Abstract
Introduction
The nuclear pore complex (NPC) is the primary gateway for transport of macromolecules between the nucleus and cytoplasm, serving as both a critical mediator and regulator of gene expression. NPCs are enormous ~120 MDa macromolecular machines embedded in the nuclear envelope, each containing ~1000 protein subunits, termed nucleoporins. Despite substantial progress in visualizing the overall shape of the NPC by cryoelectron tomography and in determining atomic resolution crystal structures of nucleoporins, the molecular architecture of the assembled NPC remains poorly understood, hindering the design of mechanistic studies that could investigate its many roles in cell biology.
Rationale
Existing cryoelectron tomographic reconstructions of the NPC remain too low in resolution to allow for de novo structure determination of the NPC or unbiased docking of nucleoporin fragment crystal structures. We sought to bridge this resolution gap by first defining the interaction network of the NPC, focusing on the evolutionarily conserved symmetric core. We developed protocols to reconstitute NPC protomers from purified, recombinant proteins, which enabled the generation of a high-resolution biochemical interaction map of the NPC symmetric core. We next determined high-resolution crystal structures of key nucleoporin interactions, providing spatial restraints for their relative orientation. Lastly, by superposing crystal structures that overlapped in sequence, we generated accurate full-length structures of the large scaffold nucleoporins. Supported by this biochemical data, we used sequential, unbiased searches to place the nucleoporin crystal structures into a previously determined cryoelectron tomographic reconstruction of the intact human NPC, thus generating a composite structure of the entire NPC symmetric core.
Results
Our analysis revealed that the inner and outer rings of the NPC utilize disparate mechanisms of interaction. While the structured coat nucleoporins of the outer ring form extensive surface contacts, the scaffold proteins of the inner ring are bridged by flexible sequences in linker nucleoporins. Our composite structure revealed a defined spoke architecture with limited cross-spoke interactions. Most nucleoporins are present in 32 copies, with notable exceptions of Nup170 and Nup188. Lastly, we observed the arrangement of the channel nucleoporins, which orient their N-termini into two sixteen-membered rings, ensuring that their N-terminal FG repeats project evenly into the central transport channel.
Conclusion
Our composite structure of the NPC symmetric core can be used as a platform for the rational design of experiments to probe NPC structure and function. Each nucleoporin occupies multiple distinct biochemical environments, explaining how such a large macromolecular complex can be assembled from a relatively small number of unique genes. Our integrated, bottom-up approach provides a paradigm for the biochemical and structural characterization of similarly large biological mega-assemblies.
Graphical Abstract
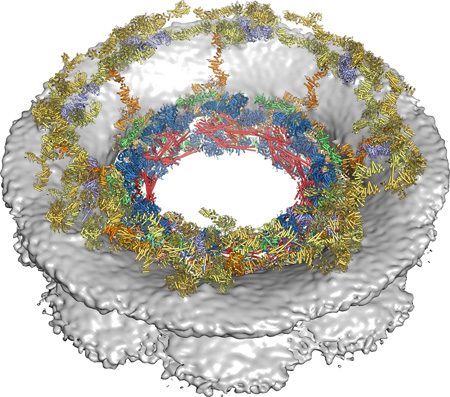
Composite structure of the nuclear pore complex symmetric core. The composite structure of the nuclear pore complex symmetric core generated by sequential, unbiased docking of nucleoporin and nucleoporin complex crystal structures into the cryoET reconstruction of the intact human NPC, viewed from above the cytoplasmic face. Nucleoporin structures are shown as colored cartoons and the nuclear envelope density is shown as a gray surface.
Summary
An integrated analysis of the symmetric core of the nuclear pore complex established its molecular architecture.
Introduction
The nuclear pore complex (NPC) is a massive molecular transport channel embedded in the nuclear envelope (1). In addition to its role as the sole mediator of bidirectional nucleocytoplasmic transport, the NPC is also involved in diverse cellular processes including transcription, mRNA maturation, and genome organization (1, 2). Despite their tremendous size (~120 MDa), NPCs are only composed of 34 different proteins (nucleoporins or nups), which assemble into eightfold-symmetric, ~1000-Å diameter pores that fuse the inner and outer nuclear membranes (Fig. 1, A and B) (1). Given their central role in cell biology, nucleoporins have been linked to a wide range of human diseases including viral infection, cancer, and neurodegenerative disease (1, 3–9). However, the structure of the NPC and the mechanisms by which it influences cellular processes remain enigmatic.
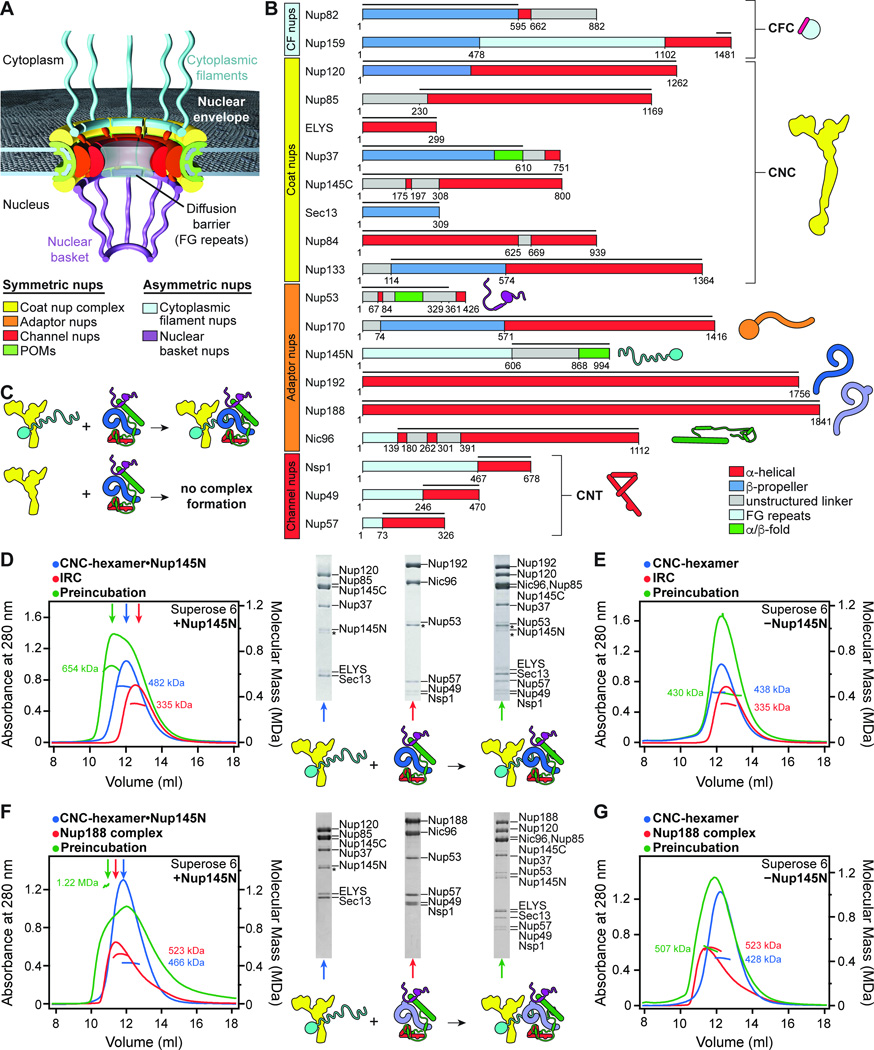
Fig. 1. Reconstitution of NPC symmetric core protomers.
(A) Cross-sectional conceptual schematic of the NPC. CNCs, colored yellow, form outer rings above the membrane on the nuclear and cytoplasmic faces. Adaptor and channel nucleoporins, colored orange and red, respectively, form concentric cylinders in the inner ring. Asymmetric nucleoporins decorate the cytoplasmic and nuclear faces of the NPC. (B) Domain organization of nucleoporins used in biochemical reconstitution experiments. Black lines indicate the construct boundaries used. Domains are drawn as boxes colored according to the legend at the bottom right for observed or predicted folds. Cartoons to the right are used throughout the text to represent the nucleoporins. Some nucleoporins form stable complexes (CNC, coat nucleoporin complex; CFC cytoplasmic filament nucleoporin complex; CNT, channel nucleoporin hetero-trimer) and are drawn as a unit. (C) Cartoon of experimental setup in panels (D–G). The CNC was preincubated with IRC/Nup188 complex in the presence or absence of Nup145N. Complex formation was only observed in the presence of Nup145N. (D, E) Reconstitution of NPC symmetric core protomers containing the CNC-hexamer and IRC is dependent on Nup145N. Identical experiments were performed in the (D) presence or (E) absence of Nup145N. Size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) profiles of the complexes in isolation (red and blue) and after preincubation (green) are shown. Representative Coomassie-stained SDS-PAGE gel slices for the peak fractions are shown with a colored arrow above the chromatogram indicating the resolved peak fraction. Measured molecular masses are indicated for each peak. Cartoons below the gel slices illustrate the respective complexes. (F, G) Reconstitution of NPC symmetric core protomers containing the CNC-hexamer and Nup188-complex is dependent on Nup145N. Identical experiments were performed in the (F) presence or (G) absence of Nup145N. Complete SDS-PAGE gels for all panels are shown in fig. S4.
Most nucleoporins are symmetrically distributed in the NPC, forming a symmetric core that is decorated by asymmetric nucleoporins on the nuclear and cytoplasmic faces (Fig. 1, A and B). Many nucleoporins contain disordered repetitive sequences enriched in phenylalanine and glycine residues called FG repeats, which collectively form a central diffusion barrier that prevents passive diffusion of macromolecules with masses greater than ~40 kDa (Fig. 1, A and B). Larger macromolecules can only traffic through the NPC with the assistance of specialized karyopherin transport factors (10).
Despite extensive efforts, the protein-protein interaction network within the NPC remains incompletely characterized, presenting a fundamental limitation to our understanding of NPC architecture. Co-purification and mass spectrometry approaches have revealed some of the strongest interactions, but provide only general spatial restraints due to their limited resolution. The most well-characterized nucleoporin interactions are those within the coat nucleoporin complex (CNC), which comprises approximately half the mass of the symmetric core and contains Nup120, Nup85, Sec13, Nup145C, Nup84, Nup133, and, in some species, Seh1, Nup37, Nup43, and ELYS (1). Structural and biochemical analyses of CNCs from multiple species reveal a highly conserved architecture in which α-helical domains form extensive interaction surfaces to assemble a large, Y-shaped complex (11–14). Recent advances have dramatically increased the resolution of cryoelectron tomographic (cryoET) reconstructions of intact NPCs, facilitating the unbiased placement of 32 copies of a ~400 kDa crystal structure of the yeast CNC into the intact human NPC (13, 14). The CNCs are arranged in pairs of concentric, eight-membered rings on both the nuclear and cytoplasmic faces of the NPC, accounting for the majority of the observed protein density in the outer rings of the NPC (13, 14).
