Abstract
Natural anion channelrhodopsins (ACRs) recently discovered in cryptophyte algae are the most active rhodopsin channels known. They are of interest both because of their unique natural function of light-gated chloride conductance and because of their unprecedented efficiency of membrane hyperpolarization for optogenetic neuron silencing. Light-induced currents of ACRs have been studied in HEK cells and neurons, but light-gated channel conductance of ACRs in vitro has not been demonstrated. Here we report light-induced chloride channel activity of a purified ACR protein reconstituted in large unilamellar vesicles (LUVs). EPR measurements establish that the channels are inserted uniformly “inside-out” with their cytoplasmic surface facing the medium of the LUV suspension. We show by time-resolved flash spectroscopy that the photochemical reaction cycle of a functional purified ACR from Guillardia theta (GtACR1) in LUVs exhibits similar spectral shifts, indicating similar photocycle intermediates as GtACR1 in detergent micelles. Furthermore, the photocycle rate is dependent on electric potential generated by chloride gradients in the LUVs in the same manner as in voltage-clamped animal cells. We confirm with this system that, in contrast to cation-conducting channelrhodopsins, opening of the channel occurs prior to deprotonation of the Schiff base. However, the photointermediate transitions in the LUVs exhibit faster kinetics. The ACR-incorporated LUVs provide a purified defined system amenable to EPR, optical and vibrational spectroscopy, and fluorescence resonance energy transfer measurements of structural changes of ACRs with the molecules in a demonstrably functional state.
Keywords: chloride channel, electron paramagnetic resonance (EPR), lipid vesicle, membrane reconstitution, photoreceptor, LUV, flash photolysis, retinal protein, chloride channel
Introduction
Channelrhodopsins, light-gated ion channels from algae, have become major neuroscience research tools (1). Optogenetic control of excitable animal cells entails two opposite actuators: induction of neuron firing by light-induced depolarization of the cell membrane and suppression of neuronal activity by light-induced hyperpolarization. Photoinduction of firing has been achieved by relatively highly conducting cation channelrhodopsins (CCRs)2 (1). Until recently, for hyperpolarization of cell membranes, low-efficiency ion pumps or chloride-conducting CCR mutants have been used (2–4). High-efficiency natural anion-conducting channelrhodopsins (ACRs) were found recently in cryptophyte algae that exhibit orders of magnitude higher sensitivity than inhibitory tools used previously (5). Their molecular mechanisms, however, remain poorly understood.
To elucidate the structural basis of channel gating, what is needed is a system in which one can measure structural changes with spectroscopic probes in the protein in functional channels. There are four requirements of such a system. (i) The system must allow measurement of light-gated channel conductance, i.e. light-dependent transmembrane ion flux. (ii) The system should contain only purified defined components and avoid the complexity of other components such as those found in electrical current measurements in animal cells, particularly transporters and other channels that potentially influence the channelrhodopsin activity measurement. (iii) The system should be amenable to optical and molecular spectroscopic probes that enable measurements of intramolecular distances: for example, fluorescent probes for energy transfer measurements or spin labels for dipolar coupling measurements. Specific labeling with probes is greatly facilitated by a purified system. (iv) In all microbial rhodopsins that translocate ions for which structures are known, including the light-driven ion pumps bacteriorhodopsin, halorhodopsin (HR), and proteorhodopsin (PR) (Govorunova et al. (29)), and most relevant here, the CCR C1C2 (6, 7), the extracellular portion of the ion channel is open in both the dark and the light, and the cytoplasmic portion of the channel is closed in the dark and opened by light. Therefore, the measurement system requires the ability to easily introduce probes specifically on the cytoplasmic side of the protein to monitor distance changes (e.g. EPR spin probes for dipolar coupling measurements or luminescence resonance energy transfer probes for energy transfer measurements).
We have succeeded in developing such a system by using large unilamellar vesicles (LUVs) and the highly conductive GtACR1. We demonstrate for the first time that purified ACR retains its light-gated channel activity in vitro. Even for the much longer studied CCR, in vitro functional activity of purified pigment was demonstrated in only one publication in black lipid films not amenable to spectroscopic analysis (8). The system we describe is amenable to optical and molecular spectroscopy.
