Abstract
E-26 transformation-specific (ETS) proteins are transcription factors directing gene expression through their conserved DNA binding domain. They are implicated as truncated forms or interchromosomal rearrangements in a variety of tumors including Ewing sarcoma, a pediatric tumor of the bone. Tumor cells express the chimeric oncoprotein EWS-FLI1 from a specific t(22;11)(q24;12) translocation. EWS-FLI1 harbors a strong transactivation domain from EWSR1 and the DNA-binding ETS domain of FLI1 in the C-terminal part of the protein. Although Ewing cells are crucially dependent on continuous expression of EWS-FLI1, its regulation of turnover has not been characterized in detail. Here, we identify the EWS-FLI1 protein as a substrate of the ubiquitin-proteasome system with a characteristic polyubiquitination pattern. Using a global protein stability approach, we determined the half-life of EWS-FLI1 to lie between 2 and 4 h, whereas full-length EWSR1 and FLI1 were more stable. By mass spectrometry, we identified two ubiquitin acceptor lysine residues of which only mutation of Lys-380 in the ETS domain of the FLI1 part abolished EWS-FLI1 ubiquitination and stabilized the protein posttranslationally. Expression of this highly stable mutant protein in Ewing cells while simultaneously depleting the endogenous wild type protein differentially modulates two subgroups of target genes to be either EWS-FLI1 protein-dependent or turnover-dependent. The majority of target genes are in an unaltered state and cannot be further activated. Our study provides novel insights into EWS-FLI1 turnover, a critical pathway in Ewing sarcoma pathogenesis, and lays new ground to develop novel therapeutic strategies in Ewing sarcoma.
Keywords: cancer biology, ETS transcription factor family, proteasome, transcription target gene, ubiquitin
Introduction
E-26 transformation-specific (ETS)2 family members are strong activators or repressors of transcription with a highly conserved ETS domain (1–3). ETS transcription factors (TFs) bind most commonly in complexes to a GGA core region to mediate gene expression (4, 5). Their main biological functions include regulation of differentiation, lineage determination of the hematopoietic system, and control of angiogenesis (6, 7). Most of the ETS family members have oncogenic potential because truncated or overexpressed ETS proteins have been linked to several cancer entities (8–11). ERG and ETV1 are frequently fused to the TMPRSS2 promoter in prostate cancer, whereas ETV1 and ETV6 are implicated in leukemia (12, 13). Like other aberrant fusion proteins, they act as drivers of uncontrolled cell growth and survival (14, 15). However, most TFs do not harbor an enzymatic pocket and are therefore difficult to target directly. Novel strategies that uncover vulnerable sites in TFs are urgently needed to develop novel targeted therapies (16).
Ewing sarcoma is a rare pediatric bone and soft tissue tumor with an aggressive behavior and prevalence to metastasize (17, 18). Its main genetic abnormalities are EWS-ETS rearrangements, among them most commonly the EWS gene on chromosome 22 fused to FLI1 on chromosome 11, which results in expression of the chimeric transcription factor EWS-FLI1 (19–21). Continuous expression of the fusion protein is crucial for tumor formation, progression, and maintenance (22, 23), and its down-regulation inhibits proliferation and reduces tumor cell growth (24–26). EWS-FLI1 is thought to function mainly as a modulator to activate and repress a wide range of target genes but also as a regulator of splicing processes or as a component of large interaction networks (27–31). However, inhibition of a single downstream target gene has not been proven effective yet for Ewing sarcoma therapy.
The turnover of most intracellular proteins is mediated via the ubiquitin-proteasome system, which triggers protein degradation (32). Several ETS proteins can be polyubiquitinated by different E3 ligases and subsequently degraded by the proteasome (33–36). Considering their high conservation, proteasomal degradation is likely to be the main mechanism of turnover for most ETS family members. Most interestingly, truncated ERG and ETV1 lack the N-terminal E3 binding domain, which results in a delayed turnover of aberrant ETS proteins (34, 36). Here, we focus specifically on the turnover of the fusion protein EWS-FLI1, which only harbors the ETS and the C-terminal domain from FLI1. The impact on the turnover of this domain fused to the N-terminal region of EWSR1 has not been investigated yet. Hence, EWS-FLI1 proteasomal turnover represents a so far uncharacterized mechanism in Ewing sarcoma tumorigenesis even though EWS-FLI1 degradation has already been linked to the lysosomal pathway (37). Moreover, the exact lysine acceptor residues for polyubiquitination have not yet been identified for any wild type or truncated ETS protein.
In the present study, we demonstrate that EWS-FLI1 is predominantly a proteasomal substrate with an unexpectedly high turnover rate mediated by polyubiquitination at a single lysine residue. Surprisingly, expression of this highly stable mutant protein in Ewing cells specifically induced subgroups of highly expressed or turnover-sensitive target genes, whereas the majority of target genes are unaltered. Hence, our study provides novel insights into EWS-FLI1 turnover, suggesting that deflecting its stability could contribute to new therapeutic concepts in Ewing sarcoma.
Results
EWS-FLI1 Turnover Is Proteasome-dependent
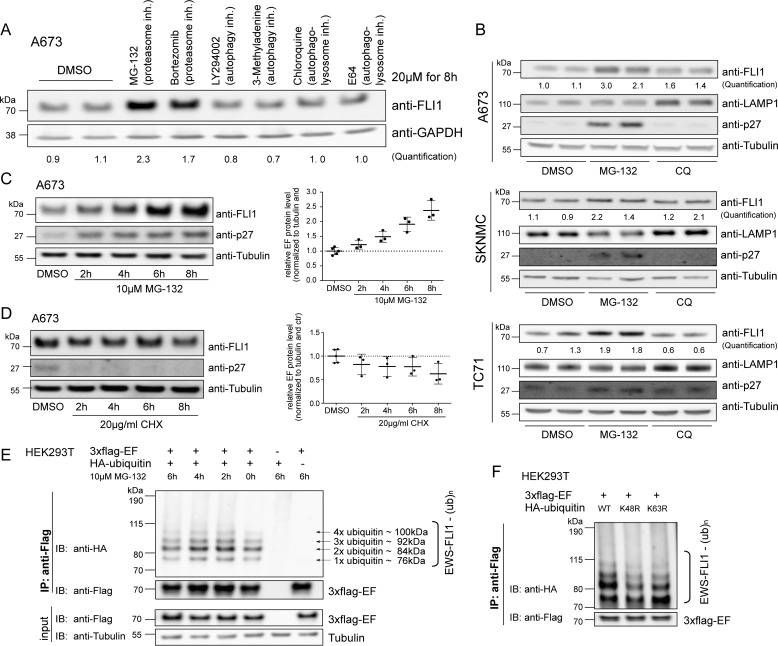
Because EWS-FLI1 expression is crucial for tumor cell survival (24–26), we were interested to analyze degradation and turnover rate of the fusion protein. To address this, we first used a panel of six different inhibitors targeting proteasomal, lysosomal, and autophagosomal protein degradation pathways. Incubation with a 20 μm concentration of the various inhibitors resulted in a 2-fold up-regulation of EWS-FLI1 protein levels only upon treatment with MG-132 and bortezomib, both inhibitors of the chymotrypsin-like proteasome activity (Fig. 1A). We next confirmed this result by selectively inhibiting the proteasomal and lysosomal degradation pathways with two compounds in three different Ewing sarcoma cell lines. Incubation with a 20 μm concentration of the proteasome inhibitor MG-132 resulted in stabilization of endogenous EWS-FLI1 fusion protein by at least 2-fold, whereas this was not the case for the lysosomal inhibitor chloroquine (Fig. 1B). p27 and LAMP1 were used as positive controls and showed up-regulation after treatment as expected. In addition, stabilization of the fusion protein in Ewing cells by MG-132 increased over a 2–8-h treatment period in a time-dependent manner (Fig. 1C). Hence, degradation of EWS-FLI1 is primarily proteasome-dependent under steady-state conditions. However, incubation with 20 μm cycloheximide (CHX) had only limited reducing effect on the endogenous fusion protein in the Ewing cells over the same 8-h time period (Fig. 1D).