In contrast to our understanding of the organization of the CNC in the intact NPC, relatively little is known about the molecular architecture of the inner ring that lines the central channel. The disordered N-terminal FG repeats of the channel nucleoporins Nup49, Nup57, and Nsp1 project into the central channel while their structured coiled-coil domains form a stable complex, termed the channel nucleoporin hetero-trimer (CNT) (15, 16). The CNT, Nic96, Nup192, and Nup145N collectively form a stable subcomplex called the inner ring complex (IRC) (15, 17). The remaining components of the symmetric core, Nup170 and Nup53, are thought to mediate interactions with the nuclear envelope, but the details of the interaction network that assembles these proteins and links them to the CNCs remains poorly defined (18, 19).
Crystal structures of many nucleoporin fragments from the symmetric NPC core have been determined, including the N-terminal domains (NTDs) of Nup192, Nup188, and Nup157; the C-terminal domain (CTD) of Nup170; the C-terminal tail domains (TAIL) of Nup192 and Nup188; the α-helical solenoid of Nic96; and the CNT bound to Nic96, CNT•Nic96R1 (15, 20–26). However, structures of full-length Nup192, Nup188, and Nup170 have remained elusive. Accurate placement of the existing structures into the intact NPC is limited by both their relatively small size and the resolution of available cryoET reconstructions. We have used an integrated approach to obtain complete atomic structures and accurate, high-resolution biochemical restraints for determining the near-atomic architecture of the NPC symmetric core.
A significant barrier preventing complete biochemical characterization of the NPC has been the difficulty of purifying significant quantities of full-length nucleoporins from a single species. To overcome this hurdle, we developed expression and purification protocols for all symmetric core nucleoporins from the thermophilic fungus Chaetomium thermophilum, which exhibit superior biochemical stability (27). By reconstituting NPC symmetric core protomers from purified proteins, we found that the interactions between flexible linker sequences and large scaffold nucleoporins drove NPC assembly. We generated a high-resolution biochemical map for these interactions and determined a series of crystal structures that revealed the structural basis for flexible linker sequence recognition by the large scaffold nucleoporins. These crystal structures enabled the construction of complete atomic structures for the ordered scaffolds of Nup170, Nup192, and Nic96. Using our biochemical restraints for validation, we performed unbiased searches to dock the atomic structures into a cryoET reconstruction of the intact human NPC with high confidence (28), thus determining a composite structure of the NPC symmetric core.
Reconstitution of NPC symmetric core protomers
We first directed our efforts towards reconstituting a soluble protomer that could recapitulate the interaction network within the assembled NPC using nucleoporins from the thermophilic fungus C. thermophilum. We used full-length proteins when possible, but FG repeats, disordered N- or C-terminal regions, or other sequences that prevented soluble protein expression were omitted (Fig. 1B; fig. S1; table S1 and S2). We first reconstituted a hetero-hexameric core CNC containing Nup120, Nup37, ELYS, Nup85, Sec13, and Nup145C, a complex analogous to the yeast CNC we previously crystallized (fig. S2, A and B) (14). This CNC hetero-hexamer was assembled with Nup84 and Nup133 to form a hetero-octameric CNC (fig. S2C). Due to the poor solubility of the intact hetero-octameric CNC, we focused our analysis on its hetero-hexameric core. Similarly, we extended our previous reconstitution of an IRC containing Nup192, Nic96, Nup145N, and the CNT by also incorporating Nup53 (fig. S3A and table S3) (15). We found that Nup188, which is evolutionarily related to Nup192, failed to incorporate into the IRC (fig. S3B). Rather, an analogous Nup188 complex formed in the absence of Nup192 (Fig. 1F). By preincubating the core CNC-hexamer with either the IRC or the Nup188 complex, we reconstituted two distinct 13-protein complexes representative of NPC symmetric core protomers (Fig. 1, C, D, and F and fig. S4, A and C).
With the ability to reconstitute NPC protomers, we sought to identify the interactions that linked the IRC or the analogous Nup188 complex to the CNC. Nup145N and Nup145C are components of the IRC/Nup188 complex and CNC, respectively, and originate from the same polypeptide chain after post-translational cleavage mediated by the Nup145N autoproteolytic domain (APD) located in the middle of the Nup145 pre-cursor polypeptide (Fig. 1B) (29). Nup145C is composed of a U-bend α-helical solenoid that is a core component of the CNC and a disordered, ~300-residue N-terminal extension (NTE) (14). Previous studies indicate that Nup145NAPD can still bind the first six N-terminal residues of Nup145C after cleavage, thus offering a possible mechanism for linking the IRC to the CNC (30). Indeed, the complexes did not interact in the absence of Nup145N (Fig. 1, E and G and fig. S4, B and D). Moreover, Nup145NAPD alone could be incorporated into the CNC (fig. S5). These results indicated that the IRC and CNC were flexibly attached via the long, intrinsically disordered sequences of Nup145N and Nup145C. Nup145NAPD also binds to the β-propeller domain of the cytoplasmic filament nucleoporin Nup82 (15). However, since this interaction was outcompeted by CNC binding (fig. S6), another interaction must play a role in retaining the cytoplasmic filaments at the cytoplasmic face of the NPC.
Symmetric core assembly is driven by flexible linkers
We next performed a systematic analysis of the molecular interaction network within the IRC and Nup188 complex, extending our previous work and additionally including Nup170 in our analysis (15). These complexes contain large scaffold domains in Nic96, Nup170, Nup188, and Nup192 as well as long, intrinsically disordered sequences in the linker nucleoporins Nup53 and Nup145N (Fig. 1B). While the CNC is primarily held together by extensive interfaces between large α-helical solenoid domains (1, 14), recent studies hint that the architectural principles of the remaining symmetric core nucleoporins are different (15, 17, 27). For example, the scaffold nucleoporin Nic96 possesses a largely unstructured NTE containing two short helical regions, Nic96R1 and Nic96R2, that are essential for IRC assembly: Nic96R1 recruits the CNT to the NPC while Nic96R2 binds Nup192 or Nup188 (15). To determine whether the structured domains of the scaffold nucleoporins interacted with each other to drive symmetric core assembly, we tested whether Nic96SOL, Nup170, Nup192, and Nup188 could form a complex, but observed no interaction (Fig. 2A and fig. S7A). Instead, complex formation was achieved only in the presence of the linker nucleoporins Nup53 and Nup145N (Fig. 2, B and C and fig. S7, B and C). Thus together with Nic96NTE, the flexible linker nucleoporins Nup53 and Nup145N are the primary driving force of IRC/Nup188 complex assembly (15).
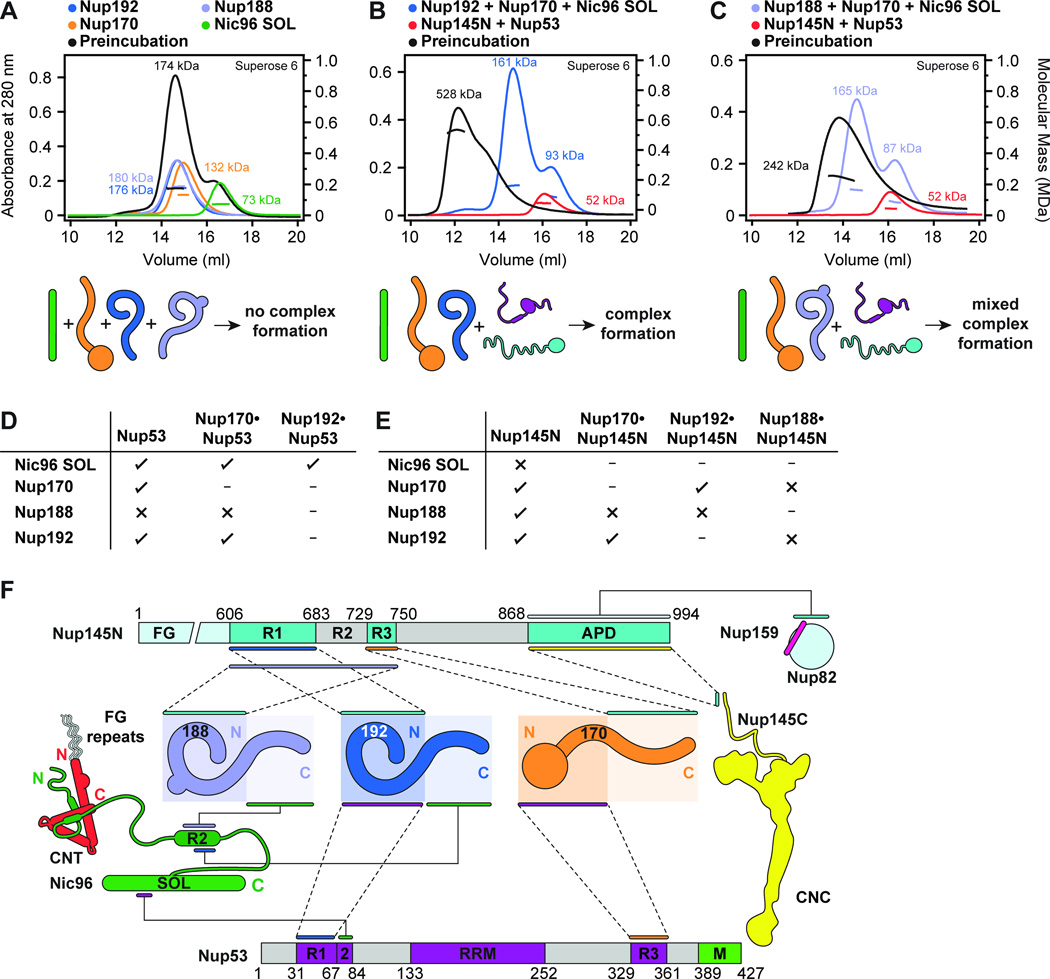
Fig. 2. Biochemical analysis of the interactions mediating NPC symmetric core assembly.
