Abstract
The thalassospiramide lipopeptides have great potential for therapeutic applications; however, their structural and functional diversity and biosynthesis are poorly understood. Here, by cultivating 130 Rhodospirillaceae strains sampled from oceans worldwide, we discovered 21 new thalassospiramide analogues and demonstrated their neuroprotective effects. To investigate the diversity of biosynthetic gene cluster (BGC) architectures, we sequenced the draft genomes of 28 Rhodospirillaceae strains. Our family-wide genomic analysis revealed three types of dysfunctional BGCs and four functional BGCs whose architectures correspond to four production patterns. This correlation allowed us to reassess the “diversity-oriented biosynthesis” proposed for the microbial production of thalassospiramides, which involves iteration of several key modules. Preliminary evolutionary investigation suggested that the functional BGCs could have arisen through module/domain loss, whereas the dysfunctional BGCs arose through horizontal gene transfer. Further comparative genomics indicated that thalassospiramide production is likely to be attendant on particular genes/pathways for amino acid metabolism, signaling transduction, and compound efflux. Our findings provide a systematic understanding of thalassospiramide production and new insights into the underlying mechanism.
Keywords: bacteria, biosynthesis, genomics, natural product, neuroprotection
Introduction
Microbial natural products are an excellent source of bioactive molecules (1). Structurally complex and diverse, these compounds have applications in many fields, including medicine, agriculture, cosmetics, and food-production/preservation (2). The thalassospiramides are a family of lipopeptides produced by marine α-proteobacteria belonging to the Rhodospirillaceae family. Fenical and co-workers (3) first characterized thalassospiramides A and B in 2007 from Thalassospira sp. CNJ-328, and then, in 2013, we reported an additional 14 thalassospiramides from Thalassospira and Tistrella strains. We found that many of these compounds display a high inhibitive potency (nanomolar levels) of human calpain protease (4), a calcium-dependent enzyme family implicated in neurological disorders (5), muscular dystrophies, cortical cataracts (6), and cancer (7). For example, inflammatory cytokines that lead to neuronal network disruption, increase intracellular calcium, and increase calpain protease activity are a possible trigger for Alzheimer's disease (8). The potent activity of thalassospiramides against calpain protease may allow them to act as neuroprotective agents and result in their therapeutic application for Alzheimer's therapy.
The thalassospiramides are hybrid non-ribosomal peptide-polyketide molecules. Unlike ribosomal peptides that are encoded directly in the genome and translated by the ribosome (9), NRPSs4 are custom-produced by a dedicated megasynthetase that acts as both the catalyst and template, incorporating a single amino acid at each enzyme module in a predictable linear fashion. In 2013, we proposed that the thalassospiramide molecules are made by two related hybrid polyketide synthase-non ribosomal peptide synthetase/NRPS pathways in Thalassospira and Tistrella strains via a highly non-canonical functioning of the megasynthetase using multimodular skipping and iteration, internal supplementation, and nonlinear substrate channeling (4). Thus, the thalassospiramides are produced by “diversity-oriented biosynthesis,” whereby the megasynthase generates a library of molecules rather than a single refined structure (10). In light of the promising bioactivity and unusual proposed biosynthesis of thalassospiramides, further investigation of the gene clusters and molecular family are valuable.
Remarkable progress in sequencing technology has advanced our understanding of particular physiological or metabolic functions in bacteria, including secondary metabolite production (11, 12). For example, gene sequence homology predicts that sioxanthin biosynthesis is present in all of the Salinispora as well as in other members of the family Micromonosporaceae (11), and the clustering of carotenoid biosynthetic genes in heterotrophic bacteria reveals that gene rearrangement is common in the production of these compounds (11). For this reason, natural product researchers have put more effort into phylogenetic analysis in the past decade. The comparison between species trees (based on ribosomal markers) and gene trees (based on biosynthetic gene sequences) with chemical character mapping is a way of identifying key functional genes and of recognizing gene loss, acquisition, and horizontal gene transfer (HGT) (13).
In this study, by integrating biochemical techniques, molecular assays, and genomic sequencing, we performed a comprehensive investigation of thalassospiramide production in the Rhodospirillaceae family. We discovered a large number of new thalassospiramides and evaluated their calpain protease-inhibitive and -neuroprotective effects. Subsequent genomic sequencing allowed us to reassess the diversity-oriented biosynthesis of thalassospiramide molecules. Further, evolutionary analysis and comparative genomics revealed implications for the biosynthetic mechanisms and future discovery of related compounds with potential clinical use.
Results
New Thalassospiramides and Their Activities
To study the breadth of thalassospiramide production, 76 Thalassospira, 29 Tistrella, 16 Oceanibaculum, two Fodinicurvata, and seven unclassified Rhodospirillaceae strains from marine environments worldwide were obtained, leading to the isolation of 21 new thalassospiramides. Fig. 1 shows the sampling locations of Thalassospira and Tistrella. Fig. 2A shows the chemical structures of 37 previously reported and newly discovered thalassospiramides. The thalassospiramides are divided into two structural groups, “A-like” and “B-like,” distinguished by the identity of the N-terminal amino acid residue. A-like molecules contain a standard amino acid residue, whereas B-like molecules have a statine-like amino acid. The thalassospiramides are further subdivided into subgroups A through J. The majority of new thalassospiramides belong to the previously described subclasses A-F, which differ in the length of the peptide chain; however, seven molecules were placed in the new subgroups H, I, and J. Thalassospiramides H, H1, H2, and H3 are truncated variants much like the C subclass and feature repeating serine or valine motifs. Thalassospiramides I and J are related to the A subgroup and possess an additional serine or valine residue, respectively. As the thalassospiramides share a 12-membered ring structure and members are amenable to “b-y” mode sequencing by tandem mass spectrometry (MS2) analysis (4), screening for new analogues and chemical structure determination of all thalassospiramides were first accomplished using high-resolution MS and MS2. Comparison between the m/z of observed MS2 fragments with diagnostic thalassospiramide fragments, such as m/z 390.19 ([M+H]+ ring), 489.26 ([M+H]+ ring+Val) or ring-modified signals m/z 414.20 ([M+H]+ ring+CH3), 503.26 ([M+H]+ ring+CH3+Val) allowed identification of all building blocks and their sequence (complete MS2 data are provided in supplemental Data S1). Additionally, we characterized at least one representative member of each new subclass thoroughly using multidimensional nuclear magnetic resonance (NMR) (supplemental Table S1).
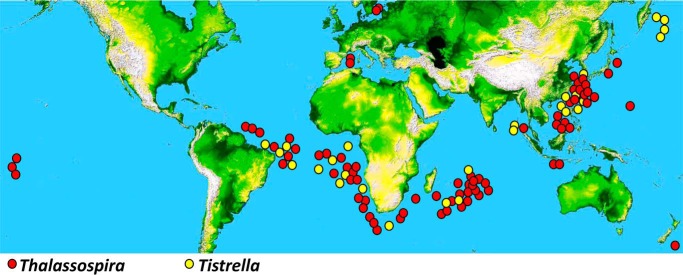
FIGURE 1.
