Abstract
Transcription factor tonicity-responsive enhancer-binding protein (TonEBP/NFAT5) is critical for osmo-adaptation and extracellular matrix homeostasis of nucleus pulposus (NP) cells in their hypertonic tissue niche. Recent studies implicate TonEBP signaling in inflammatory disease and rheumatoid arthritis pathogenesis. However, broader functions of TonEBP in the disc remain unknown. RNA sequencing was performed on NP cells with TonEBP knockdown under hypertonic conditions. 1140 TonEBP-dependent genes were identified and categorized using Ingenuity Pathway Analysis. Bioinformatic analysis showed enrichment of matrix homeostasis and cytokine/chemokine signaling pathways. C-C motif chemokine ligand 2 (CCL2), interleukin 6 (IL6), tumor necrosis factor (TNF), and nitric oxide synthase 2 (NOS2) were studied further. Knockdown experiments showed that TonEBP was necessary to maintain expression levels of these genes. Gain- and loss-of-function experiments and site-directed mutagenesis demonstrated that TonEBP binding to a specific site in the CCL2 promoter is required for hypertonic inducibility. Despite inhibition by dominant-negative TonEBP, IL6 and NOS2 promoters were not hypertonicity-inducible. Whole-disc response to hypertonicity was studied in an ex vivo organ culture model, using wild-type and haploinsufficient TonEBP mice. Pro-inflammatory targets were induced by hypertonicity in discs from wild-type but not TonEBP-haploinsufficient mice. Mechanistically, NF-κB activity increased with hypertonicity and was necessary for hypertonic induction of target genes IL6, TNF, and NOS2 but not CCL2. Although TonEBP maintains transcription of genes traditionally considered pro-inflammatory, it is important to note that some of these genes also serve anabolic and pro-survival roles. Therefore, in NP cells, this phenomenon may reflect a physiological adaptation to diurnal osmotic loading of the intervertebral disc.
Keywords: chemokine, cytokine, inflammation, NF-kappa B (NF-KB), NFAT transcription factor, NFAT5, hyperosmolarity, intervertebral disc, nucleus pulposus
Introduction
The intervertebral disc is well suited to fulfill its mechanical role in the human spine, where it permits flexion and rotation, and absorbs compressive loads (1). The matrix-rich nucleus pulposus (NP)2 at the center of the disc gives the tissue its ability to resist compression through high osmotic swelling pressure (2–4), loss of which correlates with degeneration and back pain (5). Although the high fixed charge density of the aggrecan-rich matrix allows the nucleus pulposus its water-imbibing properties, it also results in a hypertonic environment for NP cells. Importantly, tonicity of the extracellular environment fluctuates widely with diurnal cycle: water is forced out of the disc during the day when the spine is loaded and imbibed during the unloaded phase at night (6).
In mammalian cells, a key transcription factor TonEBP (NFAT5) is activated by elevated hypertonicity and promotes transcription of genes that produce or transport organic osmolytes (7). In addition, TonEBP controls transcription of several genes that are important for cell survival under hypertonic conditions independent of osmolyte accumulation (8–11). Recently, studies have shown that TonEBP participates in hypertonicity, as well as LPS-mediated induction of certain pro-inflammatory genes in macrophages and other cell types (12–16). In NP cells we have shown previously that TonEBP is important for osmoregulation and survival under hypertonic conditions (17). In addition, TonEBP regulates expression of extracellular matrix-related genes Acan and B3gat3 in NP cells and Col1 and Col2 in chondrocytes (17–19). However, little is known regarding the broader functions of TonEBP in the hypertonic niche of the NP.
The goal of this study was to elucidate whether TonEBP promotes transcription of inflammation-related genes in NP cells, even under physiologically relevant hypertonic conditions. RNA sequencing showed that TonEBP controls activities of several inflammation- and matrix turnover-related pathways. Using in vitro and ex vivo approaches and employing TonEBP haploinsufficient mice, our results demonstrate that TonEBP and NF-κB participate in activation of pro-inflammatory genes in response to hypertonic stimulus in NP cells. We hypothesize that this phenomenon reflects a physiological adaptation of NP cells to diurnal osmotic loading of the intervertebral disc and may be critical for cellular homeostasis.
Results
RNA Sequencing Reveals TonEBP as a Regulator of Pro-inflammatory Genes under Physiological Hypertonicity in NP
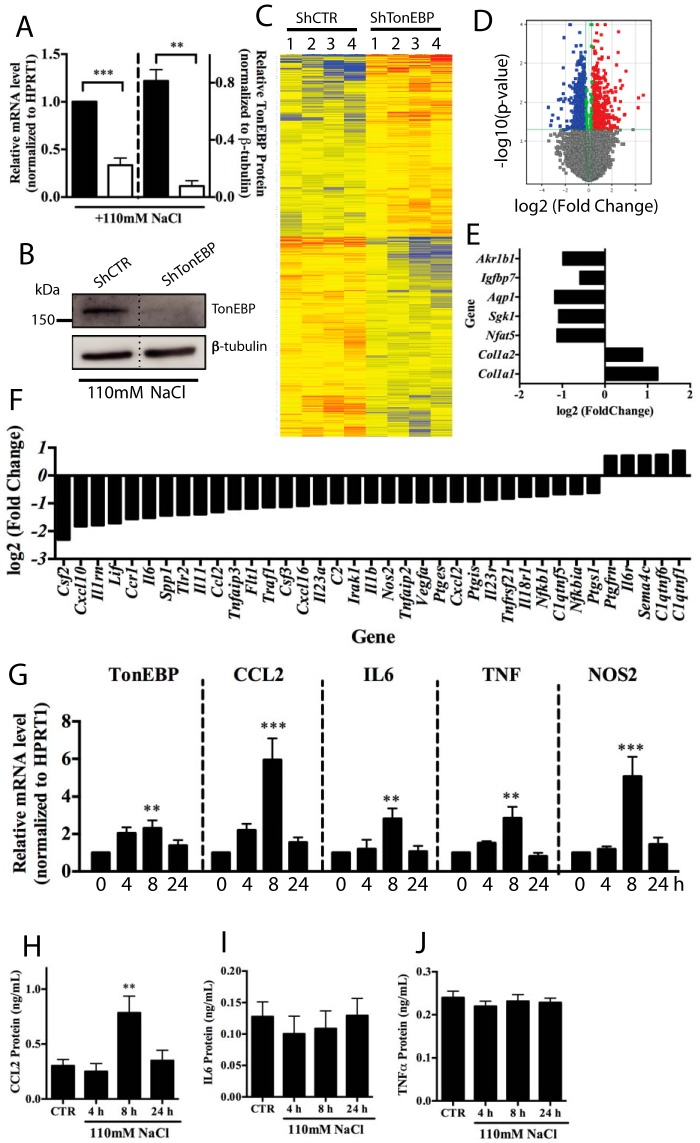
NP cells were transduced with either control (ShCTR) or TonEBP-specific (ShTonEBP) shRNA and cultured in hypertonic medium for 8 h to recapitulate the physiological state of the NP. We verified significant reduction in TonEBP mRNA (Fig. 1A) and protein levels (89.8 ± 5% decrease in protein) (Fig. 1, A and B). RNA sequencing results represented by the heat map and volcano plot (Fig. 1, C and D) depict 1140 differentially expressed transcripts (adjusted p value < 0.05), with 73 showing a log 2 (-fold change) >1.2. We validated our dataset by examining levels of known TonEBP targets Akr1b1, Igfbp7, Aqp1, Sgk1, Col1a1, and Col1a2, whose expression level changes matched previously reported data (Fig. 1E) (7–9, 11, 19). Ingenuity Pathway Analysis (IPA) software was used to identify the top pathways and functions associated with the list of differentially expressed genes (supplemental Tables 1 and 2, supplemental Fig. 1). Due to enrichment of catabolic and cytokine/chemokine-related pathways, we generated a list of all differentially expressed inflammation-related genes to investigate the role of TonEBP controlling these processes (Fig. 1F). Three genes from the list, CCL2, IL6, and NOS2, associated with disc degeneration, along with TNF, a known TonEBP target in fibroblasts (13), were further investigated.