EPR measurements with spin-labeled GtACR1 inserted into LUVs showed that the channels in LUVs are oriented nearly completely inside-out (i.e. with the cytoplasmic domain facing the medium). Measurements of flash-induced absorption changes of ACR in LUVs showed overall similarity to changes from the photocycle in detergent micelles, although rates of intermediate conversions were faster in LUVs. We definitively confirm in the LUVs the previous proposal from comparison of photocurrents in HEK cells and the photocycle in micelles (9) that opening of the channel takes place before deprotonation of the retinylidene Schiff base chromophore. Furthermore, the photocycle rate of GtACR1 in the LUVs is dependent on the membrane potential, a dependence that exists in biological membranes.
Results and Discussion
Light-induced Channel Activity in LUVs Containing GtACR1
GtACR1-inserted in LUVs were prepared with outwardly directed and inwardly directed chloride gradients. We monitored light-induced chloride fluxes by the secondary passive flow of protons (10), in this case caused by the light-induced appearance of chloride conductance. The pH-dependent fluorescent dye pyranine inside the LUV was used to monitor pH changes.
We measured the Cl− flux in LUVs that contain Cl− inside and SO42− or Asp− outside, and vice versa. Without the proton ionophore CCCP, actinic light caused relatively small changes in pH (Fig. 1), indicating low permeability of the LUV lipid membrane for protons. In the presence of protonophore CCCP, light induced a significant increase or decrease of lumenal pH depending on the direction of the chloride gradient (Fig. 1). The appearance of light-induced chloride conductance of the ACR leads to generation of a diffusion electrical potential (Δψ) across the LUV membrane. This electrical potential drives protons across the LUV membrane. Alkalization of the internal space of LUVs (i.e. the lumenal volume) took place when the chloride gradient was directed from the lumen to the exterior of LUVs, and acidification took place when the direction of the gradient was opposite, confirming that they derive from the passive chloride movement. The proton flux continues until full dissipation of the chloride gradient.
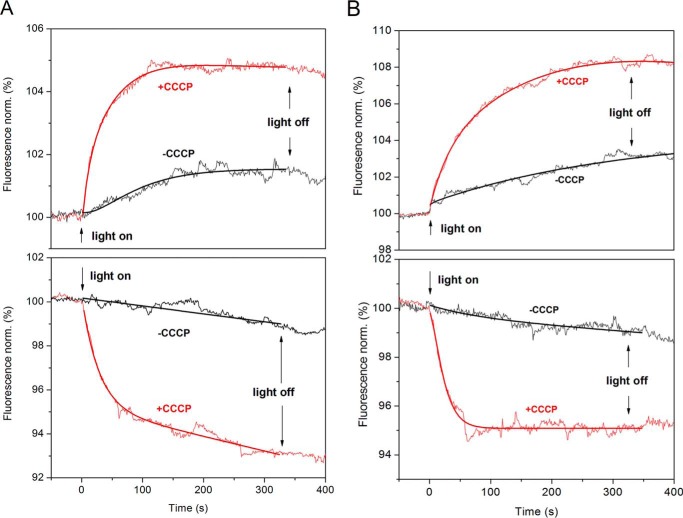
FIGURE 1.
GtACR1 function in LUVs. A and B, light-induced fluorescence changes of the pH-sensitive dye pyranine inside GtACR1-containing LUVs with Cl− inside and SO42− outside (A, top), SO42− inside and Cl− outside (A, bottom), Cl− inside and Asp− outside (B, top), and Asp− inside and Cl− outside (B, bottom). Red and black lines are with and without protonophore (CCCP), respectively. The direction of proton movement corresponds to passive chloride flux out of (A and B, top) or into (A and B, bottom) the LUVs. The fitted lines are shown on each trace. Light on and off times are indicated by arrows. norm., normalization.
In all cases, when illumination stopped, the lumenal pH did not return immediately (Fig. 1). This behavior differs from our previous results on a sodium pump rhodopsin (NaR) in LUVs, which actively transport ions (10). The passive nature of the ion movement in ACR explains the saturation of pH changes after prolonged illumination and the absence of a light-off response. Because proton flux is driven by the chloride diffusion potential, the flow stops when the chloride gradient is dissipated due to the high chloride conductance of the ACR. Closing of chloride channels upon switching off the light does not cause fast changes in Δψ.
These data offer direct evidence that purified GtACR1 acts as a light-gated Cl− channel in vesicles. The direction of pH changes corresponds to the expected direction of chloride fluxes: alkalization when chloride ions (and therefore protons) flow out of the LUV and acidification when chloride ions (and therefore protons) flow into the LUV lumen.
