Abstract
The antineoplastic agent benzyl isothiocyanate (BITC) acts by targeting multiple pro-oncogenic pathways/genes, including signal transducer and activator of transcription 3 (STAT3); however, the mechanism of action is not well known. As reported previously, BITC induced reactive oxygen species (ROS) in Panc1, MiaPaCa2, and L3.6pL pancreatic cancer cells. This was accompanied by induction of apoptosis and inhibition of cell growth and migration, and these responses were attenuated in cells cotreated with BITC plus glutathione (GSH). BITC also decreased expression of specificity proteins (Sp) Sp1, Sp3, and Sp4 transcription factors (TFs) and several pro-oncogenic Sp-regulated genes, including STAT3 and phospho-STAT3 (pSTAT3), and GSH attenuated these responses. Knockdown of Sp TFs by RNA interference also decreased STAT3/pSTAT3 expression. BITC-induced ROS activated a cascade of events that included down-regulation of c-Myc, and it was also demonstrated that c-Myc knockdown decreased expression of Sp TFs and STAT3. These results demonstrate that in pancreatic cancer cells, STAT3 is an Sp-regulated gene that can be targeted by BITC and other ROS inducers, thereby identifying a novel therapeutic approach for targeting STAT3.
Keywords: Myc (c-Myc), pancreatic cancer, reactive oxygen species (ROS), specificity protein 1 (Sp1), STAT3
Introduction
Isothiocyanates (ITCs)3 are a family of structurally related natural products that are found in cruciferous vegetables such as glucosinolates, which are hydrolyzed by the enzyme myrosinase to give the unconjugated ITCs. Cruciferous vegetables in the diet have been associated with decreased risks for human cancers and exhibit both chemoprevention and chemotherapeutic activities in animal models, and this has primarily been associated with ITCs (1–5). Sulforaphane, allyl isothiocyanate, phenethyl isothiocyanate (PEITC), and benzyl isothiocyanate (BITC) have been extensively investigated as anticancer agents, and a recent review summarizes the multiple genes and associated pathways activated by ITCs (4). Treatment of cancer cells or animal models that develop tumors with ITCs decreases cell growth, induces apoptosis, and inhibits epithelial-to-mesenchymal transition, angiogenesis, and cell cycle progression (6–16). ITCs, including BITC, exhibit a broad spectrum of activities that include induction of ROS and ROS-regulated genes, direct inhibition of genes, and formation of peptide and protein adducts (4, 5, 15–17). ITCs form adducts with both thiol and amino groups, and the formation of amino adducts forms the basis of the Edman procedure used in protein sequencing (15, 18–20). Many of these ITC-induced effects contribute to their anticancer activities; however, a recent study suggests that the alkylation of peptides by ITCs may also be responsible for allergic skin reactions to these compounds (20).
Studies in this laboratory have focused on specificity protein (Sp) transcription factors (TFs) Sp1, Sp3, and Sp4 as non-oncogene addiction genes in cancer cells, and knockdown of Sp1, Sp3, and Sp4 individually and combined inhibits cell growth, induces apoptosis, and inhibits cell migration/invasion (21). Drugs that target Sp TFs gave responses similar to those observed after Sp knockdown, and this includes decreased expression of several pro-oncogenic genes, including bcl-2, survivin, p65NFκB, β-catenin, multiple receptor tyrosine kinases, vascular endothelial growth factor (VEGF), and its receptors (22–30). We recently reported that many of the responses/genes affected by PEITC in pancreatic cancer cells were due to induction of ROS and ROS-dependent down-regulation of Sp1, Sp3, Sp4, and pro-oncogenic Sp-regulated genes (30). These results complement studies showing that hydrogen peroxide and t-butylhydroperoxide also down-regulate Sp proteins (22, 23, 27).
BITC is structurally related to PEITC, and both compounds exhibit comparable anticancer activities and modulate expression of some common genes/pathways consistent with their effects on cancer cells (6–14). It was also reported that BITC inhibits signal transducer and activator of transcription 3 (STAT3) phosphorylation and down-regulates STAT3 protein (6), and another study by the same group showed that BITC also induces ROS in pancreatic and other cancer cell lines (31–33). STAT3 is a negative prognostic factor for pancreatic and other cancers (34–40), and the functional importance of STAT3 in cancer was aptly summarized in a recent review by Frank (41). Based on these previous observations, we hypothesized that STAT3 may also be an Sp-regulated gene that can be targeted by ROS-inducing anticancer agents such as BITC, and the results of this study (41) support this hypothesis.
Results
