Abstract
Virus-host interactions play a role in many stages of the viral lifecycle, including entry. Reovirus, a model system for studying the entry mechanisms of nonenveloped viruses, undergoes a series of regulated structural transitions that culminate in delivery of the viral genetic material. Lipids can trigger one of these conformational changes, infectious subviral particle (ISVP)-to-ISVP* conversion. ISVP* formation releases two virally encoded peptides, myristoylated μ1N (myr-μ1N) and Φ. Among these, myr-μ1N is sufficient to form pores within membranes. Released myr-μ1N can also promote ISVP* formation in trans. Using thermal inactivation as a readout for ISVP-to-ISVP* conversion, we demonstrate that lipids render ISVPs less thermostable in a virus concentration-dependent manner. Under conditions in which neither lipids alone nor myr-μ1N alone promotes ISVP-to-ISVP* conversion, myr-μ1N induces particle uncoating when lipids are present. These data suggest that the pore-forming activity and the ISVP*-promoting activity of myr-μ1N are linked. Lipid-associated myr-μ1N interacts with ISVPs and triggers efficient ISVP* formation. The cooperativity between a reovirus component and lipids reveals a distinct virus-host interaction in which membranes can facilitate nonenveloped virus entry.
Keywords: conformational change, kinetics, membrane, phospholipid vesicle, reovirus, virus entry
Introduction
Host entry, a critical stage in the viral lifecycle, is necessary for incoming virus to initiate a productive infection. This step is characterized by host cell attachment, internalization, and subsequent delivery of the viral genetic material (1–3). Enveloped and nonenveloped viruses have developed numerous strategies to penetrate specific cellular compartments. Although the entry mechanisms of enveloped viruses are well established (3–5), the entry mechanisms of nonenveloped viruses are poorly understood.
Membranes play a critical role in facilitating virus entry. Enveloped viruses enter cells by fusing viral and host membranes using virally encoded fusion proteins. Membrane fusion, which culminates in the exposure of the viral genetic material, can be regulated by pH, protein-protein interactions, protein-lipid interactions, and/or lipid composition (3–6). Less is known about the role of host lipids during entry of nonenveloped viruses; however, accumulating evidence from a variety of virus systems indicates that, in addition to serving as topological barriers, specific lipids can play an active role in virus entry (7–9). Bluetongue virus, a member of Reoviridae, interacts with lysobisphosphatidic acid, a lipid enriched in the late endosomal membrane, to facilitate membrane penetration and core delivery (8). Adenovirus triggers calcium influx at the plasma membrane and lysosomal exocytosis. Released acid sphingomyelinases modify the lipid content of host membranes, which enhances adenovirus uptake and subsequent endosomal escape (7). Membranes can also promote structural transitions that are required for mammalian orthoreovirus (reovirus) entry; liposomes are sufficient to induce particle disassembly and subsequent pore formation in a lipid composition-dependent manner (9). Nonetheless, the precise mechanism by which lipids potentiate reovirus uncoating is unknown.
Particles of nonenveloped reovirus are characterized by two concentric, protein shells: the inner capsid (core) and the outer capsid. The core encapsulates 10 double-stranded RNA genome segments (10–12). Following attachment to proteinaceous or carbohydrate receptors (13–17), reovirus is internalized by receptor-mediated endocytosis (18–22). To initiate infection, particles must then undergo a series of disassembly steps. First, the σ3 outer capsid protein is digested by acid-dependent cathepsin proteases (19, 23–28). Loss of σ3 generates metastable intermediates, called infectious subviral particles (ISVPs).2 ISVPs are characterized by exposure of μ1, the cell penetration protein, on the surface of the particle (10). ISVPs can also be generated by intestinal or respiratory proteases prior to internalization into host cells (29–32). Second, metastable ISVPs undergo a structural transition to generate ISVP*s. ISVP-to-ISVP* conversion culminates in the release of μ1 N- and C-terminal fragments, myristoylated μ1N (myr-μ1N) and Φ, respectively (11, 33–39). The released fragments are thought to facilitate core delivery by generating pores within the host endosomal membrane (33, 36, 40). Many of the conformational changes that define reovirus entry can be recapitulated in vitro; ISVPs are produced by digesting purified virions with chymotrypsin (41, 42), and ISVP-to-ISVP* conversion can be induced using nonphysiological physiochemical triggers, such as heat and large, monovalent cations (35, 43–45). Lipids, a host factor that reovirus particles are likely to encounter during host entry, can also promote ISVP* formation (9).
This study investigates a mechanism for lipid- and myr-μ1N-mediated reovirus entry. Following cleavage and release from μ1, myr-μ1N may serve two functions: (i) myr-μ1N generates pores within the host endosomal membrane (33, 36, 40) and (ii) myr-μ1N induces ISVP-to-ISVP* conversion in trans (46). We propose a model in which the pore-forming activity and the ISVP*-promoting activity are linked. First, myr-μ1N promotes ISVP* formation most efficiently when lipids are present. Second, the addition of exogenous myr-μ1N to a low concentration of ISVPs alters the reaction kinetics of ISVP-to-ISVP* conversion. Third, host proteins are not required for myr-μ1N to recruit ISVPs to a lipid bilayer. Thus, membrane-embedded myr-μ1N interacts with ISVPs and induces ISVP-to-ISVP* conversion. This work reveals a heretofore unknown mechanism in which membranes can actively participate in the entry pathway of a nonenveloped virus.
Results
Susceptibility of T3D ISVPs to Lipid-mediated Thermal Inactivation Is Particle Concentration-dependent
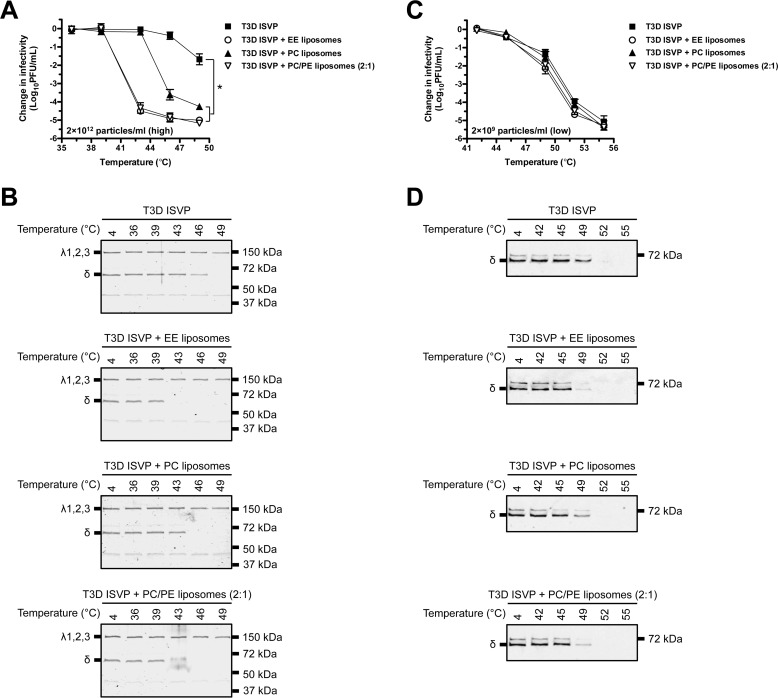
ISVPs undergo multiple structural transitions to generate ISVP*s. These conformational changes occur within an infected cell and are necessary for establishing a productive infection (35, 44, 45). Triggering ISVP-to-ISVP* conversion in vitro with heat renders the particles noninfectious (44). At high particle concentration (2 × 1012 particles/ml or 3 nm), liposomes facilitate ISVP* formation in a lipid composition-dependent manner (9). To test whether lipids also render reovirus particles noninfectious, we conducted 20-min heat inactivation experiments over a range of temperatures in the absence or presence of early endosome (EE), l-α-phosphatidylcholine (PC), or PC/l-α-phosphatidylethanolamine (PE) (2:1) liposomes. As described previously, liposomes composed of PC/PE induce ISVP-to-ISVP* conversion as efficiently as EE liposomes. Liposomes composed of PC alone can also trigger ISVP* formation, albeit to a lesser extent (9). At high particle concentration (Fig. 1A), reovirus type 3 Dearing (T3D) ISVPs were more susceptible to thermal inactivation in the presence of liposomes when compared with T3D ISVPs alone. At 43 °C, T3D ISVPs incubated with EE or PC/PE (2:1) liposomes were reduced in titer by ∼4.5 log10 units relative to control virus incubated at 4 °C. Consistent with a mechanism in which liposomes promote ISVP* formation in a lipid composition-dependent manner (9), T3D ISVPs incubated with PC liposomes were less susceptible to thermal inactivation when compared with virus incubated with EE or PC/PE (2:1) liposomes; viral titer was reduced by less than 0.5 log10 unit at 43 °C and by ∼4 log10 units at 46 °C. T3D ISVPs alone were reduced in titer by only ∼2 log10 units following incubation at 49 °C.
FIGURE 1.