FIGURE 1.
EWS-FLI1 protein turnover is proteasome-dependent. A, Western blotting analysis of EWS-FLI1 protein levels. A673 cells were treated with a 20 μm concentration of the indicated compounds for 8 h, and EWS-FLI1 protein levels were detected with anti-FLI1 antibody. Quantification represents the ratio of FLI1 over GAPDH compared with DMSO control. B, EWS-FLI1 turnover in various Ewing sarcoma cell lines. A673, SKNMC, and TC71 cells were treated with 20 μm MG-132 or chloroquine (CQ) for 10 (A673), 8 (SKNMC), and 9 h (TC71) and immunoblotted with anti-FLI1 antibody. Quantification represents the ratio of FLI1 over tubulin compared with DMSO control. C, EWS-FLI1 stabilizes in a time-dependent manner. Shown is a Western blot of A673 cells treated with 10 μm MG-132 for the indicated time points. Three independent experiments were quantified and are represented in the scatter plot with n = 3 (2–8 h) or n = 5 (DMSO); error bars represent S.D. D, half-life of endogenous EWS-FLI1 protein. Shown is a Western blot of A673 cells treated with 20 μg/ml CHX for the indicated hours. Three independent experiments were quantified with n = 3 (2–8 h) or n = 5 (DMSO); error bars represent S.D. E, EWS-FLI1 is ubiquitinated. 3xFLAG-EWS-FLI1 and HA-ubiquitin were co-expressed for 48 h in HEK293T cells and incubated with 10 μm MG-132 for the indicated hours. After immunoprecipitation of EWS-FLI1 with anti-FLAG, ubiquitination was visualized by anti-HA antibody. F, EWS-FLI1 ubiquitination consists of Lys-48-linked ubiquitin chains. 3xFLAG-EWS-FLI1 and wild type or mutant HA-ubiquitins were co-expressed for 48 h in HEK293T cells and immunoprecipitated, and ubiquitination was visualized by anti-HA antibody. inh., inhibitor; IP, immunoprecipitation; IB, immunoblotting; ub, ubiquitin; EF, EWS-FLI1.
Next, we investigated whether the fusion protein is ubiquitinated. To this end, 3xFLAG-EWS-FLI1 was co-expressed with HA-ubiquitin in HEK293T cells and immunoprecipitated after 48 h using anti-FLAG antibody. Western blotting with an anti-HA antibody revealed at least four distinct ubiquitin bands for EWS-FLI1 that could not be increased by prior treatment with MG-132 (Fig. 1E), suggesting a constant ubiquitination of the fusion protein. We next co-expressed 3xFLAG-EWS-FLI1 with wild type HA-ubiquitin or ubiquitin mutants that are deficient for Lys-48- and Lys-63-linked chains. Immunoprecipitation of the ubiquitinated 3xFLAG-EWS-FLI1 revealed mainly a decrease of the ubiquitination pattern using K48R HA-ubiquitin (Fig. 1F), indicating that this is the major, but not only, linked ubiquitin chain of the fusion protein.
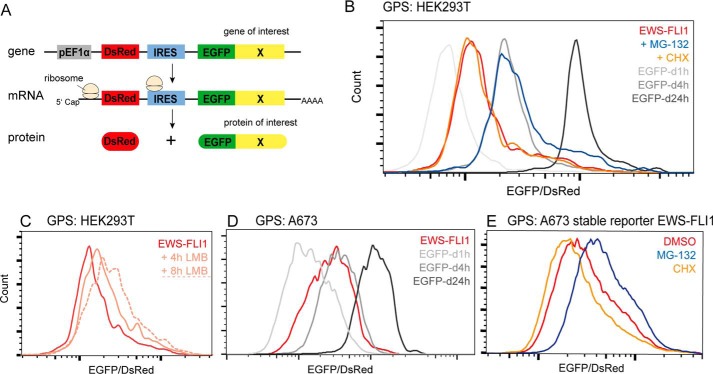
To further validate the fusion protein turnover and to overcome the limitations of classical CHX treatment to determine protein half-lives, we next used the global protein stability (GPS) approach (38) (Fig. 2A). Briefly, HEK293T cells were transduced with a reporter construct, DsRed-IRES-EGFP, in which EGFP is fused to EWS-FLI1. The ratio EGFP/DsRed was determined by FACS and represents a measure for protein stability. Known degron motifs with distinct half-lives were used as internal standards to estimate the half-life of EWS-FLI1 to be between 1 and 4 h (Fig. 2B). Incubation with MG-132 shifted the ratio closer to 4 h, confirming that EWS-FLI1 is a substrate of the proteasome system with a high turnover rate. Additionally, we treated cells with the nuclear export inhibitor leptomycin B (LMB) for 4 and 8 h. Treatment with 50 nm LMB stabilized EWS-FLI1 in a time-dependent manner (Fig. 2C), indicating that the proteasomal degradation mainly occurs in the cytosol. Similar results were obtained in transiently or stably transduced Ewing cells (Fig. 2, D and E), suggesting that the high turnover of EWS-FLI1 is mostly independent of the cellular background. Interestingly, incubation with 20 μm CHX again displayed a limited effect on the EGFP/DsRed ratio of the EWS-FLI1 reporter construct (Fig. 2, B and E), indicating that classical CHX treatment barely reflects the steady-state turnover rate of the EWS-FLI1 protein and possibly other proteins. Taken together, our results indicate that EWS-FLI1 is a polyubiquitinated protein with an unexpectedly high turnover rate in Ewing sarcoma cells.
FIGURE 2.
The EWS-FLI1 protein displays a high turnover. A, scheme illustrating GPS approach in which a reporter construct, DsRed-IRES-EGFP, is fused at the C terminus of EGFP to a protein of interest. The EGFP/DsRed ratio is determined by FACS and represents a measure for protein stability. B, EWS-FLI1 turnover measured by GPS. HEK293T were transduced for 72 h with a reporter construct fused to EWS-FLI1 or degron (d) motifs with half-lives of 1 (d1h), 4 (d4h), and 24 h (d24h) and analyzed by FACS. The reporter construct fused to EWS-FLI1 was additionally incubated with 20 μm MG-132 or 20 μg/ml CHX for 8 h or with 50 nm LMB for 4 and 8 h (dotted line) (C). D, EWS-FLI1 stability in Ewing cell line A673. GPS analysis of reporter constructs fused to EWS-FLI1 or standard degron motifs 72 h after transduction. E, A673 cells stably transfected with reporter-EWS-FLI1 construct were sorted and incubated with DMSO, 20 μm MG-132, or 20 μg/ml CHX for 8 h. d1h, d4h, and d24h, degron (d) motifs with half-lives of 1, 4, and 24 h, respectively.