FIGURE 1.
RNA sequencing of TonEBP knockdown NP cells reveals regulation of pro-inflammatory transcripts. A and B, TonEBP mRNA and protein levels decrease after transduction with TonEBP-directed shRNA. C and D, heat map (C) and volcano plot (D) depicting differentially expressed genes between control and TonEBP-shRNA transduced NP cells. E, RNA sequencing of TonEBP-silenced samples confirms decreases in TonEBP (NFAT5), along with changes in positively regulated (Akr1b1, Igfbp7, Aqp1, and Sgk1) and negatively regulated (Col1a1 and Col1a2) transcriptional targets. Log 2 (-fold change) values are shown. F, log 2 (-fold change) values for inflammation-related transcripts that are differentially expressed between control and TonEBP knockdown. G, NP cells cultured under hypertonic conditions (110 mm NaCl added) up to 24 h show increased mRNA levels of TonEBP, CCL2, IL6, TNF, and NOS2 at 8 h after stimulus. H–J, protein levels of CCL2 (H), IL6 (I), and TNFα (J) cultured under hypertonic conditions for up to 24 h. CCL2 shows significant induction at 8 h following stimulation. Levels of IL6 and TNFα do not show any change. In B, one representative Western blot image is shown; lanes between those shown were removed from the image; knockdown experiments were repeated three independent times. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Hypertonicity Causes Temporal Changes in Pro-inflammatory Gene Expression in NP
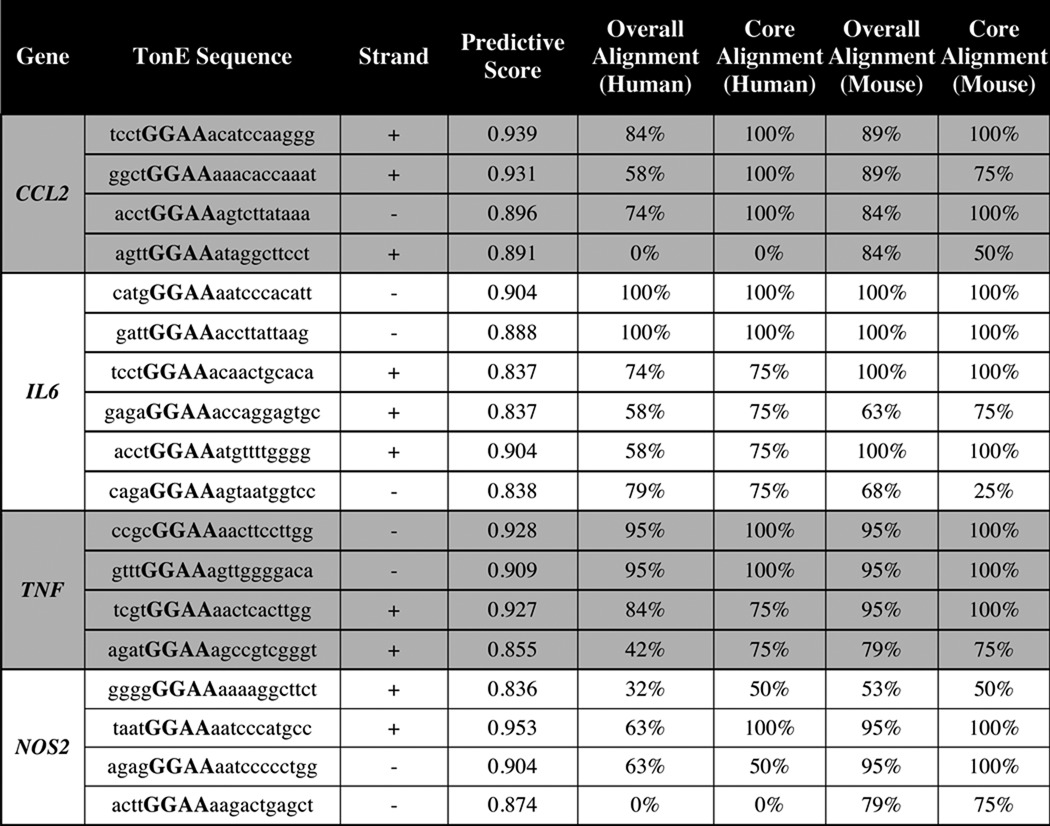
We tested whether hypertonicity induces pro-inflammatory genes by culturing NP cells under hypertonic conditions for up to 24 h. TonEBP mRNA was significantly induced by 8 h (Fig. 1G) with a concomitant induction in CCL2, IL6, TNF, and NOS2, which subsided by 24 h (Fig. 1G). CCL2 protein was also higher at 8 h after the addition of NaCl (Fig. 1H). Interestingly, levels of IL6 and TNFα protein were unaffected by hypertonicity (Fig. 1, I and J), suggesting that acute transcript increases may be required for maintenance of baseline protein expression under hypertonicity. To explore whether this regulation involved TonEBP, we analyzed the 2-kb promoter upstream of the transcription start site of these genes for potential TonEBP binding motifs (TonE). Analysis showed several predicted TonE in all promoters evaluated (Table 1). To gain an understanding of how well these TonE were conserved between species, we performed Multiz alignment of the rat, human, and mouse genomes and calculated the degree of alignment of the whole TonE (overall alignment) and alignment of the core binding sequence of the TonE (core alignment).
TABLE 1.
Prediction of TonE's in first 2-kb proximal promoters of inflammatory genes and their conservation between rat, human, and mouse genomes
CCL2, IL6, and NOS2 Promoters Are Differentially Regulated by Hypertonicity and TonEBP in NP
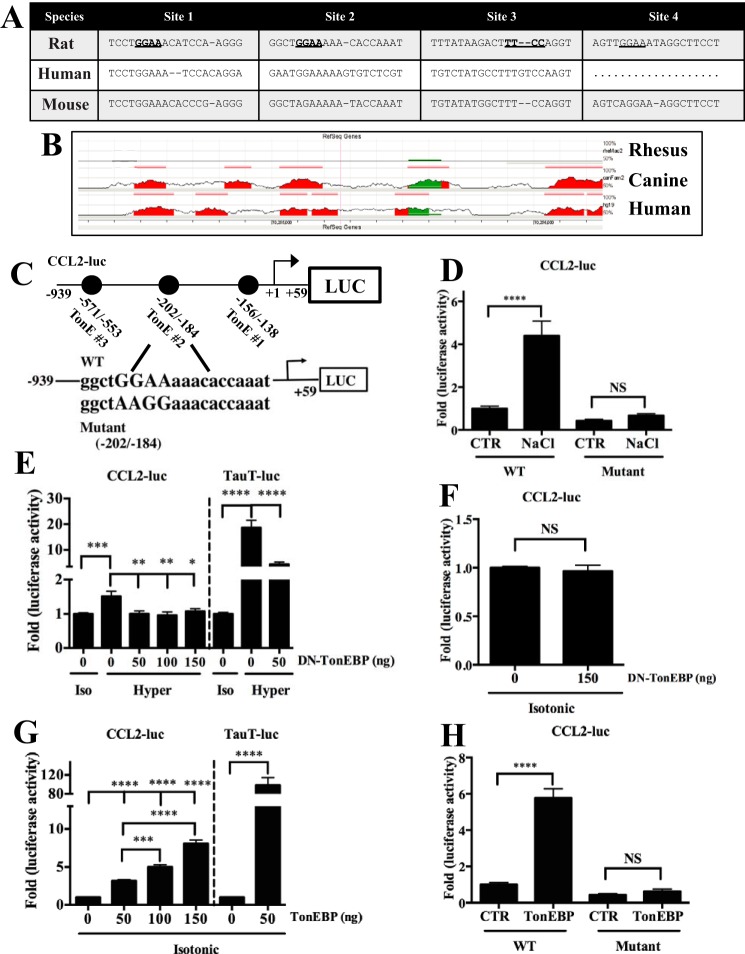
The presence of several, highly conserved predicted TonE in the CCL2 proximal promoter led us to study this promoter in more depth. We studied these TonE and overall conservation of the promoter using the Evolutionary Conserved Regions browser (Fig. 2, A and B). Based on these findings, we experimentally examined the responsiveness of the CCL2 promoter, which contains three potential TonE, to manipulation of tonicity and TonEBP levels. To test whether TonEBP bound to the promoter, we mutated TonE #2, which has a high predictive score and high core binding site conservation and is active in kidney cells (12) (Fig. 2C). Activity of the wild-type CCL2 promoter was induced by hypertonicity, and mutation of TonE #2 abolished this increase (Fig. 2D). Expression of dominant-negative TonEBP (DN-TonEBP) also inhibited hypertonic induction of the wild-type promoter (Fig. 2E), similar to activity of the taurine transporter (TauT) promoter, a well characterized TonEBP target. Under isotonic conditions, DN-TonEBP did not affect the wild-type CCL2 promoter (Fig. 2F). However, overexpression of TonEBP under isotonic conditions led to a dose-dependent increase in CCL2 promoter activity (Fig. 2G); the TonE-mutated CCL2 promoter was unresponsive (Fig. 2H).