(A) Scaffold domains of Nup192, Nup188, Nup170, and Nic96SOL do not interact with each other. SEC-MALS profiles of the individual scaffolds alone (blue, purple, orange, and green) and after their preincubation with each other (black) are shown. Measured molecular masses are indicated for each peak. (B, C) Linker nucleoporins Nup53 and Nup145N mediate scaffold nucleoporin assembly. SEC-MALS profiles of a scaffold nucleoporin mixture (blue or purple), a linker nucleoporin mixture (red), and after their preincubation with each other (black) are shown. Mixtures of scaffold nucleoporins contained either (B) Nup192 or (C) Nup188. Complete SDS-PAGE gels for all chromatograms are shown in fig. S7. (D, E) Interaction network between scaffold nucleoporins and (D) Nup53 or (E) Nup145N. Scaffold nucleoporins were tested in SEC-MALS interaction experiments for their ability to form hetero-dimeric complexes with Nup53 or Nup145N and their compatibility to also form hetero-trimeric complexes. Check marks indicate complexes that can form in SEC-MALS experiments, crosses indicate complexes that do not form, and dashes indicate complexes that were not tested. Complete SEC-MALS chromatograms and SDS-PAGE gels are shown in figs. S8 to S11. (F) Biochemical interaction map revealed by SEC-MALS interaction experiments. Minimal regions of Nup145N and Nup53 sufficient for binding to components of the NPC are depicted using colored bars and dashed lines between interacting regions. Interactions that map to the same regions on Nup145N and Nup53 do not occur simultaneously. Complete SEC-MALS chromatograms and SDS-PAGE gels are shown in figs S13 to S20. The nucleoporin schematics are according to Fig. 1B.
To further analyze the interaction network between the scaffold and linker nucleoporins, we tested which scaffolds interacted with Nup53 or Nup145N to form hetero-dimers, focusing first on the interactions within the IRC and with Nup170. Nup53 formed robust complexes with Nup192, Nic96SOL, and Nup170 (Fig. 2D and fig. S8). We previously reported that Nup145N interacts weakly with Nic96SOL (15), and here we found that Nup145N also binds to Nup192 and Nup170 (Fig. 2E and fig. S9). All of these scaffold-linker interactions are compatible, as demonstrated by the formation of hetero-trimeric complexes (Fig. 2, D and E and figs. S10 and S11). Nup192 and Nup170 can also bind to both linker nucleoporins simultaneously, indicating that the binding sites on the scaffolds are distinct (fig. S12, A and B). Indeed, we were able to reconstitute a stoichiometric hetero-tetramer composed of Nup192, Nup170, Nup53, and Nup145N (fig. S12C).
To improve the biochemical resolution of our interaction map, we next identified minimal sequence fragments of Nup53 and Nup145N sufficient for scaffold recognition. We previously mapped an interaction between a Nup53 fragment, encompassing residues 31–67 (Nup53R1), with Nup192NTD (Fig. 2F and fig. S13) (20). Here, we found an adjacent fragment containing residues 69–90 (Nup53R2) that was recognized by Nic96SOL (Fig. 2F and fig. S14, A and B). A Nup53 fragment including both of these binding sites (residues 1–90) was sufficient to link the two scaffolds into a hetero-trimeric complex (fig. S14C). We also identified a C-terminal Nup53 fragment containing residues 329–361 (Nup53R3) that interacted specifically with Nup170NTD (Fig. 2F and fig. S15). Conversely, the association between Nup170 and Nup145N mapped to Nup145N residues 729–750 (Nup145NR3) and Nup170CTD (Fig. 2F and fig. S16). Nup192 recognized a fragment of Nup145N encompassing residues 606–683 (Fig. 2F and fig. S17, A and B). Nup192NTD was sufficient for Nup145N binding (fig. S17C), but we also detected a weak interaction with Nup192CTD (fig. S17, D and E), suggesting that binding sites for Nup145N were distributed throughout Nup192. These minimal sequence fragments were specific for their binding partners (fig. S18).
While preincubation of the two linker nucleoporins with Nup192, Nup170, and Nic96SOL produced a robust pentameric complex, the analogous preincubation with Nup188 in place of Nup192 produced a mixture of species (Fig. 2, B and C and fig. S7, B and C). To understand this difference in behavior, we repeated the above analysis with Nup188 and identified a robust interaction with Nup145N whereas Nup53 binding was barely detectible (Fig. 2, D and E and figs. S8C and S9B). However, in contrast to our above results, Nup188 did not strongly bind Nup145N in the presence of Nup192 or Nup170 (Fig. 2, D and E and fig. S11). Similar to Nup192NTD, Nup188NTD was sufficient for Nup145N binding (fig. S19, A and B). However, the minimal Nup192-binding fragment of Nup145N was not sufficient for Nup188 binding (fig. S19, D and E). Instead, we only detected robust complex formation with a much longer fragment encompassing both the Nup192 and Nup170 binding sites (residues 606–750), explaining the exclusivity of their interactions (Fig. 2F and fig. S19C). We found a similar architecture for the Nup192 and Nup188 binding sites in Nic96R2. However, the Nup192 minimal binding fragment (residues 286–301) again was insufficient for Nup188 binding (fig. S20, A to C), which instead required a larger fragment (residues 274–301) (Fig. 2F and fig. S20D). Consistent with these findings, several mutations in the N-terminal region of Nic96R2 ablated Nup188 binding but had no effect on Nup192 binding (fig. S20, E to G) (15). Thus, Nup192 and Nup188 bound competitively to directly overlapping sequences in Nic96 and Nup145N, establishing the existence of a distinct Nup188 complex with an architecture analogous to the IRC.
In summary, we found that interactions between the large, ordered scaffold nucleoporins and flexible interaction motifs in Nup53, Nup145N, and Nic96NTE were the dominant driving force for assembly of the NPC symmetric core outside of the CNCs. We built a biochemical map of these interactions by identifying minimal interaction motifs, revealing that the binding sites were spatially distributed throughout the scaffold nucleoporins, but that many of the binding sites on the linker nucleoporins were adjacent or overlapping in sequence (Fig. 2F). In doing so, we identified the exclusive interactions that provide a molecular basis for the formation of two distinct complexes, the Nup192-harboring IRC and an analogous Nup188 complex. As existing crystal structures have not captured interactions between scaffold and linker nucleoporins, we used these results to identify important structural targets for determining the structural basis for this mode of interaction.
Atomic architecture of the Nup170 interaction network
We determined a crystal structure of the Nup170NTD•Nup53R3 complex at 2.1 Å resolution (Fig. 3, A and B and tables S4 and S5). In order to obtain high-resolution diffraction, we deleted residues 293–305 from the 3D4A loop of Nup170NTD (fig. S21). Nup170NTD was composed of a seven-bladed β-propeller and a C-terminal α-helical domain (Fig. 3B). An N-terminal α-helix packed against the C-terminal α-helical domain and was followed by three β-strands that formed a triple Velcro-closure against the β-propeller (Fig. 3B). Nup53 adopted an extended conformation and bound atypically to the side of the β-propeller, rather than the top, at blades 1 and 2 (Fig. 3B). The crystallized Nup53 fragment contained residues 329–361, but clear density was only observed for residues 342–355. Blade 2 of the Nup170 β-propeller deviated substantially from a canonical β-propeller blade to generate two hydrophobic pockets that accommodated Nup53 residues L346, L347, L353, and L354 (Fig. 3C). We identified several mutations in Nup170 and Nup53 that could disrupt their interaction (Fig. 3, D and E and fig. S22, A and B). Notably, we observed a complete loss of binding with mutations to Nup170 residues that are evolutionary conserved, F199, I203, and Y235, suggesting that the binding interface is evolutionarily conserved (Fig. 3, D and E; fig. S21; fig. S22, A and B).
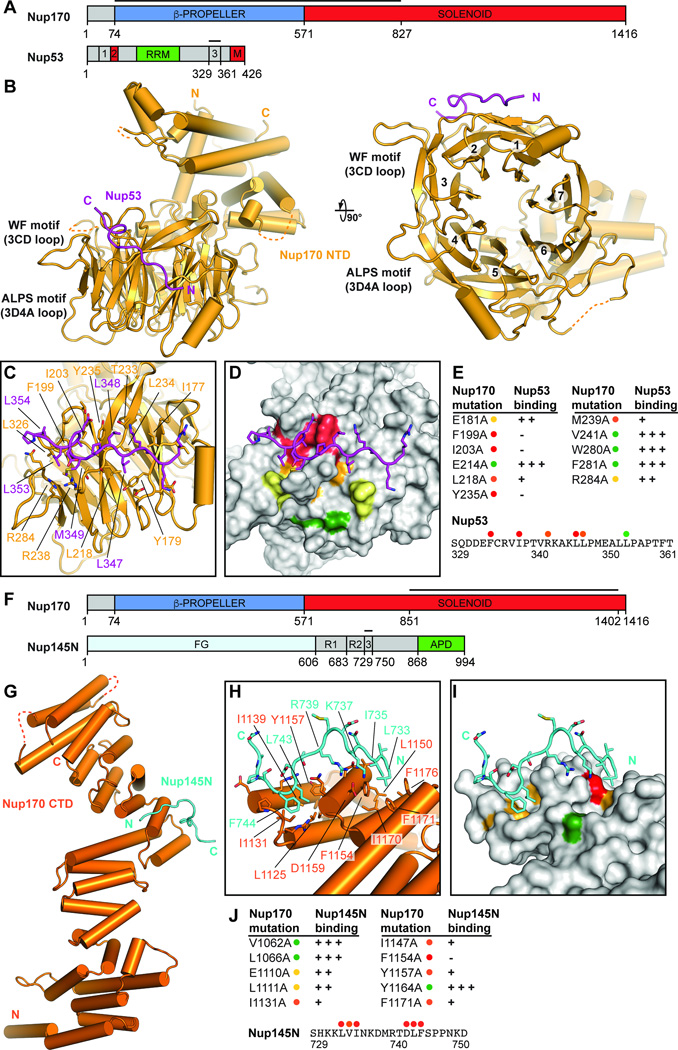
Fig. 3. Structural and biochemical analyses of the Nup170 interaction network.
(A) Domain structures of Nup170 and Nup53. Black lines indicate fragments used for crystallization. (B) Crystal structure of the Nup170NTD•Nup53R3 complex shown in cartoon representation. A 90° rotation is shown on the right. The potential WF and ALPS membrane interaction motifs are indicated. (C) Close-up view of the interaction between Nup170 and Nup53. Nup170 and Nup53 residues involved in the interaction are labeled in orange and purple, respectively. (D) Graphic summary of mutational analysis of the Nup170NTD-Nup53R3 interaction. Nup170 is shown in surface representation from the same view as in panel (C). Residues are colored in red, orange, yellow, and green to indicate mutations that had a strong, moderate, weak, or no effect on binding, respectively. (E) Tabular summary of tested Nup170 mutants and their effect on Nup53 binding; (+++) wild-type binding, (++) moderately weakened binding, (+) weak binding, and (-) no binding. Mutations in Nup53 that were tested for binding are indicated by dots above the sequence using the same color code as in panel (D). Chromatograms and representative SDS-PAGE gels are shown in fig. S22. (F) Domain structures of Nup170 and Nup145N. Black lines indicate fragments used for crystallization. (G) Crystal structure of the Nup170CTD•Nup145NR3 complex shown in cartoon representation. (H) Close-up view of the interaction between Nup170 and Nup145N. Nup170 and Nup145N residues involved in the interaction are labeled in orange and cyan, respectively. (I) Graphic summary of mutational analysis of the Nup170CTD-Nup145NR3 interaction. Nup170 is shown in surface representation from the same view as in panel (H). Coloring is according to panel (D). (J) Summary of mutational analysis of the Nup170CTD-Nup145NR3 interaction, coloring and key are same as in panel (E). Chromatograms and representative SDS-PAGE gels are shown in fig. S23.
