Abstract
The persistence of HIV in resting memory CD4+ T cells at a latent state is considered as the major barrier on the path to achieve a cure for HIV. Proteasome inhibitors (PIs) were previously reported as latency reversing agents (LRAs) but the mechanism underlying this function is yet unclear. Here we demonstrate that PIs reactivate latent HIV ex vivo without global T cell activation, and may facilitate host innate immune responses. Mechanistically, latent HIV reactivation induced by PIs is mediated by heat shock factor 1 (HSF1) via the recruitment of the heat shock protein (HSP) 90-positive transcriptional elongation factor b (p-TEFb) complex. Specifically, HSP90 downstream HSF1 gives positive feedback to the reactivation process through binding to cyclin-dependent kinase 9 (CDK9) and preventing it from undergoing degradation by the proteasome. Overall, these findings suggest proteasome inhibitors as potential latency reversing agents. In addition, HSF1/HSP90 involved in HIV transcription elongation, may serve as therapeutic targets in HIV eradication.
Keywords: cyclin-dependent kinase (CDK), Heat shock factor protein 1 (HSF1), heat shock protein 90 (Hsp90), human immunodeficiency virus (HIV), proteasome, latency reversing agent, p-TEFb
Introduction
Combined anti-retroviral therapy (cART)3 effectively prolongs the lifetime of AIDS patients and limits the spread of the virus. However, latent HIV reservoirs left post-treatment have become the primary barrier to cure HIV (1). Because latent HIV is undetectable at protein levels, it escapes the monitoring of the host immune system. To overcome this situation, a promising strategy named “shock and kill” has been proposed. It prevents new infections and the reestablishment of the latent reservoir upon the activation of the virus (2). To date, many latency reversing agents (LRAs) were discovered. Yet, besides disulfirm and panobinostat, which show good activity in vivo (3, 4), there are barely LRAs that can reduce HIV reservoirs.
Proteasome inhibitors (PIs) are in clinical use and have been demonstrated to exhibit efficient anti-cancer activity (5). Unexpectedly, bortezomib (BTZ) was reported as a bifunctional HIV antagonist. It inhibits HIV infection and also reactivates latent HIV with reduced infectivity (6). However, the mechanism of latent HIV reactivation via PIs remains to be elucidated. In addition, the use of second generation PIs, such as carfilzomib (CFZ), to reactivate latent HIV has not been reported. This is particularly important because CFZ is effective on both hematologic and solid malignancies.
It is well known that proteasome inhibition induces endoplasmic reticulum stress (ER stress), in which heat shock proteins (HSPs) and their transcription factor HSF1 widely participate (7). HSF1 has been thoroughly studied in cancer (8). In addition, it binds to the HIV 5′-long terminal repeat (LTR) and positively plays a role in HIV vital activities. Recently, we also revealed a key role for the active form of HSF1 in mediating latent HIV transcription and reactivation (9). Furthermore, HSP90 has been shown to control HIV reactivation from latency, by interacting with IKK and being involved in the degradation of IκBα and NF-κB translocation (10, 11). Recently, Joshi et al. (12) reported that inhibition of HSP90 prevents the recovery of HIV. This suggests HSP90 inhibitors as alternatives or supplementary to cART to suppress the formation of persistent HIV reservoirs (12). These studies confirm a role for HSP90 in latent HIV reactivation. However, the interplay between HSP90 and host cellular factors associated with gene transcriptional regulation requires more research. Here we investigated specifically the role of HSP90 in latent HIV reactivation under proteasome inhibition.
Toward this goal, we studied the ability of carfilzomib to reactivate latent HIV in primary CD4+ T cells from suppressive HIV+ patients, as well as in HIV latency cell models. Furthermore, the role of HSF1 in the reactivation process under proteasome inhibition was examined. We found that HSF1 was activated and it recruited the HSP90·p-TEFb complex to promote transcription elongation. Then, HSP90 was elevated and it bound to CDK9 thus preventing its degradation by ubiquitin-proteasome. Besides shedding light on the mechanism of PIs reactivation of latent HIV, this study suggests HSF1/HSP90 as potential therapeutic targets.
Results
PIs Reactivate Latent HIV in Both Latency Cell Models and Primary CD4+ T Cells
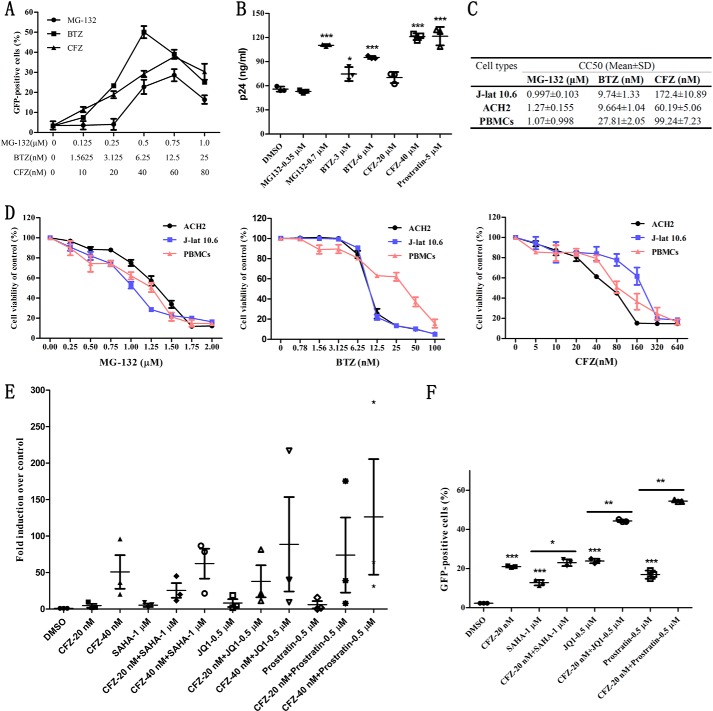
To verify the effect of PIs on latent HIV, we took J-lat 10.6 and ACH2 latency cell models as study systems. This was followed by treatment with (i) pan-proteasome inhibitor MG132, (ii) reversible proteasome inhibitor bortezomib, and (iii) irreversible proteasome inhibitor carfilzomib. J-lat 10.6 is a human Jurkat cell line integrated with a full-length HIV gene containing GFP, which allows monitoring of viral transcriptional activity (13). ACH2 is a HIV latently infected cell line with abundant secretion of infectious HIV particles under stimulation (14). After treatment with the inhibitors, the percentage of GFP-positive J-lat 10.6 cells was measured via flow cytometry (FCM). Then, the concentration of p24 in the culture supernatants of ACH2 cells was determined by enzyme-linked immunosorbent assay (ELISA). Fig. 1A shows that both BTZ and MG-132 effectively induced latent HIV LTR-driven expression of GFP. This is in agreement with a previous report (6). In addition, we found that CFZ exerted a similar activity as BTZ did. It produced the highest amount of GFP (40%) under the working concentration of 60 nm. Next, two concentrations of MG132, BTZ, and CFZ were selected to stimulate ACH2 cells. As shown in Fig. 1B, higher concentrations of PIs promoted more secretion of p24. Furthermore, the concentration of p24 in the supernatant of ACH2 cells stimulated by 40 nm CFZ was equal to that stimulated by 5 μm prostratin.
