Abstract
Fascin is an actin bundling protein that cross-links individual actin filaments into straight, compact, and stiff bundles, which are crucial for the formation of filopodia, stereocillia, and other finger-like membrane protrusions. The dysregulation of fascin has been implicated in cancer metastasis, hearing loss, and blindness. Here we identified monoubiquitination as a novel mechanism that regulates fascin bundling activity and dynamics. The monoubiquitination sites were identified to be Lys247 and Lys250, two residues located in a positive charge patch at the actin binding site 2 of fascin. Using a chemical ubiquitination method, we synthesized chemically monoubiquitinated fascin and determined the effects of monoubiquitination on fascin bundling activity and dynamics. Our data demonstrated that monoubiquitination decreased the fascin bundling EC50, delayed the initiation of bundle assembly, and accelerated the disassembly of existing bundles. By analyzing the electrostatic properties on the solvent-accessible surface of fascin, we proposed that monoubiquitination introduced steric hindrance to interfere with the interaction between actin filaments and the positively charged patch at actin binding site 2. We also identified Smurf1 as a E3 ligase regulating the monoubiquitination of fascin. Our findings revealed a previously unidentified regulatory mechanism for fascin, which will have important implications for the understanding of actin bundle regulation under physiological and pathological conditions.
Keywords: actin, cell migration, cell motility, cytoskeleton, ubiquitylation (ubiquitination), fascin, filopodia, monoubiquitination
Introduction
The compact and straight actin bundles are critical for mammalian cells to generate finger-like protrusions such as filopodia and stereocillia. These protrusions are required for diverse physiological functions including cell motility, hearing, and nutrient absorption. Fascin is a monomeric actin bundling protein essential for maximal cross-linking of actin filaments into compact and rigid bundles (1). There are three fascin isoforms (fascin-1, -2, and -3) in metazoans, with fascin-1 expressed mostly in cells with neuronal and mesenchymal lineage, fascin-2 expressed in hair cells and photoreceptor cells, and fascin-3 expressed almost exclusively in the testis (1). The dysregulation of fascin proteins has been associated with various diseases. For example, fascin-1 (hereafter referred to as fascin) is overexpressed in almost all the carcinomas (2, 3). It is believed that fascin promotes cancer metastasis by facilitating the formation of filopodia and invadopodia, which promote cancer cell motility and invasiveness (2, 4). The expression of fascin-2 is limited to the inner ear and retina (1). The loss of function mutations of fascin-2 have been correlated with hearing loss and autosomal dominant retinitis pigmentosa, presumably because of defective actin bundle structures in hair cell stereocillia and photoreceptors (5–10). Understanding the molecular mechanisms underlying fascin bundling activity and its regulation may provide new avenues to prevent cancer metastasis and to treat patients with hearing and vision loss.
We and others previously solved the X-ray crystal structure of fascin and identified residues critical for fascin bundling activity (11–14). Most of the critical fascin residues are positively charged arginine and lysine residues (11). These residues predominantly cluster at two sites, suggesting that these are the two major actin binding sites (ABS1 and ABS2)5 that cross-link actin filaments through electrostatic interactions. It has been reported that the phosphorylation of Ser39 by PKC abrogated fascin bundling activity (15). Post-translational regulation of fascin activity other than Ser39 phosphorylation is unclear. Ubiquitination controls a wide range of biological process by regulating protein degradation, interactions, localization, etc. (16). Proteins may be modified either by the monomeric ubiquitin (monoubiquitination) or the polymeric ubiquitin chain (polyubiquitination) (16, 17). Monoubiquitination has been implicated in regulating DNA repair, endocytosis, virus budding, and nuclear export (18). Fascin has been previously implicated in ubiquitin proteasome-mediated synaptic reorganization in neuronal ischemic tolerance (19). However, whether or how ubiquitination regulates fascin function is not known.
In this study we investigated the post-translational modification of fascin. We discovered that fascin was monoubiquitinated at two lysine residues in its ABS2. We further used an in vitro chemical ubiquitination method to synthesize monoubiquitinated fascin protein and investigated the effects of Lys250 monoubiquitination on fascin bundling activity, the ultrastructure of fascin bundles, and the kinetics of actin bundle formation and disassembly. Our data suggested that monoubiquitination at ABS2 inhibited fascin bundling activity by interfering the interaction between F-actin and the positively charged patch at ABS2. Our findings uncover a novel regulatory mechanism for fascin proteins and shed new light on our understanding of actin bundle regulation.
Results
Fascin Is Monoubiquitinated
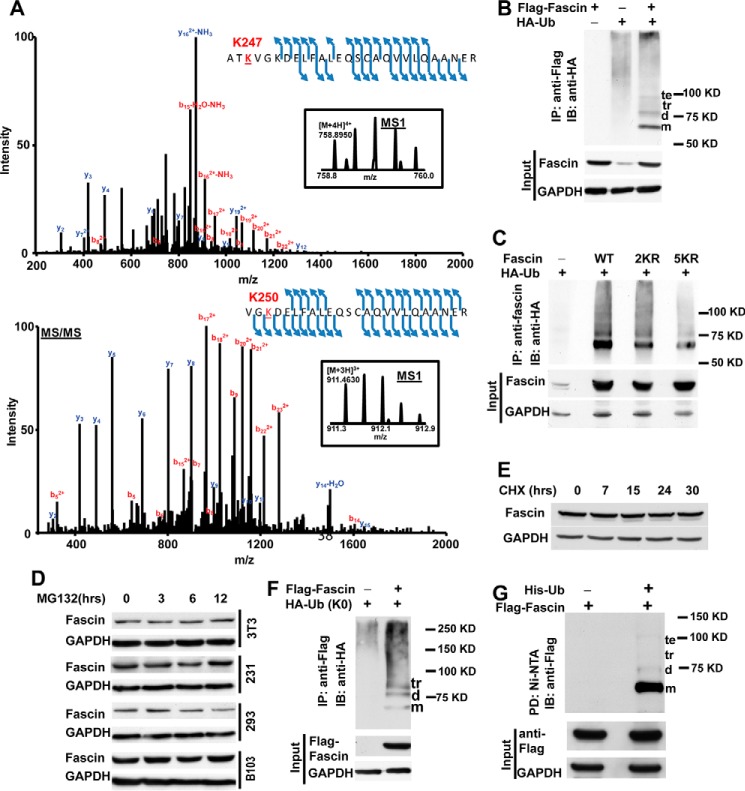
To understand molecular mechanisms that regulate fascin bundling activity, we employed mass spectrometry to identify post-translational modifications of fascin. FLAG-tagged fascin ectopically expressed in HEK293 cells was immunoprecipitated in the presence of phosphatase and pan-HDAC inhibitors, and the post-translational modifications were examined through LC-MS/MS. In addition to the previously reported phosphorylation at Ser39 (data not shown), we consistently detected fascin ubiquitination at Lys247 and Lys250 (Fig. 1A). To determine whether fascin was indeed modified by ubiquitination, HEK293 cells were co-transfected with FLAG-fascin and HA-Ub. After immunoprecipitation of FLAG-fascin, the presence of ubiquitinated fascin was probed with anti-HA antibody. As shown in Fig. 1B, ubiquitinated protein bands corresponding to mono-, di-, tri-, and tetra-Ub fascin were visible when HEK293 cells were co-transfected with HA-Ub and FLAG-fascin, but not when the cells were transfected with FLAG-fascin or HA-Ub alone, suggesting that fascin is indeed modified by ubiquitination in these cells.
FIGURE 1.
Fascin is modified by monoubiquitination. A, tandem mass spectrums showing the ubiquitination of Lys247 (upper panel) and Lys250 (lower panel), respectively. The endogenous fascin in HEK293 cells was immunoprecipitated with anti-fascin antibody, separated by SDS-PAGE, and subjected to identification of post-translational modification through mass spectrometry. B, lysates from HEK293 cells expressing FLAG-fascin and HA-Ub were immunoprecipitated with M2 anti-FLAG beads, and the presence of ubiquitinated proteins was detected with anti-HA antibody through Western blotting. C, the 2KR (K247R/K250R) and 5KR (K41R/K42R/K247R/K250R/K399R) mutations inhibited fascin ubiquitination. D, NIH3T3, MDA-MB-231, HEK293, and B103 cells were treated with proteasome inhibitor MG132 (0.5 μm) for the indicated time, and the effects of MG132 treatment on fascin protein levels in these cells were evaluated using Western blotting. E, the effect of cycloheximide (10 μm) treatment on fascin protein levels in MDA-MB-231 cells. F, FLAG-fascin was co-expressed with K0 mutant (all lysines mutated to alanines) of HA-Ub in HEK293 cells. After immunoprecipitation with M2 anti-FLAG beads, the ubiquitination of fascin by HA-Ub K0 mutant was detected with anti-HA antibody. G, ubiquitinated proteins in HEK293 cells expressing His-tagged ubiquitin were pulled down with Ni-NTA beads, and the presence of ubiquitinated fascin was detected with anti-FLAG antibody. IB, immunoblotting; IP, immunoprecipitation; m, monoubiquitinated fascin; d, diubiquitinated fascin; tr, triubiquitinated fascin; te, tetraubiquitinated fascin.
