Abstract
Lactate dehydrogenase (LDH) catalyzes the interconversion of pyruvate and lactate, which are critical fuel metabolites of skeletal muscle particularly during exercise. However, the physiological relevance of LDH remains poorly understood. Here we show that Ldhb expression is induced by exercise in human muscle and negatively correlated with changes in intramuscular pH levels, a marker of lactate production, during isometric exercise. We found that the expression of Ldhb is regulated by exercise-induced peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α). Ldhb gene promoter reporter studies demonstrated that PGC-1α activates Ldhb gene expression through multiple conserved estrogen-related receptor (ERR) and myocyte enhancer factor 2 (MEF2) binding sites. Transgenic mice overexpressing Ldhb in muscle (muscle creatine kinase (MCK)-Ldhb) exhibited increased exercise performance and enhanced oxygen consumption during exercise. MCK-Ldhb muscle was shown to have enhanced mitochondrial enzyme activity and increased mitochondrial gene expression, suggesting an adaptive oxidative muscle transformation. In addition, mitochondrial respiration capacity was increased and lactate production decreased in MCK-Ldhb skeletal myotubes in culture. Together, these results identified a previously unrecognized Ldhb-driven alteration in muscle mitochondrial function and suggested a mechanism for the adaptive metabolic response induced by exercise training.
Keywords: bioenergetics, exercise, gene regulation, mitochondria, skeletal muscle
Introduction
Muscle fitness and resistance to fatigue depend strongly on the capacity to burn the fuels, including fatty acids and glucose, to meet ATP demands (1–5). Exercise training is effective in improving muscle fitness by promoting favorable muscle metabolic reprograming including capacity for fuel burning, mitochondrial ATP production, and contraction (6–13). Conversely, many chronic diseases, including obesity, diabetes, muscular diseases, and aging, are associated with decreased muscle fitness, contributing to a vicious cycle of inactivity and further promoting the progression of chronic diseases (6–8, 11, 12, 14). Thus, a better understanding of the molecular regulatory pathways involved in the beneficial effects of exercise training on muscle fuel metabolism could yield novel therapeutic targets aimed at the prevention or treatment of diseases associated with muscle bioenergetics defects.
The molecular and cellular mechanisms of skeletal muscle adaptation to exercise training are unclear. Exercise training-induced adaptations in skeletal muscle are reflected, in part, by changes in transcriptional response and metabolite flux (1, 2, 4, 5, 11, 15, 16). Previous studies have demonstrated that the PGC-1α3 transcriptional regulatory circuit, including the nuclear receptors PPAR and ERR, is a key transducer of exercise-responsive gene expression. The PGC-1α circuit regulates a broad array of genes involved in mitochondrial biogenesis and fuel metabolism (17–25). Evidence is also emerging that manipulation of metabolic enzyme or metabolite flux in skeletal muscle can significantly affect muscle performance and resistance to fatigue (26, 27). We are just beginning to explore the physiological relevance of metabolic enzyme activation and metabolite flux alterations in regulating muscle function.
Pyruvate and lactate are critical fuel substrates of skeletal muscle particularly during exercise (15, 16, 28). A major source of pyruvate is generated by glycolysis; pyruvate can either serve as a substrate for the mitochondrial TCA cycle to fully catabolize glucose for maximal ATP production, or it can be used for lactate production through a less efficient ATP generation pathway. Another significant source of pyruvate is generated by oxidation of lactate, and exercise training is known to increase lactate oxidation in skeletal muscle (29, 30). Lactate dehydrogenase (LDH) is the key enzyme that catalyzes the interconversion of pyruvate and lactate, thereby regulating cellular pyruvate and lactate homeostasis.
LDH functions as a tetrameric complex composed of two distinct isoforms, LDH-A and LDH-B (31–34), encoded by the Ldha and Ldhb genes, respectively. LDH isoenzyme complexes are classified into LDH1 (B4), LDH2 (A1B3), LDH3 (A2B2), LDH4 (A3B1), and LDH5 (A4) based on different combination of LDH-A and LDH-B isoforms (32, 34). The LDH-A isoform is also known as the M isoform, expressed predominantly in skeletal muscle, whereas LDH-B is also referred to H isoform, is expressed primarily in the heart muscle (35). Previously studies have demonstrated that the LDH-A isoenzyme favors the reaction that converts pyruvate to lactate, whereas the LDH-B isoenzyme prefers the reverse reaction that produces pyruvate from lactate (31, 36). We have recently found that Ldhb is a glucose oxidation biomarker in skeletal muscle; the expression of Ldhb is activated by PPARβ/δ signaling and linked to the high glucose oxidative capacity in MCK-PPARβ/δ muscle (18, 37). In addition, the expression of Ldhb was also involved in PGC-1α-mediated control of lactate homeostasis in muscle (38). However, the functional significance of the Ldhb in skeletal muscle physiology is unclear.
In this study, we found that LDHB expression is induced by exercise in human muscle and negatively correlated with changes in intramuscular pH levels during isometric exercise. We also demonstrated that exercise-induced PGC-1α signaling directly drives the expression of Ldhb in skeletal muscle. We speculated that the exercise-induced Ldhb contributed to the muscle metabolic adaptations induced by exercise training. Using muscle-specific transgenic mouse lines and primary skeletal myotubes in culture, we found that chronic activation of Ldhb in skeletal muscle triggers an adaptive oxidative muscle transformation, leading to increased exercise capacity in MCK-Ldhb transgenic mice. Thus, our results identified a previously unrecognized Ldhb-driven alteration in muscle mitochondrial function and suggest a mechanism for the adaptive metabolic response induced by exercise training.
Results
Activation of LDHB Expression by Exercise Is Linked to Muscle Metabolic Parameters in Humans
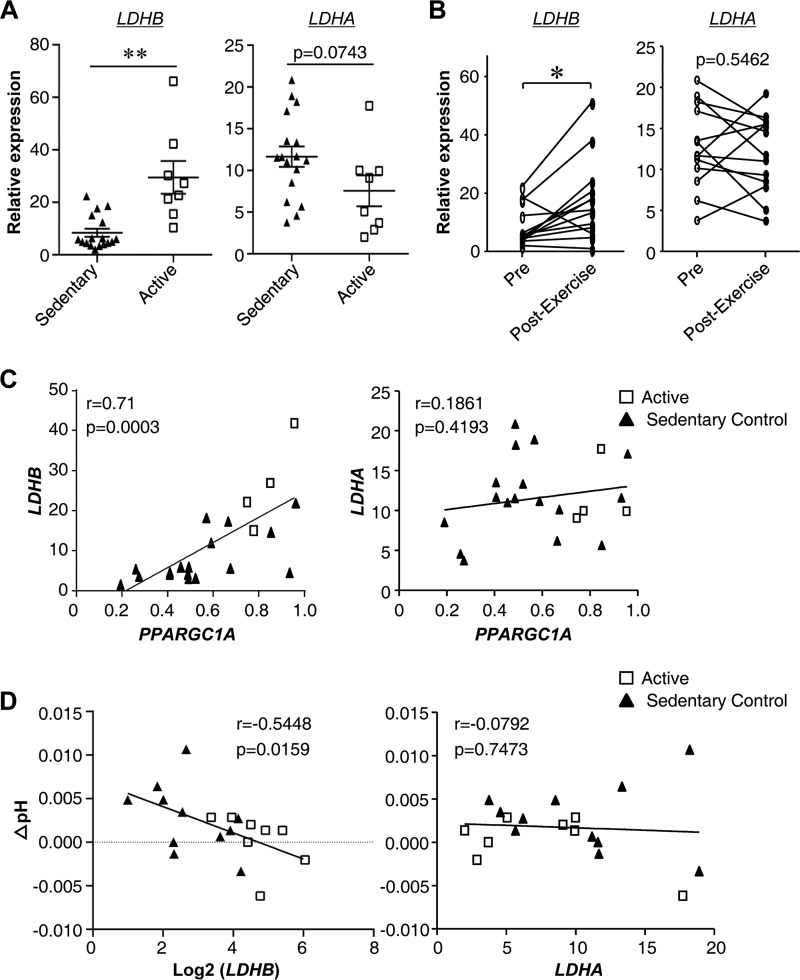
We have recently shown that LDH-B isoform (Ldhb) expression is highly correlated with skeletal muscle glucose oxidation capacity in mice (18, 37). To further explore the physiological relevance of the LDHB in humans, muscle samples from trained, active individuals and healthy sedentary controls were analyzed. Previous studies have demonstrated that the active group has higher measures of enhanced exercise performance (including VO2max and ATPmax) compared with the sedentary group (19, 39, 40). The characteristics of the human subjects are presented in Table 1. Muscle tissue from the active group exhibited higher LDHB gene expression compared with the sedentary control group (Fig. 1A). In contrast, the levels of LDHA mRNA showed a trend toward a decrease in active muscle (Fig. 1A). Additionally, we also examined the expression of LDHB in a subgroup of sedentary subjects who underwent an exercise training program. The expression levels of LDHB were significantly elevated in human muscle by exercise training (Fig. 1B). However, this induction was not observed with LDHA mRNA levels (Fig. 1B). PGC-1α is a known exercise-induced transcriptional cofactor that regulates expression of many exercise-responsive genes. As shown in Fig. 1C, there was a significant positive correlation between LDHB and PGC-1α mRNA levels in human muscle, suggesting a possible mechanism for exercise-induced Ldhb expression. Changes in intramuscular pH levels are a marker of lactate production, because lactate production indicates the generation of a proton that can be measured by the shift in resonance of inorganic phosphate. We also assessed the relationship between LDHB expression and changes in intramuscular pH levels during isometric exercise while measuring PCr recovery rate. As shown in Fig. 1D, a strong negative correlation was observed between the expression of LDHB and changes in intramuscular pH levels. This is consistent with the fact that Ldhb is the key enzyme responsible for lactate oxidation and reduction (31, 36). In contrast, LDHA expression levels did not exhibit a significant correlation with either PGC-1α levels or changes in intramuscular pH levels (Fig. 1, C and D). Together, these results demonstrate that LDHB, but not LDHA, is induced by exercise and linked to muscle metabolic parameters in humans.
