Abstract
IL-2 is the first cytokine produced when naive T helper (Th) cells are activated and differentiate into dividing pre-Th0 proliferating precursors. IL-2 expression is blocked in naive, but not activated or memory, Th cells by the transcription factor Ets-2 that binds to the antigen receptor response element (ARRE)-2 of the proximal IL-2 promoter. Ets-2 acts as an independent preinduction repressor in naive Th cells and does not interact physically with the transcription factor NFAT (nuclear factor of activated T-cells) that binds to the ARRE-2 in activated Th cells. In naive Th cells, Ets-2 mRNA expression, Ets-2 protein levels, and Ets-2 binding to ARRE-2 decrease upon cell activation followed by the concomitant expression of IL-2. Cyclosporine A stabilizes Ets-2 mRNA and protein when the cells are activated. Ets-2 silences directly constitutive or induced IL-2 expression through the ARRE-2. Conversely, Ets-2 silencing allows for constitutive IL-2 expression in unstimulated cells. Ets-2 binding to ARRE-2 in chromatin is stronger in naive compared with activated or memory Th cells; in the latter, Ets-2 participates in a change of the IL-2 promoter architecture, possibly to facilitate a quick response when the cells re-encounter antigen. We propose that Ets-2 expression and protein binding to the ARRE-2 of the IL-2 promoter are part of a strictly regulated process that results in a physiological transition of naive Th cells to Th0 cells upon antigenic stimulation. Malfunction of such a repression mechanism at the molecular level could lead to a disturbance of later events in Th cell plasticity, leading to autoimmune diseases or other pathological conditions.
Keywords: human, interleukin, T helper cells, transcription regulation, transcription repressor
Introduction
T helper (Th)3 cells are key players in controlling acquired immune responses to foreign antigens. Malfunctions of their existence, activation, and/or function lead to autoimmune diseases or to the inability of the organism to combat infectious diseases or deal with other pathological conditions. The special functions that Th cells exert presume their previous activation, which begins with the engagement of the T-cell receptor (TCR) (1). TCR-mediated stimulation and the cytokine environment where cell activation takes place influence the fate decisions of Th cells toward different Th cell lineages at the early stages of activation (2). Stimulation of TCR induces a complex downstream cascade of events in Th cells, including the activation, nuclear translocation, and coordinated function of a defined set of transcription factors, including NFAT, AP-1, Oct-1, and NF-κB (1, 3).
IL-2 is the first cytokine produced when naive Th cells are activated and differentiate into proliferating pre-Th0 precursors (4). Precise regulation of IL-2 expression is of great importance because it promotes peripheral naive Th cells to enter the S phase and divide (5). Positive and negative regulation of IL-2 gene expression requires a relatively small segment of DNA, ∼300 bp, localized immediately upstream from the transcriptional start site integrating the positive and negative action of a number of different transcriptional regulators (6, 7). Previous studies have provided significant information regarding the activation of the IL-2 gene; nevertheless, less is known about IL-2 gene repression mechanisms. IL-2 gene repression occurs at two stages, the preinduction and the postinduction, i.e. when T-cells are in the resting state and after TCR signaling when a feedback inhibition of cell activation takes place to cease the expression of IL-2 gene and terminate T-cell activation (7, 8). These known negative regulators of IL-2 expression act either directly by binding to the IL-2 promoter or indirectly to repress IL-2 transcription (7, 8).
We have previously identified an IL-2 promoter protein binding activity, present in nuclear extracts prepared from naive Th cells isolated from cord blood or adult peripheral blood but not from activated or memory Th cells, capable of repressing IL-2 gene expression (9–11). This repressor activity is exerted through the distal IL-2 purine-rich response element (PU-d) or antigen receptor response element (ARRE)-2 (−292 to −273), which is also an NFAT binding site in activated Th cells. Following naive Th cell activation, the repressor activity disappears and is replaced by a newly synthesized activator (9–11). These observations are consistent with a model where repressors bound to the IL-2 promoter during the preinduction state are replaced by activators during Th cell induction. Importantly, the preinduction repressors appear to be different from the repressors involved in turning off IL-2 transcription postinduction.
A search in public gene expression databases for DNA-binding proteins with the properties of the putative IL-2 repressor revealed that the ets-2 oncogene is a strong candidate as a repressor of IL-2 transcription in naive Th lymphocytes (12). Ets-2 is a member of the ETS family of transcription factors that bind to purine-rich DNA sequences with GGA(A/T) as a central core consensus through a highly conserved DNA binding domain (13, 14). Ets proteins function as activators or repressors of transcription in partnership with other DNA-binding proteins and coregulators and control the expression of genes involved in diverse biological functions, such as mitosis, growth, development, differentiation, apoptosis, and regulation of immunity (13–22). Ets-2 plays an important role in the maturation, proliferation, and survival of mouse thymocytes, possibly by regulating the expression of c-Myc (23). In Th2 cells, Ets-2 together with Ets-1 is responsible for the activation of IL-5 transcription (24).
The role of Ets-2 in regulating the expression of the IL-2 gene remains elusive because its core DNA binding motif GGAA exists in both the ARRE-2 and ARRE-1 of the IL-2 promoter (6, 7, 25). ARRE-1 is involved in the transcriptional activation of IL-2, and it is also bound by NFAT. In addition, there is no known connection among the biological function of Ets-2, IL-2 transcription, and the differentiation status of Th cells.
In this work, we show that Ets-2 acts as a preinduction repressor of IL-2 transcription in naive Th cells and describe its properties. We aim to show that Ets-2 expression and protein binding to the ARRE-2 of the IL-2 promoter are part of a strictly regulated process that results in a physiological transition of naive Th cells to Th0 cells upon antigenic stimulation.
Results
Ets-2 Expression in Human Peripheral Blood Mononuclear Cell (PBMC) Populations
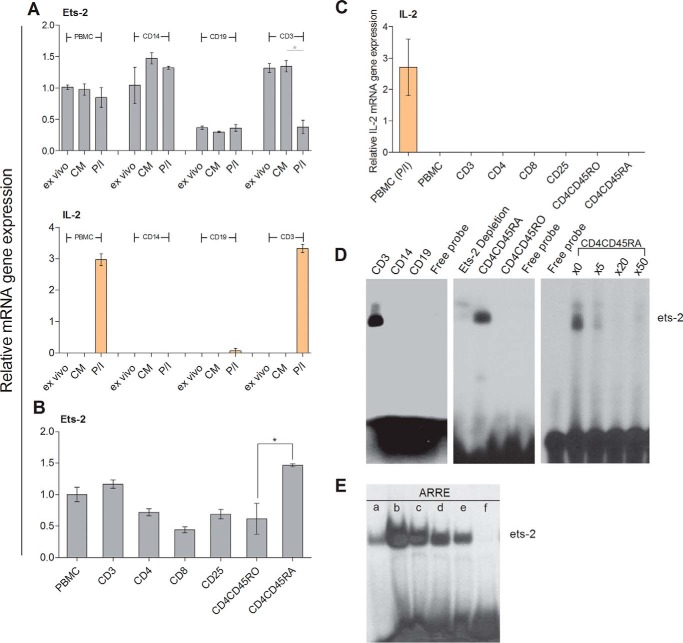
To investigate the role of Ets-2 in IL-2 transcription, we determined the levels of Ets-2 mRNA by real time PCR in PBMCs and different populations thereof isolated from healthy young adults (Fig. 1A, upper panel). Ets-2 mRNA was expressed in PBMCs ex vivo or when cultured in plain culture medium (CM) at similar levels, and its expression was slightly decreased when PBMCs were activated with the mitogens phorbol myristate acetate and ionomycin (P/I) (Fig. 1A, PBMC). Ex vivo CD14+ monocytes expressed Ets-2 mRNA at levels similar to ex vivo PBMCs, and its expression was increased when cultured ±P/I (Fig. 1A, CD14). CD19+ B-cells expressed low amounts of Ets-2 mRNA regardless of their activation status (Fig. 1A, CD19). Finally, CD3+ T-cells expressed high levels of Ets-2 mRNA that were significantly decreased following P/I stimulation (Fig. 1A, CD3).
FIGURE 1.
Ets-2 and IL-2 mRNA synthesis in human PBMC populations. A, relative Ets-2 and IL-2 mRNA levels were measured by real time PCR in PBMCs from healthy young blood donors, PBMC-derived CD14+ monocytes (CD14), CD19+ B-cells (CD19), and CD3+ T-cells (CD3). Monocytes, B-cells, and T-cells were isolated from PBMCs by negative selection using antibody-coated magnetic beads. Total RNA was isolated, and cDNA was prepared immediately after cell isolation (ex vivo) or culture of the cells in plain CM or in CM + P/I for 6 h. β2-m served as the comparative normalizer gene, and the ex vivo condition of PBMCs served as the calibrator. The results are presented as the mean values from three independent experiments; error bars represent S.E. B, relative Ets-2 mRNA levels were measured by real time PCR in PBMCs; PBMC-derived CD3+ T-cells (CD3) and different subpopulations thereof, including CD4+ Th cells (CD4), CD8+ cytotoxic T-cells (CD8), CD4+CD25+ Tegs (CD25), and CD4+CD45RO+CD25− memory Th cells (CD4CD45RO); or cord blood-derived CD4+CD45RA+CD25− naive Th cells (CD4CD45RA). The cells were isolated by negative selection in CM and processed immediately. β2-m served as the comparative normalizer gene, and the Ets-2 mRNA levels in PBMCs served as the calibrator. The results are presented as the mean values from three independent experiments; error bars represent S.E. (*, p < 0.05; Student's t test). C, IL-2 mRNA expression in different Th subpopulations was measured by real time PCR in PBMCs; PBMC-derived CD3+ T-cells (CD3) and different subpopulations thereof, including CD4+ Th cells (CD4), CD8+ cytotoxic T-cells (CD8), CD4+CD25+ Tregs (CD25), and CD4+CD45RO+CD25− memory Th cells (CD4CD45RO); or cord blood-derived CD4+CD45RA+CD25− naive Th cells (CD4CD45RA). P/I-stimulated PBMCs served as the positive control. Reverse transcribed β2-m was used as an internal control for equal loading for all samples. The results are presented as the mean values from three independent experiments; error bars represent S.E. D, nuclear protein extracts from PBMC-derived T-cells (CD3), monocytes (CD14), B-cells (CD19), memory Th cells (CD4CD45RO), cord blood-derived naive Th cells (CD4CD45RA), or cord blood-derived naive Th cells depleted of Ets-2 protein by the previous incubation of the extracts with Ets-2 antibody (Ets-2 Depletion) were analyzed for their ability to bind to a radioactive labeled oligo that encompasses the ARRE-2 binding site by EMSA. For DNA competition, the labeled ARRE-2 probe was used alone or mixed with a 5-, 20-, or 50-fold excess of unlabeled ARRE-2 oligo. The data shown are characteristic from at least three independent experiments with the same results. The purities of all isolated populations used in the experiments were tested by FACS and were >90%. E, Ets-2 binding activity to the ARRE-2. T-cell nuclear protein extracts were passed through a Protein A/G-agarose column. The eluate (a) was mixed for 1 h with 50 μg of anti-Ets-2 Ab and passed through a Protein A/G-agarose column. Bound Ets-2 was eluted and collected in 1-ml aliquots (b–e). The consecutive protein eluates (b–e) were precipitated by PEG 8000, and the pellets were redissolved in buffer. EMSAs were performed with the ARRE-2 oligonucleotide probe. f, free probe.