To determine whether Lys247 and Lys250 are required for fascin ubiquitination, we mutated the two lysine residues to arginine. The K247R/K250R double mutant (2KR) reduced the ubiquitination level by ∼50% (Fig. 1C). The additional ubiquitination at Lys41, Lys42, Lys241, Lys353, Lys399, Lys464, and Lys471 was identified on the K247R/K250R fascin mutant through mass spectrometry (data not shown), suggesting that these lysine residues were suboptimal ubiquitination sites that could be modified in the absence of Lys247 and Lys250 ubiquitination. Mutation of three additional lysine residues (5KR mutant, K41R/K42R/K247R/K250R/K399R) further reduced the levels of ubiquitinated fascin (Fig. 1C).
To investigate whether fascin protein stability is regulated by ubiquitination, we examined the effects of proteasome inhibitor MG132 on fascin protein levels in several fascin expressing cells from different lineages including NIH3T3 (murine fibroblast), B103 (rat neuroblastoma), HEK293 (human kidney), and MDA-MB-231 (breast cancer) cells. MG132 treatment had no effects on fascin protein levels in any of these cells (Fig. 1D). Furthermore, no detectable decrease in fascin protein levels was observed in MDA-MB-231 cells even 30 h after inhibition of total protein synthesis with cycloheximide (Fig. 1E). Taken together, our data suggest that fascin is a stable protein, and ubiquitination does not play a noticeable role in regulating its stability or protein expression levels.
Because our data suggested that ubiquitination may not regulate fascin protein stability, we examined the possibility that fascin might be monoubiquitinated. As shown in Fig. 1F, fascin ubiquitination was readily detectable when co-transfected with K0-HA-Ub (all lysine residues mutated to alanine). The modification by K0 mutant ubiquitin suggested that fascin could be mono- or multiubiquitinated (ubiquitination of multiple lysine residues by ubiquitin monomers). To determine the proportions of mono- and multiubiquitinated fascin, we used Ni-NTA beads to purify ubiquitinated proteins from HEK293 cells co-transfected with FLAG-fascin and His-Ub, and the presence of fascin was probed with anti-FLAG antibody. As shown in Fig. 1G, fascin purified with Ni-NTA beads were predominantly (>90%) corresponding to the monoubiquitinated band, although di-, tri-, and tetra-ubiquitinated fascin was also detectable (Fig. 1G). Similarly, we were able to observe monoubiquitinated fascin-2 when co-transfected with His-Ub (data not shown), suggesting that post-translational modification by ubiquitin is at least conserved between these two fascin isoforms. We were not able to ectopically express fascin-3 in HEK293 cells and thus not able to determine whether fascin-3 is modified by monoubiquitination. Taken together, our data suggested that both fascin and fascin-2 are post-translationally modified by monoubiquitination.
The Positively Charged Patches at Monoubiquitination Sites Are Essential for Fascin Bundling Activity
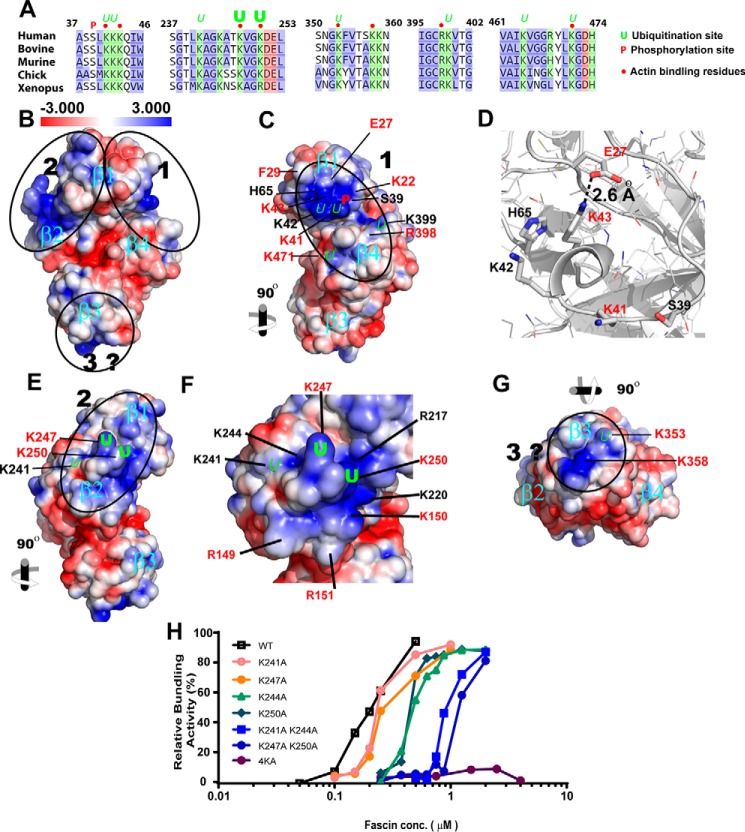
In a previous systemic alanine scan mutagenesis, we identified two major actin binding sites (ABS1 and ABS2) critical for fascin bundling activity (11). Interestingly most of the monoubiquitinated lysine residues are either required for fascin bundling activity (e.g. Lys41, Lys247, Lys250, Lys353, and Lys471), or are immediately adjacent to residues critical for bundling (e.g. Lys42 and Lys399) (Fig. 2A), indicating that monoubiquitination might be involved in the regulation of fascin-actin interaction. To understand the functional role of fascin monoubiquitination, we evaluate the structural relationship between monoubiquitination sites and electrostatic potential on the surface of solvent-accessible fascin (Fig. 2, B–E). There are two distinct patches of positive charges on fascin surface that co-localize with ABS1 and ABS2 (Fig. 2, B–D). A third positive charge patch at the tip of β-trefoil domain 3 may constitute an ancillary interaction site with F-actin (Fig. 2E). The positive charge patch at ABS1 consists of five positively charged residues Lys22, Lys41, Lys42, Lys43, and His65 from β-trefoil domain 1, with Lys41 and Lys42 being suboptimal monoubiquitination sites (Fig. 2C). Lys22, Lys41, and Lys43 have been previously shown to be essential for fascin activity. Glu27, another residue essential for fascin bundling activity, forms a 2.6 Å salt bridge with Lys43 (Fig. 2D). It is worth noting that Glu27 is the only essential residue with negative charge identified in our previous mutagenesis screening. The proximity of Glu27 to this positive charge patch and the Lys43–Glu27 salt bridge suggest that Glu27 may contribute to actin bundling by regulating the proper conformation of the ABS1 positive charge patch.
FIGURE 2.
The positively charged patches at monoubiquitination sites are required for fascin bundling activity. A, sequence alignment of fascin across species. The bold and italic Us indicate optimal and suboptimal ubiquitination sites, respectively. P indicates the PKC phosphorylation site. Red dots indicate residues critical for actin bundling activity, as identified in a previous alanine scan mutagenesis study. B, C, and E–G, surface presentation showing electrostatic properties on the solvent-accessible surface of fascin, as derived from 3LLP using PyMOL and APBS software. β1–β4 indicate the four β-trefoil domains of fascin. Residues critical for actin bundling activity are labeled in red. The ubiquitination and phosphorylation sites are labeled as in A. The molecule in B is viewed from the N- and C-terminal plane. The three positively charged patches are marked by ovals and a circle. C, the view of fascin electrostatic surface turned 90° clockwise along the y axis relative to the view in B, with residues constituting the positively charged patch at ABS1 clearly visible. D, ribbon representation showing the salt bridge between Glu27 and Lys43 at actin binding site 1. E and F, view of fascin turned 90° counterclockwise along the y axis relative to B. F shows details of the electrostatic properties in the vicinity of the 241–250 lysine-rich loop. G shows view of fascin 90° clockwise along the x axis relative to B. H, relative actin bundling activity by wild type and mutant fascin proteins at different concentrations, as determined by low speed sedimentation assays and quantified using ImageJ.