Role of BITC-induced ROS on Cell Growth, Survival, and Invasion
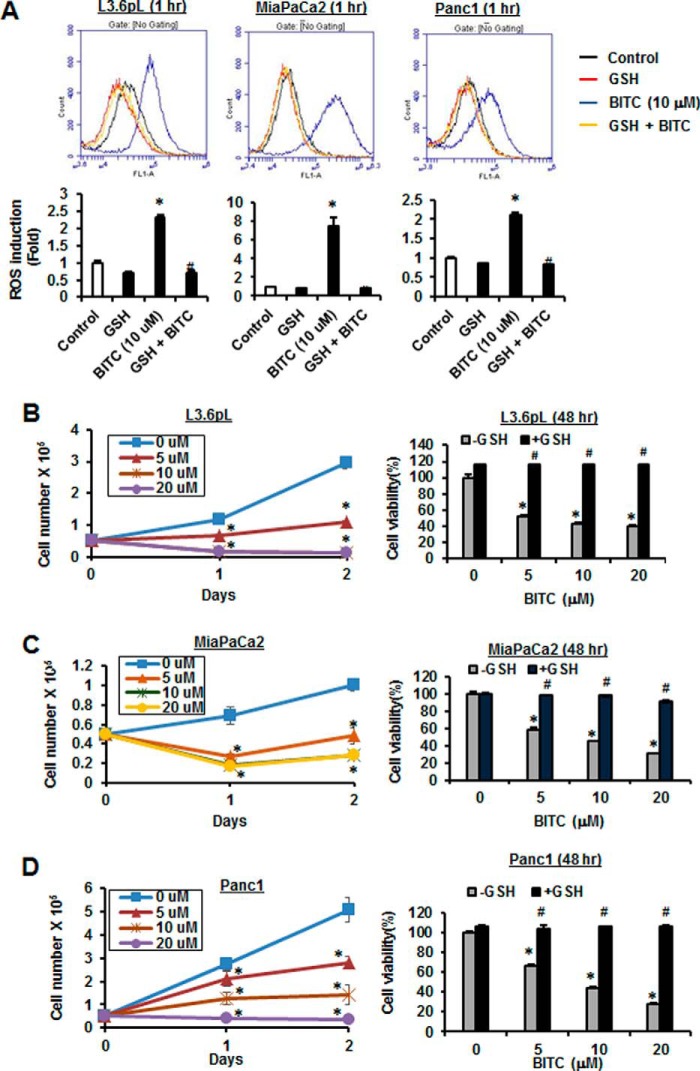
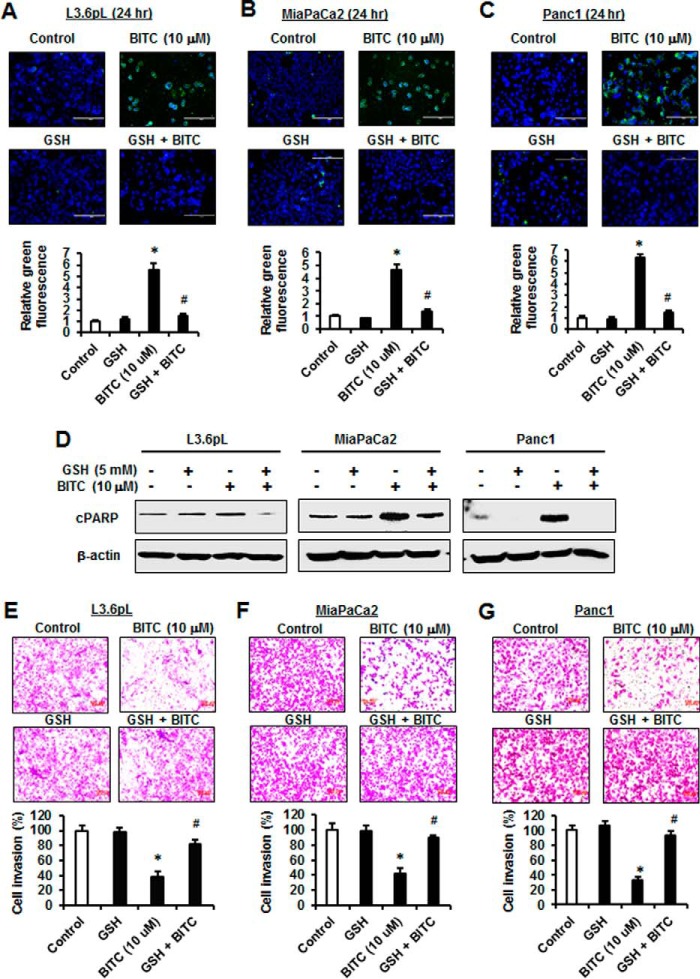
In this study, we used L3.6pL, MiaPaCa2, and Panc1 pancreatic cancer cell lines to investigate the role of ROS in mediating BITC-induced responses. Treatment of the pancreatic cancer cells with 10 μm BITC for 1 h significantly induced ROS as determined by FACS analysis using the cell-permeable CM-H2DCFDA dye, and cotreatment with GSH significantly decreased ROS induction (Fig. 1A). L3.6pL cells were treated with BITC alone resulting in growth inhibition, and after cotreatment with GSH, the growth inhibitory effects were significantly attenuated (Fig. 1B). Similar results were obtained in MiaPaCa2 (Fig. 1C) and Panc1 (Fig. 1D) cells, demonstrating that BITC-induced growth inhibition is ROS-dependent. BITC also induced annexin V staining, a marker of apoptosis, in L3.6pL, MiaPaCa2, and Panc1 cells (Fig. 2, A–C), and this response was also significantly attenuated in cells cotreated with GSH. In addition, BITC also induced PARP cleavage in L3.6pL, MiaPaCa2, and Panc1 cells (Fig. 2D); this response was also reversed after cotreatment with GSH. Fig. 2, E–G, shows that BITC (10 μm) also inhibited pancreatic cancer cell invasion in a Matrigel-coated chamber assay, and cotreatment with GSH significantly reversed this response. Thus, BITC-induced ROS plays an important role in mediating inhibition of pancreatic cancer cell growth, induction of apoptosis, and inhibition of invasion.
FIGURE 1.
BITC induces ROS and inhibits cancer cell growth. A, L3.6pL, MiaPaCa2, and Panc1 cells were pre-treated with 5 mm GSH for 30 min and then treated with vehicle DMSO (control) and 10 μm BITC alone or in combination with GSH for 1 h. ROS levels were measured by FACS using cell-permeant CM-H2DCFDA dye. L3.6pL (B), MiaPaCa2 (C), and Panc1 (D) cells were treated with different concentrations of BITC for 24 and 48 h. Cell numbers were counted by using a Coulter Z1 cell counter (left panels). Cells were pretreated with 5 mm GSH for 3 h and then treated with different concentrations of BITC for 48 h. Cell viability was measured after 48 h of treatment by MTT assay (right panels). Data represent three independent experiments and are expressed as mean ± S.E., and significant (p < 0.05) induction of ROS or growth inhibition (*) or reversal by GSH (#) is indicated.
FIGURE 2.
BITC induces apoptosis and inhibits cancer cell invasion. L3.6pL (A), MiaPaCa2 (B), and Panc1 (C) cells were pre-treated with 5 mm GSH for 3 h and then treated with DMSO (control) and 10 μm BITC alone or in combination with GSH for 24 h, and annexin V staining was determined and quantitated by fluorescence microscope. D, L3.6pL, MiaPaCa2, and Panc1 cells were treated as in A–C, and whole cell lysates were analyzed with indicated antibodies by Western blotting. L3.6pL (E), MiaPaCa2 (F), and Panc1 (G) cells were treated as in A–C, and invasion was analyzed and quantitated by using Corning BioCoat Matrigel Invasion Chamber. Data represent three independent experiments and are expressed as mean ± S.E., and significant (p < 0.05) induction of apoptosis or inhibition of invasion (*) or reversal by GSH (#) is indicated.
BITC Decreases Sp TFs and Sp-regulated STAT3 Expression
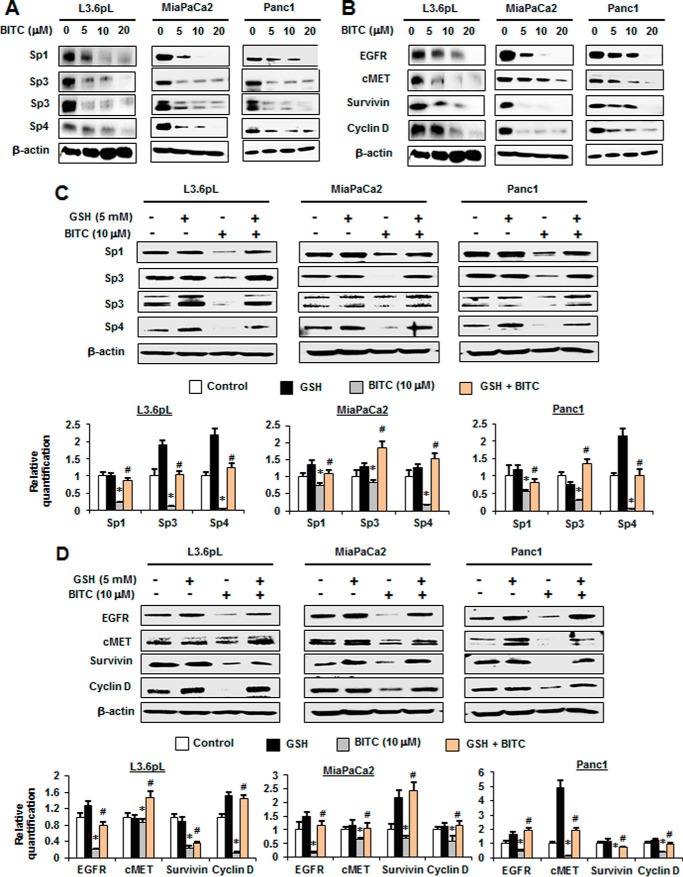
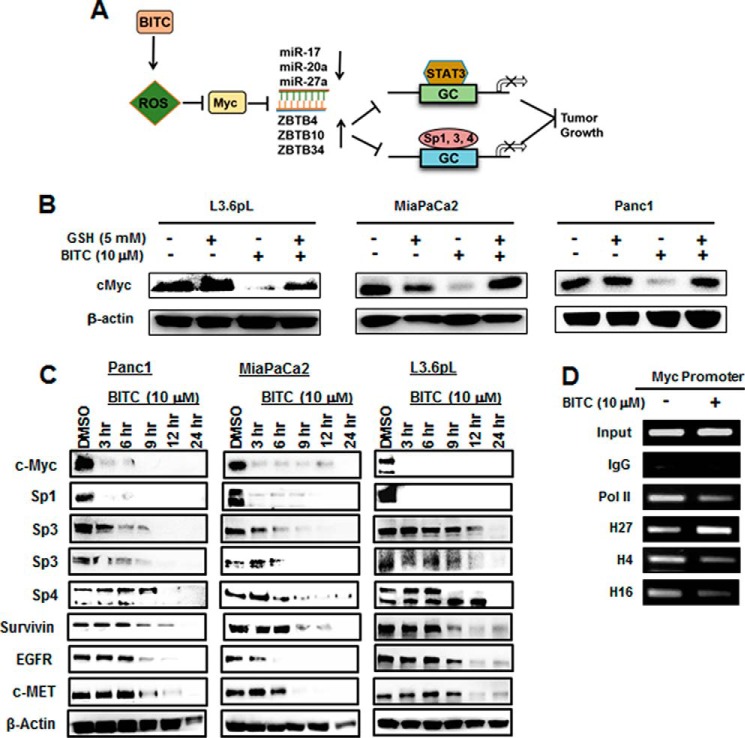
Fig. 3A illustrates the concentration-dependent effects of BITC on down-regulation of Sp1, Sp3, and Sp4 proteins in pancreatic cancer cell lines. BITC clearly decreases expression of all three Sp TFs. BITC also decreased expression of several previously identified Sp-regulated genes in L3.6pL, MiaPaCa2, and Panc1 cells, including epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (c-MET), survivin, and cyclin D1 (Fig. 3B). The role of BITC-induced ROS in mediating Sp down-regulation was also determined, and the results showed that decreased expression of Sp1, Sp3, and Sp4 after treatment with 10 μm BITC was significantly reversed after cotreatment with GSH (Fig. 3C). Moreover, similar results were observed for Sp-regulated genes EGFR, c-MET, survivin, and cyclin D1 (Fig. 3D). We also observed that BITC-induced down-regulation of Sp1, Sp3, and Sp4 in these cell lines was also inhibited after cotreatment with catalase (supplemental Fig. S1). Thus, BITC induces ROS and ROS-mediated down-regulation of Sp1, Sp3, and Sp4 in pancreatic cancer cells. Moreover, the functional effects of BITC, namely inhibition of cell growth and migration and induction of apoptosis (Figs. 1 and 2), are similar to those observed after knockdown of Sp1, Sp3, and Sp4 alone or in combination in pancreatic cancer cells (21), suggesting that these antineoplastic effects induced by BITC are due, in part, to Sp down-regulation.
