Abstract
Differentiated cells can be reprogrammed by transcription factors, and these factors that are responsible for successful reprogramming need to be further identified. Here, we show that the neuronal repressor RE1-silencing transcription factor (REST) is rich in porcine oocytes and requires for nuclear transfer (NT)-mediated reprogramming through inhibiting TGFβ signaling pathway. REST was dramatically degraded after oocyte activation, but the residual REST was incorporated into the transferred donor nuclei during reprogramming in NT embryos. Inhibition of REST function in oocytes compromised the development of NT embryos but not that of IVF and PA embryos. Bioinformation analysis of putative targets of REST indicated that REST might function on reprogramming in NT embryos by inhibiting TGFβ pathway. Further results showed that the developmental failure of REST-inhibited NT embryos could be rescued by treatment of SB431542, an inhibitor of TGFβ pathway. Thus, REST is a newly discovered transcription factor that is required for NT-mediated nuclear reprogramming.
Keywords: bioinformatics, cloning, oocyte, repressor protein, reprogramming
Introduction
Embryonic cells differentiate into all three germ layers of the body as development progresses. Once differentiated, the reversion of the differentiated state to pluripotency is strictly limited in normal development. However, experimentally the differentiated state can be returned to the pluripotent state by transcription factors (1, 2). Despite numerous attempts, the factors responsible for successful nuclear reprogramming still need to elucidate. Transcription factors maintaining the pluripotency of embryonic stem cells (ESCs)3 are called pluripotent factors, and they have an important role in nuclear reprogramming, such as Oct4, Sox2, and Nanog (3, 4). Thus, we can identify and characterize reprogramming factors by screening the pluripotent factors.
The repressor element 1 (RE1)-silencing transcription factor (REST), as a zinc finger protein, binds 21-bp RE1 sites and functions as a key negative regulator of neurogenesis, so it is also called neuron-restrictive silencer element (5). Recently, REST has been reported to induce gene expression by recruiting TET3 to the DNA for directed 5hmC generation and Nuclear SET domain-containing protein 3-mediated H3K36 trimethylation in neurons (6). Furthermore, REST has different roles in different cellular contexts, such as oncogenic and tumor-supressor functions and hematopoietic and cardiac differentiation (7, 8). In 2008, REST was proved to maintain self-renewal and pluripotency of mouse ESCs through suppression of microRNAs and believed to be a major pluripotent factor (9, 10). However, it has not been elaborated in nuclear reprogramming. Here, we provide evidence that REST plays a unique role in NT-mediated reprogramming as a supressor of the TGFβ signaling pathway in pig.
Results
Expression Pattern of REST
We first investigated the expression of REST in porcine oocytes, nuclear transfer (NT), and parthenogenetic activation (PA) embryos by real-time PCR and Western blotting analysis. Porcine fetal fibroblasts (PFFs) were used as donor cells to construct NT embryos, and REST was observed in the cells by Western blotting. Large amounts of REST mRNA and protein were stored in oocytes. After activation, REST mRNA was significantly decreased in NT and PA embryos (p < 0.001) and maintained at a low level from the four-cell to blastocyst stages (Fig. 1A), and REST protein was also degraded in one- and two-cell NT and PA embryos (Fig. 1B). We performed immunofluorescence analysis to locate REST protein in oocytes and embryos. REST was dispersed in the MII oocyte cytoplasm (Fig. 2, A and A′, n = 17), and was incorporated into transferred donor nuclei in NT embryos when the nuclei were condensed at 2 h post-NT (Fig. 2, B and B′, n = 15) and decondensed at 6 h post-NT (Fig. 2, C and C′, n = 10). In donor cells, REST was also incorporated into the nuclei (Fig. 2, D and D′). To confirm whether maternal REST could be incorporated into the transferred donor nuclei, hREST-GFP mRNA was injected into oocytes at least 2 h before NT. In control, GFP mRNA was injected and the GFP signals were dispersed in the embryos at 2 (Fig. 2, E and E′, n = 10) and 6 h (Fig. 2, G and G′, n = 12) post-NT. But in the hREST-GFP mRNA-injected embryos GFP signals were obviously observed in the transferred donor nuclei at 2 (Fig. 2, F and F′, n = 20) and 6 h (Fig. 2, H and H′, n = 16) post-NT. In in vitro fertilization (IVF) and PA embryos, maternal REST was not incorporated into the nuclei when they were condensed (Fig. 2, I and I′, n = 12; K and K′, n = 14) and incorporated with the nuclei when they were decondensed (Fig. 2, J and J′, n = 14; L and L′, n = 17). These results demonstrate that maternal REST is incorporated into transferred donor nuclei, suggesting it may function in the process of NT-mediated nuclear reprogramming.
FIGURE 1.
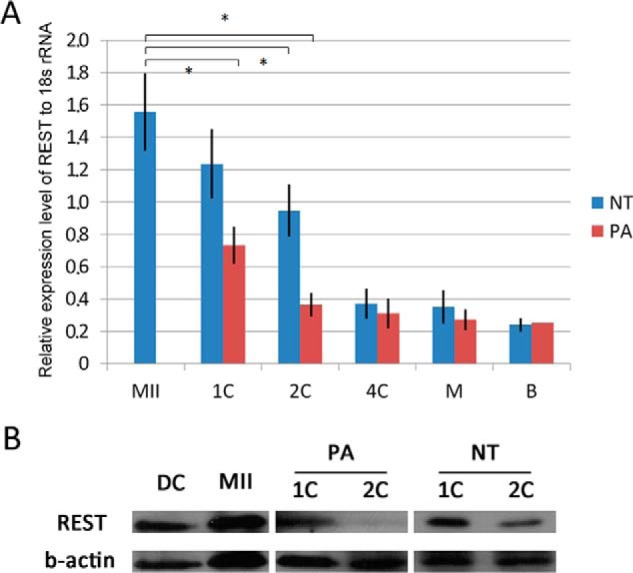
Expression pattern of REST in porcine oocytes and embryos. A, mRNA expressions of REST in porcine embryos detected by Q-PCR. Asterisk (*) indicates p < 0.001; B, protein expressions of REST in porcine oocytes, fibroblasts, and embryos checked by Western blotting. MII, MII oocytes; DC, donor cells; 1C, one-cell embryos; 2C, two-cell embryos; 4C, four-cell embryos; M, morula; B, blastocyst.