TABLE 1.
Human subject characteristics
The data represent the means ± S.E. The differences were analyzed using a two-sample t test, with a statistically significant difference defined as p < 0.05.
| Active (n = 8) | Sedentary control (n = 17) | p value | |
|---|---|---|---|
| Age (years) | 23.25 ± 1.28 | 27.63 ± 1.22 | 0.0358 |
| Weight (kg) | 75.48 ± 3.11 | 80.24 ± 2.52 | 0.2753 |
| Body-mass index (kg/m2) | 23.52 ± 1.02 | 25.69 ± 0.64 | 0.1045 |
| Fat mass (%) | 12.62 ± 1.10 | 22.42 ± 1.22 | <0.0001 |
| Fasting glucose (mg/dl) | 88.63 ± 2.05 | 92.35 ± 2.11 | 0.2838 |
| ATPmax (mm/s) | 1.01 ± 0.08 | 0.66 ± 0.03 | <0.0001 |
| Type I fiber (%) | 52.68 ± 4.71 | 29.37 ± 2.23 | <0.0001 |
| VO2 max (ml/kg/min) | 50.24 ± 1.56 | 32.41 ± 1.50 | <0.0001 |
FIGURE 1.
LDHB expression is induced by exercise in human muscle and negatively correlated with changes in intramuscular pH levels during muscle contraction. Samples from 4–8 active and 15–17 healthy sedentary controls were used for this analysis. mRNA expression levels of LDHB, LDHA, and PPARGC1A was determined by qRT-PCR. The data represent the means ± S.E. A, LDHB and LDHA expression in sedentary and active human muscle analyzed using a two-sample t test (n = 8–17). **, p < 0.01 versus sedentary controls. B, skeletal muscle LDHB and LDHA expression pre- and postexercise training of lean sedentary subjects. The differences were analyzed using paired Student's t test (n = 13). *, p < 0.05. C, Spearman correlation between LDHB and LDHA gene expression and PPARGC1A. D, Pearson correlation between LDHB and LDHA gene expression and ΔpH (changes in pH levels).
Ldhb Expression Is Regulated by Exercise-induced PGC-1α
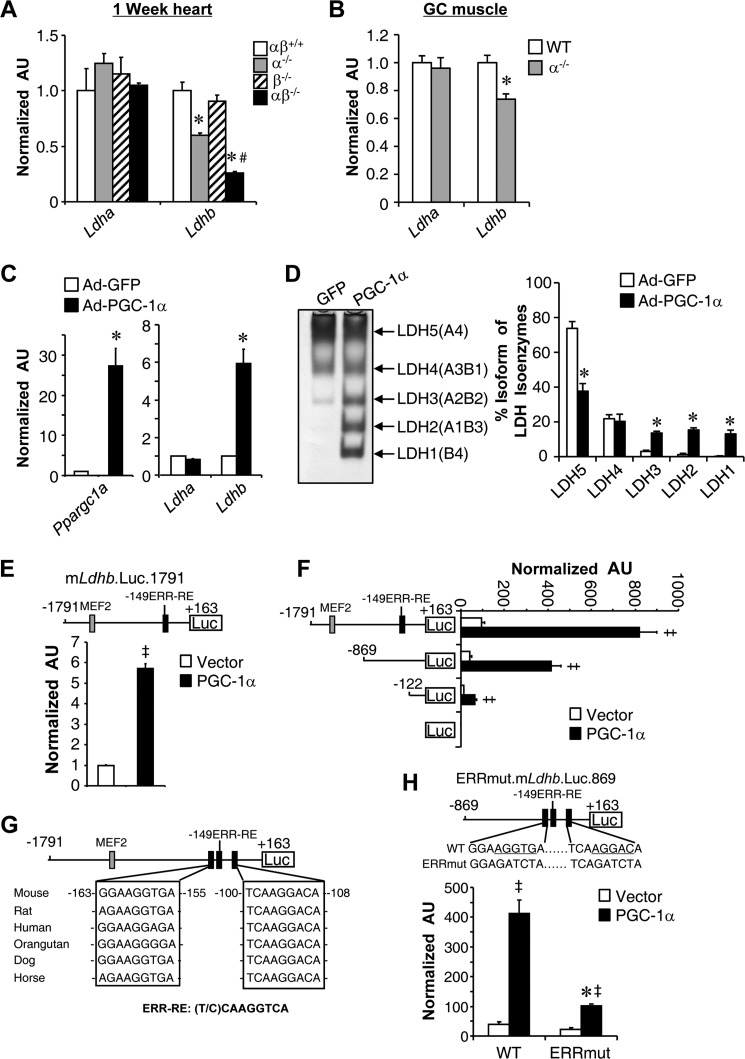
The observation that LDHB gene expression was positively correlated with PGC-1α levels in human muscle led us to explore the link between PGC-1α signaling and the expression of Ldhb. Given that Ldhb is expressed predominantly in the heart, we first conducted PGC-1 loss of function studies in mouse heart. As shown in Fig. 2A, levels of Ldhb mRNA were down-regulated in PGC-1α-deficient (PGC-1α−/−/β+/+) hearts and further reduced in PGC-1α/β deficient (PGC-1α−/−/βf/f/MCK-Cre) hearts when compared with controls (PGC-1α/β+/+). In contrast, Ldha mRNA levels were not changed in PGC-1α/β-deficient heart. Consistent with these results in the heart, disruption of PGC-1α (PGC-1α KΟ) in skeletal muscle resulted in diminished expression of Ldhb, but not Ldha (Fig. 2B). These data suggest that PGC-1α and PGC-1β function redundantly in vivo to regulate the expression of Ldhb gene. We also conducted PGC-1α gain of function in primary skeletal myotubes, in which the basal expression of Ldhb is low. Consistent with previously report (38), there was an increase in Ldhb mRNA, but not Ldha mRNA levels, in skeletal myotubes subjected to adenovirus-mediated overexpression of PGC-1α (Ad-PGC-1α) compared with a control vector (Ad-GFP) (Fig. 2C). The expected LDH isoenzyme activity shifts were confirmed by activity gel studies (Fig. 2D). We have recently shown a functional MEF2 site in the Ldhb promoter (18). Although a PGC-1α-responsive ERR site has also been described in the Ldhb promoter recently (38), we next sought to determine the precise mechanism whereby PGC-1α induces Ldhb gene transcription. Approximately 1.8 kb of the mLdhb gene promoter region containing the MEF2 and −149 ERR-RE binding sites was cloned into a PGL3 reporter vector (mLdhb.Luc.1791). Cotransfection of mLdhb.Luc.1791 with PGC-1α in C2C12 myotubes resulted in robust activation of the promoter (Fig. 2E). To map the cis-acting region conferring the PGC-1α activation, cotransfection experiments were conducted with reporter constructs containing two serial deletions of the mLdhb promoter (Fig. 2F). Both the basal and PGC-1α induced promoter activity decreased upon deletion of the promoter regions from −1791 to −869 bp containing the MEF2 sites, suggesting a role of the MEF2 site in the maximal induction of Ldhb promoter by PGC-1α. The basal promoter activity continues to decrease upon deletion the regions from −869 to −122 bp containing the previously identified −149 ERR-RE. Surprisingly, PGC-1α-mediated activation was maintained upon deletion of the −149 ERR-RE, suggesting the existence of an additional cis-regulatory element in the proximal promoter region. The analysis of the DNA sequence of Ldhb proximal promoter region identified two additional conserved putative ERR-binding sites around the previously identified −149 ERR-RE (Fig. 2G). The two new ERR sites were excellent match for an ERR-binding site (Fig. 2G). To evaluate the functionality of the newly identified ERR binding sites for PGC-1α coactivation, promoter mutational studies were next performed. Mutation of the two putative ERR response element significantly attenuated the activation of mLdhb.Luc.869 by PGC-1α (Fig. 2H). Together, these results demonstrate that multiple cis-regulatory elements, including the distal MEF2 and proximal ERR binding sites in the Ldhb promoter, contribute to the full activation of the Ldhb gene by PGC-1α.
FIGURE 2.