To estimate the pH change from the fluorescence intensity change, we measured the fluorescence intensity of free pyranine in buffer as a function of pH. In typical LUV samples, the fluorescence intensity change was +7% and −8% for Cl− outflow and inflow, which corresponds to pH changes of +0.06 and −0.07 units, respectively. These pH change values underestimate the Cl− flux because 2 mm phosphate buffer diminishes ΔpH in our system.
EPR Measurement of Insertion Orientation
In our previous study of NaR-inserted LUVs, the orientation of the pigment was evident from the direction of membrane potential changes because NaR is a unidirectional ion pump (10). Unlike in ion pumps, in ACRs, ions move in either direction through the channel depending on the sign of the gradient (5). Therefore, the orientation of the ACR molecule in LUVs membrane is not deducible from the photocurrent direction.
To detect the orientation of the GtACR1 protein in LUVs, we determined the accessibility of Cys-reactive spin labels in the LUVs with EPR spectroscopy (11). In a model structure built with the Robetta server based on the structure of C1C2 (Protein Data Bank (PDB) ID: 3UG9) (9), out of nine Cys residues in the molecule, six are located in the transmembrane helices and hence buried in the lipids; two, Cys-6 and Cys-21, are on the extracellular N-terminal region; and Cys-219 is on the loop between helices F and G facing the extracellular side (Fig. 2A). Our SDS-PAGE analysis showed that GtACR1 runs as a dimer (∼66 kDa) in the absence of β-mercaptoethanol (βME), and as a monomer (∼33 kDa) in the presence of βME (supplemental Fig. 1), indicating that GtACR1 forms a dimer, likely through one (Cys-21) or two disulfide bonds (both Cys-6 and Cys-21 are candidates for disulfide bond formation). Therefore, in the lipid environment, one or two cysteines that face the extracellular medium in the cell are expected to be exposed and available for spin labeling, and none face the cytoplasm.
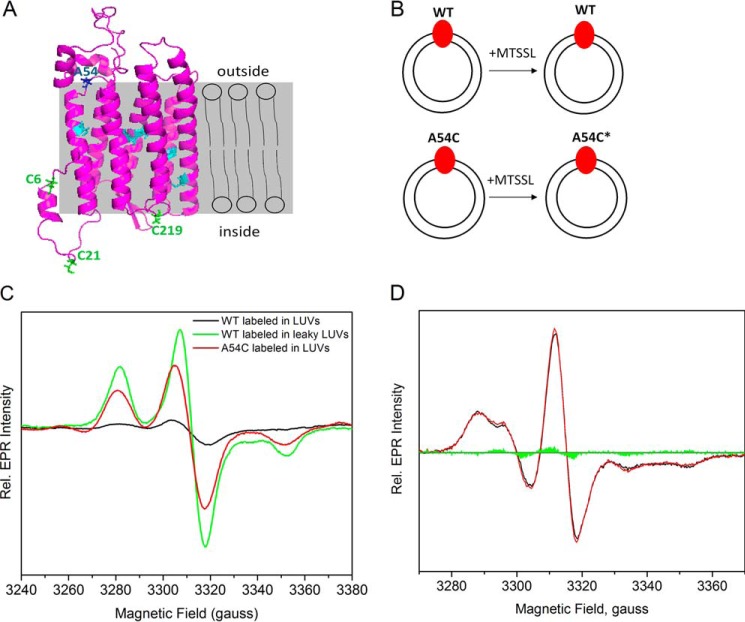
FIGURE 2.
EPR measurements of MTSSL-labeled GtACR1-inserted LUVs to detect the insertion orientation. A, in the predicted structure of GtACR1, the six cysteines in cyan are in the transmembrane helices, and the three cysteines in green face the extracellular side; Ala-54 shown in blue faces the cytoplasmic side. The labels inside and outside refer to the LUVs, of which the lipid bilayer is displayed in gray. B, GtACR1_WT-inserted LUVs (top) and GtACR1_A54C-inserted LUVs (bottom) were incubated with MTSSL. Ala-54 is predicted to face the cytoplasmic side of GtACR1 (panel A), and inserted A54C is labeled by MTSSL. The protein is colored in red, LUVs are represented as two concentric circles, and the spin label is indicated as an asterisk. C, EPR spectra of the frozen LUVs from different treatments were normalized based on their protein concentrations as described under “Experimental Procedures.” The EPR signals are shown for: labeled WT-inserted LUVs, black; labeled WT inserted in leaky LUVs, green; and labeled A54C-inserted LUVs, red. Rel., relative. D, room temperature continuous wave EPR spectra of GtACR1_A54C-inserted LUVs in the dark and light-activated states. The black and red lines are the dark and light spectra, respectively, and the green areas are the light-minus-dark difference spectrum.