The second positive charge patch at ABS2 consists of Arg149, Lys150, Arg151, Arg217, Lys220, Lys241, Lys244, Lys247, and Lys250 from β-trefoil 2, with Lys247 and Lys250 being the predominant monoubiquitination sites and Lys241 being one of the suboptimal ubiquitination sites (Fig. 2, E and F). Interestingly, the three monoubiquitinated residues locate at a lysine-rich loop that is at the center of the ABS2 positive charge patch (Fig. 2, E and F). A single lysine to alanine mutation shifted the EC50 of actin bundling activity from 0.15 μm (for WT fascin) to 0.2 μm (for K241A and K247A) or 0.4 μm (for K244A and K250A) (Fig. 2H). The EC50 further increased to 0.8–1 μm when two lysine residues were simultaneously mutated (K241A/K244A and K247A/K250A), suggesting that positive charges from this loop contribute to actin bundling accumulatively (Fig. 2H). The quadruple mutations of all the lysines in this loop lead to the complete loss of fascin activity (Fig. 2H), signifying the critical role of this monoubiquitinated loop in fascin-F-actin interaction.
Monoubiquitination Inhibits Fascin Bundling Activity
The co-localization of monoubiquitination sites with positively charged patches at actin binding sites suggests that monoubiquitination may regulate the electrostatic interaction between fascin and F-actin. On the one hand, it is possible that modification by highly positively charged ubiquitin would strengthen the fascin-actin interaction by providing additional positive charges (ubiquitin surface is highly positively charged); on the other hand, monoubiquitination may block fascin bundling activity by introducing steric hindrance to interfere with fascin-actin interaction. To understand the effects of monoubiquitination on fascin bundling activity, reconstituted actin bundling assay using purified wild type fascin and monoubiquitinated fascin would be essential.
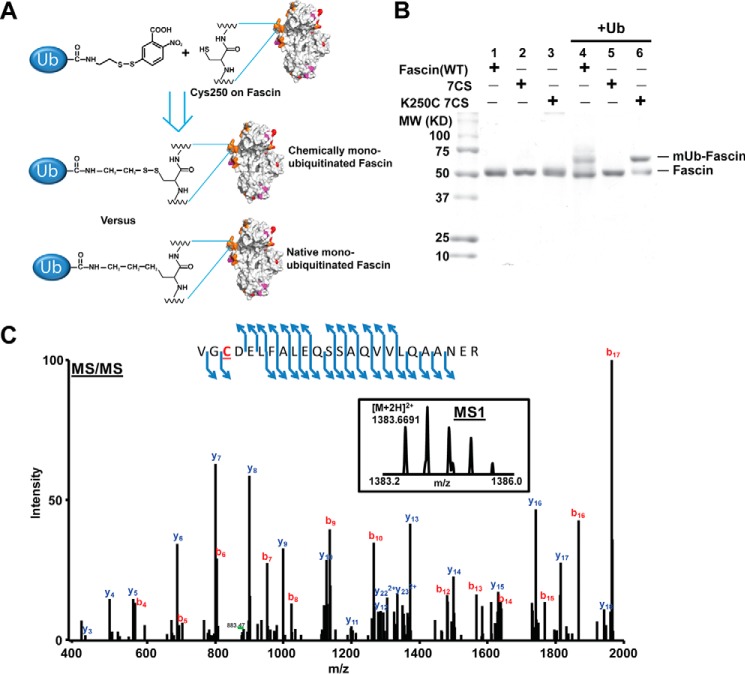
Recently, a chemical ubiquitination method has been developed to allow site-specific monoubiquitination of PCNA (20). We adapted this method to chemically monoubiquitinate fascin (Fig. 3). We decided to focus on Lys247 and Lys250 because these two sites are the predominant monoubiquitination sites and also because there is currently no report on the post-translational regulation of fascin ABS2. Recombinant His-tagged ubiquitin was expressed and purified as an intein fusion protein. After cleavage of intein with cysteamine and activation of the thiol group with DTNB, the DTNB-activated ubiquitin is able to modify cysteine by forming disulfide bond with the free thiol group (Fig. 3A). Therefore, mutation of Lys247 or Lys250 to cysteine would allow the synthesis of chemically ubiquitinated fascin that closely resembles natively monoubiquitinated fascin (Fig. 3A). Seven of the eleven cysteine residues in fascin are solvent-accessible, which explains the modification of wild type fascin by Ub-DTNB (Fig. 3B). Mutation of the seven solvent-accessible cysteines to serines (7CS mutant) abrogated the chemical ubiquitination (Fig. 3B). Next we mutated Lys247 and Lys250 to cysteine and purified recombinant K250C/7CS and K247C/7CS fascin mutants. By incubating with Ub-DTNB, we were able to obtain monoubiquitinated K250C/7CS (referred to as mUb-fascin hereafter for simplicity). The chemical ubiquitination of K247C/7CS was unsuccessful for unknown reasons. The mUb-fascin was partially purified with Ni-NTA beads (Fig. 3B), and the chemical ubiquitination of the mutant at Cys250 through a disulfide bond was confirmed through mass spectrometry (Fig. 3C).
FIGURE 3.
Chemical monoubiquitination of fascin. A, schematic illustration showing chemical immunoprecipitation of K250C/7CS fascin mutant by DTNB activated Ub-SH. The chemically monoubiquitinated fascin closely resembles the natively monoubiquitinated fascin. B, SDS-PAGE gel showing purified recombinant wild type and mutant fascin before (lanes 1–3) and after (lanes 4–6) chemical monoubiquitination. The wild type fascin could be modified by DTNB activated Ub-SH because of the presence of surface cysteines (lane 4). Mutation of the seven surface cysteines (7CS) abrogated the chemical modification (lane 5). Incubation with DTNB activated Ub-SH converts ∼70% of K250C/7CS mutant to monoubiquitinated protein (lane 6). C, tandem mass spectrum showing the modification of Cys250 in K250C/7CS mutant by chemical monoubiquitination at Cys250.
To understand the biochemical function of fascin monoubiquitination, we first used a low speed sedimentation assay to determine the effects of chemical ubiquitination on fascin bundling activity (Fig. 4, A and B). The bundling activity of the 7CS mutant was comparable with wild type fascin, with no obvious difference in actin bundling EC50, suggesting that mutation of the seven cysteines has no global effect on fascin conformation (Fig. 4, A and B). The K250C mutation slightly shifted the actin bundling EC50 from 0.15 to ∼0.25 μm, which is consistent with its localization at a positive charge patch essential for fascin bundling activity. When the K250C/7CS mutant was monoubiquitinated at Cys250, the EC50 further shifted to ∼1 μm, suggesting an over 5-fold decrease in bundling activity after monoubiquitination at the lysine-rich Lys241–Lys250 loop. Importantly, cleavage of ubiquitin from mUb-fascin with DTT was able to restore fascin bundling activity (Fig. 4A), suggesting that the inhibition of fascin activity was indeed due to the modification of Cys250 by the disulfide bond linked ubiquitin.
FIGURE 4.