FIGURE 3.
BITC down-regulates Sp1, Sp3, Sp4, and Sp-regulated genes. A and B, L3.6pL, MiaPaCa2, and Panc1 cells were treated with 0, 5, 10, and 20 μm BITC for 24 h, and whole cell lysates were analyzed for Sp1, Sp3, and Sp4 proteins (A) and Sp-regulated, pro-survival, and growth-promoting proteins by Western blottings (B). C and D, L3.6pL, MiaPaCa2, and Panc1 were pre-treated with 5 mm GSH for 3 h and then treated with 10 μm BITC alone or in combination with GSH for 24 h. The whole cell lysates were analyzed for Sp1, Sp3, and Sp4 proteins (C) and Sp-regulated, pro-survival, and growth-promoting proteins by Western blottings (D). The signals were quantitated by ImageJ software. Data represent three independent experiments and are expressed as mean ± S.E., and significant (p < 0.05) inhibition of proteins (*) or reversal by GSH (#) is indicated.
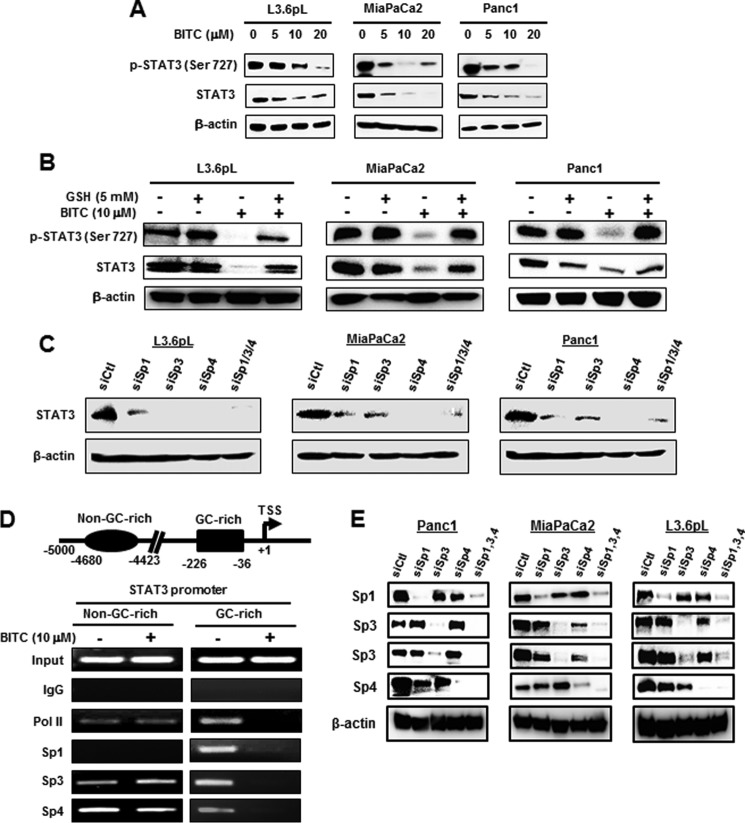
It was previously reported that BITC decreased expression of phospho-STAT3 (pSTAT3) in pancreatic cancer cells (6), and we hypothesized that STAT3 may also be an Sp-regulated gene. Treatment of L3.6pL, MiaPaCa2, and Panc1 cells with 5–20 μm BITC for 24 h and different times decreased expression of STAT3 and pSTAT3, and significant decreases were observed at the lowest concentration (Fig. 4A). The sensitivity of these cells to BITC-induced STAT3 down-regulation was similar to that observed for decreased expression of Sp TFs (Fig. 3A), suggesting that STAT3 may be an Sp-regulated gene. Moreover, BITC (10 μm)-induced STAT3 down-regulation (4-h treatment) was inhibited after cotreatment with GSH (Fig. 4B). Confirmation that STAT3 was an Sp-regulated gene was determined by RNAi, and knockdown of Sp1, Sp3, Sp4, or Sp1/3/4 (combination) (as reported previously (21); lysates from this study were used for STAT3 analysis) decreased STAT3 protein expression (Fig. 4C). The −226 to −36 proximal promoter region of the STAT3 gene contains GC-rich sequences (42), and after treatment of Panc1 cells with 10 μm BITC for 3 h, we analyzed protein interactions with the STAT3 promoter in a ChIP assay (Fig. 4D). The pol II, Sp1, Sp3, and Sp4 bound to the GC-rich region of the promoter, and BITC decreased these interactions. A ChIP assay of a distal region of the STAT3 promoter showed that pol II, Sp3, and Sp4 (not Sp1) were associated with the promoter, but BITC did not affect these interactions. Fig. 4E illustrates that knockdown of Sp1, Sp3, and Sp4 or all three combined (Sp1,3,4) is specific for the individual target except for some decrease in Sp4 in Panc1 cells transfected with siSp1 and siSp3 (21). These results provide further support that STAT3 is an Sp-regulated gene and can be targeted by BITC and other drugs that down-regulate Sp TFs.
FIGURE 4.
BITC down-regulates STAT3 and disrupts Sp binding on GC-rich STAT3 promoter. A, L3.6pL, MiaPaCa2, and Panc1 cells were treated with 0, 5, 10, and 20 μm BITC for 24 h. B, L3.6pL, MiaPaCa2, and Panc1 cells were pre-treated with 5 mm GSH for 3 h and then treated with 10 μm BITC alone or in combination with GSH for 4 h, and whole cell lysates from A and B were analyzed by Western blottings for phosphorylation of STAT3 at Ser-727 and total STAT3. C, L3.6pL, MiaPaCa2, and Panc1 cells were transfected with siRNAs for Sp1, Sp3, Sp4 and their combination of Sp1/3/4, and whole cell lysates (21) were subjected to Western blotting analysis for STAT3 expression. D, schematic diagram of the human STAT3 promoter and the positions of non-GC- and GC-rich regions are shown along with the corresponding ChIP primers spanning the regions. Panc1 cells were treated with 10 μm BITC for 3 h, and the ChIP assays were performed with control (IgG), polymerase II, Sp1, Sp3, and Sp4 antibodies on STAT3 non-GC and GC regions. E, cells were transfected with oligonucleotides target against Sp1, Sp3, and Sp4, and whole cell lysates were analyzed by Western blottings.