FIGURE 2.
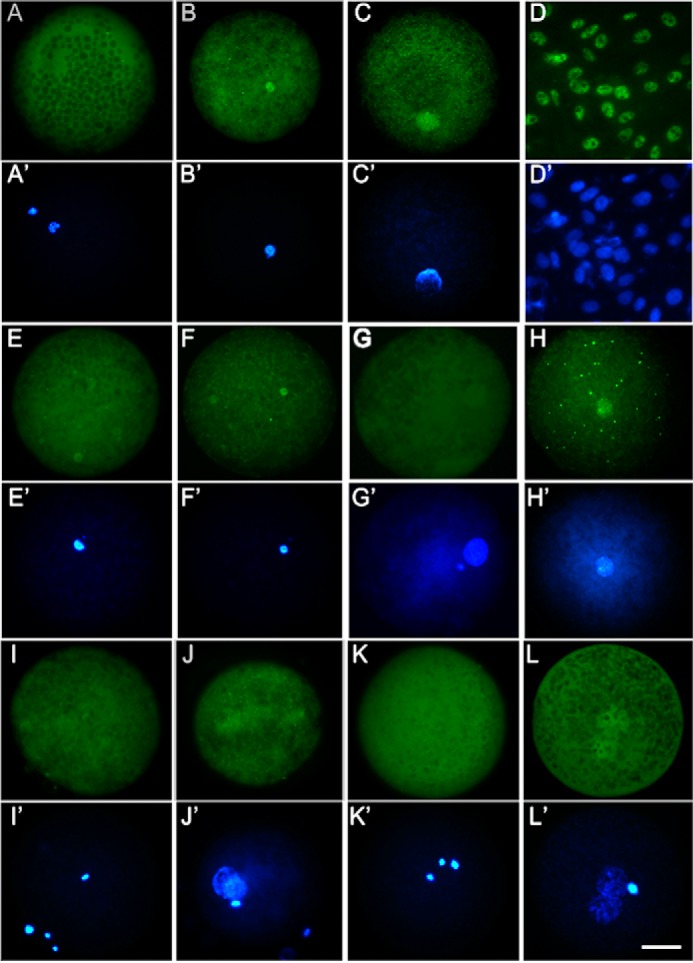
Location of REST in porcine embryos. A and A′, immunofluorescence analysis of oocyte REST; B and B′, location of REST in NT embryos at 2 h post-NT; C and C′, location of REST in NT embryos at 6 h post-NT; D and D′, expression pattern of REST in porcine fibroblasts; E and E′, detection of GFP expression pattern in NT embryos with GFP mRNA injection at 2 h post-NT; F and F′, detection of REST location in NT embryos with hREST-GFP mRNA injection at 2 h post-NT; G and G′, detection of GFP expression pattern in NT embryos with GFP mRNA injection at 6 h post-NT; H and H′, detection of REST location in NT embryos with hREST-GFP mRNA injection at 6 h post-NT; I and I′, location of REST in IVF embryos at 2 h post-fertilization; J and J′, location of REST in IVF embryos at 6 h post-fertilization; K and K′, location of REST in PA embryos at 2 h post-PA; L and L′, location of REST in PA embryos at 2 h post-PA. Green, REST or GFP; blue, DNA. Scale bar, 50 μm.
Inhibition of REST in NT Embryos
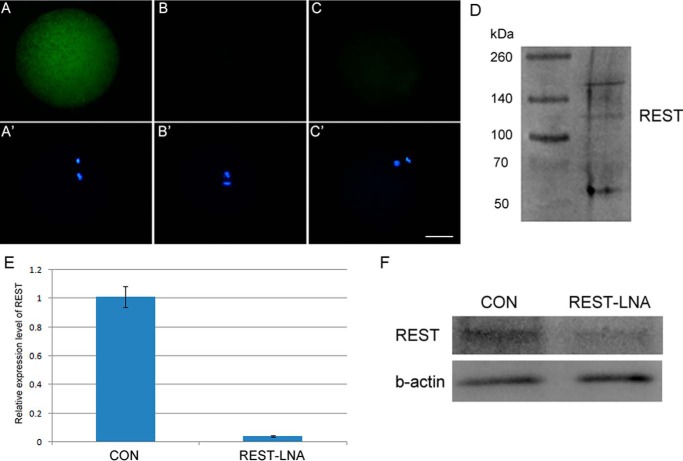
To test the role of REST in nuclear reprogramming, its function was inhibited by injection of anti-REST antibody into MII oocytes at least 2 h before NT, PA, IVF, and intracytoplasmic sperm injection (ICSI). The successful injection of the antibody used here was verified by immunofluorescence analysis (Fig. 3, A and A′, n = 15; B and B′, n = 15). In addition, no or a weak signal was detected by immunostaining the anti-REST antibody-injected oocytes at 2 h post-injection (Fig. 3, C and C′, n = 16), indicating that the injected antibody had been degraded and would not affect donor cell-derived and zygotic REST; and, effectively matching the anti-REST antibody to the porcine REST among whole oocyte proteins was verified by Western blotting (Fig. 3D). Then the in vitro developmental competency of porcine NT embryos, regarded as a stringent test of reprogramming efficiency, was examined. The rates of cleavage and cell numbers of blastocyst showed no significant difference among NT embryos with no injection (Con-NT), anti-REST antibody injection (anti-REST-NT), and IgG (IgG-NT) injection, but the proportion of anti-REST-NT embryos that developed to blastocysts was significantly lower than that of IgG-NT and Con-NT embryos (7.27 versus 20.8 and 21.53%, respectively; p < 0.05; Table 1). More anti-REST-NT embryos were arrested at the two- or four-cell stage in comparison with IgG-NT and Con-NT embryos (54.39 versus 30.33 and 28.09%, respectively; p < 0.05; Table 1). To further confirm the results, REST-specific locked nucleic acid (REST-LNA) was injected into oocytes at 33 h of in vitro maturation (IVM). Q-PCR and Western blotting analysis showed REST mRNA and protein were effectively reduced in oocytes at 42 h of IVM by REST-LNA injection (p < 0.001; Fig. 3, E and F). Consistent with anti-REST antibody, the proportion of NT embryos developed to the blastocyst stage was significantly decreased in the REST-LNA injection group (3.06 versus 17.46 and 15.37%; p < 0.05; Table 2). We also overexpressed REST by injection of hREST-GFP mRNA into oocytes, and it had no significant effect on development of NT embryos. In contrast to that in NT embryos, injections of anti-REST antibody and hREST-GFP mRNA did not affect embryonic development of IVF, ICSI, and PA embryos (Table 1). Therefore, we suggest that REST is required for successful nuclear reprogramming.