In parallel, we analyzed IL-2 expression (Fig. 1A, lower panel) and discovered a negative correlation between Ets-2 and IL-2 expression in T-cells because elevated levels of IL-2 mRNA during mitogenic activation coincided with diminished amounts of Ets-2 mRNA. Taken together, these results strongly suggest that Ets-2 expression is differentially regulated in the various PBMC populations with T-cells showing a negative correlation between Ets-2 and IL-2 expression.
To assess the levels of Ets-2 expression in distinct T-cell populations, we determined the levels of Ets-2 mRNA in ex vivo isolated T-cell populations: Ets-2 was expressed in all T-cell populations at various levels (Fig. 1B). There was a significant difference in Ets-2 mRNA levels between naive (CD4+CD45RA+CD25−) (Fig. 1B, CD4CD45RA) and memory (CD4+CD45RO+CD25−) (Fig. 1B, CD4CD45RO) Th cells with naive cells expressing Ets-2 at a significantly higher level. By comparison, cytotoxic CD3+CD8+CD25− T-cells (Fig. 1B, CD8) expressed the lowest levels of Ets-2, whereas CD4+CD25+ Tregs (Fig. 1B, CD25) expressed Ets-2 at levels similar to CD3+CD4+CD25− Th (Fig. 1B, CD4) and memory Th cells. As expected, none of the ex vivo T-cell populations expressed IL-2 (Fig. 1C).
To correlate Ets-2 mRNA synthesis with Ets-2 protein DNA binding activity, we performed electrophoretic mobility shift assays (EMSAs) using nuclear extracts prepared from PBMC-derived T-cells, monocytes, B-cells, and naive and memory Th cells using the IL-2 ARRE-2 as a probe (Fig. 1D). The results show a strong DNA binding activity when extracts from T-cells or naive Th cells were used, whereas protein extracts prepared from memory Th cells, B-cells, or monocytes had no binding activity to ARRE-2.
To test for specificity of binding activity to the ARRE-2, we performed competition assays mixing naive Th cell extracts with a 5-, 20-, or 50-fold excess of unlabeled ARRE-2 oligonucleotides. The resulting EMSA demonstrated that even at 5-fold excess of unlabeled ARRE-2 the binding activity to the probe was competed (Fig. 1D, right panel).
To verify Ets-2 protein binding to ARRE-2, we performed EMSA with Ets-2-depleted extracts from naive Th cells and demonstrated diminished binding activity (Fig. 1D, middle panel, Ets-2 Depletion). In addition, we performed EMSA with purified Ets-2 extracts isolated from peripheral blood T-cells. The protein extracts were passed through an Ets-2 antibody-binding column, and EMSAs were performed using the ARRE-2 probe mixed with consecutive protein eluates. As shown in Fig. 1E, Ets-2 protein bound to the ARRE-2 in a dose-dependent manner. Thus, Ets-2 expression correlates with DNA binding at the ARRE-2 promoter element.
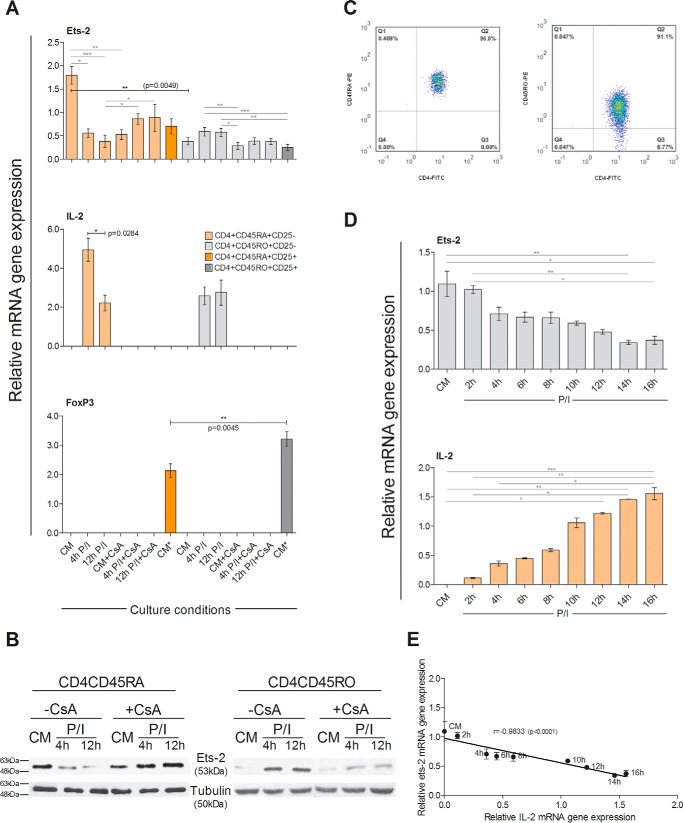
Ets-2 Expression Patterns in Th Cells under Different Culture Conditions, Effect of Cyclosporine A (CsA), and Kinetic Analysis of Ets-2 and IL-2 Expression in Naive Th Cells
Naive Th cells and Tregs, as a control, were isolated from cord blood-derived T-cells by negative and positive selection respectively. Naive Th cells were tested for their purity by flow cytometry (Fig. 2C, left panel) and were subsequently cultured ±PI ±CsA for 4 or 12 h. The levels of Ets-2, Foxp3, and IL-2 mRNA were determined by real time PCR. Ets-2 mRNA levels in naive Th cells were reduced to 31.1% at 4 h and 21.1% expression at 12 h after P/I stimulation compared with CM (−CsA) (Fig. 2A). Inversely, IL-2 mRNA was highly expressed in naive Th cells after P/I stimulation (Fig. 2A). The addition of CsA blocked IL-2 expression in naive Th cells and led to a reduction of the Ets-2 mRNA to 47.8 and 48.9% of the level of expression at 4 and 12 h after P/I activation, respectively, compared with CM (−CsA) (Fig. 2A).
FIGURE 2.
Ets-2 and IL-2 mRNA synthesis in Th cells under different culture conditions and effect of CsA. A, Ets-2, IL-2 and Foxp3 mRNA levels were measured by real time PCR in naive Th cells (CD4CD45RA) and Tregs (CD4CD25) isolated from cord blood and in memory Th cells (CD4CD45RO) and Tregs (CD4CD45ROCD25) isolated from adult PBMCs. Naive and memory Th cells were cultured ±CsA in culture medium (CM) ± P/I for 4 or 12 h. Tregs isolated from cord blood or PBMCs were cultured in CM only. Reverse transcribed β2-m was used as an internal normalizer for all samples. B, Western blotting analysis of Ets-2 protein levels in CD4CD45RA and CD4CD45RO Th cells in CM or following a 4- or 12-h induction +P/I ±CsA. Tubulin protein levels were used as an internal control for equal loading. The results shown in A and B are representatives of three independent experiments that all yielded similar results. C, FACS analysis of the isolated naive and memory Th cells (Q2) used in A and B. D, kinetic analysis of Ets-2 and IL-2 mRNA expression in naive Th cells following mitogenic stimulation. Naive (CD4+CD45RA+CD25−) Th cells were isolated from cord blood by cell sorting. The enriched cell population (>95%) was cultured with P/I for 2–16 h, and Ets-2 and IL-2 mRNA expression was assayed with real time PCR at the times indicated using β2-m as an internal normalizer in each sample. The results shown are the mean values of three independent experiments; error bars represent S.D. Statistically significant differences are indicated by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001) and were compared with two-way ANOVA (more than two groups) or Student's t test (two groups) (*, p < 0.05; **, p < 0.005). E, there was a significant negative correlation between expression levels of Ets-2 mRNA and IL-2 mRNA in naive Th cells (Spearman's r = −0.9833, p < 0.0001).
The same experiments were performed with memory Th cells and Tregs, as a control, isolated from adult PBMCs by negative and positive selection, respectively. Memory Th cells were tested for their purity by flow cytometry (Fig. 2C, right panel) and were subsequently cultured ±PI ±CsA for either 4 or 12 h. The levels of Ets-2, Foxp3, and IL-2 mRNA were determined by real time PCR analysis, which revealed distinct patterns of Ets-2 mRNA expression in memory versus naive Th cells: Ets-2 mRNA expression was ×4.7-fold lower in memory Th cells compared with naive Th cells (Fig. 2A, CM). Mitogenic stimulation of memory Th cells for 4 and 12 h induced the expression of Ets-2 mRNA ×1.5- and ×1.6-fold, respectively, compared with Ets-2 mRNA levels in unstimulated (CM) memory Th cells, whereas IL-2 mRNA was expressed normally. In contrast, mitogenic activation in the presence of CsA blocked IL-2 expression without affecting significantly Ets-2 mRNA expression (Fig. 2A).
Foxp3 was not expressed in either naive or memory Th cells under any of the culture conditions tested (Fig. 2A), whereas it was expressed in cord blood or PBMC-derived Tregs (Fig. 2A). Because Foxp3 is a known repressor of IL-2 expression in Tregs, acting through binding to the ARRE-2 and ARRE-1 of the IL-2 promoter (NFAT binding sites) (26), our results demonstrate that Ets-2 is a different suppressor of IL-2 expression from Foxp3 with the two factors acting in different types of Th cells. To note, Tregs expressed Ets-2 mRNA at different levels: adult PBMC-derived Tregs expressed 3× less Ets-2 mRNA than cord blood-derived Tregs (Fig. 2A).
