Abstract
In previous studies, we showed that the pathogenic fungus Cryptococcus neoformans (Cn) produces a specific and unique protein called antiphagocytic protein 1 (App1), which inhibits phagocytosis of Cn by alveolar macrophages (AMs). Phagocytosis of Cn by AMs occurs mainly through a complement- or Ab-mediated mechanism. Among AM receptors, complement receptor 3 (CR3) and FcRγ are the most common receptors involved in the phagocytic process. Because App1 inhibits phagocytosis of complement- but not Ab-coated erythrocytes, we investigated the role of CR3 in App1-macrophage interactions. We found that App1 binds to CR3 and if CR3 is absent from the surface of AMs, its antiphagocytic action is lost. When we investigated whether App1 would also bind to other complement receptor(s), we found that App1 does bind to complement receptor 2 (CR2) in a dose-dependent manner. In certain lymphoma cell lines, cellular proliferation is stimulated by complement through CR2, providing a potential use of App1 as a proliferation inhibitor of these cells. Initially discovered as an antiphagocytic protein regulating CR3-mediated innate immunity, App1 may also play a key role in the regulation of acquired immunity, because CR2 is mainly localized on B cells.
Antiphagocytic protein 1 (App1)4 is an antiphagocytic protein produced by the fungus Cryptococcus neoformans (Cn), an environmental human pathogen causing a life-threatening meningoencephalitis in immunocompromised patients. Upon inhalation, Cn interaction with alveolar macrophages (AMs) is the key for containment of the infection in the lung or dissemination of fungal cells through the bloodstream to the CNS. During the late 1980s and early 1990s, studies in the laboratory of B. Bolaños at the University of Puerto Rico (San Juan, Puerto Rico) identified and purified a Cn cytoplasmic factor involved in the inhibition of phagocytosis of fungal cells by mammalian macrophages. These early studies resulted in the partial isolation and purification from crude cytoplasmic extract of a 20-kDa protein. Because of such unique biological function, this protein was named App1. In recent years, we rediscovered App1 as a downstream target of the Cn sphingolipid pathway and showed that App1 was found in the culture supernatant of a Cn culture (1). We next demonstrated that App1 is transcriptionally controlled by inositol phosphoryl ceramide synthase through the production of diacylglycerol and the activating transcription factor 2 (1–3).
In our ongoing epistasis analysis to understand the mechanism(s) by which App1 inhibits phagocytosis, we produced rApp1 and created a Cn strain in which App1 was deleted (Cn Δapp1). The Cn Δapp1 is increasingly phagocytosed by macrophages compared with the Cn wild-type (WT) strain. Pharmacological treatment with increasing concentrations of rApp1 protein blocks the internalization of Cn in a dose-dependent manner (1). Because we found that rApp1 inhibits phagocytosis of complement- and not Ab-coated erythrocytes, we proposed that App1 exerts its antiphagocytic action against Cn by inhibiting complement- and not Ab-mediated phagocytosis.
The complement system is a collection of circulating and cell membrane proteins that play an important role in host defense against microbes. The most abundant complement protein in the plasma is C3. Its first cleavage product, C3b, is further degraded to iC3b, C3c, and C3dg, which serve as ligands for selective complement receptors on leukocytes (4, 5). Complement receptor 3 (CR3) is present on the surface of monocytes, macrophages, and dendritic cells, and is composed of two subunits, CD11b and CD18, and it mainly serves as the receptor for internalization of iC3b-opsonized microbes, such as Cn (reviewed in Refs. 6 and 7).
Among other complement receptors, complement receptor 2 (CR2; CD21) also binds iC3b, although its main ligands are C3d and C3dg. Instead of being localized on phagocytic cells, CR2 is mainly localized on the surface of B cells and is involved in B cell activation and differentiation (8, 9). In Burkitt's lymphoma, a non-Hodgkin lymphoma of high malignancy produced by the EBV infection of B cells, CR2 is particularly important because not only does it serve as receptor for EBV, but, through binding with its complement ligand(s), it stimulates tumor cellular proliferation (10).
In this study, we show that App1 binds to CR3 and the inhibition of phagocytosis by rApp1 is completely lost in AMs in which CR3 is absent. We show that App1 is secreted, either actively or passively, perhaps through capsule shedding. We also show that App1 binds to CR2, providing a new potential role of App1 in the adaptive immune response against Cn.
Materials and Methods
Strains, cell cultures, and growing medium
The following strains were used in this study: the Cn variety grubii serotype A strain H99 (WT), Δapp1 knockout strain, and the Δapp1Rec strain, which were created from H99 strain; Cn variety gattii serotype B strain MMRL 1336; Cn variety neoformans serotype C strain MMRL 1343; and Cn variety neoformans serotype D strain JEC21. Cn strains were routinely grown on yeast peptone dextrose medium. The Δapp1 strain was created through a double-crossover event replacing the APP1 gene open reading frame with the ADE2 gene. PCR and Southern and Western blotting were used to confirm the absence of the APP1 gene and App1 protein (1). The Δapp1Rec strain was created by reintroducing the APP1 gene back into the Δapp1 strain and specifically in the APP1 locus. This was demonstrated by Southern analysis (1). The absence of App1 in the Δapp1 strain did not have an effect on capsule formation, melanin production, or growth at 30°C, 37°C (pH 7.0 or 4.0) (1). Lysing enzyme from Tricoderma harzianum was from Sigma-Aldrich.
Chinese hamster ovary (CHO) cell lines stably transfected with human CR3 (CD18/CD11b), human CD14, or neomycin (NEO) resistance vector alone were a gift from D. Golenbock (Boston Medical Center, Boston, MA). Expression of CR3 and CD14 was determined by flow cytometry (data not shown). CHO cell lines were cultured in α MEM (Life Technologies; catalogue 12571) with 10% heat-inactivated FBS (Life Technologies; catalogue 10082-147) in G418 (final concentration 0.4 mg/ml) and 1% penicillin-streptomycin. rApp1 protein and phosphoglucose isomerase from methanosarcina (mPGI) protein were produced by D. Fedarovich in the Medical University of South Carolina Center of Biomedical Research Excellence Protein Core Facility (Charleston, SC) directed by C. Davies.
Production and purification of rApp1 and mAbs and polyclonal Ab (pAb) against App1
Large-scale production of rApp1 protein was performed as follows. The pBAD-His-App1 vector generated previously (1) was used for protein expression. Escherichia coli TOP10 cells containing pBAD-His-App1 plasmid were inoculated into 10 ml of Luria-Bertani medium supplemented with 50 μg/ml ampicillin and grown overnight at 37°C, 220 rpm. Then, 2 ml of cells was each inoculated into 5 × 1 L of Luria-Bertani medium with 50 μg/ml ampicillin and grown to an OD600 of ~0.6–0.8. The culture was induced with 20 μg/ml l-arabinose and grown overnight at 30°C, 220 rpm. Cells were collected by spinning at 4500 rpm for 10 min, and the cell pellet was frozen at −80°C for 2 h. Frozen cells were then thawed and resuspended in cold buffer A (10 mM Tris-HCl, 0.15 M NaCl, 1% Triton X-100, and 1 mM EDTA). Next, 1 mM PMSF was freshly added just before sonication on ice. The mixture was then incubated with 0.2 mg/ml lysozyme at room temperature for 1 h and then centrifuged at 10,000 rpm for 40 min at 4°C. Inclusion bodies were washed twice with 25 ml of buffer A, once with 25 ml of buffer B (10 mM Tris-HCl, 0.15M NaCl, and 1 mM EDTA), and once with 25 ml of distilled water. The inclusion bodies were incubated overnight in 10 ml of 8 M urea at 4°C on a rotary shaker. The next morning, the supernatant was centrifuged at 10,000 rpm for 30 min at 4°C, then added to 1 L of buffer C (50 mM KH2PO4, 20 mM Tris, 0.15M NaCl, 10% glycerol, and 2 mM β-ME (pH 8)) to allow for refolding of App1. The protein solution was loaded onto a pre-equilibrated (buffer D, 20 mM Tris-HCl, 0.5M NaCl, and 2 mM 2-ME (pH 7.8)) HisTrap HP 5-ml column (GE Healthcare) using a peristaltic pump P-1 (GE Healthcare). The column was washed with 100% buffer D, followed by 98% D and 2% E (20 mM Tris-HCl, 0.5 M NaCl, 0.5 M Imidazole, and 2 mM 2-ME (pH 7.8)) and then 85% D and 15% E. App1 was eluted with 40% D and 60% E. The fractions containing App1 were dialyzed twice with 1 L of buffer F (50 mM Tris-HCl/0.15 M NaCl/5% glycerol/2 mM 2-ME (pH 7.9)) at 4°C, and then concentrated.