Nup53 is anchored to the nuclear envelope by its C-terminal amphipathic helix, either directly or through an interaction with NDC1 (18, 19). Nup170 bound to Nup53R3, which is directly adjacent to this C-terminal helix, prompting us to look for features in Nup170 that could also contribute to nuclear envelope binding. We identified two motifs next to the C-terminus of Nup53R3 that would be juxtaposed with the nuclear envelope. The first was a WF motif composed of solvent exposed, evolutionarily conserved tryptophan and phenylalanine residues in the 3CD loop (Fig. 3B and fig. S22C). As tryptophan residues are enriched at membrane interfaces, the WF motif may reinforce membrane binding (31). The second motif, residing in the 3D4A loop we deleted for crystallization, is predicted to form an amphipathic helix with a striking, evolutionarily conserved absence of charged residues, a feature characteristic of amphipathic lipid packing sensing (ALPS) motifs, which are also present in Nup120 and Nup133 (fig. S22, C and D) (32, 33). The Nup170 ALPS motif contained a universally conserved proline residue on the polar face of the helix, a feature reminiscent of antimicrobial membrane destabilizing peptides (fig. S22D) (34). We propose that these additional features on Nup170 act synergistically with Nup53 binding to the nuclear envelope to help maintain the extreme membrane curvature in nuclear pores.
We next determined a crystal structure of the Nup170CTD•Nup145NR3 complex at 3.5 Å resolution, using a 2.1 Å-resolution structure of apo Nup170CTD as a search model (Fig. 3, F and G and tables S4 and S6). Nup170CTD formed an elongated α-helical solenoid containing two stacks of irregular helical pairs, arranged in a zig-zag fashion (Fig. 3G). The two stacks shared a long helix (α31) that capped the first stack and initiated the second stack. Nup145NR3 bound to a pair of deep hydrophobic pockets formed on either side of the first two helices of the second helical stack, inserting residues L733 and I735 into the first pocket and L743 and F744 into the second pocket (Fig. 3, G and H). Mutation of any of these residues completely abolished binding to Nup170 (Fig. 3, I and J and fig. S23B). Similarly, mutation of the residues that formed the hydrophobic pockets (F1171, F1154, I1131, and Y1157) strongly affected binding (Fig. 3, I and J and fig. S23A). The hydrophobic nature of both binding pockets is retained throughout eukaryotes (fig. S21), suggesting the evolutionary conservation of this interaction. Only minimal rearrangements of the binding pocket occur upon Nup145N binding (fig. S23C).
The Nup145N sequence that binds to Nup170 is also highly conserved throughout eukaryotes, and the homologous residues critical for Nup170 binding are conserved in humans (fig. S24A). During mitosis, extensive phosphorylation of hsNup98, the human homologue of Nup145N, leads to NPC and nuclear envelope disassembly (35). The most abundant mitotic phosphorylation sites in hsNup98 are at residues S608 and S612 (S741 and S745 in C. thermophilum), which flank L610 and F611 (L743 and F744 in C. thermophilum), residues we found to be critical for Nup170 binding (fig. S24, A and B) (35). To test the possibility that the interaction between Nup170 and Nup145N could be regulated by phosphorylation, we reconstituted a hsNup155•hsNup98 hetero-dimer homologous to our crystallized complex. We observed a robust interaction between hsNup155CTD and the corresponding minimal hsNup98 fragment, which was partially disrupted by a phosphomimetic mutation (S608E/S612E) (fig. S24C). As revealed by the Nup170CTD•Nup145NR3 structure, S612 (S745 in C. thermophilum) formed a hydrogen bond with N609 (D743 in C. thermophilum). Phosphorylation would therefore destabilize the conformation required to insert the critical hydrophobic residues into the binding pocket in Nup170 (fig. S24B). Disruption of this hydrogen bond by mutagenesis also completely abolished binding (Fig. 3J and fig. S23B). Thus, the Nup170-Nup145N interaction is not only highly conserved, but its disruption is also a key step in mitotic NPC disassembly in humans.
Our structures of Nup170NTD and Nup170CTD did not overlap in sequence, preventing accurate modeling of the full-length protein. Therefore, we crystallized a larger fragment of Nup170 containing the α-helical solenoids of both domains (Nup170SOL, residues 575–1402), and determined the crystal structure at 4.0 Å resolution (fig. S25A; tables S4 and S5; movie 1). While there was no conformational variability observed for the helices present in Nup170NTD, the Nup170CTD solenoid exhibited an ~20° movement resulting from a minor rearrangement of helix α27 (fig. S25A). We observed a similar conformational variability in the Nup170CTD•Nup145NR3 complex structure, where all four molecules in the asymmetric unit adopted different conformations (fig. S25B). With the structure of Nup170SOL as a template, we superposed the structures of Nup170NTD and Nup170CTD, obtaining a total of eight different conformations for full-length Nup170 (figs. S25B and S26 and movie 2).
Molecular basis for recognition of Nup53 by Nic96
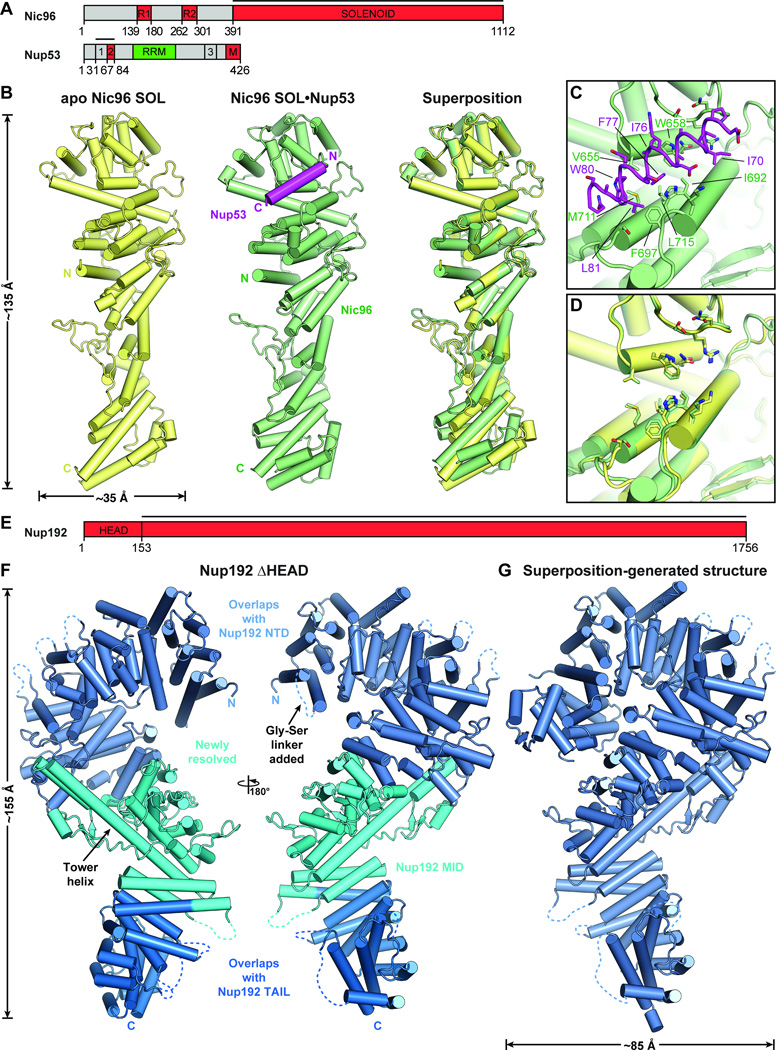
We determined crystal structures of apo Nic96SOL and a Nic96SOL•Nup53R2 complex at 3.3 and 2.65 Å resolution, respectively (Fig. 4, A and B; fig. S27; tables S4 and S7; Movie 3). Nic96SOL formed a rod-shaped molecule consisting of a U-bend α-helical solenoid with the N-terminus situated in the middle of the rod. Although residues 31–84 of Nup53 were included in the crystallization construct, only residues 67–84 were visible in the electron density. Residues 67–84 of Nup53 formed an amphipathic helix that buried its hydrophobic face into a hydrophobic groove formed by helices α14, α15, and α16 near the U-bend end of the Nic96 solenoid (Fig. 4, B and C). Consistent with our crystal structure, we found that mutations to the hydrophobic residues in this pocket disrupted binding (figs. S28 and S29). Similar to the interaction between Nup170 and Nup145N, we observed minimal conformational rearrangements upon Nup53 binding (Fig. 4D).
Fig. 4. Structural analysis of the inner ring complex nucleoporins Nic96 and Nup192.
(A) Domain structures of Nic96 and Nup53. Black lines indicate fragments used for crystallization. (B) Crystal structures of apo Nic96SOL (yellow) and the Nic96SOL•Nup53R2 complex (green and purple) and their superposition are shown in cartoon representation. See also Movie 3. (C) Close-up view of the Nup53R2-binding site in Nic96SOL. For clarity, Nup53 is shown in ribbon representation. Nic96 and Nup53 residues involved in the interaction are labeled in green and purple, respectively. (D) Close-up view of the superposition of apo and Nup53-bound structures of Nic96SOL reveals minimal conformational changes. (E) Domain structure of Nup192. A black line indicates the fragment used for crystallization. (F) Crystal structure of Nup192ΔHEAD shown in cartoon representation. A 180° rotated view is shown on the right. Regions of Nup192 that were resolved in previous crystal structures are colored in shades of blue, while the region of the protein that was not included in previous crystallographic analyses is shaded cyan. (G) Structure of full-length Nup192 generated by superposing fragment crystal structures. See also fig. S31 and Movie 4.
Structure of Nup192
Nup192, the largest symmetric core nucleoporin, forms a question-mark shape at low resolution (26). Crystal structures exist for Nup192NTD (residues 1–958) and Nup192TAIL (residues 1397–1756), but an atomic structure of the entire molecule has remained elusive (15, 20, 26). To determine the complete atomic structure of Nup192, we obtained crystals of an engineered Nup192 truncation mutant, Nup192ΔHEAD, from which we deleted the N-terminal HEAD domain (residues 1–152) and replaced a loop encompassing residues 167–184 with a short glycine-serine linker. We determined the crystal structure of Nup192ΔHEAD at 3.2 Å resolution (Fig. 4,E and F; fig. S30; tables S4 and S7).