FIGURE 3.
Anti-REST antibody injection and efficient knockdown of REST by REST-LNA. A and A′, immunostaining of anti-REST antibody injected oocytes only by secondary antibody; B and B′, immunostaining of non-injected oocytes only by secondary antibody; C and C′, immunostaining of oocytes post-2 h of anti-REST antibody injection only by secondary antibody. FITC-labeled donkey anti-rabbit IgG was used as secondary antibody, which was used to immunostain anti-REST antibody. Green, anti-REST antibody; blue, DNA. Scale bar, 50 μm. D, effectively match of anti-REST antibody to the porcine REST among whole oocyte proteins was verified by Western blotting. E, efficient knockdown of REST mRNA by REST-LNA in porcine oocytes checked by Q-PCR. Significant difference was found between the two groups by Student's t test (p < 0.001). F, efficient knockdown of REST protein in porcine oocytes checked by Western blotting analysis. CON, MII oocytes; REST-LNA, REST-LNA injected MII oocytes.
TABLE 1.
Effect of maternal REST on in vitro development of NT, IVF, ICSI, and PA porcine embryos
Note: values with different superscripts within columns denote significant differences (p < 0.05).
| Groups | Repeats | Embryos | Cleavage | Blastocyst | No. of blastocyst cells | Embryos arrested at two/four-cell stage | |
|---|---|---|---|---|---|---|---|
| % | % | ||||||
| NT | Con | 3 | 122 | 86 (71.9 ± 6.67) | 25 (21.53 ± 3.15)a | 38.33 ± 8.33 | 33 (28.07 ± 3.43)a |
| IgG | 3 | 131 | 100 (78.33 ± 7.84) | 26 (20.8 ± 2.27)a | 39.43 ± 10.56 | 40 (30.33 ± 8.33)a | |
| Anti-REST | 3 | 137 | 104 (75.52 ± 11.11) | 9 (7.27 ± 3.50)b | 32.89 ± 8.50 | 72 (54.39 ± 4.11)b | |
| Con + DMSO | 3 | 169 | 136 (81.28 ± 4.73) | 30 (18.14 ± 5.72)a | 35.25 ± 8.38 | 54 (32.48 ± 5.87)a | |
| Con + SB | 3 | 166 | 130 (79.31 ± 5.25) | 45 (28.93 ± 4.33)d | 41.26 ± 7.64 | 40 (25.74 ± 3.27)a | |
| Anti-REST + DMSO | 3 | 164 | 126 (78.26 ± 6.82) | 9 (6.48 ± 7.78)b | 30.52 ± 10.65 | 91 (56.84 ± 5.21)b | |
| Anti-REST + SB | 3 | 169 | 132 (79.10 ± 5.08) | 37 (22.94 ± 5.09)a,d | 37.41 ± 9.62 | 42 (25.25 ± 4.52)a | |
| hREST-GFP mRNA | 3 | 96 | 74 (78.13 ± 10.95) | 23 (23.09 ± 4.13)a | 35.14 ± 7.50 | 26 (27.52 ± 3.88)a | |
| GFP mRNA | 3 | 87 | 64 (75.00 ± 8.86) | 16 (19.95 ± 5.86)a | 32.25 ± 10.32 | 24 (29.32 ± 3.53)a | |
| Con | 3 | 120 | 82 (70.08 ± 6.67) | 17 (15.7 ± 4.08) | 43.00 ± 7.53 | 54 (45.26 ± 7.92) | |
| IVF | IgG | 3 | 120 | 88 (73.78 ± 5.71) | 17 (15.24 ± 4.60) | 39.33 ± 8.52 | 61 (52.64 ± 11.33) |
| Anti-REST | 3 | 120 | 85 (71.92 ± 8.48) | 16 (14.41 ± 6.86) | 41.67 ± 10.48 | 55 (47.46 ± 4.27) | |
| hREST-GFP mRNA | 3 | 120 | 78 (66.67 ± 7.86) | 16 (14.17 ± 8.17) | 37.33 ± 5.75 | 52 (43.61 ± 6.15) | |
| GFP mRNA | 3 | 120 | 77 (65.52 ± 7.24) | 15 (12.67 ± 5.70) | 44.33 ± 6.67 | 60 (49.58 ± 8.25) | |
| Con | 3 | 106 | 75 (70.65 ± 4.20) | 22 (21.55 ± 10.92) | 36.41 ± 4.21 | 27 (26.56 ± 5.25) | |
| ICSI | IgG | 3 | 99 | 77 (77.78 ± 2.39) | 19 (20.37 ± 3.27) | 34.37 ± 4.52 | 24 (25.45 ± 4.24) |
| Anti-REST | 3 | 116 | 91 (78.45 ± 6.88) | 25 (21.58 ± 2.66) | 33.33 ± 8.33 | 32 (28.24 ± 4.32) | |
| hREST-GFP mRNA | 3 | 109 | 90 (82.57 ± 5.52) | 25 (23.35 ± 8.33) | 35.48 ± 5.87 | 29 (27.43 ± 7.92) | |
| GFP mRNA | 3 | 102 | 74 (73.69 ± 7.05) | 24 (22.67 ± 4.11) | 39.39 ± 7.33 | 30 (30.26 ± 5.52) | |
| Con | 3 | 120 | 105 (89.29 ± 6.04) | 50 (42.86 ± 4.19) | 43.22 ± 5.15 | 17 (15.32 ± 3.43) | |
| PA | IgG | 3 | 120 | 110 (93.33 ± 3.43) | 55 (46.67 ± 4.33) | 43.88 ± 7.67 | 20 (17.24 ± 4.67 |
| Anti-REST | 3 | 120 | 106 (89.10 ± 7.39) | 50 (43.33 ± 5.05) | 46.67 ± 7.50 | 21 (18.61 ± 4.34) | |
| hREST-GFP mRNA | 3 | 187 | 173 (93.58 ± 7.46) | 91 (49.73 ± 7.33) | 49.48 ± 8.86 | 30 (16.92 ± 2.74) | |
| GFP mRNA | 3 | 177 | 150 (85.31 ± 7.34) | 77 (44.24 ± 8.81) | 44.93 ± 8.30 | 33 (19.33 ± 3.08 | |
TABLE 2.