Mechanism of BITC-induced Down-regulation of Sp TFs
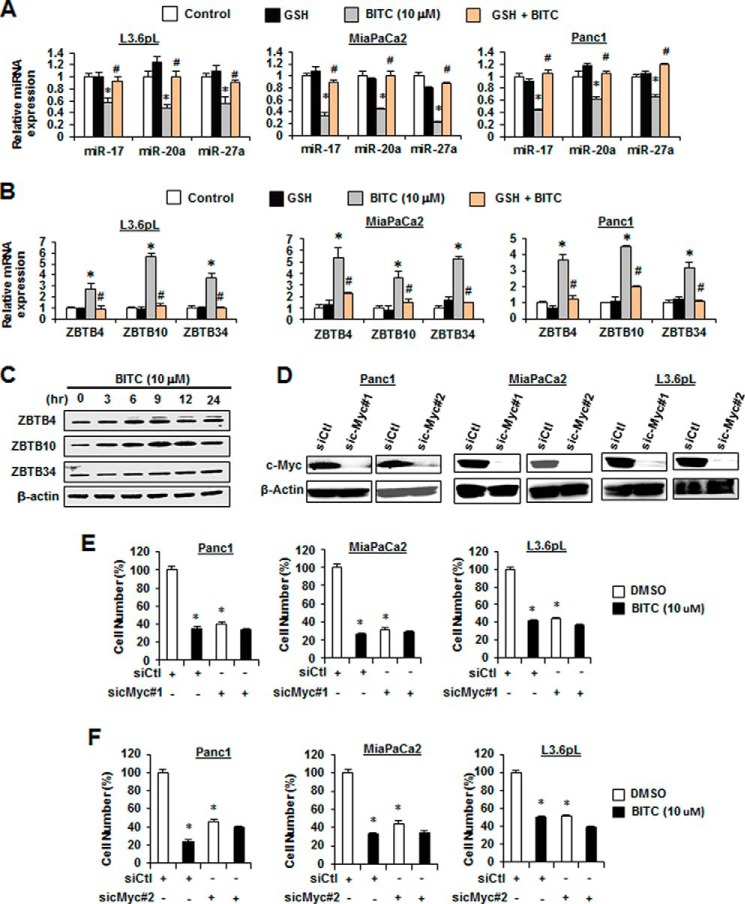
Previous studies show that ROS induces down-regulation of c-Myc and c-Myc-regulated miR-27a and miR-20a/miR-17–5p in cancer cells (30), and this results in increased expression of Sp repressor genes ZBTB10 and ZBTB4/ZBTB34, respectively (Fig. 5A). Treatment of the pancreatic cancer cells with 10 μm BITC ± GSH showed that BITC decreased c-Myc (4 h), and this response was reversed after cotreatment with GSH (Fig. 5B). We also examined the time course of BITC-induced down-regulation of Sp1, Sp3, Sp4, c-Myc, and Sp-regulated genes and observed initial rapid down-regulation of c-Myc and Sp1 as reported previously for PEITC-induced effects (Fig. 5C) (30). BITC also decreased interactions of pol II with the GC-rich proximal promoter region of the c-Myc gene (Fig. 5D), and this was accompanied by increased methylation of H3K27me3 and decreased acetylation of H4K16Ac as reported previously for ROS-induced Myc down-regulation (30). BITC (10 μm) also decreased miR-17, miR-20a, and miR-27a expression in L3.6pL, MiaPaCa2, and Panc1 cells, and this response was attenuated after cotreatment with GSH (Fig. 6A), demonstrating that BITC-mediated down-regulation of c-Myc and miR-17/miR-20a and miR-27a was ROS-dependent. BITC (10 μm) also induced ZBTB10, ZBTB4, and ZBTB34 mRNA, which was reversed after cotreatment with GSH (Fig. 6B), and induced ZBTB10, ZBTB4, and ZBTB34 protein levels in Panc1 cells at different time points over a 24-h treatment (Fig. 6C). The importance of c-Myc in mediating the effects of BITC was examined by investigating the growth inhibitory activity of BITC in the presence or absence of c-Myc, which is the key initial target of BITC-induced ROS (Fig. 5A). The results show that two oligonucleotides that knock down c-Myc (Fig. 6D) also decrease cell growth in the pancreatic cancer cell lines (Fig. 6, E and F). BITC was also a potent inhibitor of cancer cell growth in wild-type cells expressing c-Myc; however, the growth inhibitory effects of BITC were significantly attenuated in c-Myc-depleted cells (Fig. 6, E and F). These results demonstrate that BITC induces ROS-dependent down-regulation of Sp TFs through the c-Myc → miR → ZBTB axis (Fig. 5A), and this parallels previous studies showing that miR antagomirs and overexpression of ZBTB10/ZBTB4 also decrease expression of Sp TFs (21, 30).
FIGURE 5.
BITC rapidly down-regulates c-Myc. A, model of the proposed mechanism of action of BITC-induced ROS on down-regulation of STAT3 and Sp transcription factors is shown. B, L3.6pL, MiaPaCa2, and Panc1 cells were pretreated with 5 mm GSH for 3 h and then treated with 10 μm BITC for 4 h, and c-Myc protein expression was analyzed by Western blottings. The Western blottings in Figs. 4B and 5B were from the same experiment. C, L3.6pL, MiaPaCa2, and Panc1 cells were treated with 10 μm BITC alone for 3, 6, 9, 12, and 24 h, and whole cell lysates were analyzed and quantitated for c-Myc protein expression by Western blotting. D, Panc1 cells were treated with 10 μm BITC for 3 h, and ChIP assays were performed as in Fig. 4D on the Myc promoter.
FIGURE 6.
Role of BITC-induced ROS on the Myc-miR-ZBTB cascade. A and B, L3.6pL, MiaPaCa2, and Panc1 cells were pre-treated with 5 mm GSH for 3 h and then treated with 10 μm BITC alone or in combination with GSH, and after 6 h, total RNA was extracted. miR-17, miR-20a, and miR-27a (A) and ZTBT4, ZBTB10, and ZBTB34 (B) expressions were determined by real time-PCR. C, Panc1 cells were treated with 10 μm BITC for the indicated times, and ZBTB4, ZBTB10, and ZBTB34 proteins were analyzed by Western blotting. Cells were transfected with siCtl or two oligonucleotides targeting the sic-Myc#1 and sic-Myc#2 Western blottings of knockdown efficiency (D), and the effects of 10 μm BITC on proliferation of pancreatic cancer cells were determined in the presence or absence of c-Myc knockdown (E and F). Results shown in A, B, E, and F are expressed as mean ± S.E. for at least three replicate experiments, and significant (p < 0.05) changes by BITC (*) or reversal by GSH (#) are indicated in A and B. E and F, both BITC and c-Myc knockdown (sic-Myc) decrease cell proliferation (*).