EWS-FLI1 Turnover Depends on One Critical Lysine Residue
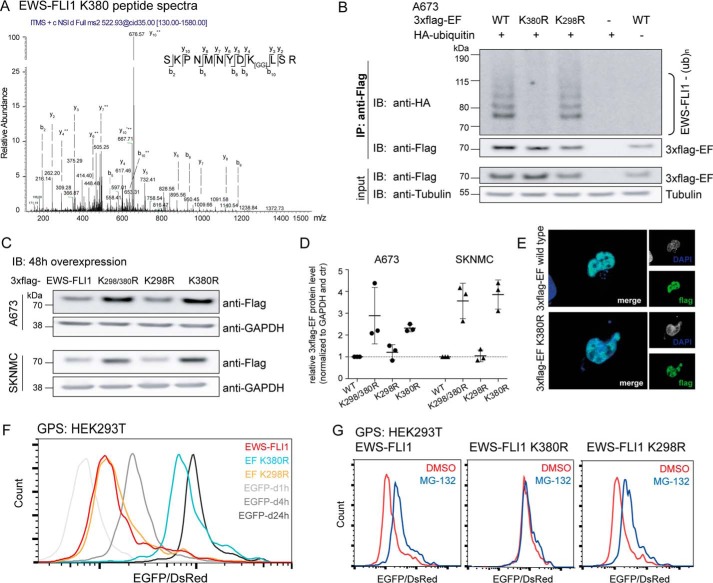
To better understand fusion protein turnover, we aimed to identify the lysine residue(s) that is important for degradation. We purified large amounts of 3xFLAG-EWS-FLI1 or a dominant-negative R386N mutant (39, 40) from HEK293T cells. The inactive mutant was included because the inability to bind DNA increases the possibility for ubiquitination. Both samples were then enzymatically digested and subjected to mass spectrometry to identify peptide shifts due to covalently attached ubiquitin. This identified two peptides with the characteristic glycine-glycine shift, including residues Lys-298 and Lys-380 (Fig. 3A and supplemental Fig. S1A). To determine whether both sites are important for fusion protein turnover, we mutated them individually to arginine. After co-expressing the mutant EWS-FLI1 with HA-ubiquitin and immunoprecipitation from A673 Ewing cells, we found that the K298R mutant was still ubiquitinated comparably with the wild type, whereas the K380R lost this characteristic pattern (Fig. 3B). In addition, transient expression of the mutants in both A673 and SKNMC Ewing cells revealed that the K380R and the double mutant K298R/K380R were 2.5- and 3.5-fold, respectively, more stable than wild type or the K298R mutant (Fig. 3, C and D). Like wild type EWS-FLI1, the K380 mutant was also still localized in the nucleus (Fig. 3E). We then investigated the role of both lysines in proteasomal turnover by GPS. The half-life of K298R was comparable with wild type EWS-FLI, whereas K380R shifted close to 24 h (Fig. 3F). Wild type EWS-FLI1 and the K298R could be stabilized by an additional incubation with MG-132; however, the EGFP/DsRed ratio of the K380R mutant did not change upon treatment (Fig. 3G). Hence, our findings suggest that Lys-380 is the major site required to regulate EWS-FLI1 protein turnover by the proteasome.
FIGURE 3.
Mass spectrometry identifies Lys-380 residue as the main ubiquitination site. A, two lysine residues were identified to be ubiquitinated by mass spectrometry. Peptide spectra with a glycine-glycine shift for residue Lys-380 (peptide spectra for the Lys-298 site in supplemental Fig. S1A). B, mutation of lysine residue Lys-380 prevents EWS-FLI1 ubiquitination. 3xFLAG-EWS-FLI1 and K298R and K380R mutants were co-expressed with HA-ubiquitin for 48 h in A673 cells, and purified ubiquitinated EWS-FLI1 was analyzed using anti-HA antibody by Western blotting. C, mutated Lys-380 residue stabilizes EWS-FLI1 protein. 3xFLAG-EWS-FLI1 and single and double mutants were transiently overexpressed for 48 h in A673 and SKNMC cells and analyzed with an anti-FLAG antibody by Western blotting. D, scatter plot for quantification for n = 3 independent experiments; error bars represent S.D. E, EWS-FLI1 mutant shows nuclear localization. 3xFLAG-EWS-FLI1 and K380R single mutant were transiently expressed for 48 h in A673 cells. Cells were fixed, stained with anti-FLAG antibody, and used for immunofluorescence (40× magnification). F, mutation of Lys-380 stabilizes EWS-FLI1 posttranslationally. HEK293T cells were transduced for 72 h with reporter constructs of EWS-FLI1, mutants K298R and K380R, and standards. EGFP/DsRed ratios were analyzed by FACS (G) after additional incubation with DMSO or 20 μm MG-132 for 8 h. IP, immunoprecipitation; IB, immunoblotting; EF, EWS-FLI1; ub, ubiquitin; d1h, d4h, and d24h, degron (d) motifs with half-lives of 1, 4, and 24 h, respectively.
EWS-FLI1 Fusion Protein and Wild Type FLI1 Share a Unique Site for Turnover
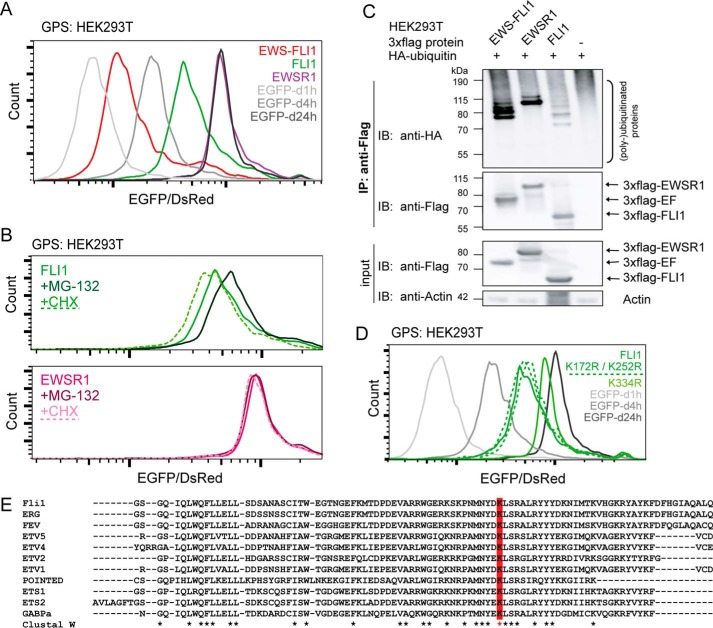
To investigate whether turnover by Lys-380 is unique to the fusion protein, we subsequently compared the stability and ubiquitination pattern with those of the full-length proteins EWSR1 and FLI1. Surprisingly, both wild type proteins were more stable with half-lives of >4 h (FLI1) and close to 24 h (EWSR1) (Fig. 4A). Although FLI1 could be stabilized by incubation with MG-132 (or destabilized by CHX), these treatments did not change the already long half-life of EWSR1 (Fig. 4B). FLI1 was clearly polyubiquitinated similarly to the fusion protein, but EWSR1 showed strong mono- and diubiquitination (Fig. 4C). Next, we mutated the previously identified lysine residue to arginine (FLI1 Lys-334). By GPS, K334R also increased its half-life toward 24 h (Fig. 4D). Additionally, mass spectrometry of purified 3xFLAG-FLI1 revealed glycine-glycine shifts for peptides containing lysines Lys-172 and Lys-252 (peptide spectra in supplemental Fig. S1, B and C). Whereas Lys-172 is located in the N-terminal part and therefore not present in the fusion protein, Lys-252 corresponds to the EWS-FLI1 Lys-298 residue. Single arginine mutants of both residues display EGFP/DsRed ratios comparable with that of wild type FLI1 (Fig. 4D, dotted lines), indicating that both fusion and full-length protein turnover is regulated by one single lysine residue. Indeed, incubation of FLI1 and K334R with MG-132 only shifted the wild type but not the K334R mutant ratio (data not shown). Interestingly, the Lys-334/Lys-380 residue is conserved within most ETS family members (Fig. 4E).
FIGURE 4.
Regulation of proteasomal turnover is conserved between EWS-FLI1 and FLI1. A, EWS-FLI1 displays fastest turnover. For GPS analysis of EWS-FLI1, EWSR1, and FLI1, reporter constructs and standards were transduced into HEK293T cells and analyzed after 72 h by FACS. B, full-length proteins EWSR1 and FLI1 were additionally treated with 20 μm MG-132 or 20 μg/ml CHX for 8 h. C, immunoprecipitation of EWS-FLI1, EWSR1, and FLI1 proteins. 3xFLAG-tagged versions were co-expressed with HA-ubiquitin for 48 h and stabilized for an additional 5 h with 20 μm MG-132. After immunoprecipitation of tag proteins, the ubiquitin pattern was analyzed by anti-HA antibody. D, mutation of conserved residue K334R stabilizes FLI1 in GPS analysis. HEK293T cells were transduced for 72 h with reporter constructs of FLI1; mutants K172R, K252R, and K334R; and standards. The EGFP/DsRed ratio was analyzed by FACS. E, turnover-dependent lysine residue is conserved within most ETS family members. Shown is a ClustalW alignment of the conserved ETS domain sequence; the Lys-334/Lys-380 residue is marked in red. IP, immunoprecipitation; IB, immunoblotting; EF, EWS-FLI1; d1h, d4h, and d24h, degron (d) motifs with half-lives of 1, 4, and 24 h, respectively. * indicates conserved amino acids upon ClustalW alignment.