For the production of App1 full-length and truncated forms, we used the pGEX-6P-3 plasmid (Amersham Biosciences), which contains the GST tag. First, a PCR fragment was generated using primers App1-BamH1-F and App1–3′-pRSET/pGEX, 5′-TTC GAA TTC TCA AAT CAT CAA TGT TCG CAG CTC-3′, using the pBAD-His-App1 plasmid as a template. The resulting 568-bp fragment was digested with BamHI and EcoRI and cloned into the BamHI- and EcoRI-restricted pGEX-6P-3 plasmid, generating pGEX-App1 (1–181) plasmid. Next, a series of plasmid carrying truncated forms of App1 proteins were generated for studying epitope recognition by Abs. Through a series of PCR and subcloning procedures, we created the following plasmids: pGEX-App1 (1–33), pGEX-App1 (33–90), pGEX-App1 (90–140), and pGEX-App1 (140–181), as indicated.
Production of CR2
Soluble CR2 was produced as follows: CR2-FH cells were added to a cell culture dish containing 10 ml of medium (made up of 500 ml of DMEM, 50 ml of dialyzed and inactivated FBS, 10 ml of glutamine synthetase expression medium supplement, 280 μl of 100 μM l-methionine sulfoximine, and 5 ml of penicillin-streptomycin). When the cells reached 90–100% confluency, they were trypsinized and split into five-cell culture dishes each with 10 ml of medium. Upon 100% confluency, the cells were trypsinized, centrifuged at 800 rpm for 5 min, and resuspended into 500 ml of medium. A total of 2 × 250 ml was then added to 2 × 235-cm2 Corning Glass CellBIND surface-expanded surface flasks and incubated for 4–7 days at 37°C, 5% CO2, and 100% relative humidity. Soluble CR2 was harvested by filtering the supernatant through a 0.22-μm Express Plus Stericup filter, then loading it onto a PBS-equilibrated HisTrap FF column containing anti-CR2 Ab using a peristaltic pump P-1. The column was washed with PBS, and the protein was eluted with 0.1 M glycine-HCl (pH 2.7). The fractions containing the protein were concentrated using an Amicon Centricon Plus-20 filter device with a 5000 Da molecular weight cutoff and the same device used for buffer exchange to PBS.
App1-CR3-binding assay
CHO cells expressing CR3 (CHO-CR3), CD14 (CHO-CD14), or empty plasmid (CHO-NEO) were a gift from D. Golenbock (Boston Medical Center, Boston, MA). CHO-CR3, CHO-CD14, or CHO-NEO were grown as monolayers in six-well culture dishes at a density of 0.025 × 104. When cells were confluent, wells were aspirated dry, washed with 10% FBS medium, and then resuspended in 1 ml of 10% FBS medium with 1 μg of rApp1/ml or with 1 μg of rApp1/ml and Ab anti-human CD11b monoclonal (Serotec) diluted 1/400. For binding studies, cells were incubated with rApp1 at 37°C for 1 h by gently shaking, and the unbound rApp1 was removed by generously washing the wells three times with 10% FBS medium. Cells were also incubated with ~500 μg of total cell protein lysate obtained from Cn WT H99 or Δapp1 mutant strain. Cell protein lysate from H99 strain would contain native App1 (nApp1) protein, whereas cell protein lysate from Δapp1 would contain all Cn proteins except App1 and was used as a negative control. Cells were incubated with protein lysate at 37°C for 1 h by gently shaking, and the unbound proteins were removed by generously washing the wells three times with 10% FBS medium.
Cells were then collected by scraping with 1 ml of cold PBS, centrifuged at 1200 rpm, and resuspended in 30 μl of 1× Laemmli sample buffer (Bio-Rad). Samples were then analyzed by a Western blot using anti-App1 4-6H Ab. For saturation experiments, the same protocol was followed with CHO-CR3 cells, with increasing concentrations of App1 from 0 to 1000 μg/ml. The resulting Western blot was semiquantified using LabWorks software to measure the total raw density of each band while subtracting the background.
Phagocytosis assay
Ex vivo phagocytosis was performed in C57BL/6J (CR3+/+) and/or B6.129S4-itgamtm1Myd/J (CR3−/−) mice. The CR3−/− are isogenic of C57BL/6J mice. Briefly, mice were anesthetized with an i.p. injection of 60 μl of xylazine/ketamine mixture, containing 95 mg of ketamine/kg body weight and 5 mg of xylazine/kg body weight. Then, Cn WT, Δapp1, or Δapp1Rec strains were inoculated intranasally. Three mice were used for each Cn strain. After 2 h, mice were euthanized by CO2 inhalation, and AMs and yeast cells were collected by broncoalveolar lavage. Cells were then centrifuged at 1200 rpm and resuspended in 500 μl of PBS, and 10 μl was plated in a standard hemocytometer for the determination of the number of yeast cells attached and/or ingested by AMs (phagocytic index). Results represent the geometric means ± SDs of the phagocytic indexes obtained from three animals per strain. In vitro phagocytosis assay was performed as follows. Fresh AMs were collected by bronchoalveolar lavage. For each mouse, ~3–5 × 105 AMs were obtained. Total cell count was assessed in a standard hemocytometer using a cell suspension diluted 1/1 with trypan blue. A total of 3 × 104 AMs was plated into each well of a 96-well microtiter plate (BD Biosciences) in 100 μl of DMEM (Invitrogen Life Technology) containing glucose with or without 10% fresh mouse serum, 1% l-glutamine, and 100 U/ml penicillin-streptomycin, and incubated at 37°C (5% CO2) for 2 h. Nonadherent AMs were removed by washing the wells with warm medium.
Cn cells were prepared, as previously described. Briefly, from a fresh overnight culture, Cn cells were washed twice with sterile PBS (pH 7.4) and resuspended in DMEM with or without 10% fresh mouse serum. Next, 6 × 105 Cn cells together with different concentration of rApp1 protein (as indicated) were added to AMs and plates were incubated at 37°C (5% CO2) for 2 h. Nonattached Cn cells were removed by washing with PBS. Cells were then fixed with ice-cold methanol and stained with Giemsa (Sigma-Aldrich). The phagocytic indexes (defined as the number of internalized yeast cells/100 macrophages) were determined for each set of experimental conditions. Eight fields per experiment were counted and averaged. Results represent the geometric means ± SDs of the phagocytic indexes obtained from three different experiments per strain.