FIGURE 2.
Activity of the CCL2 promoter is positively regulated by TonEBP in NP cells. A, TonE predicted using Genomatix MatInspector in the first 2 kb of the proximal promoter of the rat CCL2 gene were compared with the human and mouse CCL2 promoters using Multiz alignments. Sites 1 and 2 show a high degree of conservation between all species compared. B, analysis using the Evolutionary Conserved Regions Browser demonstrates sequence conservation in the 2-kb CCL2 promoter between rat, human, canine, and rhesus. ECR threshold is set at 80%. CCL2 promoter: red, intergenic regions; green, transposons and simple repeats. Height of peaks represents percentage of identity between the compared genomes. C, diagrams showing predicted TonE #1–3 in the proximal promoter of the rat CCL2 luciferase reporter construct spanning −939/+59. Both wild-type (WT) and #2 TonE-mutated (Mutant) sequences are shown. D, CCL2 promoter activity is increased in NP cells transfected with wild-type CCL2 reporter and exposed to hypertonic medium (110 mm NaCl); TonE mutant construct shows no change in activity in response to hypertonicity. E, hypertonicity (Hyper)-dependent increase in wild-type CCL2 reporter activity is abolished by DN-TonEBP even at the lowest plasmid concentration of 50 ng, similar to inhibition of TauT reporter, a known TonEBP target. F, in cells cultured under isotonic (Iso) conditions, however, expression of DN-TonEBP does not affect CCL2 promoter activity. G, expression of TonEBP at all plasmid doses induces CCL2 promoter activity, similar to the effect on TauT. H, overexpression of TonEBP results in increased activity of only the wild-type, but not the TonE-mutated CCL2 promoter. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates and 3 technical replicates per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. NS, not significant.
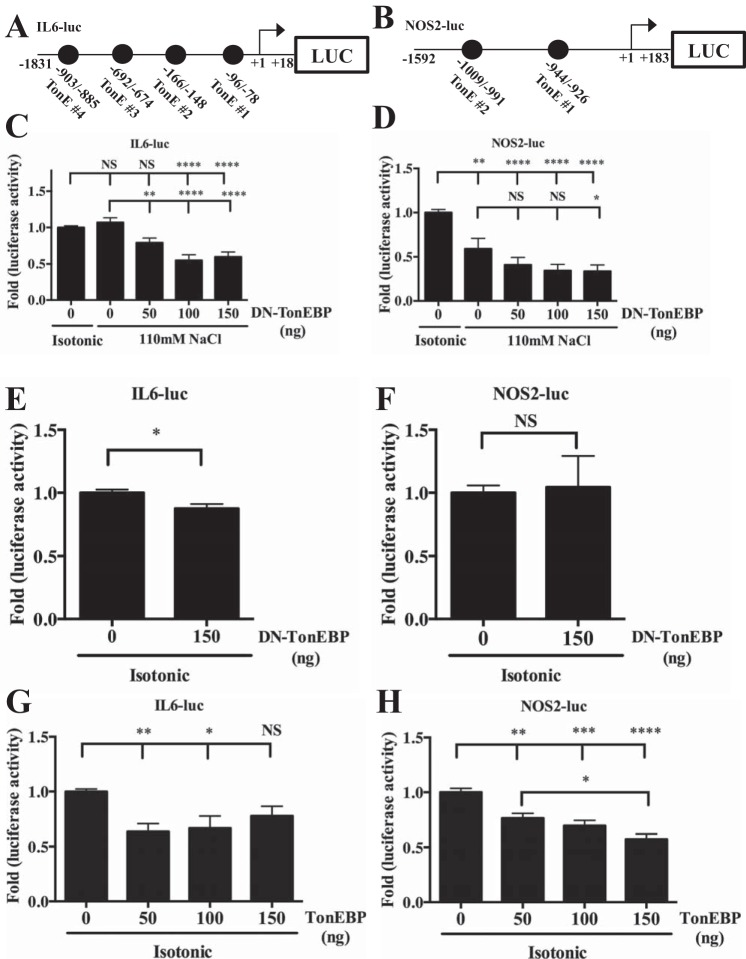
We then evaluated the activities of the proximal IL6 (Fig. 3A) and NOS2 (Fig. 3B) promoters. Unlike the CCL2 promoter, IL6 promoter activity was unaffected by hypertonicity (Fig. 3C) and NOS2 promoter activity was decreased (Fig. 3D). However, DN-TonEBP inhibited activities of both promoters under hypertonic conditions (Fig. 3, C and D). Expression of DN-TonEBP under isotonic conditions led to a slight reduction in IL6 promoter activity (Fig. 3E), but did not affect the NOS2 promoter (Fig. 3F). Surprisingly, in both cases, overexpression of TonEBP also suppressed the promoter activities (Fig. 3, G and H).
FIGURE 3.
Regulation of IL6 and NOS2 promoters by hypertonicity and TonEBP is unique in NP cells. A and B, schematics showing IL6 and NOS2 promoter luciferase constructs and their predicted TonE. C and D, treatment of NP cells with hypertonic medium has no effect on IL6 promoter activity (C) and decreases NOS2 promoter activity (D). Under hypertonic conditions, expression of DN-TonEBP suppresses activity levels of both IL6 (C) and NOS2 (D) reporters. E and F, under isotonic conditions, expression of DN-TonEBP suppresses activity of the IL6 promoter only slightly (E), but has no effect on the NOS2 promoter (F). G and H, activity levels of both IL6 (G) and NOS2 (H) promoter reporters are suppressed by overexpression of TonEBP. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates and 3 technical replicates per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. NS, not significant.