Sampling locations of the Thalassospira and Tistrella strains. In total, 76 Thalassospira strains and 27 Tistrella strains were collected.
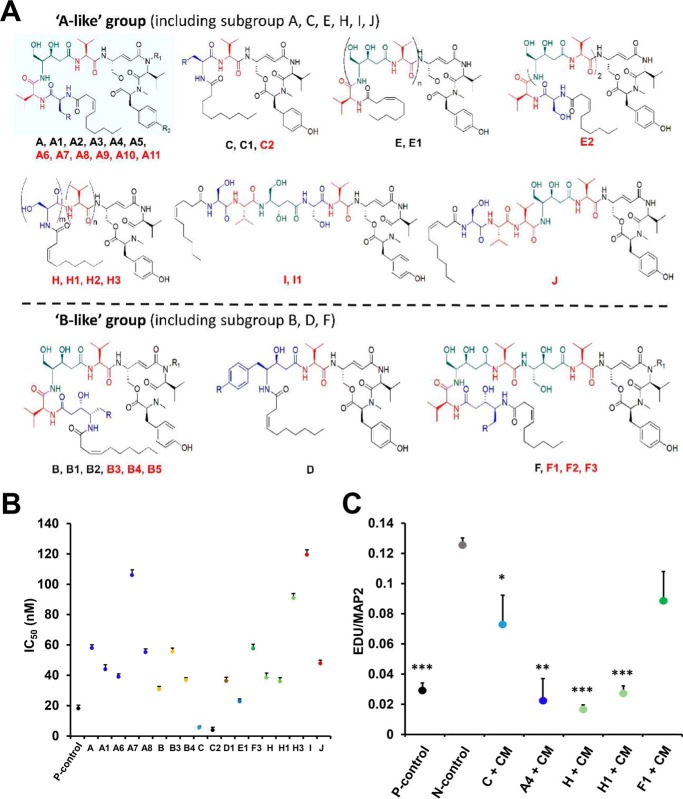
FIGURE 2.
Chemical structures and activities of thalassospiramides. A, the 21 newly discovered thalassospiramides are indicated by red text. Blue structural portions indicate N-terminal amino acids used to distinguish the A- and B-like molecules. Green portions highlight a 4-amino-3,5-dihydroxy-pentanoic acid motif, red portions indicate a common valine residue, and black portions are conserved throughout the family. Absolute configurations of these cyclopeptides are drawn on the basis of the previous report of 16 known thalassospiramides (4). Thalassospiramides A2, A3, C1, F1, H1, and I1 have saturated fatty acids at the N terminus, and A10 and A11 have fatty acids of different lengths (placement of double bond within the side chain was not determined in A11). Structures: A, r = OH, R1 = H, R2 = OH; A1, r = hydroxyPh, R1 = H, R2 = OH; A2, r = OH, R1 = H, R2 = OH, saturated; A3, r = hydroxyPh, R1 = H, R2 = OH, saturated; A4, r = iPr, R1 = H, R2 = OH; A5, r = Ph, R1 = H, R2 = OH; A6, r = hydroxyPh, R1 = CH3, R2 = OH; A7, r = Ph, R1 = CH3, R2 = OH; A8, r = OH, R1 = CH3, R2 = OH; A9, r = OH, R1 = H, R2 = OH, FA = CO(CH2)6CH3; A10, r = OH, R1 = H, R2 = OH, FA = CO(CH2)6CH3; A11, r = OH, R1 = H, R2 = OH, FA (fatty acids) = CO(CH2)6(CH = CH)CH3. C, r = hydroxyPh; C1, r = hydroxyPh, saturated; C2, r = OH. E, n = 1; E1, n = 2. H, m = 2, n = 1; H1, m = 2, n = 1, saturated; H2, m = 1, n = 1; H3, m = 1, n = 2. B, r = Ph, R1 = H; B1, r = hydroxyPh, R1 = H; B2, r = OH, R1 = CH3; B4, r = hydroxyPh, R1 = CH3; B5, r = OH, R1 = CH3. D, r = H; D1, r = OH. F, r = Ph, R1 = H; F1, r = hydroxyPh, R1 = H, saturated; F2, r = hydroxyPh, R1 = CH3; F3, r = Ph, R1 = CH3. B, in the calpain inhibitory activity assay, Z-Leu-Leu-Tyr-fluoromethylketone was used as a positive control (P-control), whereas samples without compound were defined as a negative control (N-control). All tested compounds showed significant differences from the negative control. C, the neuroprotective assay was performed using mouse primary neurons. EdU/MAP2 indicates the relative amount of DNA residing in newly replicated neuron cells and acts as a proxy for numbers of neuronal cells re-entering the cell cycle. Lower EdU/MAP2 ratios suggest neuroprotection, potentially through calpain protease inhibition. P-control, no thalassospiramide or conditioned medium added; N-control, no thalassospiramide added. All significance values are based on comparison with values shown for the negative control. Experiments were performed in triplicate. Statistics were performed using Student's t tests (*, p < 0.05; **, p < 0.01; ***, p < 0.005).
To discover new calpain inhibitors, 12 of the thalassospiramides identified in this paper were subjected to a calpain inhibition assay, exhibiting a range of potency spanning several orders of magnitude (Fig. 2B). Thalassospiramide C2, the smallest molecule, displayed the best inhibitory activity (IC50 = 1.6 nm) of the entire family. The result also hints that the molecular size of these molecules may play a role in the calpain protease inhibitory action. In light of their potency as calpain inhibitors, we examined whether thalassospiramides were also neuroprotective agents of potential use in the treatment of conditions such as Alzheimer's disease. Thalassospiramides A4, A6, A8, C, D1, H, H1, and F1 with a range of calpain inhibitory activities (A4 IC50 = 12.5 nm, A6 IC50 = 37.4 nm, A8 IC50 = 53.8 nm, C IC50 = 3.4 nm, D1 IC50 = 34.7 nm, H (H1) IC50 = 36.8 nm, and F1 IC50 = 41.3 nm) were chosen for testing via a neuroprotective assay (4, 23). Cultured primary mouse cortical neurons were exposed to inflammatory stress by adding conditioned medium from β-amyloid-stimulated THP1 cells, which are a human monocytic cell line. In the negative control, adding the conditioned medium increased neuronal cell cycling by 5-fold, indicating a neurotoxic response. Adding thalassospiramides A4, H, or H1 significantly reduced this neurotoxic response (Fig. 2C), indicating that thalassospiramides may have potent neuroprotective activities. Interestingly we found that there is not always a direct correlation between thalassospiramide behavior in the two assays; for example, thalassospiramide C possesses potent calpain inhibitory activity but only weak neuroprotective activity, thalassospiramides A6, A8, and D1 display moderate calpain inhibitory activity but show no neuroprotective activity, and the moderate calpain inhibitors H and H1 exhibit good neuroprotective activity. Clearly further studies are needed to disclose the full mechanism of the neuroprotective effects.