Embryonic development after the microinjection of LNA
Note: values with different superscripts within columns denote significant differences (p < 0.05).
| Groups | Repeats | Embryos | Blastocyst (%) | Embryos arrested at two/four-cell stage (%) | |
|---|---|---|---|---|---|
| NT | Con | 3 | 101 | 18 (17.46 ± 4.55)a | 31 (32.76 ± 5.78)a |
| REST-LNA | 3 | 98 | 3 (3.06 ± 0.12)b | 60 (60.39 ± 5.21)b | |
| Con-LNA | 3 | 102 | 16 (15.37 ± 4.72)a | 29 (27.39 ± 4.21)a | |
| Con | 4 | 160 | 21 (12.24 ± 4.08) | 74 (44.25 ± 11.78) | |
| IVF | REST-LNA | 4 | 160 | 19 (10.38 ± 4.60) | 75 (46.47 ± 11.33) |
| Con-LNA | 4 | 160 | 26 (15.82 ± 6.86) | 79 (50.35 ± 8.33) | |
| Con | 3 | 98 | 24 (24.35 ± 5.52) | 20 (22.56 ± 2.61) | |
| ICSI | REST-LNA | 3 | 86 | 19 (22.86 ± 2.98) | 23 (27.32 ± 2.22) |
| Con-LNA | 3 | 92 | 22 (25.32 ± 4,73) | 22 (24.28 ± 5.08) | |
Inhibition of REST Up-regulates TGFβ Signaling Pathway in NT Embryos
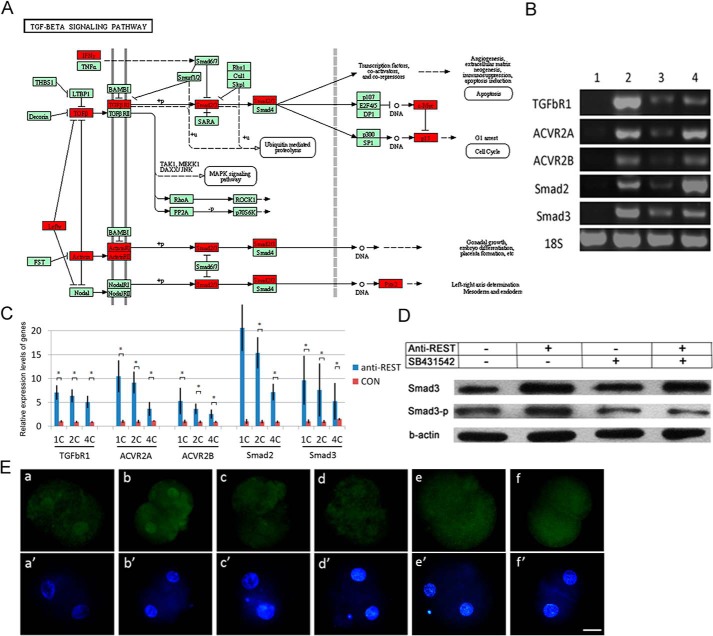
REST binds RE1 sites to repress gene expression throughout the body (11). To determine how maternal REST regulates NT-mediated nuclear reprogramming, we searched REST-targeted genes in pig. A consensus RE1 based on the sequences of 32 known RE1 elements, NT(T/C)AG(A/C)(A/G)CCNN(A/G)G(A/C)(G/S)AG, was used to screen porcine in the UCSC genome sequence database (susScr3) by using a PERL script (11). The number of putative RE1s identified in the porcine genome was 1,662, and there are 324 genes that have RE1s within 10 kb from their transcriptional start site (supplemental Table S1). Pathway analysis showed that REST putative targeted genes were widely involved in the TGFβ signaling pathway (p < 0.001, FDR = 0.1642; Fig. 4A). We therefore decided to examine expression of several key genes of the TGFβ pathway in NT embryos. By RT-PCR analysis, the expressions of TGFβR1, ACVR2A, ACVR2B, Smad2, and Smad3 were not detected in oocytes and were high in donor cell PFFs. In one-cell NT embryos, the expressions of these genes were at relatively low levels but dramatically up-regulated after REST deficiency (Fig. 4B). Consistent with RT-PCR results, Q-PCR showed that REST inhibition significantly enhanced the expressions in NT embryos from the one- to four-cell stages (Fig. 4C; p < 0.001). Moreover, we observed an increase of Smad3 and phosphorylated Smad3 (Smad3-p) expressions in two-cell REST-deficient NT embryos by Western blotting and immunofluorescence analysis (Fig. 4, D and E). The results show that inhibition of REST up-regulates the TGFβ signaling pathway in NT embryos.
FIGURE 4.
Maternal REST suppresses the TGFβ signaling pathway in porcine NT embryos. A, putative REST-targeted genes in the TGFβ signaling pathway. Putative REST targeted genes were marked by red. B, expression of REST targeted genes in TGFβ pathway checked by RT-PCR. 1, MII oocytes; 2, donor cells; 3, one-cell NT embryos; 4, anti-REST one-cell NT embryos. C, expression of REST targeted genes in the TGFβ pathway checked by Q-PCR. CON, NT embryos; anti-REST, anti-REST NT embryo. Asterisk (*) indicates p < 0.001; D, Western blotting analysis of Smad3 and Smad3-P in NT embryos; E, immunofluorescence analysis of Smad3-P in porcine two-cell NT embryos. a and a′, NT embryo (n = 14); b and b′, anti-REST NT embryo (n = 15); c and c′, NT embryo treated by SB431542 (n = 14); d and d′, anti-REST NT embryo treated by SB431542 (n = 17); e and e′, IVF embryos (n = 8); f and f′, PA embryos (n = 12). Green, Smad3-P; blue, DNA. Scale bar, 50 μm.