TonEBP Controls Hypertonic Induction and Maintenance of Pro-inflammatory Gene Expression
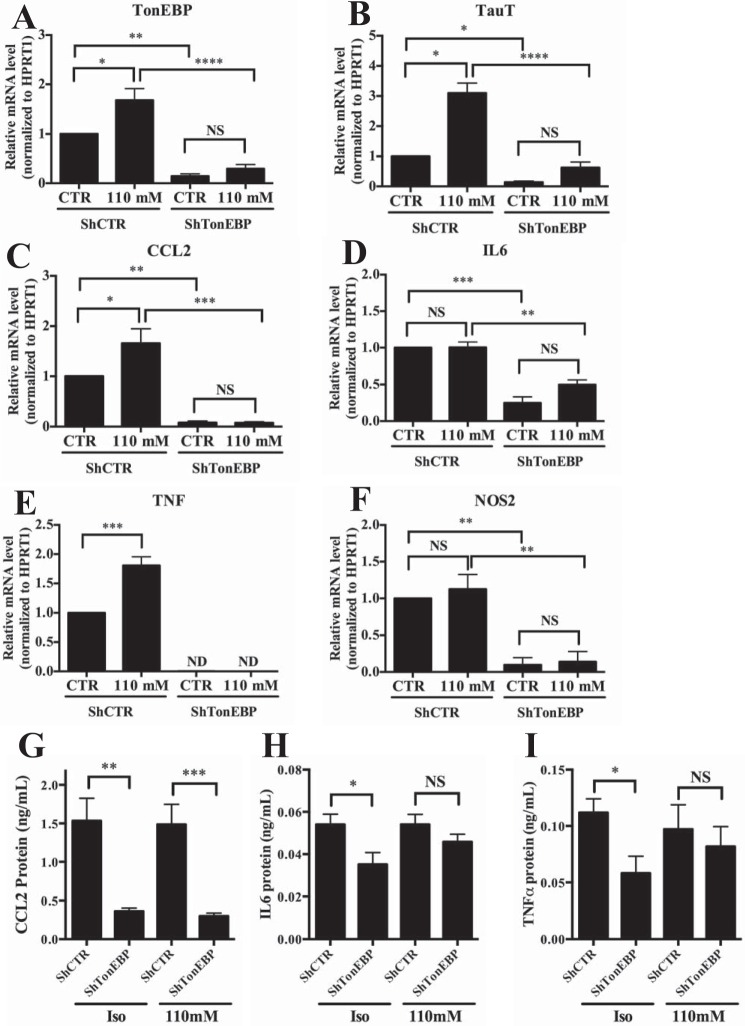
We next tested whether hypertonicity-mediated induction of pro-inflammatory genes was TonEBP-dependent by knocking down TonEBP in NP cells under isotonic or hypertonic conditions. In control cells (ShCTR), we observed hypertonicity-dependent induction in mRNAs for TonEBP, TauT, CCL2, and TNF (Fig. 4, A–C and E) but not of IL6 or NOS2 (Fig. 4, D and F). Regardless of inducibility, TonEBP knockdown was sufficient to decrease mRNA levels of all genes evaluated. We then examined protein levels of CCL2, IL6, and TNFα (Fig. 4, G–I). TonEBP silencing significantly decreased levels of CCL2 under both isotonic and hypertonic conditions. Although there was a trend of decreased IL6 and TNFα levels in TonEBP-silenced cells under hypertonicity, significant decrease was seen under isotonic conditions.
FIGURE 4.
TonEBP silencing reduces expression levels of inflammatory molecules in NP cells. A–F, irrespective of extracellular tonicity, mRNA levels of TonEBP (A), TauT (B), CCL2 (C), IL6 (D), TNF (E), and NOS2 (F) are significantly suppressed in NP cells transduced with shRNA against TonEBP (ShTonEBP) when compared with cells transduced with control shRNA (ShCTR). G–I, TonEBP-suppressed cells (ShTonEBP) also evidence significantly diminished levels of CCL2 (G), IL6 (H), and TNFα (I) protein in NP cells. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. Iso, isotonic. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. NS, not significant.
TonEBP Haploinsufficiency Prevents Hypertonicity-mediated Pro-inflammatory Gene Expression in an ex Vivo Intervertebral Disc Organ Culture Model
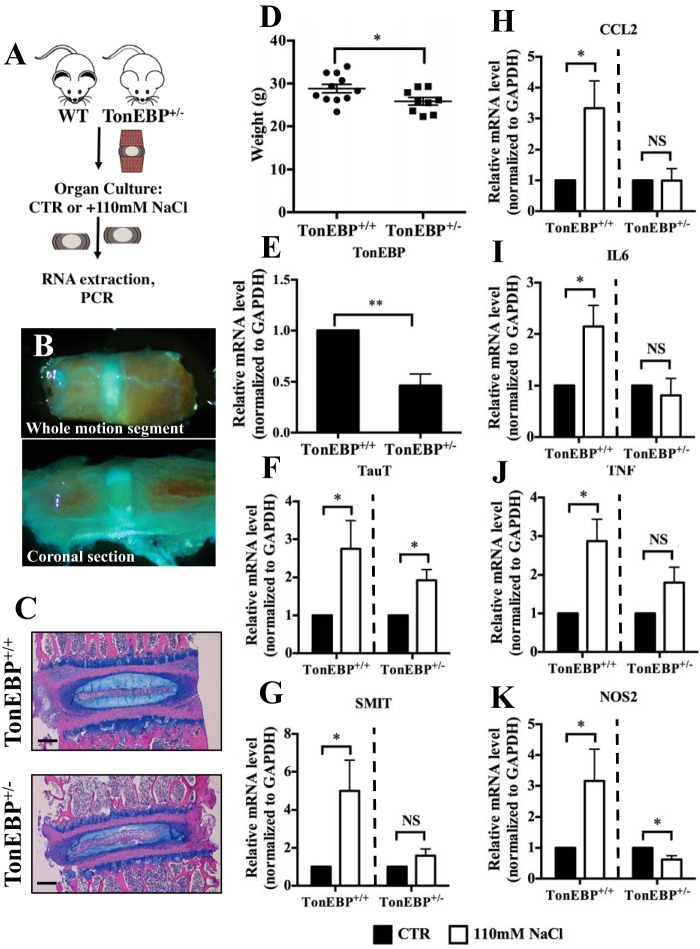
We used a whole-disc organ culture model to assess the effect of hypertonicity on inflammatory gene expression by disc cells in their native extracellular matrices, using wild-type and haploinsufficient TonEBP+/− mice (Fig. 5, A and B) (20). At this age, the overall structure, size, and health of discs from the two genotypes did not appear grossly different (Fig. 5C); weight of TonEBP+/− mice at euthanasia was slightly lower than wild-type mice (Fig. 5D). As expected, discal levels of TonEBP mRNA from haploinsufficient animals were about half of wild-type animals (Fig. 5E). Although the induction in SMIT mRNA was abolished in heterozygous animals, induction in TauT mRNA was less affected by TonEBP haploinsufficiency, indicating that some targets may be more sensitive to TonEBP modulation than others (Fig. 5, F and G). Transcript levels of CCL2, IL6, TNF, and NOS2 were induced in discs from wild-type animals. However, discs from TonEBP+/− animals failed to induce levels of any of these transcripts under hypertonicity (Fig. 5, H–K).
FIGURE 5.
TonEBP haploinsufficiency attenuates hypertonicity-mediated induction of pro-inflammatory genes in an ex vivo intervertebral disc organ culture model. A, schematic depicting the setup of the ex vivo organ culture experiments. B, representative images of intact (top) and coronally sectioned (bottom) motion segments. C, gross histological morphology of motion segments from 4-month-old (top) TonEBP+/+ and (bottom) TonEBP+/− mice following organ culture procedure (stained with H&E and Alcian blue). Scale bar, 100 μm. D, weight of TonEBP+/+ mice was only slightly higher than TonEBP+/− mice at the time of sacrifice. E, levels of TonEBP mRNA from intervertebral discs of TonEBP+/+ mice are approximately double that in discs from TonEBP+/− animals. F, mRNA level of TauT is induced by culturing motion segments in hypertonic medium (open bars), regardless of mouse genotype. G–K, mRNA levels of SMIT (G), CCL2 (H), IL6 (I), TNF (J), and NOS2 (K) in motion segments are induced by culture in hypertonic medium in wild-type animals. It is noteworthy that induction of all of these mRNAs is significantly attenuated in TonEBP haploinsufficient mice. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. For D and E, *, p ≤ 0.05, **, p ≤ 0.01. For F–K, statistical comparison is between isotonic and hypertonic groups in each genotype, *, p ≤ 0.1. Solid bars, CTR; open bars, hypertonic. NS, not significant.