FIGURE 1.
The influence of PIs on latency cell models and primary CD4+ T cells from patients. A, the percentage of GFP-positive cells was measured by FCM after treatment with MG-132, BTZ, and CFZ on J-lat 10.6 for 48 h at the indicated concentrations. B, P24 in culture supernatant of ACH2 was detected by ELISA after treatment for 48 h with PIs at the indicated concentrations. C, CC50 of PIs in three cell types used in this study. D, the cell viability of ACH2, J-lat 10.6, and PBMCs were evaluated by CCK-8 after treatment for 48 h with PIs. E, primary CD4+ T cells isolated from three suppressive HIV+ patients were co-treated for 48 h with CFZ (20 nm) and three classical LRAs (SAHA, JQ1, and prostratin), and the transcription of HIV 5′-LTR was analyzed by qPCR. Data are reported as the mean ± S.E. from three independent experiments. F, J-lat 10.6 cells were co-treated for 48 h with CFZ (20 nm) and three classical LRAs (SAHA, JQ1, and prostratin), and the percentage of GFP-positive cells was measured by FCM. The p values were defined as: *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
In view of the finding that PIs are toxic to cells, we further evaluated their effect on the cell viability of J-lat 10.6, ACH2, and peripheral blood mononuclear cells (PBMCs) using the CCK-8 assay. The CC50 of MG-132, BTZ, and CFZ in the three cell types used here were calculated as described in the legend to Fig. 1C and shown in Fig. 1D. The data reveal that MG132 displayed similar toxicity in J-lat 10.6, ACH2, and PBMCs. BTZ and CFZ behaved the same, and had CC50 of 9.74 and 172 nm for BTZ and CFZ, respectively, which are 1.6- and 4-fold higher than the working concentrations. Here, we chose 6 nm BTZ and 40 nm CFZ, which generated relatively weak toxicity, as working concentrations for further studies.
To further verify the latency reversing effect of CFZ and to evaluate its clinical value, we examined the reactivation effect of CFZ on primary CD4+ T cells from HIV-infected individuals receiving suppressive cART. Three patients who fulfilled the criteria described under ”Experimental Procedures“ were involved. The transcription of HIV 5′-LTR was analyzed by quantitative PCR (qPCR). The data revealed that the transcription of HIV LTR was dramatically up-regulated after treatment with 40 nm CFZ, but was relatively weak using 20 nm CFZ (Fig. 1E). Moreover, a synergistic effect was observed with the combination of CFZ and the representative LRAs SAHA, JQ1, and prostratin both in primary CD4+ T cells and J-lat 10.6 cells (Fig. 1, E and F). Taken together, these results demonstrate that PIs can effectively reactivate latent HIV, and also have a strong synergistic effect with classical LRAs in both the latency model and primary CD4+ T cells.
PIs Reactivate Latent HIV without Global T Cell Activation and Facilitate an Innate Immune Response
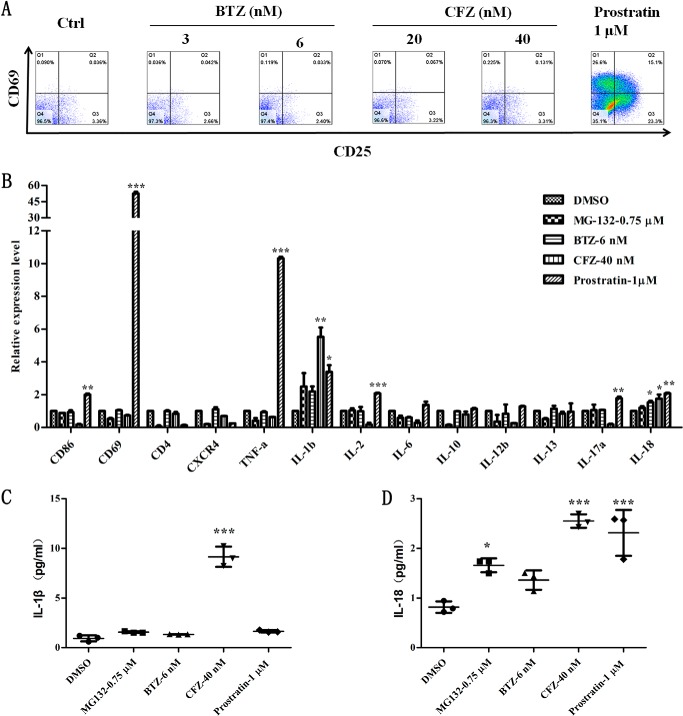
When combined with cART, activators often do not show good results in clinical trials (15). This could be attributed partially to global T cell activation and the absence of cytotoxic T lymphocyte (CTL) response (16, 17). To find out whether PIs exhibit similar traits, PBMCs isolated from human whole blood were treated with CFZ or BTZ followed by FCM detection. Interestingly, CFZ and BTZ, at two effective concentrations, did not induce the expression of activated T cell surface markers CD25 and CD69. However, prostratin reactivated latent HIV via activation of T cells (Fig. 2A) (18). Cells after treatment were followed by qPCR analysis of immune cytokines and cell surface receptors. Besides IL-1β and IL-18, there was no T cell marker or immune cytokine up-regulated under the treatment with PIs (Fig. 2B). The secretion of IL-1β and IL-18 were also observed in the cell culture supernatant (Fig. 2, C and D). In comparison, CD69, TNF-α, IL-2, and IL-17α were up-regulated under the treatment with prostratin in addition to IL-1β and IL-18. Because the transcription of genes closely related to HIV infection are inhibited, it could provide further evidence that PIs are bifunctional HIV antagonists as previously described (6). The decrease in inflammatory cytokines might imply an anti-inflammatory effect of PIs.
FIGURE 2.
PIs promote the innate immune response without causing global T cell activation. A, surface markers of T cell CD25 and CD69 of PBMCs were detected by FCM after treatment for 24 h with BTZ and CFZ. Prostratin (1 μm) was used as a positive control. B, the mRNA expression of surface markers and immune cytokines were analyzed by qPCR after treating with PIs and prostratin for 12 h. C and D, the secretion of IL-1β and IL-18 were measured by ELISA after treatment for 24 h. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
Note that the secretion of IL-1β and IL-18 triggered the host access to pyroptosis and facilitated CTL response against allogenic pathogens at the same time (19). The abundant release of pro-inflammatory cytokines resulted in inflammation-associated pathologies. In comparison, selective and slight secretion of IL-1β and IL-18 promoted the host innate immune response (20). Therefore, these results suggest that PIs not only inhibits T cell activation and HIV infection, but can also promote the innate immune response. This may trigger CTL response to assist in the elimination of latent reservoirs.