When GtACR1 were reconstituted into LUVs and then spin-labeled (Fig. 2B), the ratio between spin label and protein was only 0.2 ± 0.1 (n = 3) (Fig. 2C). This result indicated that the exposed Cys residues face the LUV lumen, preventing them from being labeled by the lipid-impermeable spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSSL). When GtACR1-LUVs were prepared in leaky or disrupted LUVs in the presence of the detergent n-dodecyl-β-d-maltopyranoside (DDM) and spin-labeled, the concentration ratio of spin label versus protein rose to 2.2 (Fig. 2C), agreeing with the modeled structure suggesting that two free cysteines were accessible. These results indicate that two cysteines not buried in the lipid face the inside of the LUVs, preventing them from being accessible to the spin label.
To confirm that the cytoplasmic surface of GtACR1 faces outward in the LUV, we introduced a cysteine mutation on Ala-54 on the loop between helices A and B on the cytoplasmic side and spin-labeled GtACR1_A54C-inserted LUVs (Fig. 2B). The concentration ratio between nitroxide and protein was 1.1 ± 0.2 (n = 3) (Fig. 2C), indicating that A54C is labeled in essentially all of the ACR protein molecules in the LUVs. Therefore, we conclude that GtACR1 has an essentially completely inside-out orientation, in which the cytoplasmic side faces the exterior medium of the LUVs (Fig. 2A), opposite to that in the cell membrane. Comparison of the labeling between A54C and WT (Fig. 2C) indicates that GtACR1 is inserted ∼93% in the inside-out orientation under our experimental conditions.
We conclude that GtACR1, like all other microbial rhodopsins that have been inserted in LUVs, namely bacteriorhodopsin (12), HR (13), PR (14), and NaR (10), is “inside-out,” i.e. the cytoplasmic side of the protein faces the medium. This orientation has been suggested to be due to the greater number of charged residues in the C-terminal region than in the N-terminal region, favoring more rapid hydrophobic N-terminal insertion into the preformed LUVs (15).
The cytoplasmic side of GtACR1 has no cysteine residue accessible to a spin label (Fig. 2A). Therefore, one can site-specifically introduce Cys residues at positions on the cytoplasmic side of the protein without endogenous Cys residues competing for probe attachment. This ability greatly facilitates future EPR measurements, as well as use of other Cys-attached probes, to map the structural changes involved in channel opening under illumination.
The EPR spectrum of A54C-inserted LUVs at ambient temperature is characteristic of a solvent-exposed nitroxide (16, 17) (Fig. 2D). We also compared the EPR line shape of A54C-inserted LUVs under illumination that activates GtACR1. The EPR spectrum in the light did not exhibit a significant difference from that in the dark, indicating similar mobility of the probe on Ala-54 in these two conditions.
Flash Photolysis of GtACR1-inserted LUVs
The photocycle of GtACR1 in detergent micelles strongly differs from those of other microbial rhodopsins (9). In particular, decay of L-like intermediates and accumulation of an M-like intermediate (a measure of the chromophore Schiff base deprotonation) are very slow. The M-like intermediate occurs long after the appearance of the conductive state. We monitored the flash-induced absorption changes of GtACR1-LUVs at three representative wavelengths (410, 460, and 590 nm) and compared the photocycle kinetics between GtACR1 in LUVs and DDM micelles (Fig. 3, A–C). The stages in the photocycle in LUVs are similar to those in the micelles. The rise and decay of the intermediates are more rapid in LUVs, with some phases ∼5-fold faster (Fig. 3, A–C). Nevertheless, the appearance of the conductive state of GtACR1 in LUVs occurs prior to deprotonation of the Schiff base, as was suggested from comparison of photocurrents in HEK cells with photocycle measurements in DDM micelles (9). This observation of a more rapid photocycle rate in LUVs than in micelles concurs with the faster M-like intermediate formation in Pichia membranes than in detergent (9). In a previous study on a CCR, CaChR1 (ChR1 from Chlamydomonas augustae), the photocycle rate of the pigment in Pichia membranes was also 3-fold faster than that in micelles (18). Furthermore, HR and PR exhibited faster photocycle kinetics in liposomes than in micelles (19, 20), suggesting that the kinetics of microbial rhodopsins are generally more rapid in the presence of lipids.