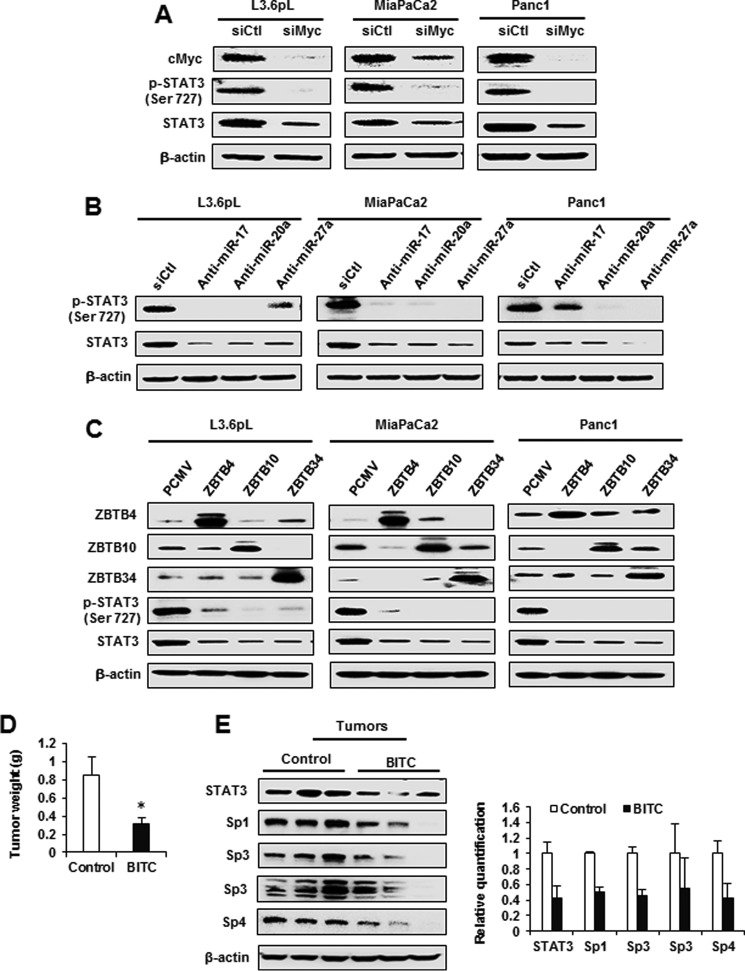
The key initial event in ROS-dependent down-regulation of Sp TFs is epigenetic down-regulation of c-Myc (30), and subsequent studies show that knockdown of c-Myc by RNA interference (siMyc) decreases Sp1, Sp3, and Sp4 expression via the pathway illustrated in Fig. 5A. The results illustrated in Fig. 7A show that knockdown of c-Myc by RNAi decreased both STAT3 and pSTAT3 proteins; moreover, transfection of miR-27a, miR-20a, and miR-17 antagomirs (Fig. 7B) also decreased STAT3 expression. Cells were also transfected with ZBTB4, ZBTB10, and ZBTB34 expression plasmids that decreased expression of STAT3 and pSTAT3 proteins (Fig. 7C). In addition, we also showed that BITC (20 mg/kg/day) inhibited tumor growth in an athymic nude mouse model bearing L3.6pL cells as xenografts (Fig. 7D). BITC did not affect body weights (supplemental Fig. S2), and Western blotting analysis of tumor lysates showed that Sp1, Sp3, Sp4, and STAT3 were decreased in tumors from BITC-treated mice (Fig. 7E). These results demonstrate that BITC-dependent repression of STAT3 is due to down-regulation of Sp TFs, and this represents a novel approach for targeting STAT3 as part of a chemotherapy regimen.
FIGURE 7.
Knockdown of c-Myc, miRNAs, and overexpression of ZBTBs down-regulates STAT3, and BITC inhibits xenograft tumor growth. A, L3.6pL, MiaPaCa2, and Panc1 cells were transfected with an siRNA targeting c-Myc, and whole cell lysates were subjected to Western blotting analysis for c-Myc, phosphorylation of STAT3 at Ser-727, and total STAT3 expression. B and C, L3.6pL, MiaPaCa2, and Panc1 cells were transfected with antago-miR-17, miR-20a, and miR-27a (B) or pCMV-ZBTB4, ZBTB10, and ZBTB34 (C) overexpression vectors. The whole cell lysates were analyzed for ZBTB4, ZBT10, and ZBTB34 phosphorylation of STAT3 at Ser-727 and total STAT3 expression. D, L3.6pL cells were injected into athymic nu/nu mice, and relative tumor weights after treatment with corn oil (control) or BITC were determined as described under “Materials and Methods.” E, protein lysates from control and BITC-treated xenograft tumor tissues were subjected to Western blotting analysis for STAT3, Sp1, Sp3, and Sp4 protein expression. Results are shown in D are expressed as mean ± S.E. for seven animals in each group, and significant (p < 0.05) changes by BITC (*) are indicated. Individual proteins were normalized to β-actin and controls (solvent-treated) for each protein were set at 1.0.
Discussion
The signal transducer and activator of transcription (STAT) family consists of seven different genes, which include STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6 (43). The tissue-specific expression and function of this family of transcription factors is highly variable with STAT2, STAT4, and STAT6 playing a role in normal cellular functions, whereas STAT1, STAT3, and STAT5 have important functions in carcinogenesis and exhibit tumor suppressor (STAT1) and oncogenic (STAT3 and STAT5) activities (44–46). Activated STAT3 (phospho-STAT3 (p-STAT3)) in particular is constitutively overexpressed in several tumor types and is a negative prognostic factor for multiple cancers (34–40). In pancreatic cancer, high expression of STAT3 is correlated with increased tumor size and stage and lymphatic metastasis (34), and the prognostic significance of microRNA-130b, which targets STAT3, was inversely correlated with STAT3 (35). The critical functional importance of STAT3 in cancer is aptly summarized in Ref. 41. It was previously reported that BITC decreased activation and expression of STAT3 in pancreatic cancer cells (6, 8), and BITC also induced ROS in some of the same cell lines (31). Several ROS-inducing anticancer agents, including PEITC, celastrol, betulinic acid, a nitro-NSAID, curcumin, histone deacetylase inhibitors, and methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate (bardoxolone-methyl), also decrease Sp1, Sp3, and Sp4 and pro-oncogenic Sp-regulated genes (22–30, 47). Based on these observations, we hypothesized that STAT3 is also an Sp-regulated gene in pancreatic cancer cells, and this is supported, in part, by the presence of Sp-binding GC-rich boxes in the 5-region of the human STAT3 gene promoter (42).
Our results confirm that BITC induces ROS in pancreatic cancer cells, and this is accompanied by ROS-dependent inhibition of pancreatic cancer cell growth, survival, and migration/invasion (Figs. 1 and 2). Moreover, like other ROS-inducing anticancer agents (22–30), BITC decreases expression of Sp1, Sp3, and Sp4 and pro-oncogenic Sp-regulated genes, including EGFR, c-MET, survivin, and cyclin D1, and similar results were observed for pSTAT3/STAT3 (Fig. 4). BITC decreased pSTAT3/STAT3, and this response was also reversed by cotreatment with the antioxidant GSH, and confirmation that STAT3 was an Sp-regulated gene was further demonstrated in Sp knockdown experiments, confirming that STAT3 was coregulated by Sp1, Sp3, and Sp4 (Fig. 4C). This was further confirmed in a ChIP assay showing that Sp1, Sp3, and Sp4 bound to the proximal GC-rich region of the STAT3 promoter, and BITC treatment resulted in loss of Sp binding (Fig. 4D).
Previous studies with PEITC in pancreatic cancer cells showed that the mechanism of ROS-dependent down-regulation involved a cascade (Fig. 5A) that was initiated by ROS-induced epigenetic down-regulation of c-Myc, which is related to migration of chromatin-modifying complexes from non-GC-rich to GC-rich promoter sequences (30, 47, 48). Both PEITC and BITC decreased pol II and the H4K16Ac activation mark on the c-Myc promoter (Fig. 5D) (30) with some differences in histone methylation marks; the subsequent effects of BITC and PEITC on miR down-regulation and induction of ZBTB genes (Sp repressors) were comparable (Fig. 5D) (30). The importance of c-Myc as a critical upstream target of BITC-induced ROS was further confirmed by showing that after c-Myc knockdown, the growth inhibitory effects of BITC were significantly attenuated (Fig. 6, E and F). These results complement previous studies on Myc knockdown in pancreatic cancer cells, which also decreased miR-27a/miR-20a and expression of Sp1, Sp3, and Sp4 proteins (30). Drug-dependent activation of ROS and the effects on c-Myc, miRs, and ZBTB transcriptional repressors (Fig. 5A) have been observed for ROS-inducing histone deacetylase inhibitors in rhabdomyosarcoma cells (47), and the miR-ZBTB-Sp component of this pathway has also been characterized for several other ROS inducers (25–28, 30, 47). Our results suggest that the BITC-induced ROS pathway illustrated in Fig. 5A significantly contributes to the anticancer activity of BITC and other ROS inducers.
The development of STAT3 inhibitors, including specific oligonucleotides, is being extensively investigated (49, 50) because of the critical role of STAT3 in carcinogenesis (41, 44–46). This study demonstrates a unique approach for targeting STAT3 by ROS-inducing anticancer agents. Moreover, we also demonstrate a relationship between c-Myc and STAT3 where ROS-dependent repression of c-Myc triggers the cascade leading to down-regulation of Sp TFs and STAT3, an Sp-regulated gene. The potential clinical applications of this approach will be dependent on the effectiveness of the drug to induce ROS and the integrity of the pathway leading to Sp down-regulation. Sp1 is a negative prognostic factor for pancreatic cancer patient survival (51), and the clinical application of BITC and similar agents would be most effective in the sub-set of patients that highly express Sp1 and other Sp TFs.
