Abstract
Hepatitis delta virus (HDV) is a satellite virus of hepatitis B virus (HBV). HDV genome encodes two forms of hepatitis delta antigen (HDAg), small HDAg (HDAg-S), which is required for viral replication, and large HDAg (HDAg-L), which is essential for viral assembly. HDAg-L is identical to HDAg-S except that it bears a 19-amino acid extension at the C terminus. Both HDAgs contain a nuclear localization signal (NLS), but only HDAg-L contains a CRM1-independent nuclear export signal at its C terminus. The nuclear export activity of HDAg-L is important for HDV particle formation. However, the mechanisms of HDAg-L-mediated nuclear export of HDV ribonucleoprotein are not clear. In this study, the host cellular RNA export complex TAP-Aly was found to form a complex with HDAg-L, but not with an export-defective HDAg-L mutant, in which Pro205 was replaced by Ala. HDAg-L was found to colocalize with TAP and Aly in the nucleus. The C-terminal domain of HDAg-L was shown to directly interact with the N terminus of TAP, whereas an HDAg-L mutant lacking the NLS failed to interact with full-length TAP. In addition, small hairpin RNA-mediated down-regulation of TAP or Aly reduced nuclear export of HDAg-L and assembly of HDV virions. Furthermore, a peptide, TAT-HDAg-L(198–210), containing the 10-amino acid TAT peptide and HDAg-L(198–210), inhibited the interaction between HDAg-L and TAP and blocked HDV virion assembly and secretion. These data demonstrate that formation and release of HDV particles are mediated by TAP and Aly.
Keywords: hepatitis virus, nuclear transport, protein translocation, viral replication, virus assembly, Aly, TAP, hepatitis delta antigen, nuclear export factor
Introduction
Hepatitis delta virus (HDV)2 is a human pathogen that is associated with fulminant hepatitis and progressive chronic liver cirrhosis upon superinfection or coinfection with hepatitis B virus (HBV) (1). Superinfection or coinfection with HBV and HDV causes severe liver disease in patients. The HDV virion is enveloped by the HBV surface antigen (HBsAg) (2, 3). Inside the HDV particle is a ribonucleoprotein (RNP) complex comprised of HDV genomic RNA of ∼1.7 kilobases and molecules of the small hepatitis delta antigen (HDAg-S, 195 residues) and large HDAg (HDAg-L, 214 residues) (4–6). The two HDAgs have the same N-terminal 195 amino acids, but HDAg-L contains an additional C-terminal 19-amino acid sequence, which is generated by posttranscriptional RNA editing (7, 8).
Host factor-mediated nucleocytoplasmic transport is critical for diverse cellular events in eukaryotes and the life cycle of viruses. In the initial stage of HDV infection, the viral genome is imported into the nucleus of the host cell through the RNA-binding activity and nuclear localization signal (NLS) of HDAgs (9), which is recognized by the NLS receptor, importin α2 (10). The viral RNA undergoes replication mediated by HDV ribozyme and host RNA polymerase in the nuclei of infected cells (11, 12). The progeny HDV RNA genome forms RNP complexes with HDAgs. In the late stage of infection, the HDV RNP is exported from the nucleus to the cytoplasm for further assembly with HBsAg. Both HDAg-S and HDAg-L contain the NLS and are mainly localized in the nucleus. HDAg-S functions as a trans-activator of HDV replication (3, 13), whereas HDAg-L is necessary for virion assembly and secretion (14, 15). The unique C-terminal domain of HDAg-L contains a CXXX motif that directs prenylation of the protein and is essential for the formation of HDV subviral particles containing HBsAg (15–18). Moreover, HDAg-L contains the chromosome region maintenance 1 (CRM1)-independent NES (198ILFPADPPFSPQS210) and triggers the nuclear export of HDV RNP essential for HDV virion morphogenesis (19). However, how host cellular proteins are involved in the HDAg-L-mediated CRM1-independent nuclear export pathways remains unclear.
In this study, our aims were to examine possible interactions between nucleus-localized HDAg-L and host factors and elucidate the importance of HDAg-L nuclear export in the life cycle of HDV. The results showed that HDAg-L formed a complex in vivo with a cellular export receptor, TAP (also known as NXF1), and an adaptor protein, Aly (also known as REF), which are responsible for RNA export (20). In metazoans, nuclear export of many mRNAs is mediated by TAP, an export receptor that cooperates with adaptor RNA-binding proteins, for example, through the combined use of an adaptor (e.g. Aly) (20). In addition, HDAg-L directly interacted via its C-terminal domain with TAP and this interaction facilitated nuclear export of HDAg-L and HDV RNA. A peptide consisting of TAT peptide, a 10-amino acid carrier peptide derived from the HIV-1 trans-activator of transcription (TAT) sequence, and residues 198–210 of HDAg-L, blocked the interaction of HDAg-L with TAP and Aly and inhibited the secretion of HDV virions. Our results demonstrate that TAP and Aly play critical roles during the processes of HDV maturation.
Results
HDAg-L Forms Complexes with the Cellular Proteins TAP and Aly
As shown in Fig. 1A, HDAg-L contains two nuclear localization signals (NLS1 and NLS2) (amino acids 35–50 and 65–77) (9, 21, 22) and a nuclear export signal (NES) (amino acids 195–214) (19), and is essential for CRM1-independent nuclear export of HDV RNA (23). Several NESs of viral proteins that pass through the CRM1-independent nuclear export pathway utilize the host nuclear export factors TAP and Aly to facilitate the morphogenesis of the virion (24–28). Therefore, possible interactions between HDAg-L and the mRNA export factors TAP and Aly were examined by co-immunoprecipitation using anti-TAP or anti-Aly antibodies from lysates of Huh7 cells transiently expressing HDAg-L or HDAg-S, which lacks the NES, as a control. As shown in Fig. 1, B and C, HDAg-L, but not HDAg-S, was co-immunoprecipitated with either TAP (B) or Aly (C). However, no co-immunoprecipitation of HDAg-L was seen using antibodies against the cellular apoptosis susceptibility protein (data not shown), which acts as a nuclear transport factor in the importin pathway in the nucleus (29). These results show that HDAg-L and TAP or Aly were present in the same complex in cells.
FIGURE 1.
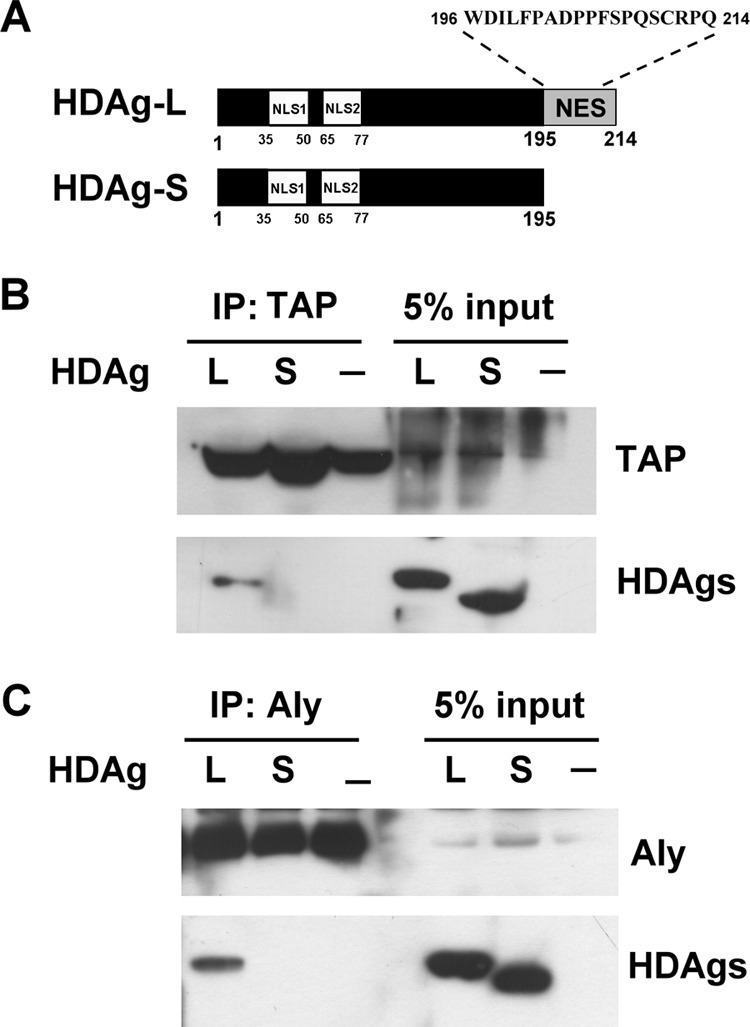
HDAg-L forms complexes with the cellular proteins TAP and Aly. A, schematic representation of HDAg-L and HDAg-S and the amino acid sequence of the C terminus of HDAg-L. The NLS and NES are shown. B and C, co-immunoprecipitation (IP) assay. Huh7 cells were left untreated or were transfected with plasmid pECE-d-BE or pECE-d-SM containing cDNAs encoding, respectively, HDAg-L and HDAg-S. At 2 days posttransfection, the cells were harvested and subjected to immunoprecipitation with anti-TAP antibodies (B), followed by Western blotting analysis with antibodies against TAP or HDAgs or with anti-Aly antibodies (C) followed by Western blotting analysis with antibodies against Aly and HDAgs.