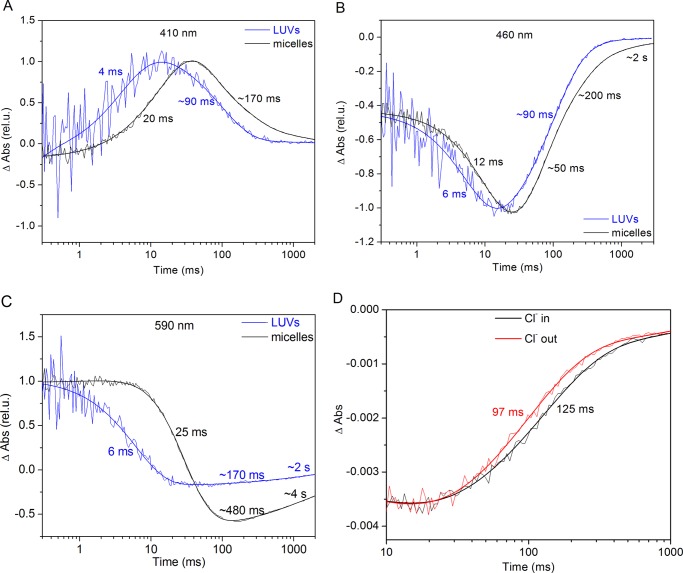
FIGURE 3.
Flash photolysis measurements of GtACR1 in LUVs and micelles. A–C, laser flash-induced absorption (Abs) changes of GtACR1 in LUVs and DDM monitored at three representing wavelengths: 410 (A), 460 (B), and 590 nm (C). The blue lines represent GtACR1-LUVs, and the black lines represent GtACR1 in DDM. The fitted lines and rates are shown on each curve. rel. u., relative units. D, laser flash-induced absorption changes of GtACR1-LUVs monitored at 500 nm with Cl− inside and Cl− outside are shown in black and red, respectively.
A positive-inside Δψ is generated in LUVs with Cl− inside under laser flash excitation with no CCCP present, and a negative Δψ is generated in LUVs with Cl− outside. We observed no difference between the absorption spectra of the two conditions, as well as between absorption of pigment in LUVs and in DDM micelles. We measured the photocycle kinetics of GtACR1 under these two different membrane potentials. Fit as first order processes, the main component of absorbance recovery at 500 nm is ∼30% faster when Cl− is outside than inside (Fig. 3D). Hence, GtACR1 under a negative potential has a faster photocycle rate than under a positive potential (Fig. 3D). The main component of channel closing of GtACR1 in HEK cells is strongly dependent on voltage, and a positive potential inside the cell membrane causes acceleration of closing (21). Because the insertion orientation of GtACR1 is opposite to that in the cell membrane, the faster photocycle decay rate under negative voltage in LUVs matches our previous result showing faster channel closing under positive voltage in HEK cells (21).
SDS-PAGE analysis on detergent-purified GtACR1 indicates that, as with C1C2 (7), GtACR1 forms a disulfide-linked dimer (supplemental Fig. 1). Because GtACR1-LUVs were prepared in the absence of any reducing agent, presumably the pigment is present as a dimer. Because GtACR1-LUVs demonstrate light-induced chloride channel activity, the dimeric form is apparently a functional state. CrChR2 (ChR2 from Chlamydomonas reinhardtii) also forms disulfide-linked dimers and has been shown to form a dimer even without disulfide bonds, indicating a dimer-stabilizing role of the disulfide linkages (22).
In summary, we have developed a system amenable to optical and molecular spectroscopic measurements of structural changes of anion channelrhodopsin with the molecules in a demonstrably functional state. GtACR1 in LUVs allows functional and spectroscopic measurements in defined lipid composition. The in vitro system developed enables investigation of channel-conductive mechanisms without perturbation by bioenergetic ion fluxes encountered in living cells. This in vitro system is amenable to molecular methods such as time-resolved flash UV-visible spectroscopy, vibrational spectroscopy, and EPR, as well as fluorescence methods such as luminescence resonance energy transfer, for characterizing light-induced protein conformational changes and atomic distance measurements under conditions in which a functional state of ACRs is confirmed.