Ldhb expression is regulated by exercise-induced PGC-1α. A, expression of the Ldhb and Ldha genes (qRT-PCR) in the hearts of PGC-1α−/−βf/f/MCK-Cremice (n = 4–7 mice/group). B, expression of the Ldhb and Ldha genes (qRT-PCR) in the GC muscle of PGC-1α KO mice (n = 10–12 mice/group). C, Ldhb, Ldha, and Ppargc1a transcript levels in myotubes harvested from muscle of WT mice and subjected to Ad-PGC-1α overexpression compared with GFP control (n = 3). D, left panel, LDH isoenzymes were separated by polyacrylamide gel electrophoresis using whole cell extracts from WT myotubes subjected to Ad-PGC-1α overexpression. A representative gel is shown. Right panel, quantification of LDH isoenzyme activity gel electrophoresis. The values represent the mean percentages (± S.E.) of total LDH activity (n = 3). E, the mLdhb.Luc.1791 promoter reporter was used in cotransfection studies in C2C12 myotubes in the presence or absence of PGC-1α (n = 3). F, results of transient transfection performed with mouse Ldhb reporter mLdhb.Luc.1791 and truncation mutants of mLdhb.Luc.859 or 122 in C2C12 myotubes in the presence or absence of PGC-1α (n = 3). G, schematic shows the putative conserved MEF2 and ERR binding sites within the Ldhb promoter regions. H, top panel, site-directed mutagenesis was used to abolish the ERR response elements. Bottom panel, the mLdhb.Luc.859 (WT) or ERRmut.mLdhb.Luc.859 promoter reporters was used in cotransfection studies in C2C12 myotubes in the presence or absence of PGC-1α (n = 3). *, p < 0.05 versus corresponding controls; #, p < 0.05 versus α−/−; ‡, p < 0.05 versus vector alone. All values represent the means ± S.E.
Activation of Ldhb Leads to Reciprocal Reduction in Ldha Level in Skeletal Muscle
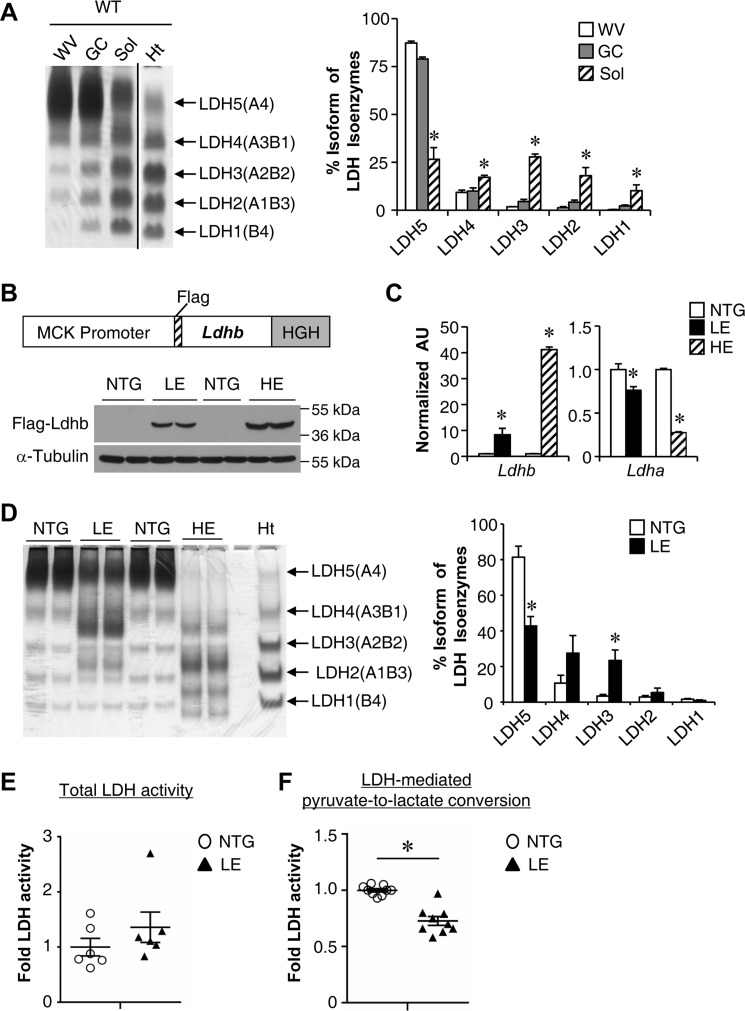
We next determined whether forced activation of Ldhb in skeletal muscle is able to affect muscle function. As an initial step, we examined the expression patterns of Ldhb in different muscle types from adult wild type mice. Consistent with the PGC-1α regulatory circuit controlling Ldhb expression, Ldhb was expressed much higher in slow fiber-dominant soleus muscle compared with fast fiber-enriched white vastus (WV) and gastrocnemius (GC) muscle (Fig. 3A). The muscle creatine kinase promoter was next used to generate skeletal muscle-specific Ldhb transgenic mice (MCK-Ldhb mice). Two independent lines of MCK-Ldhb mice with low (LE) and high (HE) levels of Ldhb overexpression were generated and characterized (Fig. 3, B and C). The MCK-Ldhb transgene transcript was expressed in a skeletal muscle-specific manner, and we observed no change in Ldhb expression in the heart (data not shown). Interestingly, real time quantitative PCR demonstrated that activation of Ldhb in skeletal muscle leads to a reciprocal suppression of the Ldha mRNA level (Fig. 3C). LDH isoenzyme assay using lactate as a substrate displayed a pronounced shift toward an Ldhb-containing isoenzyme complex in MCK-Ldhb (LE and HE) muscle (Fig. 3D). Notably, the reciprocal regulation of Ldhb and Ldha was much greater in MCK-Ldhb (HE) compared with MCK-Ldhb (LE) (Fig. 3D), suggesting a mechanism whereby Ldhb autoregulates the composition of the LDH isoenzyme complex. We subsequently focused on the MCK-Ldhb (LE) line, because of relative physiological overexpressing of Ldhb. A series of studies were next conducted to determine the effect of Ldhb overexpression on muscle LDH enzymatic activity. Whereas the total LDH activity showed an increase trend in MCK-Ldhb (LE) muscle, the enzymatic activity of LDH-mediated pyruvate to lactate conversion was significantly reduced in skeletal muscle of MCK-Ldhb (LE) compared with NTG controls (Fig. 3, E and F). These results are consistent with Ldhb catalyzing the production of lactate from pyruvate slower and less efficiently relative to Ldha, thus diverting pyruvate into mitochondria for complete oxidation (32).
FIGURE 3.
Ldhb overexpression in skeletal muscle. A, left panel, a representative LDH isoenzyme activity gel is shown. Isoenzymes were separated by polyacrylamide gel electrophoresis using whole cell extracts from WV, GC, soleus (Sol), and heart (Ht) muscle from WT mice. Note a distinct shift toward the Ldhb-containing isoenzymes LDH4, LDH3, LDH2, and LDH1 with a concomitant reduction in LDH5 (which lacks the Ldhb isoenzyme) in the soleus. Right panel, quantification of LDH isoenzyme activity gel electrophoresis (n = 3 mice). B, top panel, the schematic depicts the MCK-Ldhb construct used for transgene production. Bottom panel, representative Western blotting analysis performed with GC muscle total protein extracts prepared from NTG mice and two lines of MCK-Ldhb (LE and HE) mice using FLAG and α-tubulin (control) antibodies. C, expression of the Ldhb and Ldha genes (qRT-PCR) in the GC muscle from the indicated genotypes (n = 4–6 mice/group). D, left panel, a representative LDH isoenzyme activity gel using whole cell extracts from GC muscle from the indicated genotypes. Right panel, quantification of LDH isoenzyme activity gel electrophoresis (n = 6 mice/group). E, total LDH enzymatic activity in WV muscle of MCK-Ldhb (LE) mice (n = 6 mice/group). F, enzymatic activity of LDH-mediated pyruvate to lactate conversion in WV muscle of MCK-Ldhb (LE) mice (n = 9 mice/group). *, p < 0.05 versus corresponding controls. All values represent the means ± S.E.