The N-terminal portion of the previously unresolved middle domain of Nup192 (Nup192MID) contained three additional ARM repeats (α46-α53, residues 959–1154) that continued the superhelical solenoid we previously observed in Nup192NTD (Fig. 4F) (20). Similarly, the C-terminal portion of Nup192MID contained a HEAT repeat (α60-α61, residues 1330–1376) that extended the Nup192TAIL solenoid such that the entire protein formed a continuous HEAT/ARM repeat solenoid (Fig. 4F) (15). However, we observed an unusual insertion (residues 1155–1329) between the ARM repeats and HEAT repeat in Nup192MID containing a ~50-residue helix, α58, that reached ~75 Å from the beginning of Nup192TAIL to the C-terminus of Nup192NTD (Fig. 4F). This insertion, which we termed the Tower helix, buried several hydrophobic residues against the bottom of Nup192NTD, inducing minor rearrangements that facilitate packing of the Tower helix. While the Tower helix has only a moderate signature in secondary structure predictions, the evolutionarily-related Nup188 is also predicted to contain a similarly long, ~40 residue helix at the same location. However, previous models of either full-length Nup188 or Nup192 never anticipated the existence of the Tower helix, highlighting the importance of experimentally determining atomic resolution structures (22, 36).
Taking advantage of the extensive overlap between our crystal structure of Nup192ΔHEAD with the existing structures of Nup192NTD and Nup192TAIL, we generated the structure of full-length Nup192 by superposition (Fig. 4G and fig S31; Movie 4). Inspection of the full-length protein revealed that the first loop in the HEAD domain between α1 and α2 was close enough to contact loops in the MID domain, predominantly with polar and charged residues (Fig. 4G and fig. S31). The binding sites on Nup192 for Nup53 and Nic96, which we previously identified via mutagenesis, were located at the top and bottom of the molecule, respectively, ~140 Å apart from each other (fig. S32) (15, 20).
Architecture of the NPC symmetric core
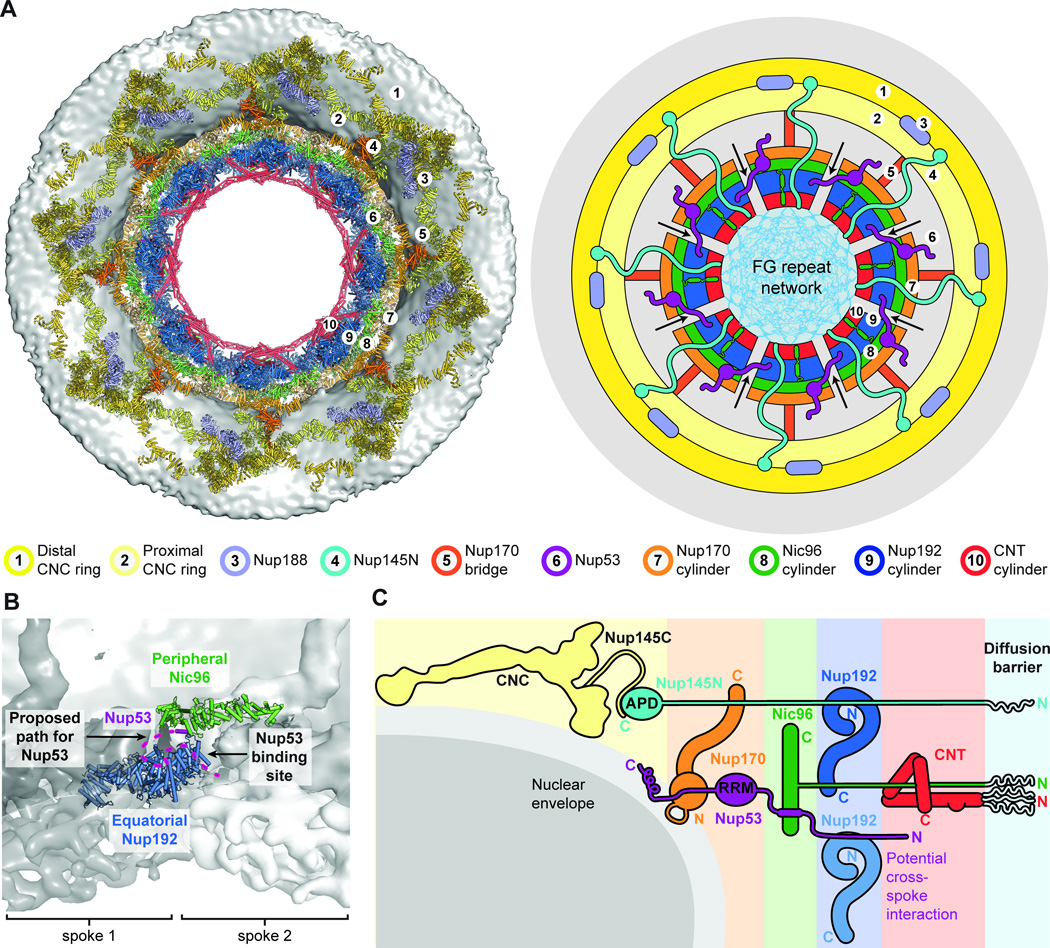
Recent advances in cryoET have produced rapidly improving reconstructions of intact NPCs, with the most recent reconstructions reporting average resolutions up to ~20 Å in the best resolved regions (13, 28, 37). We previously docked 32 copies of the yeast CNC into a ~34 Å reconstruction of the intact human NPC, taking advantage of the distinctive shape and large size of the complex (14). With the addition of the Nic96SOL•Nup53R2 structure and the full-length, superposition-generated structures of Nup192 and Nup170•Nup53R3•Nup145NR3 reported here, as well as our recently reported crystal structure of the CNT•Nic96R1 complex, accurate structures were available for essentially the entire ordered mass of the NPC symmetric core (15). The domain architectures of the symmetric core nucleoporins are highly conserved from fungi to humans (fig. S33). We successfully located these structures in the recently reported ~23 Å reconstruction of the human NPC, with the arrangement of nucleoporins validated by our biochemical restraints (28). We utilized an incremental approach to confidently place the crystal structures, starting with the largest structures possessing the most distinctive shapes, and iteratively removing the occupied density to search for subsequent structures (fig. S34). As the cryoET map possesses eightfold rotational symmetry, each unique solution defined the location and orientation of eight copies of each molecule. We first tested our approach with the yeast CNC crystal structure and found four unique placements with exceptional scores compared to 50,000 other refined placements, in excellent agreement with our previous results (fig. S35A) (14). We also readily identified the location and orientation of human Nup84CTD•Nup133CTD in unbiased searches (fig. S35B) (38). Based on previously reported biochemical data, we manually docked the Nup37, Nup43, and Nup133 β-propellers and locally optimized their fit (fig. S35C) (13, 39–41).
We next performed unbiased searches for Nup170 and Nup192 using a map from which density corresponding to the CNCs had been removed (fig. S36). As our crystallographic data indicated significant flexibility in the Nup170 solenoid, we performed searches with the eight different conformations of Nup170. These searches identified two conformations that each yielded two distinct top scoring solutions (fig. S36, A and B). Searches with full-length Nup192 revealed six unique solutions, but because Nup192 and Nup188 could not be distinguished at this resolution, we assigned two of these as Nup188 using our biochemical results and previously reported cross-linking data, as detailed in the methods (fig. S36C) (13, 42). After removing the density assigned to Nup170, Nup192, and Nup188, we successfully located four unique copies of the Nic96SOL•Nup53R2 and CNT•Nic96R1 complexes (fig. S37, A and B). Lastly, we inspected the remaining density in the inner ring in an attempt to locate the ordered domains of Nup53 and Nup145N. We determined crystal structures of Nup53RRM and Nup145NAPD•Nup145CN at 0.8 Å and 1.3 Å resolution, respectively, but could not unambiguously place them due to their small size and globular shape (fig. S38 and tables S4, S8, and S9). We attempted to generate biochemical restraints to dock Nup53RRM confidently, but were unable to find any binding partners (fig. S39). However, we did find a pair of continuous densities that readily accommodated two additional Nup170 molecules in a third distinct conformation (fig. S40A). These placements were buried in our original global search, but the conformation of this Nup170 structure still differed slightly from the remaining map density, suggesting that our crystal structures did not capture the full conformational range of Nup170 (fig. S40, B and C).
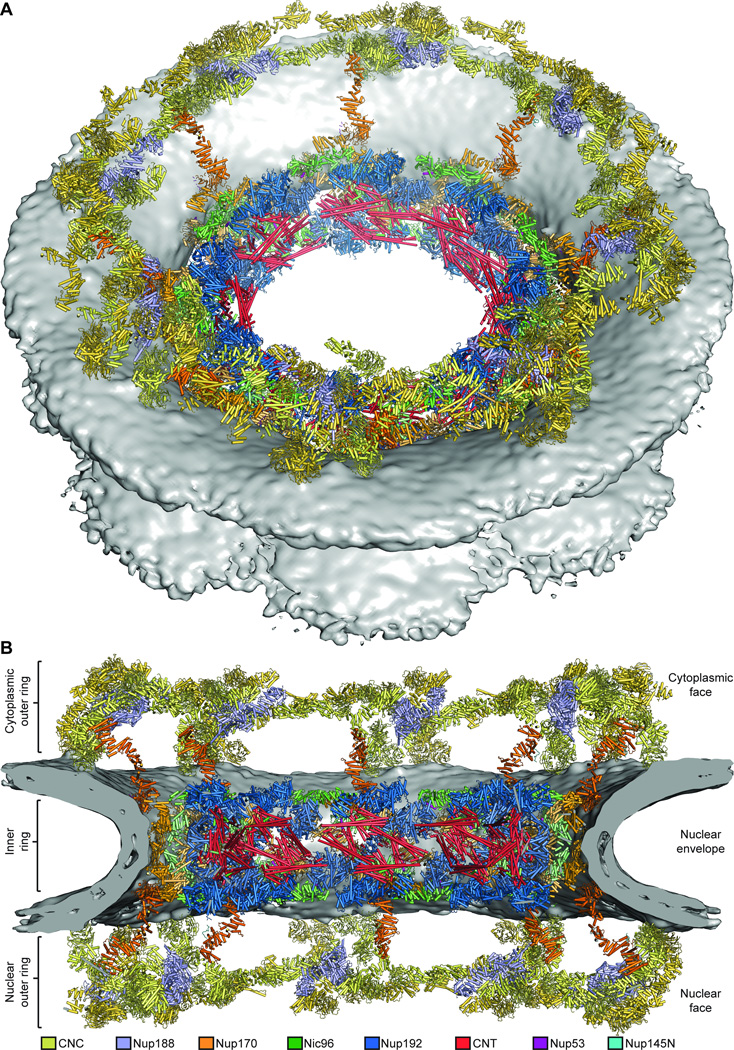
With the CNC-hexamer, Nup84CTD•Nup133CTD, Nup133NTD, Nup37NTD, Nup43, Nup188NTD, Nup188TAIL, Nup192, Nup170•Nup53R3•Nup145NR3, Nic96SOL•Nup53R2, and CNT•Nic96R1 structures placed, we acquired a composite structure accounting for nearly all of the density in the NPC symmetric core, corresponding to ~54 MDa of protein mass or ~320,000 ordered residues (Fig. 5; fig. S41; table S10; Movie 5). In total, the composite structure contained a stoichiometry of 16 copies of Nup188, 32 copies of the CNC, Nup192, Nic96, and CNT, and 48 copies of Nup170, Nup53, and Nup145N. Overall this stoichiometry is in good agreement with a previous study that used mass spectrometry to measure the relative abundances of nucleoporins in the human NPC (43).