Experimental Procedures
Cloning, Expression, and Purification of GtACR1
The opsin domain of human codon-adapted GtACR1 (residues 1–295) with a C-terminal His8 tag was subcloned into the pPIC9K vector (Invitrogen). The single colony obtained as described previously (9) was used for expression.
For protein expression and purification, the procedures were followed as described (9) with several improvements. Briefly, a starter culture was inoculated into buffered complex glycerol medium, after which the cells were transferred to 1 liter of buffered complex methanol (0.5%) medium supplemented with 5 μm all-trans-retinal (Sigma-Aldrich) and grown at 30 °C with shaking at 240 rpm. After 24 h, the cells were harvested and lysed in 100 ml of ice-cold buffer A (20 mm sodium phosphate, pH 7.4, 100 mm NaCl, 1 mm EDTA, 5% glycerol) by French press. Cell debris was removed by centrifugation at 5,000 × g for 10 min. Membrane fragments were collected by ultracentrifugation at 190,000 × g for 1 h, and then solubilized with 20 ml of buffer B (20 mm Hepes, pH 8.0, 500 mm NaCl, 5% glycerol) and 1% DDM at 4 °C for 1 h. Non-solubilized material was removed by ultracentrifugation with a TLA-100.3 rotor at 110,000 × g for 1 h. The supernatant was mixed with 2 ml of nickel-nitrilotriacetic acid resin (Qiagen) with 15 mm imidazole, incubated at 4 °C for 1 h, and loaded in a gravity flow column (bed dimensions 5 × 25 mm, Bio-Rad). After washing the resin thoroughly with 10 column volumes of buffer B containing 0.02% DDM and 40 mm imidazole, the protein was eluted with buffer B containing 0.02% DDM and 300 mm imidazole. The protein was concentrated using 4-ml Amicon® Ultra centrifugal filters with 10,000 molecular weight cut-off (Millipore) at 4 °C to about 200 μl. 100 μl of protein was further loaded into a size exclusion column using Superdex 200 10/300 GL (bed dimensions 10 × 300 mm, GE Healthcare) and eluted with buffer B with 0.02% DDM at the flow rate of 0.25 ml/min. A large portion of the protein with the peak retention volume at ∼11 ml was separated from the aggregated proteins in the void volume. The fractions of the major peak were pooled and concentrated using the centrifugal filters. The purity of GtACR1 was examined using SDS-PAGE in the presence and absence of βME.
Site-directed mutagenesis was performed with the QuikChange kit (Agilent, Santa Clara, CA) using the WT GtACR1 construct as template. We generated a structural model of GtACR1 based on the structure of C1C2 (PDB ID: 3UG9) using the homology model program at the Robetta server. Ala-54 is observed to be on the loop between helices A and B, facing the cytoplasmic side. The A54C mutant of GtACR1 was expressed and purified, as was WT. The absorption spectra of A54C matched that of GtACR1_WT, and the mutant had full light-gated Cl− channel function assessed by photoelectrophysiology measurement in HEK293 cells.
Reconstitution of GtACR1 into LUVs
Reconstitution of GtACR1 into LUVs and fluorescence measurements were based on the basic procedures established by Rigaud and co-workers (12, 23, 24). 9 mg of soy l-α-phosphatidylcholine (PC) (Avanti, Alabaster, AL) and 1 mg of 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG) (Avanti) in chloroform were mixed and dried into a thin film by Argon gas, and further dried under vacuum. Reconstitution buffer (RB) was as follows: 2 mm sodium phosphate (NaPi) and 100 mm NaCl, pH 7.4, for Cl− outflow experiment; 2 mm NaPi and 50 mm Na2SO4, pH 7.4, for Cl− inflow experiment. 1 ml of RB with 0.5 mm pyranine (Acros Organics) was added to resuspend and rehydrate the lipid film. The lipid suspension was passed through polycarbonate membranes in an extruder (Avanti) to generate LUVs. 20 μl of 10% DDM was added to LUVs and incubated for 1 h, and GtACR1 in DDM was added to the lipids/DDM at the ratio of 1:80 (w/w) and incubated for 1 h. Bio-beads (Bio-Rad) were added to the mixture three times at room temperature, and then incubated overnight at 4 °C. External pyranine was removed by size exclusion chromatography using a Sephadex G-50 column, and the LUVs were eluted with RB containing Na2SO4 for Cl− outflow, or RB containing NaCl for Cl− inflow at 4 °C. The eluate was collected for fluorescence measurements. The concentration of GtACR1 in LUVs was 0.3 ± 0.1 μm calculated from its absorbance (see below). GtACR1-LUVs were between 100 and 200 nm in size and relatively homogeneous by electron microscopy measurement. GtACR1 was reconstituted into LUVs in 2 mm NaPi buffer at pH 7.4 and 100 mm NaCl or 50 mm Na2SO4 for flash photolysis measurements, without the addition of pyranine.