Proteasome Inhibition-induced Latent HIV Reactivation Is HSF1 Dependent
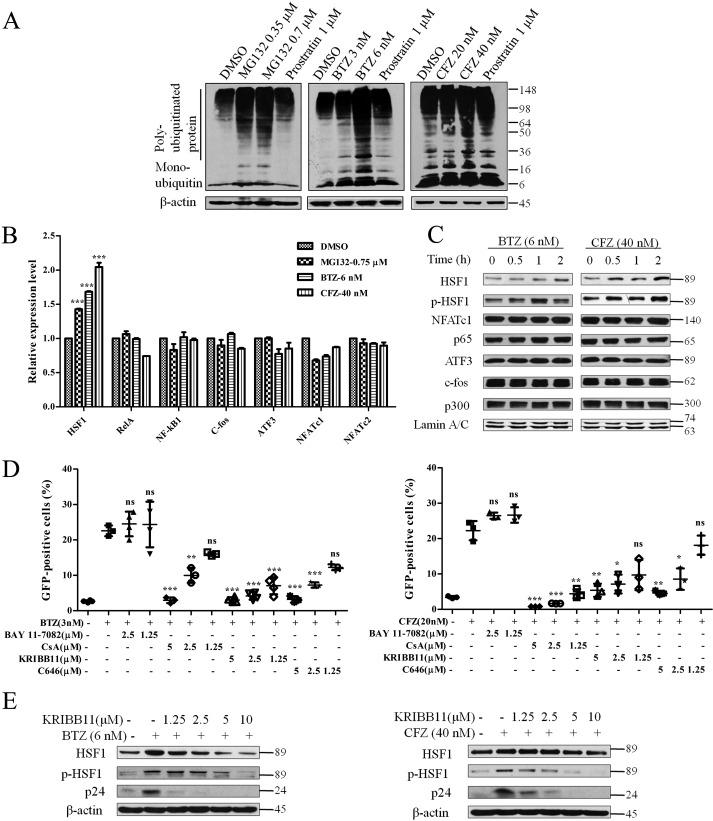
Based on the findings that PIs reactivate latent HIV, we further studied the underlying mechanism. Viral infection will influence cell response and may activate HSF1 and its downstream HSPs (21). Therefore, the J-lat 10.6 cell line, which does not produce viruses, was employed in our following research. First, we examined the extent of proteasome inhibition through detection of polyubiquitinated proteins by Western blotting. As shown in Fig. 3A, polyubiquitinated proteins obviously accumulated after the treatment with PIs. Next, we explored the nuclear transcription factors participating in this process. Various transcription factors were examined by analyzing their mRNA expression. Subunits of NF-κB (RelA and NF-κB1), T cell activation transcription factors (NFATc1 and NFATc2), and AP-1 subunits (c-fos and ATF3) were not increased when treated with PIs (Fig. 3B). In contrast, the stress protein transcription factor, HSF1, was significantly up-regulated. In addition to mRNA expression, we also investigated the protein expression of transcription regulation factors accumulated in the nucleus. In agreement with the results of mRNA expression, active HSF1 (p-HSF1) and HSF1 gradually increased, yet NFATc1, p65, ATF3, and c-fos remained unchanged (Fig. 3C). The histone acetylase p300 remained unchanged too.
FIGURE 3.
Transcription factors that participate in the latent HIV reactivation under proteasome inhibition. A, the accumulated polyubiquitinated protein was detected by Western blotting after J-lat 10.6 cells were treated with PIs and prostratin for 48 h. B, the mRNA expression of transcription regulation factors were analyzed by qPCR after treatment with PIs for 12 h. C, the accumulation of transcription factors and co-factors in the nucleus within 2 h was examined by Western blotting analysis after stimulation with 6 nm BTZ and 40 nm CFZ. D, PIs were co-treated with inhibitors on J-lat 10.6 for 48 h at the indicated concentrations. The percentage of GFP-positive cells was measured by FCM, and E, the expression of the corresponding proteins was measured by Western blotting analysis. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
Next, we carried out inhibition assays using various inhibitors to validate the results observed in the mRNA and protein expression. The data reveal that the NFAT inhibitor (CsA), HSF1 inhibitor (KRIBB11), and p300 inhibitor (C646), lowered the GFP % in a dose-dependent manner (Fig. 3D). The NF-κB inhibitor (BAY 11-7082) not only did not inhibit reactivation, but even seems to maintain the level (Fig. 3D), which is different from the latent HIV reactivation by PEP005 (15). Because HSF1 was confirmed to be a transcription factor of HIV 5′-LTR (22), we examined the protein expression of HSF1, p-HSF1 (Ser-320), and HIV capsid protein p24, under co-treatment with PIs and KRIBB11. Consistently, the proteins were up-regulated by the treatment of PIs and down-regulated by KRIBB11 (Fig. 3E). Collectively, although NFAT, AP-1, and p300 may participate in the reactivation process at basal levels, HSF1 is positively correlated with latent HIV reactivation mediated by proteasome inhibition.