Materials and Methods
Cell Lines and Reagents
The human pancreatic cancer cell lines, Panc1 and MiaPaCa2, were obtained from the American Type Culture Collection (Manassas, VA). L3.6pL cells were a generous gift from I. J. Fidler, University of Texas MD Anderson Cancer Center. These cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 1× antibiotic/antimycotic solution (Sigma). All cells were incubated in a humidified atmosphere composed of 5% CO2 at 37 °C. BITC (99%) and GSH (98% pure) were purchased from Sigma.
Measurement of ROS
ROS levels in cells were measured using cell-permeable probe CM-H2DCFDA as described in manufacturer's instructions (Life Technologies, Inc.). Briefly, cells were seeded at a density of 1.5 × 105 cells per well in 6-well plates in DMEM containing 2.5% charcoal-stripped FBS. The cells were pre-treated with GSH for 30 min and treated with vehicle (DMSO (control)), BITC alone, or a combination of BITC with GSH for 1 h. ROS levels were measured as described previously (25).
Cell Proliferation and Viability Assays
Cells were seeded at a density of 5 × 104 cells per well in 12-well plates with DMEM containing 2.5% charcoal-stripped FBS. After 24 h of plating, the cells were treated with either DMSO or different concentrations of BITC for 0, 24, and 48 h. Cells were then trypsinized and counted after 24 and 48 h using a Coulter Z1 cell counter. For cell viability assays, cells were plated in 96-well plates at a density of 3 × 103 per well. Cells were treated as described for cell proliferation assays for 48 h, and 20 μl MTT reagents (5 mg/ml) were added to each well and incubated for 3 h. After removal of the media, 200 μl of DMSO was added to each well, and absorbance was measured at 595 nm.
Measurement of Apoptosis
Cells at a density of 5 × 104 were plated in a 2-well Lab-TekTM II Chamber SlideTM (Thermo Scientific, Waltham, MA). Cells were pretreated with GSH for 3 h and treated with DMSO, BITC, or a combination of BITC with GSH for 24 h. Cells were then stained using the apoptosis assay kit according to the manufacturer's protocol (Biotium, Inc., Hayward, CA). Briefly, cells were stained with 5 μl of FITC/annexin V and Hoechst 33342 for 15 min. After three washes with 1× binding buffer, the annexin V staining was observed under a fluorescence microscope. Six randomly selected fields were selected to quantify the staining.
Cell Invasion Assay
Cells (1 × 105) with serum-free media were added to the upper surface of the pre-coated Matrigel insert, and the bottom chamber contained 10% FCS. Cells were subsequently treated with DMSO, BITC, or a combination of BITC with GSH for 24 h. The assay was carried out as described previously (52). After incubation for 24 h at 37 °C, the non-invading cells were removed from the upper surface of the insert. The cells invading to the lower surface of the insert were fixed with 4% paraformaldehyde for 10 min and stained with 0.5% crystal violet. The stained cells were counted in six randomly selected fields per insert. Cell invasion was determined using a Corning BioCoat Matrigel Invasion Chamber (Corning, Bedford, MA) according to the manufacturer's instructions.
Western Blotting Analysis
Cells were seeded at a density of 3 × 105 per well in 6-well plates and allowed to attach for 24 h. After various treatments, the whole cell lysates were subjected to Western blotting analysis. Total proteins were extracted by using RIPA lysis buffer containing 10 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 (w/v), 0.5% sodium deoxycholate, and 0.1% SDS with protease and phosphatase inhibitor mixture. The equal amounts of protein were separated in 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were incubated with primary antibodies overnight at 4 °C. The following primary antibodies were used: phospho-STAT3 (Ser-727), total STAT3, survivin, cleaved PARP, c-Met, cyclin D, and c-Myc were from Cell Signaling (Boston, MA); Sp1 and ZBTB34 were from Abcam (Cambridge, MA); and ZBTB10 was from Bethyl Laboratories Inc. (Montgomery, TX). All other antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The corresponding HRP-conjugated secondary IgG antibodies were used, and immuno-reacted proteins were detected with chemiluminescence reagent.
Chromatin Immunoprecipitation (ChIP) Assay
Panc1 cells (5 × 106) were treated with BITC for 3 h and subjected to ChIP analysis using the ChIP-IT Express magnetic chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. After treatment, cells were cross-linked with 1% formaldehyde, and then the reaction was stopped by addition of glycine. The cells were scraped with PBS and lysed with lysis buffer containing protease inhibitor mixture and PMSF. Nuclei were separated, and chromatin was sheared by sonication (10 pulses of 10 s at 25 A). The sonicated chromatins were immunoprecipitated with normal IgG (Santa Cruz Biotechnology), RNA pol II (GeneTex, Irving, CA), H3K27me3 (Abcam), H3K4me3 (Abcam), H4K16Ac (Active Motif), Sp1 (Abcam), Sp3 and Sp4 (Santa Cruz Biotechnology) with protein A-conjugated magnetic beads at 4 °C for 12 h. DNA was extracted from immunoprecipitates and subjected to PCR amplification with following primers: STAT3 promoter non-GC-rich, 5′-TATTACCCTCACTGGGTTCT-3′ (sense) and 5′-CTC CTC TAG GCT TCC TTC TAT-3′ (antisense); STAT3 promoter GC-rich, 5′-aca cgc act ggg acc tct g-3′ (sense) and 5′-ttg ttc cct cgg ctg cga c-3′ (antisense); and c-Myc promoter, 5′-GCC CTT TCC CCA GCC TTA GC-3′ (sense) and 5′-AAC CGC ATC CTT GTC CTG TGA GTA-3′ (antisense). PCR products were resolved on a 2% agarose gel in the presence of Green Glo DNA dye (Denville Scientific Inc., Holliston, MA).
Real Time Polymerase Chain Reaction (RT-PCR)
Real time PCR was used to measure expression of miR-17, miR-20a, and miR-27a and mRNA expression of ZBTB4, ZBTB10, and ZBTB34 following GSH and BITC treatment. Cells at a density of 4 × 105 were plated in a 60-mm dish, and then cells were allowed to attach for 24 h. Cells were pretreated with GSH for 3 h and treated with DMSO, BITC, or in combination of BITC with GSH for 3 h. Total RNA was extracted using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. TaqMan microRNA assays (Life Technologies, Inc.) were used to quantify the expression of miR-17, miR-20a, and miR-27a, and RNU6 was used as a control to determine relative miRNA expression. For ZBTB4, ZBTB10, and ZBTB34 mRNA expression, RNA was reverse-transcribed and amplified by using iTaq Universal SYBR Green One-Step kit (Bio-Rad). The following primers were used: TBP, 5′-TGC ACA GGA GCC AAG AGT GAA-3′ (sense) and 5′-CAC ATC ACA GCT CCC CAC CA-3′ (antisense); ZBTB4, 5′-ACC TGT GCA GGA ATT TCC AC-3′ (sense) and 5′-GAG CGG CCA AGT TAC TGA AG-3′ (antisense); ZBTB10, 5′-GCT GGA TAG TAG TTA TGT TGC-3′ (sense) and 5′-CTG AGT GGT TTG ATG GAC AGA-3′ (antisense); and ZBTB34, 5′-GCC AGC TTT CTT CAG ATG CAG TG-3′ (sense) and 5′-CTC TTC AGC ACC GAC GGT AAC A-3′ (antisense). TBP was used as a control to determine relative mRNA expression.