MCK-Ldhb Mice Exhibit Increased Exercise Performance and Enhanced Oxygen Consumption during Exercise
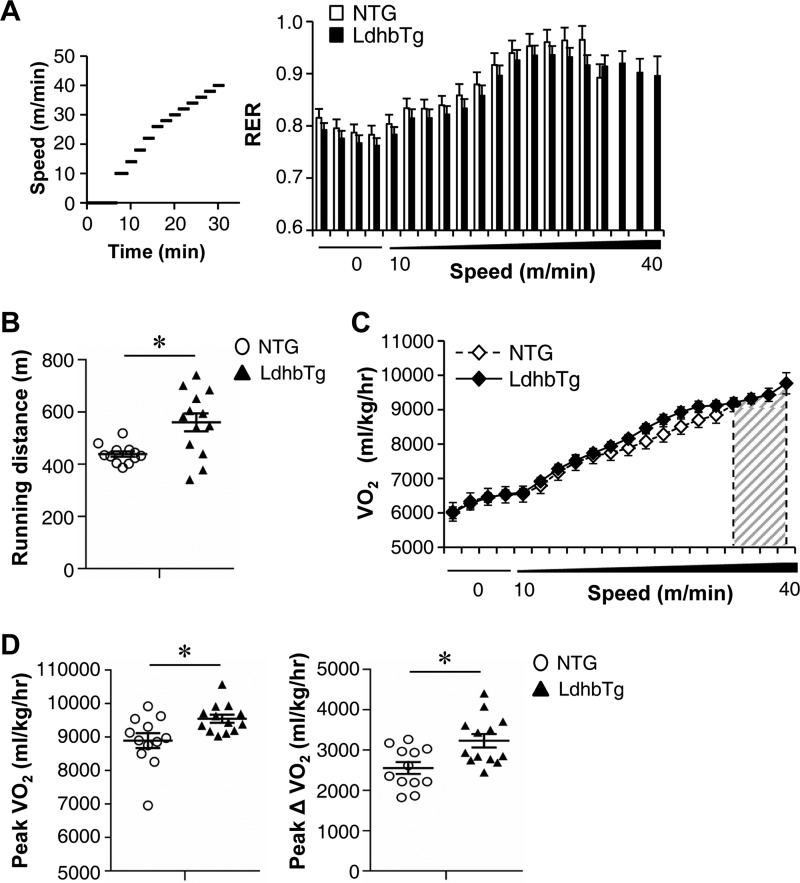
MCK-Ldhb (LE) mice appeared normal on inspection and did not exhibit an overt metabolic phenotype compared with NTG littermates on standard chow. This includes similar body weight, food intake, energy expenditure, and fasting glucose levels (data not shown). To assess the physiological effects of chronic increased Ldhb expression in skeletal muscle, exercise stress testing was conducted in MCK-Ldhb mice. The real time respiratory exchange ratio (RER) was measured during a run to exhaustion exercise protocol. Consistent with a switch to carbohydrates as the chief fuel during exercise, the RER increased with exercise in both MCK-Ldhb and in the NTG control group (Fig. 4A). Despite no change in RER levels during exercise, the MCK-Ldhb (LE) mice exercised significantly longer distance compared with the control group (Fig. 4, A and B). In addition, the MCK-Ldhb (LE) mice consumed more oxygen during the exercise period (as reflected by an increase in peak VO2) (Fig. 4, C and D). It has been shown that whole body oxygen utilization during exercise (peak ΔVO2) largely reflects changes occurring within the exercising muscle (41). Peak ΔVO2 was significantly higher in the MCK-Ldhb (LE) mice compared with NTG controls (Fig. 4D). These results demonstrate that chronic increased Ldhb expression in skeletal muscle is able to affect muscle performance.
FIGURE 4.
MCK-Ldhb mice exhibit increased exercise performance and enhanced oxygen consumption during exercise. A, left panel, schematic depicts the increments of speed over time. Right panel, RER during a graded exercise regimen as described under “Experimental Procedures” (n = 12–13 mice/group). Notably, MCK-Ldhb (LE) female mice were able to exercise at a higher speed before exhaustion. B, the scatter plots represent the mean running distance (± S.E.) in A. C, VO2 (oxygen consumption) during an exercise bout in female MCK-Ldhb (LE) and NTG mice (n = 12–13 mice/group). The gray-hatched area in the VO2 line graphs illustrate the difference in speed to exhaustion in MCK-Ldhb (LE) mice compared with NTG controls. D, peak VO2 (VO2 at the time of failure) and peak ΔVO2 (increase in oxygen consumption) are graphed. The values represent means ± S.E. (n = 12–13 mice/group). *, p < 0.05 versus NTG.
MCK-Ldhb Muscle Is Reprogrammed for Increased Capacity for Mitochondrial Oxidation
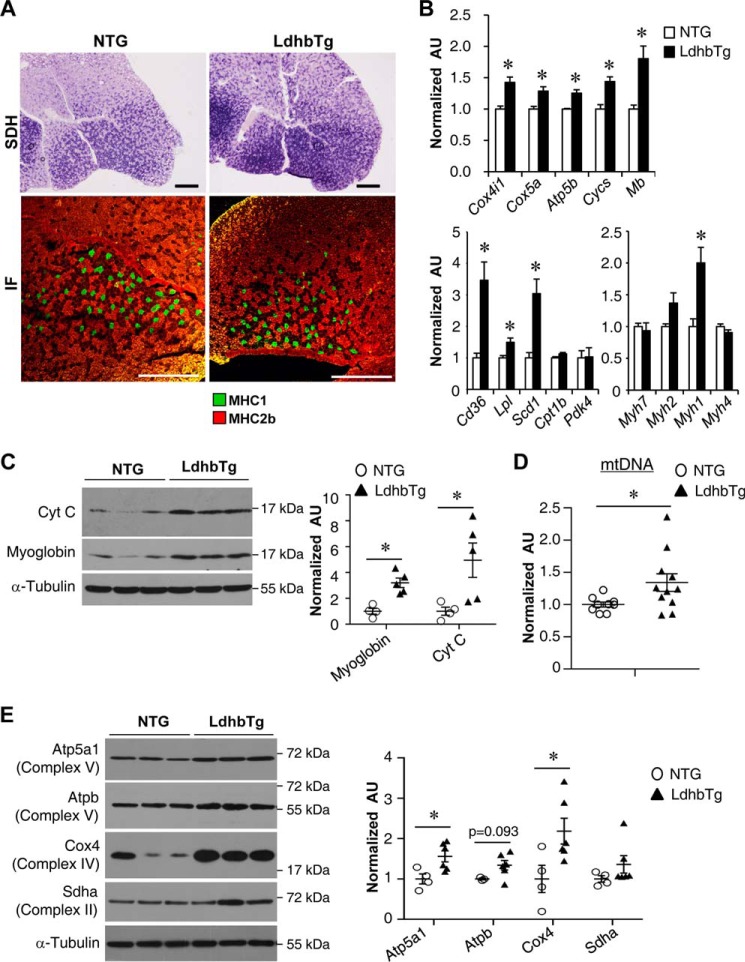
The increase in exercise capacity and oxygen consumption in MCK-Ldhb mice led us to investigate the potential impact of activating Ldhb on muscle oxidative mitochondrial activity. We first performed histochemical staining for succinate dehydrogenase (SDH), a hallmark for oxidative metabolism in skeletal muscle (42). Interestingly, the SDH enzymatic activity was higher in the GC muscle of MCK-Ldhb mice compared with their NTG littermate controls (Fig. 5A), suggesting that chronic activation of Ldhb triggers an adaptive muscle oxidative reprograming. Interestingly, however, no change in MHC1 immunofluorescence was observed in the GC muscle of MCK-Ldhb mice compared with their NTG controls (Fig. 5A). To further evaluate the effect of Ldhb in regulating mitochondrial oxidative capacity, we conducted comparative analysis of RNA isolated from GC muscle of the MCK-Ldhb mice and NTG littermate controls. Real time PCR revealed that the expression of mitochondrial oxidation genes (Cox4i1, Cox5a, Atp5b, Cycs, and Mb) was induced in the GC muscle of MCK-Ldhb mice compared with NTG controls (Fig. 5B). Moreover, we also found an increased expression of biomarker genes associated with fatty acid metabolism (Cd36, Lpl, and Scd1) in MCK-Ldhb muscle (Fig. 5B). Consistent with the fiber typing results, there was no difference in the expression of Myh7, which encodes the myosin heavy chain for type I fibers in MCK-Ldhb muscle (Fig. 5B), although the expression of the oxidative type IIx myosin gene Myh1 was increased in MCK-Ldhb muscle (Fig. 5B). The oxidative transformations in MCK-Ldhb muscle were also validated at the protein level, because the expression of the oxidative biomarkers myoglobin and cytochrome c was induced in MCK-Ldhb WV muscles compared with NTG controls (Fig. 5C). The mitochondrial DNA levels were increased in MCK-Ldhb GC muscle compared with NTG controls (Fig. 5D). In addition, Western blotting revealed significant increases in several components of the electron transport chain (e.g. Atp5a1 and Cox4) in MCK-Ldhb WV muscle (Fig. 5E). Together, these results demonstrate that chronic activation of Ldhb reprograms muscle for increased mitochondrial oxidative capacity.
FIGURE 5.
MCK-Ldhb muscle is reprogrammed for increased capacity for mitochondrial oxidation. A, top row, cross-section of the GC muscle from a 3-month-old female NTG and MCK-Ldhb (LE) stained for SDH. Bottom row, representative MHC immunofluorescence (IF) in the GC muscles of the indicated genotypes. Green, MHC1; red, MHC2b (n = 3–8 mice/group). Scale bars, 500 μm. B, expression of genes involved in mitochondrial oxidation, fuel metabolism, and contractile myosin isoforms (qRT-PCR) in GC muscle from the NTG and MCK-Ldhb (LE) mice (n = 4–6 mice/group). C, left panel, representative Western blotting analysis performed with WV muscle total protein extracts prepared from the NTG and MCK-Ldhb (LE) mice using cytochrome c, myoglobin, and α-tubulin (control) antibodies. Right panel, quantification of the myoglobin/tubulin and cytochrome c/tubulin signal ratios normalized ( = 1.0) to the NTG control (n = 4–5 mice/group). D, results of qPCR to determine mitochondrial DNA levels in GC muscle of the MCK-Ldhb (LE) mice (n = 9–11 mice/group). E, left panel, representative Western blotting analysis performed with WV muscle total protein extracts prepared from the NTG and MCK-Ldhb (LE) mice using Atp5a1, Atpb, Cox4, Sdha, and α-tubulin (control) antibodies. Right panel, quantification of the Western blot shown in the left panel. Atp5a1/tubulin, Atpb/tubulin, Cox4/tubulin, and Sdha/tubulin signal ratios were normalized (1.0) to the NTG control (n = 3–6 mice/group). *, p < 0.05 versus NTG. All values represent the means ± S.E.