HSF1 Promotes Latent HIV Reactivation through p-TEFb Recruitment
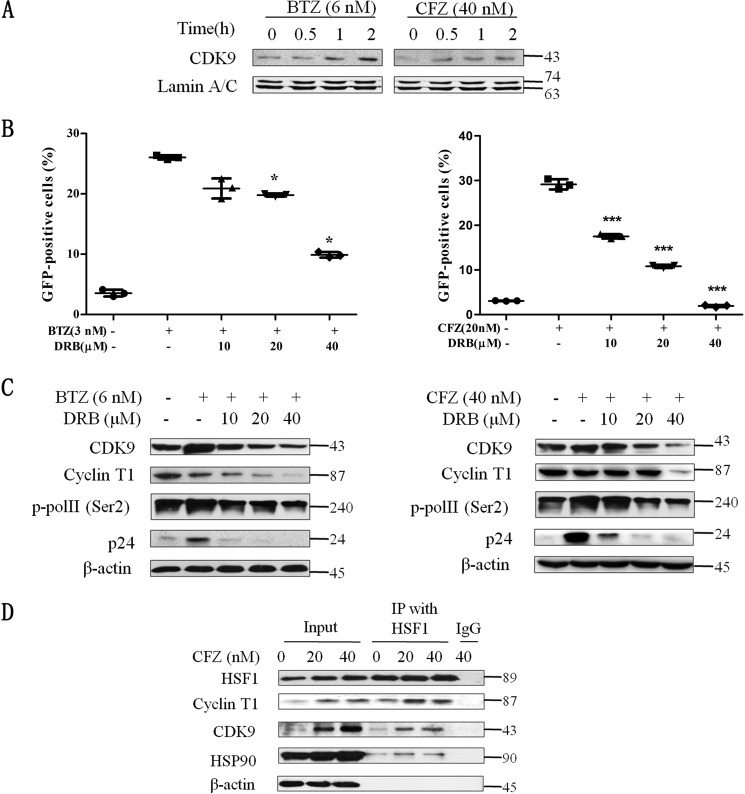
CDK9, is a subunit of the p-TEFb complex (composed of CDK9 and Cyclin T1), which promotes HIV gene transcription elongation through phosphorylating Pol II at Ser-2 and Ser-5 (23). Besides HSF1, CDK9 was also found to accumulate in the nucleus (Fig. 4A). This result indicates that transcription elongation in relationship to p-TEFb might widely participate in latent HIV reactivation induced by proteasome inhibition. In addition, the understanding that transcription elongation via p-TEFb is a rate-limiting step for latent HIV reactivation (24) strengthens our conclusion. To validate the role of p-TEFb in latency reactivation mediated by proteasome inhibition, DRB, the CDK9-specific inhibitor, was used with PIs to treat J-lat 10.6 cells. The data revealed that DRB decreased the GFP % induced by PIs in a dose-dependent manner (Fig. 4B). Simultaneously, the protein expression of p-TEFb subunits, CDK9 and cyclin T1, RNA pol II (Ser-2), as well as, HIV capsid protein p24, were detected by Western blotting analysis. In agreement with GFP %, BTZ, and CFZ elevated the expression of these proteins and DRB inhibited their expression (Fig. 4C).
FIGURE 4.
Transcriptional elongation factor p-TEFb participates in the reactivation of latent HIV under proteasome inhibition. A, the accumulation of CDK9 in the nucleus induced by PIs in 2 h was detected by Western blotting. B, PIs were co-treated with p-TEFb inhibitor (DRB) at the indicated concentrations for 48 h. Then the percentage of positive GFP cells was measured by FCM and (C) the corresponding total protein level was measured by Western blotting analysis. D, HSF1 antibody pulled down the complex of p-TEFb and HPS90 treated with CFZ at 48 h. *, p < 0.05 and ***, p < 0.001 versus control.
HSF1 was reported to promote the transcription of HIV genes by binding to the HIV 5′-LTR (22) and recruiting p-TEFb to the HSP70 promoter in colon cancer cells (25). Therefore, a set of co-immunoprecipitation experiments were performed to reveal the relationship between HSF1 and p-TEFb under proteasome inhibition conditions. As shown in Fig. 4D, CDK9 and Cyclin T1 were co-precipitated with HSF1. Moreover, HSP90 was detected in the p-TEFb complex recruited by HSF1. Taken together, these results demonstrate that p-TEFb widely participates in the latent HIV reactivation induced by proteasome inhibition, and HSF1 recruits p-TEFb to facilitate transcription elongation in the reactivation process.
HSP90 Contributes to the Reactivation Process Downstream of HSF1
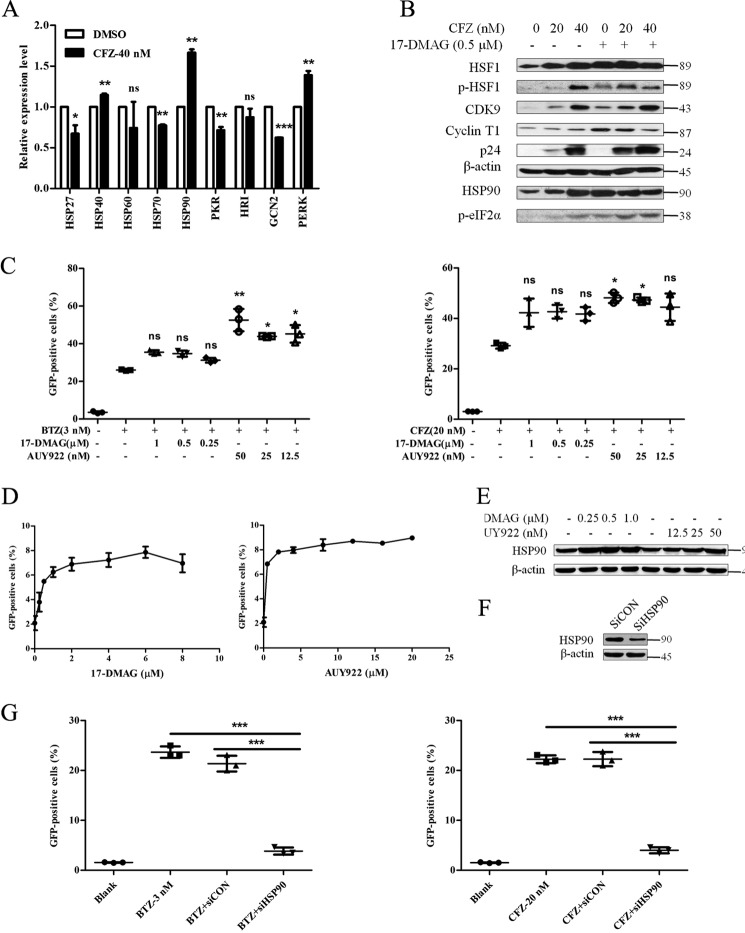
In addition to HSP90 being transcribed by HSF1, we showed that it is pulled down by HSF1. This suggests its important role in HIV latency reactivation. To further elucidate how HSP90 takes part in latency reactivation mediated by the HSF1 downstream pathway, we conducted HSP90 inhibition and HSP90 knockdown assays. First, the mRNA expression of HSPs and stress response kinases were evaluated by qPCR. As shown in Fig. 5A, only the expression of HSP90 and PKR-like endoplasmic reticulum kinase increased, whereas other HSPs (HSP27, HSP40, HSP60, and HSP70) and kinases (PKR, HRI, and GCN2) did not. These may reflect blocking of translation due to ER stress, which would be aggravated by the decreased expression of these proteins. Next, the HSP90 functional inhibitor (17-DMAG), which binds to the ATP binding site of HSP90, was co-treated with CFZ. Interestingly, 17-DMAG enhanced the expression of p24, p-HSF1, CDK9, and a cell stress marker p-eIF2α (phosphorylation of the α subunit of eukaryotic initiation factor 2) (Fig. 5B). Meanwhile, 17-DMAG maintained the GFP % induced by BTZ and CFZ (Fig. 5C). Furthermore, another HSP90 inhibitor (NVP-AUY922), which accelerates HSP90 binding to chaperones, increased GFP % as well.