Western blotting analysis (Fig. 2B) demonstrated that the changes in the quantity of the Ets-2 protein correlated with the changes in Ets-2 mRNA expression and the effect of CsA thereof, thus suggesting that regulation of Ets-2 expression in naive and memory Th cells occurs at the transcriptional level. To study further the relationship between Ets-2 and IL-2 gene expression, we performed a time course analysis. Cord blood-derived naive Th cells were activated with P/I for a time period spanning 16 h, and the relative levels of Ets-2 and IL-2 mRNA were determined by real time PCR (Fig. 2D). The results showed that Ets-2 mRNA levels began to decline at 2 h postactivation to reach minimum levels at 14 h, whereas IL-2 mRNA was evident at 2 h postactivation and increased steadily over the 16-h culture period; i.e. Ets-2 mRNA gene expression in naive Th cells correlated negatively with IL-2 mRNA gene expression (r = −0.9833) (Fig. 2E). Taken together, these results suggest that IL-2 and Ets-2 are differentially regulated in naive and memory Th cells in that only in naive Th cells does a high level of Ets-2 expression coincide with the transcriptional repression of IL-2, and IL-2 activation coincides with the disappearance of Ets-2 mRNA and protein.
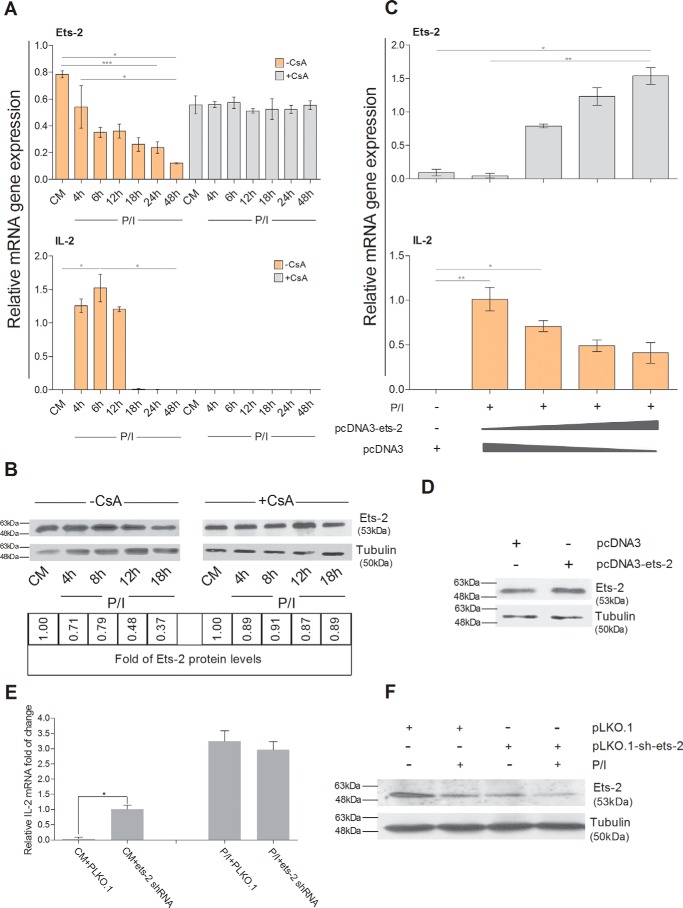
Overexpression of Ets-2 Decreases IL-2 mRNA Expression in Jurkat T-cells
To assess the direct role of Ets-2 on the transcriptional activity of the IL-2 promoter, we transfected Jurkat T-cells with an Ets-2-expressing vector (pcDNA3-ets-2) and determined the levels of IL-2 mRNA expression after P/I activation in the presence or absence of CsA.
Jurkat cells were chosen because they express high levels of IL-2 after P/I stimulation, and this activation is inhibited by CsA (27) (Fig. 3A). Jurkat cells express Ets-2 mRNA in CM, and after 4 h of mitogenic stimulation Ets-2 mRNA and protein levels begin to decrease (Fig. 3, A and B, −CsA). Furthermore, CsA stabilizes Ets-2 mRNA and protein in Jurkat cells (Fig. 3, A and B, +CsA). Thus, as is the case for naive Th cells, a negative correlation exists between Ets-2 mRNA/protein levels and IL-2 mRNA expression in Jurkat cells.
FIGURE 3.
Ets-2 and IL-2 expression in Jurkat cells. A, Jurkat cells were cultured in CM ± CsA ± P/I for a period of 4–48 h. Total RNA was isolated, cDNAs were prepared at the times indicated, and Ets-2 and IL-2 mRNA levels were measured by real time PCR using β2-m as an internal normalizer. B, Western blotting analysis of Ets-2 protein levels in Jurkat cells cultured in CM ± P/I ± CsA for a period of 4–16 h. Total protein cell extracts were prepared at the times indicated. Tubulin was used as an internal control for equal loading. Ets-2 protein levels were normalized according to tubulin levels and are also represented as -fold of expression with Ets-2 protein levels in CM ± CsA as baseline. C, Ets-2 silences IL-2 mRNA expression in Jurkat cells in a dose-dependent manner. Ets-2 and IL-2 mRNA expression was measured by real time PCR in Jurkat cells transfected with different amounts of a pcDNA3-ets-2 plasmid (0, 10, 20, and 30 μg) that overexpresses Ets-2. The amount of transfected DNA/sample was retained to 60 μg by the addition of appropriate amounts of vector pcDNA3 DNA. The cells were cultured in CM before transfection, retained for 6 h in CM after transfection to recover, and then transferred to CM + P/I for an additional 6 h. β2-m expression was used as an internal normalizer. D, Western blotting analysis to confirm expression of Ets-2 protein in transfected Jurkat cells. Jurkat cells were transfected with 20 μg of pcDNA3-ets-2 DNA for 6 h and then lysed for preparation of protein extracts. Tubulin was used as an internal control for equal loading. E, Jurkat cells were transfected with 2 μg of pLKO.1 vector or a mixture of five independent shRNA-ETS-2 clones and then cultured in CM ± P/I for 6 h. Total RNA was isolated, cDNAs were prepared, and IL-2 mRNA levels were detected by real time PCR using β2-m as an internal control. F, Western blotting analysis for verifying the silencing of Ets-2 performed with extracts prepared from transfected Jurkat cells with a mixture of the shRNA-ETS-2 clones cultured in CM ± P/I for 6 h. Tubulin was used as an internal control for equal loading. The results shown in A, C, and E are the mean values of three independent experiments; error bars represent S.D. (*, p < 0.05; **, p < 0.01; ***, p < 0.001, two-way ANOVA; *, p < 0.05, Student's t test). The results shown in B, D, and F are representative of three independent experiments.
Fig. 3C shows that expression of increasing amounts of Ets-2 caused a gradual decrease of the P/I-induced activation of the endogenous IL-2 expression. This decrease continued at a standard rate as the amount of overproduced Ets-2 increased, reaching a 60% drop when the maximum amount of the pcDNA3-ets-2 plasmid was used for transfection. Overexpression of the Ets-2 protein was verified by Western blotting (Fig. 3D).
To investigate whether silencing of the Ets-2 factor per se results in constitutive IL-2 gene expression, we transfected Jurkat cells with lentivirus vectors containing short hairpin RNA (shRNA) clones to silence the endogenous Ets-2 mRNA. The cells were cultured ±P/I for 6 h. Reduction of the Ets-2 protein was verified by Western blotting analysis (Fig. 3F), and IL-2 mRNA expression was assayed by real time PCR. The results (Fig. 3E) show that when Ets-2 expression is inhibited in unstimulated Jurkat cells, i.e. in the absence of functional NFAT (CM + ets-2 shRNA), low expression of IL-2 mRNA ensues, confirming that Ets-2 acts as an independent repressor of IL-2 expression. Conversely, in P/I-stimulated Jurkat cells, the reduction of Ets-2 expression by shRNAs does not affect IL-2 expression. Taken together, these data confirm the negative causality between Ets-2 and IL-2 expression and that Ets-2 acts as a preinduction repressor of IL-2 transcription.
Ets-2 Represses the Activation of the IL-2 Promoter through the ARRE-2
Because the putative repressor of IL-2 expression works through the ARRE-2 of the IL-2 promoter, we next sought to determine whether Ets-2 represses IL-2 transcription via the ARRE-2.
To this end, we cotransfected P/I-stimulated Jurkat cells with increasing amounts of the pcDNA3-ets-2 expression vector together with (i) a CAT reporter plasmid bearing the human proximal IL-2 promoter (−300/+1), (ii) a CAT reporter plasmid bearing the human proximal IL-2 promoter (−300/+1) with a mutation in the ARRE-2 (the GGAA Ets-2 binding site was changed to CTAA), (iii) a CAT reporter bearing four copies of the ARRE-2, (iv) a CAT reporter bearing four mutant copies of the ARRE-2, or (v) a CMV-CAT plasmid as a control. The results (Fig. 4) show that high level expression of Ets-2 resulted in a significant reduction of expression of both the IL-2-CAT (Fig. 4A) and 4xARRE-CAT (Fig. 4C) reporters in a dose-dependent manner but had no effect on the expression of IL-2-CAT reporter with the mutated ARRE-2 (Fig. 4B), 4xmutantARRE-CAT (Fig. 4D), and CMV-CAT (Fig. 4E). The above results are in accordance with the EMSA results depicted in Fig. 1, D and E, and show that the repression of the IL-2 gene expression is mediated through the binding of the Ets-2 protein to the ARRE-2 promoter element.
FIGURE 4.