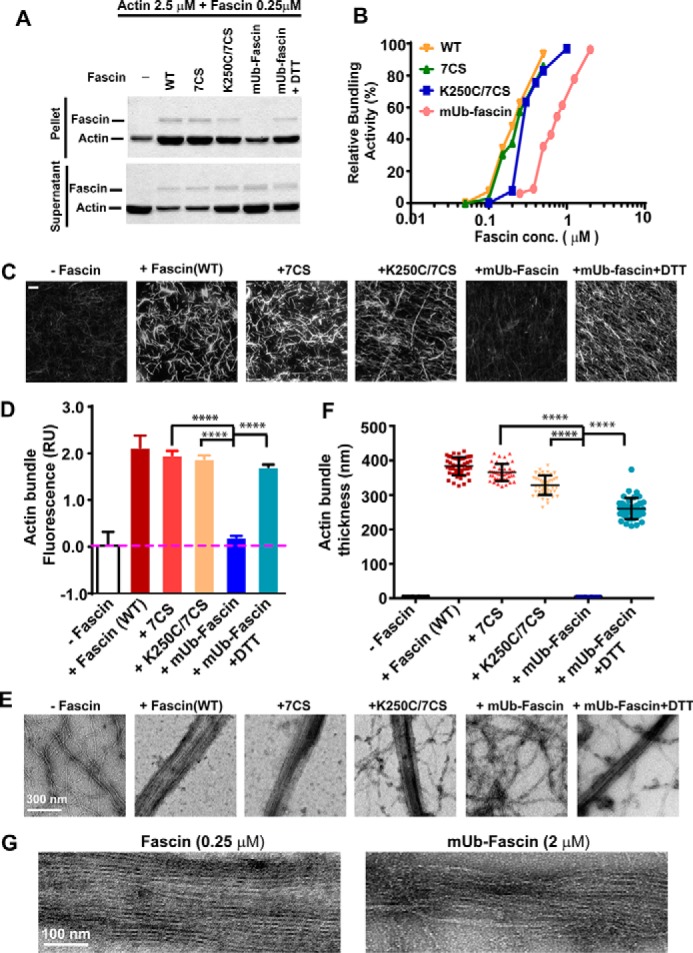
Monoubiquitination inhibits fascin bundling activity. A, the effects of fascin mutations and monoubiquitination on actin bundling activity as determined by low speed sedimentation assay. The increased amount of actin in pellet and the decrease in supernatant indicate the cross-linking of actin into bundles. B, F-actin was incubated with different concentrations of wild type, mutant, or mUb-fascin at the indicated concentrations. The individual actin filaments and F-actin bundles were separated by low speed sedimentation. The relative bundling activity was quantified by densitometry measurement of SDS-PAGE gels. C and E, F-actin (2.5 μm) was incubated with fascin proteins (0.25 μm) for 1 h at room temperature, and the formation of actin bundles was visualized using fluorescence microscopy (C, after staining with Alexa 488-phalloidin) or with TEM (E, after negative staining with uranyl acetate). D, quantitation of fluorescence intensity of fluorescent micrographs from C. F, measurement of actin bundle width from E. G, TEM micrograph showing details of actin bundles cross-linked by wild type fascin (0.25 μm) or mUb-fascin (2 μm). ****, p < 0.0001 as determined by two-tailed two sample t test.
Next, we used two complementary assays to further evaluate the effects of monoubiquitination on fascin bundling activity (Fig. 4, C–F). To visualize actin filaments and bundles directly under a fluorescent microscope, F-actin incubated with or without wild type, mutant, or mUb-fascin was stained with phalloidin. As shown in Fig. 4C, wild type fascin, 7CS, and K250C/7CS mutants induced the formation of thick and bright actin bundles. The bundling activity of K250C/7CS was abrogated after chemical ubiquitination (Fig. 4, C and D). To evaluate the effects of monoubiquitination on the ultrastructure of F-actin bundles, we used transmission electron microscopy (TEM) to visualize the actin bundles cross-linked by wild type, mutant, or mUb-fascin (Fig. 4, E–G). Incubation with 0.5 μm wild type fascin or 7CS mutant completely cross-linked all the actin filaments into tight and straight bundles with an average diameter of 383.5 ± 26.2 nm (Fig. 4, D and E). The K250C mutation slightly decreased the bundle thickness to 328.8 ± 28.3 nm and resulted in incomplete bundling of actin filaments (Fig. 4, D and E). No actin bundle was observed when F-actin was incubated with 0.5 μm mUb-fascin. The abrogation of actin bundling activity by chemical ubiquitination could be at least partially rescued by treatment with DTT (average bundle thickness, 260.6 ± 30.3 nm) (Fig. 4, D and E), which is consistent with the nature of the disulfide bond-linked chemical ubiquitination. It is worth noting that although mUb-fascin was able to cross-link F-actin at a saturating 2 μm, the bundles by Ub-fascin were less densely packed and disordered (Fig. 4G), suggesting inherently defective bundling by mUb-fascin even at high concentrations.
Monoubiquitination Regulates the Dynamic of Fascin Bundle Assembly and Disassembly
To evaluate the effects of monoubiquitination on fascin bundling dynamics, we used light scattering to monitor the cross-linking of F-actin (Fig. 5A). Incubation of F-actin with wild type fascin led to increase in light scattering, suggesting the increase in bundle size after cross-linking of actin filaments. The increase in light scattering quickly reached a plateau at ∼15 min for wild type fascin and the 7CS mutant (Fig. 5A). Interestingly, there was a 10–15-min lag in actin bundling when actin filaments were incubated with 0.5 μm K250C/7CS mutant, although the increase in light scattering eventually reached a plateau comparable with that of wild type and 7CS mutant fascin after 60 min. No increase in light scattering was observed when actin filaments were incubated with 0.5 μm mUb-fascin. At the saturating bundling concentration for mUb-fascin (2 μm), there was a delay in actin bundling similar to that of 0.5 μm K250C mutant (Fig. 5A).
FIGURE 5.
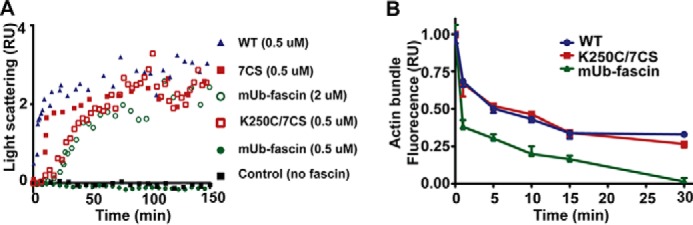
The effects of monoubiquitination on fascin bundling dynamics. A, light scattering data showing the effects of monoubiquitination and mutations on fascin bundle formation dynamics. The increase in F-actin light scattering after incubation with fascin suggested the cross-linking of actin filaments and the increase in bundle size. B, the effects of monoubiquitination on actin bundle disassembly dynamics measured via phalloidin fluorescent staining.
Next, we investigated the effects of monoubiquitination on fascin bundle disassembly. The actin bundles were cross-linked by incubation with wild type fascin, K250C mutant, or mUb-fascin at saturating concentrations. The bundles were separated from unbound fascin proteins by centrifugation. The disassembly was initiated by resuspending the bundles in actin polymerization solution and monitored by the decrease in fluorescence. As shown in Fig. 5B, the fluorescence of wild type fascin and K250C/7CS bundles decreased by ∼50% at 5 min, indicating the disassembly of actin bundles. In contrast, fluorescence of Ub-fascin bundles decreased by more than 50% within 1 min, indicating an 5-fold increase in disassembly rates. Taken together, our data suggested that perturbation of the positive charge patches at the actin binding sites of fascin will affect both the avidity and kinetics of the interaction between fascin and F-actin.
It is well established that fascin promotes cell migration in an actin bundling dependent manner. To determine the effects of monoubiquitination on the cellular function of fascin, we expressed wild type fascin or 2KR (K247R/K250R) mutant in DLD-1 colon cancer cells (Fig. 6A). The lysine to arginine mutations prevent monoubiquitination at these sites without perturbing the positive charges at the actin binding sites. The ectopic expression of wild type fascin in DLD-1 cells promoted the numbers of migrated cells by ∼2-fold (Fig. 6, B and C). The ectopic expression of 2KR mutant further promoted the migration of DLD-1 cells when compared with wild type fascin (Fig. 6, B and C). Taken together, our data suggested that monoubiquitination inhibits the pro-migratory activity of fascin.
FIGURE 6.
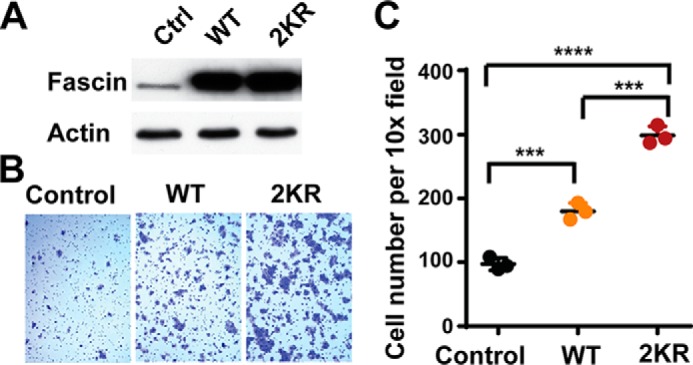
The effects of monoubiquitination on fascin-mediated cell migration. A, the protein levels of fascin in DLD-1 cells stably expressing control vector (Ctrl), wild type fascin, and 2KR fascin mutant. B and C, representative images (B) and quantitation (C) of cell migration assay showing that mutation of Lys247 and Lys250 to arginine (2KR) increased the pro-migration activity of fascin in DLD-1 cells.