Fig. 5. Architecture of the NPC symmetric core.
Composite structure of the NPC symmetric core generated by docking nucleoporin and nucleoporin complex crystal structures into the cryoET reconstruction of the intact human NPC (EMD-3103). The density corresponding to the nuclear envelope is shown as a gray surface. Proteins are color-coded according to the legend at the bottom. (A) View from above the cytoplasmic face and (B) a cross-sectional view from within the transport channel. Details are discussed in the text. See also figs. S34 to S37; S40; Movie 5.
Spoke architecture
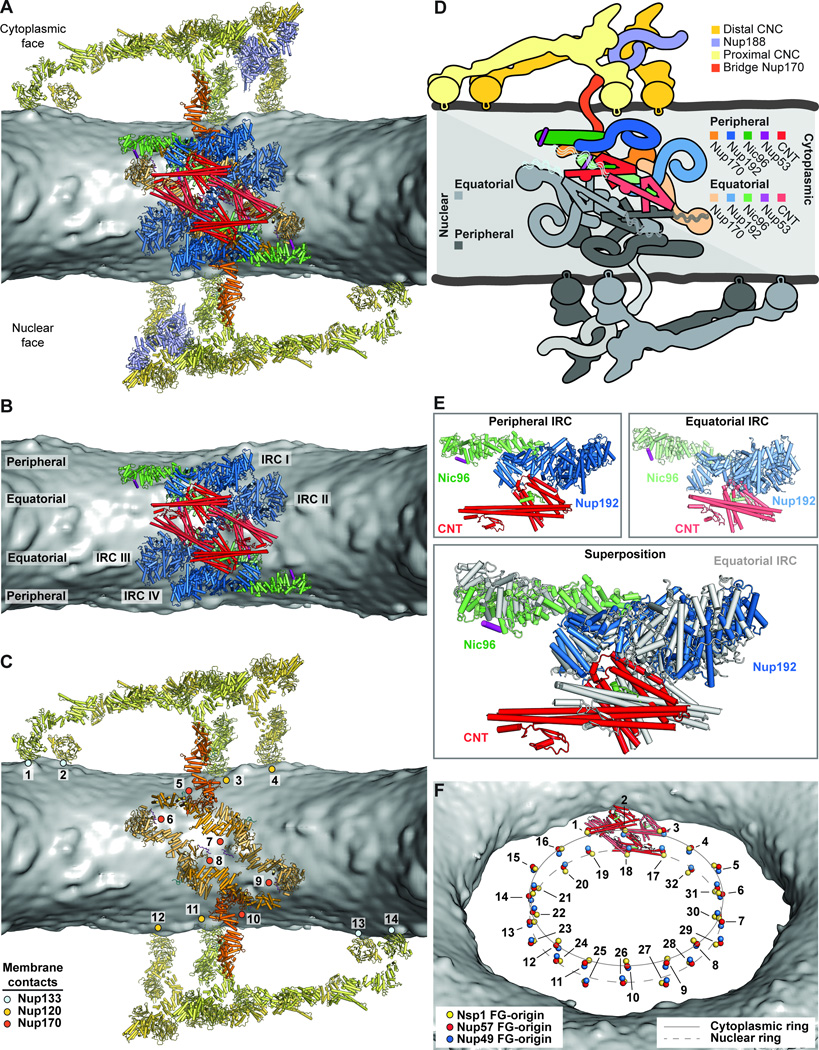
The symmetric core of the NPC consisted of eight spokes related by an eight-fold rotational axis of symmetry perpendicular to the nuclear envelope (Fig. 5A). Outer rings resided above the nuclear and cytoplasmic faces of the nuclear envelope, while an inner ring was embedded in the pore and spanned the nuclear envelope (Fig. 5B). When viewed from the cytoplasm, the nucleoporins formed distinct cylinders, with the CNTs lining the transport channel, surrounded by successive cylinders formed by Nup192, Nic96, Nup170, and the CNCs (Fig. 5A). Nic96NTE and the linker nucleoporins Nup53 and Nup145N spanned these cylinders. Only C8 rotational symmetry was applied to generate the cryoET reconstruction, yet we observed an additional two-fold axis of symmetry in our composite structure relating the nuclear and cytoplasmic sides within each spoke (Fig. 6, A to D) (28).
Fig. 6. Architecture of the NPC spoke.
(A) A single NPC symmetric core spoke is shown in cartoon representation from the same cross-sectional view as in Fig. 5B. Different shades of colors are used to indicate biochemically distinct dockings of the same protein. See also Movie 6. (B) Same view as in panel (A), but with Nup170 and the nucleoporins from the outer ring removed to highlight the organization of the four inner ring complexes (IRCs). (C) The membrane coat of the NPC. Nup192 and Nic96 molecules and CNTs have been removed for clarity. Contacts between the nuclear envelope and the ALPS motifs in Nup120, Nup133, and Nup170 are indicated by dots. (D) Schematic of the NPC spoke. The proteins corresponding to the nuclear side of the spoke are colored in gray, demonstrating the two-fold rotational symmetry relating the cytoplasmic and nuclear halves of each spoke. (E) Equatorial and peripheral IRCs adopt similar conformations. Identical views of the cytoplasmic peripheral IRC, the cytoplasmic equatorial IRCs, and their superposition are shown. For clarity, the equatorial IRC is colored in gray in the superposition. (F) Organization of FG repeats in the inner ring. Colored spheres indicate the positions of the N-termini of the three channel nucleoporins for all 32 CNT copies in the inner ring. Despite four distinct CNT positions in each spoke, the N-termini for the cytoplasmic and nuclear complexes are arranged in approximately the same plane. Thus, while not possessing true 16-fold symmetry, the CNT N-termini approximately form two 16-membered rings, which are indicated by solid and dashed lines. Notably, the FG repeats of the 16-membered CNT rings project circumferentially into the central transport channel with opposite directionality; cytoplasmic CNT ring (counterclockwise), nuclear CNT ring (clockwise). The CNT molecules for a single spoke are shown in cartoon representation to indicate the directionality of the FG repeats.
Each inner ring spoke contained four copies of the IRC (Fig. 6B and Movie 6). Our results provided spatial restraints for Nic96R1 and Nic96R2, allowing us to trace the path of Nic96NTE, which emerges from the middle of Nic96SOL, to its binding sites on Nup192 and the CNT. We refer to these four distinct IRCs as nuclear peripheral, nuclear equatorial, cytoplasmic peripheral, and cytoplasmic equatorial IRCs in the following text (Fig. 6B). The nuclear and cytoplasmic equatorial IRCs were related to each other directly by the two-fold rotational axis of symmetry, as were the nuclear and cytoplasmic peripheral IRCs. Unexpectedly, the subunits in the equatorial and peripheral IRCs were in approximately the same relative orientation, which was readily apparent upon superposition (Fig. 6E). Because the subunits were placed independently, this surprising symmetry was an emergent property of the composite structure. The docking reveals that the CNT and Nup192 are in close proximity, suggesting that additional weaker interactions orient the CNTs in the fully assembled NPC. We observed additional knobs of density adjacent to the Nup57 α/β domains of each CNT, which were unexplained by our composite structure (fig. S37D). When we superposed structures of CNT fragments from Xenopus laevis onto our docked CNT molecules (16), we found that the metazoan-specific ferredoxin-like domain of Nup57 accounted for these extra knobs of density (fig. S37, C and D), further validating our composite structure.
The mechanism by which FG repeats in the central transport channel form a diffusion barrier and facilitate transport remains controversial, partly because the stoichiometry and orientation of the FG repeats remain unknown. Our composite structure revealed a total of 32 CNTs in the inner ring, which would project 96 distinct polypeptide chains in clusters of three into the central channel (Fig. 6F). The orientation of the CNTs suggested that the FG repeats would emanate circumferentially towards the adjacent spoke rather than pointing radially towards the center of the channel. Unexpectedly, the N-termini of the peripheral and equatorial CNTs were evenly spaced and roughly planar such that they approximately formed two 16-membered rings (Fig. 6F).
In the outer rings, each spoke contained two CNCs on either face, which we refer to as proximal and distal CNCs based on their distance from the inner ring. The orientations of Nup133 relative to the CNC core differed slightly between the distal and proximal CNCs, but yielded the same overall architecture, as each pair of distal and proximal CNCs formed an arch over the nuclear envelope (fig. S42). The outer rings also contained a Nup188 molecule on either face (fig. S43). The majority of the CNC components were ~100 Å above the membrane, and the only contacts with the membrane were made by the β-propeller domains of Nup120 and Nup133 through their ALPS motifs (Fig. 6C). Similarly, the IRCs did not make direct contacts with the membrane, but instead were surrounded by a network of Nup170 molecules that formed the outermost layer of the inner ring (Fig. 6C). Each spoke contained three distinct pairs of Nup170 molecules, which we refer to as equatorial, peripheral, and bridging Nup170 molecules. The equatorial pair occupied alternating orientations equatorially along the surface of the nuclear envelope (Fig. 6C and fig. S36A). The resolution of the cryoET reconstruction was high enough for these molecules that the central holes of the β-propellers were readily visible at higher contour levels (fig. S36A). The bridging pair of Nup170 molecules bridged the inner ring to the outer CNC rings, via a contact with Nup120 (Fig. 6C and figs. S36B and S43). The peripheral pair of Nup170 molecules, which had a weaker quality of fit and was identified only after placing all other symmetric core components, contacted both the equatorial and bridging Nup170 molecules (Fig. 6C and fig. S40). The equatorial Nic96 molecules contacted multiple Nup170 molecules, effectively bridging the nuclear and cytoplasmic networks. Notably, the ALPS and WF motifs we identified in Nup170NTD and the C-terminal amphipathic helix of Nup53 were oriented directly adjacent to the nuclear envelope (Fig. 6C). Thus, Nup170 and Nic96 constitute a membrane coat for the inner ring analogous to the CNCs in the outer ring.