App1-CR2-binding assay: ELISA
The assay was performed in a 96-well microtiter plate (Maxisorp NUNC). First, the wells were coated with 50 μl/well of 5 μg/ml extracellular domain of mouse CR2 in coating buffer (5 mM Na2CO3, 35 mM NaHCO3 (pH 9.6)) and incubated overnight at 4°C. The plate was then blocked with 2% BSA in PBS, incubated 2 h at 37°C, and then washed three times with PBS/0.1% Tween 20 (PBST). A total of 1 μg/50 μl/well rApp1 in 1% BSA and 1× PBS was added and incubated overnight at 4°C. After three washes with PBST, 50 μl of anti-App1 mAb/well, diluted 1/20 with 1% BSA/PBS, was added and incubated 2 h at 37°C. The plate was washed three times with PBST and incubated with 50 μl of mouse secondary Ab anti-IgG HRP/well (Jackson ImmunoResearch Laboratories) diluted 1/5000 with 1% BSA/PBS, for 2 h at 37°C. After three washes with PBST, the color was developed with 50 μl/well 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich). The reaction was then stopped with 50 μl of 2 M H2SO4, and the plate was read at 450 nm with a VersaMax plate reader. For App1-iC3b-binding assay, 8 μg/50 μl/well human iC3b (Complement Technology) diluted in 1% BSA/1× PBS was incubated 20 min with CR2 coated on the bottom of the plate, and then rApp1 (1 μg/50 μl/well) was added and incubated overnight at 4°C. For competition assay, different dilutions of mAb 4-6H and 3-11F were preincubated with 1 μg of rApp1 for 1 h at 37°C, and then added to the CR2-coated wells and incubated overnight at 4°C. Plates were then processed as described above.
Immunoprecipitation of App1
A starter culture of H99 was prepared in yeast peptone dextrose (10 ml) and incubated for 24 h at 30°C, 250 rpm. A 1/40 dilution in yeast nitrogen base was then conducted, and the culture was incubated 24 h at 30°C, 250 rpm. The culture was centrifuged at 2800 rpm, and the cells were washed three times in 0.5 M NaCl/50 mM EDTA, then resuspended in sterile water and incubated 1 h at 37°C, with occasional mixing. Following incubation, the cells were centrifuged at 2800 rpm and resuspended in spheroplastic solution (1 ml) consisting of 1 M sorbitol, 0.1 M sodium citrate (pH 5.8), and 0.01 M EDTA with or without 10 mg of lysing enzyme. Cells were incubated for 3.5 h at 37°C with occasional mixing. The sample was then centrifuged at 1000 rpm for 10 min, and the resulting supernatant was centrifuged again for 20 min at 10,000 × g at 4°C. The supernatant was carefully transferred to a sterile tube and placed on ice. A total of 50 μl of protein G-Sepharose resin slurry/1 ml of supernatant was added to preclear the lysate. The sample was rocked at 4°C for 1 h, and then centrifuged at 10,000 × g at 4°C for 1 min. The supernatant was transferred to a sterile tube and placed on ice. Anti-App1 pAb 545 (1/50 dilution) and 50 μl/ml protein G-Sepharose resin slurry were added to the supernatant, and the sample was rocked at 4°C for 24 h. Following centrifugation at 10,000 × g, 4°C for 1 min, the supernatant was removed and the resin was washed twice with spheroplastic solution (without lysing enzyme) before being used for SDS-PAGE and Western blot.
Capture ELISA
The assay was performed in a 96-well microtiter plate (Maxisorp; Nunc). First, the wells were coated with 50 μl/well mAb 4-6H at 1/20 dilution in coating buffer (5 mM Na2CO3, 35 mM NaHCO3 (pH 9.6)) and incubated overnight at 4°C. The plate was then blocked with 2% BSA in PBS, incubated 2 h at 37°C, and then washed three times with PBST. Varying concentrations of rApp1 in 1% BSA in PBS were used for standard curve. A total of 50 μl of medium (treated or untreated with lysing enzyme) was added and incubated overnight at 4°C. After three washes with PBST, 50 μl of anti-App1 pAb was diluted 1/5,000 with 1% BSA/PBS, added to each well, and incubated 2 h at 37°C. The plate was washed three times with PBST and incubated with 50 μl of rabbit secondary Ab anti-IgG HRP per well (Jackson ImmunoResearch Laboratories) diluted 1/20,000 with 1% BSA/PBS, for 2 h at 37°C. After three washes with PBST, the color was developed with 50 μl/well 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich). The reaction was then stopped with 50 μl of 2 M H2SO4, and the plate was read at 450 nm with a VersaMax plate reader. The experiments were performed twice.
Statistics
Statistical analysis was performed using two-tailed Student's t test.
Results
Production and characterization of anti-App1 Abs
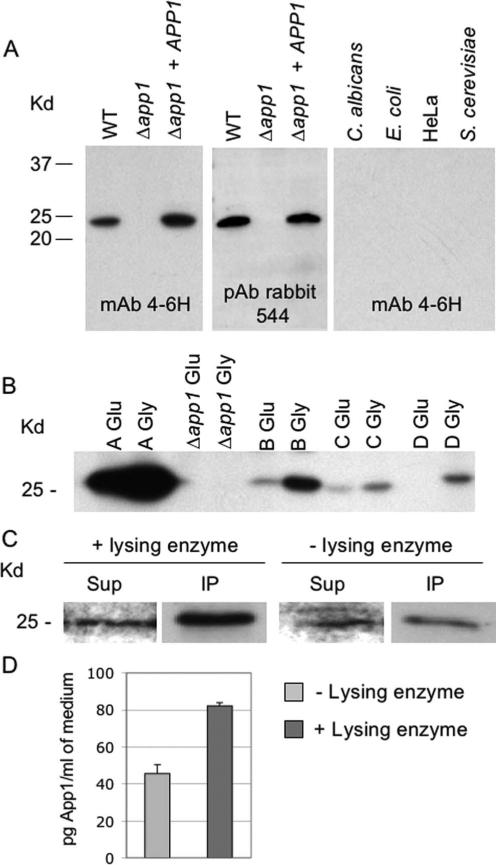
To study the antiphagocytic function of App1 in vivo and in vitro, we produced anti-App1 mAb and pAb (Fig. 1). These tools were necessary to study the function of App1 at the molecular level. We produced two mouse mAb (mAb 4-6H and mAb 3-11F) and two rabbit pAb (pAb 544 and pAb 545). Fig. 1A illustrates that mAb 4-6H and pAb 544 react with App1 produced by WT Cn or Δapp1 + APP1 reconstituted strain, but not with Δapp1 strain. App1 appears to be a Cn-specific protein because it is not found in Candida albicans, E. coli, the cervical human cancer HeLa cell line, or Saccharomyces cerevisiae. Comparing the production of App1 among different serotypes of Cn, App1 is mainly produced by Cn serotype A strain compared with other serotypes. Interestingly, its production is significantly up-regulated when cells are grown in presence of glycerol compared with glucose as the sole carbon source (Fig. 1B). These results suggest that App1 is a Cn-specific protein, which is differentially expressed by different Cn serotypes.
FIGURE 1.