Novel Thalassospiramide Production Patterns and Biosynthetic Gene Cluster (BGC) Architectures
We originally reported two thalassospiramide production patterns that were each directly correlated with a gene cluster variant (4). Thalassospira sp. TrichSKD10 and Thalassospira sp. CNJ-328 produce both A-like and B-like thalassospiramides (pattern A, B-like) and contain a complete version of the gene cluster, whereas Tistrella mobilis KA081020–065 and Tistrella bauzanensis TIO7329 contain a contracted version of the gene cluster and produce only A-like thalassospiramides (pattern A-like) (4). This study has a far larger sample size, providing a comprehensive picture of the production patterns across the Rhodospirillaceae family and hinting at more variants of the gene cluster. Intriguingly, two Thalassospira mesophila strains produced only thalassospiramide subclasses A and C and several newly identified A-like thalassospiramides (H, I, and J subclasses); this new pattern is denoted “Anew-like” (supplemental Table S1). Of the 29 Tistrella strains (26 T. mobilis and three T. bauzanensis), only 11 produced thalassospiramides, and all adhered to the A-like pattern (supplemental Table S2). Another new pattern of thalassospiramide production, consisting of both A-like and trace amounts of B-like molecules, denoted as “A, weak B-like,” was detected from Oceanibaculum pacificum 1A02656, the sole producing strain of the 16 Oceanibaculum strains. Seven representative Thalassospira and Tistrella strains were grown under varied culture conditions (supplemental Table S3), and the production patterns for each strain remained consistent (supplemental Table S4).
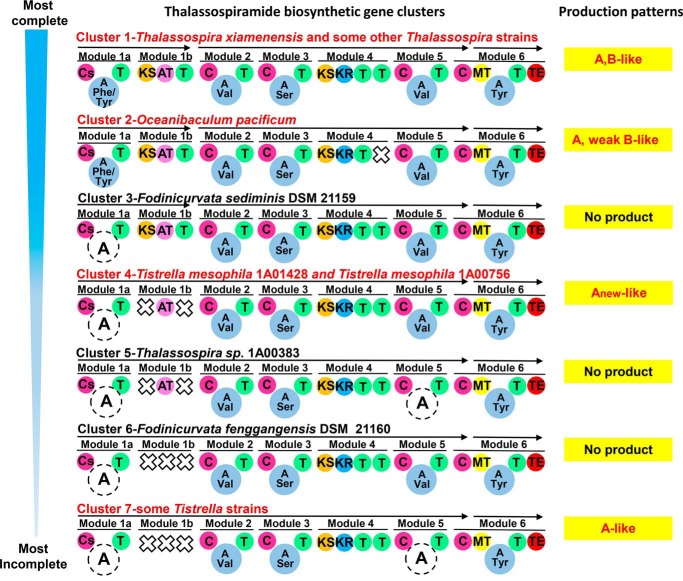
To investigate additional BGC architectures, we performed genome sequencing and bioinformatics to uncover all the clusters. The genomes of 28 Rhodospirillaceae strains were sequenced using Illumina next-generation technology and uploaded to the National Center for Biotechnology Information (NCBI) database (see supplemental Table S5 for further information and accession numbers). These included 11 strains with the production pattern A, B-like, one strain with the pattern A-like, two strains with the pattern Anew-like, one strain with the pattern A, weak B-like, as well as 13 randomly selected strains with no thalassospiramide production. All sequenced genomes had >98% completeness. In addition, 27 publically available Rhodospirillaceae genomes were obtained from the NCBI database. All 55 genome sequences were analyzed using bioinformatics software. Every strain that produced thalassospiramide molecules had some version of the putative BGC, and a total of seven different architectures for the biosynthetic pathway were identified (Fig. 3). The BGC comprises six modules, although A domains in modules 1a and 5 were found to be truncated in certain strains. The truncated A domain in module 1a is shown in more detail in supplemental Fig. S1. The presence of gene clusters 1, 2, 4, and 7 in particular strains, along with their individual pathway architectures, corresponded to the observed compound production patterns A, B-like; A, weak B-like; Anew-like; and A-like; respectively.
FIGURE 3.
Seven putative thalassospiramide BGCs in the Rhodospirillaceae family. Bacterial strains and molecule production patterns are specified after the cluster name. Truncated domains (A domains in modules 1a and 5; for a detailed description of these truncated domains see supplemental Fig. S1) are denoted by a dotted circle, and missing domains are denoted by a cross. Cs, starter condensation; C, condensation; A, adenylation; T, thiolation; KS, ketosynthase; AT, acyltransferase; KR, ketoreductase; MT, methyltransferase; TE, thioesterase.
Evolution of Thalassospiramides BGCs
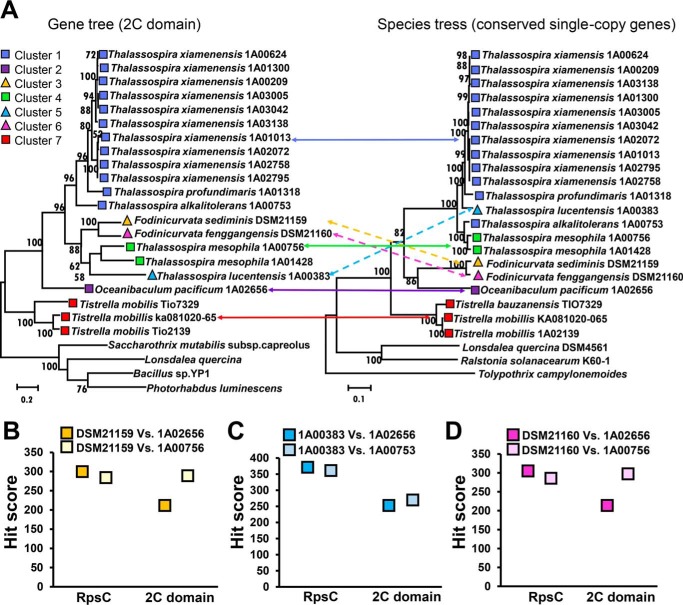
To investigate the relationships between the seven BGCs further, we undertook several phylogenetic comparisons. First, separate functional gene trees were built using maximum likelihood investigating domains that were present in all versions of the pathway and that would be functional based on our hypothesis for the biosynthetic steps. All trees displayed similar architectures, suggesting that the thalassospiramide BGCs are a cohesive unit and not arising from different sources. As the C domains in modules 2 and 6 and the A domain in module 6 tend to be more conserved (indicated by protein alignment), the trees for these three domains are shown in Fig. 4A and supplemental Figs. S2 and S3. Second, we built a species tree of sequenced strains using a concatenation of 28 essential genes (Fig. 4A). The species and gene trees were directly compared, and, somewhat surprisingly, in all cases the gene tree was highly similar to the corresponding species tree with respect to the four functional clusters (1, 2, 4, and 7). By contrast, the relative position of the dysfunctional clusters (3, 5, and 6) in the species tree and the gene tree were not consistent. We propose that clusters 3 and 6 are introduced into Fodinicurvata by HGT, as clusters 3 and 6 are closely related to cluster 1, despite the strain itself being more distantly related to Thalassospira xiamenensis on the basis of essential genes; a similar situation was observed for cluster 2. Protein sequences of the ribosomal protein S3 (RpsC) and the 2C domain from the strains Fodinicurvata sediminis DSM21159 (cluster 3), Thalassospira lucentensis 1A00383 (cluster 5), and Fodinicurvata fenggangensis DSM21160 (cluster 6) were searched against homologues in other strains using the Basic Local Alignment Search Tool (BLAST) to confirm their relative phylogenetic distances. For example, based on the hit scores of RpsC protein pairs, DSM21159 was closer to O. pacificum 1A02656 than to T. mesophila 1A00756; by contrast, hit scores of the 2C domain sequences indicated a higher similarity between DSM21159 and 1A00756 than between DSM21159 and 1A02656 (Fig. 4, B–D).