Together, our findings indicate that, despite differences in turnover, stability of both EWS-FLI1 and wild type FLI1 is regulated by the same critical conserved residue in the ETS domain.
EWS-FLI1 Target Gene Expression Is Subdivided into Three Major Groups
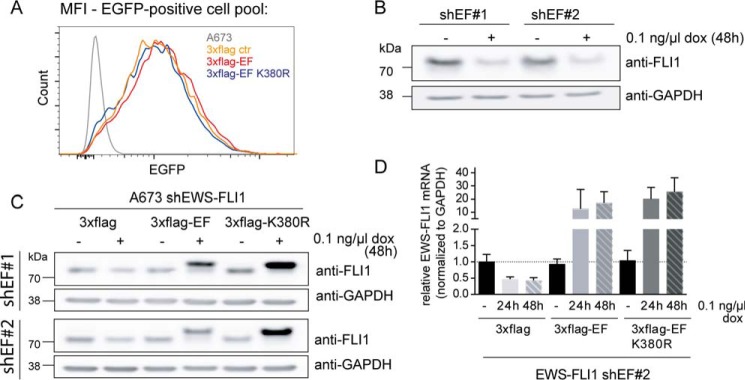
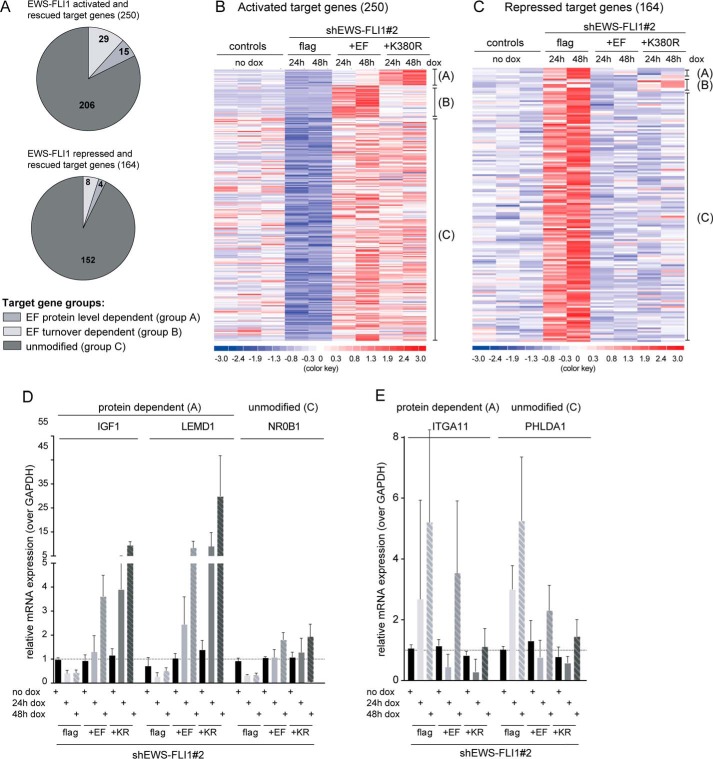
Modulation of EWS-FLI1 target gene signature is one of the best studied mechanisms in Ewing sarcoma. EWS-FLI1 activates a wide range of target genes like NKX2.2, NR0B1, and IGF1 (30, 31, 41–43) but also has repressive functions as shown for IGFBP3, PHLDA1, and lysyl oxidase (44–46). Consequently, this is thought to be critical for oncogenic properties as depletion of the fusion protein results in growth inhibition and cellular senescence (24, 47). To better understand the effect of EWS-FLI1 stability on the target gene signature, we established inducible exchange cell lines that were capable of knocking down the endogenous EWS-FLI1 while simultaneously expressing 3xFLAG-EWS-FLI1 or the K380R mutant. We first sorted cells expressing these FLAG-tagged overexpression constructs by flow cytometry for the same low EGFP expression (the selection marker of the pInducer21 vector) to obtain a homogeneously expressing population (Fig. 5A). We then subsequently transduced each of these clonal pools with two different shRNA constructs against the 3′-UTR of EWS-FLI1 (Fig. 5B). Upon doxycycline induction for 48 h, endogenous EWS-FLI1 was exchanged with ectopic wild type or mutant 3xFLAG-EWS-FLI1versions (Fig. 5C). As observed before, the K380R mutant stabilized the protein posttranslationally by at least 4-fold (Fig. 5C), whereas EWS-FLI1 mRNA levels were comparable as shown for shEF2 (Fig. 5D; not shown for shEF1-based cell lines). We then extracted total RNA for microarray expression profiling and analyzed the EWS-FLI1 target gene signature for differential gene expression (supplemental Fig. S2). A total of 497 probe sets were down-regulated by at least 1.5-fold due to EWS-FLI1 depletion of which 250 could be rescued to at least normal levels by induction of EWS-FLI1 wild type (Fig. 6, A and B). Only if the EWS-FLI1 target genes were modulated and recovered in the paired comparison of EWS-FLI1 induction and knockdown at both 24 and 48 h were they were considered as rescued. This pattern was similar for repressed target genes as 333 probe sets were at least 1.5-fold up-regulated of which 164 could be repressed again by ectopic EWS-FLI1 (Fig. 6, A and C). However, we were most interested in the target genes that were further modulated with stabilized protein levels. Surprisingly, the large majority of these genes appeared to be unmodified and displayed no difference in target gene expression between wild type and mutant EWS-FLI1. However, two subgroups were modulated differentially by the mutated fusion protein. One subgroup seems to be protein-sensitive as 15 individual genes displayed higher RNA levels by at least 1.5-fold upon expression of stabilized EWS-FLI1 protein. From the other subgroup, 29 rescued candidates were fusion protein turnover-sensitive and could only partly be modulated by the mutant in comparison with wild type EWS-FLI1 (Fig. 6, A and B, and supplemental Table S2). Among the protein-sensitive candidates, IGF1 and long non-coding RNA EWSAT1 have already been characterized (41, 48), whereas most other target genes have not been implicated in EWS-FLI1 oncogenesis. Selected target genes from the protein-dependent and unmodified group were validated by quantitative RT-PCR for two different shEWS-FLI1 sequences (Fig. 6D, shown for shEF2). Although the exact mechanism of EWS-FLI1 gene repression is not fully understood, the pattern of target gene modulation resembled that of activated target genes. The majority of repressed genes were unmodified, but here also two subgroups differentially modulated repression upon expression of stabilized protein levels (Fig. 6, C and E, and supplemental Table S2).
FIGURE 5.
Generation of exchange Ewing cell lines. A, EGFP (selection marker) sorting of pInducer21 vector-transduced cells. A673 cells with 3xFLAG, 3xFLAG-EWS-FLI1, or 3xFLAG-K380R mutant were sorted for the same low EGFP expression. After sorting, all cell pools were analyzed by FACS for EGFP and plotted for corresponding mean fluorescence intensity (MFI). B, shEF sequences of shRNA against the 3′-UTR of EWS-FLI1 are sufficient to down-regulate EWS-FLI1 protein. Shown is a Western blot of A673 cells with shEF constructs 1 and 2 after incubation with 0.1 ng/μl doxycycline for 48 h. C, inducible EWS-FLI1 exchange cell lines. Double-transduced A673 cells were incubated with 0.1 ng/μl doxycycline for 48 h. Endogenous and exogenous EWS-FLI1 protein levels were analyzed by Western blotting using anti-FLI1 antibody, and mRNA levels were analyzed by quantitative RT-PCR (D) with n = 4; error bars represent 95% confidence intervals. dox, doxycycline; EF, EWS-FLI1; ctr, control.