Liposomes facilitate thermal inactivation of T3D ISVPs in a virus concentration-dependent manner. A and C, lipid-mediated thermal inactivation curves. T3D ISVPs at 2 × 1012 particles/ml (A) or 2 × 109 particles/ml (C) were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes for 20 min at the indicated temperatures. The change in infectivity relative to samples incubated at 4 °C was determined by plaque assay. Data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent replicates for each reaction condition). B and D, liposome-mediated ISVP* formation. T3D ISVPs at 2 × 1012 particles/ml (B) or 2 × 109 particles/ml (D) were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes for 20 min at the indicated temperatures. Each reaction was then treated with trypsin for 30 min on ice. Following digestion, equal particle numbers from each reaction were analyzed by SDS-PAGE. In panel B, the gels were Coomassie Brilliant Blue-stained. In panel D, the gels were analyzed for the presence of the μ1 δ fragment by Western blotting (n = 3 independent replicates for each reaction condition, one representative experiment is shown).
As a consequence of ISVP* formation, the reovirus outer capsid protein, μ1, reorganizes into a protease-sensitive conformation (35). After incubating T3D ISVPs in the absence or presence of liposomes under conditions identical to Fig. 1A, this structural transition was assayed by determining the susceptibility of the δ fragment (a product of μ1 cleavage during ISVP-to-ISVP* conversion) to trypsin digestion (Fig. 1B) (35, 44). Consistent with the thermal inactivation curves shown in Fig. 1A, inclusion of EE, PC, or PC/PE (2:1) liposomes reduced the temperature that was required to render δ protease-sensitive.
At the high particle concentration used in Fig. 1, A and B (2 × 1012 particles/ml or 3 nm), ISVP* formation induced by heat does not follow first-order kinetics (34, 35, 46–48). In contrast, ISVP-to-ISVP* conversion at low particle concentration (e.g. 2 × 109 particles/ml or 0.003 nm) has been shown to approximate a first-order reaction (44, 46). As such, by measuring the rate of infectivity loss at low particle concentration, three thermodynamic parameters can be calculated: enthalpy of activation (ΔH‡), entropy of activation (ΔS‡), and free energy of activation (ΔG‡) (44, 49). To test the hypothesis that lipids facilitate ISVP* formation by lowering the energy barrier of the reaction, T3D ISVPs (2 × 109 particles/ml or 0.003 nm) were incubated for 20 min over a range of temperatures in the absence or presence of EE, PC, or PC/PE (2:1) liposomes (Fig. 1C). At low particle concentration (i.e. conditions that are more likely to arise during infection), we expected ISVP-to-ISVP* conversion to occur most efficiently when lipids were present. Nonetheless, T3D ISVPs alone and T3D ISVPs incubated with liposomes were equally susceptible to thermal inactivation at 42, 45, and 55 °C. At 49 and 52 °C, virus incubated with liposomes was reduced in titer to a greater extent than virus alone; however, the difference in infectivity was less than 1 log10 unit. Similar results were obtained using the trypsin sensitivity assay (Fig. 1D). At 49 °C, the μ1 δ fragment was only marginally more sensitive to trypsin digestion in virus incubated with liposomes when compared with virus alone.
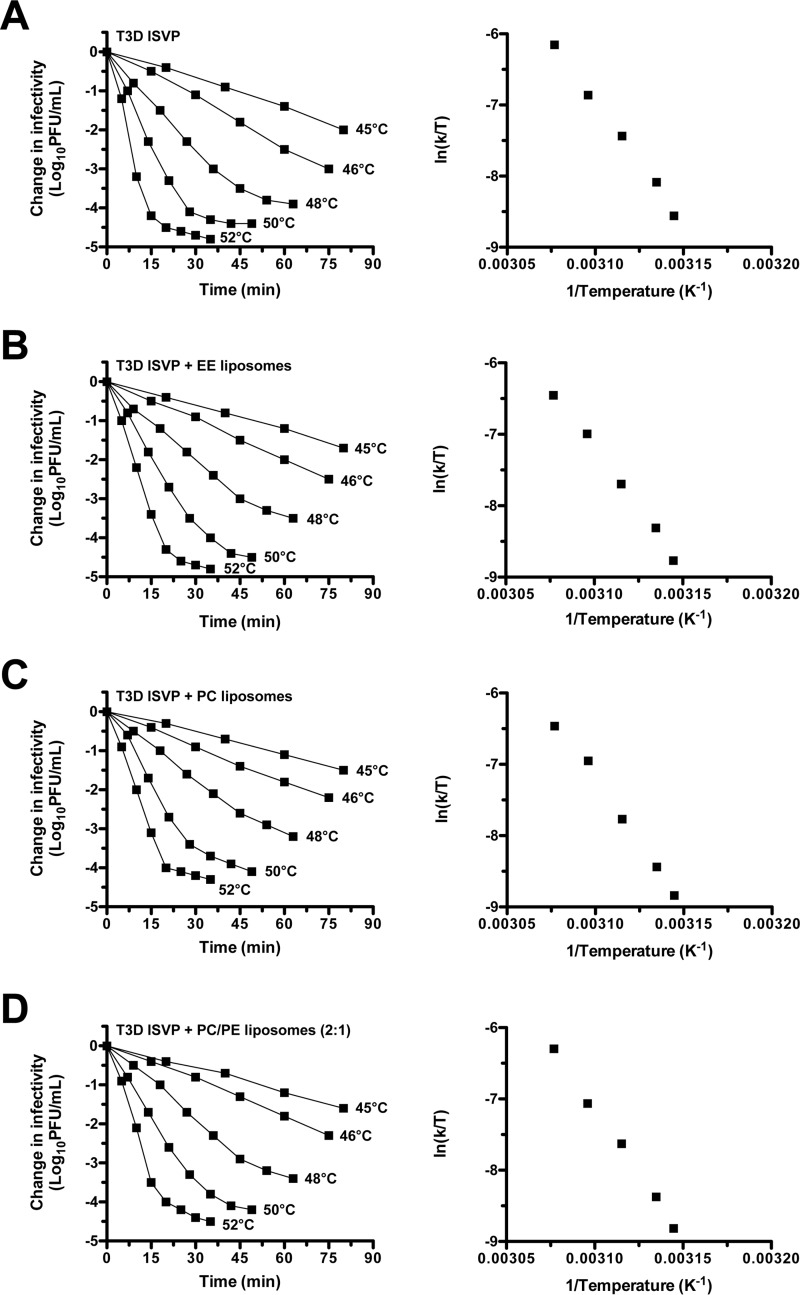
To further quantify the effects of lipids on ISVP-to-ISVP* conversion (i.e. loss of infectivity), kinetic analyses were performed. T3D ISVPs (2 × 109 particles/ml or 0.003 nm) were incubated in the absence or presence of EE, PC, or PC/PE (2:1) liposomes at constant temperatures. Under each reaction condition, change in infectivity decreased linearly with respect to time (Fig. 2, plots on the left side of each panel); liposomes did not alter the first-order kinetics of ISVP* formation at low particle concentration. The slopes of the inactivation curves were used to generate Arrhenius plots (Fig. 2, plots on the right side of each panel) (44). ΔH‡ and ΔS‡ were then calculated from the slope and the y-intercept of the Arrhenius plots using the Eyring absolute equation (49). ΔG‡ was calculated for each reaction condition at 48 °C using the following formula: ΔG‡ = ΔH‡ − T ΔS‡ (Table 1) (44, 49). The thermodynamic values derived from these reactions were not significantly different in the absence or presence of liposomes. These results demonstrate that lipids promote T3D ISVP thermal inactivation (i.e. ISVP-to-ISVP* conversion) only under conditions in which the particle concentration is high.
FIGURE 2.
Liposomes do not lower the energy barrier to thermally inactivate T3D ISVPs. A–D, plots on the left side of each panel, kinetics of lipid-mediated thermal inactivation. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes at the indicated temperatures. At the indicated time points, the change in infectivity relative to samples incubated at 4 °C was determined by plaque assay (n = 2 independent replicates for each reaction condition, one experiment is shown). A–D, plots on the right side of each panel, Arrhenius plots of lipid-mediated thermal inactivation. The rate constant, k, was determined for each thermal inactivation curve as described previously (44). The rate constants were plotted as ln(k/T) versus the reciprocal of the incubation temperature (T) in degrees Kelvin (n = 2 independent replicates for each reaction condition, one experiment is shown).
TABLE 1.
Thermodynamic values for T3D ISVP (2 × 109 particles/ml or 0.003 nm) thermal inactivation
| Reaction | Calculated value ± 95% confidence intervala |
ΔG‡b | |
|---|---|---|---|
| ΔH‡ | ΔS‡ | ||
| kJ mol−1 | kJ mol−1 K−1 | kJ mol−1 | |
| T3D ISVP | 286 ± 10 | 0.589 ± 0.020 | 94 |
| T3D ISVP + EE liposomes | 282 ± 10 | 0.583 ± 0.020 | 95 |
| T3D ISVP + PC liposomes | 297 ± 13 | 0.629 ± 0.019 | 95 |
| T3D ISVP + PC/PE liposomes (2:1) | 302 ± 12 | 0.644 ± 0.020 | 95 |
a Calculated by fitting the Eyring absolute equation to the data in Fig. 2.
b Calculated from ΔH‡ and ΔS‡ at 48 °C (321 K).