FIGURE 4.
Phylogenetic analysis of Rhodospirillaceae strains. A, trees based on functional genes (2C domains, left) and 28 concatenated conserved single-copy genes (right) are compared. Protein BLAST analysis using thalassospiramide A and C domains identified the NRPS proteins in the bacteria Tolypothrix, Lonsdalea, and Ralstonia; therefore, genes encoding these proteins are serving as the out groups. Bootstrap values based on 1000 replicates are shown at the nodes. Squares with different colors distinguish bacterial strains with functional gene clusters for thalassospiramide biosynthesis, whereas triangles with different colors distinguish strains with dysfunctional gene clusters. Solid lines indicate consistency between the gene tree and the species tree, whereas dashed lines indicate inconsistency between the two trees, likely because of HGT events. B–D, protein sequences of the ribosomal protein S3 (RpsC) and the 2C domain from the strains DSM21159, 1A00383, and DSM21160 were compared by protein BLAST to sequences in other strains to confirm their relative phylogenetic distances.
Genome-wide Functions Associated with Thalassospiramide Production
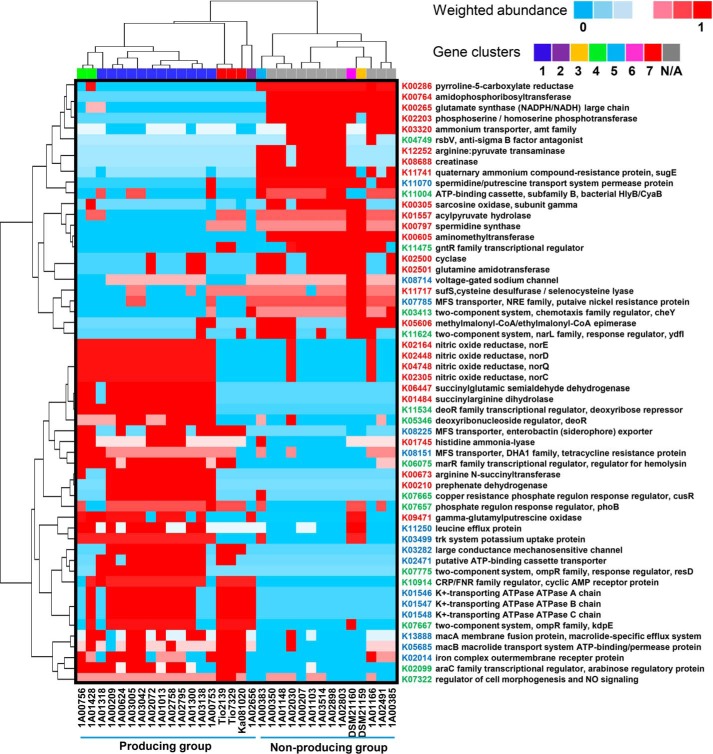
Linking HGT to functional loss of BGCs suggested that particular metabolic functions beyond the BGCs may influence thalassospiramide production. Comparative genomics were performed between 18 genomes of thalassospiramide-producing bacteria (Thalassospira, Tistrella, and Oceanibaculum) and 14 genomes of non-producing bacteria (Thalassospira and Tistrella), which in total revealed 108 significantly changed Kyoto Encyclopedia of Genes and Genomes (KEGG) genes (Student's t test, p < 0.001; supplemental Data S2). A large proportion of these genes were classified under the categories of amino acid metabolism (24 genes), signal transduction (18 genes), and compound transport (14 genes) (Fig. 5). Remarkably, based on genes within these three categories, the thalassospiramide-producing and -non-producing bacteria formed two distinct groups. Two genes for glutamate biosynthesis via arginine, succinylarginine dihydrolase (astB and K01484), and succinyl glutamic semialdehyde dehydrogenase (astE, K06447), were present in nearly all genomes of thalassospiramide-producing bacteria but absent in the genomes of all non-producing bacteria. Similarly, a gene involved in l-tyrosine production, prephenate dehydrogenase (K00210), was only found in thalassospiramide-producing bacteria.
FIGURE 5.
Genes differentiating the thalassospiramide-producing and -non-producing bacteria. Genes were identified based on comparison between 18 genomes of thalassospiramide-producing bacteria and 14 genomes of non-producing bacteria using Student's t test (p < 0.001). A large proportion of these KEGG-annotated genes belong to the categories of amino acid metabolism (KEGG number in red), signal transduction (KEGG number in green), and compound transport (KEGG number in blue). All of the 108 significantly changed genes are listed in supplemental Table S2.
In addition to amino acid metabolic pathways, the producing and non-producing bacteria could also be differentiated by a number of genes for signal transduction. For example the pho family regulator (K07657) was highly enriched in the genomes of thalassospiramide-producing bacteria (Fig. 5). The thalassospiramide-producing bacteria were also equipped with a number of specific transporters for compound transport, such as the mac family transporters (K13888 and K05685), which involve efflux of macrolide antibiotics (14), and the major facilitator superfamily (MFS) proteins (K08225 and K08151), which are involved in symport, antiport, and uniport of various substrates, including antibiotics (15). Pathway reconstruction confirmed the presence of specific amino acid metabolism functions for the thalassospiramide-producing bacteria, which could be exemplified by comparison of two closely related strains, 1A00753 and 1A00383 (supplemental Fig. S4). Moreover, the alignment of genomes of these two strains suggested that particular genes involved in amino acid metabolism, carbohydrate transport, and signal transduction are lost from strain 1A00383 (supplemental Fig. S5).