Colocalization of HDAg-L with Endogenous TAP and Aly in the Nucleus
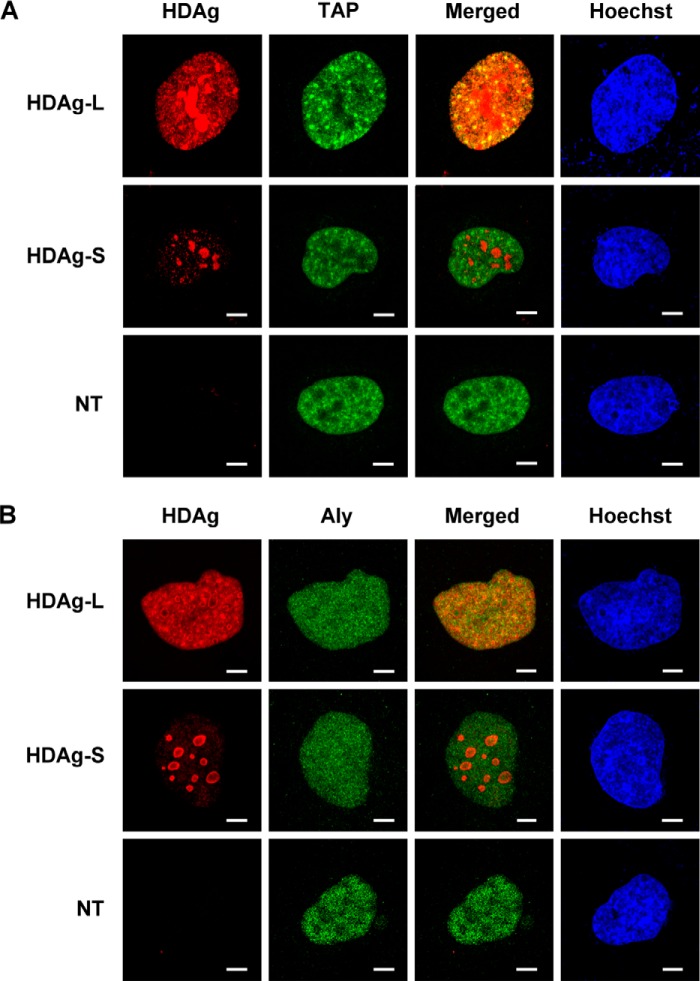
To determine the relative distribution of HDAgs, TAP, and Aly in mammalian cells, Huh7 cells were transiently transfected with plasmids encoding wild-type HDAgs, then expression of HDAgs was examined by confocal microscopy. Consistent with our previous findings (30), wild-type HDAg-L and HDAg-S were both localized in the nucleus (Fig. 2, A and B). Double labeling with monoclonal antibodies against TAP (Fig. 2A) or Aly (Fig. 2B) showed that HDAg-L colocalized with TAP or Aly throughout the nucleus, whereas HDAg-S was mainly localized in the nucleolus and was not colocalized with TAP or Aly. These results show that HDAg-L, but not HDAg-S, colocalizes with TAP and Aly in the same cellular compartment in the nucleus.
FIGURE 2.
Subcellular localization of HDAg-L, HDAg-S, TAP, and Aly in Huh7 cells. Huh7 cells were left untreated or were transfected with plasmid pECE-d-BE or pECE-d-SM containing cDNA encoding, respectively, HDAg-L or HDAg-S, then the subcellular localization of HDAg-L or HDAg-S and either TAP (A) or Aly (B) was examined by immunofluorescence staining with antibodies against HDAg, TAP, or Aly and confocal microscopy. NT, non-transfected. The bars on the images represent 20 μm.
Identification of TAP as an HDAg-L-interacting Protein and Mapping of the HDAg-L-binding Domain in TAP
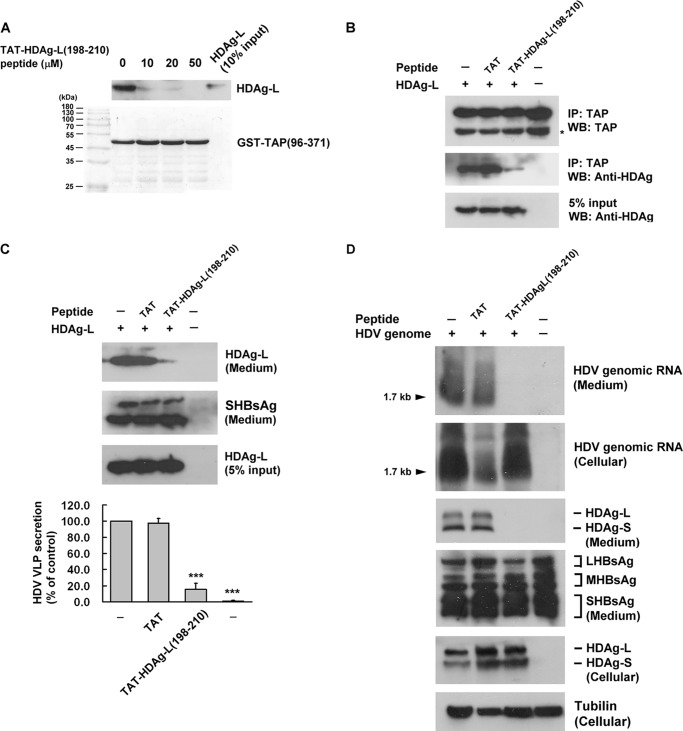
To examine the interaction between TAP and HDAg-L, GST-TAP(1–619) was overexpressed and treated with PreScission Protease to cleave between GST and TAP, then glutathione-Sepharose was added to remove GST and PreScission Protease to obtain purified full-length TAP(1–619) protein (Fig. 3A). The interaction between TAP and HDAg-L was analyzed in a GST pulldown assay using a GST-HDAg-L(198–210) fusion protein or GST precoupled to glutathione-Sepharose and purified TAP(1–619) protein, and the results showed that TAP bound in vitro to GST-HDAg-L(198–210), but not GST (Fig. 3B), indicating that the C-terminal domain of HDAg-L interacts directly with TAP(1–619). To determine what domains of TAP interact with HDAg-L, a series of GST-TAP fusion proteins precoupled to glutathione-Sepharose beads (20) were tested for their ability to pulldown full-length HDAg-L transiently expressed in Huh7 cells. As shown in Fig. 3C, the fusion protein GST-TAP(96–371), containing amino acids 96–371 of TAP, but none of the other TAP domains (amino acids 188–619, 188–550, 371–619, 371–550, or 551–619) was able to pulldown HDAg-L. None of the GST-TAP fusion proteins was able to pulldown HDAg-S (data not shown). This result shows that HDAg-L binds to amino acids 96–371 of TAP.
FIGURE 3.
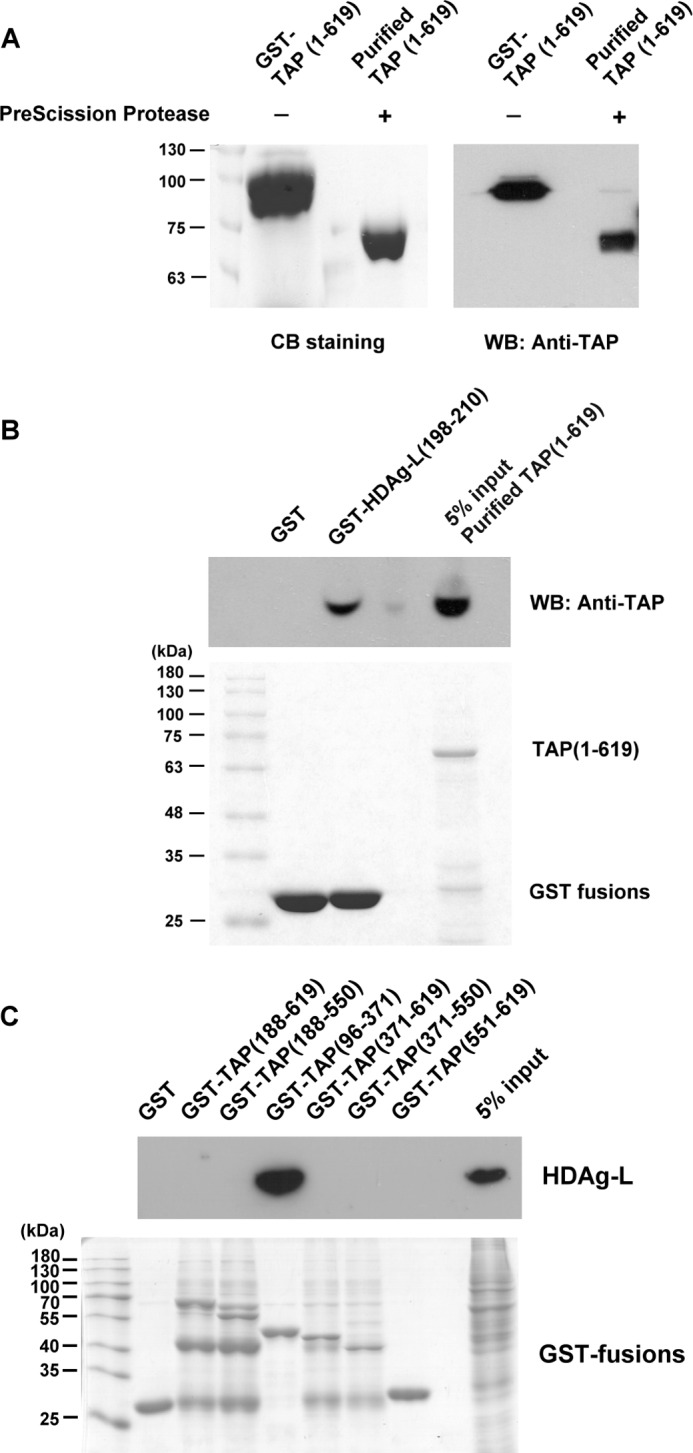
Identification of TAP as an HDAg-L-interacting protein and mapping of the HDAg-L-binding domain in TAP. A, purification of TAP(1–619). GST-TAP(1–619) was incubated with PreScission Protease and then the glutathione-Sepharose beads were added to removed the cleaved GST, uncleaved GST-TAP(1–619), and the PreScission Protease (also a GST fusion protein). Following cleavage reactions, the purified proteins in the supernatant were detected by Coomassie Blue staining (left panel) or the Western blotting analysis was performed using antibodies against TAP (right panel). The positions of the molecular weight markers are shown on the left. B, GST pulldown assay with purified TAP(1–619) and GST-HDAg-L(198–210). The GST pulldown assay was performed with 100 μg of GST-HDAg-L(198–210) fusion protein precoupled to glutathione-Sepharose beads and 100 μg of purified TAP(1–619). 5% of input purified TAP(1–619) was used as control. Following GST pulldown, Western blotting analysis was performed using antibodies against TAP (top panel) or the pulled down proteins were detected by Coomassie Blue staining (bottom panel). The positions of the molecular weight markers are shown on the left. C, mapping of the HDAg-L-binding domain in TAP. The GST pulldown assay was performed with various GST-TAP fusion proteins precoupled to glutathione-Sepharose beads and lysates prepared from Huh7 cells transiently expressing HDAg-L. Following GST pulldown, Western blotting analysis was performed using antibodies against HDAg or Aly (top panel) or the pulled down proteins were detected by Coomassie Blue staining (bottom panel). The positions of the molecular weight markers are shown on the left.