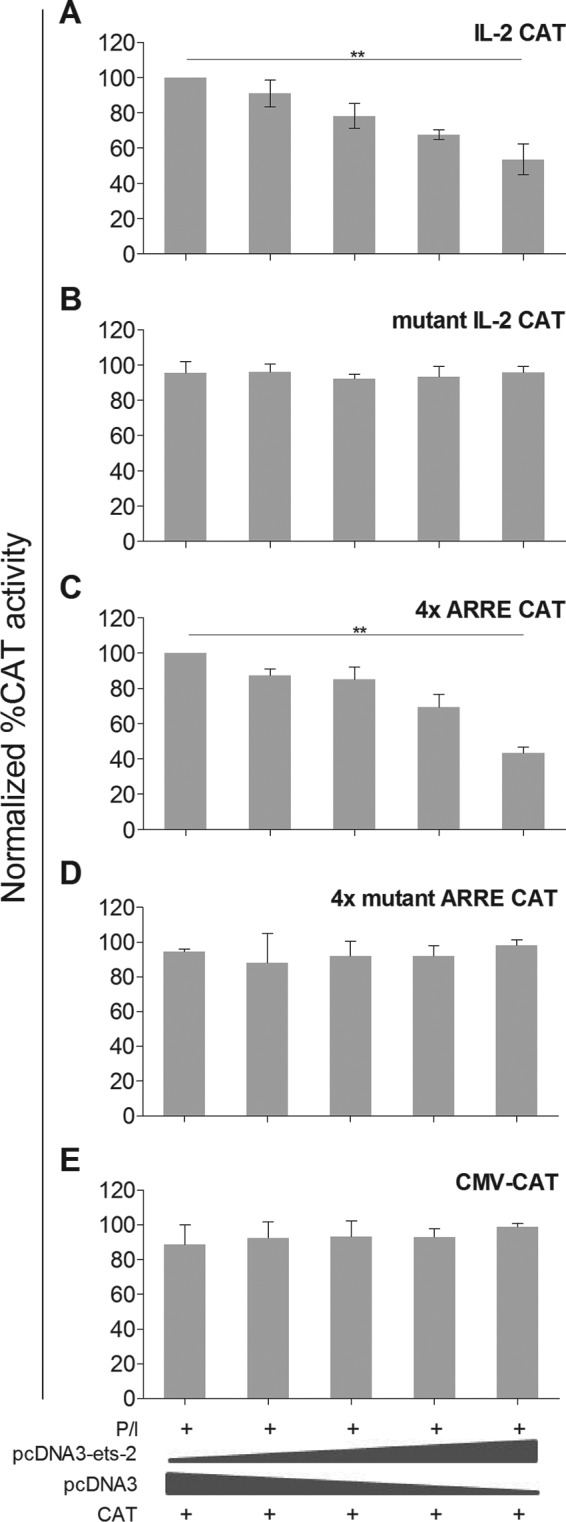
Ets-2 silences IL-2 mRNA expression through the ARRE-2. A, CAT enzyme expression driven by a 301-bp IL-2 upstream promoter region (IL-2 CAT). B, CAT enzyme expression driven by a 301-bp IL-2 upstream promoter region but with the ARRE-2 mutated (GGAA to CTAA) (mutant IL-2 CAT). C, four repeats of the ARRE-2 of the IL-2 promoter (4x ARRE CAT). D, four mutant repeats of the ARRE-2 of the IL-2 promoter (4x mutant ARRE CAT); in C and D, the ARRE-2 repeats were placed upstream of a minimal promoter of herpesvirus thymidine kinase. E, CMV promoter as a control (CMV-CAT). CAT assays were performed in extracts from Jurkat cells transfected with different amounts of pcDNA3-ets-2 DNA (0, 5, 10, 20, and 40 μg) that overexpressed Ets-2 and a constant amount (20 μg) of each CAT reporter. The amount of transfected DNA/sample was retained to 60 μg by the addition of appropriate amounts of vector pcDNA3 DNA. The cells were cultured in CM before transfection, retained for 6 h in CM after transfection to recover, and then transferred to CM + P/I for an additional 6 h. The results in A–E show the percentage of relative CAT activity from three independent experiments with the highest CAT activity in each panel used as baseline; error bars represent S.E. (**, p < 0.01, two-way ANOVA).
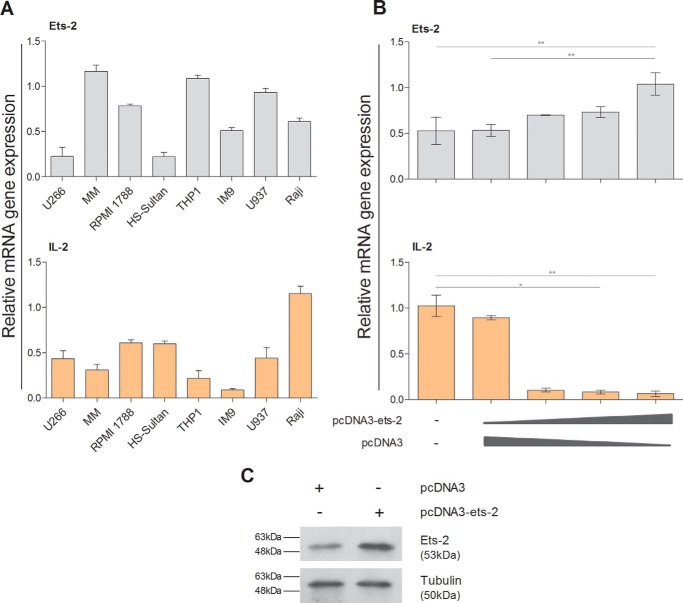
Ets-2 Is a General Repressor of IL-2 Expression
To test whether Ets-2 alone can repress IL-2 expression, i.e. in a non-T-cell environment, we searched for cell lines that express IL-2 mRNA constitutively and investigated whether overexpression of Ets-2 reduces their IL-2 mRNA levels. We found that the leukemic cell lines U266, RPMI 1788, Sultan, THP1, IM9, U937, Raji, and multiple myeloma (MM) cells express the endogenous Ets-2 and IL-2 genes at varied levels with MM and THP1 cells showing the highest constitutive expression of Ets-2 and Raji cells showing the highest constitutive expression for IL-2 (Fig. 5A).
FIGURE 5.
Ets-2 is a general repressor of IL-2. A, Ets-2 and IL-2 mRNA levels in different leukemic cell lines measured by RT-PCR. All cells were cultured in CM. Reverse transcribed β2-m from each cell line was used as an internal control. B, Ets-2 and IL-2 mRNA levels were measured by real time PCR in Raji cells transfected with varying amounts of pcDNA3-ets-2 plasmid (0, 10, 20, and 30 μg). The amount of transfected DNA/sample was retained to 60 μg by the addition of appropriate amounts of vector pcDNA3 DNA. Cells were kept in CM after transfection for 6 h. β2-m was used as an internal control. Error bars represent S.E. (*, p < 0.05; **, p < 0.01, two-way ANOVA). C, Western blotting analysis to confirm overexpression of Ets-2 protein in transfected Raji cells. Raji cells were transfected with 20 μg of pcDNA3-ets-2 DNA or pcDNA3 and lysed 6 h post-transfection, and total protein extracts were prepared. Tubulin was used as an internal control for equal loading. The results shown in A, B, and C are representative of three independent experiments.
Raji cells were chosen to assess whether overexpression of Ets-2 protein can down-regulate IL-2 expression levels. To this end, they were transfected with increasing amounts of the Ets-2 expression vector pcDNA3-ets-2, and the levels of the endogenous IL-2 mRNA were determined by real time PCR. The levels of overexpressed Ets-2 were determined by Western blotting (Fig. 5C). The results show that overexpression of Ets-2 in Raji cells blocks the synthesis of endogenous IL-2 mRNA (Fig. 5B). Therefore, Ets-2 can also act as a repressor of IL-2 expression in a heterologous system, such as the B-cell line Raji, suggesting that it is a general repressor of IL-2 expression.
Chromatin Immunoprecipitation (ChIP) Analysis to Assay the Physiological Interactions between Ets-2 and IL-2 Promoter in Living Cells
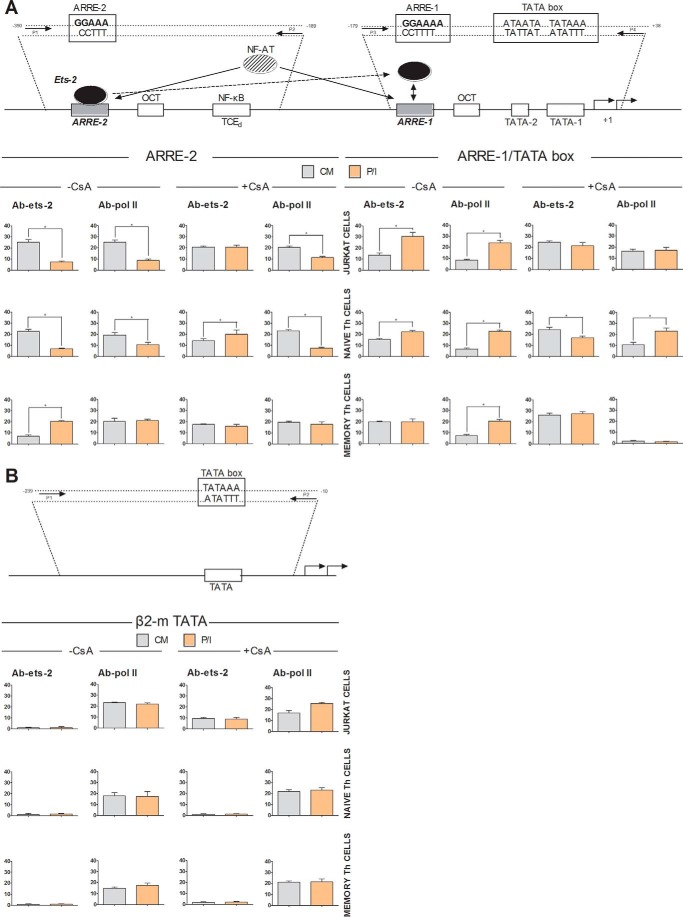
To assay the physiological interactions between Ets-2 and IL-2 promoter in vivo, we performed ChIP analysis using chromatin prepared from Jurkat cells, cord blood-derived naive Th cells, and adult PBMC-derived and in vitro expanded memory Th cells cultured in the presence or absence of P/I or CsA for 6 h. Cross-linked chromatin was subjected to immunoprecipitation using antibodies against human Ets-2 and POLII or a negative control antibody to IgG; the association of Ets-2 or POLII with the IL-2 promoter was detected by real time PCR using specific sets of primers. We examined Ets-2 binding at the ARRE-2 and the region of the core IL-2 promoter that includes the TATA box and ARRE-1 (that also contains Ets-2/NFAT binding sequences) (Fig. 6A). A chromatin region that encompasses the human β2-m gene promoter, including the TATA element, was chosen as a negative control for Ets-2 binding (Fig. 6B).
FIGURE 6.
