. 93, 12251–12255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Linder M., Salovuori I., Ruohonen L., and Teeri T. T. (1996) Characterization of a double cellulose-binding domain: synergistic high affinity binding to crystalline cellulose. J. Biol. Chem. 271, 21268–21272 [DOI] [PubMed] [Google Scholar]
- 59. Hildén L., Väljamäe P., and Johansson G. (2005) Surface character of pulp fibres studied using endoglucanases. J. Biotechnol. 118, 386–397 [DOI] [PubMed] [Google Scholar]
- 60. Ghattyvenkatakrishna P. K., Alekozai E. M., Beckham G. T., Schulz R., Crowley M. F., Uberbacher E. C., and Cheng X. (2013) Initial recognition of a cellodextrin chain in the cellulose-binding tunnel may affect cellobiohydrolase directional specificity. Biophys. J. 104, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koivula A., Kinnari T., Harjunpää V., Ruohonen L., Teleman A., Drakenberg T., Rouvinen J., Jones T. A., and Teeri T. T. (1998) Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 429, 341–346 [DOI] [PubMed] [Google Scholar]
- 62. Taylor C. B., Payne C. M., Himmel M. E., Crowley M. F., McCabe C., and Beckham G. T. (2013) Binding site dynamics and aromatic-carbohydrate interactions in processive and non-processive family 7 glycoside hydrolases. J. Phys. Chem. B 117, 4924–4933 [DOI] [PubMed] [Google Scholar]
- 63. Jana S., Hamre A. G., Wildberger P., Holen M. M., Eijsink V. G., Beckham G. T., Sørlie M., and Payne C. M. (2016) Aromatic-mediated carbohydrate recognition in processive Serratia marcescens chitinases. J. Phys. Chem. B 120, 1236–1249 [DOI] [PubMed] [Google Scholar]
- 64. Colussi F., Sørensen T. H., Alasepp K., Kari J., Cruys-Bagger N., Windahl M. S., Olsen J. P., Borch K., and Westh P. (2015) Probing substrate interactions in the active tunnel of a catalytically deficient cellobiohydrolase (Cel7). J. Biol. Chem. 290, 2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ståhlberg J., Johansson G., and Pettersson G. (1991) A new model for enzymatic hydrolysis of cellulose based on the two-domain structure of cellobiohydrolase I. Nat. Biotechnol. 9, 286–290 [Google Scholar]
- 66. Bhikhabhai R., Johansson G., and Pettersson G. (1984) Isolation of cellulolytic enzymes from Trichoderma reesei QM 9414. J. Appl. Biochem. 6, 336–345 [PubMed] [Google Scholar]
- 67. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., and Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414: analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 68. Sipos B., Benko Z., Dienes D., Réczey K., Viikari L., and Siika-aho M. (2010) Characterisation of specific activities and hydrolytic properties of cell-wall-degrading enzymes produced by Trichoderma reesei Rut C30 on different carbon sources. Appl. Biochem. Biotechnol. 161, 347–364 [DOI] [PubMed] [Google Scholar]
- 69. Velleste R., Teugjas H., and Väljamäe P. (2010) Reducing end-specific fluorescence labeled celluloses for cellulase mode of action. Cellulose 17, 125–138 [Google Scholar]
- 70. Kostylev M., and Wilson D. (2013) Two-parameter kinetic model based on a time-dependent activity coefficient accurately describes enzymatic cellulose digestion. Biochemistry 52, 5656–5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Horn S. J., and Eijsink V. G. H. (2004) A reliable reducing end assay for chito-oligosaccharides. Carbohydr. Polym. 56, 35–39 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.