Lipids Enhance the ISVP-to-ISVP*-promoting Activity of Preconverted ISVP* Supernatant
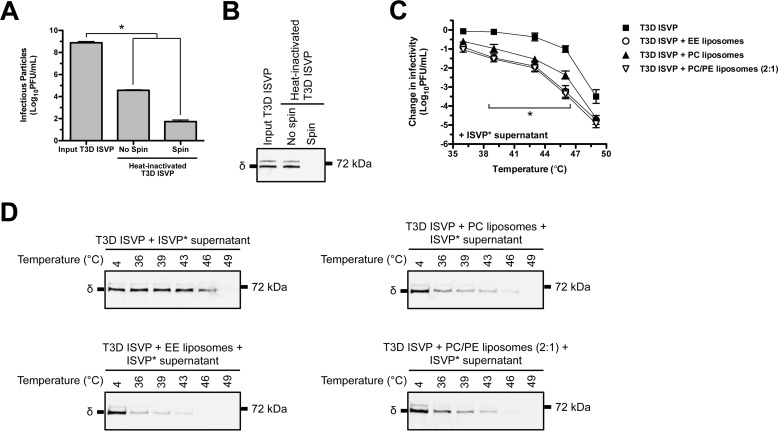
The observation that liposomes promote thermal inactivation at high particle concentration, but not at low particle concentration, suggests that one or more of the reaction products interacts with one or more of the input components to drive the reaction forward (i.e. the reaction does not follow first-order kinetics). ISVP-to-ISVP* conversion culminates in the release of at least three reovirus-derived fragments: myr-μ1N, Φ, and σ1 (11, 33–39). Among these, myr-μ1N is sufficient to generate 4–10-nm pores in target membranes (e.g. liposomes and bovine RBCs) (33, 36, 40) and to induce ISVP* formation in trans; however, its promoting activity is also enhanced by Φ (46). To determine whether the released fragments can facilitate lipid-mediated thermal inactivation, T3D ISVPs (2 × 109 particles/ml or 0.003 nm) supplemented with the supernatant of preconverted ISVP*s were incubated for 20 min over a range of temperatures in the absence or presence of EE, PC, or PC/PE (2:1) liposomes. ISVP* supernatant was generated by heating input T3D ISVPs for 5 min at 52 °C (36, 46). The reactions were then centrifuged to pellet particles, and the supernatant was transferred to tubes containing target ISVPs. The supernatant (spin) was analyzed by plaque assay (Fig. 3A) and by Western blotting (Fig. 3B) to confirm the clearance of input virus. As expected, virus incubated with ISVP* supernatant, which contains released myr-μ1N, Φ, and σ1 (36), was more susceptible to thermal inactivation than virus incubated without ISVP* supernatant (compare Figs. 1C and 3C). Furthermore, the released fragments functioned more efficiently in the presence of liposomes. At 46 °C, virus incubated with ISVP* supernatant and EE liposomes was reduced in titer by ∼3 log10 units, whereas virus incubated with ISVP* supernatant alone was reduced in titer by ∼1 log10 unit.
FIGURE 3.
The supernatant of preconverted ISVP*s cooperates with liposomes to facilitate thermal inactivation of T3D ISVPs. A and B, generation of ISVP* supernatant. Input T3D ISVPs at 2 × 1012 particles/ml were incubated at 52 °C for 5 min. The heat-inactivated virus (No spin) was centrifuged to pellet particles. The supernatant (Spin) was immediately transferred to tubes containing target T3D ISVPs for thermal inactivation reactions. Aliquots of the no spin and spin reactions were analyzed for residual infectivity by plaque assay (A) and for the presence of the μ1 δ fragment by Western blotting (B). In panel A, data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent preparations of ISVP* supernatant were analyzed). C, ISVP* supernatant- and liposome-mediated thermal inactivation curves. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes and ISVP* supernatant for 20 min at the indicated temperatures. The change in infectivity relative to samples incubated at 4 °C was determined by plaque assay. Data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent replicates for each reaction condition). D, ISVP* supernatant- and liposome-mediated ISVP* formation. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes and ISVP* supernatant for 20 min at the indicated temperatures. Each reaction was then treated with trypsin for 30 min on ice. Following digestion, equal particle numbers from each reaction were analyzed by SDS-PAGE. The gels were analyzed for the presence of the μ1 δ fragment by Western blotting (n = 3 independent replicates for each reaction condition, one representative experiment is shown).
The thermal inactivation results were confirmed using the trypsin sensitivity assay (Fig. 3D). In the presence of liposomes, ISVP* supernatant induced ISVP-to-ISVP* conversion more efficiently (i.e. triggered reorganization of the μ1 δ fragment at lower temperatures) than ISVP* supernatant that was incubated with virus alone. These results suggest that lipids potentiate the in trans promoting activity of one or more of the released fragments.
Lipids Enhance the ISVP-to-ISVP*-promoting Activity of Released myr-μ1N
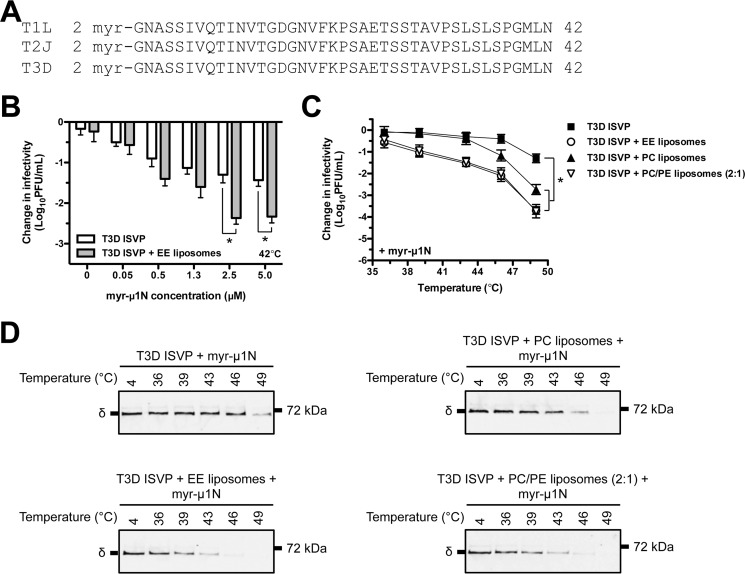
Small, virally encoded peptides are thought to play an essential role during cell entry of many nonenveloped viruses. myr-μ1N, a μ1-derived pore-forming peptide, is conserved across the prototype strains (e.g. T1L (reovirus type 1 Lang), T2J (reovirus type 2 Jones), and T3D) of all three mammalian orthoreovirus serotypes (Fig. 4A). Moreover, aqua and avian reoviruses each encode a peptide that is similar, but not identical, in sequence to myr-μ1N (39, 50–53).
FIGURE 4.
myr-μ1N cooperates with liposomes to facilitate thermal inactivation of T3D ISVPs. A, amino acid sequence alignments of the reovirus myr-μ1N peptide. T1L, reovirus type 1 Lang; T2J, reovirus type 2 Jones. B, dose-dependent analysis of myr-μ1N-mediated thermal inactivation. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with myr-μ1N (concentrations shown in figure) in the absence (white bars) or presence (gray bars) of EE liposomes for 20 min at 42 °C. The change in infectivity relative to samples incubated at 4 °C was determined by plaque assay. Data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent replicates for each reaction condition). C, myr-μ1N- and liposome-mediated thermal inactivation curves. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes and 2.5 μm myr-μ1N for 20 min at the indicated temperatures. The change in infectivity relative to samples incubated at 4 °C was determined by plaque assay. Data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent replicates for each reaction condition). D, myr-μ1N- and liposome-mediated ISVP* formation. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes and 2.5 μm myr-μ1N for 20 min at the indicated temperatures. Each reaction was then treated with trypsin for 30 min on ice. Following digestion, equal particle numbers from each reaction were analyzed by SDS-PAGE. The gels were analyzed for the presence of the μ1 δ fragment by Western blotting (n = 3 independent replicates for each reaction condition, one representative experiment is shown).