FIGURE 6.
Differential regulation of target gene expression by EWS-FLI1 protein levels. A, analysis of microarray expression data of A673 exchange cell lines reveals three subgroups (A–C) of target gene regulation. Shown is a pie chart distribution for EWS-FLI1-activated (upper) and -repressed (lower) target genes and their differential modulation of stabilized over wild type EWS-FLI1. B and C, heat map of activated-rescued (B) and repressed-rescued (C) target gene signatures. The comparison of stabilized over wild type EWS-FLI1 target genes suggested three distinct groups. D and E, validation of selected activated (D) or repressed (E) target genes by quantitative RT-PCR based on RNA extracted from shEF2 exchange cell lines with n = 4; error bars represent 95% confidence intervals. dox, doxycycline; EF, EWS-FLI1; KR, K380R.
Taken together, our results indicate that 82% of activated and 93% of repressed target genes have unaltered levels and cannot be further increased by higher EWS-FLI1 protein levels. Only subsets of the target gene signature are differentially modulated, implying that stability of EWS-FLI1 directs these important subgroups or primarily modulates other mechanisms.
Discussion
Here, we have demonstrated that EWS-FLI1 is a polyubiquitinated substrate that is primarily degraded by the proteasome. The high turnover of the fusion protein is controlled by one critical lysine residue located in the conserved ETS domain. By exchanging the wild type EWS-FLI1 with its turnover-deficient mutant, we could show that protein stabilization differentially modulates two subsets of target genes whose expression could either be enhanced (protein-dependent) or only partially rescued (turnover-dependent). However, the majority of target genes are in an unmodified state and independent of protein stabilization.
Attachment of ubiquitins to a substrate triggers a variety of outcomes, including degradation and signaling depending on the ubiquitin acceptor site and ubiquitin chain linkage (32). Here, we identified the fusion protein EWS-FLI1 as a substrate of the proteasome system in a time- and dose-dependent manner. The previously suggested lysosomal degradation route (37) possibly contributes to turnover but appears less relevant compared with the proteasomal pathway under steady-state conditions.
During the last decades, a variety of different technologies have been developed to dissect and understand ubiquitin-mediated processes (49). GPS profiling was initially established to identify novel proteasomal or cullin-RING ligase substrates (38, 50, 51). Thus, we applied this approach to analyze EWS-FLI1 turnover. Determination of the EGFP/DsRed ratio for the fusion protein and known standard reporter constructs confirmed EWS-FLI1 turnover as proteasome-dependent and mapped the half-life to be between 1 and 4 h. Like most potent transcription factors, EWS-FLI1 also displays a fast turnover (52, 53). Using leptomycin B, we could show that EWS-FLI1 is indeed exported out of the nucleus for degradation, suggesting that the fusion protein tightly maintains the balance of its cytosolic proteolysis. The apparent exclusive nuclear localization might therefore be due to its rapid cytosolic degradation.
However, incubation of cells with the translation synthesis inhibitor CHX barely reflected the high turnover observed in the GPS under steady-state conditions. Using this assay, others have already suggested EWS-FLI1 as a stable protein (37). In contrast, by using antisense oligodeoxynucleotides (26) and RNAi approaches, it is known that the fusion protein can be efficiently down-regulated within a short 24-h time frame, which would not be possible with a highly stable protein. Therefore, the discrepancy between the GPS/RNAi data and CHX treatment remains. However, complete inhibition of protein synthesis by CHX also affects other components of the ubiquitin system such as E2 conjugating enzymes or E3 ligases. Possibly, if a required E3 ligase is unstable and immediately depleted, EWS-FLI1 turnover might be prolonged as seen here with classical CHX treatment.
Although EWS-FLI1 has been shown to be posttranslationally modified (54–56), ubiquitination remained unknown. Here, we have demonstrated that the fusion protein is polyubiquitinated with a clear and distinct pattern, which supports the concept of a constant turnover. To better understand how the ubiquitinated fusion protein is degraded, we used affinity-purified EWS-FLI1 for mass spectrometry and searched for glycine-glycine modifications in the fragmented peptide sequence. From two possible ubiquitin acceptor sites, only mutation of the Lys-380 clearly abolished ubiquitination and posttranslationally stabilized the protein. Although we focused on EWS-FLI1 degradation, we do not exclude that this critical lysine may also trigger non-proteolytic signaling via alternative linked polyubiquitin chains. It is also worth mentioning here that we have analyzed our mass spectrometry data for occurrence of glycine-glycine shifts of the ubiquitin moieties. We have identified peptide shifts mainly for Lys-48- but also Lys-63-linked ubiquitins (data not shown), confirming our findings that Lys-48-linked chains are highly abundant but not the exclusive EWS-FLI1 ubiquitination pattern. Other ubiquitin-linked chains or mixed chains might also be relevant. Furthermore, mutation of this residue may also inhibit other modifications such as sumoylation. Acetylation of this residue has not been detected in contrast to the Lys-298 site (55).
The comparison of EWS-FLI1 with its full-length proteins revealed that the polyubiquitin pattern is similar to that of FLI1. Indeed, mutation of the corresponding lysine residue in the ETS domain posttranslationally stabilizes both proteins. As the Lys-380/Lys-334 residue is highly conserved among most ETS family members, it might contribute to proteasomal degradation in other ETS proteins and their fusion proteins. Most interestingly, EWS-FLI1 displays a higher turnover compared with its full-length proteins EWSR1 and FLI1. In contrast, the ETS proteins ERG and ETV1, which are fused to the TMPRSS2 promoter in prostate cancer, stabilize the protein and confer a physiological advantage to cancer cells (33, 34, 36). The truncated protein versions display increased stability due to loss of the N-terminal E3 ligase motif. However, EWS-FLI1 is not a truncated version of FLI1; instead the N-terminal domain is dominated by EWSR1 with a stronger transactivation domain. If the turnover and subsequent E3 ligase binding of ETS members are conserved, EWS-FLI1 might possibly interact with E3 ligases via its EWSR1 domain, giving rise to a new regulatory mechanism. However, the basic concept that truncated oncogenic TFs are more stable than their wild type counterparts (36) might not apply for fusion proteins consisting of two unrelated protein domains. Whether this is indeed a general paradigm or rather specific for EWSR1- related fusion proteins needs to be further elucidated.
We then deciphered the role of EWS-FLI1 turnover in transcriptional regulation of target genes. To investigate the influence of stabilized EWS-FLI1 on target gene expression, we established an inducible Ewing cell system that allowed depleting endogenous EWS-FLI1 protein while simultaneously expressing the wild type fusion protein or a turnover-deficient mutant. This system largely circumvents interference with endogenous fusion protein, enabling us to study the dynamic behavior of posttranslationally stabilized EWS-FLI1 protein on transcriptional regulation. Surprisingly, around 85% of both activated and repressed target genes are in an unaltered state, including well characterized target genes such as NR0B1 and STEAP1 (30, 57).
Although our EWS-FLI1 wild type overexpression constructs are around 2-fold above the endogenous EWS-FLI1 levels, it is possible that cofactors expressed at endogenous levels or interacting proteins limit further modulation. The knowledge of the EWS-FLI1 interactome is continuously expanding (28, 29); however, which cofactors or interacting proteins are most important or influence the fusion protein in a context-dependent matter still remain unsolved. Nevertheless, longer induction of wild type or mutated ectopic EWS-FLI1 up to 96 h induced cell death irrespective of the construct (data not shown).
Besides the majority of unmodified target genes, two other subgroups display a different behavior. 15 activated target genes are more highly expressed upon expression of stabilized EWS-FLI1 protein levels. This group includes protein-coding genes such as IGF1 (41) and LEMD1 and RNA-coding genes such as EWSAT1 (48). Interestingly, EWSAT1 was also observed to be elevated upon higher EWS-FLI1 expression in a different experimental setting (48), suggesting that our approach reflects a global pattern. Furthermore, 29 repressed target genes are less expressed upon expression of stabilized EWS-FLI1. None of these turnover-sensitive target genes have yet been characterized, although there are a few interesting candidates such as anaplastic lymphoma kinase and TP63.