RNA Interference, Antagomir, and Overexpression Assays
Small interfering RNAs (siRNAs) for c-Myc were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). siRNAs for Sp1, Sp3, and Sp4 were purchased from Sigma. Antagomirs for miR-17, miR-20a, and miR-27a were obtained from Ambion (Austin, TX). Cells were seeded at a density of 6 × 104 per well in 6-well plates in DMEM with 2.5% charcoal-stripped FBS without antibiotic and left to attach for 24 h. The siRNAs, antagomirs, or pCMV-ZBTB4, ZBT10, and ZBTB34 overexpression vectors were transfected with Lipofectamine 2000 according to the manufacturer's instructions. After 72 h of transfection, cells were harvested and used for subsequent analysis. The Sp knockdown studies in Panc1, L3.6pL, and MiaPaCa2 cells and effects on Sp1, Sp3, and Sp4 expression have been reported (21), and the lysates were used for determining STAT3 and pSTAT3.
Xenograft Study
Female athymic nu/nu mice of 4–6 weeks old were purchased from Harlan Laboratories (Houston, TX). L3.6pL cells were harvested and suspended at a concentration of 1 × 106 cells in 100 μl of DMEM with ice-cold Matrigel (1:1 ratio) and injected subcutaneously into either side of the flank area of nude mice. Seven days after tumor cell inoculation, mice were randomized and treated every other day with either vehicle (corn oil) or BITC (20 mg/kg body weight) in a volume of 100 μl injected intraperitoneally. All mice were weighed once a week over the course of treatment to monitor changes in body weight. The xenografted tumors were relatively deep, and we were unable to determine accurate tumor volumes. All animals were sacrificed after 21 days of treatment, and tumor weights were determined. All animal studies were carried out according to the procedures approved by the Texas A&M University Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical significance of differences between the groups was determined by Student's t test. The results are presented with three independent experiments as mean with standard error at 95% confidence intervals. Results were considered statistically significant at a p value of less than 0.05.
Author Contributions
S. S. conceived and designed the research and wrote the paper. R. K., I. J., K. K., and E. H. performed the research, analyzed the data, and prepared the figures.
Supplementary Material
This work was supported by National Institutes of Health Grant P30-ES023512 (to S. S.), a College of Veterinary Science and Biomedical Sciences postdoctoral research grant (to R. K.), and a Texas AgriLife Research and Sid Kyle endowment. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1 and S2.
- ITC
- isothiocyanate
- BITC
- benzyl isothiocyanate
- ROS
- reactive oxygen species
- TF
- transcription factor
- PEITC
- phenethyl isothiocyanate
- EGFR
- EGF receptor
- CM-H2DCFDA
- 5-(and 6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester
- pol II
- polymerase II
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PARP
- poly(ADP-ribose) polymerase.
References
- 1. Higdon J. V., Delage B., Williams D. E., and Dashwood R. H. (2007) Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 55, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta P., Kim B., Kim S. H., and Srivastava S. K. (2014) Molecular targets of isothiocyanates in cancer: recent advances. Mol. Nutr. Food Res. 58, 1685–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen J. H., Kristal A. R., and Stanford J. L. (2000) Fruit and vegetable intakes and prostate cancer risk. J. Natl. Cancer Inst. 92, 61–68 [DOI] [PubMed] [Google Scholar]
- 4. Singh S. V., and Singh K. (2012) Cancer chemoprevention with dietary isothiocyanates mature for clinical translational research. Carcinogenesis 33, 1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan N., and Mukhtar H. (2015) Dietary agents for prevention and treatment of lung cancer. Cancer Lett. 359, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahu R. P., and Srivastava S. K. (2009) The role of stat-3 in the induction of apoptosis in pancreatic cancer cells by benzyl isothiocyanate. J. Natl. Cancer Inst. 101, 176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu C., Shen G., Chen C., Gélinas C., and Kong A. N. (2005) Suppression of NF-κB and NF-κB-regulated gene expression by sulforaphane and PEITC through IκBα, IKK pathway in human prostate cancer PC-3 cells. Oncogene 24, 4486–4495 [DOI] [PubMed] [Google Scholar]
- 8. Boreddy S. R., Sahu R. P., and Srivastava S. K. (2011) Benzyl isothiocyanate suppresses pancreatic tumor angiogenesis and invasion by inhibiting HIF-α/VEGF/RHO-GTPases: pivotal role of stat-3. PLoS ONE 6, e25799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J. H., Xu C., Keum Y. S., Reddy B., Conney A., and Kong A. N. (2006) Inhibition of EGFR signaling in human prostate cancer PC-3 cells by combination treatment with β-phenylethyl isothiocyanate and curcumin. Carcinogenesis 27, 475–482 [DOI] [PubMed] [Google Scholar]
- 10. Yang M. D., Lai K. C., Lai T. Y., Hsu S. C., Kuo C. L., Yu C. S., Lin M. L., Yang J. S., Kuo H. M., Wu S. H., and Chung J. G. (2010) Phenethyl isothiocyanate inhibits migration and invasion of human gastric cancer ags cells through suppressing mapk and NF-κB signal pathways. Anticancer Res. 30, 2135–2143 [PubMed] [Google Scholar]
- 11. Lai K. C., Huang A. C., Hsu S. C., Kuo C. L., Yang J. S., Wu S. H., and Chung J. G. (2010) Benzyl isothiocyanate (BITC) inhibits migration and invasion of human colon cancer HT29 cells by inhibiting matrix metalloproteinase-2/-9 and urokinase plasminogen (upa) through PKC and MAPK signaling pathway. J. Agric. Food Chem. 58, 2935–2942 [DOI] [PubMed] [Google Scholar]
- 12. Srivastava S. K., Xiao D., Lew K. L., Hershberger P., Kokkinakis D. M., Johnson C. S., Trump D. L., and Singh S. V. (2003) Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits growth of PC-3 human prostate cancer xenografts in vivo. Carcinogenesis 24, 1665–1670 [DOI] [PubMed] [Google Scholar]
- 13. Xiao D., Vogel V., and Singh S. V. (2006) Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by bax and bak. Mol. Cancer Ther. 5, 2931–2945 [DOI] [PubMed] [Google Scholar]
- 14. Kim S. H., Nagalingam A., Saxena N. K., Singh S. V., and Sharma D. (2011) Benzyl isothiocyanate inhibits oncogenic actions of leptin in human breast cancer cells by suppressing activation of signal transducer and activator of transcription 3. Carcinogenesis 32, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trachootham D., Zhou Y., Zhang H., Demizu Y., Chen Z., Pelicano H., Chiao P. J., Achanta G., Arlinghaus R. B., Liu J., and Huang P. (2006) Selective killing of oncogenically transformed cells through a ros-mediated mechanism by β-phenylethyl isothiocyanate. Cancer Cell 10, 241–252 [DOI] [PubMed] [Google Scholar]
- 16. Tang L., and Zhang Y. (2005) Mitochondria are the primary target in isothiocyanate-induced apoptosis in human bladder cancer cells. Mol. Cancer Ther. 4, 1250–1259 [DOI] [PubMed] [Google Scholar]
- 17. Nakamura Y., Ohigashi H., Masuda S., Murakami A., Morimitsu Y., Kawamoto Y., Osawa T., Imagawa M., and Uchida K. (2000) Redox regulation of glutathione S-transferase induction by benzyl isothiocyanate: correlation of enzyme induction with the formation of reactive oxygen intermediates. Cancer Res. 60, 219–225 [PubMed] [Google Scholar]
- 18. Edman P., and Henschen A. (1975) in Protein Sequence Determination (Needleman S. B., ed) pp. 232–279, Springer-Verlag, Berlin [Google Scholar]
- 19. Rydberg P., Lüning B., Wachtmeister C. A., Eriksson L., and Törnqvist M. (2002) Applicability of a modified Edman procedure for measurement of protein adducts: mechanisms of formation and degradation of phenylthiohydantoins. Chem. Res. Toxicol. 15, 570–581 [DOI] [PubMed] [Google Scholar]