Production of anti-App1 mAb and pAb. A, Western blot analysis of App1 production in Cn WT Δapp1, Δapp1 + APP1 reconstituted strains, C. albicans, E. coli, cervical human cancer HeLa cell line, and S. cerevisiae using mAb 4-6H and pAb 544. B, Production of App1 by different Cn serotypes (A–D), as indicated, grown in glucose (Glu) or glycerol (Gly) as a sole carbon source. C, App1 protein is found in the medium and can be immunoprecipitated using pAb 544. Treatment with lysing enzyme increases the amount of App1 in the medium. Sup, supernatant; IP, immunoprecipitation. D, Capture ELISA of App1 present in the medium before (−) and after (+) treatment with lysing enzyme.
To exert its antiphagocytic function, Cn App1 should be either secreted and/or exposed on the surface of Cn (capsule/cell wall). Because we already found that App1 is present in the supernatant (1), we wondered whether digesting Cn cells with lysing enzyme would enrich its presence in the medium. Lysing enzyme from T. harzianum hydrolyzes poly(1–3)-glucose of the yeast glucan-releasing cell wall and capsule materials in the medium. Unexpectedly, we found that upon treatment with lysing enzyme, App1 protein is enriched in the medium compared with untreated cells (Fig. 1, C and D), suggesting that App1 may interact with capsule and/or cell wall materials.
App1 binds to CHO cells expressing CR3
Because in our previous studies we showed that rApp1 blocks the phagocytosis of complement- and not Ab-coated erythrocytes, we reasoned that App1 would inhibit the complement-mediated phagocytosis of Cn. Among complement receptors, CR3 (CD11b/CD18) on the surface of AMs plays a major role in the attachment and internalization of many fungi, including C. albicans (11), Blastomyces dermatitidis (12), and Cn (13–15). Thus, we decided to focus our attention on CR3.
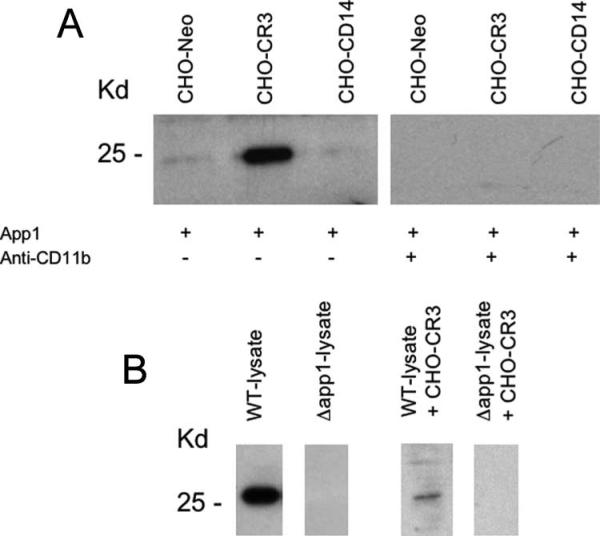
To begin studying the App1-CR3 interaction, we tested whether App1 would interact with a CHO cell line expressing human CR3 (CHO-CR3). As negative controls, we used CHO cells expressing CD14 (CHO-CD14) or an empty vector (CHO-NEO). The expression of the indicated receptor on CHO cells was confirmed and quantified by flow cytometry analysis (data not shown). rApp1 was incubated with CHO-CR3, CHO-CD14, and CHO-NEO, as indicated in Fig. 2A. Cells were then washed and collected, and membrane proteins were loaded for SDS-PAGE and Western blot analysis using anti-App1 mAb 4-6H. As illustrated in Fig. 2A, rApp1 binds to CHO-CR3, but not to CHO-CD14 or CHO-NEO.
FIGURE 2.
rApp1 and nApp1 bind to CHO-CR3. A, rApp1 was incubated with CHO expressing CR3 (CHO-CR3), CHO expressing CD14 (CHO-CD14), or CHO expressing empty vector (CHO-Neo). rApp1 specifically binds to CHO-CR3, but not to CHO-CD14 or CHO-Neo cells. The addition of anti-CD11b Ab, which binds to CD11b (one of the two subunits of CR3), completely abolished App1 binding. B, Lysate protein extracts from WT strain (WT-lysate) containing nApp1 or lysate protein extracts from Δapp1 strain (Δapp1-lysate) were incubated with CHO-CR3 (WT-lysate + CHO-CR3 or Δapp1-lysate + CHO-CR3). nApp1 binds to CR3.
As mentioned above, CR3 is comprised of two subunits: CD11b and CD18. Thus, we wondered whether App1 would bind to CD11b and/or CD18. As shown in Fig. 2A, coincubation with anti-CD11b Ab totally abrogates the interaction between rApp1 and CHO-CR3. Coincubation with anti-CD18 Ab did not inhibit rApp1 binding to CHO-CR3 (data not shown). These results suggest that rApp1 may bind to the CD11b subunit of CR3.
To demonstrate that Cn nApp1 binds to CR3, we incubated the cell lysate obtained from Cn WT or Δapp1 strain with CHO-CR3 cell line. After incubation, total proteins were extracted and analyzed by Western blot using mAb 4-6H. As illustrated in Fig. 2B, nApp1 was detected only when total proteins obtained from Cn WT, and not Δapp1, were incubated with CHO-CR3, indicating that nApp1 interacts with CR3.
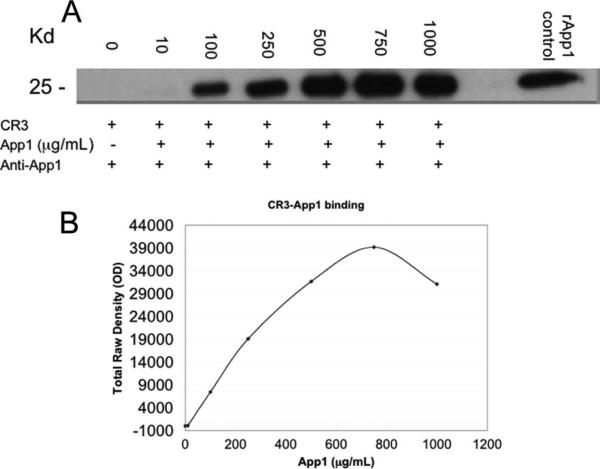
To further prove the binding between App1 and CR3, we tested whether the binding was saturable. CHO cells expressing CR3 were incubated with increasing amounts of App1. As shown in Fig. 3, we found that the binding between App1 and CR3 is saturable, as the binding curve plateaus. Proving the binding is saturable further proves the App1 and CR3 interaction.
FIGURE 3.
The binding of CR3 and rApp1 is saturable. A, CHO-CR3 cells were incubated with increasing concentrations of rApp1 from 0 to 1000 μg/ml. The cells were washed and lysed, and proteins were extracted and run on a Western blot. rApp1 was also run as a positive control for App1 on the Western blot. B, The bands from the Western blot were semi-quantified using LabWorks software to measure the total raw density of each band while subtracting the background. Data are representative of three independent experiments.