FIGURE 5.
HSP90 positively regulates the latent HIV reactivation induced by PIs. A, J-lat 10.6 cells were treated with CFZ (40 nm) for 12 h and the mRNA expression of HSPs and stress kinases were detected by qPCR. B, total protein indicated in the figure was measured after co-treatment with CFZ and 17-DMAG for 48 h on J-lat 10.6 cells. C, the percentage of GFP-positive cells was measured by FCM after co-treatment for 48 h with PIs and HSP90 inhibitors. D, the percentage of GFP-positive cells was measured by FCM after treatment with NVP-AUY922 and 17-DMAG on J-lat 10.6 for 48 h. E, the expression of HSP90 was detected by Western blotting analysis after treatment for 48 h with NVP-AUY922 and 17-DMAG on J-lat 10.6. F, the knockdown of HSP90 was verified by detecting the expression of HSP90 after electroporating siRNA into J-lat 10.6 cells for 48 h. G, the percentage of GFP-positive cells was measured by FCM after treating with PIs on HSP90-knockdown cells for 48 h. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
Therefore, the independent influence of 17-DMAG and NVP-AUY922 was examined. The data reveal that both inhibitors could slightly reactivate latent HIV (Fig. 5D). Moreover, they up-regulated the expression of HSP90 (Fig. 5E). This is in agreement with a previous study showing that HSP90 inhibitors induced HSP90 expression (26). Consequently, HSP90 was knocked down to confirm its function in latency reactivation. The effect of HSP90 knockdown was then detected by Western blotting analysis (Fig. 5F). After HSP90 knockdown, J-lat cells were treated with BTZ and CFZ as usual. In comparison to a control group, HSP90 knockdown significantly impaired GFP % induced by either BTZ or CFZ (Fig. 5G).
Overall, we conclude that the amount of HSP90 is in direct correlation with the extent of latent HIV reactivation mediated by proteasome inhibition. Presumably, the recruitment of p-TEFb by HSF1 plays a leading role in latent HIV reactivation, and the generated HSP90 gives positive feedback to HSF1.
HSP90 and CDK9 Synergistically Enhance the Function of HSF1
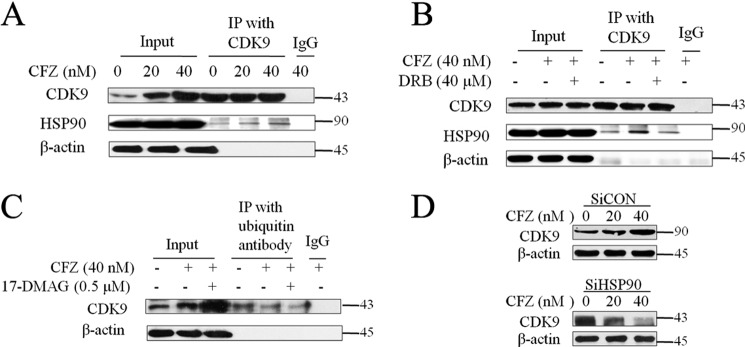
We demonstrated that HSP90 is an important factor in reactivating latent HIV mediated by HSF1. Therefore, we further explored how it works with p-TEFb. Here we show that HSP90 is involved in the complex of p-TEFb recruited by HSF1. Other studies reported that the HSP90·Cdc37 complex interacts with p-TEFb and influences the activity of CDK9 (27). To determine the relationship between HSP90 and CDK9 under proteasome inhibition, we showed a direct interaction between HSP90 and CDK9. As shown in Fig. 6A, CDK9 pulled down HSP90 in a dose-dependent manner. In addition, the CDK9 inhibitor, DRB, hampered the interaction between CDK9 and HSP90 (Fig. 6B). These results demonstrate that the binding of HSP90 to CDK9 is closely related to the activity of CDK9. In other words, we believe that the binding of HSP90 enhances the activity of CDK9 on reversing HIV latency.
FIGURE 6.
The relationship between HSP90 and CDK9 under proteasome inhibition. A and B, J-lat 10.6 cells were treated with CFZ alone or co-treated with the CDK9 inhibitor (DRB) for 48 h. HSP90 was detected by Western blotting analysis after being pulled down by CDK9 antibodies. C, ubiquitylated CDK9 was detected through pulling down CDK9 by polyubiquitin antibody after co-treatment with CFZ and 17-DMAG for 48 h. D, HSP90-knockdown cells were treated with CFZ for 48 h, and the expression of CDK9 was detected by Western blotting analysis. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
We further investigated how proteasome inhibition aids the positive potency of HSP90. Our results revealed that polyubiquitinated proteins were accumulated in J-lat cells under treatment with PIs (Fig. 3A). Therefore, we analyzed the ubiquitination modification of CDK9. The data revealed that the CDK9 pulled down by the polyubiquitin antibody did not increase. Concomitantly, total CDK9 increased (Fig. 6C). Moreover, lower expression of HSP90 due to HSP90 knockdown decreased CDK9 expression induced by CFZ, despite the fact that CFZ increased CDK9 expression in the control group (Fig. 6D). These results demonstrate that HSP90 facilitates the latent HIV reactivation under proteasome inhibition by preventing CDK9 from degradation by ubiquitin-proteasome. Thus it enhances the influence of HSF1, which recruits p-TEFb to function.
Discussion
LRAs operate primarily via three pathways: chromatin/HDAC, PKC/NF-κB, and RNA pol II/p-TEFb. They are responsible for chromatin deconstruction, transcription initiation, and the transcription elongation step, respectively. Usually, NFAT, AP-1, and NF-κB are involved in the HIV 5′-LTR transcription regulation domain (27). In the present study, proteasome inhibition reactivated latent HIV without causing NF-κB activation. Instead, HSF1 works as a hub. It was first reported in 2011 as a transcription factor for HIV genes (22). However, its role in HIV infection is not yet fully known even though HSPs were already reported to interact with HIV proteins Nef and Tat in the transcription process (28, 29). Here, we revealed the role of HSF1 in reversing HIV latency under proteasome inhibition, where HSF1 can recruit p-TEFb. This indicates that HSF1 works on both HIV gene transcription and elongation steps.
We found that PKR-like endoplasmic reticulum kinase and p-eIF2α respond under proteasome inhibition (Fig. 5, A and B). This indicates that proteasome inhibition causes ER stress in HIV latency cells. We therefore propose PIs as stress/HSF1 modulators, a novel class of LRAs, which is different from PKC modulators (30), HDACis (31), and p-TEFb activators (32). PKC modulators have synergistic effects with HDACis and p-TEFb activators (15, 24). Similarly, carfilzomib, the representative compound of stress/HSF1 modulators, shows synergistic effects with prostratin, SAHA, and JQ1, the representative compounds of the other three pathways (Fig. 1, E and F). Also, in our study HSP90 inhibitors (expression inducers) synergize with stress/HSF1 modulators.