UV-visible Spectroscopy and Laser Flash Photolysis
Absorption spectra of detergent-extracted GtACR1 and GtACR1-LUVs were collected using a Cary 4000 spectrophotometer (Varian, Palo Alto, CA) equipped with an integrating sphere. Absorption spectra of the LUVs were obtained by subtracting a baseline from the logarithmic scale of absorbance in Origin 7.0 (OriginLab, Northampton, MA) to eliminate light scattering following an established method (25). The protein concentration in LUVs was determined from absorbance at 515 nm divided by the extinction coefficient of 45,000 m−1 cm−1, a commonly used first approximation for microbial rhodopsins (26, 27).
Light-induced absorption changes were measured with a laboratory-constructed crossbeam apparatus as described previously (9). Excitation flashes (532 nm, 6 ns, up to 40 mJ) were provided by a Surelite I Nd-YAG laser (Continuum). Signals were amplified by a low-noise current amplifier and digitized with a GaGe Octopus digitizer board (model CS8327, DynamicSignals), with maximum sampling rate of 50 MHz. The time interval between excitation flashes was 15 s, and up to 100 sweeps were averaged for each wavelength. Data analysis was performed with pClamp 10.2 (Molecular Devices) and Origin-Pro 7 (OriginLab) software. Logarithmic filtration of the data was performed by using the GageCon program (kindly provided by L. S. Brown, University of Guelph, Ontario, Canada). Flash-induced absorption changes at four representative wavelengths were recorded: 410, 460, 590, and 500 nm. The absorption increase at 410 nm monitors the deprotonation of the retinylidene Schiff base. The absorption changes at 590 and 460 nm are associated with the biphasic decay of the earliest intermediate in the photochemical reaction cycle, a red-shifted species characteristic of microbial rhodopsins called K, and formation of the subsequent blue-shifted L-like intermediate, which in ACRs represents the conductive state, respectively (9). The absorption change at 500 nm near the absorption maximum of the unphotolyzed state of GtACR1 monitored the overall photocycle of the pigment.
Light-induced Ion Transport in GtACR1-inserted LUVs Monitored with Fluorescence
GtACR1-LUVs were placed into a quartz cuvette with constant stirring, and the fluorescence intensity was measured using a QuantaMaster model QM3-SS (Photon Technology International, Edison, NJ) fluorometer at 5 °C. The fluorescence excitation and emission for pyranine were set at 450 and 500 nm, respectively, and the fluorescence intensity of pyranine increased as pH increased (28). Actinic illumination of vesicles was performed with a halogen-tungsten light source (Ushio, Cypress, CA) equipped with a heat filter and a long-pass orange filter (transmitting 530–700 nm) through a flexible light guide whose aperture was positioned above the cuvette. An interference filter transmitting 500 ± 20 nm was placed along the emission path to remove further stray and diffuse light. The excitation and emission slits were adjusted to obtain near-maximal fluorescence intensity. Protonophore CCCP (0.5 μm in ethanol) was used.
Spin Labeling and EPR Measurement of GtACR1-inserted LUVs
GtACR1 in detergent micelles and LUVs were labeled with spin label (MTSSL, Santa Cruz Biotechnology, Dallas, TX) in our measurements. GtACR1 was exchanged into buffer C (10 mm MES, 100 mm NaCl, pH 6.2) with 0.02% DDM and subsequently incorporated into LUVs in buffer C. GtACR1_WT-LUVs and A54C-LUVs were labeled with MTSSL using a molar ratio of GtACR1:MTSSL = 1:20 overnight at 4 °C or room temperature for 5 h. Unreacted MTSSL was removed by dialysis in a 1-ml Slide-A-LyzerTM G2 dialysis cassette (Thermo Fisher) four times against 1 liter of buffer C at 4 °C for 2–3 days. GtACR1-LUVs were concentrated by pelleting with centrifugation at 190,000 × g and resuspending in buffer C. In a separate experiment, GtACR1-LUVs were prepared in the presence of DDM, and the resulting LUVs were incubated with MTSSL. The unreacted spin label was removed by extensive dialysis as mentioned against buffer C with 0.02% DDM. All GtACR1-LUVs were then frozen and stored in liquid N2 before EPR measurements.