Up-regulation of TGFβ Pathway by Inhibition of REST Is Involved in Reprogramming Failure
To determine whether up-regulation of the TGFβ pathway in REST-deficient NT embryos leads to the failure of NT-mediated nuclear reprogramming, a specific TGFβ pathway inhibitor, SB431542, was used. We found 0.1 μm SB431542 treatment for 12 h post-activation had no negative effect on development of PA embryos (Table 3), so NT embryos were treated as the method. SB431542 treatment could dramatically decrease Smad3 and Smad3-p in normal and REST-deficient NT embryos detected by Western blotting and immunofluorescence analysis (Fig. 4, D and E), indicating the TGFβ pathway was efficiently inhibited by SB431542. Then, we asked whether TGFβ pathway inhibition in REST-deficient NT embryos can rescue the failure of nuclear reprogramming. As expected, SB431542 treatment successfully rescued the embryonic development to blastocysts (anti-REST + DMSO-NT versus Con + DMSO-NT and anti-REST + SB-NT, 6.48 versus 18.14 and 22.94%, respectively; p < 0.05; Table 1). Furthermore, the development of NT embryos was significantly enhanced by SB431542 treatment (Con + DMSO-NT versus Con + SB-NT, 18.14 versus 28.93%, respectively; p < 0.05; Table 1). These results indicate that the failure of NT-mediated reprogramming in REST-deficient NT embryos can, at least to some extent, be attributed to TGFβ pathway up-regulation and TGFβ pathway may block nuclear reprogramming. In induced pluripotent stem (iPS) cells technology, inhibition of TGFβ pathway promotes reprogramming through inducing Nanog (12). Here, we also found that the expressions of Nanog in one- and two-cell NT embryos were remarkably enhanced after TGFβ pathway inhibition checked by Q-PCR and Western blotting analysis (p < 0.001; Fig. 5, A and B). Taken together, our results indicate the REST repressing TGFβ pathway regulates NT-mediated reprogramming (Fig. 5C).
TABLE 3.
Effect of SB431542 with different concentrations on in vitro development of porcine PA embryos
Note: values with different superscripts within columns denote significant differences (p < 0.05).
| Groups | Repeats | Embryos | Cleavage | Blastocyst |
|---|---|---|---|---|
| % | ||||
| Con. | 3 | 121 | 98 (79.91 ± 6.36)a | 32 (27.64 ± 7.09)a |
| 1 μm | 3 | 119 | 69 (56.38 ± 3.19)b | 16 (14.53 ± 7.81)b |
| 0.5 μm | 3 | 124 | 84 (66.94 ± 5.48)c | 22 (18.42 ± 4.53)b |
| 0.1 μm | 3 | 131 | 96 (74.48 ± 5.42)a,c | 34 (26.41 ± 4.19)a |
FIGURE 5.
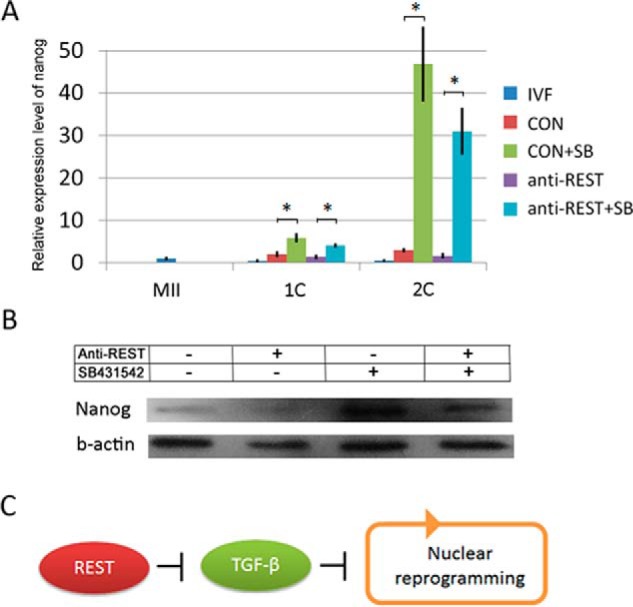
TGFβ signaling pathway represses expression of Nanog in porcine NT embryos. A, mRNA expression pattern of Nanog in porcine embryos checked by Q-PCR. MII, MII oocytes; IVF, IVF embryos; CON, NT embryos; CON + SB, NT embryo treated by SB431542; anti-REST, anti-REST NT embryos; anti-REST + SB, anti-REST NT embryo treated by SB431542. Asterisk (*) indicates p < 0.001; B, Western blotting analysis of Nanog in porcine NT embryos at the two-cell stage. C, proposed model of maternal REST on nuclear reprogramming. REST promotes nuclear reprogramming by suppressing TGFβ signaling pathway.
Discussion
Differentiated cell nuclei can be reprogrammed to a pluripotent state by NT into oocytes, iPS technology, and cell fusion with ESCs (2, 3, 13, 14). NT mediated-reprogramming has been proven to be the most efficient way (15), and complex transcription factors are significant in the process. Therefore, identification and characterization of these factors will provide us important information on nuclear reprogramming. In the study, we found that REST was rich in oocytes and required for NT-mediated reprogramming through inhibiting TGFβ pathway in pig.
REST is a zinc finger protein, and represses neuronal gene transcription in nonneuronal cells (16–18). Mice that lack REST exhibit malformations in the developing nervous system and die by embryonic day 11.5 of embryogenesis. However, these mice appear normal until embryonic day 9.5 (16). This is consistent with our observations that REST deficiency has no obvious effect on early embryonic development of IVF and PA embryos. REST was found to incorporate into the condensed and decondensed transferred donor nuclei in one-cell NT embryos; and REST inhibition by anti-REST antibody and REST-LNA injections remarkably decreased the in vitro developmental competency of NT embryos. So, we believe that REST is required for NT-mediated reprogramming. REST can bind RE1 sites to repress gene expression (5). In porcine genome, 1,662 RE1s and 324 corresponding genes were identified. These numbers are comparable with human and mouse (11). Pathway analysis showed that TGFβ signaling pathway could be suppressed by REST. Confirming that, up-regulation of the TGFβ pathway was observed in REST-deficient NT embryos, revealing TGFβ pathway is suppressed by REST in NT embryos.