Ets-2 binding to the endogenous IL-2 loci by ChIP analysis. A, ChIP analysis in Jurkat cells, CD4+CD45RA+CD25− naive Th cells (CD4CD45RA), and CD4+CD45RO+CD25− in vitro generated memory Th cells (CD4CD45RO) by real time PCR. The schematic representation of the human IL-2 promoter shows the relative positions of selected transcription factors, the ARRE-2 and ARRE-1, the TATA box, and the transcriptional initiation start site (+1). The positions of the primer pairs used for the PCR amplifications are indicated by arrows. The P1-P2 primer pair was used to amplify the region mapping the ARRE-2 and produced a 200-bp fragment. The P3-P4 primer pair was used to amplify the region mapping the TATA box and the ARRE-1 and produced a 140-bp fragment. The cells were cultured in CM ± P/I ± CsA for 6 h. They were then harvested, and their extracts were prepared for ChIP analysis. ChIP was carried out using antibodies to Ets-2 and POLII (and IgG for background determination). Before immunoprecipitation, the cross-linked chromatin templates of the samples were equalized by UV measurements. Real time PCR on cross-linked chromatin from cells cultured in CM or P/I before immunoprecipitation was used as a control to calculate the baseline chromatin amount for each PCR condition and set of primers. The results represent the DNA enrichment as a percentage of immunoprecipitated chromatin (IP) for every condition and set of primers relative to corresponding chromatin input (according to the equation 100 × ((Ct IP/Ct input) − (Ct IgG/Ct input) where Ct is the cycle at which the threshold line is crossed). B, to control for the accuracy of the ChIP assays and for the specificity of the antibody and the amplified regions, real time PCR was performed to detect the TATA region of the β2-m gene promoter (β2-m TATA). The schematic representation of the human β2-m promoter shows the TATA box, the transcriptional initiation start site (+1), and the positions of the primer pairs used for the PCR amplifications as indicated by the arrows. The results shown are the mean values of three independent experiments; error bars represent S.E. (*, p < 0.05, Student's t test). The control antibody to IgG failed to precipitate chromatin.
The results show that in unstimulated Jurkat and naive Th cells Ets-2 is bound to the region encompassing the ARRE-2. P/I activation led to the departure of Ets-2 from the ARRE-2 and the binding of POLII to the core promoter (TATA box), a result consistent with the activation of IL-2 in both Jurkat and naive Th cells (Fig. 6A, −CsA). The inability to detect significant Ets-2 binding to ARRE-2 in activated naive Th or Jurkat cells (6-h P/I) coincides with the decrease of Ets-2 mRNA and Ets-2 protein (Figs. 2 and 3).
The opposite was observed in memory Th cells where the ARRE-2 region was amplified in activated (P/I) compared with resting cells (CM) (Fig. 6A, −CsA). The binding of Ets-2 to ARRE-2 in P/I-activated memory Th cells coincided with the increase of Ets-2 mRNA (Fig. 2A) and protein (Fig. 2B) in these cells.
In contrast, Ets-2 binding to the IL-2 TATA/ARRE-1 region was increased in stimulated Jurkat and naive Th cells, whereas in memory Th cells Ets-2 was bound almost equally to the TATA/ARRE-1 region in both CM and P/I conditions (Fig. 6A, −CsA). The strength of the PCR amplification in the ChIP assays coincided with the amounts of Ets-2 mRNA and Ets-2 protein shown in Figs. 2 and 3.
Incubation of cells with CsA resulted in stabilization of Ets-2 binding to ARRE-2 even after P/I activation (Fig. 6A, +CsA). These results are consistent with the stabilization of Ets-2 mRNA and protein shown in Figs. 2 and 3 (+CsA). Furthermore, CsA blocked the binding of the Ets-2 protein to the TATA/ARRE-1 region. Because P/I-dependent NFAT activation of IL-2 expression is blocked in the presence of CsA, we conclude that the decrease of the binding status of Ets-2 to the ARRE-2 promoter element is a prerequisite for the transcriptional activation of IL-2.
Using an antibody to human POLII, similar results were obtained for both Jurkat and naive Th cells (Fig. 6A, −CsA). Before transcriptional induction (cells cultured in CM), POLII was bound to the ARRE-2 region, whereas after P/I induction POLII binding was detected at the IL-2 TATA/ARRE-1 region. The presence of POLII at the ARRE-2 in resting conditions may reflect its association with a factor(s) that binds to this element in a “ready to use state.” The subsequent presence of POLII in the TATA/ARRE-1 region in P/I conditions is in agreement with the transcriptional activation of the IL-2 gene. In contrast, in memory Th cells, POLII binds equally well to the ARRE-2 region in both resting and activated cells (Fig. 6A, −CsA). In agreement with the results for Jurkat and naive Th cells, transcriptional induction coincides with the interaction of POLII with the TATA/ARRE-1 region. Blocking the transcriptional activation pathway of the IL-2 gene by CsA also blocked binding of POLII to the core promoter (Fig. 6A, +CsA).
Control ChIP assays using the Ets-2 antibody revealed no binding of Ets-2 to the β2-m gene promoter around the TATA box under all conditions tested and in all cell types, a result that validates the specificity of our experiments (Fig. 6B). In contrast, using antibody to POLII, the genomic region around the TATA box of β2-m promoter was precipitated under all culture conditions at equal amounts per cell type, a result consistent with the expression properties of the β2-m gene (Fig. 6B).
Taken together, the results show that in naive Th and Jurkat cells Ets-2 binds to the chromatin region that encompasses the ARRE-2 of the IL-2 promoter when the IL-2 gene is not transcriptionally active. Induction of the IL-2 gene is accompanied with reduced binding of Ets-2 to the ARRE-2 and, at the same time, increased binding to the ARRE-1. In contrast, in memory Th cells Ets-2 binds more strongly to the ARRE-2 in P/I conditions and equally to the ARRE-1 region under CM or P/I conditions. Therefore, Ets-2 is a preinduction repressor of IL-2 transcription in naive Th cells acting through the ARRE-2 of the IL-2 promoter. Furthermore, a movement of Ets-2 binding from ARRE-2 to ARRE-1 is revealed when naive Th cells are activated. This movement constitutes ARRE-2 acting as a cis element necessary for the transcriptional activation of IL-2 in memory Th cells.
Ets-2 and NFAT Do Not Interact Physically
ARRE-2 is a binding site for both Ets-2 and NFAT factors through overlapping DNA sequences. NFAT binding is necessary for the transcriptional activation of IL-2 in Th cells. We maintain that, upon cell activation, Ets-2 must be released from the ARRE-2, which will then be occupied by NFAT. A question that remains open is whether this replacement involves the physical interaction between the two factors. To answer this, we performed coimmunoprecipitation assays with extracts prepared from Jurkat cells cultured in CM ± P/I for 6 h. The results (Fig. 7A) show that by using human anti-Ets-2 antibody as the fist antibody we were unable to detect any physical interactions between Ets-2 and NFAT-1 or NFAT-2 in both unstimulated and stimulated cells. Parallel experiments confirmed the functional integrity of NFAT, showing that NFAT interacted with c-Fos (28, 29) (Fig. 7B). Therefore, Ets-2 must depart from ARRE-2 to permit NFAT binding upon cell activation, and during this process there is no physical interaction between Ets-2 and NFAT.
FIGURE 7.
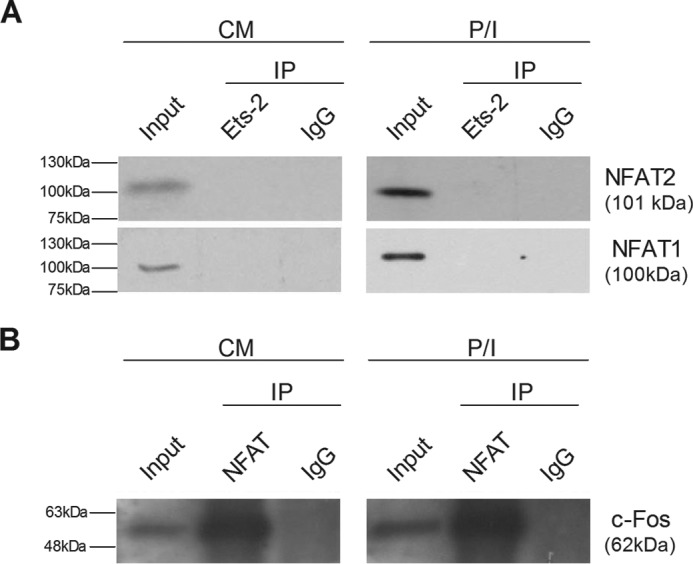
Ets-2 and NFAT do not interact physically. A, Jurkat cells were cultured in CM or P/I, harvested, and subjected to immunoprecipitation (IP) with a human anti-Ets-2 antibody. Detection of interactions of Ets-2 with NFAT-1 or NFAT-2 was carried out by Western blotting analysis with the corresponding specific antibodies. B, as a positive control, extracts from the same cells were immunoprecipitated with a human anti-NFAT antibody for the detection of the interaction of NFAT with c-Fos by Western blotting analysis. An anti-human IgG antibody was used as a negative control to confirm the specificity of the immunoprecipitations.
Discussion
A major question in immunology and immunopathology is the nature of the molecular mechanisms that underlie the developmental transition of naive (uncommitted) Th cells to the status of proliferating Th0 precursors, to Th0 immature effector cells, and then to the various Th lineages (2, 4). Although the role of NFAT and other transcription factors is well documented when induction of Th cells takes place (6, 7), little is known about the molecules and the mechanism that restrict the induction of the response pathway to the antigen in naive effectors. The repression mechanism that controls IL-2 transcription in naive Th cells is possibly part of a general mechanism that controls the developmental changes at the molecular level. Data from our previous work (9–12) have pointed to a mechanism in which the pivotal role should be attributed to a molecule or molecules that exist and/or function as suppressors of IL-2 transcription in naive Th cells but disappear and/or change their function in activated or memory Th cells. Analogous patterns of expression have been documented for already identified suppressors of Th cell activation or IL-2 transcription; for example, the expression of Foxj1, which controls the transcriptional expression of IκB, is eliminated when Th cells are activated (30). The same expression mode has been shown for the transcriptional repressors of IL-2 expression DREAM (31) and Tob (32). The product of the ets-2 oncogene is a molecule that fulfils a similar role (13) but with a different molecular and biological function to Tob and DREAM according to the present work that shows that Ets-2 acts as a preinduction repressor of IL-2 expression in naive Th cells. Preinduction repression mechanisms also play a significant role in the transcriptional expression of IFN-β (33). Such mechanisms possibly safeguard gene expression from spatiotemporal aberrant induction or, more importantly, from stochastic events that may lead single cells to express genes ectopically and/or out of time with disastrous results for cell fate or function.