Increased Mitochondrial Function in MCK-Ldhb Skeletal Muscle
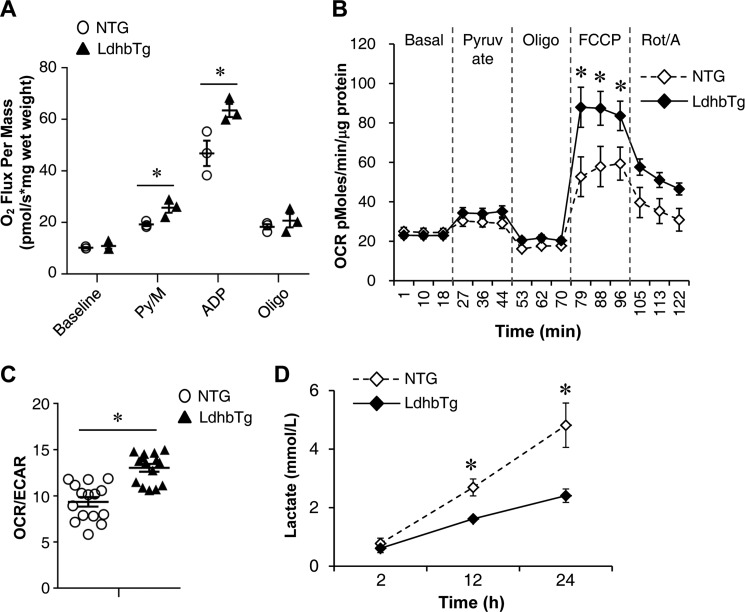
Mitochondrial respiration rates were determined in the extensor digital longus muscle of the MCK-Ldhb (LE) mice and corresponding NTG controls. Consistent with the oxidative transformations in MCK-Ldhb muscle, pyruvate-driven state 3 respiration rates were significantly higher in MCK-Ldhb muscle compared with the NTG controls (Fig. 6A). To directly determine the effects of activating Ldhb on muscle mitochondrial function, oxygen consumption rates (OCRs) were also measured in primary myotubes isolated from MCK-Ldhb skeletal muscle. As shown in Fig. 6B, activation of Ldhb significantly stimulated the OCR in the presence of the uncoupler FCCP, a sign of enhanced mitochondrial function. We also determined the extracellular acidification rate (ECAR) (a measure of glycolysis) along with OCRs in these cells. Ldhb overexpression significantly induced OCR/ECAR ratio, indicative of a shift toward more oxidative phosphorylation for cellular energy production (Fig. 6C). Consistent with the aerobic metabolism in MCK-Ldhb myotubes, the rate of lactate production decreased in myotubes isolated from MCK-Ldhb muscle compared with NTG controls (Fig. 6D). These results demonstrate that chronic activation of Ldhb in muscle cells promotes a shift toward a more oxidative phenotype, which is consistent with the phenotypic changes observed in MCK-Ldhb mice.
FIGURE 6.
Increased mitochondrial function in MCK-Ldhb skeletal muscle. A, mitochondrial respiration rates were determined from the extensor digital longus muscle of the indicated genotypes using pyruvate/malate as substrate. Pyruvate/malate (Py/M)-stimulated, ADP-dependent respiration, oligomycin-induced (Oligo), and the respiratory control ratio are shown (n = 3 mice/group). B, OCRs in primary mouse myotubes isolated from the NTG and MCK-Ldhb (LE) mice. Basal OCR was first measured, followed by administration of 10 mm sodium pyruvate, 2 μm oligomycin (to inhibit ATP synthase), uncoupler FCCP (2 μm), or rotenone/antimycin (Rot/A) (1 μm) as indicted. C, OCR/ECAR ratio using pyruvate as substrate indicates a shift in cellular energy production to oxidative phosphorylation (n = 3 separate experiments done with 5 biological replicates). D, lactate concentrations in culture medium from primary mouse myotubes isolated from the NTG and MCK-Ldhb (LE) mice (n = 3). *, p < 0.05 versus NTG controls. All values represent the means ± S.E.
Discussion
Mitochondrial oxidative metabolism and energy production are critical for muscle performance. Exercise is known to be the best medicine for many chronic illnesses including obesity, diabetes, muscular diseases, and aging, by promoting favorable metabolic and structural adaptations to improve muscle fitness (6–13). Delineation of the molecular regulatory pathways involved in the beneficial effects of exercise training on muscle fuel metabolism has implications for new therapeutic approaches for many human diseases associated with muscle bioenergetics defects. Herein, we discover a novel mechanism for exercise-induced metabolic changes in skeletal muscle. Our results support the following conclusions: 1) LDHB expression is induced by exercise in human muscle and negatively correlated with changes in intramuscular pH levels during muscle contraction; 2) exercise-induced PGC-1α signaling directly regulates the transcription of the Ldhb gene by coactivating multiple cis-regulatory elements in the Ldhb promoter; and 3) chronic activation of Ldhb triggers a secondary mitochondrial oxidative metabolism program in skeletal muscle. We therefore identify a previously unrecognized Ldhb-driven alteration in muscle mitochondrial function and suggest a mechanism for the adaptive metabolic response induced by exercise training.
Previous studies have established that the PGC-1α transcriptional regulatory circuit, including nuclear receptors PPAR and ERR, is a key transducer of skeletal muscle adaptation to exercise by directly regulates the expression of genes involved in mitochondrial fuel metabolism (17–25). We have recently shown that Ldhb is a downstream target of PPARβ/δ signaling (18, 37). In addition, the PGC-1α-ERR axis has also been implicated in the regulation of Ldhb expression (38). In the present study, our data suggest that Ldhb is a regulator of mitochondrial function that acts downstream of the PGC-1α/nuclear receptor regulatory circuit. Exercise training induces the expression of both PGC-1α and Ldhb in muscle. It is likely that the Ldhb-driven alteration of skeletal muscle mitochondrial function contributes the broad effect of exercise PGC-1α signaling on muscle metabolic adaptations.
The observed role of Ldhb in regulating muscle mitochondrial function was surprising, given that Ldhb is a glycolytic enzyme responsible for lactate oxidation and reduction (31, 36). Several lines of evidence presented here support the conclusion that chronic activation of Ldhb triggers a secondary beneficial muscle metabolic reprograming, in addition to regulating pyruvate/lactate homeostasis. First, MCK-Ldhb mice are able to run longer during exercise. Second, diverse aspects of aerobic metabolism, including the capacity for oxygen consumption during exercise, muscle SDH activity, and mitochondrial respiration, are increased in MCK-Ldhb muscle compared with NTG controls. Third, a broad array of mitochondrial metabolism genes and oxidative biomarkers are induced in MCK-Ldhb muscle compared with NTG controls.
Whereas our results provide significant evidence that increased Ldhb activity affects muscle mitochondrial function, the molecular basis of this finding was not fully delineated in this study. Interestingly, we do not see significant changes in oxidative biomarkers in younger MCK-Ldhb muscle (6 weeks old) compared with NTG controls. This could relate to the age difference. Alternatively, the secondary mitochondrial gene programs triggered by chronic activation of Ldhb are not manifest as early in 6-week-old muscle because other postnatal programs are dominant during the first several weeks after birth (42). Ldhb has recently been shown to locate outside of the mitochondrial matrix (43), and lactate oxidation was shown to regulate mitochondrial oxidative gene expression in muscle cells (44). It is possible that an alteration in pyruvate/lactate oxidation in MCK-Ldhb muscle triggers the mitochondrial oxidative gene expression. It is also intriguing to speculate that Ldhb can serve as a unique glycolytic enzyme that could directly affect muscle metabolic gene expression. Consistent with this later notion, the glycolytic enzyme complex SASEME has recently been shown to sense glucose metabolism and directly regulate chromatin modifications (45). Whether such mechanisms are relevant to our study remains to be determined; future studies aimed at assessing the mechanism whereby muscle metabolic reprograming is altered in MCK-Ldhb muscle will likely require metabolic profiling and genome-wide chromatin survey.
We found that the LDH isoenzyme complex was dynamically regulated in human and mouse skeletal muscle. The induced LDHB expression during exercise is consistent with previous reports that exercise training increases lactate oxidation in human muscle (29, 30, 46, 47). Our data suggest there were both PGC-1α-dependent and independent mechanisms that regulate the LDH isoenzyme composition. First, our study reveals an intriguing autoregulatory loop whereby Ldhb directly controls LDH isoenzyme complex, given that activation of Ldhb leads to reciprocal reduction in Ldha level in skeletal muscle. Second, our data also demonstrated that Ldhb expression is regulated by exercise-induced PGC-1α, providing a mechanism for exercise-induced Ldhb expression. Although a PGC-1α-responsive ERR binding site (−149 ERR-RE) has been previously described (38), the functionality of the Ldhb gene promoter has not been fully characterized. Using a robust Ldhb promoter reporter assay in C2C12 myotubes, we were able to identify two additional functional ERR-REs in the Ldhb promoter. In addition, consistent with our previous report (18), our data also support the involvement of the distal MEF2 site for both the basal and full activation of the Ldhb promoter by PGC-1α. The involvement of the MEF2 site is also corroborated by the fact that slow oxidative muscle fibers with higher MEF2 activity are enriched in Ldhb, whereas the fast glycolytic muscle fibers have the opposite effect.