Because the abovementioned genes (Fig. 5) show significant differences between the thalassospiramide producing and non-producing groups, we conjectured that these genes co-occur with the BGCs across all bacterial genomes. Co-occurrence analysis of all genes in the 32 genomes using a Spearman's correlation coefficient of >0.8 and statistically significant p value of <0.01 revealed a strong correlation between thalassospiramides BGCs and 16 genes, including succinyl arginine dihydrolase (K01484), succinyl glutamic semialdehyde dehydrogenase (K06447), arginine N-succinyl transferase (K00673), deoR family transcriptional regulator, deoxyribose operon repressor (K11534), K+-transporting ATPase A chain (K01546), K+-transporting ATPase B chain (K01547), K+-transporting ATPase C chain (K01548), monosaccharide-transporting ATPase (K10820), and large conductance mechanosensitive channel (K03282) (supplemental Fig. S6). Notably, all of these genes are absent in DSM21159, 1A00383, and DSM21160, the strains with clusters 3, 5, and 6, respectively.
Discussion
This study describes a remarkable diversity-oriented biosynthetic system for the production of 37 thalassospiramides within the Rhodospirillaceae family. We discovered new thalassospiramides and demonstrated that the thalassospiramides have neuroprotective activities (Fig. 2) and are worth exploring in the context of human Alzheimer's disease and other neurodegenerative diseases. The identification of several extremely promising calpain protease inhibitors and neuroprotective agents from the thalassospiramides, where small structural changes have large effects on molecule potency, emphasizes the importance of the complex diversity-oriented biosynthesis.
Our family-wide genome sequencing effort revealed seven BGCs wherein clusters 2, 3, 4, 5, and 6 are newly identified in this report. The pathways differ at biosynthetic modules 1a, 1b, 4, and 5, with newly identified clusters lacking certain functional domains in comparison with cluster 1 (Fig. 3). On a cursory analysis of the pathways, clusters 2–6 appear to lie along a continuum of “completeness” between cluster 1 and 7. Regarding the functional BGCs 1, 2, 4, and 7, the more complete the cluster is, the more diverse the molecules it produces, motivating us to reassess the biosynthetic mechanism based on the compound structures, production patterns, and gene cluster architectures.
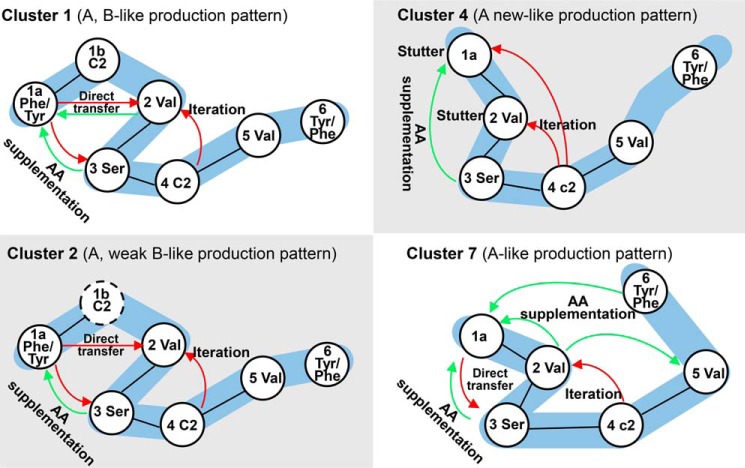
Several of the new thalassospiramides, including C2 and F1, are simply additional examples of previously described biosynthetic mechanisms involving in trans amino acid supplementation and adenylation domain promiscuity, respectively. However, the majority of the new molecules result from new instances of non-canonical biosynthesis, thereby expanding our understanding of this biosynthetic system. The first example is that of the new thalassospiramides A6, A7, A8, B3, B4, B5, F2, and F3. They are made by cluster 1-containing Thalassospira strains and have structures very similar to known thalassospiramides, with the addition of a methyl group on the valine nitrogen of the core cyclic peptide. However, based upon protein BLAST analysis, there is no methyltransferase domain in module 5, indicating that this methyl group might result from the in trans activity of an external transferase. Second, clusters 1, 2, and 4 produce thalassospiramide A9, in which the presence of a phenylalanine residue in the core cyclic portion of the peptide suggests that the A domain in module 6 can activate both Tyr and Phe in a similar manner to module 1a. Third, the new A-like thalassospiramide groups H, I, and J are synthesized by cluster 4. A single module stuttering is proposed for the production of thalassospiramides H, H1, H2, H3, and J, where either module 1a or module 2 is used in a repeated fashion. Finally and most intriguingly, the thalassospiramides I and I1 can be produced by the iterative use of modules 1 through 4. This observation is particularly interesting as cluster 4 also produces thalassospiramide A through iterative use of modules 2–4. A summary of the putative biosynthetic mechanisms of gene clusters 1, 2, 4, and 7, including module skipping and stuttering, multimodular iteration, in trans amino acid activation, and substrate channeling, is represented in Fig. 6. The array of unusual mechanisms used by this family of homologous pathways to produce molecular diversity is without parallel.
FIGURE 6.
Representation of the proposed biosynthetic pathways of gene clusters 1, 2, 4, and 7. The white ribbons represent the linear sequences of the gene products, black solid lines identify the modules found within the same protein, the black dashed line suggests that the module has no function, red arrows show the movement of the growing peptide chain along the megasynthase, and green arrows show the movement of amino acids from supplementing the A domain to the T domain.
The proposed biosynthetic mechanism cannot explain why the dysfunctional clusters 3, 5, and 6 do not produce thalassospiramides while they appear to contain the necessary modules; however, our evolutionary analysis comparing the seven BGCs has revealed a potential explanation. We propose that, being the most complete and common, cluster 1 is likely to be the ancestral pathway inherited vertically by the Thalassospira and that clusters 2, 4, and 7 could have arisen through module/domain losses (Figs. 3 and 4), whereas clusters 3, 5, and 6 have arisen through HGT events (Fig. 4). Therefore, successful thalassospiramide production might require additional metabolic pathways that are deficient in the host strains of clusters 3, 5, and 6. This hypothesis is consistent with the deduced biosynthetic mechanisms that suggest that in trans activity of an unknown external transferase is involved. To test this hypothesis further, we performed comparative genomics.