The E3 Ubiquitin Ligase Smurf1 Promotes Fascin Monoubiquitination
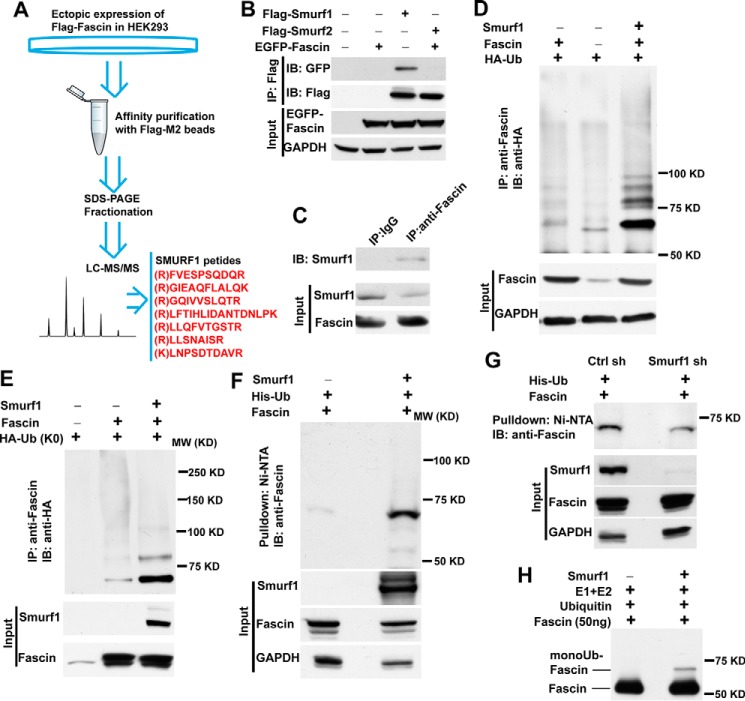
To determine the E3 ubiquitin ligase-mediating fascin monoubiquitination, we used FLAG-M2 beads to pull down fascin, and the proteins associated with fascin were fractionated by SDS-PAGE and identified through mass spectrometry. Interestingly, six peptides corresponding to the E3 ubiquitin ligase were identified through LC-MS/MS (Fig. 7A). EGFP-fascin was co-immunoprecipitated by Smurf1, but not its analog Smurf2, suggesting that fascin specifically interacted with Smurf1 (Fig. 7B). To determine whether Smurf1 interacted with endogenous fascin, the fascin protein in HEK293 cells was immunoprecipitated by anti-fascin antibody, and the co-immunoprecipitated Smurf1 was detected by Western blotting. As shown in Fig. 7C, anti-fascin antibody, but not the control IgG, precipitated Smurf1, indicating the interaction between the two endogenous proteins. The ectopic expression of Smurf1 together with wild type or K0 ubiquitin in HEK293 cells significantly increased the amount of mono- and multiubiquitinated fascin (Fig. 7, D–F), whereas Smurf1 knockdown in HEK293 cells decreased the amount of monoubiquitinated fascin by ∼50%, suggesting that Smurf1 is at least partially responsible for catalyzing fascin monoubiquitination (Fig. 7G). To determine whether fascin might be able to directly mediate fascin monoubiquitination, we purified recombinant Smurf1 and conducted an in vitro ubiquitination assay. Purified fascin was incubated with ubiquitin, E1, and E2 ligase and with or without recombinant Smurf1. As shown in Fig. 7H, a mono-Ub fascin band was detectable when Smurf1 was present in the in vitro ubiquitination reaction, suggesting that Smurf1 directly catalyzes the monoubiquitination of fascin.
FIGURE 7.
The E3 ubiquitin ligase Smurf1 promotes fascin monoubiquitination. A, schematic diagram showing identification of Smurf1 as an fascin interacting protein through FLAG affinity purification and LC-MS/MS. B, co-immunoprecipitation of EGFP-fascin by Smurf1, but not Smurf2. HEK293 cells were transiently transfected with EGFP-fascin and with or without FLAG-Smurf1 or FLAG-Smurf2. The FLAG-tagged proteins were precipitated with anti-FLAG M2 beads, and the presence of EGFP-fascin was detected by Western blotting. C, co-immunoprecipitation of endogenous fascin and Smurf1. Fascin protein in the HEK293 cell lysate was precipitated with anti-fascin antibody, and the co-precipitated Smurf1 was detected by Western blotting. D, lysates from HEK293 cells were immunoprecipitated with anti-fascin antibody, and the presence of ubiquitinated proteins was detected with anti-HA antibody through Western blotting. E, fascin was co-expressed with K0 mutant (all lysine mutated to alanine) of HA-Ub and with or without Smruf1 in HEK293 cells. After immunoprecipitation with anti-fascin antibody, the ubiquitination of fascin by HA-Ub K0 mutant was detected with anti-HA antibody. F, ubiquitinated proteins in HEK293 cells expressing His-tagged ubiquitin were pulled down with Ni-NTA beads, and the presence of ubiquitinated fascin was detected with anti-fascin antibody. G, the effect of Smurf1 knockdown on the levels of monoubiquitinated fascin. H, purified fascin protein (50 ng) was incubated with ubiquitin, E1, and E2 ubiquitin ligase in the presence or absence of recombinant Smurf1. The presence of fascin and mono-Ub fascin was detected by Western blotting with anti-fascin antibody. IB, immunoblotting; IP, immunoprecipitation.
Discussion
Because individual actin filaments are insufficient to overcome compressive forces from the plasma membrane (21), the cross-linking of F-actin into compact and rigid bundles is essential to generate finger-like protrusions such as filopodia. In filopodia fascin undergoes rapid cycles of association and dissociation, with t½ <10 s (4, 21). Even in the highly stable mechanosensory stereocillia in hair cells, fascin-2 undergoes rapid turnover with t½ several orders of magnitude faster than any other known hair cell bundling proteins (22). Interestingly, in A375 melanoma cells the turnover of fascin in invadopodia was ∼7 times slower than fascin in filopodia (4). The differential fascin turnover speeds in the cell suggest active regulation of fascin dynamics in the actin cytoskeleton. Indeed, the dissociations of phosphomimetic mutants of fascin and fascin-2 from actin bundles in filopodia and stereocilia were much faster than wild type proteins (4, 22), suggesting a role for post-translational modification in fascin and fascin-2 turnover. However, paradoxically, the constitutively active mutations of the PKC phosphorylation sites on fascin and fascin-2 (S39A for fascin and S38A for fascin-2) did not have a noticeable effect on their fast turnover in filopodia and stereocilia. It is possible that the turnover of fascin is regulated by other post-translational regulation in addition to PKC phosphorylation.
Here we report that fascin and fascin-2 are modified by monoubiquitination. The monoubiquitination sites of fascin locate at a lysine-rich 241–250 loop within the ABS2 of fascin. The four lysine residues in this loop are critical for fascin bundling activity, as demonstrated by the mutagenesis and actin bundling assay. Interestingly, the 241–250 loop locates at the center of the positive charge patch at the ABS2. Although single mutation of these lysine residues to alanine or cysteine only moderately reduced bundling activity, the double and quadruple mutations drastically inhibited fascin bundling, suggesting that these residues cumulatively contribute to fascin-actin interaction.
Taking advantage of a recently developed chemical ubiquitination approach (20), we were able to achieve site-specific ubiquitination of fascin mimicking native monoubiquitination. Using in vitro actin bundling assays, we demonstrated that monoubiquitination at the ABS2 remarkably reduced fascin bundling activity. Notably, although at high concentration (2 μm) mUb-fascin was able to bundle F-actin, the bundles were less compact and inherently disordered. Moreover, monoubiquitination significantly slowed the initiation of cross-linking and accelerated the disassembly of actin bundles, suggesting that monoubiquitination modulates fascin activity and turnover in actin bundles. The quick turnover of fascin may help to relieve local stresses constantly generated inside filopodial actin bundles, which might be essential to maintain the integrity of filopodia (21). Monoubiquitination regulates a wide range of biological process by promoting or inhibiting protein-protein interactions (17). This could be achieved either by modifying specific residues or a defined domain in substrates (16). Interestingly, most of the monoubiquitinated residues in fascin are confined to its actin binding sites. It is possible that the positively charged patches in the actin binding sites are critical for both the interaction with F-actin and the recognition by the ubiquitin ligase(s). Importantly, many of the ubiquitinated lysine residues are either directly involved in fascin-actin interaction or are immediately adjacent to residues critical for fascin bundling activity, suggesting that monoubiquitination of these residues will introduce steric hindrance inhibiting the interaction between fascin binding sites and F-actin.