In summary, we have identified exercise-induced Ldhb as a novel regulator of mitochondrial oxidative metabolism in skeletal muscle. Future studies aimed at the mechanisms involved in triggering the adaptive responses could yield new therapeutic targets aimed at the prevention or treatment of diseases associated with muscle bioenergetics defects.
Experimental Procedures
Animal Studies
All animal studies were conducted in strict accordance with the institutional guidelines for the humane treatment of animals and were approved by the institutional animal care and use committees at the Model Animal Research Center of Nanjing University.
Generation of MCK-Ldhb Mice
To generate mice with muscle-specific Ldhb overexpression, a cDNA encoding the mouse Ldhb gene was cloned into the EcoRV site downstream of the mouse MCK gene promoter (kind gift of E. N. Olson, University of Texas Southwestern). The transgene was linearized with XhoI and SacII digestion and microinjected into C57BL/6J embryos by the transgenic mouse facility at the Model Animal Research Center of Nanjing University. Transgenic mice were identified by PCR amplification of a 722-bp product using primers specific for Ldhb (5′-CAGACAATGACAGTGAGAACTGGAAGGAGG) and the human growth hormone poly(A) component of the MCK construct (5′-ATTGCAGTGAGCCAAGATTGTGCCACTGCA). Two independent lines were generated, exhibiting low and high levels of transgenic expression (LE and HE). Unless specifically indicated, the results described here were generated using the low expressing MCK-Ldhb line (LE), compared with corresponding NTG controls. Of note, the majority of the phenotypic characterization of these mice was performed in female MCK-Ldhb mice. In addition, several readouts, including oxidative biomarkers, were similarly induced in male MCK-Ldhb mice. The PGC-1α/βf/f/MCK-Cre mice have been described previously (48).
Human Studies
After signing the informed written consent approved by the Pennington Biomedical Research Center ethical review board, patients were enrolled in clinical trial performed at the Pennington Biomedical Research Center (Baton Rouge, LA). Volunteers qualified for the study (ACTIV; Clinicaltrials.gov ID NCT00401791) if they ranged in age from 20 to 40, had a body-mass index of 20–30 kg/m2, were non-diabetic, were taking no medications, and were otherwise healthy. After baseline testing, 13 nonobese sedentary subjects participated in an exercise training protocol consisting of alternating day sessions of a progressive 30–60-min interval protocol (75–85% maximum aerobic capacity (VO2 max)) and a 50-min aerobic protocol (70% VO2 max), both performed on a stationary bicycle. Subjects exercised on 13 days of a 3-week period. Details on subject characteristics and procedures have been described previously (19, 39, 40). Briefly, after an overnight fast and local anesthesia, skeletal muscle was collected from the vastus lateralis muscle, cleaned, and mounted for fiber typing or flash frozen in liquid nitrogen for RNA isolation.
Mouse Studies
Mice were acclimated (run for 9 min at 10 m/min followed by 1 min at 20 m/min) to the treadmill for 2 consecutive days prior to the experimental protocol. RERs during exercise were determined as described previously (18). Briefly, mice were placed in an enclosed treadmill attached to the Comprehensive Laboratory Animal Monitoring System (Columbus Instruments) for 15 min at a 0° incline and 0 m/min. The mice were then challenged with 2-min intervals of increasing speed at a 0° incline. The increasing speeds used in the protocol were 10, 14, 18, 22, 26, 28, 30, 32, 34, 36, 38, and 40 m/min. The protocol was performed until exhaustion; running distance was derived by calculating the treadmill speed and running time. The measurements were collected before the exercise challenge and throughout the challenge, and peak VO2 was measured at the time of failure.
Mitochondrial Respiration Studies
Mitochondrial respiration rates were measured in saponin-permeabilized extensor digital longus muscle fibers with pyruvate/malate as substrate as described previously (37). In brief, the muscle fibers were separated and transferred to BIOPS buffer. The muscle fibers bundles were then permeabilized with 50 μg/ml saponin in BIOPS solution. Measurement of oxygen consumption in permeabilized muscle fibers was performed in buffer Z at 37 °C and in the respiration chambers of an Oxygraph 2K (Oroboros Inc., Innsbruck, Austria). Following measurement of basal, pyruvate (10 mm)/malate (5 mm) respiration, maximal (ADP-stimulated) respiration was determined by exposing the mitochondria to 4 mm ADP. Uncoupled respiration was evaluated following addition of oligomycin (1 μg/ml). Respiration rates were determined and normalized to fiber bundle wet weight using Datlab 5 software (Oroboros Inc.), and the data are expressed as pmol O2 s−1 mg wet weight−1.
Histologic Analyses
Muscle tissue was frozen in isopentane that had been cooled in liquid nitrogen. SDH and immunofluorescence staining was performed as previously described (24).
RNA Analyses
Quantitative RT-PCR was performed as described previously, with modifications (18, 19). Briefly, total RNA was extracted from mouse muscle or primary myotubes using RNAiso Plus (Takara Bio). The purified RNA samples were then reverse transcribed using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio). Real time quantitative RT-PCR was performed using the ABI Prism Step-One system with SYBR® Premix Ex TaqTM (Takara Bio). Specific oligonucleotide primers for target gene sequences are listed below. Arbitrary units of target mRNA were corrected to the expression of 36b4. For mouse gene, the following primers were used: 36b4, 5′-ATCCCTGACGCACCGCCGTGA, 5′-TGCATCTGCTTGGAGCCCACGT; Cox4i1, 5′-TACTTCGGTGTGCCTTCGA, 5′-TGACATGGGCCACATCAG; Cox5a, 5′-TTAAATGAATTGGGAATCTCCAC, 5′-GTCCTTAGGAAGCCCATCG; Atp5b, 5′-GCAGGGACAGCAGACTGG, 5′-GCCATCCATAGCAATAGTTCTGA; Scd1, 5′-TTCCCTCCTGCAAGCTCTAC, 5′-CAGAGCGCTGGTCATGTAGT; Cpt1b, 5′-GAGTGACTGGTGGGAAGAATATG, 5′-GCTGCTTGCACATTTGTGTT; Mb, 5′-CCGGTCAAGTACCTGGAGTT, 5′-TGAGCATCTGCTCCAAAGTC; Pdk4, 5′-CCGCTGTCCATGAAGCA, 5′-GCAGAAAAGCAAAGGACGTT; Cycs, 5′-ACCAAATCTCCACGGTCTGTT, 5′-GGATTCTCCAAATACTCCATCAG; Lpl, 5′-TTTGTGAAATGCCATGACAAG, 5′-CAGATGCTTTCTTCTCTTGTTTGT; Ldha, 5′-TGCCTACGAGGTGATCAAGCT, 5′-GCACCCGCCTAAGGTTCTTC; Ldhb, 5′-AGTCTCCCGTGCATCCTCAA, 5′-AGGGTGTCCGCACTCTTCCT; Ppargc1a, 5′-CGGAAATCATATCCAACCAG, 5′-TGAGAACCGCTAGCAAGTTTG; Ppargc1b, 5′-TCCAGAAGTCAGCGGCCT, 5′-CTGAGCCCGCAGTGTGG; Esrra, 5′-AGGAGTACGTCCTGCTG, 5′-CCTCAGCATCTTCAATG; Esrrb, 5′-ACGGCTGGATTCGGAGAAC, 5′-TCCTGCTCAACCCCTAGTAGATTC; Esrrg, 5′-TGACTTGGCTGACCGAG, 5′-CCGAGGATCAGAATCTCC; Ppara, 5′-ACTACGGAGTTCACGCATGTG, 5′-TTGTCGTACACCAGCTTCAGC; Ppard, 5′-GTATGCGCATGGGACTCAC, 5′-GTCTGAGCGCAGATGGACT; Myh7, 5′-GCCAACTATGCTGGAGCTGATGCCC, 5′-GGTGCGTGGAGCGCAAGTTTGTCATAAG; Myh1, 5′-GGCAGCAGCAGCTGCGGAAGCAGAGTCTGG, 5′-GAGTGCTCCTCAGATTGGTCATTAGC; Myh2, 5′-GGCACAAACTGCTGAAGCAGAGGC, 5′-GGTGCTCCTGAGGTTGGTCATCAGC; Myh4, 5′-GAGCTACTGGATGCCAGTGAGCGC, 5′-CTGGACGATGTCTTCCATCTCTCC. For human gene: LDHB,5′-GATGGATTTTGGGGGAACAT, 5′-AACACCTGCCACATTCACAC; and PPARGC1A, 5′-TGAGAGGGCCAAGCAAAG, 5′-ATAAATCACACGGCGCTCTT.