The Nuclear Export Mutant HDAg-L-P205A Cannot Interact with TAP
The C-terminal domain spanning amino acids 198–210 of HDAg-L contains the NES, which directs nuclear export of HDAg-L to HBsAg in the cytoplasm via a CRM1-independent pathway (19). To determine whether the NES of HDAg-L was also responsible for the interaction with TAP in the cytoplasm, GST-HDAg-L(198–210), a fusion protein containing GST and amino acids 198–210 of HDAg-L, and GST-HDAg-L(198–210)-P205A, in which Pro205 of HDAg-L was replaced by alanine, were purified and tested in a pulldown assay of Huh7 cell lysate. As shown in Fig. 4A, when precoupled to glutathione-Sepharose beads, GST-HDAg-L(198–210) was able to pulldown endogenous TAP and this effect was abolished when the P205A mutation was introduced into the NES of HDAg-L. To further study the role of Pro205 of HDAg-L in the interaction with TAP in vivo, a plasmid encoding full-length HDAg-L with the P205A mutation was transiently transfected into Huh7 cells and a co-immunoprecipitation assay performed on the cell lysates using anti-TAP antibodies. As shown in Fig. 4B, wild-type HDAg-L was co-immunoprecipitated with endogenous TAP, whereas HDAg-L-P205A was not. These results show that Pro205 in the NES of HDAg-L is critical for the interaction of HDAg-L with TAP.
FIGURE 4.
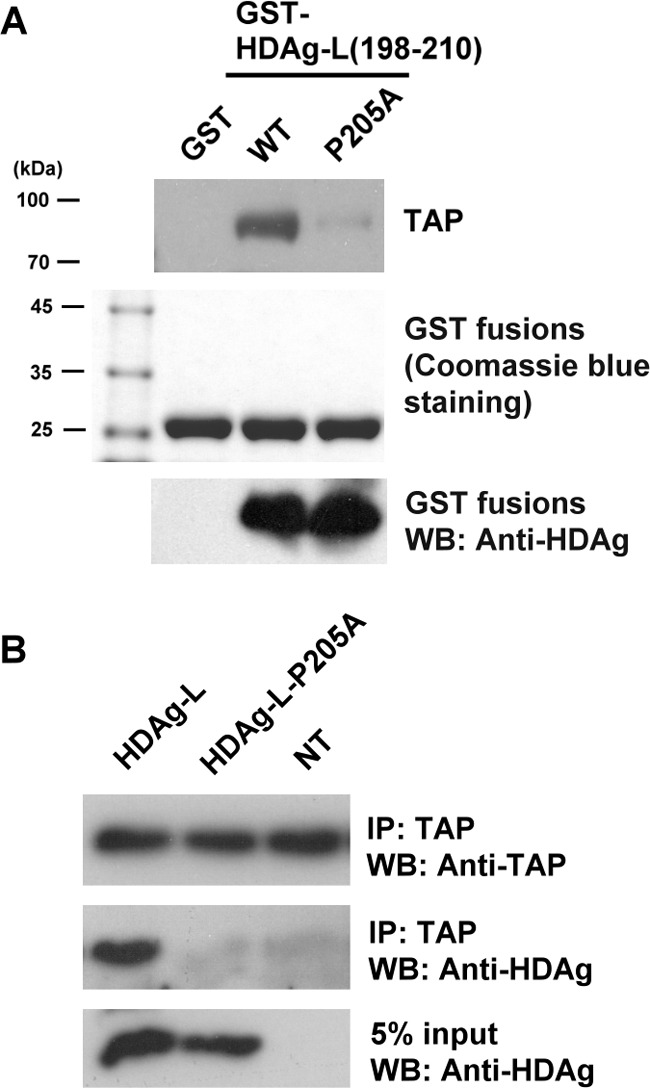
The nuclear export mutant HDAg-L-P205A cannot bind to TAP. A, GST pulldown assay with TAP and GST fusion proteins containing either HDAg-L(198–210) or the P205A mutant. The GST pulldown assay was performed with either GST-HDAg-L(198–210) or the mutant precoupled to glutathione-Sepharose beads and lysates were prepared from Huh7 cells. Following GST pulldown, Western blotting analysis was performed using antibodies against TAP and HDAg (top and bottom panels) or the pulled down proteins were detected by Coomassie Blue staining (middle panel). The positions of the molecular weight markers are shown on the left. B, Huh7 cells were transfected with plasmid pECE-d-BE or pECE-d-BE(P205A) containing cDNA encoding, respectively, HDAg-L and HDAg-L-P205A, then, at 2 days posttransfection, were harvested and the cell lysates were subjected to immunoprecipitation (IP) with anti-TAP antibodies, followed by Western blotting analysis with antibodies against TAP or HDAgs. NT, non-transfected.
An NLSs-deleted Mutant of HDAg-L Fails to Interact with TAP
HDAg-L is a nucleocytoplasmic shuttling protein, and, in addition to the NES, contains two NLSs between amino acids 35 and 77. To test whether HDAg-L interacted with TAP in the cytoplasm, the construct HDAg-L-d35/88 lacking amino acid residues 35–88 was used to examine the relative distribution of HDAg-L-d35/88 and endogenous TAP in Huh7 cells transiently expressing HDAg-L-d35/88 using immunofluorescent antibodies and confocal microscopy. Fig. 5A shows that endogenous TAP was exclusively localized in the nucleus, whereas HDAg-L-d35/88 was localized in the cytoplasm. We then examined the association of HDAg-L-d35/88 with TAP in vivo by transfecting Huh7 cells with plasmids encoding wild-type HDAg-L and HDAg-L-d35/88 and performing a co-immunoprecipitation assay on the cell lysates using anti-TAP antibodies. As shown in Fig. 5B, wild-type HDAg-L, but not HDAg-L-d35/88, could be co-immunoprecipitated with endogenous TAP. These results show that the NLS of HDAg-L is required for it to be present in the same nuclear compartment with TAP and facilitates HDAg-L to bind TAP.
FIGURE 5.
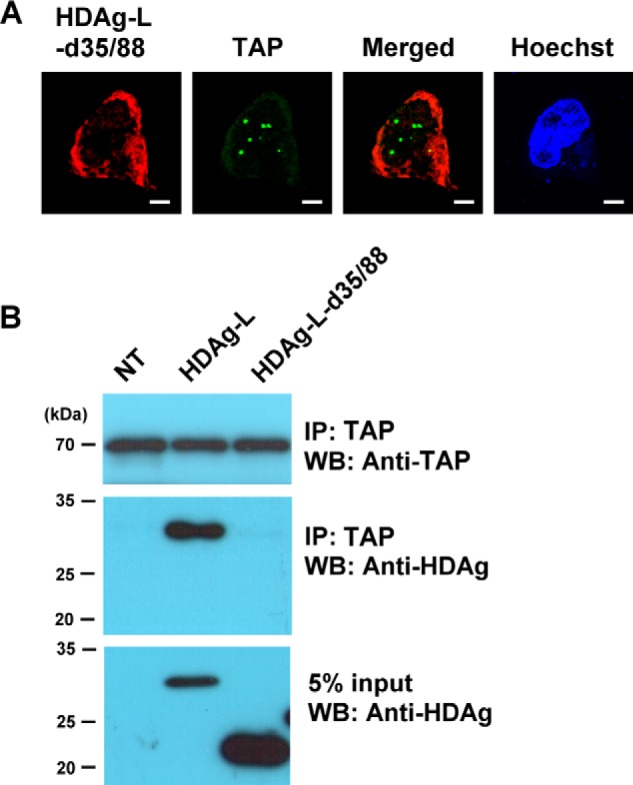
The HDAg-L NLS deletant mutant does not bind to TAP. A, Huh7 cells were transfected with plasmid pECEL-d35/88, then, at 2 days posttransfection, the subcellular localization of HDAg-L-d35/88 and TAP was examined by immunofluorescence staining with antibodies against HDAg and TAP using confocal microscopy. The bars on the images represent 20 μm. B, Huh7 cells were transfected with plasmid pECE-d-BE, coding for full-length HDAg-L, or pECEL-d35/88 lacking residues 35–88, then, at 2 days posttransfection, were harvested and the cell lysate was subjected to immunoprecipitation (IP) with anti-TAP antibodies, followed by Western blotting (WB) analysis with antibodies against TAP or HDAgs. The positions of the molecular weight markers are shown on the left. NT, non-transfected.