The ISVP-to-ISVP*-promoting activity of myr-μ1N has been described previously (46). myr-μ1N cleavage and release were required for ISVP* supernatant to retain promoting activity and for ISVP* formation to occur in a virus concentration-dependent manner. To determine whether lipids enhance myr-μ1N-mediated ISVP* formation, T3D ISVPs (2 × 109 particles/ml or 0.003 nm) supplemented with a synthetic myr-μ1N peptide were incubated for 20 min at 42 °C in the absence or presence of EE liposomes (Fig. 4B). As expected, myr-μ1N induced particle inactivation in a concentration-dependent manner. Promoting activity plateaued at 2.5 μm peptide, which corresponds to the release of all 600 copies of myr-μ1N from each particle in ∼3 × 1012 particles/ml (46). Moreover, T3D ISVPs incubated with 2.5 μm myr-μ1N and EE liposomes were reduced in titer by ∼2.5 log10 units, whereas virus incubated with 2.5 μm peptide alone was reduced in titer by ∼1.5 log10 units. We obtained similar results when the myr-μ1N peptide was incubated with a different prototype reovirus strain, T1L (data not shown).
Next, T3D ISVPs (2 × 109 particles/ml or 0.003 nm) supplemented with 2.5 μm myr-μ1N were incubated for 20 min over a range of temperatures in the absence or presence of EE, PC, or PC/PE (2:1) liposomes. T3D ISVPs incubated with myr-μ1N and liposomes were reduced in titer to a greater extent than virus incubated with myr-μ1N alone (Fig. 4C). Moreover, T3D ISVPs incubated with myr-μ1N and PC liposomes inactivated less efficiently than virus incubated with myr-μ1N and EE or PC/PE (2:1) liposomes. The thermal inactivation results were confirmed using the trypsin sensitivity assay (Fig. 4D). In the presence of liposomes, myr-μ1N induced ISVP* formation at a lower temperature than myr-μ1N incubated with virus alone. Together, these data demonstrate that lipids promote ISVP-to-ISVP* conversion by maximizing the in trans promoting activity of released myr-μ1N.
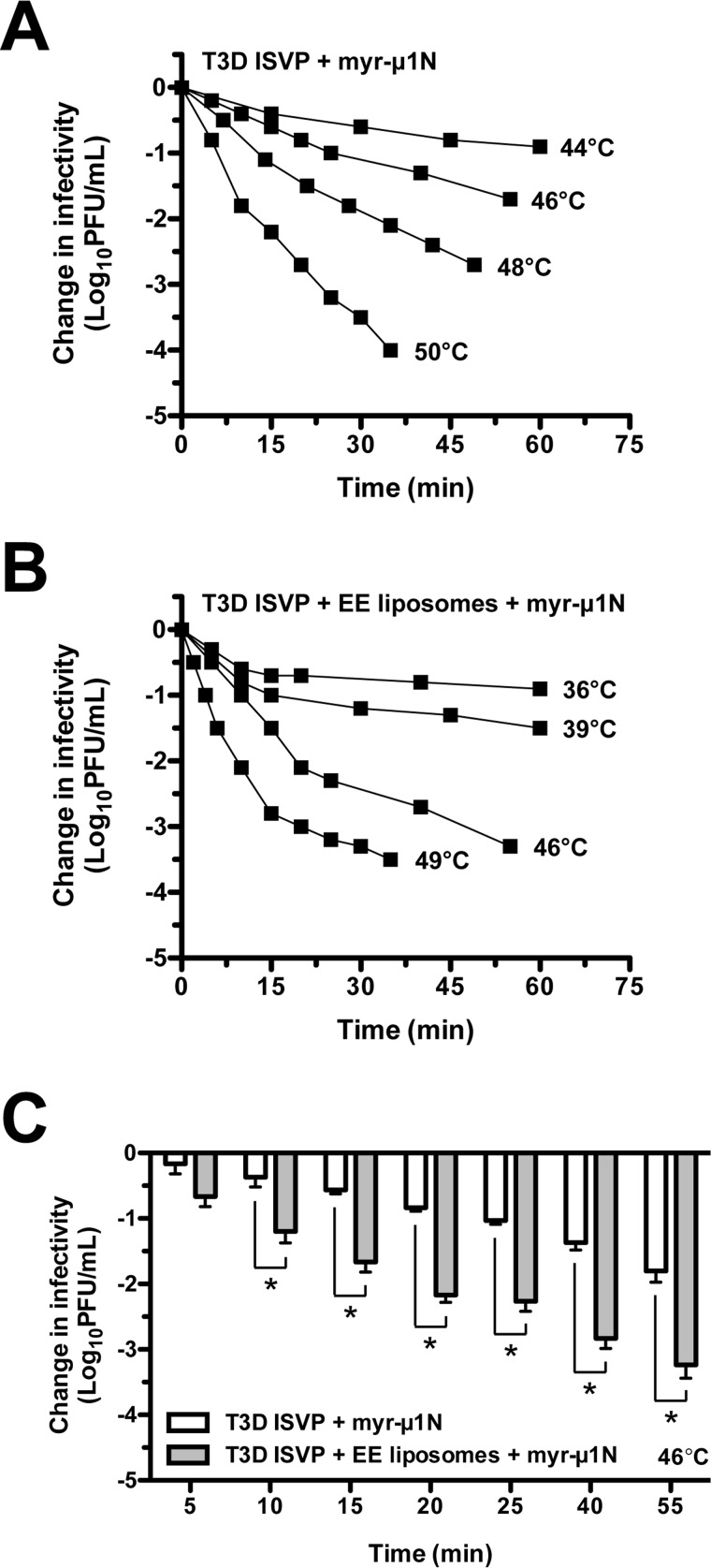
Addition of Exogenous myr-μ1N Alters the Reaction Kinetics of T3D ISVP Thermal Inactivation
At high particle concentration, ISVP-to-ISVP* conversion does not follow first-order kinetics (34, 35, 46–48). Given the observation that myr-μ1N promotes thermal inactivation (Fig. 4) (46), we hypothesized that a low concentration of T3D ISVPs incubated with a high concentration of the trans-activating factor myr-μ1N would lose infectivity following non-first-order kinetics. T3D ISVPs (2 × 109 particles/ml or 0.003 nm) supplemented with 2.5 μm myr-μ1N were incubated in the absence or presence of EE liposomes at constant temperatures. When liposomes were included in the reactions, the decrease in infectivity was biphasic: an initial, fast phase and a second, slower phase (Fig. 5B). The reactions did not obey first-order kinetics. Thus, the addition of myr-μ1N to the virus-liposome mixtures was sufficient to alter the kinetics of T3D ISVP thermal inactivation. In the absence of liposomes, the decrease in infectivity was biphasic at higher temperatures (Fig. 5A, see 50 °C curve), but more linear at lower temperatures (Fig. 5A, see 44 °C curve). Consistent with the results shown in Fig. 4, EE liposomes also enhanced the promoting activity of myr-μ1N (Fig. 5C). At 46 °C, T3D ISVPs incubated with myr-μ1N and EE liposomes were reduced in titer by 1.5 log10 units at ∼15 min, whereas T3D ISVPs incubated with myr-μ1N alone were reduced in titer by 1.5 log10 units at ∼55 min. Together, these results provide direct, experimental evidence that released myr-μ1N is responsible for the particle concentration-dependent mechanism of ISVP-to-ISVP*conversion (i.e. the addition of exogenous myr-μ1N changes a first-order reaction into a non-first-order reaction) (46).
FIGURE 5.
myr-μ1N alters the reaction kinetics of T3D ISVP thermal inactivation. A and B, kinetics of myr-μ1N- and lipid-mediated thermal inactivation. T3D ISVPs at 2 × 109 particles/ml were incubated in virus storage buffer supplemented with 2.5 μm myr-μ1N in the absence (A) or presence (B) of EE liposomes at the indicated temperatures. At the indicated time points, the change in infectivity relative to samples incubated at 4 °C was determined by plaque assay (n = at least 2 independent replicates for each reaction condition, one experiment is shown). C, the results from panels A and B at 46 °C are plotted on a bar graph. Data are presented as mean ± S.D., *, p ≤ 0.01 (n = 3 independent replicates for each reaction condition).
ISVP* Supernatant and myr-μ1N Can Recruit T3D ISVPs to Liposomes
Our results demonstrate that lipids enhance the in trans promoting activity of myr-μ1N. Following ISVP-to-ISVP* conversion, released myr-μ1N and Φ are sufficient to form size-selective pores (4–10 nm in diameter) within target membranes (e.g. liposomes and bovine RBCs) (33, 36, 40). Due to its membrane-targeting property (40), myr-μ1N may induce ISVP* formation most efficiently when lipid-associated. When formed within RBCs, myr-μ1N-generated pores can recruit reovirus ISVP*s (33, 36); however, it is unclear whether RBC surface proteins are required for this interaction. Moreover, because these experiments induced ISVP-to-ISVP* conversion with heat, it is unknown whether pores can recruit ISVPs, in addition to ISVP*s, to membranes. When compared with ISVP*s, ISVPs are substantially less hydrophobic and present different portions of μ1 on the surface of the particle (35).