As seen from the data presented in this work, the ets-2 gene is expressed at lower levels in ex vivo or resting memory Th cells compared with ex vivo or resting naive Th cells. This difference in the transcriptional expression rate of the ets-2 gene shows an alternative role or a different need for the ets-2 gene product in naive and post-naive Th cells.
The expression of ets-2 follows a completely different pattern in naive versus memory Th cells because, upon cell stimulation, it declines in naive cells and increases in memory cells. Nevertheless, when memory Th cells are stimulated, the amount of the Ets-2 mRNA is significantly lower compared with that of naive Th cells. The same is evident when NFAT binding, which is necessary for the expression of the IL-2 gene, is blocked by CsA. The level of Ets-2 mRNA in memory Th cells is significantly lower than in naive Th cells when both types of cells are stimulated in the presence of CsA. This implies that Ets-2 mRNA expression in naive and memory Th cells is independent of the presence of IL-2 transcriptional activators; i.e. a transcriptional repression mechanism exists in naive Th cells that implicates the expression levels of Ets-2 as a key event in the transition of Th cells from naive to memory. The observation that Ets-2 mRNA and protein levels are significantly decreased in stimulated memory Th cells and remain low in resting memory Th cells is in concert with EMSA results showing that the binding of the repressor to the ARRE-2 disappears in extracts from stimulated naive Th cells or that the repressor does not bind to ARRE-2 in resting memory Th cells (Refs. 10 and 11 and this work).
Our combined results from shRNA, ChIP, PCR, EMSA, and transfection experiments demonstrate a significant inhibitory potential for the Ets-2 protein in naive Th cells via its binding to the ARRE-2 of the IL-2 promoter. The binding activity of Ets-2 protein on the ARRE-2 of IL-2 promoter disappears as soon as the naive Th cells are activated. In addition, overexpression of exogenous Ets-2 in Jurkat cells leads to a decrease of endogenous IL-2 transcription and of IL-2 promoter or ARRE-2 expression-driven reporter genes in a specific and dose-dependent manner, demonstrating that the amount of Ets-2 is of critical importance for its silencing activity (Fig. 4, A and C). Conversely, overexpression of exogenous Ets-2 in Jurkat cells does not affect the expression of reporter plasmids driven by the mutant ARRE-2 either in the context of the IL-2 promoter or as a multiple mutant Ets-2 binding target site in mutant ARRE-2 expression-driven reporter genes (Fig. 4, B and D). Reversely, silencing of Ets-2 in resting Jurkat T-cells results in the transient expression of IL-2 (Fig. 3E). These observations reinforce the notion that Ets-2 binding to ARRE-2 and Ets-2 availability are key prerequisites to Ets-2 repression function. To note, the silencing activity of Ets-2 on IL-2 expression is not restricted to T-cells because overexpression of Ets-2 in the B leukemic cell line Raji, which expresses high levels of IL-2 mRNA constitutively, inhibits the expression of the endogenous IL-2 gene.
Our results from the ChIP assays suggest a different role of Ets-2 in naive and activated naive Th cells versus memory Th cells. Ets-2 is not only expressed at lower levels in resting memory Th cells compared with naive Th cells but also shows a reverse cooperation with the ARRE-2 in memory Th cells versus naive Th cells when the cells are activated. When memory Th cells are quiescent, Ets-2 binds to the ARRE-1 of IL-2 promoter, and this binding is not significantly altered when memory Th cells are activated. At the same time, when memory Th cells are activated, Ets-2 also binds to the ARRE-2. In contrast, upon transcriptional activation, Ets-2 seems to move from ARRE-2 to the ARRE-1 region of the human IL-2 promoter in naive Th and Jurkat cells, a transition that is part of the process during which Ets-2 aborts the role of IL-2 preinduction repressor. This movement does not take place in memory Th cells, probably because a cooperation of Ets-2 with the ARRE-1 has been established during their prior activation as naive cells. The movement of Ets-2 from the ARRE-2 to the ARRE-1 after naive Th cell activation is possibly fixed to a permanent status in memory Th cells, reflecting a mechanism of fast transcriptional response when memory Th cells encounter their cognate antigen.
The binding of Ets-2 to the ARRE-1 in activated naive and quiescent memory Th cells and the change in Ets-2 mRNA expression levels are critical events for aborting the role of a repressor. Abortion of the repressor function of Ets-2 is in agreement with other reports that attribute a dual role of transcriptional repressor and/or activator to Ets-2 (21) or that show the engagement of Ets-2 in the activation and subsequent transcription of IL-2 (34).
These findings fit well with the current understanding of the role of the transcription factors of the ETS family in controlling the expression of a wide variety of genes in hemopoietic and other cell types (13–15). Ets factors share a common DNA binding motif, the ets domain, which has been shown to be a variation of the winged helix-turn-helix motif (35). Ets-1, Ets-2, and other members of the same family make contacts with a 20-bp sequence containing a central GGAA motif, which also exists in the IL-2 promoter ARRE-2 and ARRE-1 cis elements.
We propose that Ets-2 is highly expressed in naive Th cells and binds preferentially to the GGAA motif of the ARRE-2, blocking the expression of IL-2. Because active NFAT is not present in resting cells, direct binding of Ets-2 to the ARRE-2 of the IL-2 promoter provides a physical barrier to the basic transcriptional apparatus alone or together with factors, such as chromatin modulators, transcription factors, or the chromatin 3D structure per se. Ets-2 must depart from ARRE-2 to permit NFAT binding upon cell activation. Accordingly, coimmunoprecipitation experiments (Fig. 7) showed that there is no physical interaction between Ets-2 and NFAT.
Upon mitogenic stimulation of naive Th cells, Ets-2 does not bind to the ARRE-2 any longer. In contrast, it binds to the ARRE-1 that has been shown to be important for IL-2 transcriptional activation (6, 36). The subsequent lower expression of Ets-2 in memory Th cells renders it unable to act as a repressor, so it no longer has an inhibitory effect on IL-2 expression. Therefore, when it is necessary to produce IL-2 mRNA again, Th cells can perform this faster because they do not have to overcome the Ets-2 barrier. In a sense, Ets-2 expression levels may act as an epigenetic memory key, which marks the transition from the naive to the memory status of Th cells. In this context, the tight preinduction repression control of IL-2 mRNA in naive Th cells is no longer necessary.
We suggest that Ets-2 expression and protein binding to the ARRE-2 of the IL-2 promoter are part of a strictly regulated process that results in a physiological transition of naive Th cells to Th0 cells upon antigenic stimulation. Malfunction of such a preinduction repression mechanism at the molecular level could lead to the disturbance of later events in Th cell plasticity, leading in turn to autoimmune diseases (37) or other pathological conditions.
Experimental Procedures
Cells
Cord blood (CB) samples (10–15 ml) of healthy full term neonates (Neonatal Intensive Care Unit, St. Andrew's General Hospital, Patras, Greece) and peripheral blood samples (10–30 ml) from healthy adults (Faculty of Medicine and Blood Transfusion Center, Patras University Hospital, Patras, Greece) were collected in heparinized tubes. When specifically stated, 200–300 ml of blood from underweighted blood collections were used (Blood Transfusion Center). The cell lines used were IM9 and RPMI 1788 (Epstein-Barr virus-transformed B lymphoblastoid cells), HS-Sultan and Raji (Burkitt's lymphoma), U266 (plasmacytoma) (European Collection of Cell Cultures, Salisbury, Wiltshire, UK), Jurkat (T-cell acute lymphoblastic leukemia), THP-1 (acute monocytic leukemia), U937 (histiocytic lymphoma) (American Type Culture Collection, Manassas, VA), and MM cells derived from a patient with MM IgG/κ type stage IIIA and kept in culture for 9 months without the addition of exogenous growth factors.
Cell Isolation, Phenotyping, and Cultures
Mononuclear cells were isolated from blood by centrifugation over a Ficoll-Paque gradient (Biochrom AG, Berlin, Germany) and washed four times with ice-cold RPMI 1640 culture medium (Gibco). For the isolation of pure cell populations from PBMCs or CB mononuclear cells, all the procedures were carried out at 4 °C. The isolated cell populations were phenotyped and used when their purity was >90%. Flow cytometric acquisition and analysis were performed on at least 10,000 acquired events per sample using a Coulter EPICS-XL-MCL cytometer. The data were analyzed using FlowJo version 7.5 software (Tree Star Inc., Ashland, OR).
CD3+ T-cells, CD19+ B-cells and CD14+ monocytes were isolated using antibody-coated magnetic bead kits for the isolation of untouched human T-cells, B-cells, or monocytes from PBMCs at a concentration of 106 cells/ml, two beads/cell (Dynabeads®, Invitrogen, Biotech ASA, Oslo, Norway). The antibodies used for FACS analysis were the mouse anti-human monoclonal antibodies (mAbs) CD3-PE-Cy5 (Beckman Coulter, France), CD19-FITC (Beckman Coulter), and CD14-FITC (Becton Dickinson Biosciences/Pharmingen). T helper (CD4+) and T cytotoxic (CD8+) cells were separated from CD3+ T-cells with the same method and phenotyped with the mAbs CD4-FITC (Beckman Coulter) and CD8-FITC (Beckman Coulter). From the isolated CD4+ T-cells, the CD25+ cells were removed by positive selection using the appropriate Dynabeads and stored frozen for further use. Naive Th cells (CD45RA+) were CB CD4+CD25− T-cells and were phenotyped with the mAb CD45RA-PE (Becton Dickinson Biosciences).
Memory Th cells (CD45RO+) were generated in vitro as follows. PBMCs from 200–300 ml of blood were cultured (2 × 106 cells/ml) with the mitogens phorbol myristate acetate (20 ng/ml) and ionomycin (2 μm) (P/I) overnight, washed two times with RPMI 1640 medium, and cultured in fresh plain CM (RPMI 1640 medium containing 10% fetal bovine serum and 1% penicillin/streptomycin) for 3 days. The CM was then replaced by fresh CM + P/I. This procedure was repeated three times. At the end of the culture period, the CM was changed, and the cells were cultured for 4 days. CD3+ T-cells and CD4+ T-cells were isolated by negative selection. CD25+ cells were then removed by positive selection and stored frozen. The remaining cells were phenotyped with the mAb CD45RO-PE (BD Biosciences). All types of cells were cultured in CM (106/ml) ± P/I ± CsA (1 μg/ml; kindly provided by Novartis, Basel, Switzerland) for 6 h unless indicated otherwise.