The antiphagocytic action of App1 depends on the presence of CD11b
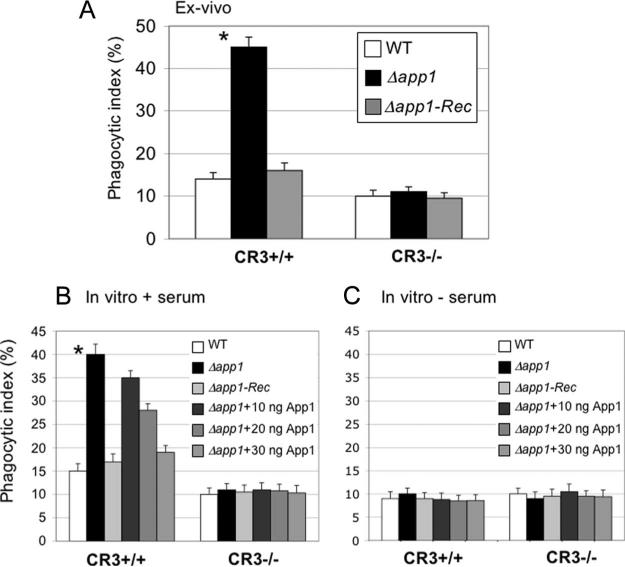
To establish the role of CD11b in Cn phagocytosis, we sought to perform an ex vivo phagocytosis using CR3+/+ WT (C57BL/6J) and CR3−/− mice in which CD11b is deleted (Jackson B6.129S4-itgamtm1Myd/J). The CR3−/− mouse model is an isogenic model of C57BL/6J. Thus, we measured the phagocytic index of Cn WT, Δapp1, and Δapp1 + APP1 (Δapp1Rec) strains after 2 h of lung infection. As shown in Fig. 4A, CR3+/+ AMs readily phagocytosed Cn Δapp1 compared with WT or Δapp1Rec strain. Interestingly, when CD11b is absent (CR3−/−), the Δapp1 strain is phagocytosed by AMs similarly to the WT or the Δapp1Rec strains. These results suggest that CD11b is required to efficiently internalize Δapp1.
FIGURE 4.
The antiphagocytic action of App1 is lost when CD11b subunit is absent in AMs. A, Ex vivo phagocytosis of Cn WT, Δapp1, and Δapp1 + APP1 (Δapp1Rec) strains after 2 h of intranasal inoculation of C57BL/6J (CR3+/+) or B6.129S4-itgamtm1Myd/J (CR3−/−) mice, in which the CD11b (and not the CD18) subunit is absent. Phagocytosis of Cn Δapp1 strain increases in CR3+/+ AMs compared with Cn WT (*, p < 0.05), whereas it does not in CR3−/− AMs. B, Treatment with rApp1 restores WT phenotype in CR3+/+ AMs, whereas rApp1 treatment does not affect phagocytosis when CD11b subunit is absent from the surface of AMs (CR3−/−) (*, p < 0.05). C, Treatment with rApp1 has no effect on phagocytosis when Cn cells are not opsonized with serum regardless of the presence of CR3.
Next, we tested the effect of rApp1 on Cn phagocytosis by CR3+/+ and CR3−/− AMs in vitro. Whereas treatment with rApp1 decreases phagocytosis in CR3+/+ AMs of Cn Δapp1 in a dose-dependent manner, treatment with rApp1 did not affect the internalization of Cn WT, Δapp1, or Δapp1Rec strains by AMs lacking CD11b (Fig. 4B), and this effect is lost when Cn cells are not opsonized with serum (Fig. 4C). These results suggest that App1 exerts its antiphagocytic action through the CD11b subunit of CR3 by blocking the interaction of iC3b on the surface of Cn with CR3. As expected, the overall phagocytosis of Cn WT strain in CR3−/− was decreased compared with CR3+/+, although not completely abolished, because Cn is also phagocytosed through receptors other than complement (7, 16).
App1 binds to CR2 in vitro, and this binding is inhibited by iC3b
The results showing that App1 binds to CR3 raise the question of whether it also binds to other complement receptor(s). Thus, in addition to CR3, we studied App1-CR2 interaction. We selected CR2 because of the following: 1) the secondary structure of App1 is similar to C3d, which is the natural ligand of CR2; and 2) App1 contains an acidic pocket between aa 33 and 90 that shows high similarity to the amino acids found in C3d required for binding to CR2.
CR2 is a complement receptor mainly found on B cells, although it can also be present on a subset of T and dendritic cells (17). CR2 principally binds C3d, but can also bind iC3b and C3dg complement ligands. Notably, the molecular interaction among C3, C3d, and CR2 has been recently solved (18, 19), and the amino acids of the ligand interacting with the receptor have been identified (20, 21).
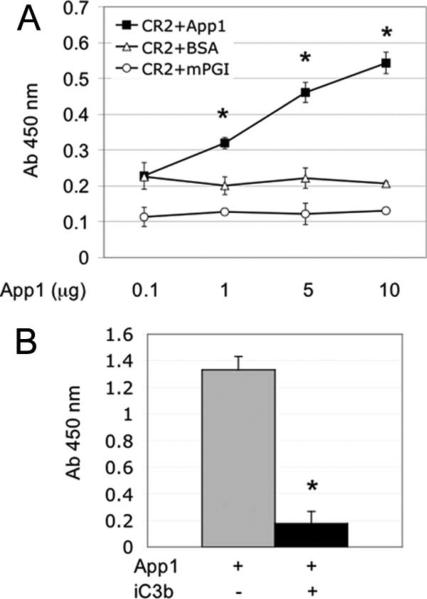
Thus, we examined the App1-CR2 interaction using an ELISA. The CR2 N-terminal short-consensus repeats responsible for the C3d-CR2 binding (short consensus repeat 1–4, residues 1–257 of mature protein, GenBank M35684) were expressed and purified, as described in Materials and Methods. The N terminus mouse rCR2 was absorbed onto an ELISA plate, rApp1 was added, and rCR2-rApp1 interaction was examined using mAb against App1. As shown in Fig. 5A, we found a dose- and time-dependent interaction between rApp1 and rCR2, whereas no interaction was observed between CR2 and BSA or CR2 and mPGI, a His-tagged control protein used to ensure that the effect seen for rApp1 is specific. Importantly, preincubation of an equimolar concentration iC3b completely abolished the binding of rApp1 to rCR2 (Fig. 5B). These results suggest that, in addition to CR3, rApp1 interacts also with CR2.
FIGURE 5.
App1 binds to CR2. A, CR2 recombinant protein was coated onto an ELISA plate, and App1, mPGI, or BSA was added at different concentrations, as indicated. Binding was detected using mAb 4-6H anti-App1. Significant binding of rApp1 to CR2 is observed at 1, 5, and 10 μg (*, CR2-App1 vs CR2-BSA; p < 0.05). B, Coincubation of App1 and iC3b (in equimolar concentrations) to CR2 blocks App1-CR2 binding almost completely (*, p < 0.001).
App1 epitope recognition by anti-App1 Abs
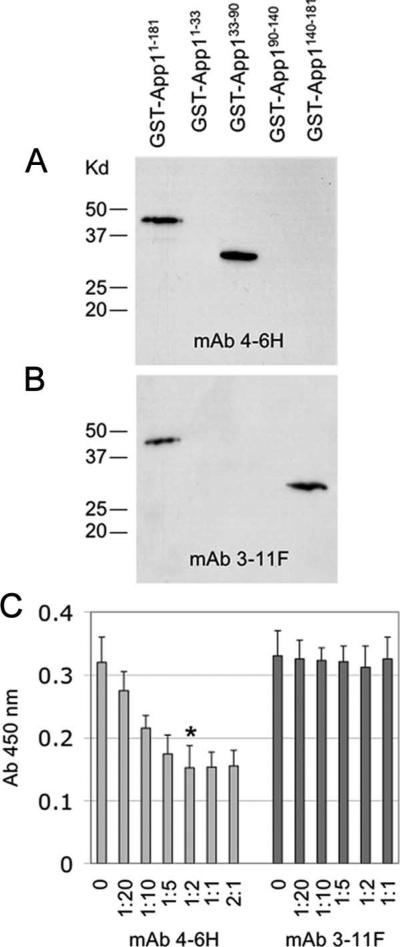
Both mAb and pAb anti-App1 were tested for App1 epitope recognition. The rApp1 full-length and different App1-truncated forms were loaded on a SDS-PAGE, and Western blot was performed using either mAb 4-6H (Fig. 6A) or 3-11F (Fig. 6B), or pAb rabbit 544 (data not shown). The results show that mAb 4-6H recognizes a domain present within 33–90, whereas mAb 3-11F recognizes a domain present within 140–181. Polyclonal 544 and 545 recognizes all domains, as expected (data not shown).