Knockdown of TAP or Aly Inhibits Nuclear Export of HDAg-L
We have previously demonstrated an essential role of HDAg-L in nuclear export and that this nuclear export activity is essential for HDAg-L to form HDV virions with HBsAg (4). To examine the importance of TAP and Aly in HDAg-L nuclear export, plasmids expressing specific shRNAs directed against TAP or Aly were transfected into Huh7 cells, then Western blotting analysis was performed to analyze the effectiveness of the shRNAs in down-regulating the expression of endogenous TAP or Aly, using nontransfected cells as controls. Fig. 6A shows that TAP and Aly shRNAs were, respectively, highly efficient in reducing levels of endogenous TAP and Aly. The nuclear export of HDAg-L was then examined in the TAP- and Aly-down-regulated cells by immunofluorescence staining. We have previously demonstrated that HDAg-L is a nuclear protein, but relocalizes to the cytoplasm in the presence of HBsAg (19). The staining patterns of HDAg-L in the presence of HBsAg were classified into three types, type I (nucleolus stained), type II (both nucleolus and nucleoplasm stained), and type III (nucleolus, nucleoplasm, and cytoplasm stained). Fig. 6B shows that, in non-silenced cells, in the presence of HBsAg, wild-type HDAg-L was found in both the nucleus and cytoplasm (type III pattern) in about 41% of transfected cells. After treatment with shRNA specific for TAP or Aly, transport of HDAg-L to the cytoplasm dropped from 41 to 6 or 9%, respectively, indicating a critical role of both TAP and Aly in the nuclear export of HDAg-L.
FIGURE 6.
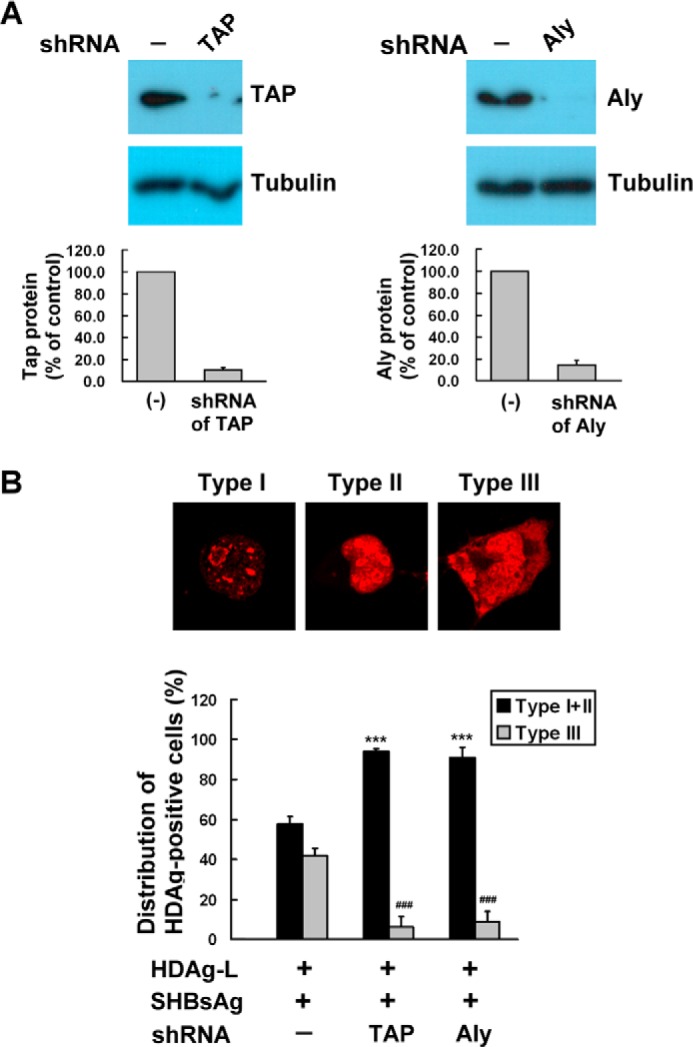
Knockdown of TAP and Aly inhibits nuclear export of HDAg-L. A, Huh7 cells were left untreated or were transfected with plasmid expressing TAP- or Aly-targeted shRNAs, then were harvested at 2 days posttransfection and analyzed by Western blotting analysis with antibodies against TAP and Aly. The top panel shows a typical result and the bottom panel the quantification of TAP and Aly expressed as the mean ± S.D. for three independent experiments. B, cellular distribution of HDAgs in the presence of TAP- or Aly-targeted shRNAs. Huh7 cells were transfected with HDAgs (L) alone or together with small HBsAg in the absence of presence of TAP or Aly shRNA, then, at 72 h posttransfection, immunofluorescence staining was performed using anti-HDAg antibodies. The top panels show the three HDAg staining patterns: type I, nucleolus; type II, both nucleolus and nucleoplasm; type III, nucleolus, nucleoplasm, and cytoplasm. The bottom panel shows the statistical analysis in which fields each containing at least 100 HDAg-positive cells were randomly selected and the number of cells with each type of HDAg staining pattern counted, and calculated as a percentage of the total number of the HDAg-positive cells in the same field. The results shown are the mean ± S.D. for three independent experiments. ***, p < 0.001 versus type I + type II control. ###, p < 0.001 versus type III control.
Assembly and Release of HDV Virions Is Decreased by shRNA-mediated Down-regulation of TAP and Aly
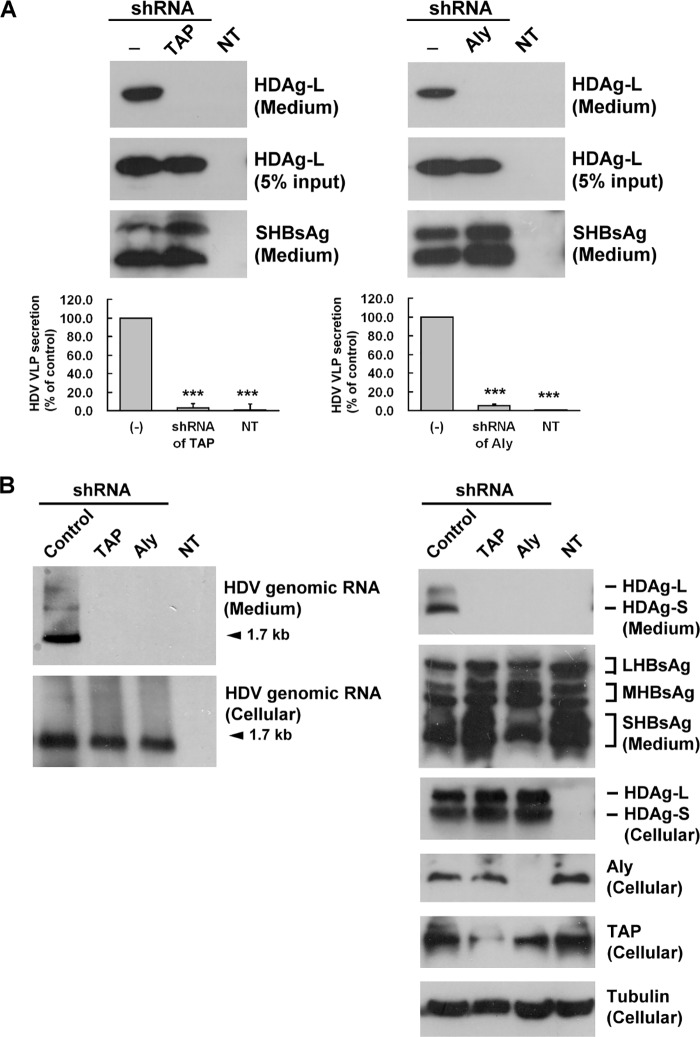
Both HDAgs have to form RNPs with the viral genomic RNA in the nucleus, then HDAg-L mediates HDV RNP export out of the nucleus to assemble with HBsAg in the cytoplasm to generate HDV virion, which is then secreted out of the host cell. To examine whether the association of HDAg-L with TAP and Aly contributes to the packaging of HDV virus-like particles (VLPs), Huh7 cells were cotransfected with plasmids encoding HDAg-L and HBsAg with or without TAP or Aly shRNA encoding plasmids to see if HDV VLPs could be packaged and secreted into the culture medium. At 4 days posttransfection, HDV VLPs were isolated from the culture medium; HDV VLP levels were represented by HDAg-L levels. As shown in Fig. 7A, using shRNA specific for TAP (left panel) or Aly (right panel), there was a significant decrease in HDV VLP levels in the culture medium to, respectively, about 3.3 or 5.2% of control levels, with no effect on HBsAg levels. In addition, to generate the biologically relevant HDV virions, the HBV cell line, HepG2.2.15 cells, which stably express the HBV genome, were cotransfected with plasmid pSVD2 encoding HDV genomic RNA, which could undergo viral replication. To examine the effect of TAP or Aly shRNA, the release of HDV virions was detected in transfected HepG2.2.15 cells. At 7 days posttransfection, viral proteins and RNA were isolated from both culture cells and culture medium. As shown in Fig. 7B, HDV genomic RNA was analyzed by Northern blotting, and HDAg, HBsAg, TAP, and Aly proteins were analyzed by Western blotting. Using shRNA specific for TAP or Aly showed a significant decrease in HDV genomic RNA (1.7 kb) and HDAg protein levels in the culture medium, with no effect on large HBsAg (LHBsAg), middle HBsAg (MHBsAg), and small HBsAg (SHBsAg) levels. TAP or Aly shRNA had no effect on the cellular HDV genomic RNA and cellular HDAg protein levels. These results show that shRNA-mediated knockdown of TAP or Aly inhibits the release of HDV virions containing HDV genomic RNA, but not HDV gene replication.