Inter-spoke interactions
We next searched for interfaces mediating interactions between spokes. Several potential interactions have been identified in previous studies, including one between Nup133NTE and Nup120NTD (40). Our composite structure revealed four additional interactions between CNC components that could link adjacent spokes: (1) between the neighboring proximal Nup84 and distal Nup85, (2) between the proximal Nup133 α-helical solenoid and distal Nup120 α-helical solenoid, (3) between the proximal Nup133 β-propeller and proximal Nup120 β-propeller, and (4) between the distal Nup133 β-propeller and distal Nup120 β-propeller (fig. S43, A to C). The space between the proximal and distal CNCs contained density that readily accommodated a Nup188 molecule. Nup188 recognized a special niche generated in the CNC inter-spoke interface, bridging four CNC molecules by potentially making contacts with: (5) the distal Sec13, (6) the distal Nup85, (7) the proximal Nup43, and (8) the proximal Nup133 of a neighboring spoke (fig. S43, B and C). The Nic96R2 binding site in Nup188TAIL was not occluded by any additional density (fig. S43B). Due to the absence of strong density in this region, we cannot determine whether only Nup188 or entire an Nup188 complex would be anchored to the outer rings at this site. However, any flexibly tethered components of the NPC, including the remainder of the Nup188 complex, may not be clearly visible in cryoET reconstructions.
Inspection of the interaction surfaces also provided a molecular explanation for how two CNC rings are assembled. Nup133 and Nup170 bind to distinct Nup120 molecules via overlapping interfaces on the proximal and distal Nup120 molecules, respectively, effectively capping the CNCs on either side (Figs. 43, D and E). These results are in agreement with a common evolutionary origin for Nup120, Nup133, and Nup170, which all possess similar domain architectures, contact the nuclear envelope via ALPS motifs, and interact with each other at inter-spoke interfaces. The U-bend solenoid nucleoporins, Nup85, Nup145C, Nup84, and Nic96, bridge these inter-spoke interfaces to form a continuous membrane-bending coat. These results also support the protocoatomer hypothesis of a common evolutionary origin for vesicle coats and the NPC coat, wherein the extant nucleoporins derived from an ancient membrane coat containing these protein folds (44).
We could only identify a single interaction that would analogously link the inner ring spokes, which would be mediated by an interaction between Nup53R1 and Nup192 (Fig. 7). In our composite structure, peripheral Nic96 molecules oriented Nup53R2 directly adjacent to the Nup53-binding site on equatorial Nup192 molecules from a neighboring spoke (Fig. 7B). Our biochemical mapping experiments identified adjacent binding sites for Nic96 and Nup192 on Nup53 (Fig. 2F) and that a fragment containing both these binding sites could bridge the two nucleoporins (fig. S14C). Thus, binding of Nup53R1 in trans to a Nup192 molecule from a neighboring spoke would link the inner ring spokes.
Fig. 7. Cylindrical organization of the NPC.
(A) A top view of the composite structure of the NPC symmetric core viewed from the cytoplasm is shown (left). Coloring is according to Fig. 5. Schematic representation on the right illustrates the distinct concentric cylinders observed in the symmetric core. Cartoons of the linker nucleoporins and the unstructured NTE of Nic96 are shown to indicate how they span across multiple cylinders. Arrows indicate gaps between the inner ring spokes, which represent proposed paths for inner nuclear membrane protein transport. (B) Only a single contact is observed between the two adjacent inner ring spokes. A close-up view of the inter-spoke interface in the inner ring is shown in an orientation similar to Fig. 5A. Two adjacent spokes are depicted as gray and white surfaces, on the left and right, respectively. An equatorial Nup192 molecule (spoke 1) and a peripheral Nic96 molecule bound to Nup53 (spoke 2) are shown in cartoon representation. The Nup53 binding site on Nup192 is labeled and the proposed path of the peptide is drawn as a purple dashed line. (C) A schematic representation illustrating the concentric cylinder organization of the symmetric core shown from the side. The order and arrangement of the binding sites for the linker nucleoporins observed in the composite structure are depicted. The proposed inter-spoke interaction between Nup53 and Nup192 is drawn at the bottom.
An open question regarding nucleocytoplasmic transport has been the mechanism of inner nuclear membrane (INM) protein transport through the NPC, particularly for INM proteins with large globular nuclear domains. Peripheral channels on the order of ~100 Å have been proposed as routes for INM transport (45), but we observed no such channels through the inner ring in either our composite structure or the cryoET reconstruction (Fig. 5). Given the dense packing within each spoke, traffic of INM proteins through a spoke would require significant disruption of NPC structure (Fig. 7A). Rather, the most likely path through the inner ring would be at the inter-spoke interfaces where Nup53R1 and Nup192 interact (Fig. 7). The CNCs form an ~100-Å arch above this interface, providing an uninterrupted path to the inner ring and possibly explaining the previously observed upper limit for the size of nuclear domains (Fig. 6A) (46). However, the channel at the inner ring was much smaller, suggesting that rearrangements at the inter-spoke interface may be necessary to traffic large nuclear domains. Thus, our composite structure of the NPC symmetric core enables rational design of experiments to further understand the mechanism of INM protein import.
A flexible linker mediates CNC oligomerization
While reconstituting the CNCs, we noticed that assembly of the CNC-octamer spontaneously generated a separate solution phase (fig. S44A). Similar phase transitions were previously seen in other systems with multiple binding valencies, suggesting that the oil droplet formation we observed resulted from oligomerization of the CNC (47). We previously identified an interaction between Nup133NTE and Nup120 that would mediate head-to-tail CNC ring formation consistent with the composite structure of the NPC (40). Removal of the unstructured Nup133NTE completely ablated both oil droplet and complex formation, suggesting that this flexible interaction serves as a driving force for CNC ring formation (fig. S44). We also found that Nup170 could be incorporated into this separate solution phase and that this incorporation was ablated by C-terminal truncation, which was consistent with the interaction between the proximal Nup120 and bridging Nup170 molecules we observed in our composite structure (fig. S44A).
Conservation of NPC architecture
Our results establish the principles that drive the assembly of nucleoporins in the NPC. While the outer rings assemble largely via structurally rigid interaction surfaces, inner ring assembly is primarily driven by flexible linker sequences within Nup53, Nup145N, and Nic96NTE. This dichotomy may reflect the different roles of the respective complexes. The outer rings provide a structural scaffold for the NPC and given their location above the plane of the nuclear envelope, their assembly would not be affected dramatically by the dynamic generation of membrane curvature during fusion of the inner and outer nuclear membranes. In contrast, the proteins in the inner ring occupy an environment that only exists after membrane fusion, likely necessitating conformational flexibility over the course of NPC assembly.
The importance of these flexible interactions in the NPC is highlighted by their evolutionary conservation despite poor overall sequence conservation in the linker nucleoporins Nup53 and Nup145N. The overall folds of the scaffold proteins are well-conserved in S. cerevisiae (fig. S45) and furthermore, point mutations in the binding pockets of S. cerevisiae Nic96, Nup170, and Nup157 also disrupted their interaction with linker sequences (figs. S46 to S48). A complete understanding of the interaction network in S. cerevisiae has been partially intractable because of the genetic redundancy that arises from several gene duplications (Nup170/Nup157, Nup53/Nup59, and Nup145N/Nup100/Nup116). We found that the paralogs mostly retained the ability to form these interactions, but did not detect an interaction between scNup188 and any of the Nup145N paralogs (figs. S46 to S50).
Our structural data also highlighted the evolutionary conservation of nucleoporin structure and their interactions. While crystal structures of the human scaffold nucleoporins have not been determined, previous comparisons of the fungal CNC with low-resolution reconstructions of the human CNC suggest a conserved architecture (13, 14). Superposition of the structures of Nup53RRM and Nup145NAPD with their human homologues also revealed that their folds were identical (fig. S38) (30). In addition, the mechanism of interaction between Nup170 and Nup145N is conserved in humans, and we found that phosphomimetic mutations weakened the interaction between hsNup98 and hsNup155. Several other phosphorylation sites have been identified in Nup98, many of which potentially overlap with scaffold binding sites (35). Therefore, phosphorylation could also regulate other key interactions, including those that occur between Nup145N and Nup188, Nup192, and the CNC. We note that Nup53 is similarly phosphorylated in a cell-cycle dependent manner, and thus phosphorylation of both linker nucleoporins would be an effective means to disassemble the entire inner ring of the NPC (48).
Conclusions
Determining the molecular details of nucleocytoplasmic transport has been a longstanding challenge, at least in part due to an incomplete understanding of the architecture and biochemistry of the NPC itself. We have used purified, recombinant proteins to systematically characterize the nucleoporin interaction network and determine atomic resolution structures of nucleoporin complexes. This approach was crucially complemented by recent advances in cryoET reconstructions (28). Using the results of our divide-and-conquer approach, we were able to dock the available crystal structures into a cryoET reconstruction of the human NPC, yielding a composite structure for the entire NPC symmetric core. This union of bottom-up and top-down approaches offers a paradigm for determining the architectures of similarly complex macromolecular assemblies.
Our composite structure differs dramatically from the previously reported computational models not only in relative and absolute stoichiometry, but also in overall architecture (49). These discrepancies highlight the complexities that must be accommodated when attempting a holistic, computational approach. We observed a remarkable degree of symmetry in the structure of the NPC, which explains how such a limited vocabulary of proteins can generate such a large macromolecular structure. Most nucleoporins also occupy multiple, distinct biochemical environments. Nup170 offers a dramatic example of this property, as biochemically distinct versions of Nup170 are either buried in the inner ring or are exposed in the bridge between the inner and outer rings. Similarly, Nup120 utilizes overlapping, exclusive interfaces to contact Nup170 and Nup133. Due to this diversification of nucleoporin function, the NPC can be encoded by a relatively small number of genes. The gene duplications of nucleoporins in S. cerevisiae may reflect the gradual separation of these distinct functions into several genes. Nup170 appears to also adopt different conformations in each of its distinct biochemical environments, which is consistent with the wide conformational range we observed in crystal structures. It is possible that the different conformations are the result of different mechanical forces acting on Nup170 at each position.