FIGURE 6.
mAb 4-6H interacts with App1 differently than mAb 3-11F. A, Western blot analysis showing that mAb 4-6H interacts with an epitope within aa 33 and 90, whereas B, mAb 3-11F interacts with App1 within aa 140–181. C, Preincubation of increasing concentrations of mAb 4-6H, but not mAb 3-11F, to 1 μg of App1 partially inhibits App1-CR2 interaction. *, 1:2 vs 0, p < 0.05.
Because of these difference between mAb 4-6H and mAb 3-11F, we wondered whether a preincubation of rApp1 with its mAb would interfere with CR2 binding. Interestingly, preincubation with anti-App1 mAb 4-6H inhibited rApp1-rCR2 interaction by 50%, whereas incubation with anti-App1 mAb 3-11F did not show any inhibition of App1-CR2 interaction (Fig. 6C). These results suggest that rApp1 binds to CR2 and possibly, but not exclusively, with a domain within aa 33–90.
Discussion
Cn is a facultative intracellular pathogen and, as such, it has the ability to survive intra- and extracellularly. Whereas the identification of specific fungal factor(s) or molecule(s) that allows Cn to survive intracellularly is the focus of intensive investigations, little is known about the factors that would allow Cn to escape phagocytosis and survive extracellularly, such as in alveolar space, bloodstream, and extracellular tissues. The production of the polysaccharide capsule provides effective protection to extracellular yeast cells against AMs and other host immune cells, because the capsule material negatively affects diverse aspects of the host immune response (22, 25). In addition to the polysaccharide capsule, Cn produces certain glycosphingolipid(s), such as glucosylceramide, which is required for its growth in extracellular environments characterized by high CO2 and alkaline pH (26) and to transport Cn polysaccharide outside fungal cells (27).
In recent years, we identified a Cn factor (App1) that specifically regulates the internalization of Cn by AMs in vitro and ex vivo, and, most likely, also during the infection because App1 was found in sera of AIDS patients affected with cryptococcosis (1). When we investigated whether App1 would regulate pathogenicity of Cn in murine models of cryptococcosis, we found the intriguing result that, compared with Cn WT strain, Δapp1 mutant is hypervirulent in immunocompromised mouse models lacking T and NK cells (Tgε26), but hypovirulent in complement-deficient model (A/Jcr) (1) or immunocompetent mouse models, such as CBA/J and C57BL/6 (data not shown). We hypothesized that the App1 phenotype is due to the unique ability of the protein to modulate phagocytosis. If more Cn cells will be internalized by AMs (lower production of App1) and AMs cannot effectively kill them, phagocytosis can be considered an opportunity of the fungus to cause disease. Under this condition, the host could benefit from a switch of fungal cells from the intra- to the extracellular environment by specifically delivering App1 in the infected lung.
In this work, we studied how App1 blocks phagocytosis of Cn, and the results led us to explore whether App1 would bind to complement receptors other than CR3, such as CR2. CR3 is an opsonic receptor present on the surface of phagocytic cells. It recognizes complement fragment iC3b deposited on microbial surface, such as Cn, and it regulates phagocytosis. CR3 is comprised of two subunits, CD11b, which binds iC3b, and fungal β glucan and CD18, which bind, in the case of Cn, glucuronoxylomannan, which is the major component of the Cn polysaccharide capsule. CR2 is found on the surface of B and follicular dendritic cells (28), and in many subsets of T cells, including CD4+ and CD8+ (29, 30). CR2 can bind many complement fragments, such as C3b, iC3b, C3dg, and C3d (6, 31, 32). It is also called CD21 and, instead of being involved in the regulation of phagocytosis, CR2 plays a key role in the enhancement of humoral immune responses (33, 34) by linking the binding of specific ligands to signal transduction events mediated by other members of the CD21/CD19 complex on B lymphocytes (35, 36). Expression of CR2 is also required on follicular dendritic cells for robust humoral immune response, probably by mediating Ag trapping or cell-cell interactions in lymphoid organs (17).
The interaction of App1 with CR3 is shown by using CHO cells expressing human CR3. App1 interacts with CHO-CR3, but not with CHO-CD14 or CHO-NEO cells. The total absence of this interaction when anti-CD11b Ab is coincubated suggests that App1 binds to CD11b. There is still a possibility that App1 binds to CD18 and that inhibition of anti-CD11b is due to steric hindrance.
To add more proof of the binding between App1 and CR3, we show that binding of rApp1 to CR3 is saturable and that nApp1 also binds to CR3. Moreover, the fact that App1 exerts its effect through CR3 is supported by the total lack of its antiphagocytic effect when AMs from CR3−/− mice lacking CD11b subunit are used for assessing phagocytosis of Cn. Fig. 4 clearly shows that presence of App1 in WT and reconstituted cells blocks phagocytosis in CR3+/+, but not in CR3−/− AMs. In fact, loss of App1 (Δapp1) dramatically increases phagocytosis of Cn by CR3-positive, but not by CR3-negative cells. This suggests the following: 1) it is the activity of App1 that, in WT Cn, makes CR3-mediated phagocytosis marginal compared with phagocytosis mediated by other processes (compare phagocytic indexes of WT Cn in CR3+/+ vs CR3−/−); and 2) App1 is extremely effective in inhibiting CR3-mediated phagocytosis because only ~4% of phagocytic index of WT Cn is due to CR3 (difference between WT Cn in CR3+/+ and CR3−/−), whereas ~35% of phagocytic index of Δapp1 is due to CR3 (difference between Cn Δapp1 in CR3+/+ vs CR3−/−). Moreover, this phagocytic effect of App1 through CR3 is further confirmed by the requirement of serum, and therefore complement, for App1 antiphagocytic activity. These results suggest that the App1 produced by Cn, either the one found in the medium and/or the one associated with the cell wall/capsule, interacts with CD11b on the surface of AMs, and this interaction prevents the binding of iC3b-opsonized Cn cells and, thus, phagocytosis.
In addition to CR3, App1 appears to interact with CR2. The interaction of App1 with CR2 in vitro is shown by ELISA using rCR2. We hypothesize that App1 interacts with CR3 or CR2 possibly through the acidic binding motifs present in App1 protein within aa 33 and 90. This hypothesis is supported by the following observations: 1) the secondary structure of App1 is similar to the secondary structure of complement ligands of CR2 and CR3; 2) the acidic domain of C3d interacting with CR2 is similar to an acidic domain found within 33–90; and 3) App1-CR2 interaction is partially inhibited by mAb 4-6H, which recognizes 33–90, but not with 3-11F, which recognizes 140–181. It is interesting, however, that mAb 4-6H does not totally abolish the interaction of App1 with CR2, suggesting that App1 interacts with CR2 with an additional epitope that is not recognized by mAb 4-6H. The competition studies showing that the App1-CR3 interaction is abolished by anti-CD11b, but not anti-CD18 Abs suggest that App1 and iC3b bind to the same domain of the CD11b subunit of CR3. The possibility that App1 and iC3b bind to the same domain is indirectly supported by the fact that iC3b competes with App1 for CR2. Further structural studies are warranted to determine the physical interaction between App1 and CR3 or CR2.