FIGURE 7.
Interference with assembly and release of HDV virions in cells expressing TAP- or Aly-targeted shRNAs. A, Huh7 cells were cotransfected with plasmid pECE-C-ES encoding HBsAg and pECE-d-BE encoding HDAg-L in the presence or absence of TAP- or Aly-targeted shRNAs. Four days posttransfection, protein lysates were prepared from the transfected cells and HDV VLPs were collected from the culture medium. The HDV VLPs were subjected to Western blotting analysis with antibodies against HDAg or HBsAg (top panel) and the packaging activity calculated and normalized to HDAg-L in the absence of TAP- and Aly-targeted shRNAs (bottom panel). The results are the mean ± S.D. for three independent experiments. ***, p < 0.001 versus control. B, HepG2.2.15 cells were cotransfected with plasmid pSVD2 expressing a dimeric HDV RNA in the presence or absence of TAP- or Aly-targeted shRNAs or pLKO.1 control plasmid. Seven days posttransfection, protein lysates and RNA were prepared from the transfected cells and HDV virions were collected from the culture medium. A DIG-labeled HDV antigenomic RNA transcribed in vitro from plasmid pD3 was used as a probe to perform Northern blot analysis (left panel). Rabbit antiserum specific for HDAgs, Aly, TAP, and tubulin and goat polyclonal antibodies specific to HBsAg were used to perform Western blotting analysis (right panel) as indicated. NT, non-transfection.
The Cell-permeable Peptide TAT-HDAg-L(198–210) Inhibits the HDAg-L/TAP Interaction and Release of HDV Virions
TAT peptide is cell-permeable and is widely used as a therapeutic agent in many diseases by coupling it to a peptide or protein, allowing it to be transported into the cell (31). We therefore examined whether a peptide consisting of TAP peptide fused to residues 198–210 of HDAg-L, TAT-HDAg-L(198–210), interfered with the direct interaction between TAP and HDAg-L and could be a possible therapeutic agent for HDV infection. First, the effect of TAT-HDAg-L(198–210) on the interaction between HDAg-L and TAP(96–371) was examined in a competition assay in vitro by performing a GST pulldown assay with glutathione-Sepharose beads and a lysate of GST-TAP(96–371) and HDAg-L-expressing Huh7 cells in the presence or absence of different concentrations (0, 10, 20, or 50 μm) of TAT-HDAg-L(198–210). As shown in Fig. 8A, at all concentrations tested, TAT-HDAg-L(198–210) significantly reduced the amount of HDAg-L protein bound to GST-TAP(96–371). In subsequent studies, 10 μm TAT-HDAg-L(198–210) was used. In an in vivo study, Huh7 cells cotransfected with pECE-C-ES expressing HBsAg, and pECE-d-BE expressing HDAg-L were cultured for 2 days in the presence or absence of 10 μm TAT-HDAg-L(198–210), then TAP in the cell lysates was immunoprecipitated and the protein in the precipitates analyzed for HDAg by Western blotting. Fig. 8B shows that complex formation between HDAg-L and TAP was blocked by TAT-HDAg-L(198–210). Interestingly, as shown in Fig. 8C, in the presence of TAT-HDAg-L(198–210), HDV virion levels detected by the presence of HDAg-L in the culture medium were only 15.3% of those seen in the absence of the peptide, whereas control TAT peptide had no inhibitory effect. Furthermore, HepG2.2.15 cells were cotransfected with plasmid pSVD2 encoding a dimeric form of HDV genomic RNA, with or without TAT-HDAg-L(198–210). As shown in Fig. 8D, HDV virions and cell lysates were analyzed for HDV genomic RNA by Northern blotting and for HDAg and HBsAg proteins by Western blotting. Using TAT-HDAg-L(198–210) showed a significant decrease in HDV genomic RNA (1.7 kb) and HDAg protein levels in the culture medium, with no effect on large HBsAg (LHBsAg), middle HBsAg (MHBsAg), and small HBsAg (SHBsAg) levels. TAT-HDAg-L(198–210) had no effect on the cellular HDV genomic RNA and cellular HDAg protein levels. These results show that TAT-HDAg-L(198–210) inhibits both the HDAg-L/TAP interaction and the assembly and secretion of HDV virions.
FIGURE 8.
Effect of the cell-permeable peptide TAT-HDAg-L(198–210) on the interaction between HDAg-L and TAP and on the release of HDV virions. A, competition between HDAg-L(198–210) and GST-TAP(96–371) for binding to HDAg-L. The GST pulldown competition assay was performed using GST-TAP(96–371) generated in bacteria, HDAg-L was prepared from transfected Huh7 cells and various amounts of HDAg-L(198–210). The top panel shows Western blotting (WB) analysis with antibodies against HDAgs and the bottom panel shows Coomassie Blue staining. B, effect of TAT-HDAg-L(198–210) on the binding of HDAg-L to TAP. Huh7 cells cotransfected with plasmids pSVD2 expressing a dimeric HDV RNA, pECE-C-ES expressing HBsAg, and pECE-d-BE expressing HDAg-L were cultured for 2 days in the presence or absence of 10 μm TAT-HDAg-L(198–210) or TAT peptide as control. They were then harvested and cell lysates were subjected to immunoprecipitation (IP) with anti-TAP antibodies followed by Western blotting analysis with antibodies against TAP or HDAgs. C, effects of TAT-HDAg-L(198–210) on the assembly of HDV virions. Huh7 cells were cotransfected with plasmid pSVD2 expressing a dimeric HDV RNA, pECE-C-ES encoding HBsAg, and pECE-d-BE encoding HDAg-L and incubated for 4 days with or without 10 μm TAT-HDAg-L(198–210) with TAT peptide as control. RNA and protein lysates were then prepared from the transfected cells and HDV virions were collected from the culture medium. Top panel, HDV virions subjected to Western blotting analysis with antibodies against HDAg or HBsAg. Bottom panel, HDV packaging activity calculated and normalized to HDAg-L in the absence of peptide. The results are the mean ± S.D. for three independent experiments. ***, p < 0.001 versus control. D, HepG2.2.15 cells were cotransfected with plasmid pSVD2 expressing a dimeric HDV RNA in the presence or absence of TAT-HDAg-L(198–210). Seven days posttransfection, protein lysates and RNA were prepared from the transfected cells and HDV virions were collected from the culture medium. Northern blot analysis was performed to detect cellular and secreted HDV genomic RNA. Western blotting analysis was performed to detect cellular and secreted HDAgs and HBsAg as indicated.