Most interestingly, the activity of TFs can be influenced by their own turnover as shown for estrogen receptor α or c-MYC (58, 59). It was recently shown that the proteasomal turnover of c-MYC is required for full activity. Lysine-mutated c-MYC failed to induce tumorigenesis as inhibitory complexes could not be removed during target gene activation (58), whereas other TFs are independent of ongoing degradation for their transcriptional activity (60). Surprisingly, EWS-FLI1 induced distinct behavior in three subgroups of target genes. Speculatively, the two small subgroups of EWS-FLI1 protein-sensitive and turnover-sensitive target genes might be of greater importance, and their dynamics might trigger different response pathways in Ewing sarcoma oncogenicity. Although it is not yet clear how these subgroups are defined at their regulatory regions, possible driver target genes might be found in these two subgroups. Alternatively, because most target gene levels were unaltered, oncogenic properties might also depend on additional mechanisms such as interacting proteins and/or influence on the splicing machinery.
Taken together, turnover regulation of the major oncogenic driver EWS-FLI1 represents an important regulatory mechanism in Ewing sarcoma pathogenesis. Interference with fusion protein degradation might not only be a novel therapeutic strategy in Ewing sarcoma treatment but might also be applicable to other ETS-based fusion proteins.
Experimental Procedures
Cell Lines and Reagents
HEK293T cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), 2 mmol/liter glutamine (BioConcept, Allschwil, Switzerland), and 100 units/ml penicillin/streptomycin (ThermoFisher Scientific AG, Reinach, Switzerland) at 37 °C in 5% CO2. Ewing sarcoma cell lines A673, SKNMC, and TC71 were cultured in RPMI 1640 medium as described above. Additionally, dishes were precoated with 0.2% gelatin (Sigma-Aldrich). All cell lines have been tested and found to be mycoplasma-negative. The following reagents were used: bortezomib (Selleckchem, Houston, TX), chloroquine, cycloheximide, DMSO, doxycycline, E64, leptomycin B (all Sigma-Aldrich), LY294002, MG-132 (Millipore, Billerica, MA), and 3-methyladenine (ApexBio, Houston, TX).
Plasmids and Cloning
The coding sequences for human EWS-FLI1, FLI1, and EWS were subcloned into the NotI site of pCMV-3xFLAG vector (Sigma-Aldrich). For global protein stability, the coding sequences of DsRed-IRES-EGFP, d24-EGFP, d4-EGFP, and d1-EGFP (Addgene catalog numbers 41941, 41944, 41943, and 41942) were cloned into the EcoRI site of pR-EF1 (Cellecta Inc., Mountain View, CA). All cDNAs (EWS-FLI1, FLI1, EWS, and mutants) were inserted into the BstBI site at the 3′-end of DsRed-IRES-EGFP.
3xFLAG-EWS-FLI1 cDNA was cloned into the SpeI-BamHI sites of pInducer21 ORF (Addgene catalog number 46948). The pRSIT-U6Tet-shRNA-PGKTetRep-2A-GFP-2A-puro vector with shRNAs against EWS-FLI1 was purchased from Cellecta Inc. with the following target sequences: shEF1, 5′-ATAGAGGTGGGAAGCTTATAA-3′ (described previously in Ref. 31); shEF2, 5′-CGTCATGTTCTGGTTTGAGAT-3′ (designed against the C-terminal FLI1 part according to RefSeq accession number NM_002017.4).
Cloning was performed by In-Fusion HD Cloning (Clontech) according to the manufacturer's protocol. All mutations were introduced using site-directed mutagenesis. Detailed information of plasmids, cloning, and mutagenesis primers can be found in supplemental Table ST1. All clones were verified by sequencing.
Transient Transfection
For HEK293T cells, complete DMEM was mixed with PolyethylenimineMax (Polyscience, Cham, Switzerland) and plasmids for 15 min and then added to the cells for 48 h. For Ewing sarcoma cells, JetPrime (Polyplus Transfections, Illkirch, France) reagent was used according to the manufacturer's instruction in antibiotic-free RPMI 1640 medium.
Virus Production and Transduction
HEK293T cells were transfected with cDNA vectors and pMDL, pREV, and pVSV with JetPrime according to the manufacturer's instruction in antibiotic-free DMEM. Medium was replaced 24 h after transfection, and virus was harvested after an additional 48 h. Viral supernatant was cleared by centrifugation, filtered, and concentrated if necessary (Amicon® Ultra 15 ml, Millipore). Ewing sarcoma cells were infected with the viral supernatant supplemented with 10 μg/ml Polybrene (Sigma-Aldrich) for 16–18 h.
Global Protein Stability Assay and Flow Cytometry
Lentivirus from HEK293T cells transfected with pR-EF1-DsRed-IRES-d1/d4/d24/EGFP-cDNA, pMDL, pREV, and pVSV was used to transduce HEK293T or Ewing sarcoma cells as described to about 5–10% DsRed-positive cells. Cells were harvested 72h after transduction; incubated with cycloheximide, leptomycin B, or MG-132 for 4–8 h as indicated; and analyzed by FACS. Cells were resuspended in PBS and filtered using a 40-μm cell strainer (BD Biosciences). FACS analysis was performed on a FACS CantoTM II cytometry system (BD Biosciences), and data were analyzed by FlowJo software (Treestar Inc., Ashland, OR). At least 50,000 cells were recorded for each experiment. Cell sorting was done with a FACS AriaTM III cytometry system (BD Biosciences).
Cell Lysis, Western Blotting, and Antibodies
Cells were harvested and lysed in standard lysis buffer (50 mm Tris/HCl, 150 mm NaCl, 50 mm NaF, 5 mm Na4P2O7, 1 mm Na3VO4, 10 mm β-glycerol phosphate, 1% Triton X-100 with protease inhibitor mixture Complete Mini® (Sigma-Aldrich)). Lysates were sonicated and cleared by centrifugation. Proteins were separated by 4–12 or 10% BisTris NuPAGE precast gels (ThermoFisher Scientific AG) and transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked with 5% milk powder in PBS with 0.2% Tween 20, and primary antibodies were incubated overnight at 4 °C followed by a 2-h incubation with HRP-linked secondary antibody at room temperature. Proteins were detected by chemiluminescence using ECL detection reagent (GE Healthcare) or SuperSignalTM Western blotting reagent (ThermoFisher Scientific AG). Quantification of blots was performed using ImageJ (version 1.46r). The following commercial antibodies were used: anti-actin (dilution, 1:1000; Cell Signaling Technology), anti-FLAG (clone M2; 1:1000; Sigma-Aldrich), anti-FLI1 (1:1000; MyBiosource LLC, San Diego, CA), anti-GAPDH (1:1000; Cell Signaling Technology), anti-HA (1:1000; Millipore), anti-LAMP1 (1:500; Developmental Studies Hybridoma Bank, Iowa City, IA), anti-p27 (1:1000 (Cell Signaling Technology) and 1:200 (ThermoFisher Scientific AG)) and anti-Tubulin (1:40,000; Sigma-Aldrich).
Immunoprecipitation of Ubiquitinated Proteins
Cells were transfected for 48 h and treated with 10 μm MG-132 prior to lysis in ubiquitin lysis buffer (2% SDS, 150 mm NaCl, 10 mm Tris/HCl, 2 mm Na3VO4, 50 mm NaF with Complete Mini protease inhibitor mixture), boiled for 10 min, and sonicated. Lysates were diluted 1:10 in dilution buffer (150 mm NaCl, 10 mm Tris/HCl, 2 mm EDTA, 1% Triton X-100), incubated for 30 min at 4 °C, and cleared by centrifugation for 30 min at maximum speed. Immunoprecipitation was performed using anti-FLAG antibody (Sigma-Aldrich) coupled to Dynabeads Protein G (ThermoFisher Scientific AG). Lysates were incubated for 1 h at 4 °C, washed three times, eluted at room temperature using 3xFLAG peptide (Sigma-Aldrich), and prepared for Western blotting analysis. An extended description for detection of EWS-FLI1 ubiquitination sites by mass spectrometry can be found in supplemental Fig. S1.