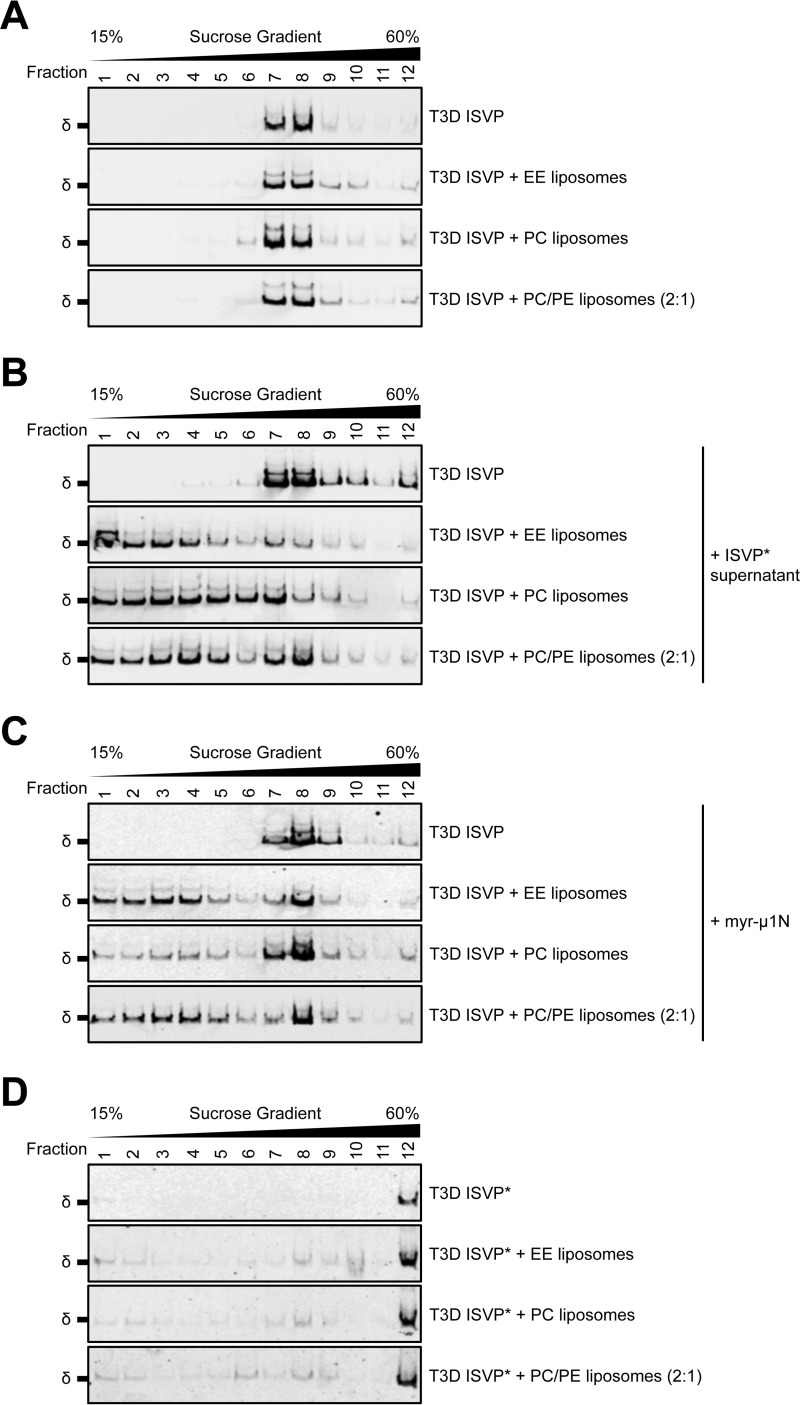
To determine whether myr-μ1N can recruit reovirus to liposomes (i.e. membranes that lack host proteins), T3D ISVPs were incubated with EE, PC, or PC/PE (2:1) liposomes that were pretreated with the supernatant of preconverted ISVP*s or with 2.5 μm myr-μ1N. The reactions were applied to sucrose gradients and sedimented by ultracentrifugation. When T3D ISVPs were mixed with liposomes alone (i.e. lacking ISVP* supernatant or myr-μ1N pretreatment), the μ1 δ fragment was observed predominantly in fractions 7 and 8, similar to virus alone (Fig. 6A). Thus, virus-liposome interactions were not detected. In contrast, when T3D ISVPs were mixed with liposomes that were preincubated with ISVP* supernatant (Fig. 6B) or with myr-μ1N (Fig. 6C), the μ1 δ fragment was observed predominantly in fractions 1–8; virus-liposome interactions prevented T3D ISVPs from fully sedimenting through the gradient. Furthermore, ISVP*s display a unique sedimentation profile. T3D ISVPs were converted to ISVP*s with heat in the absence or presence of liposomes. Following ultracentrifugation, the majority of the μ1 δ fragment was observed in fraction 12 (Fig. 6D). Together, these results suggest that myr-μ1N alone can recruit T3D ISVPs to membranes. Thus, we conclude that the pore-forming activity and the ISVP*-promoting activity of myr-μ1N are linked. Lipids facilitate ISVP-to-ISVP* conversion (Figs. 4 and 5) by increasing the efficiency of ISVP-myr-μ1N interactions.
FIGURE 6.
ISVP* supernatant and myr-μ1N can recruit T3D ISVPs to liposomes. A–D, T3D ISVPs at 2 × 1012 particles/ml were incubated in virus storage buffer supplemented with EE, PC, or PC/PE (2:1) liposomes for 10 min on ice (A–C) or for 20 min at 49 °C (D). When indicated, ISVP* supernatant (B) or 2.5 μm myr-μ1N (C) was included in the reactions. The samples were then applied to the top of sucrose gradients and sedimented by ultracentrifugation. Fractions were collected from the top of the gradients. Equal volumes from each fraction were analyzed by SDS-PAGE. The gels were analyzed for the presence of the μ1 δ fragment by Western blotting (n = 3 independent replicates for each reaction condition, one representative experiment is shown).
Discussion
There is an increasing body of evidence that host lipids can play an active role in multiple stages of nonenveloped virus entry, including internalization and pore formation (7–9). For reovirus, membranes facilitate particle uncoating (i.e. ISVP-to-ISVP* conversion) in a lipid composition-dependent manner (9). In this work, we explore a potential mechanism by which lipids cooperate with reovirus to promote viral entry. Using thermal inactivation as a readout for ISVP-to-ISVP* conversion, we show that liposomes facilitate this conformational change at high particle concentration, but not at low particle concentration (Figs. 1 and 2). The concentration dependence is due to myr-μ1N (Figs. 4 and 5), a virally encoded peptide that is released from reovirus during entry (11, 33–39) and promotes particle inactivation through a positive-feedback loop (46). Notably, myr-μ1N triggers ISVP-to-ISVP* conversion most efficiently when liposomes are present (Figs. 4 and 5). Moreover, host proteins are not required for myr-μ1N to recruit ISVPs to membranes (Fig. 6). Finally, under conditions in which thermal inactivation approximates first-order kinetics, we show that lipids alone do not affect the free energy of activation (Fig. 2 and Table 1); however, the first-order behavior is altered by the inclusion of exogenous myr-μ1N (Fig. 5). This observation further supports the conclusion that myr-μ1N functions as an in trans promoting factor to induce ISVP-to-ISVP* conversion (46). Together, our results provide experimental evidence that lipids can facilitate reovirus entry by interacting with, and enhancing the ISVP*-promoting activity of, myr-μ1N.
At high particle concentration, reovirus ISVPs are susceptible to lipid-mediated ISVP-to-ISVP* conversion (Fig. 1). Even within the limited volume of an endosome, the virus concentration used in these experiments (2 × 1012 particles/ml or 3 nm) cannot be achieved biologically. There are at least two non-mutually exclusive strategies to overcome this limitation: (i) perform the reaction under conditions in which the concentration of input components can be reduced without affecting output and (ii) sequester the input components within a confined space. At low particle concentration (2 × 109 particles/ml or 0.003 nm), inclusion of liposomes alone or myr-μ1N alone only marginally enhanced ISVP-to-ISVP* conversion (Figs. 1, 2, and 4). In contrast, inclusion of lipids and myr-μ1N significantly reduced the thermal energy that was needed to inactivate ISVPs (Figs. 4 and 5). Thus, myr-μ1N, in cooperation with lipids, can promote ISVP-to-ISVP* conversion under conditions that are more relevant to a bona fide infection (i.e. low particle concentration). Following cleavage and release from μ1, myr-μ1N, which alone can perforate liposomes and bovine RBCs, is expected to oligomerize within host membranes (33, 36, 40). Thus, it is presumed that a high local concentration of myr-μ1N is generated at the site of pore formation. Moreover, lipid-associated myr-μ1N recruits ISVPs to membranes (Fig. 6). Together, these observations suggest that membranes can act as a scaffold to bring two input components into close proximity. High local concentrations of lipid-associated myr-μ1N and ISVPs increase the efficiency of ISVP-to-ISVP* conversion.