Mitochondrial DNA Analyses
Genomic/mitochondrial DNA was measured as described previously (37). Mitochondrial DNA content was determined by SYBR Green analysis (Takara Bio). The levels of NADH dehydrogenase subunit 1 (mitochondrial DNA) were normalized to the levels of lipoprotein lipase (genomic DNA).
Antibodies and Immunoblotting Studies
Antibodies directed against MHC1 (BA-D5) and MHC2b (BF-F3) were purchased from the Developmental Studies Hybridoma Bank; antibodies directed against cytochrome c (bs1089) and α-tubulin (bs1699) antibody were from Bioworld; anti-myoglobin (sc-25607) was from Santa Cruz; anti-FLAG M2 (F1804) was from Sigma; anti-Atpb (ab14730) was from Abcam; anti-Atp5a1 (14676–1-AP), anti-Cox4 (11242–1-AP), and anti-Sdha (14865–1-AP) were from Proteintech. Western blotting studies were performed as previously described (18, 19).
LDH Isoenzyme Analysis and Activity Assay
LDH isoenzyme patterns were determined as previously described (18). Five major LDH isoenzymes are found, because Ldhb polypeptide has more acidic amino acid residues than the Ldha polypeptide; thus the electrophoretic mobilities of the LDH isoenzymes migrate toward the positive electrode end as follows: LDH 1 > LDH 2 > LDH 3 > LDH 4 > LDH 5. Briefly, primary skeletal myotubes or mouse skeletal muscle were homogenized in a solution of 0.9% NaCl, 5 mm Tris-HCl, pH 7.4, and the lysates were centrifuged for 30 min at 15,000 × g to remove the cellular debris. 100 μg of protein/lane was loaded onto a 6% nondenaturing polyacrylamide gel. Following electrophoresis, the gel was placed in 10 ml of staining solution containing 0.1 m sodium lactate, 1.5 mm NAD, 0.1 m Tris-HCl (pH 8.6), 10 mm NaCl, 5 mm MgCl2, 0.03 mg/ml phenazinmethosulphate, and 0.25 mg/ml nitro blue tetrazolium. Protein extracted from mouse heart served as a positive control. Total LDH activity and specific LDH activities (pyruvate to lactate conversions) were determined using the LDH assay kit (Nanjing Jiancheng Bioengineering Institute, A020-2) and (Beijing Leagene Biotechnology, TE0155) according to the manufacturer's protocol, respectively. Changes in absorbance were determined with a VersaMax ELISA microplate reader (Molecular Devices) at 450 nm for total LDH activity and 340 nm for LDH-mediated pyruvate to lactate conversion assay.
Lactate Concentration Measurement
Cell culture medium was collected. Lactate concentration was then determined with a VersaMax ELISA Microplate Reader (Molecular Devices) using the lactic acid assay kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer's protocol.
Cell Culture
Primary muscle cells were isolated from skeletal muscles as previously described (18, 19). For differentiation, the cells were washed with PBS, refed with 2% horse serum/DMEM differentiation medium, and refed daily. The cells were then differentiated for 3 days prior to harvest. Primary myoblasts were infected with an adenovirus overexpressing GFP or PGC-1α as previously described (18, 19). 12 h postinfection, the cells were induced to differentiation for 3 days.
Oxygen Consumption Measurements
Cellular OCRs were measured using the XF24 analyzer (Seahorse Bioscience Inc.) per the manufacturer's protocol. The basal OCR was first measured in XF assay medium without sodium pyruvate, followed by administration of 10 mm sodium pyruvate. Uncoupled respiration was evaluated following the addition of oligomycin (2 μm) to inhibit ATP synthase by addition of the uncoupler FCCP (2 μm) and then followed by the addition of rotenone/antimycin (1 μm). Immediately after measurement, total protein levels were measured with the Micro BCA protein assay kit (Thermo Scientific) for data correction.
Cell Transfection and Luciferase Reporter Assays
pcDNA3.1 and pcDNA3.1-PGC-1α vectors have been described previously (19). The mouse Ldhb gene promoter deletion series was generated by PCR amplification from C57BL/6J genomic DNA followed by cloning into the pGL3 Basic luciferase reporter plasmid using BglII and MluI sites. The following 5′ primers were used: 5′-CTGGCTGACCTAGATCTCCGTTTC (mLdhb.Luc.1791), 5′-TGGATGAGACAAAGATCTAAGAATGTGG (mLdhb.Luc.869), and 5′-GAGAGATCTTGCACACTCCAGCCTTG (mLdhb.Luc.122). The same 3′ primer was used with all constructs: 5′-ACAACACACGCGTTGATGTTCAG. Site-directed mutagenesis was performed using complementary oligonucleotides as follows (with mutated nucleotides shown in lowercase): 5′-GTGCCTCAGCGGAgatctACCTCTAACTTTAG (ERRmut#1) and 5′-AAAGTTAGAGGTagatcTCCGCTGAGGCA (ERRmut#2). C2C12 cells were obtained from the American Type Culture Collection and were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1,000 units/ml penicillin, and 100 μg/ml streptomycin. Transient transfections in C2C12 cells were performed using Attractene transfection reagent (Qiagen) as per the manufacturer's protocol. Briefly, 350 ng of reporter was cotransfected with 100 ng of nuclear receptor expression vectors and 25 ng of CMV promoter-driven Renilla luciferase to control for transfection efficiency. The cells were harvested 48 h after transfection. The luciferase assay was performed using Dual-Glo (Promega) according to the manufacturer's recommendations. All transfection data are presented as the means ± S.E. for at least three separate transfection experiments.
Statistical Analyses
All mouse and cell studies were analyzed by Student's t test or one-way analysis of variance coupled to a Fisher's least significant difference post hoc test when more than two groups were compared. The data represent the means ± S.E., with a statistically significant difference defined as a value of p < 0.05. Statistical analyses in human studies were performed using JMP 9.0.0 (SAS Institute Inc.), and values are presented as means ± S.E. Gene expression levels in human studies were analyzed using the Spearman correlation or Pearson correlation test. Significant differences were defined as p < 0.05.
Author Contributions
X. L. and L. L. contributed equally to this work and performed most of the experiments with assistance from T. F., Q. Z., D. Z., L. X., J. L., Y. K., H. X., F. Y., and L .L., whereas R. B. V. and D. P. K. contributed reagents and provided scientific insight and discussion. S. R. S. was the principal investigator responsible for the clinical studies. Z. G. provided oversight of the study including experimental design and data interpretation and wrote the manuscript. All authors reviewed and contributed to the manuscript.
Acknowledgments
We give special thanks to Dr. Jiansheng Kang (Shanghai Institute for Biological Sciences), Dr. Bin Lv (Wenzhou Medical University), and Dr. Su-neng Fu (Tsinghua University) for ETC antibodies.