- 20. Karlsson I., Samuelsson K., Ponting D. J., Törnqvist M., Ilag L. L., and Nilsson U. (2016) Peptide reactivity of isothiocyanates–implications for skin allergy. Sci. Rep. 6, 21203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hedrick E., Cheng Y., Jin U. H., Kim K., and Safe S. (2016) Specificity protein (sp) transcription factors sp1, sp3 and sp4 are non-oncogene addiction genes in cancer cells. Oncotarget 7, 22245–22256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jutooru I., Chadalapaka G., Sreevalsan S., Lei P., Barhoumi R., Burghardt R., and Safe S. (2010) Arsenic trioxide downregulates specificity protein (sp) transcription factors and inhibits bladder cancer cell and tumor growth. Exp. Cell Res. 316, 2174–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jutooru I., Chadalapaka G., Lei P., and Safe S. (2010) Inhibition of NFκB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J. Biol. Chem. 285, 25332–25344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pathi S. S., Lei P., Sreevalsan S., Chadalapaka G., Jutooru I., and Safe S. (2011) Pharmacologic doses of ascorbic acid repress specificity protein (sp) transcription factors and sp-regulated genes in colon cancer cells. Nutr. Cancer 63, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chintharlapalli S., Papineni S., Lei P., Pathi S., and Safe S. (2011) Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (sp) transcription factors. BMC Cancer 11, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pathi S. S., Jutooru I., Chadalapaka G., Sreevalsan S., Anand S., Thatcher G. R., and Safe S. (2011) Gt-094, a no-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microrna-27a: Zbtb10-specificity protein pathway. Mol. Cancer Res. 9, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jutooru I., Chadalapaka G., Abdelrahim M., Basha M. R., Samudio I., Konopleva M., Andreeff M., and Safe S. (2010) Methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate decreases specificity protein transcription factors and inhibits pancreatic tumor growth: role of microrna-27a. Mol. Pharmacol. 78, 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chadalapaka G., Jutooru I., and Safe S. (2012) Celastrol decreases specificity proteins (sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis 33, 886–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chadalapaka G., Jutooru I., Burghardt R., and Safe S. (2010) Drugs that target specificity proteins downregulate epidermal growth factor receptor in bladder cancer cells. Mol. Cancer Res. 8, 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jutooru I., Guthrie A. S., Chadalapaka G., Pathi S., Kim K., Burghardt R., Jin U. H., and Safe S. (2014) Mechanism of action of phenethylisothiocyanate and other reactive oxygen species-inducing anticancer agents. Mol. Cell. Biol. 34, 2382–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sahu R. P., Zhang R., Batra S., Shi Y., and Srivastava S. K. (2009) Benzyl isothiocyanate-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of MAPK in human pancreatic cancer cells. Carcinogenesis 30, 1744–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu X., Zhu Y., Yan H., Liu B., Li Y., Zhou Q., and Xu K. (2010) Isothiocyanates induce oxidative stress and suppress the metastasis potential of human non-small cell lung cancer cells. BMC Cancer 10, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C. L., Huang A. C., Yang J. S., Liao C. L., Lu H. F., Chou S. T., Ma C. Y., Hsia T. C., Ko Y. C., and Chung J. G. (2011) Benzyl isothiocyanate (BITC) and phenethyl isothiocyanate (PEITC)-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of caspase-3, mitochondria dysfunction and nitric oxide (NO) in human osteogenic sarcoma u-2 os cells. J. Orthop. Res. 29, 1199–1209 [DOI] [PubMed] [Google Scholar]
- 34. Huang C., Huang R., Chang W., Jiang T., Huang K., Cao J., Sun X., and Qiu Z. (2012) The expression and clinical significance of pstat3, vegf and vegf-c in pancreatic adenocarcinoma. Neoplasma 59, 52–61 [DOI] [PubMed] [Google Scholar]
- 35. Zhao G., Zhang J. G., Shi Y., Qin Q., Liu Y., Wang B., Tian K., Deng S. C., Li X., Zhu S., Gong Q., Niu Y., and Wang C. Y. (2013) Mir-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting stat3. PLoS ONE 8, e73803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frank D. A. (2013) Transcription factor stat3 as a prognostic marker and therapeutic target in cancer. J. Clin. Oncol. 31, 4560–4561 [DOI] [PubMed] [Google Scholar]
- 37. Xiong H., Du W., Wang J. L., Wang Y. C., Tang J. T., Hong J., and Fang J. Y. (2012) Constitutive activation of stat3 is predictive of poor prognosis in human gastric cancer. J. Mol. Med. 90, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 38. Alvarez J. V., Mukherjee N., Chakravarti A., Robe P., Zhai G., Chakladar A., Loeffler J., Black P., and Frank D. A. (2007) A stat3 gene expression signature in gliomas is associated with a poor prognosis. Transl. Oncogenomics 2, 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M. X., Bi X. Y., Huang Z., Zhao J. J., Han Y., Li Z. Y., Zhang Y. F., Li Y., Chen X., Hu X. H., Zhao H., and Cai J. Q. (2015) Prognostic role of phospho-stat3 in patients with cancers of the digestive system: a systematic review and meta-analysis. PLoS ONE 10, e0127356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee J., Kang W. K., Park J. O., Park S. H., Park Y. S., Lim H. Y., Kim J., Kong J., Choi M. G., Sohn T. S., Noh J. H., Bae J. M., Kim S., Lim D. H., Kim K. M., and Park C. K. (2009) Expression of activated signal transducer and activator of transcription 3 predicts poor clinical outcome in gastric adenocarcinoma. APMIS 117, 598–606 [DOI] [PubMed] [Google Scholar]
- 41. Frank D. A. (2007) Stat3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 251, 199–210 [DOI] [PubMed] [Google Scholar]
- 42. Kato K., Nomoto M., Izumi H., Ise T., Nakano S., Niho Y., and Kohno K. (2000) Structure and functional analysis of the human stat3 gene promoter: alteration of chromatin structure as a possible mechanism for the upregulation in cisplatin-resistant cells. Biochim. Biophys. Acta 1493, 91–100 [DOI] [PubMed] [Google Scholar]
- 43. Ihle J. N. (2001) The stat family in cytokine signaling. Curr. Opin. Cell Biol. 13, 211–217 [DOI] [PubMed] [Google Scholar]
- 44. Yu H., Lee H., Herrmann A., Buettner R., and Jove R. (2014) Revisiting stat3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 [DOI] [PubMed] [Google Scholar]
- 45. He G., and Karin M. (2011) Nf-κb and stat3–key players in liver inflammation and cancer. Cell Res. 21, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu H., Pardoll D., and Jove R. (2009) Stats in cancer inflammation and immunity: a leading role for stat3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hedrick E., Crose L., Linardic C. M., and Safe S. (2015) Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Mol. Cancer Ther. 14, 2143–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Hagan H. M., Wang W., Sen S., Destefano Shields C., Lee S. S., Zhang Y. W., Clements E. G., Cai Y., Van Neste L., Easwaran H., Casero R. A., Sears C. L., and Baylin S. B. (2011) Oxidative damage targets complexes containing DNA methyltransferases, sirt1, and polycomb members to promoter cpg islands. Cancer Cell 20, 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hong D., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Zhou T., Schmidt J., Jo M., Lee S. J., Yamashita M., Hughes S. G., Fayad L., Piha-Paul S., et al. (2015) Azd9150, a next-generation antisense oligonucleotide inhibitor of stat3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 7, 314ra185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong A. L., Soo R. A., Tan D. S., Lee S. C., Lim J. S., Marban P. C., Kong L. R., Lee Y. J., Wang L. Z., Thuya W. L., Soong R., Yee M. Q., Chin T. M., Cordero M. T., Asuncion B. R., Pang B., et al. (2015) Phase I and biomarker study of opb-51602, a novel signal transducer and activator of transcription (stat) 3 inhibitor, in patients with refractory solid malignancies. Ann. Oncol. 26, 998–1005 [DOI] [PubMed] [Google Scholar]
- 51. Jiang N. Y., Woda B. A., Banner B. F., Whalen G. F., Dresser K. A., and Lu D. (2008) Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 17, 1648–1652 [DOI] [PubMed] [Google Scholar]
- 52. Kim K., Chadalapaka G., Lee S. O., Yamada D., Sastre-Garau X., Defossez P. A., Park Y. Y., Lee J. S., and Safe S. (2012) Identification of oncogenic microrna-17–92/zbtb4/specificity protein axis in breast cancer. Oncogene 31, 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.