Quantitative Real Time PCR
For RT-PCR analysis, the cells were cultured for 6 h or for the times indicated. Real time quantitative PCR was performed using the Mx3000PTM Quantitative PCR System thermal cycler and MxProTM software (Stratagene Corp., La Jolla, CA). β2-m expression served as the normalizer gene. All measurements were done in triplicate. The following primers were used: for ets-2, 5′-CTCGTGTGTCTCAACCATCTT-3′ and 5′-CGCTCTGTGCCTCAGAATAG-3′, yielding a 112-bp PCR product; for il-2, 5′-CTCACCAGGATGCTCACATTTA-3′ and 5′-CCTCCAGAGGTTTGAGTTCTTC-3′, yielding a 97-bp PCR product; for foxp3, 5′-GAGCTGCCTACAGTGCCCCTA-3′ and 5′-CATTTGCCAGCAGTGGGTAG-3′, yielding a 150-bp PCR product; and for β2-m, 5′-TCGCGCTACTCTCTCTTTCT-3′ and 5′-TTTCCATTCTCTGCTGGATGAC-3′, yielding an 88-bp PCR product. The RT-PCRs were performed in a total volume of 20 μl containing 1 μl of cDNA, a 200 nm concentration of each primer pair, 10 μl of 2× KAPA SYBR Fast qPCR mixture, and 0.4 μl of 6-Carboxyl-X-Rhodamine low (KAPA SYBR FAST qPCR kit, Kapa Biosystems Inc., Woburn, MA). The optimal real time thermocycler conditions were 95 °C for 15 min followed by 40 cycles of 95 °C for 30 s to denature the cDNA, 58 °C for 30 s for annealing, and 72 °C for 30 s for extension.
DNA Plasmids
An Ets-2 expression vector, pcDNA3-ets-2, was generated by inserting a PCR-derived HindIII/BamHI fragment of a human ets-2 cDNA into a HindIII/BamHI-digested pcDNA3 plasmid (kindly provided by Dr. I. Talianidis, Alexander Fleming Biomedical Sciences Research Center Institute of Molecular Biology and Genetics, Athens, Greece). As a PCR template for the production of the ets-2 cDNA fragment, total human cDNA was mixed with Vent polymerase (New England Biolabs GmbH, Frankfurt am Main, Germany) and the specific ets-2 cloning primers 5′-CCCAAGCTTGGCAGGATGAATGATTTCGG and 5′-CGGGATCCTCAGTCCTCCGTGTCG. The sequence of pcDNA3-ets-2 plasmid was verified by automated sequencing analysis (Minotech, Crete, Greece). The target genes used were IL-2-CAT, which contains 301 bp of the 5′-flanking sequences of the human IL-2 gene (spanning nucleotides −300/+1) linked to the CAT gene; mutant IL-2-CAT, which contains 301 bp of the 5′-flanking sequences of the human IL-2 gene (spanning nucleotides −300/+1) with a mutated ARRE-2 (GGAA to TCAA) linked to the CAT gene; 4xARRE-CAT containing four copies of the ARRE-2 of the IL-2 promoter (−292/−264) linked to a truncated inactive thymidine kinase promoter (−270/+56); 4xmutantARRE-CAT containing four mutated copies of the ARRE-2 of the IL-2 promoter linked to a truncated inactive thymidine kinase promoter; and CMV-CAT (25) as a control. The mutant IL-2-CAT plasmid was constructed by inverse PCR using the wild-type human IL-2-CAT plasmid as a template mixed with Pfu polymerase (Thermo Scientific) and the specific mutant inverse PCR primers 5′-CCTTAAAGAAAGGACTAAAAACTGTTTC-3′ and 5′-GAAACAGTTTTTAGTCCTTTCTTTAAGG-3′. It was then subjected to digestion with DpnI for the removal of parental methylated and hemimethylated DNA; the derived plasmids were used for Escherichia coli transformation and were further subjected to sequencing analysis for the verification of the mutation.
Cell Transfections, shRNAs, and CAT Assays
Jurkat and Raji cells were transfected using Lipofectamine LTX DNA transfection reagents (Invitrogen, Life Technologies). The cells were transfected with various amounts of pcDNA3-ets-2 in the presence and absence of the target genes as indicated (see “Results”). When required, pcDNA3 was used to make up for the same amount of DNA per experimental point. For the experiments investigating the effect of shRNA on ets-2, five independent clones in pLKO.1 vectors containing shRNA to human Ets-2 (Mission shRNA, Sigma-Aldrich) were mixed and used for transfection of Jurkat cells as indicated (see “Results”). The transfected cells were allowed to recover in CM (with 5% FCS) overnight and were cultured for a further 6 h ±P/I as indicated. They were then processed either for CAT analysis, PCR, or Western immunoblotting as indicated. CAT assays were performed as described (25) using 40 μg of cell lysate per experimental point.
Immunoprecipitations
For immunoprecipitation experiments, Jurkat cells (107 cells per experimental point) were cultured in CM ± P/I for 6 h and then lysed in a buffer containing 50 mm Tris-HCl (pH 7.6), 150 mm NaCl, 5 mm EDTA, 5 mm MgCl2, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and Complete EDTA-free protease inhibitor mixture tablets (Roche Applied Science). The cell lysates were centrifuged at 13,000 rpm for 10 min at 4 °C, and after removal of 10% of the total volume of the supernatants to be used as input in the subsequent Western analysis, the remaining supernatants were precleared with 50 μl of Dynabeads Protein G per experimental point overnight. Immunoprecipitations were carried out for 3 h at 4 °C with 25 μl (5 μg) of human anti-Ets-2 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) or human anti-NFAT (Santa Cruz Biotechnology Inc.) antibody previously bound to 50 μl of Dynabeads Protein G per experimental point at 4 °C for 3 h. Immunocomplexes were washed three times with the lysis buffer and resuspended in 2× sample buffer. The immunoprecipitated proteins were analyzed by SDS-PAGE followed by Western immunoblotting with antibodies to human NFAT-1, NFAT-2, c-Fos (Santa Cruz Biotechnology Inc.), and IgG (BD Biosciences) as indicated (see “Results”).
SDS-PAGE and Western Immunoblotting
106 cells per experimental point were washed two times with PBS, collected by centrifugation at 4 °C for 1 min, and lysed in 20 μl of radioimmune precipitation assay buffer containing protease inhibitors (Sigma-Aldrich). After 20-min incubation on ice, the cell debris was removed by centrifugation, and the protein concentrations of the supernatants were estimated by the Bradford assay (Sigma-Aldrich). A volume that contained 10 μg of protein was mixed with an equal volume of 2× SDS sample buffer, and the samples were heated at 100 °C for 2 min. Total proteins were analyzed by SDS-PAGE followed by Western immunoblotting. The levels of Ets-2 protein were determined using a rabbit polyclonal Ab to human Ets-2 (Santa Cruz Biotechnology Inc.). The same membranes were reprobed with a mouse mAb to tubulin (Upstate, Biotechnology UBI, Lake Placid, NY). As secondary Abs, HRP-conjugated goat anti-rabbit IgG (Upstate) and goat anti-mouse IgG (Upstate) were used. Immunoreactive bands were visualized by the ECL LumiGLO detection kit (Upstate).
Protein Purification and EMSA
Nuclear protein extracts from CD3+, CD14+, CD19+, CD4+CD45RA+CD25−, and CD4+CD45RO+CD25− cells (5 × 106 cells per experimental point) were prepared as described previously (25). Ets-2 depletion was carried out using a human anti-Ets-2 antibody (Santa Cruz Biotechnology Inc.) as described previously (12). For Ets-2 protein purification, all the procedures were carried out at 4 °C. Ets-2 was isolated from a total of 5 mg of T-cell nuclear protein extracts divided in five aliquots of 1 mg. Each protein aliquot was diluted 5× in KIB buffer (20 mm HEPES (pH 7.6), 100 mm KCl, 0.02 mm EDTA, and 0.5 mm 1,4-dithio-dl-threitol) and passed through a Protein A/G-agarose column. The eluate was mixed for 1 h with 50 μg of Ets-2 Ab (Santa Cruz Biotechnology Inc.) and then passed through a Protein A/G-agarose column. Bound Ets-2 was eluted with ImmunoPure Gentle Ag/Ab Elution Buffer (Thermo Scientific, Pierce) and collected in 1-ml aliquots. For EMSA, the proteins were precipitated by 10% (w/v) PEG 8000, and the pellets were redissolved in KIB buffer. EMSA were performed as described (25, 37). The oligonucleotide probe used was the AREE-2 sequence of the human IL-2 promoter, AAGAAAGGAGGAAAAACTGTTT (37).
ChIP Assays
Cord blood CD4+CD45RA+CD25− naive Th cells, in vitro generated CD4+CD45RO+CD25− memory Th cells, and Jurkat cells were cultured in CM for 6 h ±P/I, washed one time with RPMI 1640 medium, resuspended in CM (5 × 106 cells/ml), and fixed by the addition of formaldehyde to a 1.1% final concentration. Fixed cells were used at a concentration of 2 × 107 cells per experimental point. ChIP assays were performed as described (38). For each immunoprecipitation, 10 μg of chromatin was used. The antibodies used were anti-Ets-2 (sc-351) and anti-POLII (sc-9001); an anti-human IgG antibody (sc-2027, Santa Cruz Biotechnology Inc.) was used as a negative control. The levels of DNA at target genomic sites were quantitatively measured by real time PCR using the KAPA SYBR FAST qPCR kit Master Mix (2×) Universal (Kapa Biosystems) and the Mx3000P Quantitative PCR System thermal cycler and analyzed with MxPro software. The primers used for the ARRE-2 Ets-2 binding site on the IL-2 promoter were 5′-CTTGCTGTTGTCCACCAC-3′ and 5′-TGGATGTAGGTGAAATCCC-3′, yielding a 201-bp PCR product. The primers used for the TATA/ARRE-1 region on the IL-2 promoter were 5′-TCTTTGGGGGTTTAAAGAAATT C-3′ and 5′-AGGAGTTGAGGTTACTGTGAG-3′, yielding a 217-bp PCR product. The primers used for the β2-m promoter were 5′-CGCCGATGTACAGACAGCAAA-3′, 5′-TGCTGTCAGCTTCAGGAATG-3′, yielding a 229-bp PCR product. The program was set to 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s; this was followed by a dissociation program. The normalized (to input) immunoprecipitation values are expressed as percentage of input.