TonEBP Controls Expression of Select Inflammatory Targets through p65/NF-κB under Hypertonicity
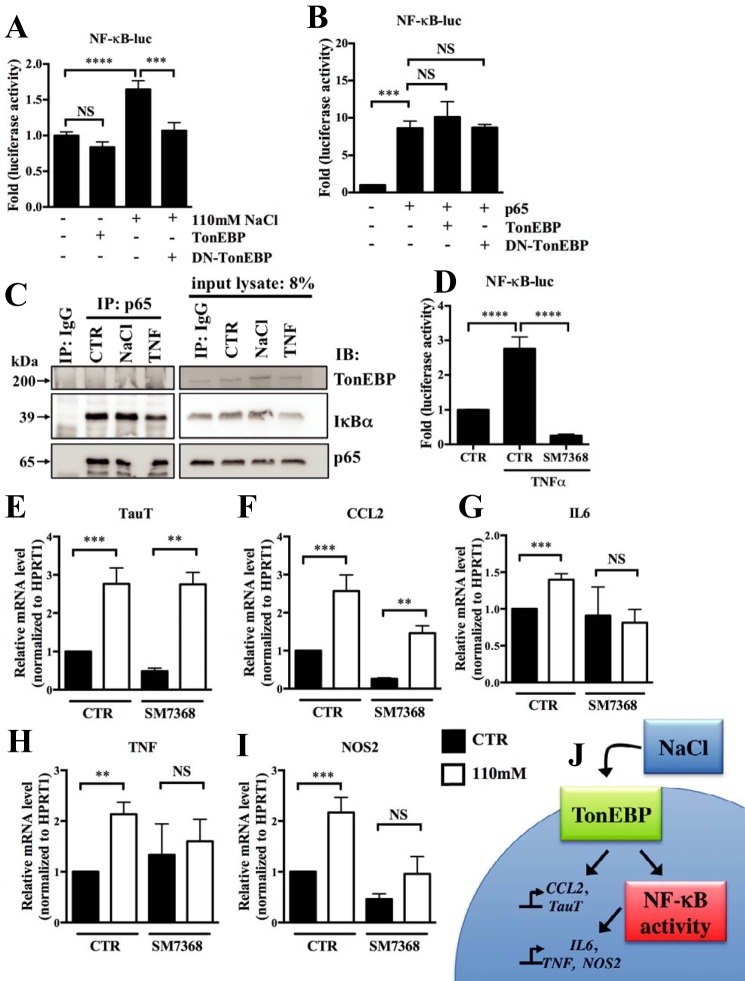
Because the NF-κB pathway is a common regulator of the pro-inflammatory genes studied here, we investigated the relationship between hypertonicity, TonEBP, and NF-κB signaling in NP cells. Hypertonicity increased activity of the NF-κB-responsive reporter, and this induction was blocked by DN-TonEBP. Interestingly, under isotonic conditions, TonEBP overexpression had no effect on NF-κB activity (Fig. 6A) and p65-mediated induction of NF-κB activity was not affected by overexpression of TonEBP or DN-TonEBP (Fig. 6B). Because both TonEBP and p65 are Rel family members and undergo homo/heterodimerization to promote transcription, we investigated their interaction. Immunoprecipitations failed to show association between these proteins, regardless of tonicity or presence of TNFα (Fig. 6C), whereas we were able to detect interaction between p65 and its known interacting protein, IκBα. We then tested whether NF-κB signaling contributed to hypertonic induction of pro-inflammatory targets using SM7368, an inhibitor that blocks TNFα-dependent NF-κB reporter activity (Fig. 6D). Despite inhibiting NF-κB activation, TauT (Fig. 6E) and CCL2 (Fig. 6F) were induced by hypertonicity. In contrast, SM7368 blocked hypertonic induction of IL6 (Fig. 6G), TNF (Fig. 6H), and NOS2 (Fig. 6I).
FIGURE 6.
Hypertonic induction of pro-inflammatory genes requires activity of the NF-κB pathway. A, activity of the NF-κB reporter is increased in cells cultured under hypertonicity (110 mm NaCl added), and induction is blocked by co-transfection with DN-TonEBP. Reporter activity is unaffected by TonEBP overexpression alone. B, p65-mediated induction of NF-κB reporter activity is not affected by expression of either TonEBP or DN-TonEBP under isotonic conditions. C, immunoprecipitation (IP) of p65 in NP cells following stimulation with NaCl (110 mm) or 50 ng/ml TNFα. TonEBP was not immunoprecipitated with p65 under either condition. IκBα is used as a positive control, showing successful immunoprecipitation of p65 with its interacting partners. D, TNFα-mediated induction of the NF-κB reporter activity is blocked by treatment with inhibitor SM7368 (5 μm). E–I, induction in mRNA levels of TauT (E) and CCL2 (F) by hypertonicity (open bars) occurs despite treatment with NF-κB inhibitor SM7368, whereas induction of IL6 (G), TNF (H), and NOS2 (I) is completely abolished. J, schematic representation of TonEBP-mediated control of pro-inflammatory genes under hypertonic stimulus. For E–I, solid bars, CTR; open bars, hypertonic. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. Co-immunoprecipitation of TonEBP and p65 was attempted in six independent experiments. For A, B, and D, 3 technical replicates were performed per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. NS, not significant.
Discussion
NP cells reside in a hypertonic environment within the disc, the severity of which fluctuates with daily activity (6). The transcription factor TonEBP plays a pro-survival role in the NP under hypertonic conditions via regulation of canonical osmotic response genes (3, 17) while also regulating matrix synthesis and tissue hydration genes (17, 18, 21–23). The present study was aimed at determining whether TonEBP promotes inflammation in response to hypertonicity, as has been reported in other cell types.
RNA sequencing and subsequent investigation showed that TonEBP maintained CCL2 mRNA and protein and IL6 and TNFα mRNA expression levels. Although TonEBP maintained IL6 and TNFα protein levels under isotonic conditions, this was not the case under hypertonicity. It is possible that under hypertonicity another factor compensates for TonEBP absence to maintain IL6 and TNFα protein levels at the post-transcriptional stage. Previous reports have demonstrated hypertonic induction of CCL2, IL6, and TNF (12, 24), which was TonEBP-dependent in some instances. On the other hand, hypertonicity suppressed LPS-mediated IL6 production in macrophages (16). These results suggest that the TonEBP-mediated response to hypertonicity is likely cell-type specific. Timing of this response may also depend on cell type, as we detected only a very transient induction in mRNA levels of these targets.
We have shown that the CCL2 gene is induced by hypertonicity and that this induction requires the action of TonEBP on a highly conserved TonE. Our result is in agreement with a previous study in kidney cells, which showed a lack of hypertonic response after deleting this TonE (12). Although they were not inducible by hypertonicity, maintenance of the IL6 and NOS2 promoters under hypertonic conditions required TonEBP. In addition, overexpression studies demonstrated that precise control of TonEBP levels was crucial in sustaining promoter activities. These studies suggest that, in NP cells, control of IL6 and NOS2 transcription by TonEBP may be unique. In LPS-treated mouse embryonic fibroblasts, TonEBP, indeed, bound to the region containing predicted TonE in IL6 and NOS2 promoters, pointing to context- and cell type-specific differences in the mechanism by which TonEBP controls these genes (15). Induction may also involve post-transcriptional mechanisms, including increased mRNA stability via osmo-sensitive micro RNAs (25).