Membranes facilitate particle inactivation in a lipid composition-dependent manner; liposomes that contain phosphatidylcholine and phosphatidylethanolamine trigger ISVP* formation most efficiently (9). Given the membrane-targeting potential of myr-μ1N (33, 36, 40), we speculate that certain lipids, or certain lipid formulations, position myr-μ1N to interact with target ISVPs. The basis for lipid specificity is unclear; however, factors such as membrane fluidity or rigidity, composition of fatty acid tails, and/or composition of lipid head groups may influence pore formation, and thus, orientation of the inserted peptide. Furthermore, the properties that affect the conformation of embedded myr-μ1N and the properties that affect pore formation are likely not separable. The myr-μ1N peptide contains two regions of alternating, hydrophobic residues. When mutations are made within these regions, the capacity of myr-μ1N to generate pores is reduced (40); however, whether these mutated peptides can interact with each other has not been demonstrated. As such, hydrophobic forces are likely important for oligomerization of myr-μ1N and for myr-μ1N-lipid interactions. Moreover, it is unknown whether ISVPs interact exclusively with pore-associated myr-μ1N. Thermal inactivation is induced by myr-μ1N in a dose-dependent manner (Fig. 4). Thus, it is possible that ISVPs interact with multiple, pore-associated peptides to promote particle conversion.
We propose a model in which lipids enhance the ISVP*-promoting activity of membrane-embedded myr-μ1N (Fig. 7). Reovirus ISVPs may exist in reversible equilibrium with a conformationally altered form, called ISVPΔ (54). The capsids of nonenveloped viruses can exhibit structural flexibility (55, 56). Capsid “breathing” transiently and reversibly exposes normally internal peptides on the surface of the particle. ISVPΔ likely represents a flexible form of the ISVP. It was suggested that thermal energy induces spontaneous ISVPΔ-to-ISVP* conversion (46). ISVP* formation releases myr-μ1N, Φ, and σ1 (11, 33–39). myr-μ1N, which can perforate liposomes in the absence Φ or σ1 (33, 36, 40), is predicted to interact with the endosomal membrane. When RBCs are incubated with ISVP* supernatant or with myr-μ1N, pores that are capable of recruiting ISVP*s are formed (33, 36). In the absence of host proteins, myr-μ1N can also recruit ISVPs to membranes (Fig. 6). Finally, specific lipids (e.g. phosphatidylcholine and phosphatidylethanolamine) enhance the ISVP*-promoting activity of myr-μ1N (Figs. 4 and 5). Together, these results are consistent with myr-μ1N-lipid interactions serving two complementary functions: recruiting ISVPs to the endosomal membrane and augmenting myr-μ1N-mediated ISVP-to-ISVP* conversion. Thus, lipids function within a positive-feedback loop by artificially raising the concentration of myr-μ1N at the site of pore formation and by positioning myr-μ1N to interact with ISVPs more effectively. Nonetheless, questions regarding this model remain. The myr-μ1N binding site on target ISVPs, including the stoichiometry of the interaction (i.e. the minimum number of myr-μ1N peptides that is needed to convert a single ISVP), is currently unknown. The stability of the myr-μ1N-ISVP interaction at physiological conditions is also unknown. As discussed earlier, reovirus ISVPs can undergo “structural breathing” to form a conformationally altered intermediate (i.e. ISVPΔ) (54). This structural transition may facilitate ISVP* formation by exposing internal epitopes that interact with lipid-associated myr-μ1N (46, 54). Next, if membranes only position myr-μ1N to interact with ISVPs, the initial trigger of ISVP-to-ISVP* conversion is still unclear. Partial conversion, which releases a limited quantity of myr-μ1N, may occur at 37 °C (35, 46). Thus, the endosomal membrane would be in position to amplify the ISVP*-promoting activity. These questions are the subject of ongoing investigations.
FIGURE 7.
Model for lipid-mediated ISVP-to-ISVP* conversion. See “Discussion” for details. This figure was modified from research originally published in Proceedings of the National Academy of Sciences (Agosto, M. A., Myers, K. S., Ivanovic, T., and Nibert, M. L. (2008) A positive-feedback mechanism promotes reovirus particle conversion to the intermediate associated with membrane penetration. Proc. Natl. Acad. Sci. U.S.A. 105, 10571–10576). ©2008 National Academy of Sciences, U.S.A.
A significant question remaining is whether lipids cooperate with myr-μ1N to facilitate reovirus entry into host cells. At high particle concentration, PC liposomes promote ISVP-to-ISVP* conversion (Fig. 1) (9). Thus, we attempted to enzymatically deplete PC from cells with phospholipases. PC constitutes a large proportion of the plasma and endosomal membranes (40–50% of the phospholipid content) (57). Not surprisingly, reducing PC levels was severely cytotoxic, which precluded us from assessing its impact on viral entry (data not shown). Another question is whether lipid-associated myr-μ1N can induce ISVP-to-ISVP* conversion under conditions that are likely to arise during a bona fide infection. As described earlier, high local concentrations of myr-μ1N and ISVPs may be generated on the endosomal membrane. At low particle concentration (e.g. a single ISVP within an endosome), the inclusion of lipids alone or myr-μ1N alone only marginally enhanced ISVP* formation (Figs. 1, 2, and 4). In contrast, this conformational change does occur when ISVPs are incubated with liposomes and myr-μ1N (Figs. 4 and 5). Moreover, lipids reduce the amount of myr-μ1N that is needed to inactivate ISVPs (Fig. 4). Together, these results suggest that the in vitro mechanism of myr-μ1N-mediated ISVP-to-ISVP* conversion might be biologically relevant. Lipids facilitate entry by enhancing the ISVP*-promoting activity of myr-μ1N.
Viruses utilize related pathways to penetrate the host and deliver their genetic material. In most cases, enveloped viruses mediate fusion between viral and host membranes, whereas the role of lipids during entry of nonenveloped viruses is an active area of research. Our work reveals that reovirus can usurp host membranes to amplify its own triggering activity. Thus, reovirus overcomes a limiting environment in which common promoting factors (e.g. virus-receptor interactions and low pH) do not facilitate the structural transitions required for entry.
Experimental Procedures
Cells and Viruses
Spinner-adapted murine L929 (L) cells were grown at 37 °C in Joklik's minimal essential medium (Lonza, Walkersville, MD) supplemented with 5% FBS (Life Technologies), 2 mm l-glutamine (Invitrogen), 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 25 ng/ml amphotericin B (Sigma-Aldrich). T3D was generated by plasmid-based reverse genetics (58, 59).
Virus Purification
T3D virions were propagated and purified as described previously (45, 60). Briefly, L cells infected with second- or third-passage reovirus stocks were lysed by sonication. Virus particles were extracted from lysates using Vertrel-XF specialty fluid (DuPont) (61). The extracted particles were layered onto 1.2–1.4-g/cm3 CsCl step gradients. The gradients were then centrifuged at 187,000 × g for 4 h at 4 °C in an SW 41 Ti rotor (Beckman Coulter). Bands corresponding to purified virus particles (∼1.36 g/cm3) (62) were isolated and dialyzed into virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl). Following dialysis, the particle concentration was determined by measuring optical density of the purified virus stocks at 260 nm (A260; 1 unit at A260 = 2.1 × 1012 particles/ml) (62).
Generation of ISVPs
T3D virions (2 × 1012 particles/ml or 4 × 1012 particles/ml) were digested with 200 μg/ml TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone)-treated chymotrypsin (Worthington Biochemical) in a total volume of 100 μl of virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl) for 20 min at 32 °C (41, 42). After 20 min, the reaction mixtures were incubated on ice for 20 min and quenched by the addition of 1 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich). The generation of ISVPs was confirmed by SDS-PAGE and Coomassie Brilliant Blue staining.
Liposome Preparation
The lipids used in this study (PC from chicken egg, sphingomyelin (SM) from porcine brain, cholesterol (Chl) from ovine wool, PE from chicken egg, l-α-phosphatidylserine (PS) from porcine brain, and lysobisphosphatidic acid (LBPA)) were purchased from Avanti Polar Lipids (Alabaster, AL). All lipids were dissolved in chloroform and stored at −20 °C. Prior to liposome preparation, the lipids were dried under a stream of argon gas. 1 mm liposomes were prepared by resuspending the dried lipids in 250 μl of virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl) and passing the resuspension (31 times) through an Avanti Mini Extruder with a 0.1-μm-pore size polycarbonate membrane (Avanti Polar Lipids). The liposome compositions used in this study were PC alone and PC/PE (2:1 molar ratio). The composition of early endosome liposomes was Chl/PC/PE/SM/PS/LBPA (50:26:13:5:5:1 molar ratio) (9, 57).
Plaque Assays
Plaque assays to determine infectivity were performed as described previously (44, 60). Briefly, control or heat-treated virus samples were diluted in PBS supplemented with 2 mm MgCl2. L cell monolayers in 6-well plates (Greiner Bio-One, Kremsmünster, Austria) were infected with 250 μl of diluted virus for 1 h at room temperature. Following the viral attachment incubation, the monolayers were overlaid with 4 ml of serum-free medium 199 (Sigma-Aldrich) supplemented with 1% Bacto Agar (BD Biosciences), 10 μg/ml TLCK-treated chymotrypsin (Worthington Biochemical), 2 mm l-glutamine (Invitrogen), 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 25 ng/ml amphotericin B (Sigma-Aldrich) The infected cells were incubated at 37 °C, and plaques were counted 5 days after infection.