Author Contributions
A. M. conceived and coordinated the study. I. P., I. A., M. A., T. G., and A. M. performed the experiments. A. M., I. P., and T. G. wrote the paper. D. T. critically revised the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. E. Farri-Kostopoulou, St. Andrew's General Hospital of Patras, for providing the cord blood samples; our students, colleagues, and the staff of the Blood Transfusion Center of Patras University Hospital for the peripheral blood samples; Dr. E. Mandouvalou for help with the ChIP assays; and Dr. P. Sakellaraki and Dr. P. Spadidea for help with flow cytometry and Western blotting.
This work was co-supported by the European Union (European Social Fund) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework-Research Funding Program Heracleitus II Grant D.276.001.020 and K. Karatheodoris Grant B407, University of Patras (to A. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- Th
- T helper
- NFAT
- nuclear factor of activated T-cells
- TCR
- T-cell receptor
- ARRE
- antigen receptor response element
- PBMC
- peripheral blood mononuclear cell
- CM
- culture medium
- P/I
- phorbol myristate acetate and ionomycin
- Treg
- regulatory T cell
- CsA
- cyclosporine A
- CAT
- chloramphenicol acetyltransferase
- MM
- multiple myeloma
- POLII
- polymerase II
- β2-m
- β2-microglobulin
- CB
- cord blood
- PE
- phycoerythrin
- qPCR
- quantitative PCR
- Ab
- antibody
- ANOVA
- analysis of variance
- ETS
- E26 transformation-specific
- DREAM
- downstream regulatory element antagonist modulator
- ChIP
- chromatin immunoprecipitation
- OCT
- octamer-binding transcription factor
- TCEd
- distal T-cell element.
References
- 1. Smith-Garvin J. E., Koretzky G. A., and Jordan M. S. (2009) T cell activation. Annu. Rev. Immunol. 27, 591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamane H., and Paul W. E. (2013) Early signaling events that underlie fate decisions of naive CD4+ T cells toward distinct T-helper cell subsets. Immunol. Rev. 252, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naito T., Tanaka H., Naoe Y., and Taniuchi I. (2011) Transcriptional control of T-cell development. Int. Immunol. 23, 661–668 [DOI] [PubMed] [Google Scholar]
- 4. Travers P., Janeway C. A., Walport M., and Shlomchik M. J. (2001) Immunobiology: the Immune System in Health and Disease, p. 323, Garland Publishing, New York [Google Scholar]
- 5. Firpo E. J., Koff A., Solomon M. J., and Roberts J. M. (1994) Inactivation of a Cdk2 inhibitor during interleukin 2-induced proliferation of human T lymphocytes. Mol. Cell. Biol. 14, 4889–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Serfling E., Avots A., and Neumann M. (1995) The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim. Biophys. Acta 1263, 181–200 [DOI] [PubMed] [Google Scholar]
- 7. Bunting K., Wang J., and Shannon M. F. (2006) Control of interleukin-2 gene transcription: a paradigm for inducible, tissue-specific gene expression. Vitam. Horm. 74, 105–145 [DOI] [PubMed] [Google Scholar]
- 8. Liu J. O. (2005) The yins of T cell activation. Sci. STKE 2005, re1. [DOI] [PubMed] [Google Scholar]
- 9. Mouzaki A., Weil R., Muster L., and Rungger D. (1991) Silencing and trans-activation of the mouse IL-2 gene in Xenopus oocytes by proteins from resting and mitogen-induced primary T-lymphocytes. EMBO J. 10, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mouzaki A., Rungger D., Tucci A., Doucet A., and Zubler R. H. (1993) Occurrence of a silencer of the interleukin-2 gene in naive but not in memory resting T helper lymphocytes. Eur. J. Immunol. 23, 1469–1474 [DOI] [PubMed] [Google Scholar]
- 11. Mouzaki A., and Rungger D. (1994) Properties of transcription factors regulating interleukin-2 gene transcription through the NFAT binding site in untreated or drug-treated naive and memory T-helper cells. Blood 84, 2612–2621 [PubMed] [Google Scholar]
- 12. Argyropoulos C., Nikiforidis G. C., Theodoropoulou M., Adamopoulos P., Boubali S., Georgakopoulos T. N., Paliogianni F., Papavassiliou A. G., and Mouzaki A. (2004) Mining microarray data to identify transcription factors expressed in naive resting but not activated T lymphocytes. Genes Immun. 5, 16–25 [DOI] [PubMed] [Google Scholar]
- 13. Oikawa T., and Yamada T. (2003) Molecular biology of the Ets family of transcription factors. Gene 303, 11–34 [DOI] [PubMed] [Google Scholar]
- 14. Hollenhorst P. C., McIntosh L. P., and Graves B. J. (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gallant S., and Gilkeson G. (2006) ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. 54, 149–163 [DOI] [PubMed] [Google Scholar]
- 16. Lopez R. G., Carron C., Oury C., Gardellin P., Bernard O., and Ghysdael J. (1999) TEL is a sequence-specific transcriptional repressor. J. Biol. Chem. 274, 30132–30138 [DOI] [PubMed] [Google Scholar]
- 17. Mavrothalassitis G., and Ghysdael J. (2000) Proteins of the ETS family with transcriptional repressor activity. Oncogene 19, 6524–6532 [DOI] [PubMed] [Google Scholar]
- 18. Irvin B. J., Wood L. D., Wang L., Fenrick R., Sansam C. G., Packham G., Kinch M., Yang E., and Hiebert S. W. (2003) TEL, a putative tumor suppressor, induces apoptosis and represses transcription of Bcl-XL. J. Biol. Chem. 278, 46378–46386 [DOI] [PubMed] [Google Scholar]
- 19. Wolvetang E. J., Wilson T. J., Sanij E., Busciglio J., Hatzistavrou T., Seth A., Hertzog P. J., and Kola I. (2003) ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum. Mol. Genet. 12, 247–255 [DOI] [PubMed] [Google Scholar]
- 20. Sussan T. E., Yang A., Li F., Ostrowski M. C., and Reeves R. H. (2008) Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature 451, 73–75 [DOI] [PubMed] [Google Scholar]
- 21. Baker K. M., Wei G., Schaffner A. E., and Ostrowski M. C. (2003) Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278, 17876–17884 [DOI] [PubMed] [Google Scholar]
- 22. Anderson M. K., Hernandez-Hoyos G., Diamond R. A., and Rothenberg E. V. (1999) Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126, 3131–3148 [DOI] [PubMed] [Google Scholar]
- 23. Zaldumbide A., Carlotti F., Pognonec P., and Boulukos K. E. (2002) The role of the Ets2 transcription factor in the proliferation, maturation, and survival of mouse thymocytes. J. Immunol. 169, 4873–4881 [DOI] [PubMed] [Google Scholar]
- 24. Blumenthal S. G., Aichele G., Wirth T., Czernilofsky A. P., Nordheim A., and Dittmer J. (1999) Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J. Biol. Chem. 274, 12910–12916 [DOI] [PubMed] [Google Scholar]
- 25. Mouzaki A., Doucet A., Mavroidis E., Muster L., and Rungger D. (2000) A repression-derepression mechanism regulating the transcription of human immunodeficiency virus type 1 in primary T cells. Mol. Med. 6, 377–390 [PMC free article] [PubMed] [Google Scholar]
- 26. Marson A., Kretschmer K., Frampton G. M., Jacobsen E. S., Polansky J. K., MacIsaac K. D., Levine S. S., Fraenkel E., von Boehmer H., and Young R. A. (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emmel E. A., Verweij C. L., Durand D. B., Higgins K. M., Lacy E., and Crabtree G. R. (1989) Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science 246, 1617–1620 [DOI] [PubMed] [Google Scholar]
- 28. Jia Y. Y., Lu J., Huang Y., Liu G., Gao P., Wan Y. Z., Zhang R., Zhang Z. Q., Yang R. F., Tang X., Xu J., Wang X., Chen H. Z., and Liu D. P. (2014) The involvement of NFAT transcriptional activity suppression in SIRT1-mediated inhibition of COX-2 expression induced by PMA/ionomycin. PLoS One 9, e97999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamanaka Y., Abu-Amer W., Foglia D., Otero J., Clohisy J. C., Abu-Amer Y. (2008) NFAT2 is an essential mediator of orthopedic particle-induced osteoclastogenesis. J. Orthop. Res. 26, 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin L., Spoor M. S., Gerth A. J., Brody S. L., and Peng S. L. (2004) Modulation of Th1 activation and inflammation by the NF-κB repressor Foxj1. Science 303, 1017–1020 [DOI] [PubMed] [Google Scholar]
- 31. Savignac M., Pintado B., Gutierrez-Adan A., Palczewska M., Mellström B., and Naranjo J. R. (2005) Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 24, 3555–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tzachanis D., Freeman G. J., Hirano N., van Puijenbroek A. A., Delfs M. W., Berezovskaya A., Nadler L. M., and Boussiotis V. A. (2001) Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182 [DOI] [PubMed] [Google Scholar]
- 33. Senger K., Merika M., Agalioti T., Yie J., Escalante C. R., Chen G., Aggarwal A. K., and Thanos D. (2000) Gene repression by coactivator repulsion. Mol. Cell 6, 931–937 [DOI] [PubMed] [Google Scholar]
- 34. Bhat N. K., Thompson C. B., Lindsten T., June C. H., Fujiwara S., Koizumi S., Fisher R. J., and Papas T. S. (1990) Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: regulatory role for the protooncogene ETS1. Proc. Natl. Acad. Sci. U.S.A. 87, 3723–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shore P., Whitmarsh A. J., Bhaskaran R., Davis R. J., Waltho J. P., and Sharrocks A. D. (1996) Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 16, 3338–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson C. B., Wang C. Y., Ho I. C., Bohjanen P. R., Petryniak B., June C. H., Miesfeldt S., Zhang L., Nabel G. J., Karpinski B., and Leiden J. M. (1992) cis-Acting sequences required for inducible interleukin-2 enhancer function bind a novel Ets-related protein, Elf-1. Mol. Cell. Biol. 12, 1043–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouzaki A., Theodoropoulou M., Gianakopoulos I., Vlaha V., Kyrtsonis M. C., and Maniatis A. (2002) Expression patterns of Th1 and Th2 cytokine genes in childhood idiopathic thrombocytopenic purpura (ITP) at presentation and their modulation by intravenous immunoglobulin G (IVIg) treatment: their role in prognosis. Blood 100, 1774–1779 [PubMed] [Google Scholar]
- 38. Lomvardas S., and Thanos D. (2002) Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110, 261–271 [DOI] [PubMed] [Google Scholar]