Comparison between the genomes of thalassospiramide-producing and -non-producing bacteria identified KEGG genes for amino acid metabolism (such as genes for glutamine and tyrosine biosynthesis), signal transduction (such as the pho family), and compound efflux (such as the MFS transporters) (Fig. 5). Although there is no evidence that glutamate is directly involved in thalassospiramide biosynthesis, glutamate metabolism is at the center of several amino acid metabolic pathways, and it affects the biosynthesis of a variety of bacterium-sourced natural products (16, 17). Enhancement of the tyrosine biosynthetic pathway can increase the production of salidrosides in engineered Escherichia coli (18). The biosynthetic steps to form the thalassospiramides directly involve tyrosine, suggesting potential linkages between tyrosine metabolic pathways and thalassospiramide biosynthesis. Genome mining efforts indicate that secondary metabolites in microbes are under tight regulation, silencing many biosynthetic gene clusters under laboratory conditions (19, 20). The pho family regulators control antibiotic biosynthesis in Streptomyces (21). Thus, the presence or absence of particular regulators in thalassospiramide-producing bacteria suggests their potential roles in controlling thalassospiramide production. In Streptomyces, modifying the MFS transporters can increase the production of secondary metabolites by up to 10-fold (21). In addition, a remarkable degree of evolutionary relatedness has been demonstrated between the transporters and biosynthetic enzymes (22). Currently, the transporters for thalassospiramide secretion have not yet been confirmed experimentally; however, the non-random distribution of transporters in the thalassospiramide-producing and non-producing bacterium genomes indicates their potential impact.
To conclude, the thalassospiramide production patterns and functional BGC architectures are consistent within the Rhodospirillaceae family, allowing a reassessment of the biosynthetic mechanisms. Particular genes/pathways for amino acid metabolism, signal transduction, and molecule transport may have important functions driving thalassospiramide production, emphasizing that molecule production via diversity-oriented biosynthesis is also impacted by genes/pathways beyond the BGCs. This study is an excellent example of the application of integrative biochemical and genomic approaches in discovery of genes and mechanisms within natural product biosynthesis.
Experimental Procedures
Fermentation, Secondary Metabolite Extraction, and LC/MS Analysis
Information on the 130 strains used for compound production examination is listed in supplemental Table S6. Each bacterial strain was cultured on solid media (yeast extract, 2 g liter−1; agar, 16 g liter−1; sea salt, 35 g liter−1; 37 °C) for 5–7 days, and then a single colony was inoculated into a 50-ml liquid culture of GYTP medium (glucose, 10 g liter−1; yeast extract, 2 g liter−1; tryptone, 2.5 g liter−1; peptone, 2.5 g liter−1; sea salt, 35 g liter−1) and grown with shaking for 3 days at 28 °C. The cultures were extracted twice with an equal volume of ethyl acetate. The organic layers were combined and dried, and the solvent was removed in vacuo. Extracts were analyzed by reverse-phase ultra-performance liquid chromatography-mass spectrometry (Waters 1.7-μm BEH C18 column, 0.25 ml min−1, gradient from 5% to 95% CH3CN with 0.1% formic acid, 30 min).
Isolation and Characterization of Thalassospiramides
Bacterial strains were cultured in GYTP medium (40 × 1 liter) at 28 °C for 4 days. The cultures were extracted twice with an equal volume of ethyl acetate. The organic layers were combined and dried, and the solvent was removed in vacuo. The extracts were subjected to flash C18 chromatography, and elution with 30%, 60%, 90%, and 100% aqueous methanol yielded four fractions. New thalassospiramides were purified from these fractions by reverse-phase HPLC (Phenomenex Luna 5-μm C18, 250 × 100 mm, 100 Å, 3.0 ml min−1) with an isocratic method of 55% CH3CN in water. Electrospray ionization TOF high-resolution MS analysis was conducted using a microTOF mass spectrometer (Bruker Daltonics GmbH) in positive ion-scanning mode with a mass range of 100–2000 Da and a voltage of 4.5 kV. MS2 analysis was performed using an LTQ Velos dual pressure ion trap mass spectrometer (Thermo Fisher Scientific). 1D NMR and 2D NMR spectra were obtained using a 500-MHz Varian Inova spectrometer. All samples were dissolved in methanol-d4, and chemical shifts were reported in parts per million relative to TMS.
Calpain Inhibitory Activity Assay
As described previously (4, 23), the calpain inhibitory activity assay was performed using the calpain activity assay kit (BioVision) with Z-Leu-Leu-Tyr-fluoromethylketone used as a positive control. Briefly, the compounds were dissolved in DMSO and diluted in methanol to a series of concentrations. The assays were performed in 96-well plates, initiated by mixing 85 μl of extraction buffer with 0.1 units HCAN1 (BioVision). Subsequently, 10 μl of reaction buffer, 1 μl of test compound, and 5 μl of calpain substrate (Ac-LLY-AFC) were added to each well. The plate was then subjected to incubation at 37 °C for 1 h. The test samples were then recorded under an excitation wavelength of 400 nm and emission wavelength of 505 nm. Samples without compound were defined as a negative control. Three replicates were performed to generate an average value.
Cultures of Mouse Primary Neurons
Embryonic cortical neurons were harvested from C57BL/6 E16.5 mouse embryos by established methods. On day 16.5 of gestation, pregnant mice were sacrificed and their embryos removed. Embryonic cerebral cortices were dissected into PBS containing glucose, minced with forceps, and digested with 0.25% trypsin-EDTA (Sigma-Aldrich) for 10 min at 37 °C. After digestion, tissues were rinsed in DMEM (Thermo Fisher Scientific) with 10% FBS (Sigma-Aldrich) to inactivate the trypsin, rinsed a second time in Neurobasal medium (Gibco), and triturated to produce a single-cell suspension. The cells were then plated onto poly-l-lysine-coated glass coverslips at a final density of 45,000 cells/well. The cells were then grown in Neurobasal medium supplemented with B27 (Gibco), 2 mM GlutaMAX (Gibco), 100 units ml−1 penicillin (Gibco), and 100 μg ml−1 streptomycin (Gibco). Cultures were maintained at 37 °C in 5% CO2. Compound treatments were not initiated until 14 days in vitro.
Neuroprotective Assay by Mouse Primary Neurons
Human THP-1 cells (an immortalized human monocyte cell line, ATCC), were grown at 37 °C in a 5% CO2 atmosphere in RPMI 1640 culture medium (Gibco) containing 10% heat-inactivated FBS, 5 × 10−5 m β-mercaptoethanol (Sigma-Aldrich), 1% sodium pyruvate (Sigma-Aldrich), 100 units/ml penicillin (Gibco), and 100 μg ml−1 streptomycin (Gibco). To create the THP-1 conditioned medium, 4 μl of a 2 mm fibrillar Aβ25–35 (Tocris) suspension was added to 24-well plates and allowed to air-dry. THP-1 cells were subsequently plated on the Aβ-coated wells at a density of 20,000/well in Neurobasal medium. After 48 h, the medium was harvested and centrifuged to remove cells and other debris and then used immediately to treat primary neurons as described previously (24).