TGFβ pathway has been implicated in the development and maintenance of various organs (19, 20), and is necessary for the maintenance of self-renewal and pluripotency of both human and mouse ESCs (21). During embryonic development, the pathway is believed to play critical roles in the specification of cell identities in embryonic and extra embryonic lineages of the post-implantation embryo (19, 22–24). Before implantation, embryonic phenotypes for loss-of-function mutation of the pathway are not detected (20). Transcriptome sequencing and analyzing pig embryos in vivo and in vitro also show that the TGFβ pathway is not active well before maternal zygotic transition at the four- to eight-cell stages (25). Those data suggest the function of the TGFβ pathway is suppressed during early embryonic development. In the study, activation of the TGFβ pathway at a certain level was detected in NT embryos, but not in IVF and PA embryos, and the activation in NT embryos could be attributed to donor cell PFFs in which the TGFβ pathway activated. Here, we proposed that REST was required to silence the TGFβ pathway in NT embryos, and in IVF and PA embryos, the pathway was unactivated and, in the regard, REST was not needed. So, the function of REST is only necessary for NT embryos, not for IVF and PA embryos. In addition, inhibition of REST up-regulated the TGFβ signaling pathway in NT embryos. So, we believe inhibition of the TGFβ pathway by REST may be involved in successful nuclear reprogramming.
To test the point, SB431542 was used to treat NT embryos. SB431542 treatment could successfully rescue the development failure of REST-deficient NT embryos and improve developmental potential of normal NT embryos. The results reveal that the TGFβ pathway may have a negative effect on NT-mediated reprogramming. It has been demonstrated that TGFβ pathway inhibition can replace Sox2 and promote the completion of iPS reprogramming through induction of the reprogramming factor Nanog (12). Coincidentally, high level Nanog expression was observed in the TGFβ pathway-inhibited NT embryos. Previous reports have been shown that inhibition of the TGFβ pathway by SB431542 increases Bmp signaling (26) and Bmp signaling induces Nanog expression (27). The cross-talk between TGFβ and Bmp signaling may result in Nanog induction. We conclude that inhibition of the TGFβ pathway improves NT-mediated reprogramming perhaps by up-regulation of Nanog.
So far, many studies have focused on identification of reprogramming factors (28–35). In the study, we demonstrate that REST acts as a repressor of the TGFβ pathway and is critical for NT-mediated nuclear reprogramming, and inhibition of the TGFβ pathway by SB431542 treatment promotes the reprogramming efficiency in pig. In addition to better understanding the detailed mechanism of how TGFβ pathway inhibition contributes to increased reprogramming efficiency, whether or not our observation can be generally applied to other animal species warrants future investigation. The simplicity of SB431542 treatment during NT makes the testing of our approach worthwhile. If so, SB431542 treatment has the potential to enhance cloning efficiency in a broad range of mammalian species, including humans. Our method could hold great promise for human therapeutic cloning (36).
Experimental Procedures
hREST-GFP mRNA in Vitro Transcription and Plasmid Construction
pEGFP-C1 and hREST-GFP (RG211570, Origene) plasmids were linearized before in vitro transcription. RNA synthesis and poly(A) tailing were carried out with a MEGA script T7 Kit (Ambion, Carlsbad, CA) according to the manufacturer's instructions.
Oocyte and Embryo Manipulations
Before NT, IVF, ICSI, and PA, 10 picoliters of 1 mg/ml of anti-REST antibody (ab21635, Abcam), 100 ng/μl of GFP and hREST-GFP mRNAs solution were injected into matured oocytes. After injection, oocytes were kept for at least 2 h before manipulations, which allows the antibody to bind endogenous REST. Moreover, 10 picoliters of 10 μm REST-LNA (Locked Nucleic Acid, Exiqon) was injected into porcine oocytes with the first polar body collected at 33 h of IVM (35), and the oocytes matured at 42 h were used for NT, ICSI, and IVF. The procedure for porcine NT, PA, and IVF has been described previously (35). After fusion, 0.1 μm SB431542 was used to treat NT embryos for 12 h. Cumulus cell-free oocytes were directly activated by the same parameters as for the somatic cell nuclear transfer procedure to produce PA embryos.
ICSI was performed by using an inverted microscope (Olympus IX71, Olympus Optical Co. Ltd.) with a piezoactuated micromanipulator (PMAS-CT150; Prime Tech Ltd, Tsuchiura, Japan). A 100-ml drop of HEPES-M199 containing 0.5% (v/v) FBS and a 20-ml drop of 4% (w/v) polyvinylpyrrolidone (Mr 360,000; Sigma) were placed in a 35-mm dish and covered with mineral oil. Next, 20–30 oocytes were placed in the 100-ml drop and the sperm suspension was transferred to the polyvinylpyrrolidone drop. The oocyte was positioned with a holding pipette so that the first polar body was at the 6 or 12 o'clock positions. A single sperm was injected into the cytoplasm with a micropipette. Activation of ICSI zygotes was induced with 2DC pulses of 1.2 kV/cm for 30 ms on a BTX Elector-Cell Manipulator 2001 (BTX, San Diego, CA).
The embryos were cultured in porcine zygote medium-3 at 39 °C in 5% CO2 in air. The cleavage and blastocyst rates were assessed at 48 and 156 h after activation, and the number of blastocyst cells was examined by nuclear staining with 5 μg/ml of Hoechst 33342.
RE1 Database Construction
A search was performed of the porcine genome GenBank formatted DNA sequence flat files (downloaded from UCSC susScr3 version) by using a PERL script constrained by a core 17 nucleotide regular expression pattern. This pattern represents an RE1 consensus sequence, derived from alignment of 32 known RE1 sequences containing degeneracies reflecting known variations (12). The search output and corresponding annotations or external references (SWISS-PROT Version 40.43 and TREMBL Version 22.13 protein sequence databases) were used to assign gene description and determination of annotated genes within 100 kb on either strand.
Q-PCR Analysis
Total RNA was extracted using the PureLink TM Micro-to-Midi System (Invitrogen) according to the manufacturer's instructions, and reverse transcription was used to generate cDNAs using the Prime Script TM RT Reagent kit (TaKaRa). Real-time PCR was performed using SYBR Premix Ex TaqTM (TaKaRa) and the 7500 Real-time PCR System (Applied Biosystems). The reaction parameters were 95 °C for 30 s followed by 40 two-step cycles of 95 °C for 5 s and 60 °C for 34 s. All the primer pairs used for PCR amplification are shown in Table 4. Ct values were calculated using Sequence Detection System software (Applied Biosystems), and the amount of target sequence normalized to the reference sequence was calculated as 2−▵▵Ct.
TABLE 4.