Generation of ISVP* Supernatant
The supernatant of preconverted ISVP*s was generated as described previously (36, 46). Briefly, input T3D ISVPs (2 × 1012 particles/ml or 3 nm) were incubated at 52 °C for 5 min. The heat-inactivated virus (no spin) was then centrifuged at 16,000 × g for 10 min at 4 °C to pellet particles. The supernatant (spin) was immediately transferred to tubes containing target T3D ISVPs for thermal inactivation reactions (Fig. 3) or for ISVP-liposome coflotation experiments (Fig. 6B). To ensure that intact T3D ISVPs did not contaminate the transferred ISVP* supernatant, aliquots of the no spin and spin reactions were analyzed for residual infectivity by plaque assay and for the presence of the μ1 δ fragment by Western blotting using a rabbit, α-reovirus polyclonal antibody (63).
Synthetic myr-μ1N
The myr-μ1N peptide was synthesized by and purchased from ABI Scientific (Sterling, VA). The peptide comprised amino acids 2–42 of T3D μ1 and was modified with an N-terminal N-myristoyl group (myr). To improve solubility, the peptide contained six additional amino acids (RGKGRC) at its C terminus (40). Purified peptide was dissolved in 100% dimethylformamide. When the myr-μ1N peptide was added to thermal inactivation (Figs. 4 and 5) or ISVP-liposome coflotation (Fig. 6C) reactions, the final dimethylformamide concentration, including the no peptide control, was adjusted to 0.25%.
Thermal Inactivation and Trypsin Sensitivity Assays
For experiments shown in Figs. 1, 3, and 4, T3D ISVPs (2 × 1012 particles/ml (3 nm) or 2 × 109 particles/ml (0.003 nm)) were incubated in the absence or presence of 1 mm EE, PC, or PC/PE (2:1) liposomes for 20 min at the indicated temperatures in a Bio-Rad S1000 thermal cycler. When specified, ISVP* supernatant (15 μl of supernatant per 30 μl of total reaction volume) (Fig. 3) or myr-μ1N peptide (concentrations shown in Fig. 4B or 2.5 μm in Fig. 4, C and D) was included. Before adding virus, ISVP* supernatant or myr-μ1N was mixed with liposomes, vortexed briefly, and incubated on ice for 5 min. The total volume of each reaction was 30 μl in virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl). For each reaction condition, an aliquot was also incubated for 20 min at 4 °C. Following incubation, 10 μl of each reaction was diluted into 40 μl of ice-cold virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl), and infectivity was determined by plaque assay. The change in infectivity at a given temperature (T) was calculated using the following formula: log10(PFU/ml)T − log10(PFU/ml)4 °C. Under each reaction condition at high particle concentration (2 × 1012 particles/ml or 3 nm), the titers of the 4 °C control samples were between 5 × 109 and 5 × 1010 PFU/ml. Under each reaction condition at low particle concentration (2 × 109 particles/ml or 0.003 nm), the titers of the 4 °C control samples were between 5 × 106 and 5 × 107 PFU/ml. The remaining 20 μl of each reaction was treated with 0.08 mg/ml trypsin (Sigma-Aldrich) for 30 min on ice. Following digestion, equal particle numbers from each reaction were solubilized in reducing SDS sample buffer and analyzed by SDS-PAGE. In Fig. 1B, the gels were Coomassie Brilliant Blue-stained and imaged on an Odyssey imaging system (LI-COR, Lincoln, NE). In Figs. 1D, 3D, and 4D, the reaction mixtures were analyzed for the presence of the μ1 δ fragment by Western blotting using a rabbit, α-reovirus polyclonal antibody (63).
For the experiments shown in Figs. 2 and 5, T3D ISVPs (2 × 109 particles/ml or 0.003 nm) were incubated in the absence or presence of 1 mm EE, PC, or PC/PE (2:1) liposomes for the indicated amounts of time at the indicated temperatures in a water bath. When specified, 2.5 μm myr-μ1N peptide was included. Before adding virus, myr-μ1N was mixed with liposomes, vortexed briefly, and incubated for 5 min on ice. The total volume of each reaction was 2 ml in virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl). For each reaction condition, an aliquot was also incubated at 4 °C. At the indicated time points, 10 μl of each reaction was diluted into 40 μl of ice-cold virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl), and infectivity was determined by plaque assay. The change in infectivity at a given temperature (T) was calculated using the following formula: log10(PFU/ml)T − log10(PFU/ml)4 °C. Under each reaction condition, the titers of the 4 °C control samples were between 5 × 106 and 5 × 107 PFU/ml. The rate constant, k, was determined for each thermal inactivation curve as described previously (44). To generate Arrhenius plots, the rate constants were plotted as ln(k/T) versus the reciprocal of the incubation temperature (T) in degrees Kelvin. The thermodynamic values shown in Table 1 were calculated using the Eyring absolute equation as described previously (44, 49). ΔG‡ was calculated at 48 °C (321 K) using the following formula: ΔG‡ = ΔH‡ − T ΔS‡.
ISVP-Liposome Coflotation Assay
T3D ISVPs (2 × 1012 particles/ml or 3 nm) were incubated in the absence or presence of 1 mm EE, PC, or PC/PE (2:1) liposomes for 10 min on ice (Fig. 6, A–C) or for 20 min at 49 °C (Fig. 6D). When indicated, liposomes were pretreated with ISVP* supernatant (10 μl of supernatant per 100 μl of total reaction volume) (Fig. 6B) or with 2.5 μm myr-μ1N peptide (Fig. 6C) for 1 min at 37 °C. All reactions were performed in virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl). The samples were then applied to the top of 15–60% linear sucrose gradients (mass/volume in virus storage buffer (10 mm Tris, pH 7.4, 15 mm MgCl2, 150 mm NaCl)) and sedimented at 77,000 × g for 2 h at 4 °C in a SW 41 Ti rotor (Beckman Coulter). Following ultracentrifugation, 1-ml fractions were collected from the top of the gradients and solubilized in reducing SDS sample buffer. Equal volumes from each fraction were analyzed by SDS-PAGE. The gels were analyzed for the presence of the μ1 δ fragment by Western blotting using a rabbit, α-reovirus polyclonal antibody (63).
Statistical Analyses
Unless stated otherwise, the reported values represent the mean of three independent, biological replicates. Error bars indicate the S.D. p values were calculated using Student's t test (two-tailed, unequal variance assumed).
Author Contributions
A. J. S. and P. D. designed the study; A. J. S. performed the experiments; A. J. S. and P. D. analyzed the data; and A. J. S. and P. D. wrote the manuscript.
Acknowledgments
We thank Karl Boehme, Tuli Mukhopadhyay, Kevin Sokoloski, Adam Zlotnick, and members of our laboratory for helpful suggestions and reviews of the manuscript.
This work was supported by NIAID, National Institutes of Health Grant 1R01AI110637 (to P. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ISVP
- infectious subviral particle
- myr-μ1N
- myristoylated μ1N
- EE
- early endosome
- PC
- l-α-phosphatidylcholine
- PE
- l-α-phosphatidylethanolamine
- PS
- l-α-phosphatidylserine
- Chl
- cholesterol
- SM
- sphingomyelin
- LBPA
- lysobisphosphatidic acid
- TLCK
- Nα-p-tosyl-l-lysine chloromethyl ketone
- L cells
- L929 cells.