Biochemical diversification of proteins within the same protein complex also generates enormous challenges for computationally modeling the structure of the NPC and similar complexes, as distance restraints such as crosslinks that are valid in one biochemical environment may be violated in another. This challenge is exacerbated by the possibility of flexibly tethered domains or nucleoporins, such as those in the Nup188 complex. Our results also highlight the confounding effect generated by the flexible linker nucleoporins Nup53 and Nup145N, which occupy binding sites that span the entirety of the inner ring, rather than a single globular volume. The structure of the NPC could be used as a template for the development of methods that can accommodate these additional complexities.
Our composite structure of the NPC provides a rich platform for contextualizing previous results, not all of which can be commented upon here. The structure also permits the rational design of new experiments to not only further validate the structure, but also begin structure-function interrogation of the NPC. Our biochemical results built a map of the strongest and most conserved interactions, but our composite structure clearly indicates that many additional interactions can occur in the context of the assembled NPC. However, a structural understanding of the entire NPC at single residue resolution still requires several advances. Successful placement of crystal structures of fungal nucleoporins into a cryoET reconstruction of the intact human NPC highlights not only the evolutionary conservation of NPC structure, but also the need for structural characterization of human nucleoporins and further improvement of the resolution of cryoET reconstructions to improve the accuracy of the composite structure of the NPC. Improved resolution in cryoET reconstructions where secondary structure elements can be visualized would enable flexible fitting of high-resolution crystal structures. We were not able to dock any of the asymmetric components of NPC into the composite structure, due to the small size of extant structures and the absence of high-quality restraints (fig. S41). A similar approach to the one used here will be necessary to extend our analysis to the many proteins in the cytoplasmic filaments and nuclear basket that interact directly with transport factors and are implicated in human disease. Lastly, high-resolution structural characterization of the remaining interactions, especially those involving flexible linker sequences, remains critical to building a truly complete structure of the NPC, as these interactions may never be resolved in cryoET reconstructions.
Supplementary Material
Movie 1. Generation of the Nup170•Nup53R3•Nup145NR3 complex structure by superposition. A 360° rotation of the crystal structure of Nup170SOL followed by a demonstration of the process used for generating the full-length structure by superposition. Colors are the same as in fig. S25.
Movie 2. Morph animation of observed Nup170 conformations. The eight different conformations observed in the superposition-generated structures of full-length Nup170 are animated in morph movies. The animation pauses at each of the eight experimentally observed conformations of Nup170, colored the same as in fig. S25.
Movie 3. Structure of the Nic96SOL•Nup53R2 complex. A 360° rotation of the crystal structure of Nic96SOL •Nup53R2. Colors are the same as in Fig. 4.
Movie 4. Generation of the structure of full-length Nup192 by superposition. A 360° rotation of the crystal structure of Nup192ΔHEAD followed by a demonstration of the process used for generating the full-length structure by superposition. Colors are the same as in fig. S31.
Movie 5. Overview of the composite structure of the NPC symmetric core. The structures are shown as docked into the cryoET reconstruction of the intact human NPC, oriented with the cytoplasmic face on top and the nuclear face on bottom. The nuclear envelope and density corresponding to protein density were colored in gray and white, respectively. Crystal structures of symmetric core nucleoporins are first shown in cartoon representation, colored according to Fig. 5, then without the cryoET reconstruction density, and finally in surface representation.
Movie 6. Rotating structure of a single spoke of the NPC symmetric core. The crystal structures are shown as docked into the cryoET reconstruction of the intact human NPC, oriented with the cytoplasmic face on top and the nuclear face on bottom. The nuclear envelope is colored in gray. Crystal structures of symmetric core nucleoporins are shown in cartoon representation, colored according to Fig. 6.
Acknowledgments
We thank Jee-Young Mock, Stefan Petrovic and Alina Patke for critical reading of the manuscript. We thank Martin Beck for sharing the cryoET reconstruction of the intact human NPC prior to publication, Ed Hurt for providing material and discussions, and Camille Bayes, Slava Butkovich, Christopher Frick, Antoine Koehl, Hanna Kuberczyk, Roland Tran, and Stephan Zimmerman for experimental support. We also acknowledge Jens Kaiser and the scientific staff of the Stanford Synchrotron Radiation Laboratory (SSRL) Beamline 12-2, the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA) at the Advanced Photon Source (APS), and the Advanced Light Source (ALS) beamline 8.2.1 for their support with X-ray diffraction measurements. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program for their support of the Molecular Observatory at the California Institute of Technology (Caltech). The operations at the SSRL, ALS, and APS are supported by the U.S. Department of Energy and the National Institutes of Health (NIH). GM/CA has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). D.H.L and A.M.D. were supported by a NIH Research Service Award (5 T32 GM07616). D.H.L. was also supported by an Amgen Graduate Fellowship through the Caltech-Amgen Research Collaboration. T.S. was supported by a Postdoctoral Fellowship of the Deutsche Forschungsgemeinschaft. F.M.H. was supported by a Ph.D. fellowship of the Boehringer Ingelheim Fonds. Y.F. was supported by a visiting Ph.D. student scholarship from the China Scholarship Council (CSC). A.H. was supported by Caltech startup funds, the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award of the Edward Mallinckrodt Jr. Foundation, a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research, a Camille-Dreyfus Teacher Scholar Award, NIH grant R01-GM111461, and is a Heritage Principal Investigator of the Heritage Medical Research Institute. The coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 5HAX (Nup170NTD•Nup53R3), 5HAY (Nup170CTD), 5HAZ (Nup170CTD), 5HB0 (Nup170CTD•Nup145NR3), 5HB1 (Nup170SOL), 5HB2 (Nic96SOL), 5HB3 (Nic96SOL•Nup53R2), 5HB4 (Nup192ΔHEAD), 5HB5 (Nup145NAPD), 5HB6 (Nup145NAPD•Nup145CN), 5HB7 (Nup53RRM, space group P212121), 5HB8 (Nup53RRM, space group P3121). A PyMol session file containing the composite structure of the NPC symmetric core can be obtained from our webpage (http://ahweb.caltech.edu). This work is dedicated to the memory of Mandy Hoelz.
Footnotes
Supplementary Materials:
Materials and Methods
Figs. S1–S59
Tables S1–S10
References (50–67)
The authors declare no financial conflicts of interest.
References and Notes
- 1.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 2.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Sage V, Mouland AJ. Viral subversion of the nuclear pore complex. Viruses. 2013;5:2019–2042. doi: 10.3390/v5082019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol. Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woerner AC, et al. Cytoplasmic protein aggregates interfere with nucleo-cytoplasmic transport of protein and RNA. Science. 2015;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 7.Kaneb HM, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jovicic A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 11.Thierbach K, et al. Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure. 2013;21:1672–1682. doi: 10.1016/j.str.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 2015;22:425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bui KH, von Appen A, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 14.Stuwe T, Correia AR, et al. Architecture of the nuclear pore complex coat. Science. 2015;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuwe T, Bley CJ, Thierbach K, Petrovic S, et al. Architecture of the fungal nuclear pore inner ring complex. Science. 2015;350:56–64. doi: 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D. Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science. 2015;350:106–110. doi: 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- 17.Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol. 2015;22:774–781. doi: 10.1038/nsmb.3084. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhardt N, Redolfi J, Antonin W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci. 2014;127:908–921. doi: 10.1242/jcs.141739. [DOI] [PubMed] [Google Scholar]
- 19.Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA. 2014;111:2530–2535. doi: 10.1073/pnas.1311081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo HS, Blus BJ, Jankovic NZ, Blobel G. Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc. Natl. Acad. Sci. USA. 2013;110:16450–16455. doi: 10.1073/pnas.1316607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen KR, et al. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J. Biol. Chem. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrader N, et al. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J. Biol. Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- 26.Sampathkumar P, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 28.von Appen A, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teixeira MT, et al. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodel AE, et al. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 31.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 32.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 33.Kim SJ, et al. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol. Cell. Proteomics. 2014;13:2911–2926. doi: 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez DI, Lee TH, Sani MA, Aguilar MI, Separovic F. Proline facilitates membrane insertion of the antimicrobial peptide maculatin 1.1 via surface indentation and subsequent lipid disordering. Biophys J. 2013;104:1495–1507. doi: 10.1016/j.bpj.2013.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurell E, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Flemming D, et al. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 2012;177:99–105. doi: 10.1016/j.jsb.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Eibauer M, et al. Structure and gating of the nuclear pore complex. Nat. Commun. 2015;6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilokapic S, Schwartz TU. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2012;109:15241–15246. doi: 10.1073/pnas.1205151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo HS, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc. Natl. Acad. Sci. USA. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C, et al. Crystal structure of human nuclear pore complex component NUP43. FEBS Lett. 2015;589:3247–3253. doi: 10.1016/j.febslet.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Kim DI, et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ori A, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katta SS, Smoyer CJ, Jaspersen SL. Destination: inner nuclear membrane. Trends Cell Biol. 2014;24:221–229. doi: 10.1016/j.tcb.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 46.Ungricht R, Kutay U. Establishment of NE asymmetry-targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol. 2015;34:135–141. doi: 10.1016/j.ceb.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lusk CP, et al. Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic. 2007;8:647–660. doi: 10.1111/j.1600-0854.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- 49.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Generation of the Nup170•Nup53R3•Nup145NR3 complex structure by superposition. A 360° rotation of the crystal structure of Nup170SOL followed by a demonstration of the process used for generating the full-length structure by superposition. Colors are the same as in fig. S25.
Movie 2. Morph animation of observed Nup170 conformations. The eight different conformations observed in the superposition-generated structures of full-length Nup170 are animated in morph movies. The animation pauses at each of the eight experimentally observed conformations of Nup170, colored the same as in fig. S25.
Movie 3. Structure of the Nic96SOL•Nup53R2 complex. A 360° rotation of the crystal structure of Nic96SOL •Nup53R2. Colors are the same as in Fig. 4.
Movie 4. Generation of the structure of full-length Nup192 by superposition. A 360° rotation of the crystal structure of Nup192ΔHEAD followed by a demonstration of the process used for generating the full-length structure by superposition. Colors are the same as in fig. S31.
Movie 5. Overview of the composite structure of the NPC symmetric core. The structures are shown as docked into the cryoET reconstruction of the intact human NPC, oriented with the cytoplasmic face on top and the nuclear face on bottom. The nuclear envelope and density corresponding to protein density were colored in gray and white, respectively. Crystal structures of symmetric core nucleoporins are first shown in cartoon representation, colored according to Fig. 5, then without the cryoET reconstruction density, and finally in surface representation.
Movie 6. Rotating structure of a single spoke of the NPC symmetric core. The crystal structures are shown as docked into the cryoET reconstruction of the intact human NPC, oriented with the cytoplasmic face on top and the nuclear face on bottom. The nuclear envelope is colored in gray. Crystal structures of symmetric core nucleoporins are shown in cartoon representation, colored according to Fig. 6.