Further insights into inflammatory gene regulation came from ex vivo organ culture studies performed using haploinsufficient TonEBP mice. This organ culture preserves the native cell-matrix and cell-cell interactions in all disc compartments. Therefore, these organ culture studies confirmed that hypertonicity-mediated induction of pro-inflammatory genes required TonEBP and was not due to a stress response evoked by a sudden change in tonicity. Canonical TonEBP targets TauT and SMIT displayed differing sensitivities to TonEBP levels, suggesting that TonEBP preferentially activates transcription of some targets, such as TauT, over others.
Because NF-κB controls expression of many inflammatory genes, we investigated potential cross-talk between TonEBP and RelA. The effect of hypertonicity on NF-κB activity appears to be cell type-specific, with reports of both inductive (26) and repressive (27) effects. In NP cells, NF-κB activity was controlled in a TonEBP-dependent manner under hypertonic conditions. TonEBP modulation did not affect p65-dependent NF-κB activity under isotonic conditions, indicating that hypertonicity produces a permissive environment for cross-talk between TonEBP and NF-κB signaling. However, the lack of TonEBP immunoprecipitation with p65 showed that cross-talk does not involve physical interaction. Interestingly, in other cells, these proteins have been shown to interact at the immediate onset of hypertonic stimulation (26) and in response to LPS treatment (28). Interestingly, NF-κB activity was required for hypertonic induction of only a subset of the studied targets; TauT and CCL2 were refractory to inhibition. These results suggest that, under hypertonic conditions, cross-talk between TonEBP and NF-κB controls a subset of targets, whereas some targets are controlled by TonEBP alone.
These results were compelling because inflammation and extracellular matrix content are hallmarks of spondyloarthritis and disc degeneration, a major cause of degenerative spondylolisthesis and spinal instability (29–31). Specifically, levels of CCL2, IL6, TNF, and NOS (32–34) are linked to disc degeneration. However, it is counterintuitive that physiological loading of the healthy disc would activate an inflammatory program. It is, therefore, more likely that the acute nature of CCL2 induction may be tied to diurnal loading of the disc, considering that genes controlling circadian rhythm are essential for disc homeostasis (35, 36). It is noteworthy that in other cell types, CCL2 promotes survival (37), proliferation (38), and phosphorylation of Akt, ERK, and STAT3 (39), molecules critical for NP function (22, 40–43). Therefore, it is feasible that in the NP, the moderate, transient increase in CCL2 elicited by the hypertonic milieu serves a physiological function.
In summary, hypertonic induction of traditionally pro-inflammatory genes is seen in various cell types. However, the timing of the response to hypertonicity and the mechanism by which TonEBP promotes transcription of select target genes such as IL6 and NOS2 are unique in NP cells. These differences might explain how the responses are finely tuned in a context- and cell type-dependent fashion to promote homeostatic maintenance of NP health. However, it is important to note that dysregulation of TonEBP could also potentially promote inflammation.
Experimental Procedures
Isolation and Treatment of NP Cells
Rat NP cells were isolated using a method previously described by Risbud et al. (44). Collection of animal tissues for cell isolation was approved by Thomas Jefferson University's Institutional Animal Care and Use Committee (IACUC). Cells were maintained in DMEM with 10% FBS and antibiotics. For hypertonic culture, 110 mm NaCl was added to medium.
Plasmids and Reagents
Luciferase reporter plasmids were provided by Drs. Kojima (12) (CCL2-luc), Atreya (45) (IL6-luc), Ito (46) (TauT-luc), and Taubman (47) (NF-κB-luc). NOS2-luc (plasmid 19296) (48), p65 (plasmid 20012) (49), psPAX2 (plasmid 12260), and pMD2G (plasmid 12259) were from Addgene. As transfection control, pRL-TK (Promega) was used. Lentiviral ShTonEBP (TRCN0000020019) and control ShRNA pLKO.1 were from Sigma. TonEBP+/+ and haploinsufficient TonEBP+/− were from Dr. Kwon (20).
Lentiviral Studies
HEK-293T cells in 10-cm plates (1.3 × 106 cells/plate) were transfected with 9 μg of either lentiviral ShCTR (pLKO.1) or ShTonEBP plasmids, plus 6 μg of psPAX2 and 3 μg of pMD2.G. After 16 h, medium was removed and replaced with DMEM with 5% FBS. Lentiviral particles were harvested at 48 and 60 h after transfection and concentrated using PEG solution. NP cells were transduced with medium containing viral particles and 8 μg/ml Polybrene. Cells and conditioned medium were collected 5 days after transduction.
RNA Sequencing
Illumina TruSeq Stranded Total RNA Sample Prep with Ribo-Zero was used to prepare the library. Libraries were chemically denatured and applied to an Illumina HiSeq v4 single read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR Cluster Kit v4-cBot. After transfer of the flow cell to an Illumina HiSeq 2500, a 50-cycle single-read sequence run was performed (HiSeq SBS Kit v4). For data analysis, Rn5 Ensembl annotations (Build 75) were downloaded and converted to genePred format. Reads were aligned to the transcriptome reference index using NovoAlign (v2.08.01), allowing up to 50 alignments for each read. Read counts were generated using the USeq Defined Region Differential Seq application and used in DESeq2 to measure the differential expression between each condition, controlling for sample preparation batch. For IPA, differentially expressed gene lists were used as input to identify related pathways, diseases, and networks.
Real-time Quantitative RT-PCR
For in vitro assays, total DNA-free RNA was extracted from NP cells using RNeasy mini columns (Qiagen), and cDNA was made using EcoDry premix (Clontech). For ex vivo assays, RNA was isolated using TRIzol (Thermo Fisher) and treated with DNA-free DNase treatment kit (Ambion). cDNA and gene-specific primers (Integrated DNA Technologies, Coralville, IA) were added to SYBR Green master mixture, and mRNA expression was quantified using the Step-One Plus System (Applied Biosystems).
Western Blotting
Cells were placed on ice following treatment and washed with ice-cold PBS. Buffers included 1× protease inhibitor cocktail (Roche Applied Science), NaF (4 mm), Na3VO4 (20 mm), NaCl (150 mm), β-glycerophosphate (50 mm), and DTT (0.2 mm). Total cell proteins were resolved on 10% SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline, Tween 20 and incubated overnight at 4 °C in blocking buffer with rabbit anti-TonEBP (1:1000, Novus, catalogue number NB120-3446, lot Q1220667), rabbit anti-p65 (1:1000, Cell Signaling, catalogue number D14E12, lot 8), mouse anti-IκBα (1:1000, Cell Signaling, catalogue number 4814), or mouse anti-β-tubulin antibody (1:2000, Developmental Studies Hybridoma Bank (DSHB), catalogue number E-7). Specificity of the TonEBP antibody is evidenced by loss of signal with TonEBP-specific knockdown (Fig. 1B) Immunolabeling was detected with ECL reagent. Densitometric analysis (ImageQuant) was performed by first normalizing protein-of-interest levels to the housekeeping protein (β-tubulin) and then normalizing to the experimental control group.
ELISA
Conditioned medium was filtered (0.45 μm) and supplemented with 1× protease inhibitor cocktail (Roche Applied Science). ELISA was performed using Mini ELISA Kits (PeproTech).
Bioinformatic Analysis of Promoters and TonE Prediction
Promoter sequences were downloaded from the UCSC Genome Table Browser. MatInspector (Genomatix) was used to identify predicted TonEBP binding sites with a score cutoff of 0.8. The Ensembl browser was used for Multiz alignments of TonE predicted in the rat promoter against human and mouse. The ECR Browser was used to visually represent evolutionary conservation between the human, canine, and rhesus CCL2 promoters.