References
- 1. Smith A. E., and Helenius A. (2004) How viruses enter animal cells. Science 304, 237–242 [DOI] [PubMed] [Google Scholar]
- 2. Tsai B. (2007) Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23, 23–43 [DOI] [PubMed] [Google Scholar]
- 3. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kielian M. (2014) Mechanisms of virus membrane fusion proteins. Annu. Rev. Virol. 1, 171–189 [DOI] [PubMed] [Google Scholar]
- 5. Weissenhorn W., Dessen A., Calder L. J., Harrison S. C., Skehel J. J., and Wiley D. C. (1999) Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 6. White J. M., Delos S. E., Brecher M., and Schornberg K. (2008) Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luisoni S., Suomalainen M., Boucke K., Tanner L. B., Wenk M. R., Guan X. L., Grzybek M., Coskun Ü., and Greber U. F. (2015) Co-option of membrane wounding enables virus penetration into cells. Cell Host Microbe 18, 75–85 [DOI] [PubMed] [Google Scholar]
- 8. Patel A., Mohl B. P., and Roy P. (2016) Entry of bluetongue virus capsid requires the late endosome-specific lipid lysobisphosphatidic acid. J. Biol. Chem. 291, 12408–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snyder A. J., and Danthi P. (2015) Lipid membranes facilitate conformational changes required for reovirus cell entry. J. Virol. 90, 2628–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dryden K. A., Wang G., Yeager M., Nibert M. L., Coombs K. M., Furlong D. B., Fields B. N., and Baker T. S. (1993) Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122, 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., Ji Y., Zhang L., Harrison S. C., Marinescu D. C., Nibert M. L., and Baker T. S. (2005) Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Å resolution. Structure 13, 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dermody T. S., Parker J. S. L., and Sherry B. (2013) Orthoreoviruses. in Fields Virology, 6th Ed., pp. 1304–1346, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 13. Barton E. S., Connolly J. L., Forrest J. C., Chappell J. D., and Dermody T. S. (2001) Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 14. Chappell J. D., Duong J. L., Wright B. W., and Dermody T. S. (2000) Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 74, 8472–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gentsch J. R., and Pacitti A. F. (1985) Effect of neuraminidase treatment of cells and effect of soluble glycoproteins on type 3 reovirus attachment to murine L cells. J. Virol. 56, 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gentsch J. R., and Pacitti A. F. (1987) Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology 161, 245–248 [DOI] [PubMed] [Google Scholar]
- 17. Paul R. W., Choi A. H., and Lee P. W. (1989) The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology 172, 382–385 [DOI] [PubMed] [Google Scholar]
- 18. Borsa J., Morash B. D., Sargent M. D., Copps T. P., Lievaart P. A., and Szekely J. G. (1979) Two modes of entry of reovirus particles into L cells. J. Gen Virol 45, 161–170 [DOI] [PubMed] [Google Scholar]
- 19. Borsa J., Sargent M. D., Lievaart P. A., and Copps T. P. (1981) Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111, 191–200 [DOI] [PubMed] [Google Scholar]
- 20. Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M. L., and Kirchhausen T. (2004) Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118, 591–605 [DOI] [PubMed] [Google Scholar]
- 21. Maginnis M. S., Forrest J. C., Kopecky-Bromberg S. A., Dickeson S. K., Santoro S. A., Zutter M. M., Nemerow G. R., Bergelson J. M., and Dermody T. S. (2006) β1 integrin mediates internalization of mammalian reovirus. J. Virol. 80, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maginnis M. S., Mainou B. A., Derdowski A., Johnson E. M., Zent R., and Dermody T. S. (2008) NPXY motifs in the β1 integrin cytoplasmic tail are required for functional reovirus entry. J. Virol. 82, 3181–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baer G. S., and Dermody T. S. (1997) Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71, 4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang C. T., and Zweerink H. J. (1971) Fate of parental reovirus in infected cell. Virology 46, 544–555 [DOI] [PubMed] [Google Scholar]
- 25. Sturzenbecker L. J., Nibert M., Furlong D., and Fields B. N. (1987) Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dermody T. S., Nibert M. L., Wetzel J. D., Tong X., and Fields B. N. (1993) Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67, 2055–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ebert D. H., Deussing J., Peters C., and Dermody T. S. (2002) Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277, 24609–24617 [DOI] [PubMed] [Google Scholar]
- 28. Silverstein S. C., Astell C., Levin D. H., Schonberg M., and Acs G. (1972) The mechanisms of reovirus uncoating and gene activation in vivo. Virology 47, 797–806 [DOI] [PubMed] [Google Scholar]
- 29. Amerongen H. M., Wilson G. A., Fields B. N., and Neutra M. R. (1994) Proteolytic processing of reovirus is required for adherence to intestinal M cells. J. Virol. 68, 8428–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bass D. M., Bodkin D., Dambrauskas R., Trier J. S., Fields B. N., and Wolf J. L. (1990) Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 64, 1830–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bodkin D. K., Nibert M. L., and Fields B. N. (1989) Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63, 4676–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nygaard R. M., Golden J. W., and Schiff L. A. (2012) Impact of host proteases on reovirus infection in the respiratory tract. J. Virol. 86, 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agosto M. A., Ivanovic T., and Nibert M. L. (2006) Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borsa J., Long D. G., Sargent M. D., Copps T. P., and Chapman J. D. (1974) Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology 4, 171–188 [DOI] [PubMed] [Google Scholar]
- 35. Chandran K., Farsetta D. L., and Nibert M. L. (2002) Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76, 9920–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivanovic T., Agosto M. A., Zhang L., Chandran K., Harrison S. C., and Nibert M. L. (2008) Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J. 27, 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nibert M. L., Odegard A. L., Agosto M. A., Chandran K., and Schiff L. A. (2005) Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345, 461–474 [DOI] [PubMed] [Google Scholar]
- 38. Nibert M. L., Schiff L. A., and Fields B. N. (1991) Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65, 1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Odegard A. L., Chandran K., Zhang X., Parker J. S., Baker T. S., and Nibert M. L. (2004) Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78, 8732–8745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang L., Agosto M. A., Ivanovic T., King D. S., Nibert M. L., and Harrison S. C. (2009) Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J. Virol. 83, 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Borsa J., Copps T. P., Sargent M. D., Long D. G., and Chapman J. D. (1973) New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J. Virol. 11, 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Joklik W. K. (1972) Studies on the effect of chymotrypsin on reovirions. Virology 49, 700–715 [DOI] [PubMed] [Google Scholar]
- 43. Borsa J., Sargent M. D., Long D. G., and Chapman J. D. (1973) Extraordinary effects of specific monovalent cations on activation of reovirus transcriptase by chymotrypsin in vitro. J. Virol. 11, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Middleton J. K., Severson T. F., Chandran K., Gillian A. L., Yin J., and Nibert M. L. (2002) Thermostability of reovirus disassembly intermediates (ISVPs) correlates with genetic, biochemical, and thermodynamic properties of major surface protein μ1. J. Virol. 76, 1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarkar P., and Danthi P. (2010) Determinants of strain-specific differences in efficiency of reovirus entry. J. Virol. 84, 12723–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Agosto M. A., Myers K. S., Ivanovic T., and Nibert M. L. (2008) A positive-feedback mechanism promotes reovirus particle conversion to the intermediate associated with membrane penetration. Proc. Natl. Acad. Sci. U.S.A. 105, 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Borsa J., Long D. G., Copps T. P., Sargent M. D., and Chapman J. D. (1974) Reovirus transcriptase activation in vitro: further studies on the facilitation phenomenon. Intervirology 3, 15–35 [DOI] [PubMed] [Google Scholar]
- 48. Agosto M. A., Middleton J. K., Freimont E. C., Yin J., and Nibert M. L. (2007) Thermolabilizing pseudoreversions in reovirus outer-capsid protein μ1 rescue the entry defect conferred by a thermostabilizing mutation. J. Virol. 81, 7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pollard E. C. (1953) The Physics of Viruses, pp. 103–119, Academic Press, New York [Google Scholar]
- 50. Attoui H., Fang Q., Mohd Jaafar F., Cantaloube J. F., Biagini P., de Micco P., and de Lamballerie X. (2002) Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol 83, 1941–1951 [DOI] [PubMed] [Google Scholar]
- 51. Liemann S., Chandran K., Baker T. S., Nibert M. L., and Harrison S. C. (2002) Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X., Tang J., Walker S. B., O'Hara D., Nibert M. L., Duncan R., and Baker T. S. (2005) Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 343, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X., Jin L., Fang Q., Hui W. H., and Zhou Z. H. (2010) 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Borsa J., Sargent M. D., Kay C. M., and Oikawa K. (1976) Circular dichroism of intermediate subviral particles of reovirus: elucidation of the mechanism underlying the specific monovalent cation effects on uncoating. Biochim. Biophys. Acta 451, 619–627 [DOI] [PubMed] [Google Scholar]
- 55. Li Q., Yafal A. G., Lee Y. M., Hogle J., and Chow M. (1994) Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 68, 3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lewis J. K., Bothner B., Smith T. J., and Siuzdak G. (1998) Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. U.S.A. 95, 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., and Gruenberg J. (1998) A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392, 193–197 [DOI] [PubMed] [Google Scholar]
- 58. Kobayashi T., Antar A. A., Boehme K. W., Danthi P., Eby E. A., Guglielmi K. M., Holm G. H., Johnson E. M., Maginnis M. S., Naik S., Skelton W. B., Wetzel J. D., Wilson G. J., Chappell J. D., and Dermody T. S. (2007) A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kobayashi T., Ooms L. S., Ikizler M., Chappell J. D., and Dermody T. S. (2010) An improved reverse genetics system for mammalian orthoreoviruses. Virology 398, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Furlong D. B., Nibert M. L., and Fields B. N. (1988) σ1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mendez I. I., Hermann L. L., Hazelton P. R., and Coombs K. M. (2000) A comparative analysis of Freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90, 59–67 [DOI] [PubMed] [Google Scholar]
- 62. Smith R. E., Zweerink H. J., and Joklik W. K. (1969) Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39, 791–810 [DOI] [PubMed] [Google Scholar]
- 63. Wetzel J. D., Chappell J. D., Fogo A. B., and Dermody T. S. (1997) Efficiency of viral entry determines the capacity of murine erythroleukemia cells to support persistent infections by mammalian reoviruses. J. Virol. 71, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]