The primer list
| Gene | Primer sequence (5′-3′) | Length | Accession number |
|---|---|---|---|
| bp | |||
| 18S rRNA | F: TCCAATGGATCCTCGCGGAA | 149 | NR002170 |
| R: GGCTACCACATCCAAGGAAG | |||
| REST | F: GAGGCGGAGTCTGAGGAGCAG | 192 | GU991112.1 |
| R: GTGGTCGTAGCGGTTGGTGTTG | |||
| ACVR2A | F: AGGTGCTATACTTGGTAGATCAGAAACTC | 187 | NM_001204765.1 |
| R: CAGCCAACAACCTTGCTTCACTA | |||
| ACVR2B | F: CACGGGAGTGCATCTACTACAACGCC | 165 | NM_001005350.1 |
| R: CGTCTAGCCAGCAGCCCTTCTTCAC | |||
| ACVR1B | F: GAGTTATGAGGCGCTGCGGGTG | 112 | NM_001195322.1 |
| R: GCTGAGCTGGGACAGGGTCTTCTTG | |||
| TGFBR1 | F: AGTAAGACATGATTCAGCCACAGATACAA | 172 | NM_001038639.1 |
| R: AGCTATTTCCCAGAATACTAAGCCCATT | |||
| Smad2 | F: GCTGCTCTTCTGGCTCAGTCCG | 123 | NM_001256148.1 |
| R: TACTGTCTGCCTTCGGTATTCTGCTC | |||
| Smad3 | F: CAGCGACCACCAGATGAACCACAG | 145 | NM_214137.1 |
| R: CTCGTAGTAGGAGATGGAGCACCAGAAG | |||
| Nanog | F: CCTCCATGGATCTGCTTATTC | 118 | AY596464 |
| R: CATCTGCTGGAGGCTGAGGT |
Western Blotting
The procedure for Western blotting analysis has been described previously (11). Oocytes or embryos without zona pellucida were transferred to cold 40 mm sodium phosphate, pH 7.6, containing 50 mm NaCl, 50 μm sodium orthovanadate, 10 mm sodium fluoride, 20 μm MG132, 2 μm matrix metalloprotease inhibitor III (444264, Calbiochem), and 1% protease inhibitor mixture III (539134, Calbiochem). Homogenization was carried out with a Tekmar homogenizer by three 15-s bursts with a minute cooling between. Homogenates were centrifuged for 1 h at 100,000 × g. The supernatant solutions are referred to as “soluble” fractions. The pellets were suspended in 0.2 ml of complete buffer containing 1% ASB-14 and were mixed every 15 min for 2 h with Radnoti glass pestles (Unitek, Monrovia, CA). After centrifugation for 1 h at 100,000 × g, the supernatants, referred to as “membrane extracts” were removed, and the pellets were discarded. About 50 embryos of each soluble and membrane extract for each gene were separated by lithium dodecyl sulfate-polyacrylamide gel electrophoresis on 4–12% BisTris NuPAGE gels (the gels have been run under the same experimental conditions) and transferred to PVDF membranes (Invitrogen); nonspecific binding was blocked by overnight incubation in 1% casein in PBS at room temperature. Antibodies against REST (ab21635, Abcam), Smad3 (ab40854, Abcam), Smad3-p (ab118825, Abcam), and Nanog (500-P236, Peprotech) were used, and β-actin (A1978, Sigma) served as a loading control. After a 2-h incubation at room temperature with secondary antibodies, protein bands were detected by enhanced chemiluminescence with the RPN2108 kit (Amersham Biosciences) and BioMax Light film (Eastman Kodak Co.).
Immunofluorescence Analysis
Oocytes and embryos without zonae pellucidae were washed twice in PBS, then fixed in freshly prepared 4% paraformaldehyde in PBS, permeabilized in 1% Triton X-100 in PBS, and left in blocking solution (1% BSA in PBS) for 1 h. For immunolabeling, the embryos were incubated overnight with anti-REST (ab21635, Abcam), anti-Nanog (500-P236, Peprotech), or anti-Smad3-p (ab118825, Abcam) antibodies; washed three times, and incubated for 1 h with secondary antibody FITC-labeled donkey anti-mouse IgG (A21202, Invitrogen) diluted 1:1000 with blocking solution. Immunofluorescence of injected oocytes and one-cell NT embryos without anti-REST antibody (only secondary antibody) was used to analyze REST antibody injection and degradation. Samples were washed and counterstained with 5 μg/ml of Hoechst 33342. Fluorescence was detected and imaged using a Nikon fluorescence microscope.
Statistical Analysis
Statistical analysis was performed using SPSS 13.0 for MicroSoftTM Windows. Data are shown as the mean ± S.D. One-way analysis of variance was used to assess any differences between groups. The Duncan method was used for pairwise comparisons followed by a Bonferroni correction. p < 0.05 (two-tailed) was considered statistically significant.
Author Contributions
Z. H. L. and Q. R. K. designed and conceived the experiments; B. T. X., Q. R. K., and H. Z. conducted the oocyte and embryo manipulations; Q. R. K. and T. Q. H. conducted molecular experiments; J. Y. L. contributed to bioinformation analysis; R. Y. W. conducted the cell manipulations. Z. H. L. and Q. R. K. wrote and all authors reviewed the manuscript.
Supplementary Material
Acknowledgment
We gratefully acknowledge Dr. Jilong Liu in Huanan Agricultural University for proteomic analysis.