The competition between App1 and iC3b, as well as the binding of App1 to CR2 and CR3, which are expressed on a variety of cells, can lead to numerous effects during Cn infection, as follows. 1) The inhibition of AMs’ phagocytosis by App1 can affect the initiation of the infection in the lung and potentially the dissemination from the lung to other organs, because AMs may transport Cn cells outside the lung (1, 37). This possibility is supported by the observation that App1 is highly produced at low glucose concentrations (Fig. 1). 2) The inhibition of CR3-mediated phagocytosis by App1 on other CR3+ phagocytic cells other than AMs, such as dendritic cells (38, 39) or other tissue-specific macrophages, such as Kupffer cells (40), may imply a role of App1 in different tissues/organs during the infection. This possibility is supported by the observation that App1 is found in sera of AIDS patients during cryptococcosis, suggesting that its effect goes beyond the function exerted in the lung (1). 3) The inhibition of the CR3-dependent cytotoxic activity of NK cells (41) by App1 may add another mean of interference from App1 on the host defense mechanisms. This possibility is supported by the observation that Cn Δapp1 mutant is hypervirulent in a mouse model lacking T and NK cells (data not shown). 4) Finally, the potential binding of App1 to CR2 (and CR3) on dendritic cells can result in alteration of Ag trafficking or cell-cell interaction in lymphoid organs (17), whereas the binding of App1 to CR2 on B cells may alter Ab production. This possibility is supported by the fact that macrophages overloaded with Cn Δapp1 mutant is readily transported in lymphoid organs in the mediastinal area (1). Thus, in addition to a role on innate immunity through the regulation of phagocytosis, the existence of an App1-CR2 interaction may suggest a novel role for App1 in the adaptive host immune response against Cn.
In previous work, we found that the Cn Δapp1 mutant is hypervirulent in immunodeficient hosts due to the increased phagocytosis, followed by the inability of the host macrophage to mature and kill the pathogen (1). Because Cn replicates faster intracellularly than extracellularly in condition of immunodeficiency, the infection will progressive more rapidly. Thus, it is envisioned that delivery of App1 protein to the lung of immunodeficient individuals susceptible to develop cryptococcosis would be beneficial and would significantly decrease fungal burden. Whether the beneficial effect of App1 on AMs through CR3 will be hampered by the potential detrimental effects on dendritic or B cells through CR2 awaits further characterization. Once the structural analysis between CR2, CR3, and App1 will be performed, a possibility will be to create small App1 peptide(s) that could specifically bind one, but not the other complement receptor. Thus, structural studies of App1 with CR3 and CR2 will provide further insights on this potential and exciting therapeutic strategy against cryptococcosis.
The fact that App1 binds CR2 opens up the possibility of many applications for this protein. CR2 has been noted to be highly expressed in certain lymphoma cell lines, and the proliferation of these cells is stimulated through complement fragments. The potential clinical implications for App1 in modulating CR2 signaling is exciting because CR2 is not only involved in tumor proliferation mediated by the Epstein-Barr viral infection, but also in the regulation of autoimmunity disorders, such as lupus erythematous and arthritis rheumatoid (reviewed in Ref. 42). Thus, the possibility to modulate CR2 function in such disorders to ameliorate symptoms and signs of lupus erythematous and arthritis rheumatoid using App1 is particularly intriguing.
In conclusion, we found that a fungal protein involved in the regulation of phagocytosis of Cn by AMs interacts with CR3 and CR2, and this interaction has a profound effect in fungal physiopathology and has potential for applications in cancer and autoimmunity disorders.
Acknowledgments
We thank all members of Del Poeta's and Dr. Luberto's laboratories for helpful and constructive discussion. We are particularly grateful to Dr. Dzimitry Fedarovich for providing assistance and expertise in producing App1 and mPGI recombinant proteins in the Medical University of South Carolina Center of Biomedical Research Excellence Protein Core directed by Dr. Christopher Davies.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
M.D.P. is supported by National Institutes of Health Grants AI56168 and AI71142. C.L. is supported by National Institutes of Health Grant RR17677 Project 6 from the Centers of Biomedical Research Excellence Program of the National Center for Research Resources, and from National Science Foundation/EPSCoR Grant EPS-0132573. M.D.P. is a Burroughs Wellcome New Investigator in Pathogenesis of Infectious Diseases.
Abbreviations used in this paper: App1, antiphagocytic protein 1; AM, alveolar macrophage; CHO, Chinese hamster ovary; Cn, Cryptococcus neoformans; CR2, complement receptor 2; CR3, complement receptor 3; mPGI, phosphoglucose isomerase from methanosacrina; nApp1, native App1; NEO, neomycin; pAb, polyclonal Ab; PBST, PBS/0.1% Tween 20; WT, wild type.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Luberto C, Martinez-Marino B, Taraskiewicz D, Bolanos B, Chitano P, Toffaletti DL, Cox GM, Perfect JR, Hannun YA, Balish E, Del Poeta M. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Invest. 2003;112:1080–1094. doi: 10.1172/JCI18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mare L, Iatta R, Montagna MT, Luberto C, Del Poeta M. APP1 transcription is regulated by IPC1-DAG pathway and is controlled by ATF2 transcription factor in Cryptococcus neoformans. J. Biol. Chem. 2005;280:36055–36064. doi: 10.1074/jbc.M507285200. [DOI] [PubMed] [Google Scholar]
- 3.Tommasino N, Villani M, Qureshi A, Henry J, Luberto C, Del Poeta M. Atf2 transcription factor binds to the APP1 promoter in Cryptococcus neoformans: stimulatory effect of diacylglycerol. Eukaryot. Cell. 2008;7:294–301. doi: 10.1128/EC.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walport MJ. Complement: second of two parts. N. Engl. J. Med. 2001;344:1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 5.Walport MJ. Complement: first of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 6.Kozel TR. Activation of the complement system by pathogenic fungi. Clin. Microbiol. Rev. 1996;9:34–46. doi: 10.1128/cmr.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitz SM. Receptor-mediated recognition of Cryptococcus neoformans. Jpn. J. Med. Mycol. 2002;43:133–136. doi: 10.3314/jjmm.43.133. [DOI] [PubMed] [Google Scholar]
- 8.Carroll MC. The role of complement in B cell activation and tolerance. Adv. Immunol. 2000;74:61–88. doi: 10.1016/s0065-2776(08)60908-6. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MC. The complement system in B cell regulation. Mol. Immunol. 2004;41:141–146. doi: 10.1016/j.molimm.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Hatzfeld A, Fischer E, Levesque JP, Perrin R, Hatzfeld J, Kazatchkine MD. Binding of C3 and C3dg to the CR2 complement receptor induces growth of an Epstein-Barr virus-positive human B cell line. J. Immunol. 1988;140:170–175. [PubMed] [Google Scholar]