Over the years we have performed fascin Western blotting using various cancer cells and fibroblasts with high fascin levels (including fascin Western blotting experiments in Fig. 1D), and we rarely see fascin bands corresponding to mono- or multiubiquitinated fascin using anti-fascin antibody. Therefore it is likely only a small fraction of fascin is modified by mono- or multiubiquitination. However, this should not be interpreted as fascin monoubiquitination has no physiological relevance. Indeed, mutation of the two ubiquitination sites to arginine (2KR mutant) enhances cell migration, suggesting that monoubiquitination is an important inhibitory mechanism for fascin-mediated cell migration. Although for some proteins (e.g. SETDB1 and PCNA) it is important to have a significant fraction modified by monoubiquitination, there are also plenty of examples where only a small fraction of proteins is modified to regulate their physiological functions. For instance, there is convincing evidence that monoubiquitination inhibits the function of p53 and Smad4 by promoting nuclear export (p53) or preventing the formation of transcriptional complexes (Smad4), despite the fact that only very small fractions of p53 and Smad4 are monoubiquitinated (23, 24). This is probably because monoubiquitination of these proteins may be activated only in some circumstances or in some subcellular compartments. Monoubiquitination is a highly dynamic process regulated by ubiquitin ligases and deubiquitinases (18). Our data suggested that fascin monoubiquitination is promoted by the E3 ubiquitin ligase Smurf1. Interestingly, the localization of Smurf1 to filopodia and lamellipodia has been previously reported (25). It is possible that filopodial Smurf1 ubiquitinylates fascin to control filopodium flexibility and/or retraction. Future investigation into this area is warranted.
The actin cytoskeleton in metastatic cancer cells is dysregulated to promote cell motility, invasiveness and metastatic dissemination. In recent years, fascin has emerged as a prominent driver for cancer metastasis. The overexpression of fascin has been reported in almost all the carcinomas (2, 14, 26, 27), and fascin overexpression uniformly correlates with aggressive clinical course, metastatic progression, and poor prognosis (14, 28–33). The transcriptional regulation of fascin in cancer has been extensively studied, with inflammatory cytokines, hypoxia, epithelial to mesenchymal transcription factors, and Wnt/β-catenin pathways, among others, being implicated in the up-regulation of fascin levels in various cancers (33–38). In contrast, little is known about the post-translational regulation of fascin other than the Ser39 phosphorylation by PKC. Paradoxically, many PKC agonists such as phorbol 12-myristate 13-acetate (PMA) are well known carcinogens. Therefore how PKC phosphorylation of fascin contributes to cancer progression remained to be determined.
Materials and Methods
Ubiquitination Assay
HEK293 or 293T cells were co-transfected with plasmids encoding FLAG-fascin, HA-Ub, and Smurf1 using the PEI method as previously described (39). The cells were harvested 36–48 h post transfection and lysed on ice with radioimmune precipitation assay buffer supplemented with 1% SDS and protease inhibitors. After brief sonication, the lysates were heated at 97 °C for 8 min and centrifuged at 12,000 × g for 10 min. The supernatant was incubated with M2 anti-FLAG beads (Sigma) overnight at room temperature. The beads were washed three times and subjected to SDS-PAGE. The Western blotting was performed with anti-HA antibody.
To isolate ubiquitinated fascin with Ni-NTA beads, 293T cells were co-transfected with plasmids encoding FLAG-fascin and His-Ub using PEI. The cells were scraped from the plate and resuspended in 1 ml of PBS at ∼24–32 h post transfection. One-fifth of the cells were lysed in SDS loading buffer for determination of input. The rest of the cells were lysed in buffer A. After sonication and centrifugation, the supernatant was incubated with 30 μl of Ni-NTA beads at room temperature for 4–16 h. The beads were washed subsequently with buffer A, buffer B, buffer C with 0.2 Triton X-100, and then buffer C alone for 5 min each. The protein was eluted with 50 μl of elution buffer for 20 min and subjected with Western blotting using anti-FLAG antibody. The buffers were: A, 6 m guanidinium HCl, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris, pH 8.0, 10 mm imidazole, 10 mm β-mercaptoethanol; B, 8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris, pH 8.0, 10 mm imidazole, 10 mm β-mercaptoethanol; C, 8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris, pH 6.3, 10 mm imidazole, 10 mm β-mercaptoethanol; and elution buffer, 200 mm imidazole, 0.15 m Tris-Cl, pH 6.7, 30% glycerol, 0.72 mm β-mercaptoethanol, 5% SDS.
F-Actin Polymerization
G-Actin powder was purchased from Cytoskeleton (catalog no. AKL99-C). F-Actin was polymerized by incubating G-actin (25 μm) in actin polymerization buffer (20 mm Tris, pH 8, 100 mm KCl, 1 mm ATP, 2 mm MgCl2) for 60 min at room temperature. The actin filaments were centrifuged at 20,000 × g for 30 min at 4 °C to remove any protein aggregates.
Purification of Wild Type and Mutant Fascin Proteins
The wild type and mutant fascin proteins (7CS: C19S/C61S/C89S/C260S/C226S/C397S/C456S; K247C/7CS, K250C/7CS, K241A, K244A, K247A, K250A, K241A/K244A, K247A/K250A, and K241A/K244A/K247A/K250A) were expressed as recombinant GST fusion protein and purified as described previously (11, 14). Escherichia coli BL21 (DE3) cells expressing fascin proteins were cultured at 37 °C in LB medium (1 liter) containing 50 μg/ml kanamycin. The protein expression was induced with 0.1 mm isopropyl β-d-thiogalactopyranoside when the A600 of the culture reached 0.8–1.0. After overnight incubation at 16 °C, the cells were harvested and resuspended in GST buffer A (50 mm Tris, 150 mm NaCl, 1% Triton, 0.1 mm PMSF, 5 mm EDTA, pH 8). The cells were lysed by sonication, and the lysate was centrifuged at 40,000 g for 30 min at 4 °C. The supernatant was incubated with 1 ml of glutathione-agarose beads (Thermo catalog no. 16100) at 4 °C for 2 h. After washing with GST wash buffer (50 mm Tris, 150 mm NaCl, pH 8) five times, the beads were resuspended in thrombin buffer (50 mm Tris, 150 mm NaCl, 1 mm CaCl2, pH 7.6) containing with 30 unit thrombin (Sigma, catalog no. T9326) and incubated overnight at 4 °C. The cleaved fascin protein in the supernatant was concentrated using a 30-kDa centrifugal filter (Amicon catalog no. Ultra-15 ml) to over 8 mg/ml. The aliquoted proteins were frozen with liquid nitrogen and stored at −80 °C.
Chemical Monoubiquitination of Fascin
Chemical ubiquitination of fascin was carried out using a protocol previously described for PCNA (20) with modifications. To purify His-ubiquitin-intein-CBD fusion protein, BL21 (DE3) cells were transformed with pTYB1-Ubiquitin plasmid (a generous gift from Zhihao Zhuang, University of Delaware). The cells were cultured at 37 °C in LB medium until A600 reached 0.8–1.0. The protein expression was induced with 0.1 mm isopropyl β-d-thiogalactopyranoside at 16 °C for 18 h. The cells were harvested by centrifugation, resuspended in lysis buffer (20 mm Tris, 200 mm NaCl, 1 mm EDTA, 5% glycerol, pH 7.5), and lysed by sonication. The lysate was centrifuged at 40,000 × g for 30 min at 4 °C. The supernatant was incubated with 4 ml of chitin resin for 2 h at 4 °C. After washing five times with 50 ml of high salt wash buffer (20 mm Tris, 1 m NaCl, 1 mm EDTA, 5% glycerol, pH 7.5) and five times with 50 ml of low salt wash buffer (50 mm Tris, 100 mm NaCl, pH 8.5), the His-ubiquitin was cleaved by incubation with 10 ml of cleavage buffer (50 mm Tris, 100 mm NaCl, 100 mm cysteamine, pH 8.5) overnight at 4 °C. The resin was eluted with another 10 ml of cleavage buffer. His-Ub-SH was then concentrated with a 10-kDa Centrifugal Filter (Amicon catalog no. Ultra-15 ml).
To remove free cysteamine, the samples were applied to HiTrap desalting column (GE Healthcare catalog no. 11-0003-29) and eluted with ligation buffer (20 mm Tris, 50 mm NaCl, 1 mm EDTA, pH 7.0). The fractions containing His-Ub-SH were combined and concentrated to 400 μl. To activate the thiol group, His-Ub-SH was incubated with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (4.8 mg, 12 μm) in 50 mm sodium phosphate, pH 7.5, at room temperature for 30 min. After desalting, ubiquitin protein fractions were with a 10-kDa centrifugal filter. To chemically monoubiquitinate fascin, K250C/7CS fascin was mixed with the DTNB activated His-Ub-SH (250 μl, 0.6 μmol) at 1:4 molar ratios and incubated at room temperature for 2 h. Aliquots were removed at various time points and quenched by flash freezing with liquid N2. The reaction products at various time points were analyzed in a 10% denaturing non-reducing SDS-PAGE gel. Excess His-ubiquitin was removed with a 50-kDa centrifugal filter and concentrated to ≥5 mg/ml.