The Role of TAP and Aly in Viral Assembly and Release under Condition of HDV Infection
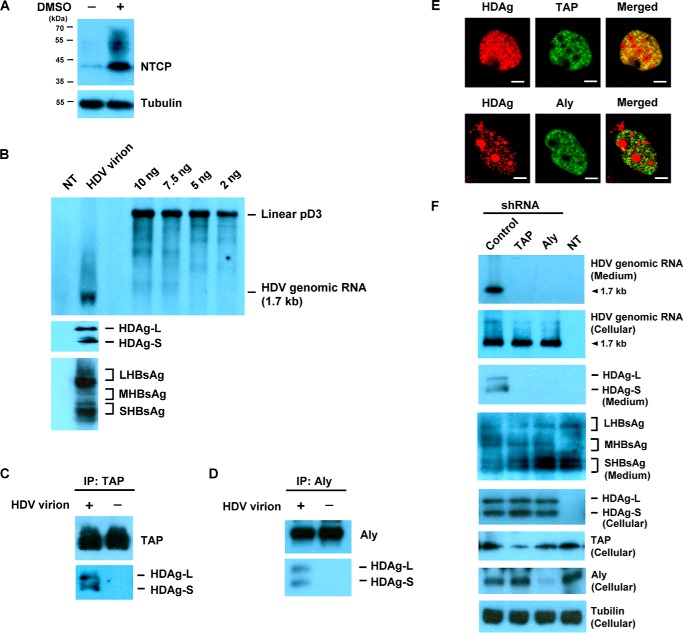
The human sodium taurocholate cotransporting polypeptide (NTCP), a multiple transmembrane transporter predominantly expressed in the liver, is a functional receptor for HBV and HDV (32). HuS-E/2 is an immortalized cell line derived from human primary hepatocytes, and has been reported to be susceptible to HBV infection (33). To observe whether the interaction between HDAgs, TAP, and Aly was biologically relevant to HDV infection, HuS-E/2 cells were used in the following HDV infection experiments. After DMSO treatment, expression of NTCP was significantly increased in HuS-E/2 cells (Fig. 9A). Large amounts of HDV infectious virion was prepared and the HDV particles were confirmed to contain HDV genomic RNA, HDAgs, and envelope proteins by performing Northern blot analysis and Western blotting analysis (Fig. 9B). Next, HDV virions were used to infect DMSO-treated HuS-E/2 cells, and the cell lysate was collected 7 days later. The interactions between HDAgs and TAP or Aly were examined by co-immunoprecipitation using anti-TAP or anti-Aly antibodies. As shown in Fig. 9, C and D, HDAgs were co-immunoprecipitated with either TAP (C) or Aly (D). To determine the relative distribution of HDAgs, TAP, and Aly in HDV-infected cells, expression of HDAgs was examined by confocal microscopy. Double labeling with monoclonal antibodies against TAP or Aly showed that HDAgs colocalized with TAP or Aly throughout the nucleus (Fig. 9E). To examine the effect of TAP or Aly shRNA under the condition of HDV infection, DMSO-treated HuS-E/2 cells were co-transfected with plasmid encoding TAP shRNA or Aly shRNA and plasmid p1.3HBcl encoding HBsAgs for viral assembly. Then HDV were infected in the transfected cells and the release of HDV virions was detected at 7 days post-infection. Viral proteins and RNA were isolated from both culture cells and culture medium. As shown in Fig. 9F, HDV genomic RNA were analyzed by Northern blotting, and HDAg, HBsAg, TAP, and Aly proteins were analyzed by Western blotting. Using shRNA specific for TAP or Aly showed a significant decrease in HDV genomic RNA (1.7 kb) and HDAg protein levels in the culture medium, with no effect on HBsAg protein levels. TAP or Aly shRNA had no effect on the cellular HDV genomic RNA and cellular HDAgs. Taken together, these results show that DMSO-differentiated HuS-E/2 cells are susceptible to HDV infection, and TAP and Aly play an essential role in the assembly and release of HDV virions in HDV-infected cells.
FIGURE 9.
The role of TAP and Aly in viral assembly and release under conditions of HDV infection. A, Western blotting analysis of NTCP after DMSO treatment in HuS-E/2 cells. DMSO-differentiated HuS-E/2 cells were harvested and the cell lysate was subjected to Western blotting analysis with antibodies against NTCP. B, HDV virions were prepared and subjected to Northern blot analysis with a DIG-labeled probe (top panel) and Western blotting analysis with antibodies against HDAg or HBsAg (bottom panel). C and D, co-immunoprecipitation assay. DMSO-differentiated HuS-E/2 cells were left untreated or were infected with HDV. At 7 days post-infection, the cells were harvested and subjected to immunoprecipitation (IP) with anti-TAP antibodies (C), followed by Western blotting analysis with antibodies against TAP or HDAgs or with anti-Aly antibodies (D) followed by Western blotting analysis with antibodies against Aly and HDAgs. E, DMSO-differentiated HuS-E/2 cells were infected with HDV. At 7 days post-infection, cells were examined by immunofluorescence staining with antibodies against HDAg, TAP, or Aly and confocal microscopy. The bars on the images represent 20 μm. F, DMSO-differentiated HuS-E/2 cells were co-transfected with plasmid encoding TAP shRNA or Aly shRNA and plasmid p1.3HBcl encoding HBsAgs for 2 days. Then, cells were infected with HDV. Seven days post-infection, protein lysates and RNA were prepared from the infected cells and HDV virions were collected from the culture medium. Northern blot analysis was performed to detect secreted and cellular HDV genomic RNA. Western blotting analysis was performed to detect cellular TAP, Aly, HDAgs, and tubulin and secreted HDAgs and HBsAgs as indicated. NT, non-infection.
Discussion
In this study, we examined whether the cellular RNA export factors TAP and Aly were involved in the process by which HDAg-L regulates the CRM1-independent nuclear export pathway, and, in particular, in HDV morphogenesis. Our results showed that TAP is an HDAg-L-interacting protein. Site-directed mutagenesis of Pro205 of HDAg-L to alanine resulted in loss of the ability to bind to TAP, loss of RNP nuclear export activity, and inability to support the assembly of HDV. The reason may be partially explained by the change of the tertiary structure of HDAg-L induced by the mutation, thus preventing the interaction. Use of TAP shRNA or Aly shRNA resulted in inhibition of secretion of HDV virions. We therefore conclude that the interactions between HDAg-L, TAP, and Aly are essential steps in HDV assembly.
Although TAP exhibits RNA-binding activity by itself in vitro (34, 35), it predominantly selects its mRNA cargoes through interactions with adaptor RNA-binding proteins. The protein Aly directly binds to TAP and recruits it to mRNA cargoes. Accordingly, we speculate that the interaction between HDAg-L, TAP, and Aly described here is essential in guiding HDV RNP assembly and release. Our data support the hypothesis that HDAg-L functions like adaptor RNA-binding proteins and facilitates nuclear export of HDV genomic RNA through the TAP-mediated nuclear export pathway, which ensures fast and specific virion formation regulated by several accessory factors, including Aly.
TAP has a modular domain organization, including binding sites for adaptor RNA-binding proteins and FG-nucleoporins (34, 36). A fragment of TAP consisting of the N-domain (residues 96–371), which is known to be involved in adaptor binding with Aly (37) or SRP20 (38), was found to bind to HDAg-L. Whether HDAg-L competes with the interaction between TAP and Aly or SRP20 requires elucidation. Further truncation analyses or site-directed mutagenesis also needs to be performed to determine the amino acid residues of TAP that are important for its interaction with either Aly or HDAg-L. In addition, p15 and the co-adaptor Thoc5 are necessary for the TAP-mediated nuclear export of cellular mRNA (20) and it remains to be determined whether these proteins have functional roles in the TAP-mediated nuclear export of HDAg-L.
The nuclear-cytoplasmic transport of proteins and RNAs across the nuclear envelope occurs through channels formed by macromolecular structures known as nuclear pore complexes (39), which span the double membrane of the nuclear envelope and act as a gateway for macromolecular trafficking. A general theme in the nuclear export of proteins is that specialized export receptors recognize cargoes with specific export signals. A well known example is the recognition of the classical leucine-rich NES by the CRM1 exportin (40–42). The host NES-interacting protein (NESI), which interacts with lamin A/C and nucleoporins in the nuclear envelope, has been reported to facilitate the nuclear export of HDAg-L (43). It will be an interesting issue to elucidate the association between NESI protein and TAP-/Aly-mediated nuclear export pathway. In addition, it has been reported that HDV RNAs could be exported to the cytoplasm immediately after synthesis and processing without HDAg-L (44). It is possible that knockdown of Aly and TAP interfere with secretion of virus particles but do not block egress of HDV RNA from the nucleus.
Several viral proteins utilize the host nuclear export factors TAP and Aly to pass through the CRM1-independent nuclear export pathway. The Epstein-Barr virus early protein EB2 contains a CRM1-independent arginine-rich NES that promotes nucleocytoplasmic export by binding Aly (24) and TAP (25). Efficient ICP27-mediated nuclear export of herpes simplex virus 1 RNA requires TAP (26, 27). Both HDAg-L (19) and the human cytomegalovirus transactivator protein pUL69 (28) contain a proline-rich NES. pUL69 functions as a viral nuclear RNA export factor and interacts with the cellular DEXD/H-box RNA helicase UAP56, which recruits Aly and thus indirectly the TAP-p15 export receptor (45). These results are in agreement with our finding that the viral nucleoprotein HDAg-L promotes HDV RNA export by interacting with TAP and Aly.
HDV genotype I is more widely spread throughout the world than genotype II and is more often associated with severe outcomes (46). The level of farnesylation of genotype II HDAg-L is similar to that of genotype I HDAg-L (47). However, the packaging activity of genotype II is lower than that of genotype I (47, 48). The relatively low assembly efficiency of HDV genotype II results in the milder disease phenotype of HDV genotype II (48). On the other hand, HDV genotype III is only found in the north of South America, and genotype III HDAg-L shows significant divergence from genotypes I and II HDAg-L (49). Amino acid sequences show that the clathrin box-like domain found in genotypes I and II is not present in the C terminus of genotype III HDAg-L. In our study, we used genotype I HDAg-L to study the nuclear export pathway of HDV and found that nuclear export and virus assembly of HDV genotype I is TAP-/Aly-mediated, but the molecular mechanisms involved in HDV genotypes II and III assembly remain to be elucidated.