This work was supported by Ministry of Science and Technology of China 973 Program Grant 2015CB856300, National Natural Science Foundation of China Grants 31471110 and 81400821, Natural Science Foundation of Jiangsu Province Grant BK20140600 (to Z. G.), and National Institutes of Health Grant RO1DK045416 (to D. P. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PGC
- PPARγ coactivator
- PPAR
- peroxisome proliferator-activated receptor
- LDH
- lactate dehydrogenase
- ERR
- estrogen-related receptor
- MCK
- muscle creatine kinase
- WV
- white vastus
- GC
- gastrocnemius
- LE
- low level of Ldhb overexpression
- HE
- high level of Ldhb overexpression
- NTG
- nontransgenic
- RER
- respiratory exchange ratio
- SDH
- succinate dehydrogenase
- OCR
- oxygen consumption rate
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- ECAR
- extracellular acidification rate
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Burke L. M., and Hawley J. A. (1999) Carbohydrate and exercise. Curr. Opin. Clin. Nutr. Metab. Care 2, 515–520 [DOI] [PubMed] [Google Scholar]
- 2. Coggan A. R. (1991) Plasma glucose metabolism during exercise in humans. Sports Med. 11, 102–124 [DOI] [PubMed] [Google Scholar]
- 3. Hargreaves M. (2004) Muscle glycogen and metabolic regulation. Proc. Nutr. Soc. 63, 217–220 [DOI] [PubMed] [Google Scholar]
- 4. Hawley J. A. (2002) Adaptations of skeletal muscle to prolonged, intense endurance training. Clin. Exp. Pharmacol. Physiol. 29, 218–222 [DOI] [PubMed] [Google Scholar]
- 5. Holloszy J. O., Kohrt W. M., and Hansen P. A. (1998) The regulation of carbohydrate and fat metabolism during and after exercise. Front. Biosci. 3, D1011–D1027 [DOI] [PubMed] [Google Scholar]
- 6. Booth F. W., and Thomason D. B. (1991) Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol. Rev. 71, 541–585 [DOI] [PubMed] [Google Scholar]
- 7. Neufer P. D., Bamman M. M., Muoio D. M., Bouchard C., Cooper D. M., Goodpaster B. H., Booth F. W., Kohrt W. M., Gerszten R. E., Mattson M. P., Hepple R. T., Kraus W. E., Reid M. B., Bodine S. C., Jakicic J. M., et al. (2015) Understanding the cellular and molecular mechanisms of physical activity-induced health benefits. Cell Metab. 22, 4–11 [DOI] [PubMed] [Google Scholar]
- 8. Rowe G. C., Safdar A., and Arany Z. (2014) Running forward: new frontiers in endurance exercise biology. Circulation 129, 798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zierath J. R., and Hawley J. A. (2004) Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2, e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan Z., Okutsu M., Akhtar Y. N., and Lira V. A. (2011) Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J. Appl. Physiol. 110, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egan B., and Zierath J. R. (2013) Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184 [DOI] [PubMed] [Google Scholar]
- 12. Holloszy J. O., and Coyle E. F. (1984) Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 56, 831–838 [DOI] [PubMed] [Google Scholar]
- 13. Liu J., Liang X., and Gan Z. (2015) Transcriptional regulatory circuits controlling muscle fiber type switching. Sci. China Life Sci. 58, 321–327 [DOI] [PubMed] [Google Scholar]
- 14. Mujika I., and Padilla S. (2001) Muscular characteristics of detraining in humans. Med. Sci. Sports Exerc. 33, 1297–1303 [DOI] [PubMed] [Google Scholar]
- 15. Hagberg J. M., Seals D. R., Yerg J. E., Gavin J., Gingerich R., Premachandra B., and Holloszy J. O. (1988) Metabolic responses to exercise in young and older athletes and sedentary men. J. Appl. Physiol. 65, 900–908 [DOI] [PubMed] [Google Scholar]
- 16. Cairns S. P. (2006) Lactic acid and exercise performance: culprit or friend? Sports Med. 36, 279–291 [DOI] [PubMed] [Google Scholar]
- 17. Arany Z., He H., Lin J., Hoyer K., Handschin C., Toka O., Ahmad F., Matsui T., Chin S., Wu P. H., Rybkin I. I., Shelton J. M., Manieri M., Cinti S., Schoen F. J., et al. (2005) Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 1, 259–271 [DOI] [PubMed] [Google Scholar]
- 18. Gan Z., Burkart-Hartman E. M., Han D. H., Finck B., Leone T. C., Smith E. Y., Ayala J. E., Holloszy J., and Kelly D. P. (2011) The nuclear receptor PPARβ/δ programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 25, 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gan Z., Rumsey J., Hazen B. C., Lai L., Leone T. C., Vega R. B., Xie H., Conley K. E., Auwerx J., Smith S. R., Olson E. N., Kralli A., and Kelly D. P. (2013) Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Invest. 123, 2564–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narkar V. A., Downes M., Yu R. T., Embler E., Wang Y. X., Banayo E., Mihaylova M. M., Nelson M. C., Zou Y., Juguilon H., Kang H., Shaw R. J., and Evans R. M. (2008) AMPK and PPARδ agonists are exercise mimetics. Cell 134, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Narkar V. A., Fan W., Downes M., Yu R. T., Jonker J. W., Alaynick W. A., Banayo E., Karunasiri M. S., Lorca S., and Evans R. M. (2011) Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 13, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rangwala S. M., Wang X., Calvo J. A., Lindsley L., Zhang Y., Deyneko G., Beaulieu V., Gao J., Turner G., and Markovits J. (2010) Estrogen-related receptor γ is a key regulator of muscle mitochondrial activity and oxidative capacity. J. Biol. Chem. 285, 22619–22629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., and Evans R. M. (2004) Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zechner C., Lai L., Zechner J. F., Geng T., Yan Z., Rumsey J. W., Collia D., Chen Z., Wozniak D. F., Leone T. C., and Kelly D. P. (2010) Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 12, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., and Spiegelman B. M. (2002) Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 26. Seiler S. E., Koves T. R., Gooding J. R., Wong K. E., Stevens R. D., Ilkayeva O. R., Wittmann A. H., DeBalsi K. L., Davies M. N., Lindeboom L., Schrauwen P., Schrauwen-Hinderling V. B., and Muoio D. M. (2015) Carnitine acetyltransferase mitigates metabolic inertia and muscle fatigue during exercise. Cell Metab. 22, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muoio D. M., Noland R. C., Kovalik J. P., Seiler S. E., Davies M. N., DeBalsi K. L., Ilkayeva O. R., Stevens R. D., Kheterpal I., Zhang J., Covington J. D., Bajpeyi S., Ravussin E., Kraus W., Koves T. R., et al. (2012) Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 15, 764–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juel C., and Halestrap A. P. (1999) Lactate transport in skeletal muscle: role and regulation of the monocarboxylate transporter. J. Physiol. 517, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jorfeldt L., and Wahren J. (1970) Human forearm muscle metabolism during exercise: V. quantitative aspects of glucose uptake and lactate production during prolonged exercise. Scand. J. Clin. Lab. Invest. 26, 73–81 [DOI] [PubMed] [Google Scholar]
- 30. Omachi A., and Lifson N. (1956) Metabolism of isotopic lactate by the isolated perfused dog gastrocnemius. Am. J. Physiol. 185, 35–40 [DOI] [PubMed] [Google Scholar]
- 31. Cahn R. D., Zwilling E., Kaplan N. O., and Levine L. (1962) Nature and development of lactic dehydrogenases: the two major types of this enzyme form molecular hybrids which change in makeup during development. Science 136, 962–969 [DOI] [PubMed] [Google Scholar]
- 32. Dawson D. M., Goodfriend T. L., and Kaplan N. O. (1964) Lactic dehydrogenases: functions of the two types rates of synthesis of the two major forms can be correlated with metabolic differentiation. Science 143, 929–933 [DOI] [PubMed] [Google Scholar]
- 33. Draoui N., and Feron O. (2011) Lactate shuttles at a glance: from physiological paradigms to anti-cancer treatments. Dis. Model. Mech. 4, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Hall G. (2000) Lactate as a fuel for mitochondrial respiration. Acta Physiol. Scand. 168, 643–656 [DOI] [PubMed] [Google Scholar]
- 35. Markert C. L., Shaklee J. B., and Whitt G. S. (1975) Evolution of a gene: multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science 189, 102–114 [DOI] [PubMed] [Google Scholar]
- 36. Pesce A., McKay R. H., Stolzenbach F., Cahn R. D., and Kaplan N. O. (1964) The comparative enzymology of lactic dehydrogenases: I. properties of the crystalline beef and chicken enzymes. J. Biol. Chem. 239, 1753–1761 [PubMed] [Google Scholar]
- 37. Liu J., Liang X., Zhou D., Lai L., Xiao L., Liu L., Fu T., Kong Y., Zhou Q., Vega R. B., Zhu M. S., Kelly D. P., Gao X., and Gan Z. (2016) Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 8, 1212–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Summermatter S., Santos G., Pérez-Schindler J., and Handschin C. (2013) Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc. Natl. Acad. Sci. U.S.A. 110, 8738–8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Costford S. R., Bajpeyi S., Pasarica M., Albarado D. C., Thomas S. C., Xie H., Church T. S., Jubrias S. A., Conley K. E., and Smith S. R. (2010) Skeletal muscle NAMPT is induced by exercise in humans. Am. J. Physiol. Endocrinol. Metab. 298, E117–E126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bajpeyi S., Pasarica M., Moro C., Conley K., Jubrias S., Sereda O., Burk D. H., Zhang Z., Gupta A., Kjems L., and Smith S. R. (2011) Skeletal muscle mitochondrial capacity and insulin resistance in type 2 diabetes. J. Clin. Endocrinol. Metab. 96, 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Knight D. R., Poole D. C., Schaffartzik W., Guy H. J., Prediletto R., Hogan M. C., and Wagner P. D. (1992) Relationship between body and leg VO2 during maximal cycle ergometry. J. Appl. Physiol. 73, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 42. Schiaffino S., and Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 [DOI] [PubMed] [Google Scholar]
- 43. Elustondo P. A., White A. E., Hughes M. E., Brebner K., Pavlov E., and Kane D. A. (2013) Physical and functional association of lactate dehydrogenase (LDH) with skeletal muscle mitochondria. J. Biol. Chem. 288, 25309–25317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hashimoto T., Hussien R., Oommen S., Gohil K., and Brooks G. A. (2007) Lactate sensitive transcription factor network in L6 cells: activation of MCT1 and mitochondrial biogenesis. FASEB J. 21, 2602–2612 [DOI] [PubMed] [Google Scholar]
- 45. Li S., Swanson S. K., Gogol M., Florens L., Washburn M. P., Workman J. L., and Suganuma T. (2015) Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell 60, 408–421 [DOI] [PubMed] [Google Scholar]
- 46. Apple F. S., and Rogers M. A. (1986) Skeletal muscle lactate dehydrogenase isozyme alterations in men and women marathon runners. J. Appl. Physiol. 61, 477–481 [DOI] [PubMed] [Google Scholar]
- 47. Hittel D. S., Kraus W. E., Tanner C. J., Houmard J. A., and Hoffman E. P. (2005) Exercise training increases electron and substrate shuttling proteins in muscle of overweight men and women with the metabolic syndrome. J. Appl. Physiol. 98, 168–179 [DOI] [PubMed] [Google Scholar]
- 48. Martin O. J., Lai L., Soundarapandian M. M., Leone T. C., Zorzano A., Keller M. P., Attie A. D., Muoio D. M., and Kelly D. P. (2014) A role for peroxisome proliferator-activated receptor γ coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ. Res. 114, 626–636 [DOI] [PMC free article] [PubMed] [Google Scholar]