To remove the disulfide bond linked ubiquitin, Ub-fascin was incubated with 1 mm DTT at 25 °C. The complete removal of ubiquitination was analyzed with 10% non-reducing SDS-PAGE. Same concentration of DTT used in low speed sedimentation assay, electron microscopy analysis, and fluorescence microscopy analysis to rescue the bundling activity mUb-fascin.
Low Speed Sedimentation Actin Bundling Assay
Low speed sedimentation assays were carried out as we described previously (11, 14). 2.5 μm F-Actin was mixed with wild type, mutant fascin, or Ub-fascin for 1 h at room temperature to allow the formation of actin bundles. The mixtures were centrifuged for 20 min at 11,000 × g at 4 °C to precipitate actin bundles. The supernatant was carefully separated from the pellet, and the amount of actin and fascin in the supernatant and pellet was analyzed with 10% SDS-PAGE.
To calculate the relative actin bundling activity, the amount of actin in pellet and supernatant was quantified by densitometry measurement of Coomassie Blue-stained SDS-PAGE gels. The relative bundling activity was calculated using the formula B(f) = (P(f) − P(0))/(P(f) + S(f) − P(0)), where B(f) is the relative bundling activity of fascin protein at a given concentration f, and P(f), P(0), and S(f) are the optical density of SDS-PAGE actin bands in the pellet (P(f) and P(0)) or supernatant (S(f)) when fascin concentration is f or 0, respectively.
Electron Microscopy Analysis of Fascin Bundles
Bundles were prepared by mixing F-actin with fascin proteins and incubating for 1 h at room temperature. The samples were then applied to carbon-coated 300 mesh copper grids (EMS, catalog no. CF300-CU). 5 μl of the samples were applied on to grids for 1 min before removal of the solution. The samples were then negatively stained with 10 μl of 1% (w/v) uranyl acetate for 1 min and dried at room temperature overnight. A JEM-1400 transmission electron microscope (JEOL) was used to visualize actin filaments and fascin bundles at an accelerating voltage of 80 kV and a magnification of 80,000×–200,000×. Images were captured on an Orius wide field side mount device camera (Gatan). The width of bundles and the number of filaments of per bundle were measured using ImageJ software. Fifty or more bundles were analyzed.
Fluorescence Microscopy Analysis of Fascin Bundles
F-actin was mixed with recombinant wild type or mutant fascin protein in actin polymerization and incubated at room temperature for 60 min. Actin was then labeled by adding 1% Alexa 488-phalloidin (Invitrogen, catalog no. A12379) to the mixtures. The samples were applied to a coverslip that was freshly coated with 1 mg/ml of poly-d-lysine. After 1 min of incubation, the coverslips were mounted on slides, and the images were taken using the Zeiss automated upright fluorescent microscope and AxioVision microscopy software (Carl Zeiss Vision, Inc., San Diego, CA). Three randomly selected fields (63× objectives) were photographed, and the fluorescence density was analyzed using ImageJ software. For Fascin-Actin bundle dissociation assay, after mixing F-Actin with Fascin at room temperature for 60 min, samples were centrifuged for 30 min at 15,000 × g at 4 °C. After removing the supernatants, the pellet was resuspended with actin polymerization buffer and incubated at 37 °C water bath. At different time points, actin bundles were removed from the water bath and labeled by adding 1% Alexa 488-phalloidin to the mixtures. The samples were applied to a coverslip that was freshly coated with 1 mg/ml of poly-d-lysine. After 1 min of incubation, the coverslips were mounted, and the images were acquired and analyzed as previously described.
Static Light Scattering Assay
Static light scattering (SLS) assays were performed as previously described (40). A Zetasizer Nano S (Malvern Instruments, Worchestershire, UK) dynamic light scattering unit with a 4-milliwatt HeNe laser and an avalanche photodiode detector was used to determine SLS intensities. Bundling activity of actin was monitored via changes in SLS intensities with incubation period. A volume of 25 μl of F-actin (5 μm) and 25 μl of fascin (1.0–4.0 μm) was mixed at a 1:1 ratio prior to measurements. During measurements, scattering intensities were averaged over a 3-min sample period. Bundling kinetics was reported as fractional enhancement in SLS intensity (I − I0)/I0 over its starting intensity, I0.
Analysis by LC-MS/MS Mass Spectrometry
The gel band containing target protein was excised and treated with Tris(2-carboxy-ethyl) phosphine hydrochloride and iodoacetamide. Trypsin in-gel digestion was carried on at 37 °C overnight. The extracted peptides were analyzed by LC-MS/MS. A nanoflow liquid chromatograph (U3000; Dionex, Sunnyvale, CA) coupled to an electrospray ion trap mass spectrometer (LTQ-Orbitrap; Thermo, San Jose, CA) was used for tandem mass spectrometry peptide sequencing experiments. Five tandem mass spectra were collected in a data-dependent manner following each survey scan. The MS scans were performed in Orbitrap to obtain accurate peptide mass measurement, and the MS/MS scans were performed in linear ion trap using 60 s of exclusion for previously sampled peptide peaks. Sequest I and Mascot II searches were performed against the Swiss-Prot human database downloaded on February 10, 2009. Two trypsin missed cleavages were allowed, the precursor mass tolerance was 1.08 Da. MS/MS mass tolerance was 0.8 Da. Dynamic modifications included carbamidomethylation (Cys), oxidation (Met), and ubiquitination (Gly-Gly addition on Lys). Both the Mascot and Sequest search results were summarized in Scaffold 2.0.
In Vitro Ubiquitination Assay
Ubiquitin activating enzyme E1 (catalog no. BML-UW9410-0050) was from Enzo Life Sciences. UbcH7/UBE2L3 (catalog no. E2-640-050) and ubiquitin (U-100H-10M) were from R&D Systems. Recombinant GST-Smurf1 was cloned into pGEX-4T1 vector, expressed in E. coli BL21/DE3, and purified with glutathione beads. 100 ng of E1, E2, and purified recombinant Smurf1 and 20 ng of ubiquitin was incubated with 500 ng of fascin in ubiquitination reaction buffer (50 mm Tris-HCl, pH 7.4, 10 mm Mgcl2, 5 mm ATP, and 2 mm DTT) at 30 °C for 60 min. 10% of the reaction product was separated on 10% SDS-PAGE gel and subjected to Western blotting with anti-fascin antibody.
Author Contributions
S. Y. conceived the study and wrote the manuscript. S. Y., S. Lin, S. Lu, and J. S. designed the study and analyzed the data. M. Mulaj and M. Muschol designed the experiments and analyzed the data in Fig. 5A. S. Lin, S. Lu, J. S., M. Mulaj, and T. K. performed the experiments. B. F. performed and analyzed all the LC-MS/MS experiments. J. H. and L. W. revised the manuscript.
Acknowledgments
We thank Zhihao Zhuang for pTYB1-ubiquitin; Jaya Padmanabhan for B103 cells; Jiandong Chen for valuable advice; and Bin Fang and the Proteomic Core at the Moffitt Cancer Center for help with mass spectrometry.
This work is supported in part by National Institutes of Health Grant R01CA175741 and National Natural Science Foundation of China Grants 81572618 and 31671448. The core facilities at the Moffitt Cancer Center are supported in part by the Cancer Center Support Grant P30 CA076292 from the National Cancer Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ABS
- actin binding site
- Ub
- ubiquitin
- mUb
- monoubiquitinated
- DTNB
- 5,5-dithio-bis-(2-nitrobenzoic acid)
- Ni-NTA
- nickel-nitrilotriacetic acid
- TEM
- transmission electron microscopy
- PCNA
- proliferating cell nuclear antigen
- PMA
- phorbol 12-myristate 13-acetate
- SLS
- static light scattering.