Importantly, stress/HSF1 modulators have many advantages because they reactivate latent HIV without global T cell activation. They selectively promote the release of innate immune cytokines in addition to their ability to suppress viral infection. This also suggests the involvement of NF-κB inhibition. An interesting question is how proteasome inhibition causes selective generation of IL-1β and IL-18, but suppresses other inflammatory cytokines such as TNF-α, IL-6, and others. A possible explanation is that IL-1β and IL-18 are cleaved by caspase-1 and then released into the extracellular domain where pyroptosis takes place (33). The accumulation of misfolded proteins due to ER stress results in cell pyroptosis as well as NF-κB inhibition. Note that NF-κB has a central role in inflammatory responses by generating most of the inflammatory cytokines (34). In light of these results, the data regarding IL-1β and IL-18 imply that additional transcription initiation factors could be hijacked when the proteasome is inhibited. The activation of HSF1 may partially account for that. Nevertheless, more evidence is needed.
From a therapeutic point of view, PIs did not induce T cell activation and selectively promoted the generation of innate immune cytokines. There is a possibility that a PI will influence antigen presentation (35) and therefore may prevent it from triggering the immune system to clear latent HIV reservoirs. This should be taken into consideration when assessing the feasibility of LRAs.
Anderson et al. (11) suggested that HSP90 can control the reactivation of HIV from latency through interacting with inhibitors of IKK. Joshi et al. (12) reported that inhibition of HSP90 prevents HIV rebound. Joshi et al. (12) suggested HSP90 inhibitors as alternatives or supplementary to cART to suppress rebound viremia from persistent HIV reservoirs. In our study, we suggested that HSP90 serves as a safeguard to CDK9, playing a positive role in latent HIV reactivation. Interestingly, HSP90 inhibitors 17-DMAG and AUY-922, used in our study, up-regulated HSP90 expression and slightly reactivated latent HIV at the same time. Our study is in line with previous reports and strengthens the viewpoint that HSP90 is likely to be a target for HIV cure. Nevertheless, the first question raised is whether this phenomenon is due to HSP90 binding to CDK9. The second question is why reactivation induced by proteasome inhibitors is enhanced by 17-DMAG and AUY-922, whereas reactivation induced by TNF-α or TPA are inhibited (11). Regarding the significance of activating or inhibiting latent HIV, this may depend on the strategy of reactivation or suppression latent HIV. Nevertheless, the detailed mechanism of latent HIV reactivation, as well as the suggestion that HSP90 is a target for HIV cure needs further studies.
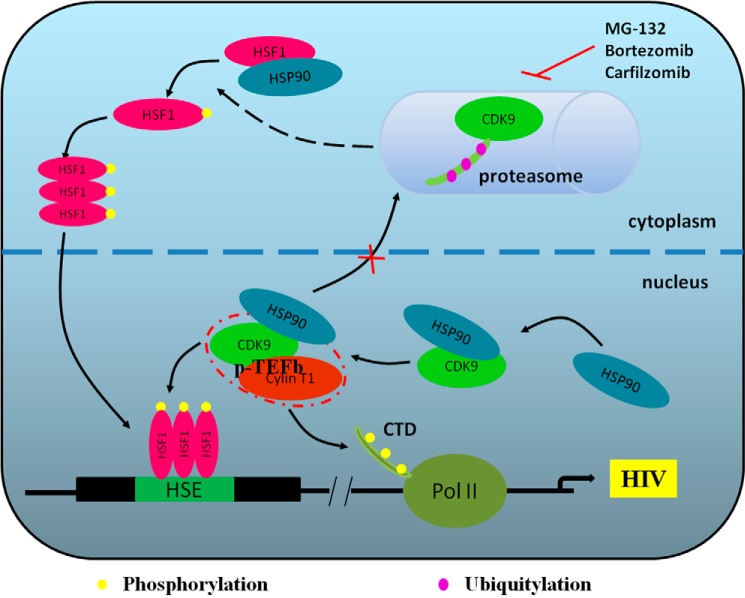
In summary, we suggest a mechanism for latent HIV reactivation, in which HSF1 recruits p-TEFb during ER stress and its downstream factor HSP90 leads to a positive feedback (Fig. 7). This underlines the observation that proteasome inhibitors reactivate latent HIV without global T cell activation, and highlights HSF1/HSP90 as targets for the therapy of HIV.
FIGURE 7.
An overview of the full story. Proteasome inhibition induces the phosphorylation of HSF1 and its translocation into the nucleus. In addition to trimeric HSF1 binding to HSE, which initiates HIV transcription, HSF1 recruits p-TEFb to phosphorylate RNA pol II thus promoting transcription elongation. The reactivation process is facilitated by generating HSP90 that binds to CDK9 to prevent it from degradation by the proteasome, thereby giving positive feedback to HSF1.
Experimental Procedures
Reagents
Primary antibodies and secondary antibodies were purchased from Cell Signaling Technology (Beverly, MA) and Santa Cruz Biotechnology (Santa Cruz, CA). Conjugated antibodies were purchased from BD Biosciences (San Jose, CA). Compounds and inhibitors were from Sigma and Merck Calbiochem (Darmstadt, Germany).
Cell Culture
The whole peripheral blood provided by Guangzhou Blood Center was from healthy donors, and PBMCs were isolated by Ficoll density gradient centrifugation using Histopaque-1077 (Sigma). The patient peripheral blood was collected in The Eighth People's Hospital of Guangzhou (Guangzhou, China) with informed consents, and the experiment was approved by the Ethical Committee of The Eighth People's Hospital of Guangzhou and performed in accordance with relevant guidelines and regulations. HIV-infected patients were selected based on sustained plasma viral load suppression (plasma viral loads were <20 copies/ml for >12 months and CD4 count >350 cells/μl), and their essential information was not shown. The primary CD4+ T cells were isolated by EasySep kit (STEMCELL Technologies Inc., Vancouver, BC, Canada) according to the manufacturer's instructions for PBMCs and the purity was >90%. J-lat 10.6, ACH2 (were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program), and PBMCs were maintained in RPMI1640 (Gibco) supplemented with 10% fetal bovine serum (Gibco) at 37 °C with 5% CO2.
Cell Viability Assay
Cell viability was evaluated by a CCK-8 kit (Dojindo Molecular Technologies, Inc., Japan). J-lat 10.6, ACH2 cells, and PBMCs plated as 1 × 106 cells per well in a 96-well plate were incubated with drugs or co-treated with different inhibitors for 48 h. Then 10 μl of CCK-8 reagent was added to 100 μl of cell culture mixture and incubated for an additional 4 h. Optical density (OD) was recorded at a wavelength of 450 nm on a microplate reader (TECAN, Swiss). The influence of inhibitors on cell viability was not shown.