HDV can dramatically worsen liver disease in patients coinfected or superinfected with HBV (50). HDV infection also increases the risk for HCC and mortality in patients with compensated cirrhosis type B (51), but currently there is no effective medical therapy for HDV-related chronic hepatitis. In this study, we have provided several lines of evidence that HDAg-L interacts with the host factor TAP in a highly specific manner. Whether HDV infection can disrupt TAP-mediated host mRNA transport awaits further clarification. TAP is important for mRNA nuclear export in mammalian cells, and TAP depletion results in high nuclear accumulation of poly(A)+ RNA (20). In human HeLa and monkey CV1 cells stably expressing replication-competent HDV, FACS analysis revealed a progressive decline in the percentage of HDAg-positive cells that was due to growth disadvantage, rather than apoptosis (52). Cirrhosis development without nodular regeneration is observed in liver biopsies of HDV patients (53). Based on these results, the inhibitory effects of nuclear HDAg-L on the biological functions of TAP/Aly would be expected to impair liver regeneration, resulting in progressive chronic liver disease. Furthermore, understanding how the HDAg-L/TAP/Aly interaction is involved in the life cycle of HDV will allow us to determine the biological roles of HDV in human liver diseases. Our study also suggests that TAP or Aly may be new molecular targets for treating HDV infection.
Experimental Procedures
Plasmids
pGEX-HDAg-L(198–210) and pGEX-HDAg-L(198–210)-P205A
Plasmid pGEX-HDAg-L(198–210) has been described previously (54). For the construction of plasmid pGEX-HDAg-L(198–210)-P205A, a cDNA fragment coding for amino acid residues 198–210 of HDAg-L in which proline 205 had been mutated to alanine was generated by annealing two synthetic oligonucleotides representing the two strands of HDAg-L(198–210), with EcoRI/SalI recognition sequences added to the two ends. This cDNA fragment was then cloned into the EcoRI/SalI sites of plasmid pGEX-6P-1 (GE Healthcare Biosciences).
pECE-d-BE, pECE-d-SM, pECE-d-BE(P205A), pECEL-d35/88, pSVD2, pD3, and pECE-C-ES
Plasmid pECE-d-BE, encoding HDAg-L, and pECE-d-SM, encoding HDAg-S (21), and pECE-d-BE(P205A), encoding mutant HDAg-L in which proline 205 has been replaced by alanine, and pECEL-d35/88, encoding HDAg-L lacking amino acid residues 35–88 (30), and pSVD2, containing a dimeric form of HDV genomic RNA under control of the simian virus 40 early promoter (15), and pD3, containing a trimeric form of HDV genomic RNA, and pECE-C-ES, encoding HBsAg (15), have been described previously.
Plasmids Encoding GST-TAP Fusion Proteins
Plasmids encoding GST fusion proteins containing amino acids 1–619, 96–371, 188–550, 188–619, 371–550, 371–619, or 551–619 of TAP protein (20) were kindly provided by Dr. Jun Katahira, Osaka University, Japan.
pLKO.1-shTAP, pLKO.1-shAly, and pLKO.1
These plasmids were obtained from the National RNAi Core Facility (Academia Sinica, Taiwan). pLKO.1-shTAP was transcribed to shRNA 5′-CGCGAACGAUUUCCCAAGUUA-3′ for TAP RNA interference and pLKO.1-shAly was transcribed to shRNA 5′-GAACUCUUUGCUGAAUUUGGA-3′ for Aly RNA interference. Vector pLKO.1 was used as control.
pcDNA3.0-HA-LHBsAg, pcDNA3.0-HA-MHBsAg, and pcDNA3.0-HA-SHBsAg
Plasmids pcDNA3.0-HA-LHBsAg, pcDNA3.0-HA-MHBsAg, and pcDNA3.0-HA-SHBsAg containing cDNA fragments encoding large, middle, and small HBsAg, respectively, were generated as described previously (33).
p1.3HBcl
p1.3HBcl, which contains a 1.3-fold HBV genome of the ayw subtype on a modified pUC13 vector backbone, in which the transcription of pregenomic RNA is controlled by the core promoter and enhancer I and II regulatory elements of the virus, has been described previously (55).
Antibodies and Reagents
Rabbit polyclonal antibodies against HDAg were generated as described previously (9). Rabbit polyclonal antibodies against TAP and Aly were from Genetex and goat polyclonal antibodies against HBsAg from Dako. Mouse monoclonal antibodies against TAP and Aly were purchased from Sigma, whereas mouse monoclonal antibody against tubulin was purchased from Millipore. Goat polyclonal antibodies against NTCP were purchased from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used for Western blotting, whereas Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies (Invitrogen) were used for immunofluorescent staining.
Cell Lines and DNA Transfection
Huh7 cells and HepG2.2.15 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 100 nm nonessential amino acids (all from Gibco). DNA transfection was performed using cationic liposomes (Invitrogen) according to the manufacturer's directions. HuS-E/2 cells were grown as described previously (56).
Synthetic Peptides TAT and TAT-HDAg-L(198–210)
The cell-permeable TAT peptide is a 10-amino acid carrier peptide from the HIV-1 TAT sequence (48GRKKRRQRRR57) (57). Peptide TAT-HDAg-L(198–210) consists of this a 10-amino acid peptide, HDAg-L(198–210) (ILFPADPPFSPQS), and two proline residues inserted as a spacer between TAT peptide and HDAg-L(198–210) to allow for maximal flexibility (58).
Purification of TAP(1–619) and GST Pulldown Assay
Expression and purification of recombinant fusion proteins and the GST pulldown assay were performed as described previously (54). To purified TAP(1–619), GST-TAP(1–619) was cleaved by PreScission Protease (GE Healthcare) between the GST moiety and the TAP(1–619) according to the manufacturer's directions. Because the protease is fused to GST, both GST and the protease were easily removed from cleavage reactions using glutathione-SepharoseTM 4B. For the GST pulldown competition assay, the test GST fusion protein was first incubated for 4 h at 4 °C with glutathione-Sepharose 4B beads in the presence of increasing amounts of TAT-HDAg-L(198–210), then the mixture was incubated for 18 h at 4 °C with a lysate of Huh7 cells transiently expressing HDAg-L. The beads were then precipitated, extensively washed with PBS containing 1% Triton X-100, and the bound proteins resolved by SDS-PAGE.
Immunofluorescence Staining, Co-immunoprecipitation, and Western Blotting Analysis
Immunofluorescence staining, co-immunoprecipitation, and Western blotting analysis were performed as previously described (19).
Harvesting of HDV Virions and Determination of Packaging Activity
To determine the packaging activity of HDAg-L and HBsAg under various conditions, HDV virions were collected from the culture supernatant of HepG2.2.15 cells at 7 days posttransfection or Huh7 cells at 4 days posttransfection as described previously (19). Protein lysates or viral RNA prepared from the HDV virions were then subjected to Western blotting analysis and Northern blot analysis, respectively.
Preparation of HDV Virions and HDV Infection of Cell Culture
To generate infectious HDV virions, Huh7 cells were transfected with plasmids pSVD2, pcDNA3.0-HA-LHBsAg, pcDNA3.0-HA-MHBsAg, and pcDNA3.0-HA-SHBsAg. HDV virions were collected from the culture supernatant at 6 days posttransfection. For HDV infection, HuS-E/2 cells were differentiated by incubation with 2% DMSO for 10 days, as described previously (33).
RNA Isolation and Northern Blot Analysis
Total RNA was isolated from cultured cells and HDV virions using TRIzol® reagent (Invitrogen). RNase-free DNase I (MDBio Inc.) was used to remove cellular or viral genomic DNA and the reaction was terminated by heating at 70 °C for 10 min. To detect HDV genomic RNA in transfected cells and HDV virions, a digoxigenin (DIG)-labeled HDV trimeric antigenomic RNA was transcribed in vitro from plasmid pD3. Northern blot analysis was performed using a DIG-labeled probe according to the procedures described by the manufacturer (Roche Biochemicals).
Statistical Analysis
All values are expressed as the mean ± S.D. of the results from at least three separate experiments. One-way analysis of variance followed by Dunnett's multiple comparison test was used to compare differences among groups of samples. Asterisks indicate that the values were significantly different from the control (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author Contributions
H. C. H. conducted most of the experiments and analyzed the results. C. P. L. conducted the immunofluorescence staining. H. K. L. and M. F. C. conducted the Northern blot analysis. Y. H. L. and Y. C. L. conducted experiments on virus packaging activity. C. H. conceived the idea for the project and wrote the paper.
Acknowledgments
We thank Dr. Jun Katahira (Department of Biochemistry, Graduate School of Medicine, Osaka University, Osaka, Japan) for providing plasmids encoding the GST-TAP fusion proteins. We thank Kunitada Shimotohno (Kyoto University, Japan) for providing the HuS-E/2 cells.
This work was supported by Grants MOST104-2320-B-077-003-MY3 and MOST103-2313-B-134-001-MY3 from the Ministry of Science and Technology, Taiwan and Grants MOHW105-NRICM-M-315-000106 and MM10501-0274 from the National Research Institute of Chinese Medicine, Ministry of Health and Welfare, Taiwan. The authors declare that they have no conflicts of interest with the contents of this article.
- HDV
- hepatitis delta virus
- HBV
- hepatitis B virus
- HDAg-L
- large hepatitis delta antigen
- HDAg-S
- small hepatitis delta antigen
- HBsAg
- HBV surface antigen
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- RNP
- ribonucleoprotein
- CRM1
- chromosome region maintenance 1
- TAT
- trans-activator of transcription
- VLP
- virus-like particle
- NTCP
- sodium taurocholate cotransporting polypeptide
- DIG
- digoxigenin.