Immunofluorescence and Microscopy
A673 cells were seeded on cover slides for 24 h and transiently transfected with FLAG-tagged plasmids for an additional 48 h. After fixing with 4% paraformaldehyde (Carl Roth, Arlesheim, Switzerland), cells were permeabilized and stained with anti-FLAG antibody (1:300) in 4% horse serum (Sigma-Aldrich) and 0.1% Triton X-100 in PBS overnight. Fluorescent secondary antibody (Alexa Fluor 488 anti-mouse; Sigma-Aldrich) in PBS with 4% horse serum was applied for 1 h. Cover slides were fixed on objective glass with DAPI Vectashield® mounting medium (Vector laboratories Inc., Burlingame, CA) and analyzed by an Axioskop 2 MOT Plus microscope (Carl Zeiss Microscopy LLC, Thornwood, NY).
Quantitative PCR and Microarray Analysis
Total RNA was extracted from Ewing sarcoma cells using a Qiagen RNeasy kit (Qiagen Instruments AG, Hombrechtikon, Switzerland). cDNA synthesis was carried out using a High-Capacity Reverse Transcription Kit (Applied Biosystems by ThermoFisher Scientific AG). Quantitative PCR was performed using TaqMan gene expression master mix (ThermoFisher Scientific AG) and assays on demand (Applied Biosystems by ThermoFisher Scientific AG). A complete list can be found in supplemental Table ST1. Replicate values were pooled and represented as geometric mean values with a 95% confidence interval. Microarray expression analysis with total RNA was performed using GeneChip® Human Gene 2.0 ST Array (Atlas Biolabs, Berlin, Germany). Data are accessible under Gene Expression Omnibus (GEO) accession number GSE81018.
Microarray Data Analysis
The raw microarray data were processed and normalized using the robust multiarray average algorithm (61). Differential expression was computed using the Bioconductor package limma providing a moderated t test that is adapted to a low number of replicates (62). Differential expression results were filtered based on -fold change and p values. All computations were done using R/Bioconductor. An extended description for target gene grouping can be found in supplemental Fig. S2.
Author Contributions
M. E. G., B. W. S., and F. K. N. designed the study and wrote the paper. M. E. G. and F. P. designed, performed, and analyzed the experiments. L. A. L.-G. performed and analyzed the mass spectrometry experiments. L. H. provided help with flow cytometry and sorted stable cell lines. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by Swiss National Science Foundation Grant 31003A-144177. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2 and Tables ST1 and S2.
- ETS
- E-26 transformation-specific
- EWS
- Ewing sarcoma protein
- TF
- transcription factor
- ERG
- ETS-related gene
- CHX
- cycloheximide
- GPS
- global protein stability
- IRES
- internal ribosome entry site
- EGFP
- enhanced GFP
- LMB
- leptomycin B
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Hollenhorst P. C., McIntosh L. P., and Graves B. J. (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laudet V., Hänni C., Stéhelin D., and Duterque-Coquillaud M. (1999) Molecular phylogeny of the ETS gene family. Oncogene 18, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 3. Wasylyk B., Hahn S. L., and Giovane A. (1993) The Ets family of transcription factors. Eur. J. Biochem. 211, 7–18 [DOI] [PubMed] [Google Scholar]
- 4. Li R., Pei H., and Watson D. K. (2000) Regulation of Ets function by protein-protein interactions. Oncogene 19, 6514–6523 [DOI] [PubMed] [Google Scholar]
- 5. Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., and Graves B. J. (1992) Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 6, 975–990 [DOI] [PubMed] [Google Scholar]
- 6. Lelièvre E., Lionneton F., Soncin F., and Vandenbunder B. (2001) The Ets family contains transcriptional activators and repressors involved in angiogenesis. Int. J. Biochem. Cell Biol. 33, 391–407 [DOI] [PubMed] [Google Scholar]
- 7. Sharrocks A. D. (2001) The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 8. Davidson B., Reich R., Goldberg I., Gotlieb W. H., Kopolovic J., Berner A., Ben-Baruch G., Bryne M., and Nesland J. M. (2001) Ets-1 messenger RNA expression is a novel marker of poor survival in ovarian carcinoma. Clin. Cancer Res. 7, 551–557 [PubMed] [Google Scholar]
- 9. Golub T. R., Barker G. F., Bohlander S. K., Hiebert S. W., Ward D. C., Bray-Ward P., Morgan E., Raimondi S. C., Rowley J. D., and Gilliland D. G. (1995) Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 92, 4917–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petrovics G., Liu A., Shaheduzzaman S., Furusato B., Sun C., Chen Y., Nau M., Ravindranath L., Chen Y., Dobi A., Srikantan V., Sesterhenn I. A., McLeod D. G., Vahey M., Moul J. W., and Srivastava S. (2005) Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 24, 3847–3852 [DOI] [PubMed] [Google Scholar]
- 11. Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., and Chinnaiyan A. M. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- 12. Kumar-Sinha C., Tomlins S. A., and Chinnaiyan A. M. (2008) Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer 8, 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seth A., and Watson D. K. (2005) ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer 41, 2462–2478 [DOI] [PubMed] [Google Scholar]
- 14. Mertens F., Antonescu C. R., Hohenberger P., Ladanyi M., Modena P., D'Incalci M., Casali P. G., Aglietta M., and Alvegård T. (2009) Translocation-related sarcomas. Semin. Oncol. 36, 312–323 [DOI] [PubMed] [Google Scholar]
- 15. Mitelman F., Johansson B., and Mertens F. (2007) The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 [DOI] [PubMed] [Google Scholar]
- 16. Hagenbuchner J., and Ausserlechner M. J. (2016) Targeting transcription factors by small compounds—current strategies and future implications. Biochem. Pharmacol. 107, 1–13 [DOI] [PubMed] [Google Scholar]
- 17. Bernstein M., Kovar H., Paulussen M., Randall R. L., Schuck A., Teot L. A., and Juergens H. (2006) Ewing's sarcoma family of tumors: current management. Oncologist 11, 503–519 [DOI] [PubMed] [Google Scholar]
- 18. Esiashvili N., Goodman M., and Marcus R. B. Jr. (2008) Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: surveillance epidemiology and end results data. J. Pediatr. Hematol. Oncol. 30, 425–430 [DOI] [PubMed] [Google Scholar]
- 19. Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., Aurias A., and Thomas G. (1992) Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165 [DOI] [PubMed] [Google Scholar]
- 20. May W. A., Gishizky M. L., Lessnick S. L., Lunsford L. B., Lewis B. C., Delattre O., Zucman J., Thomas G., and Denny C. T. (1993) Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc. Natl. Acad. Sci. U.S.A. 90, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. May W. A., Lessnick S. L., Braun B. S., Klemsz M., Lewis B. C., Lunsford L. B., Hromas R., and Denny C. T. (1993) The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol. 13, 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bailly R. A., Bosselut R., Zucman J., Cormier F., Delattre O., Roussel M., Thomas G., and Ghysdael J. (1994) DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 14, 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun B. S., Frieden R., Lessnick S. L., May W. A., and Denny C. T. (1995) Identification of target genes for the Ewing's sarcoma EWS/FLI fusion protein by representational difference analysis. Mol. Cell. Biol. 15, 4623–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kovar H., Aryee D. N., Jug G., Henöckl C., Schemper M., Delattre O., Thomas G., and Gadner H. (1996) EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 7, 429–437 [PubMed] [Google Scholar]