Flow Cytometry Analysis
After treatment, J-lat 10.6 cells were collected to calculate the percentage of GFP-positive cells and determine the level of HIV gene expression by flow cytometry through FITC channel (FACS Canto II, BD Biosciences, San Jose, CA). PBMCs (1 × 106) were treated with drugs for 24 h and collected to incubate with CD25-PE and CD69-APC antibodies (BD Biosciences) at 4 °C for 30 min in the dark to perform surface staining. Cells were washed to eliminate fluorescence background interference, and then 5 × 104 cells were collected to analyze T cell activation markers through dual channels. Fluorescence value and dot plots were analyzed by FlowJo 7.6 software (Treestar, San Carlos, CA).
qPCR
J-lat 10.6 cells or isolated primary CD4+ T cells were incubated with PIs or co-treated with LRAs for different time intervals. Total RNA was extracted using TRIzol (Invitrogen) and chloroform, then precipitated with isopropyl alcohol. RNA reverse transcription to cDNA was done according to the procedure of the PrimeScript RT reagent kit (TAKARA, Japan) with a gDNA Eraser (TAKARA) to remove genome DNA. Quantitative PCR was performed using a SYBR Select Master Mix (Applied Biosystems, Foster City, CA) on the 7500 Real-time PCR System (Applied Biosystems) using a standard two-step procedure (denatured: 95 °C/15 s, annealed/extended: 60 °C/1 min, 40 cycles). The 2−ΔΔCT method was adopted to analyze the relative expression levels referred to the GAPDH gene.
ELISA
J-lat 10.6 cells (2 × 106) were treated with drugs for 24 h and the supernatants were obtained by centrifugation followed by removal of the cell precipitation. Then 50-μl supernatants were used to detect the secretion of IL-1β and IL-18 by ELISA according to the ELISA kit protocol (eBioscience, San Diego, CA).
Protein Extraction for Western Blotting Analysis
After treatment, cells were lysed in RIPA lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 1 mm EDTA, 1% Triton X-100, 0.25% sodium deoxycholate, 0.1% SDS) The supernatants were collected as a whole protein extract. The nucleoprotein was extracted using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer's protocol. Protein samples were stored at −80 °C or directly used for Western blotting analysis.
Electroporation of siRNA
The control siRNA and three duplexes of human-HSP90AA1 siRNAs were purchased from RiboBio Co., Ltd. (Guangzhou, China). J-lat 10.6 cells were collected and re-suspended with Opti-MEM (Invitrogen) at 5 × 106 cells/ml. siRNAs were added to 400 μl of cell suspension to a final concentration of 20 nm. The mixture was transferred into a 0.4-cm electroporation cuvette before incubating at room temperature for 15 min. Then the cells were electroporated with a Bio-Rad Gene Pulser Xcell using the following conditions: voltage, 140 V; capacitance, 1000 microfarads. The mixture was added to 1.6 ml of preincubated complete medium prior to incubation on ice for 5 min. After 48 h, a fraction of cells were collected to conducting Western blotting analysis or subjected to other experiments.
Immunoprecipitation (IP) and Co-IP
J-lat 10.6 cells (5 × 106) were treated with PIs for 24 h. Protein extracts were obtained by adding 300 μl of IP lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100). The supernatants were obtained by centrifugation followed by adding 1 μg of mouse or rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) and incubating with shaking for 30 min at 4 °C. Then 30 μl of protein A/G plus agarose (Santa Cruz Biotechnology) was added to the mixture and incubated for additional 30 min at 4 °C. Next, the mixture was centrifuged at 2500 rpm for 5 min to obtain supernatants. After protein quantification of the supernatants, 2 μg of primary antibodies (Santa Cruz Biotechnology) were added to 300–500 μg of total protein and incubated with shaking overnight at 4 °C. All the inputs were made with 20 μg of protein. The next day, 30 μl of protein A/G plus agarose was added to the mixture and incubated for an additional 4 h at 4 °C. The precipitation was then washed for three times with IP lysis buffer including 0.1% PMSF and re-suspended with loading buffer, then denaturated at 100 °C. Finally, the samples were analyzed by Western blotting and detected with relevant antibodies.
Statistical Analysis
All the experiments described were independently repeated at least three times to verify reproducibility. The data were analyzed using GraphPad Prism 5.0 software (San Diego, CA), and the error bars were presented as mean ± S.D. based on at least three independent experiments. The statistical analysis was performed using one-way analysis of variance with Dunnett's test. p values were defined as *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author Contributions
X.-Y. P. designed the experiments and drafted the manuscript, W. Z. participated in the discussion, C.-Y. W. operated the PBMC experiments, J. L., and X.-Y. Z. analyzed the data, R.-X. R., T.-R. X., and K. W. performed the FCM analysis, Y. S. discussed and modified the manuscript, S.-W. L designed and mastered the project process.
Acknowledgments
We thank Dr. Shibo Jiang and Huanzhang Zhu at Fudan University, China, for kindly gifting many essential cells and reagents. We also thank Yoel Klug for assistance in editing the manuscript.
This work was supported by Natural Science Foundation of China Grant 31370781, National S&T Key Special Foundation Grant 2014ZX09509001-004, and Natural Science Foundation of Guangdong Province Grant S2011020005207 (to S. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- cART
- combined anti-retroviral therapy
- LRA
- latency reversing agent
- PI
- proteasome inhibitor
- BTZ
- bortezomib
- CFZ
- carfilzomib
- ER
- endoplasmic reticulum
- HSF
- heat shock factor
- LTR
- long terminal repeat
- FCM
- flow cytometry
- GFP
- green fluorescent protein
- PBMC
- peripheral blood mononuclear cell
- CCK-8
- cell counting kit-8
- qPCR
- quantitative PCR
- CTL
- cytotoxic T lymphocyte
- IP
- immunoprecipitation
- HDACis
- histone deacetylase inhibitors
- DRB
- 5,6-dichlorobenzimidazole
- SAHA
- suberoylanilide hydroxamic acid
- 17-DMAG
- 17-dimethylaminoethylamino-17-demethoxygeldanamycin.