Transfections and Dual-Luciferase Assay
Cells were transferred to 48-well plates (2 × 104 cells/well) 1 day prior to transfection. To measure the effects of hypertonicity, cells were transfected with 250 ng of CCL2, IL6, or NOS2 reporters and 250 ng of pRL-TK plasmid and cultured in isotonic or hypertonic conditions. For gain- and loss-of-function studies, FLAG-TonEBP, FLAG-DN-TonEBP, or backbone plasmid (50–150 ng) was co-transfected with reporters and pRLTK. In all experiments, plasmids were premixed with the transfection reagent, Lipofectamine 2000 (Invitrogen). 48 h after transfection, cells were harvested, and firefly and Renilla luciferase activities were measured using the Dual-LuciferaseTM reporter assay (Promega) and a luminometer (TD-20/20, Turner Designs).
Site-directed Mutagenesis
Site-directed mutagenesis of the rat CCL2 promoter was performed according to the manufacturer's protocol, using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs). Primers used for CCL2 promoter mutants are (mutated TonE underlined): forward, 5′-AGTAGTGGCTAAAGGAAACACCAAATTCC-3′; reverse, 5′-GGGAGCAAATGAAGCTGC-3′. Mutations were verified by sequencing (Applied Biosystems 3730 DNA Sequencer).
Ex Vivo Disc Organ Culture and Histology
4-month-old mice were sacrificed according to IACUC guidelines. Whole spines were carefully dissected en bloc, and extraneous tissues were removed. For each experimental group, lumbar and caudal motion segments from a single mouse were pooled together. A total of 20 mice were used (11 TonEBP+/+, 9 TonEBP+/−). Motion segments were equilibrated overnight in DMEM. 16 h later, fresh medium with or without 110 mm NaCl was added and cultured for 8 h. After treatment, some motion segments were stored in RNAlater (Ambion), and vertebrae and endplates were removed using a dissecting microscope (Zeiss Stemi 305, imaged with Axiocam ERc 5s). Discs were snap-frozen and pulverized (BioSpec BioPulverizer) before RNA isolation. Undissected motion segments were fixed for 48 h in 4% paraformaldehyde, decalcified in 12.5% EDTA, and embedded in paraffin. Sagittal sections (7 μm) were deparaffinized, rehydrated through graded ethanol, and stained with Alcian blue, eosin, and hematoxylin. Sections were visualized using a Zeiss Axio Imager A2 and imaged with Axiocam 105 color camera and N-Achroplan 5× objective.
Statistics
All experiments were performed at least three times. For quantitative measurements, results are presented as the mean ± S.E. Differences between groups were assessed by analysis of variance and Student's t test using GraphPad Prism Software. p values < 0.05 were considered significant for in vitro experiments; p values < 0.1 were considered statistically significant for ex vivo organ culture experiments, as noted in legends.
Author Contributions
Z. I. J., I. M. S., and M. V. R. conceived the study. Z. I. J. conducted the experiments, analyzed data, and wrote the manuscript. I. M. S. designed the study, wrote the manuscript, and secured funding. M. V. R. designed experiments, interpreted results, secured funding, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants AR055655 and AR064733 (to M. V. R.) and by National Institutes of Health Grant T32 AR052273 (to I. M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables 1 and 2 and supplemental Fig. 1.
The sequence data reported in this paper have been submitted to the GEO Database under GEO Accession Number GSE86552.
- NP
- nucleus pulposus
- TonE
- tonicity-responsive enhancer element
- DN-TonEBP
- dominant-negative TonEBP
- luc
- luciferase
- CTR
- control.
References
- 1. Inoue N., and Espinoza Orías A. A. (2011) Biomechanics of intervertebral disk degeneration. Orthop. Clin. North Am. 42, 487–499, vii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kiani C., Chen L., Wu Y. J., Yee A. J., and Yang B. B. (2002) Structure and function of aggrecan. Cell Res. 12, 19–32 [DOI] [PubMed] [Google Scholar]
- 3. Johnson Z. I., Shapiro I. M., and Risbud M. V. (2014) Extracellular osmolarity regulates matrix homeostasis in the intervertebral disc and articular cartilage: Evolving role of TonEBP. Matrix Biol. 40, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eyre D. R., and Muir H. (1977) Quantitative analysis of types I and II collagens in human intervertebral discs at various ages. Biochim. Biophys. Acta 492, 29–42 [DOI] [PubMed] [Google Scholar]
- 5. Borthakur A., Maurer P. M., Fenty M., Wang C., Berger R., Yoder J., Balderston R. A., and Elliott D. M. (2011) T1ρ magnetic resonance imaging and discography pressure as novel biomarkers for disc degeneration and low back pain. Spine 36, 2190–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsumura Y., Kasai Y., Obata H., Matsushima S., Inaba T., and Uchida A. (2009) Changes in water content of intervertebral discs and paravertebral muscles before and after bed rest. J. Orthop. Sci. 14, 45–50 [DOI] [PubMed] [Google Scholar]
- 7. Burg M. B., Kwon E. D., and Kültz D. (1997) Regulation of gene expression by hypertonicity. Annu. Rev. Physiol. 59, 437–455 [DOI] [PubMed] [Google Scholar]
- 8. Chen S., Grigsby C. L., Law C. S., Ni X., Nekrep N., Olsen K., Humphreys M. H., and Gardner D. G. (2009) Tonicity-dependent induction of Sgk1 expression has a potential role in dehydration-induced natriuresis in rodents. J. Clin. Invest. 119, 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lanaspa M. A., Andres-Hernando A., Li N., Rivard C. J., Cicerchi C., Roncal-Jimenez C., Schrier R. W., and Berl T. (2010) The expression of aquaporin-1 in the medulla of the kidney is dependent on the transcription factor associated with hypertonicity, TonEBP. J. Biol. Chem. 285, 31694–31703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hollborn M., Vogler S., Reichenbach A., Wiedemann P., Bringmann A., and Kohen L. (2015) Regulation of the hyperosmotic induction of aquaporin 5 and VEGF in retinal pigment epithelial cells: involvement of NFAT5. Mol. Vis. 21, 360–377 [PMC free article] [PubMed] [Google Scholar]
- 11. Lee S. D., Choi S. Y., Lim S. W., Lamitina S. T., Ho S. N., Go W. Y., and Kwon H. M. (2011) TonEBP stimulates multiple cellular pathways for adaptation to hypertonic stress: organic osmolyte-dependent and -independent pathways. Am. J. Physiol. Renal Physiol. 300, F707–F715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kojima R., Taniguchi H., Tsuzuki A., Nakamura K., Sakakura Y., and Ito M. (2010) Hypertonicity-induced expression of monocyte chemoattractant protein-1 through a novel cis-acting element and MAPK signaling pathways. J. Immunol. 184, 5253–5262 [DOI] [PubMed] [Google Scholar]
- 13. Esensten J. H., Tsytsykova A. V., Lopez-Rodriguez C., Ligeiro F. A., Rao A., and Goldfeld A. E. (2005) NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone. Nucleic Acids Res. 33, 3845–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. López-Rodríguez C., Aramburu J., Jin L., Rakeman A. S., Michino M., and Rao A. (2001) Bridging the NFAT and NF-κB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 15, 47–58 [DOI] [PubMed] [Google Scholar]
- 15. Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., Del Val M., Aramburu J., and López-Rodríguez C. (2012) Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 209, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim N.-H., Hong B.-K., Choi S. Y., Moo Kwon H., Cho C.-S., Yi E. C., and Kim W.-U. (2013) Reactive oxygen species regulate context-dependent inhibition of NFAT5 target genes. Exp. Mol. Med. 45, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai T.-T., Danielson K. G., Guttapalli A., Oguz E., Albert T. J., Shapiro I. M., and Risbud M. V., (2006) TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc. J. Biol. Chem. 281, 25416–25524 [DOI] [PubMed] [Google Scholar]