- 11.Forsyth CB, Plow EF, Zhang L. Interaction of the fungal pathogen Candida albicans with integrin CD11b/CD18: recognition by the I domain is modulated by the lectin-like domain and the CD18 subunit. J. Immunol. 1998;161:6198–6205. [PubMed] [Google Scholar]
- 12.Newman SL, Chaturvedi S, Klein BS. The WI-1 antigen of Blastomyces dermatitidis yeasts mediates binding to human macrophage CD11b/CD18 (CR3) and CD14. J. Immunol. 1995;154:753–761. [PubMed] [Google Scholar]
- 13.Cross CE, Collins HL, Bancroft GJ. CR3-dependent phagocytosis by murine macrophages: different cytokines regulate ingestion of a defined CR3 ligand and complement-opsonized Cryptococcus neoformans. Immunology. 1997;91:289–296. doi: 10.1046/j.1365-2567.1997.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong ZM, Murphy JW. Cryptococcal polysaccharides bind to CD18 on human neutrophils. Infect. Immun. 1997;65:557–563. doi: 10.1128/iai.65.2.557-563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taborda CP, Casadevall A. CR3 (CD11b/CD18) and CR4 (CD11c/ CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity. 2002;16:791–802. doi: 10.1016/s1074-7613(02)00328-x. [DOI] [PubMed] [Google Scholar]
- 16.Levitz SM, Tabuni A. Binding of Cryptococcus neoformans by human cultured macrophages: requirements for multiple complement receptors and actin. J. Clin. Invest. 1991;87:528–535. doi: 10.1172/JCI115027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y, Xu C, Fu YX, Holers VM, Molina H. Expression of complement receptors 1 and 2 on follicular dendritic cells is necessary for the generation of a strong antigen-specific IgG response. J. Immunol. 1998;160:5273–5279. [PubMed] [Google Scholar]
- 18.Szakonyi G, Guthridge JM, Li D, Young K, Holers VM, Chen XS. Structure of complement receptor 2 in complex with its C3d ligand. Science. 2001;292:1725–1728. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- 19.Clemenza L, Isenman DE. Structure-guided identification of C3d residues essential for its binding to complement receptor 2 (CD21). J. Immunol. 2000;165:3839–3848. doi: 10.4049/jimmunol.165.7.3839. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert HE, Eaton JT, Hannan JP, Holers VM, Perkins SJ. Solution structure of the complex between CR2 SCR 1–2 and C3d of human complement: an x-ray scattering and sedimentation modelling study. J. Mol. Biol. 2005;346:859–873. doi: 10.1016/j.jmb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Hannan JP, Young KA, Guthridge JM, Asokan R, Szakonyi G, Chen XS, Holers VM. Mutational analysis of the complement receptor type 2 (CR2/CD21)-C3d interaction reveals a putative charged SCR1 binding site for C3d. J. Mol. Biol. 2005;346:845–858. doi: 10.1016/j.jmb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Feldmesser M, Rivera J, Kress Y, Kozel TR, Casadevall A. Antibody interactions with the capsule of Cryptococcus neoformans. Infect. Immun. 2000;68:3642–3650. doi: 10.1128/iai.68.6.3642-3650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casadevall A, Cassone A, Bistoni F, Cutler JE, Magliani W, Murphy JW, Polonelli L, Romani L. Antibody and/or cell-mediated immunity, protective mechanisms in fungal disease: an ongoing dilemma or an unnecessary dispute? Med. Mycol. 1998;36:95–105. [PubMed] [Google Scholar]
- 24.Vecchiarelli A. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 2000;38:407–417. doi: 10.1080/mmy.38.6.407.417. [DOI] [PubMed] [Google Scholar]
- 25.Murphy JW. Immunological down-regulation of host defenses in fungal infections. Mycoses. 1999;42(Suppl. 2):37–43. [PubMed] [Google Scholar]
- 26.Rittershaus PC, Kechichian TB, Allegood J, Merrill AHJ, Hennig M, Luberto C, Del Poeta M. Glucosylceramide is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T, Takeda J, Hong K, Kozono H, Sakai H, Inoue K. Monoclonal antibodies to mouse complement receptor type 1 (CR1): their use in a distribution study showing that mouse erythrocytes and platelets are CR1-negative. J. Immunol. 1988;140:3066–3072. [PubMed] [Google Scholar]
- 29.Kaya Z, Afanasyeva M, Wang Y, Dohmen KM, Schlichting J, Tretter T, Fairweather D, Holers VM, Rose NR. Contribution of the innate immune system to autoimmune myocarditis: a role for complement. Nat. Immunol. 2001;2:739–745. doi: 10.1038/90686. [DOI] [PubMed] [Google Scholar]
- 30.Kaya Z, Tretter T, Schlichting J, Leuschner F, Afanasyeva M, Katus HA, Rose NR. Complement receptors regulate lipopolysaccharide-induced T-cell stimulation. Immunology. 2005;114:493–498. doi: 10.1111/j.1365-2567.2004.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holers VM. Complement receptors. Year Immunol. 1989;4:231–240. [PubMed] [Google Scholar]
- 32.Dierich MP, Schulz TF, Eigentler A, Huemer H, Schwable W. Structural and functional relationships among receptors and regulators of the complement system. Mol. Immunol. 1988;25:1043–1051. doi: 10.1016/0161-5890(88)90136-8. [DOI] [PubMed] [Google Scholar]
- 33.Molina H, Holers VM, Li B, Fung Y, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, Chaplin DD. Markedly impaired humoral immune response in mice deficient in complement receptors 1 and 2. Proc. Natl. Acad. Sci. USA. 1996;93:3357–3361. doi: 10.1073/pnas.93.8.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahearn JM, Fischer MB, Croix D, Goerg S, Ma M, Xia J, Zhou X, Howard RG, Rothstein TL, Carroll MC. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity. 1996;4:251–262. doi: 10.1016/s1074-7613(00)80433-1. [DOI] [PubMed] [Google Scholar]
- 35.Fearon DT, Carter RH. The CD19/CR2/TAPA-1 complex of B lymphocytes: linking natural to acquired immunity. Annu. Rev. Immunol. 1995;13:127–149. doi: 10.1146/annurev.iy.13.040195.001015. [DOI] [PubMed] [Google Scholar]
- 36.Tedder TF, Zhou LJ, Engel P. The CD19/CD21 signal transduction complex of B lymphocytes. Immunol. Today. 1994;15:437–442. doi: 10.1016/0167-5699(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 37.Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 2002;186:522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- 38.Romani L, Bistoni F, Puccetti P. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr. Opin. Microbiol. 2003;6:338–343. doi: 10.1016/s1369-5274(03)00081-x. [DOI] [PubMed] [Google Scholar]
- 39.Romani L, Montagnoli C, Bozza S, Perruccio K, Spreca A, Allavena P, Verbeek S, Calderone RA, Bistoni F, Puccetti P. The exploitation of distinct recognition receptors in dendritic cells determines the full range of host immune relationships with Candida albicans. Int. Immunol. 2004;16:149–161. doi: 10.1093/intimm/dxh012. [DOI] [PubMed] [Google Scholar]
- 40.Yan J, Vetvicka V, Xia Y, Hanikyrova M, Mayadas TN, Ross GD. Critical role of Kupffer cell CR3 (CD11b/CD18) in the clearance of IgM-opsonized erythrocytes or soluble β-glucan. Immunopharmacology. 2000;46:39–54. doi: 10.1016/s0162-3109(99)00157-5. [DOI] [PubMed] [Google Scholar]
- 41.Vetvicka V, Hanikyrova M, Vetvickova J, Ross GD. Regulation of CR3 (CD11b/CD18)-dependent natural killer (NK) cell cytotoxicity by tumor target cell MHC class I molecules. Clin. Exp. Immunol. 1999;115:229–235. doi: 10.1046/j.1365-2249.1999.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erdei A, Prechl J, Isaak A, Molnar E. Regulation of B-cell activation by complement receptors CD21 and CD35. Curr. Pharm. Design. 2003;9:1849–1860. doi: 10.2174/1381612033454351. [DOI] [PubMed] [Google Scholar]