To test the neuroprotective behavior of the thalassospiramides, the compounds were assayed at 1 μg ml−1. After 24-h pretreatment, half of the culture medium in each well was removed and replaced with conditioned medium diluted 1:3 in Neurobasal medium to reach a final concentration of 12.5%. Cell cycle activity in the neuronal cultures was monitored by the incorporation of 10 μm 5-ethynyl-2′-deoxyuridine. After 24 h, the cells were rinsed in buffer and fixed with 4% paraformaldehyde in 0.1 m PBS for 20 min at room temperature followed by repeated rinsing in PBS.
The 5-ethynyl-2′-deoxyuridine (EdU) staining was performed using Click-iT chemistry according to the instructions of the manufacturer (Life Technologies). For immunostaining, nonspecific binding staining was blocked with 1% BSA (Sigma-Aldrich) and 0.1% Tween 20. Cells were then incubated overnight at 4 °C with chicken anti-microtubule-associated protein 2 (MAP2) antibody (Abcam) diluted 1:5000 in blocking buffer Phosphate Buffered Saline Tween-20 (PBST). Cells were subsequently rinsed in PBS and then incubated with secondary antibody (A488-labeled donkey anti-chicken antibody, 1:500 in PBST) for 1 h at room temperature. Cells were counterstained with DAPI (Sigma-Aldrich) for 5 min. The fluorescently labeled cultures were mounted in Hydromount and analyzed on a fluorescence microscope (Olympus DP80). All assays were run in triplicate.
DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
DNA extraction was performed as described previously (25). Briefly, the cultured bacterial cells were pelleted by centrifugation at 4000 × g for 10 min and then lysed with lysozyme, proteinase K, and 10% SDS. Total DNA was extracted using the AllPrep DNA/RNA mini kit (Qiagen) following the descriptions of the manufacturer. PCR amplification was carried out using Phusion DNA polymerase (New England Biolabs) and the 16S rRNA gene primer pair8F/1492R in a thermal cycler (Bio-Rad) following these steps: initial denaturation at 98 °C for 30 s, 26 cycles of 98 °C for 10 s, 60 °C for 10 s and 72 °C for 15 s, and a final extension at 72 °C for 5 min. The 16S rRNA genes were sequenced in the Sanger sequencing platform before being annotated online at the NCBI website.
Genome Sequencing, Assembly, and Analysis
Genomic sequencing and analysis were performed using methods described in our previous study (26). The draft genomes were sequenced using the Illumina Hi-seq 2000 platform (Shanghai South Gene Technology Co.). Assembly was performed using SPAdes Genome Assembler 3.6.1 on our local server (27). The specified K values 21, 31, 41, 51, 61, 71, and 81 were used under the “careful” and “pe” options. The optional output after assembly was selected based on N50 and N90 of the contiguous sequences. The completeness and contamination of the genomes were estimated using CheckM (28). Gene prediction was performed with Glimmer v3.0 (29), and gene annotation was done by searching the KEGG (30) database. Adenylation domain specificity was predicted using Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) (31) and NRPS predictor 2 (32), and the condensation and ketosynthase domain functions were analyzed with NaPDoS (33). Non-random co-occurrence patterns of KEGG genomes and the thalassospiramide BGCs were constructed according to methods described in a previous study (34). The statistical analyses were performed in the R studio environment (R 2.13) using the vegan, igraph, and hmisc packages. The cutoff for network generation was a Spearman's correlation coefficient of >0.8 and a statistically significant p value of <0.01. The results were visualized in Gephi (version 0.8.2).
Phylogenetic Analyses
Single gene alignment of nearly full-length (>1300 bp) 16S rRNA genes was conducted in Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 (35) using the Muscle algorithm with the following parameters: gap open penalty of −50, unweighted pair group method with arithmetic mean clustering method, and minimum diagonal length of 24. Gblocks (36) analysis was used to eliminate less informative sites in the alignments. The construction of maximum likelihood trees of 16S rRNA genes was conducted using MEGA version 6.0 with the Tamura-Nei model and the nearest neighbor interchange method with 1000 bootstrap replicates. The construction of maximum parsimony trees of 16S rRNA genes was performed using the subtree-pruning-regrafting method with 1000 bootstrap replicates.
The biosynthetic domains 2C, 6C, and 6A of all predicted thalassospiramide clusters were aligned in MEGA version 6.0 using the Muscle algorithm with the following parameters: gap open penalty of −2.9, gap extension penalty of 0, a hydrophobicity multiplier 1.2, unweighted pair group method with arithmetic mean clustering method, and minimum diagonal length of 24. Alignments of the functional gene trees were subjected to Gblocks analysis to remove less informative sites. Maximum likelihood trees were built using the Jones-Taylor-Thornton model and nearest neighbor interchange method with 1000 bootstrap replicates. Maximum parsimony trees were built using the site rate ranking (SRR) method with 1000 bootstrap replicates. Automated Phylogenomic Inference Application (AMPHORA) (37) was used to predict 31 conserved single-copy genes (tsf, smpB, rpsS, rpsM, rpsK, rpsJ, rpsI, rpsE, rpsC, rpsB, rpoB, rpmA, rplT, rplS, rplP, rplN, rplM, rplL, rplK, rplF, rplE, rplD, rplC, rplB, rplA, pyrG, pgk, nusA, infC, frr, and dnaG). Subsequently, 28 genes (rplL, rplT, and rpmA could not be detected in all genomes) were used to construct the tree. The same parameters used in the construction of the functional gene trees were used.
Statistical Analyses
Significant differences between the effects of the tested compounds and the negative controls in calpain inhibitory and neuroprotective assays were assessed using Student's t tests. Genes with significantly changed numbers in the 18 genomes of thalassospiramide-producing bacteria and the 14 genomes of non-producing bacteria were also identified using Student's t test (p < 0.001).
Author Contributions
P. Y. Q. designed the experiments. W. Z., L. L., and A. C. R. carried out most data analyses. Z. S. and Q. L. provided the samples. Z. L., Y. X., B. Z., and K. H. performed the experiments. L. L., W. Z., A. R., B. S. M., and P. Y. Q. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Nadine Ziemert for discussions regarding the phylogenetic approaches, Prof. William Fenical for providing the strain Thalassospira sp. CNJ-328, Prof. Elizabeth Mann for providing the strain Thalassospira sp. TrichSKD10, and Pui Shuen Wong for performing the MS analysis.
This work was supported by grants from the China Ocean Mineral Resources Research and Development Association (DY135 to P. Y. Q.), the Natural Sciences and Engineering Research Council of Canada (NSERC-DG to A. C. R.), and the National Institutes of Health (GM97509 to B. S. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Data S1 and S2, Figs. S1–S6, and Tables S1–S6.