References
- 1. Kureishy N., Sapountzi V., Prag S., Anilkumar N., and Adams J. C. (2002) Fascins, and their roles in cell structure and function. Bioessays 24, 350–361 [DOI] [PubMed] [Google Scholar]
- 2. Hashimoto Y., Skacel M., and Adams J. C. (2005) Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int. J. Biochem. Cell Biol. 37, 1787–1804 [DOI] [PubMed] [Google Scholar]
- 3. Machesky L. M., and Li A. (2010) Fascin: invasive filopodia promoting metastasis. Commun. Integr. Biol. 3, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li A., Dawson J. C., Forero-Vargas M., Spence H. J., Yu X., König I., Anderson K., and Machesky L. M. (2010) The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shin J. B., Longo-Guess C. M., Gagnon L. H., Saylor K. W., Dumont R. A., Spinelli K. J., Pagana J. M., Wilmarth P. A., David L. L., Gillespie P. G., and Johnson K. R. (2010) The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J. Neurosci. 30, 9683–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perrin B. J., Strandjord D. M., Narayanan P., Henderson D. M., Johnson K. R., and Ervasti J. M. (2013) β-Actin and fascin-2 cooperate to maintain stereocilia length. J. Neurosci. 33, 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin-Jones J., and Burnside B. (2007) Retina-specific protein fascin 2 is an actin cross-linker associated with actin bundles in photoreceptor inner segments and calycal processes. Invest. Ophthalmol. Vis. Sci. 48, 1380–1388 [DOI] [PubMed] [Google Scholar]
- 8. Yokokura S., Wada Y., Nakai S., Sato H., Yao R., Yamanaka H., Ito S., Sagara Y., Takahashi M., Nakamura Y., Tamai M., and Noda T. (2005) Targeted disruption of FSCN2 gene induces retinopathy in mice. Invest. Ophthalmol. Vis. Sci. 46, 2905–2915 [DOI] [PubMed] [Google Scholar]
- 9. Wada Y., Abe T., Itabashi T., Sato H., Kawamura M., and Tamai M. (2003) Autosomal dominant macular degeneration associated with 208delG mutation in the FSCN2 gene. Arch. Ophthalmol. 121, 1613–1620 [DOI] [PubMed] [Google Scholar]
- 10. Wada Y., Abe T., Takeshita T., Sato H., Yanashima K., and Tamai M. (2001) Mutation of human retinal fascin gene (FSCN2) causes autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 42, 2395–2400 [PubMed] [Google Scholar]
- 11. Yang S., Huang F. K., Huang J., Chen S., Jakoncic J., Leo-Macias A., Diaz-Avalos R., Chen L., Zhang J. J., and Huang X. Y. (2013) Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 288, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., and Dominguez R. (2011) Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286, 30087–30096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedeh R. S., Fedorov A. A., Fedorov E. V., Ono S., Matsumura F., Almo S. C., and Bathe M. (2010) Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein. J. Mol. Biol. 400, 589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen L., Yang S., Jakoncic J., Zhang J. J., and Huang X. Y. (2010) Migrastatin analogues target fascin to block tumour metastasis. Nature 464, 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., and Matsumura F. (1997) Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 16. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 17. Husnjak K., and Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 [DOI] [PubMed] [Google Scholar]
- 18. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 19. Meller R., Thompson S. J., Lusardi T. A., Ordonez A. N., Ashley M. D., Jessick V., Wang W., Torrey D. J., Henshall D. C., Gafken P. R., Saugstad J. A., Xiong Z. G., and Simon R. P. (2008) Ubiquitin proteasome-mediated synaptic reorganization: a novel mechanism underlying rapid ischemic tolerance. J. Neurosci. 28, 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J., Ai Y., Wang J., Haracska L., and Zhuang Z. (2010) Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 6, 270–272 [DOI] [PubMed] [Google Scholar]
- 21. Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., and Borisy G. G. (2006) Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hwang P., Chou S. W., Chen Z., and McDermott B. M. Jr. (2015) The stereociliary paracrystal is a dynamic cytoskeletal scaffold in vivo. Cell Reports 13, 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., and Gu W. (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 24. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., et al. (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 [DOI] [PubMed] [Google Scholar]
- 25. Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., and Wrana J. L. (2003) Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 26. Adams J. C. (2004) Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16, 590–596 [DOI] [PubMed] [Google Scholar]
- 27. Hashimoto Y., Skacel M., Lavery I. C., Mukherjee A. L., Casey G., and Adams J. C. (2006) Prognostic significance of fascin expression in advanced colorectal cancer: an immunohistochemical study of colorectal adenomas and adenocarcinomas. BMC Cancer 6, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hashimoto Y., Shimada Y., Kawamura J., Yamasaki S., and Imamura M. (2004) The prognostic relevance of fascin expression in human gastric carcinoma. Oncology 67, 262–270 [DOI] [PubMed] [Google Scholar]
- 29. Yoder B. J., Tso E., Skacel M., Pettay J., Tarr S., Budd T., Tubbs R. R., Adams J. C., and Hicks D. G. (2005) The expression of fascin, an actin-bundling motility protein, correlates with hormone receptor-negative breast cancer and a more aggressive clinical course. Clin. Cancer Res. 11, 186–192 [PubMed] [Google Scholar]
- 30. Pelosi G., Pasini F., Fraggetta F., Pastorino U., Iannucci A., Maisonneuve P., Arrigoni G., De Manzoni G., Bresaola E., and Viale G. (2003) Independent value of fascin immunoreactivity for predicting lymph node metastases in typical and atypical pulmonary carcinoids. Lung Cancer 42, 203–213 [DOI] [PubMed] [Google Scholar]
- 31. Puppa G., Maisonneuve P., Sonzogni A., Masullo M., Chiappa A., Valerio M., Zampino M. G., Franceschetti I., Capelli P., Chilosi M., Menestrina F., Viale G., and Pelosi G. (2007) Independent prognostic value of fascin immunoreactivity in stage III-IV colonic adenocarcinoma. Br. J. Cancer 96, 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pelosi G., Pastorino U., Pasini F., Maissoneuve P., Fraggetta F., Iannucci A., Sonzogni A., De Manzoni G., Terzi A., Durante E., Bresaola E., Pezzella F., and Viale G. (2003) Independent prognostic value of fascin immunoreactivity in stage I nonsmall cell lung cancer. Br. J. Cancer 88, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao X., Gao S., Ren H., Sun W., Zhang H., Sun J., Yang S., and Hao J. (2014) Hypoxia-inducible factor-1 promotes pancreatic ductal adenocarcinoma invasion and metastasis by activating transcription of the actin-bundling protein fascin. Cancer Res. 74, 2455–2464 [DOI] [PubMed] [Google Scholar]
- 34. Sun J., He H., Pillai S., Xiong Y., Challa S., Xu L., Chellappan S., and Yang S. (2013) GATA3 transcription factor abrogates Smad4 transcription factor-mediated fascin overexpression, invadopodium formation, and breast cancer cell invasion. J. Biol. Chem. 288, 36971–36982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun J., He H., Xiong Y., Lu S., Shen J., Cheng A., Chang W. C., Hou M. F., Lancaster J. M., Kim M., and Yang S. (2011) Fascin protein is critical for transforming growth factor β protein-induced invasion and filopodia formation in spindle-shaped tumor cells. J. Biol. Chem. 286, 38865–38875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li A., Morton J. P., Ma Y., Karim S. A., Zhou Y., Faller W. J., Woodham E. F., Morris H. T., Stevenson R. P., Juin A., Jamieson N. B., MacKay C. J., Carter C. R., Leung H. Y., Yamashiro S., et al. (2014) Fascin is regulated by slug, promotes progression of pancreatic cancer in mice, and is associated with patient outcomes. Gastroenterology 146, 1386–1396.e1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanda Y., Kawaguchi T., Kuramitsu Y., Kitagawa T., Kobayashi T., Takahashi N., Tazawa H., Habelhah H., Hamada J., Kobayashi M., Hirahata M., Onuma K., Osaki M., Nakamura K., Kitagawa T., et al. (2014) Fascin regulates chronic inflammation-related human colon carcinogenesis by inhibiting cell anoikis. Proteomics 14, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 38. Vignjevic D., Schoumacher M., Gavert N., Janssen K. P., Jih G., Laé M., Louvard D., Ben-Ze'ev A., and Robine S. (2007) Fascin, a novel target of β-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 67, 6844–6853 [DOI] [PubMed] [Google Scholar]
- 39. Yang S., Zhang J. J., and Huang X. Y. (2012) Mouse models for tumor metastasis. Methods Mol. Biol. 928, 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mulaj M., Foley J., and Muschol M. (2014) Amyloid oligomers and protofibrils, but not filaments, self-replicate from native lysozyme. J. Am. Chem. Soc. 136, 8947–8956 [DOI] [PMC free article] [PubMed] [Google Scholar]