References
- 1. Rizzetto M. (1983) The delta agent. Hepatology 3, 729–737 [DOI] [PubMed] [Google Scholar]
- 2. Bonino F., Heermann K. H., Rizzetto M., and Gerlich W. H. (1986) Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58, 945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai M. M. (1995) The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64, 259–286 [DOI] [PubMed] [Google Scholar]
- 4. Chang M. F., Baker S. C., Soe L. H., Kamahora T., Keck J. G., Makino S., Govindarajan S., and Lai M. M. (1988) Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 62, 2403–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiner A. J., Choo Q. L., Wang K. S., Govindarajan S., Redeker A. G., Gerin J. L., and Houghton M. (1988) A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62, 594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo G. X., Chao M., Hsieh S. Y., Sureau C., Nishikura K., and Taylor J. (1990) A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polson A. G., Bass B. L., and Casey J. L. (1996) RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380, 454–456 [DOI] [PubMed] [Google Scholar]
- 8. Polson A. G., Ley H. L 3rd, Bass B. L., and Casey J. L. (1998) Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18, 1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang M. F., Chang S. C., Chang C. I., Wu K., and Kang H. Y. (1992) Nuclear localization signals, but not putative leucine zipper motifs, are essential for nuclear transport of hepatitis delta antigen. J. Virol. 66, 6019–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chou H. C., Hsieh T. Y., Sheu G. T., and Lai M. M. (1998) Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol. 72, 3684–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Macnaughton T. B., Wang Y. J., and Lai M. M. (1993) Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J. Virol. 67, 2228–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., and Taylor J. (1988) Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 62, 2674–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuo M. Y., Chao M., and Taylor J. (1989) Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63, 1945–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang F. L., Chen P. J., Tu S. J., Wang C. J., and Chen D. S. (1991) The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. U.S.A. 88, 8490–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang M. F., Chen C. J., and Chang S. C. (1994) Mutational analysis of delta antigen: effect on assembly and replication of hepatitis delta virus. J. Virol. 68, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glenn J. S., Watson J. A., Havel C. M., and White J. M. (1992) Identification of a prenylation site in delta virus large antigen. Science 256, 1331–1333 [DOI] [PubMed] [Google Scholar]
- 17. Hwang S. B., and Lai M. M. (1993) Isoprenylation mediates direct protein-protein interactions between hepatitis large delta antigen and hepatitis B virus surface antigen. J. Virol. 67, 7659–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Komla-Soukha I., and Sureau C. (2006) A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 80, 4648–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee C. H., Chang S. C., Wu C. H., and Chang M. F. (2001) A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 276, 8142–8148 [DOI] [PubMed] [Google Scholar]
- 20. Katahira J., Inoue H., Hurt E., and Yoneda Y. (2009) Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 28, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang M. F., Chen C. H., Lin S. L., Chen C. J., and Chang S. C. (1995) Functional domains of delta antigens and viral RNA required for RNA packaging of hepatitis delta virus. J. Virol. 69, 2508–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alves C., Freitas N., and Cunha C. (2008) Characterization of the nuclear localization signal of the hepatitis delta virus antigen. Virology 370, 12–21 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y. H., Chang S. C., Huang C., Li Y. P., Lee C. H., and Chang M. F. (2005) Novel nuclear export signal-interacting protein, NESI, critical for the assembly of hepatitis delta virus. J. Virol. 79, 8113–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hiriart E., Farjot G., Gruffat H., Nguyen M. V., Sergeant A., and Manet E. (2003) A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278, 335–342 [DOI] [PubMed] [Google Scholar]
- 25. Juillard F., Hiriart E., Sergeant N., Vingtdeux-Didier V., Drobecq H., Sergeant A., Manet E., and Gruffat H. (2009) Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J. Virol. 83, 12759–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson L. A., Li L., and Sandri-Goldin R. M. (2009) The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83, 6335–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson L. A., and Sandri-Goldin R. M. (2009) Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83, 1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lischka P., Rosorius O., Trommer E., and Stamminger T. (2001) A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20, 7271–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kutay U., Bischoff F. R., Kostka S., Kraft R., and Görlich D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071 [DOI] [PubMed] [Google Scholar]
- 30. Huang C., Chang S. C., Yang H. C., Chien C. L., and Chang M. F. (2009) Clathrin-mediated post-Golgi membrane trafficking in the morphogenesis of hepatitis delta virus. J. Virol. 83, 12314–12324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rizzuti M., Nizzardo M., Zanetta C., Ramirez A., and Corti S. (2015) Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today 20, 76–85 [DOI] [PubMed] [Google Scholar]
- 32. Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., Fu L., Song M., Chen P., Gao W., Ren B., Sun Y., Cai T., Feng X., Sui J., and Li W. (2012) Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1, e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang H. C., Chen C. C., Chang W. C., Tao M. H., and Huang C. (2012) Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol. 86, 9443–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katahira J., Strässer K., Podtelejnikov A., Mann M., Jung J. U., and Hurt E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18, 2593–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liker E., Fernandez E., Izaurralde E., and Conti E. (2000) The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 19, 5587–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bachi A., Braun I. C., Rodrigues J. P., Panté N., Ribbeck K., von Kobbe C., Kutay U., Wilm M., Görlich D., Carmo-Fonseca M., and Izaurralde E. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6, 136–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodrigues J. P., Rode M., Gatfield D., Blencowe B. J., Carmo-Fonseca M., and Izaurralde E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang Y., Yario T. A., and Steitz J. A. (2004) A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 101, 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wente S. R. (2000) Gatekeepers of the nucleus. Science 288, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 40. Wen W., Meinkoth J. L., Tsien R. Y., and Taylor S. S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473 [DOI] [PubMed] [Google Scholar]
- 41. Fornerod M., Ohno M., Yoshida M., and Mattaj I. W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 42. Stade K., Ford C. S., Guthrie C., and Weis K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 43. Huang C., Jiang J. Y., Chang S. C., Tsay Y. G., Chen M. R., and Chang M. F. (2013) Nuclear export signal-interacting protein forms complexes with lamin A/C-Nups to mediate the CRM1-independent nuclear export of large hepatitis delta antigen. J. Virol. 87, 1596–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Macnaughton T. B., and Lai M. M. (2002) Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76, 3928–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas M., Lischka P., Müller R., and Stamminger T. (2011) The cellular DEXD/H-box RNA-helicases UAP56 and URH49 exhibit a CRM1-independent nucleocytoplasmic shuttling activity. PloS One 6, e22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu J. C., Choo K. B., Chen C. M., Chen T. Z., Huo T. I., and Lee S. D. (1995) Genotyping of hepatitis D virus by restriction-fragment length polymorphism and relation to outcome of hepatitis D. Lancet 346, 939–941 [DOI] [PubMed] [Google Scholar]
- 47. O'Malley B., and Lazinski D. W. (2005) Roles of carboxyl-terminal and farnesylated residues in the functions of the large hepatitis delta antigen. J. Virol. 79, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu S. C., Syu W. J., Sheen I. J., Liu H. T., Jeng K. S., and Wu J. C. (2002) Varied assembly and RNA editing efficiencies between genotypes I and II hepatitis D virus and their implications. Hepatology 35, 665–672 [DOI] [PubMed] [Google Scholar]
- 49. Wu J. C., Chiang T. Y., and Sheen I. J. (1998) Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 79, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 50. Saracco G., Macagno S., Rosina F., and Rizzetto M. (1988) Serologic markers with fulminant hepatitis in persons positive for hepatitis B surface antigen. A worldwide epidemiologic and clinical survey. Ann. Intern. Med. 108, 380–383 [DOI] [PubMed] [Google Scholar]
- 51. Fattovich G., Giustina G., Christensen E., Pantalena M., Zagni I., Realdi G., and Schalm S. W. (2000) Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B: the European Concerted Action on Viral Hepatitis (Eurohep). Gut 46, 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang D., Pearlberg J., Liu Y. T., and Ganem D. (2001) Deleterious effects of hepatitis delta virus replication on host cell proliferation. J. Virol. 75, 3600–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rizzetto M., Verme G., Recchia S., Bonino F., Farci P., Aricò S., Calzia R., Picciotto A., Colombo M., and Popper H. (1983) Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen: an active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern Med. 98, 437–441 [DOI] [PubMed] [Google Scholar]
- 54. Huang C., Chang S. C., Yu I. C., Tsay Y. G., and Chang M. F. (2007) Large hepatitis delta antigen is a novel clathrin adaptor-like protein. J. Virol. 81, 5985–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chou Y. C., Jeng K. S., Chen M. L., Liu H. H., Liu T. L., Chen Y. L., Liu Y. C., Hu C. P., and Chang C. (2005) Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. J. Virol. 79, 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aly H. H., Watashi K., Hijikata M., Kaneko H., Takada Y., Egawa H., Uemoto S., and Shimotohno K. (2007) Serum-derived hepatitis C virus infectivity in interferon regulatory factor-7-suppressed human primary hepatocytes. J. Hepatol. 46, 26–36 [DOI] [PubMed] [Google Scholar]
- 57. Vivès E., Brodin P., and Lebleu B. (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 58. Bonny C., Oberson A., Negri S., Sauser C., and Schorderet D. F. (2001) Cell-permeable peptide inhibitors of JNK: novel blockers of β-cell death. Diabetes 50, 77–82 [DOI] [PubMed] [Google Scholar]