This work was support by National Natural Science Foundation of China Grant 31470079, National Basic Research Program of China Grant 2011CBA01006, the Fund for Fostering Talents in Basic Science of the National Natural Science Foundation of China Grant J1210069, “Academic Backbone” Project of Northeast Agricultural University Grant 15XG19, and Doctoral Research Foundation of Northeast Agricultural University Grants 2009RC56 and 2012RCB06. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- ESC
- embryonic stem cells
- NT
- nuclear transfer
- RE1
- repressor element 1
- REST
- RE1-silencing transcription factor
- PA
- parthenogenetic activation
- PFF
- porcine fetal fibroblasts
- IVF
- in vitro fertilization
- ICSI
- intracytoplasmic sperm injection
- IVM
- in vitro maturation
- LNA
- locked nucleic acid
- Q-PCR
- quantitative PCR
- iPS
- induced pluripotent stem
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Gurdon J. B., Elsdale T. R., and Fischberg M. (1958) Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182, 64–65 [DOI] [PubMed] [Google Scholar]
- 2. Wilmut I., Schnieke A. E., McWhir J., Kind A. J., and Campbell K. H. (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 4. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 5. Ooi L., and Wood I. C. (2007) Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8, 544–554 [DOI] [PubMed] [Google Scholar]
- 6. Perera A., Eisen D., Wagner M., Laube S. K., Künzel A. F., Koch S., Steinbacher J., Schulze E., Splith V., Mittermeier N., Müller M., Biel M., Carell T., and Michalakis S. (2015) TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 11, 283–294 [DOI] [PubMed] [Google Scholar]
- 7. Negrini S., Prada I., D'Alessandro R., and Meldolesi J. (2013) REST: an oncogene or a tumor suppressor? Trends Cell Biol. 23, 289–295 [DOI] [PubMed] [Google Scholar]
- 8. Tokoyoda K., and Radbruch A. (2012) Signals controlling rest and reactivation of T helper memory lymphocytes in bone marrow. Cell. Mol. Life Sci. 69, 1609–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kagalwala M. N., Singh S. K., and Majumder S. (2008) Stemness is only a state of the cell. Cold Spring Harb. Symp. Quant. Biol. 73, 227–234 [DOI] [PubMed] [Google Scholar]
- 10. Ding N., Zhou H., Esteve P. O., Chin H. G., Kim S., Xu X., Joseph S. M., Friez M. J., Schwartz C. E., Pradhan S., and Boyer T. G. (2008) Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell 31, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruce A. W., Donaldson I. J., Wood I. C., Yerbury S. A., Sadowski M. I., Chapman M., Göttgens B., and Buckley N. J. (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. U.S.A. 101, 10458–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ichida J. K., Blanchard J., Lam K., Son E. Y., Chung J. E., Egli D., Loh K. M., Carter A. C., Di Giorgio F. P., Koszka K., Huangfu D., Akutsu H., Liu D. R., Rubin L. L., and Eggan K. (2009) A small-molecule inhibitor of TGF-β signaling replaces Sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tada M., Morizane A., Kimura H., Kawasaki H., Ainscough J. F., Sasai Y., Nakatsuji N., and Tada T. (2003) Pluripotency of reprogrammed somatic genomes in embryonic stem hybrid cells. Dev. Dyn. 227, 504–510 [DOI] [PubMed] [Google Scholar]
- 14. Tada M., Takahama Y., Abe K., Nakatsuji N., and Tada T. (2001) Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 [DOI] [PubMed] [Google Scholar]
- 15. Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., et al. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z. F., Paquette A. J., and Anderson D. J. (1998) NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142 [DOI] [PubMed] [Google Scholar]
- 17. Qureshi I. A., and Mehler M. F. (2010) Impact of nuclear organization and dynamics on epigenetic regulation in the central nervous system: implications for neurological disease states. Ann. N.Y. Acad. Sci. 1204, E20–E37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qureshi I. A., Gokhan S., and Mehler M. F. (2010) REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle 9, 4477–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robertson E. J. (2014) Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin. Cell Dev. Biol. 32, 73–79 [DOI] [PubMed] [Google Scholar]
- 20. Papanayotou C., and Collignon J. (2014) Activin/Nodal signalling before implantation: setting the stage for embryo patterning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, pii. 21030539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park K. S. (2011) Tgf-β family signaling in embryonic stem cells. Int. J. Stem Cells 4, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossant J., and Tam P. P. (2009) Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713 [DOI] [PubMed] [Google Scholar]
- 23. Shen M. M. (2007) Nodal signaling: developmental roles and regulation. Development 134, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 24. Takaoka K., Yamamoto M., and Hamada H. (2011) Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nat. Cell Biol. 13, 743–752 [DOI] [PubMed] [Google Scholar]
- 25. Cao S., Han J., Wu J., Li Q., Liu S., Zhang W., Pei Y., Ruan X., Liu Z., Wang X., Lim B., and Li N. (2014) Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 15, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu R. H., Sampsell-Barron T. L., Gu F., Root S., Peck R. M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R., and Thomson J. A. (2008) NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suzuki A., Raya A., Kawakami Y., Morita M., Matsui T., Nakashima K., Gage F. H., Rodríguez-Esteban C., and Izpisúa Belmonte J. C. (2006) Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kikyo N., Wade P. A., Guschin D., Ge H., and Wolffe A. P. (2000) Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289, 2360–2362 [DOI] [PubMed] [Google Scholar]
- 29. Jullien J., Astrand C., Halley-Stott R. P., Garrett N., and Gurdon J. B. (2010) Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl. Acad. Sci. U.S.A. 107, 5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamada H., Van Thuan N., Reed P., Nelson D., Katoku-Kikyo N., Wudel J., Wakayama T., and Kikyo N. (2006) Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell. Biol. 26, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wen D., Banaszynski L. A., Liu Y., Geng F., Noh K. M., Xiang J., Elemento O., Rosenwaks Z., Allis C. D., and Rafii S. (2014) Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111, 7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyamoto K., Nagai K., Kitamura N., Nishikawa T., Ikegami H., Binh N. T., Tsukamoto S., Matsumoto M., Tsukiyama T., Minami N., Yamada M., Ariga H., Miyake M., Kawarasaki T., Matsumoto K., and Imai H. (2011) Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc. Natl. Acad. Sci. U.S.A. 108, 7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hansis C., Barreto G., Maltry N., and Niehrs C. (2004) Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr. Biol. 14, 1475–1480 [DOI] [PubMed] [Google Scholar]
- 34. Gonda K., Fowler J., Katoku-Kikyo N., Haroldson J., Wudel J., and Kikyo N. (2003) Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell. Biol. 5, 205–210 [DOI] [PubMed] [Google Scholar]
- 35. Kong Q., Xie B., Li J., Huan Y., Huang T., Wei R., Lv J., Liu S., and Liu Z. (2014) Identification and characterization of an oocyte factor required for porcine nuclear reprogramming. J. Biol. Chem. 289, 6960–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tachibana M., Amato P., Sparman M., Gutierrez N. M., Tippner-Hedges R., Ma H., Kang E., Fulati A., Lee H. S., Sritanaudomchai H., Masterson K., Larson J., Eaton D., Sadler-Fredd K., Battaglia D., et al. (2013) Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.