References
- 1. Bruner K. M., Hosmane N. N., and Siliciano R. F. (2015) Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 23, 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deeks S. G. (2012) HIV: shock and kill. Nature 487, 439–440 [DOI] [PubMed] [Google Scholar]
- 3. Søgaard O. S., Graversen M. E., Leth S., Olesen R., Brinkmann C. R., Nissen S. K., Kjaer A. S., Schleimann M. H., Denton P. W., Hey-Cunningham W. J., Koelsch K. K., Pantaleo G., Krogsgaard K., Sommerfelt M., Fromentin R., et al. (2015) The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog. 11, e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olesen R., Vigano S., Rasmussen T. A., Søgaard O. S., Ouyang Z., Buzon M., Bashirova A., Carrington M., Palmer S., Brinkmann C. R., Yu X. G., Østergaard L., Tolstrup M., and Lichterfeld M. (2015) Innate immune activity correlates with CD4 T cell-associated HIV-1 DNA decline during latency-reversing treatment with panobinostat. J. Virol. 89, 10176–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moreau P., Richardson P. G., Cavo M., Orlowski R. Z., San Miguel J. F., Palumbo A., and Harousseau J. L. (2012) Proteasome inhibitors in multiple myeloma: 10 years later. Blood 120, 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller L. K., Kobayashi Y., Chen C. C., Russnak T. A., Ron Y., and Dougherty J. P. (2013) Proteasome inhibitors act as bifunctional antagonists of human immunodeficiency virus type 1 latency and replication. Retrovirology 10, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Egger L., Madden D. T., Rhême C., Rao R. V., and Bredesen D. E. (2007) Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell Death Differ 14, 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scherz-Shouval R., Santagata S., Mendillo M. L., Sholl L. M., Ben-Aharon I., Beck A. H., Dias-Santagata D., Koeva M., Stemmer S. M., Whitesell L., and Lindquist S. (2014) The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 158, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pan X. Y., Zhao W., Zeng X. Y., Lin J., Li M. M., Shen X. T., and Liu S. W. (2016) Heat shock factor 1 mediates latent HIV reactivation. Sci. Rep. 6, 26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roesch F., Meziane O., Kula A., Nisole S., Porrot F., Anderson I., Mammano F., Fassati A., Marcello A., Benkirane M., and Schwartz O. (2012) Hyperthermia stimulates HIV-1 replication. PLoS Pathog. 8, e1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson I., Low J. S., Weston S., Weinberger M., Zhyvoloup A., Labokha A. A., Corazza G., Kitson R. A., Moody C. J., Marcello A., and Fassati A. (2014) Heat shock protein 90 controls HIV-1 reactivation from latency. Proc. Natl. Acad. Sci. U.S.A. 111, E1528–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joshi P., Maidji E., and Stoddart C. A. (2016) Inhibition of heat shock protein 90 prevents HIV rebound. J. Biol. Chem. 291, 10332–10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jordan A., Bisgrove D., and Verdin E. (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clouse K. A., Powell D., Washington I., Poli G., Strebel K., Farrar W., Barstad P., Kovacs J., Fauci A. S., and Folks T. M. (1989) Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 142, 431–438 [PubMed] [Google Scholar]
- 15. Jiang G., Mendes E. A., Kaiser P., Wong D. P., Tang Y., Cai I., Fenton A., Melcher G. P., Hildreth J. E., Thompson G. R., Wong J. K., and Dandekar S. (2015) Synergistic reactivation of latent HIV expression by ingenol-3-angelate, PEP005, targeted NF-κB signaling in combination with JQ1 induced p-TEFb activation. PLoS Pathog. 11, e1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang H. C., Shen L., Siliciano R. F., and Pomerantz J. L. (2009) Isolation of a cellular factor that can reactivate latent HIV-1 without T cell activation. Proc. Natl. Acad. Sci. U.S.A. 106, 6321–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng K., Pertea M., Rongvaux A., Wang L., Durand C. M., Ghiaur G., Lai J., McHugh H. L., Hao H., Zhang H., Margolick J. B., Gurer C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., et al. (2015) Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Korin Y. D., Brooks D. G., Brown S., Korotzer A., and Zack J. A. (2002) Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 76, 8118–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen I. Y., and Ichinohe T. (2015) Response of host inflammasomes to viral infection. Trends Microbiol 23, 55–63 [DOI] [PubMed] [Google Scholar]
- 20. Smale S. T. (2010) Selective transcription in response to an inflammatory stimulus. Cell 140, 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borsa M., Ferreira P. L., Petry A., Ferreira L. G., Camargo M. M., Bou-Habib D. C., and Pinto A. R. (2015) HIV infection and antiretroviral therapy lead to unfolded protein response activation. Virol. J. 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rawat P., and Mitra D. (2011) Cellular heat shock factor 1 positively regulates human immunodeficiency virus-1 gene expression and replication by two distinct pathways. Nucleic Acids Res. 39, 5879–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itzen F., Greifenberg A. K., Bösken C. A., and Geyer M. (2014) Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 42, 7577–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Budhiraja S., and Rice A. P. (2013) Reactivation of latent HIV: do all roads go through P-TEFb? Future Virol. 8, 10.2217/fvi.13.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon Y. J., Kim J. A., Shin K. D., Shin D. S., Han Y. M., Lee Y. J., Lee J. S., Kwon B. M., and Han D. C. (2011) KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J. Biol. Chem. 286, 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sreedhar A. S., Soti C., and Csermely P. (2004) Inhibition of Hsp90: a new strategy for inhibiting protein kinases. Biochim. Biophys. Acta 1697, 233–242 [DOI] [PubMed] [Google Scholar]
- 27. Chan J. K., and Greene W. C. (2011) NF-κB/Rel: agonist and antagonist roles in HIV-1 latency. Curr. Opin. HIV AIDS 6, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar M., and Mitra D. (2005) Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 280, 40041–40050 [DOI] [PubMed] [Google Scholar]
- 29. Wheeler D. S., Dunsmore K. E., and Wong H. R. (2003) Intracellular delivery of HSP70 using HIV-1 Tat protein transduction domain. Biochem. Biophys. Res. Commun. 301, 54–59 [DOI] [PubMed] [Google Scholar]
- 30. Mehla R., Bivalkar-Mehla S., Zhang R., Handy I., Albrecht H., Giri S., Nagarkatti P., Nagarkatti M., and Chauhan A. (2010) Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS ONE 5, e11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bartholomeeusen K., Fujinaga K., Xiang Y., and Peterlin B. M. (2013) Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J. Biol. Chem. 288, 14400–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z., Guo J., Wu Y., and Zhou Q. (2013) The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 41, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jorgensen I., and Miao E. A. (2015) Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 265, 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mitchell S., Vargas J., and Hoffmann A. (2016) Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 8, 227–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu H. T., Janßen L., Lawand M., Kim J., Perez-Arroyo A., Culina S., Gdoura A., Burgevin A., Cumenal D., Fourneau Y., Moser A., Kratzer R., Wong F. S., Springer S., and van Endert P. (2014) Endoplasmic reticulum targeting alters regulation of expression and antigen presentation of proinsulin. J. Immunol. 192, 4957–4966 [DOI] [PubMed] [Google Scholar]