All sequenced genomes have been deposited in the NCBI database and can be accessed through the numbers listed in supplemental Table S5.
- NRPS
- non-ribosomal peptide synthetase
- HGT
- horizontal gene transfer
- BGC
- biosynthetic gene cluster
- NCBI
- National Center for Biotechnology Information
- BLAST
- Basic Local Alignment Search Tool
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- MFS
- major facilitator superfamily
- EdU
- 5-ethynyl-2′-deoxyuridine
- MS2
- tandem mass spectrometry.
References
- 1. Newman D. J., and Cragg G. M. (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cotter P. D., Hill C., and Ross R. P. (2005) Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788 [DOI] [PubMed] [Google Scholar]
- 3. Oh D. C., Strangman W. K., Kauffman C. A., Jensen P. R., and Fenical W. (2007) Thalassospiramides A and B, immunosuppressive peptides from the marine bacterium Thalassospira sp. Org. Lett. 9, 1525–1528 [DOI] [PubMed] [Google Scholar]
- 4. Ross A. C., Xu Y., Lu L., Kersten R. D., Shao Z., Al-Suwailem A. M., Dorrestein P. C., Qian P. Y., and Moore B. S. (2013) Biosynthetic multitasking facilitates thalassospiramide structural diversity in marine bacteria. J. Am. Chem. Soc. 135, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saatman K. E., Creed J., and Raghupathi R. (2010) Calpain as a therapeutic target in traumatic brain injury. Neurotherapeutics 7, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morton J. D., Lee H. Y., McDermott J. D., Robertson L. J., Bickerstaffe R., Jones M. A., Coxon J. M., and Abell A. D. (2013) Amacrocyclic calpain inhibitor slows the development of inherited cortical cataracts in a sheep model. Invest. Ophthalmol. Vis. Sci. 54, 389–395 [DOI] [PubMed] [Google Scholar]
- 7. Leloup L., and Wells A. (2011) Calpains as potential anti-cancer targets. Expert Opin. Ther. Targets 15, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herrup K., and Yang Y. (2007) Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 8, 368–378 [DOI] [PubMed] [Google Scholar]
- 9. Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., et al. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meiser P., Weissman K. J., Bode H. B., Krug D., Dickschat J. S., Sandmann A., and Müller R. (2008) DKxanthene biosynthesis: understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism. Chem. Biol. 15, 771–781 [DOI] [PubMed] [Google Scholar]
- 11. Richter T. K., Hughes C. C., and Moore B. S. (2015) Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway. Environ. Microbiol. 17, 2158–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmerk C. L., Welander P. V., Hamad M. A., Bain K. L., Bernards M. A., Summons R. E., and Valvano M. A. (2015) Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ. Microbiol. 17, 735–750 [DOI] [PubMed] [Google Scholar]
- 13. McDaniel L. D., Young E., Delaney J., Ruhnau F., Ritchie K. B., and Paul J. H. (2010) High frequency of horizontal gene transfer in the oceans. Science 330, 50–50 [DOI] [PubMed] [Google Scholar]
- 14. Kobayashi N., Nishino K., and Yamaguchi A. (2001) Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marger M. D., and Saier M. H. Jr. (1993) A major superfamily of transmembrane facilitators that can catalyze uniport, symport, and antiport. Trends Biochem. Sci. 18, 13–20 [DOI] [PubMed] [Google Scholar]
- 16. Zhang K., Li H., Cho K. M., and Liao J. C. (2010) Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine. Proc. Natl. Acad. Sci. U.S.A. 107, 6234–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kudo F., Miyanaga A., and Eguchi T. (2014) Biosynthesis of natural products containing β-amino acids. Nat. Prod. Rep. 31, 1056–1073 [DOI] [PubMed] [Google Scholar]
- 18. Bai Y., Bi H., Zhuang Y., Liu C., Cai T., Liu X., Zhang X., Liu T., and Ma Y. (2014) Production of salidroside in metabolically engineered Escherichia coli. Sci. Rep. 4, 6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brakhage A. A. (2013) Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32 [DOI] [PubMed] [Google Scholar]
- 20. Liu G., Chater K. F., Chandra G., Niu G., and Tan H. (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niu G., Li L., Wei J., and Tan H. (2013) Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem. Biol. 20, 34–44 [DOI] [PubMed] [Google Scholar]
- 22. Janeček Š., and Gabriško M. (2016) Remarkable evolutionary relatedness among the enzymes and proteins from the α-amylase family. Cell Mol. Life Sci. 6, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu L., Meehan M. J., Gu S., Chen Z., Zhang W., Zhang G., Liu L., Huang X., Dorrestein P. C., Xu Y., Moore B. S., and Qian P. Y. (2015) Mechanism of action of thalassospiramides, a new class of calpain inhibitors. Sci. Rep. 5, 8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Q., Combs C., Cannady S. B., Geldmacher D. S., and Herrup K. (2000) β-Amyloid activated microglia induce cell cycling and cell death in cultured cortical neurons. Neurobiol. Aging 21, 797–806 [DOI] [PubMed] [Google Scholar]
- 25. Zhang W., Wang Y., Tian R. M., Bougouffa S., Yang B., Cao H. L., Zhang G., Wong Y. H., Xu W., Batang Z., Al-Suwailem A., Zhang X. X., and Qian P. Y. (2014) Species sorting during biofilm assembly by artificial substrates deployed in a cold seep system. Sci. Rep. 4, 06647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang W., Wang Y., Bougouffa S., Tian R., Cao H., Li Y., Cai L., Wong Y. H., Zhang G., Zhou G., Zhang X., Bajic V. B., Al-Suwailem A., and Qian P. Y. (2015) Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool. Environ. Microbiol. 17, 4089–4104 [DOI] [PubMed] [Google Scholar]
- 27. Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., and Pevzner P. A. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., and Tyson G. W. (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delcher A. L., Bratke K. A., Powers E. C., and Salzberg S. L. (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanehisa M., and Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., Lee S. Y., Fischbach M. A., Müller R., Wohlleben W., Breitling R., Takano E., and Medema M. H. (2015) antiSMASH 3.0: a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Röttig M., Medema M. H., Blin K., Weber T., Rausch C., and Kohlbacher O. (2011) NRPS predictor 2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ziemert N., Podell S., Penn K., Badger J. H., Allen E., and Jensen P. R. (2012) The natural product domain seeker NaPDos: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7, e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freilich S., Zarecki R., Eilam O., Segal E. S., Henry C. S., Kupiec M., Gophna U., Sharan R., and Ruppin E. (2011) Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2, 589. [DOI] [PubMed] [Google Scholar]
- 35. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 37. Wu M., and Eisen J. A. (2008) A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.