- 25. Tanaka K., Iwakuma T., Harimaya K., Sato H., and Iwamoto Y. (1997) EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. J. Clin. Investig. 99, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toretsky J. A., Connell Y., Neckers L., and Bhat N. K. (1997) Inhibition of EWS-FLI-1 fusion protein with antisense oligodeoxynucleotides. J. Neurooncol. 31, 9–16 [DOI] [PubMed] [Google Scholar]
- 27. Petermann R., Mossier B. M., Aryee D. N., Khazak V., Golemis E. A., and Kovar H. (1998) Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 17, 603–610 [DOI] [PubMed] [Google Scholar]
- 28. Selvanathan S. P., Graham G. T., Erkizan H. V., Dirksen U., Natarajan T. G., Dakic A., Yu S., Liu X., Paulsen M. T., Ljungman M. E., Wu C. H., Lawlor E. R., Üren A., and Toretsky J. A. (2015) Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 112, E1307–E1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toretsky J. A., Erkizan V., Levenson A., Abaan O. D., Parvin J. D., Cripe T. P., Rice A. M., Lee S. B., and Uren A. (2006) Oncoprotein EWS-FLI1 activity is enhanced by RNA helicase A. Cancer Res. 66, 5574–5581 [DOI] [PubMed] [Google Scholar]
- 30. Kinsey M., Smith R., and Lessnick S. L. (2006) NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol. Cancer Res. 4, 851–859 [DOI] [PubMed] [Google Scholar]
- 31. Smith R., Owen L. A., Trem D. J., Wong J. S., Whangbo J. S., Golub T. R., and Lessnick S. L. (2006) Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 9, 405–416 [DOI] [PubMed] [Google Scholar]
- 32. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 33. An J., Ren S., Murphy S. J., Dalangood S., Chang C., Pang X., Cui Y., Wang L., Pan Y., Zhang X., Zhu Y., Wang C., Halling G. C., Cheng L., Sukov W. R., et al. (2015) Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol. Cell 59, 904–916 [DOI] [PubMed] [Google Scholar]
- 34. Gan W., Dai X., Lunardi A., Li Z., Inuzuka H., Liu P., Varmeh S., Zhang J., Cheng L., Sun Y., Asara J. M., Beck A. H., Huang J., Pandolfi P. P., and Wei W. (2015) SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell 59, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji Z., Degerny C., Vintonenko N., Deheuninck J., Foveau B., Leroy C., Coll J., Tulasne D., Baert J. L., and Fafeur V. (2007) Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 26, 395–406 [DOI] [PubMed] [Google Scholar]
- 36. Vitari A. C., Leong K. G., Newton K., Yee C., O'Rourke K., Liu J., Phu L., Vij R., Ferrando R., Couto S. S., Mohan S., Pandita A., Hongo J. A., Arnott D., Wertz I. E., et al. (2011) COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 [DOI] [PubMed] [Google Scholar]
- 37. Elzi D. J., Song M., Hakala K., Weintraub S. T., and Shiio Y. (2014) Proteomic Analysis of the EWS-Fli-1 interactome reveals the role of the lysosome in EWS-Fli-1 turnover. J. Proteome Res. 13, 3783–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yen H. C., Xu Q., Chou D. M., Zhao Z., and Elledge S. J. (2008) Global protein stability profiling in mammalian cells. Science 322, 918–923 [DOI] [PubMed] [Google Scholar]
- 39. Liang H., Mao X., Olejniczak E. T., Nettesheim D. G., Yu L., Meadows R. P., Thompson C. B., and Fesik S. W. (1994) Solution structure of the ets domain of Fli-1 when bound to DNA. Nat. Struct. Biol. 1, 871–875 [DOI] [PubMed] [Google Scholar]
- 40. Welford S. M., Hebert S. P., Deneen B., Arvand A., and Denny C. T. (2001) DNA binding domain-independent pathways are involved in EWS/FLI1-mediated oncogenesis. J. Biol. Chem. 276, 41977–41984 [DOI] [PubMed] [Google Scholar]
- 41. Riggi N., Suvà M. L., Suvà D., Cironi L., Provero P., Tercier S., Joseph J. M., Stehle J. C., Baumer K., Kindler V., and Stamenkovic I. (2008) EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 68, 2176–2185 [DOI] [PubMed] [Google Scholar]
- 42. Kinsey M., Smith R., Iyer A. K., McCabe E. R., and Lessnick S. L. (2009) EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing's sarcoma. Cancer Res. 69, 9047–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tirado O. M., Mateo-Lozano S., Villar J., Dettin L. E., Llort A., Gallego S., Ban J., Kovar H., and Notario V. (2006) Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing's sarcoma cells. Cancer Res. 66, 9937–9947 [DOI] [PubMed] [Google Scholar]
- 44. Boro A., Prêtre K., Rechfeld F., Thalhammer V., Oesch S., Wachtel M., Schäfer B. W., and Niggli F. K. (2012) Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing's sarcoma. Int. J. Cancer 131, 2153–2164 [DOI] [PubMed] [Google Scholar]
- 45. Prieur A., Tirode F., Cohen P., and Delattre O. (2004) EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 24, 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sankar S., Bell R., Stephens B., Zhuo R., Sharma S., Bearss D. J., and Lessnick S. L. (2013) Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 32, 5089–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsunobu T., Tanaka K., Nakamura T., Nakatani F., Sakimura R., Hanada M., Li X., Okada T., Oda Y., Tsuneyoshi M., and Iwamoto Y. (2006) The possible role of EWS-Fli1 in evasion of senescence in Ewing family tumors. Cancer Res. 66, 803–811 [DOI] [PubMed] [Google Scholar]
- 48. Marques Howarth M., Simpson D., Ngok S. P., Nieves B., Chen R., Siprashvili Z., Vaka D., Breese M. R., Crompton B. D., Alexe G., Hawkins D. S., Jacobson D., Brunner A. L., West R., Mora J., et al. (2014) Long noncoding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J. Clin. Investig. 124, 5275–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williamson A., Werner A., and Rape M. (2013) The Colossus of ubiquitylation: decrypting a cellular code. Mol. Cell 49, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., and Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yen H. C., and Elledge S. J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 [DOI] [PubMed] [Google Scholar]
- 52. Kodadek T., Sikder D., and Nalley K. (2006) Keeping transcriptional activators under control. Cell 127, 261–264 [DOI] [PubMed] [Google Scholar]
- 53. Muratani M., and Tansey W. P. (2003) How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 54. Bachmaier R., Aryee D. N., Jug G., Kauer M., Kreppel M., Lee K. A., and Kovar H. (2009) O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing's sarcoma. Oncogene 28, 1280–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schlottmann S., Erkizan H. V., Barber-Rotenberg J. S., Knights C., Cheema A., Uren A., Avantaggiati M. L., and Toretsky J. A. (2012) Acetylation increases EWS-FLI1 DNA binding and transcriptional activity. Front Oncol 2, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klevernic I. V., Morton S., Davis R. J., and Cohen P. (2009) Phosphorylation of Ewing's sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochem. J. 418, 625–634 [DOI] [PubMed] [Google Scholar]
- 57. Grunewald T. G., Diebold I., Esposito I., Plehm S., Hauer K., Thiel U., da Silva-Buttkus P., Neff F., Unland R., Müller-Tidow C., Zobywalski C., Lohrig K., Lewandrowski U., Sickmann A., Prazeres da Costa O., et al. (2012) STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol. Cancer Res. 10, 52–65 [DOI] [PubMed] [Google Scholar]
- 58. Jaenicke L. A., von Eyss B., Carstensen A., Wolf E., Xu W., Greifenberg A. K., Geyer M., Eilers M., and Popov N. (2016) Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol. Cell 61, 54–67 [DOI] [PubMed] [Google Scholar]
- 59. Reid G., Hübner M. R., Métivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., and Gannon F. (2003) Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11, 695–707 [DOI] [PubMed] [Google Scholar]
- 60. Yao J., Munson K. M., Webb W. W., and Lis J. T. (2006) Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 [DOI] [PubMed] [Google Scholar]
- 61. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., and Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 62. Gentleman R. (2005) Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer Science+Business Media, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.