- 18. Hiyama A., Gajghate S., Sakai D., Mochida J., Shapiro I. M., and Risbud M. V. (2009) Activation of TonEBP by calcium controls β1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J. Biol. Chem. 284, 9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Windt A. E., Haak E., Das R. H. J., Kops N., Welting T. J. M., Caron M. M. J., van Til N. P., Verhaar J. A. N., Weinans H., and Jahr H. (2010) Physiological tonicity improves human chondrogenic marker expression through nuclear factor of activated T-cells 5 in vitro. Arthritis Res. Ther. 12, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Go W. Y., Liu X., Roti M. A., Liu F., and Ho S. N. (2004) NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. U.S.A. 101, 10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gajghate S., Hiyama A., Shah M., Sakai D., Anderson D. G., Shapiro I. M., and Risbud M. V. (2009) Osmolarity and intracellular calcium regulate aquaporin2 expression through TonEBP in nucleus pulposus cells of the intervertebral disc. J. Bone Miner. Res. 24, 992–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsai T.-T., Guttapalli A., Agrawal A., Albert T. J., Shapiro I. M., and Risbud M. V. (2007) MEK/ERK signaling controls osmoregulation of nucleus pulposus cells of the intervertebral disc by transactivation of TonEBP/OREBP. J. Bone Miner. Res. 22, 965–974 [DOI] [PubMed] [Google Scholar]
- 23. Hiyama A., Gogate S. S., Gajghate S., Mochida J., Shapiro I. M., and Risbud M. V. (2010) BMP-2 and TGF-β stimulate expression of β1,3-glucuronosyl transferase 1 (GlcAT-1) in nucleus pulposus cells through AP1, TonEBP, and Sp1: role of MAPKs. J. Bone Miner. Res. 25, 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ueno M., Shen W.-J., Patel S., Greenberg A. S., Azhar S., and Kraemer F. B. (2013) Fat-specific protein 27 modulates nuclear factor of activated T cells 5 and the cellular response to stress. J. Lipid Res. 54, 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang W., Liu H., Wang T., Zhang T., Kuang J., Luo Y., Chung S. S. M., Yuan L., and Yang J. Y. (2011) Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res. 39, 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roth I., Leroy V., Kwon H. M., Martin P.-Y., Féraille E., and Hasler U. (2010) Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-κB activity. Mol. Biol. Cell. 21, 3459–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wright F. L., Gamboni F., Moore E. E., Nydam T. L., Mitra S., Silliman C. C., and Banerjee A. (2014) Hyperosmolarity invokes distinct anti-inflammatory mechanisms in pulmonary epithelial cells: evidence from signaling and transcription layers. PLoS ONE. 9:e114129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H. H., Sanada S., An S. M., Ye B. J., Lee J. H., Seo Y.-K., Lee C., Lee-Kwon W., Küper C., Neuhofer W., Choi S. Y., and Kwon H. M. (2016) LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding. Sci. Rep. 6, 24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Satomi K., Hirabayashi K., Toyama Y., and Fujimura Y. (1992) A clinical study of degenerative spondylolisthesis: radiographic analysis and choice of treatment. Spine 17, 1329–1336 [DOI] [PubMed] [Google Scholar]
- 30. Risbud M. V., and Shapiro I. M. (2014) Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 10, 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schleich C., Müller-Lutz A., Matuschke F., Sewerin P., Sengewein R., Schmitt B., Ostendorf B., Wittsack H.-J., Stanke K., Antoch G., and Miese F. (2015) Glycosaminoglycan chemical exchange saturation transfer of lumbar intervertebral discs in patients with spondyloarthritis. J. Magn. Reson. Imaging. 42, 1057–1063 [DOI] [PubMed] [Google Scholar]
- 32. Watanabe T., Kato S., Sato K., and Nagata K. (2005) Nitric oxide regulation system in degenerative lumbar disease. Kurume Med. J. 52, 39–47 [DOI] [PubMed] [Google Scholar]
- 33. Phillips K. L. E., Chiverton N., Michael A. L. R., Cole A. A., Breakwell L. M., Haddock G., Bunning R. A. D., Cross A. K., and Le Maitre C. L. (2013) The cytokine and chemokine expression profile of nucleus pulposus cells: implications for degeneration and regeneration of the intervertebral disc. Arthritis Res. Ther. 15, R213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altun I. (2016) Cytokine profile in degenerated painful intervertebral disc: variability with respect to duration of symptoms and type of disease. Spine J. 16, 857–861 [DOI] [PubMed] [Google Scholar]
- 35. Dudek M., Yang N., Ruckshanthi J. P., Williams J., Borysiewicz E., Wang P., Adamson A., Li J., Bateman J. F., White M. R., Boot-Handford R. P., Hoyland J. A., and Meng Q.-J. (2016) The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration. Ann. Rheum. Dis. 10.1136/annrheumdis-2016-209428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suyama K., Silagi E. S., Choi H., Sakabe K., Mochida J., Shapiro I. M., and Risbud M. V. (2016) Circadian factors BMAL1 and RORα control HIF-1α transcriptional activity in nucleus pulposus cells: implications in maintenance of intervertebral disc health. Oncotarget 7, 23056–23071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fang W. B, Jokar I., Zou A., Lambert D., Dendukuri P., and Cheng N. (2012) CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J. Biol. Chem. 287, 36593–36608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Q., Sun W., Liao Y., Zeng H., Shan L., Yin F., Wang Z., Zhou Z., Hua Y., and Cai Z. (2015) Monocyte chemotactic protein-1 promotes the proliferation and invasion of osteosarcoma cells and upregulates the expression of AKT. Mol. Med. Rep. 12, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ji W.-T., Chen H.-R., Lin C.-H., Lee J.-W., and Lee C.-C. (2014) Monocyte chemotactic protein 1 (MCP-1) modulates pro-survival signaling to promote progression of head and neck squamous cell carcinoma. PLoS ONE 9, e88952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Risbud M. V., Guttapalli A., Albert T. J., and Shapiro I. M. (2005) Hypoxia activates MAPK activity in rat nucleus pulposus cells: regulation of integrin expression and cell survival. Spine 30, 2503–2509 [DOI] [PubMed] [Google Scholar]
- 41. Risbud M. V., Fertala J., Vresilovic E. J., Albert T. J., and Shapiro I. M. (2005) Nucleus pulposus cells upregulate PI3K/Akt and MEK/ERK signaling pathways under hypoxic conditions and resist apoptosis induced by serum withdrawal. Spine 30, 882–889 [DOI] [PubMed] [Google Scholar]
- 42. Cheng C.-C., Uchiyama Y., Hiyama A., Gajghate S., Shapiro I. M., and Risbud M. V. (2009) PI3K/AKT regulates aggrecan gene expression by modulating Sox9 expression and activity in nucleus pulposus cells of the intervertebral disc. J. Cell. Physiol. 221, 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z., Shen J., Wu W. K. K., Yu X., Liang J., Qiu G., and Liu J. (2012) Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS ONE 7, e53176. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., and Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1α under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 45. Zenker S., Panteleev-Ivlev J., Wirtz S., Kishimoto T., Waldner M. J., Ksionda O., Tybulewicz V. L. J., Neurath M. F., and Atreya I. (2014) A key regulatory role for Vav1 in controlling lipopolysaccharide endotoxemia via macrophage-derived IL-6. J. Immunol. 192, 2830–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ito T., Fujio Y., Hirata M., Takatani T., Matsuda T., Muraoka S., Takahashi K., and Azuma J. (2004) Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 382, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu J., and Taubman M. B. (2010) Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-κB-dependent pathway. J. Biol. Chem. 285, 8927–8935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lowenstein C. J., Alley E. W., Raval P., Snowman A. M., Snyder S. H., Russell S. W., and Murphy W. J. (1993) Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon γ and lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 90, 9730–9734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanjabi S., Williams K. J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., and Smale S. T. (2005) A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 19, 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.