EPR spectra were recorded with a Bruker EMX spectrometer at 115 K or ambient temperature. Data analyses and spectral integrations were conducted using the WinEPR program furnished with the EMX system (11). The following parameters were used for the EPR measurements: frequency, 9.3 GHz; microwave power, 1 milliwatt (115 K) and 10 milliwatt (ambient temperature); modulation frequency, 100 kHz; modulation amplitude, 2 or 4 G (115 K) and 2 G for ambient temperature EPR; and time constant, 0.16 s. The spin densities of the frozen samples were calculated based on the double integrations of the EPR signals and compared with that of a 1 mm Cu2+ standard (11).
Ambient temperature EPR spectra of LUVs were recorded with samples placed in 50-μl capillary tubes. Illumination of the sample was achieved with the same light source equipped with a light guide and a heat and long-pass yellow filter through the front window of the resonator.
Author Contributions
H. L., O. A. S., G. W., and J. L. S. designed the study and wrote the paper. H. L. expressed and purified GtACR1, and reconstituted it into vesicles. H. L. and O. A. S. performed the flash photolysis measurements, and G. W. performed the EPR measurements. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Ah-Lim Tsai for use of an EPR instrument, Dr. Heidi Vitrac for discussions on LUV preparation, and E. Cassie Lane for technical support.
This work was supported by National Institutes of Health Grant R01GM027750 (to J. L. S.) and Endowed Chair Number AU-0009 from the Robert A. Welch Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. 1.
- CCR
- cation channelrhodopsin
- Δψ
- membrane electric potential
- LUV
- large unilamellar vesicle
- GtACR1
- Guillardia theta ACR1
- ACR
- anion channelrhodopsin
- NaR
- sodium pump rhodopsin
- HR
- halorhodopsin
- PR
- proteorhodopsin
- DDM
- n-dodecyl-β-d-maltopyranoside
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazine
- MTSSL
- S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanethiosulfonate
- NaPi
- sodium phosphate
- βME
- β-mercaptoethanol
- RB
- reconstitution buffer.
References
- 1. Deisseroth K. (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow B. Y., Han X., Dobry A. S., Qian X., Chuong A. S., Li M., Henninger M. A., Belfort G. M., Lin Y., Monahan P. E., and Boyden E. S. (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wietek J., Wiegert J. S., Adeishvili N., Schneider F., Watanabe H., Tsunoda S. P., Vogt A., Elstner M., Oertner T. G., and Hegemann P. (2014) Conversion of channelrhodopsin into a light-gated chloride channel. Science 344, 409–412 [DOI] [PubMed] [Google Scholar]
- 4. Berndt A., Lee S. Y., Ramakrishnan C., and Deisseroth K. (2014) Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344, 420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Govorunova E. G., Sineshchekov O. A., Janz R., Liu X., and Spudich J. L. (2015) Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Volz P., Krause N., Balke J., Schneider C., Walter M., Schneider F., Schlesinger R., and Alexiev U. (2016) Light and pH-induced changes in structure and accessibility of transmembrane helix B and its immediate environment in channelrhodopsin-2. J. Biol. Chem. 291, 17382–17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato H. E., Zhang F., Yizhar O., Ramakrishnan C., Nishizawa T., Hirata K., Ito J., Aita Y., Tsukazaki T., Hayashi S., Hegemann P., Maturana A. D., Ishitani R., Deisseroth K., and Nureki O. (2012) Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feldbauer K., Zimmermann D., Pintschovius V., Spitz J., Bamann C., and Bamberg E. (2009) Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. U.S.A. 106, 12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sineshchekov O. A., Li H., Govorunova E. G., and Spudich J. L. (2016) Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc. Natl. Acad. Sci. U.S.A. 113, E1993–E2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H., Sineshchekov O. A., da Silva G. F., and Spudich J. L. (2015) In vitro demonstration of dual light-driven Na+/H+ pumping by a microbial rhodopsin. Biophys. J. 109, 1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu G., Rogge C. E., Wang J. S., Kulmacz R. J., Palmer G., and Tsai A. L. (2007) Oxyferryl heme and not tyrosyl radical is the likely culprit in prostaglandin H synthase-1 peroxidase inactivation. Biochemistry 46, 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rigaud J. L., Bluzat A., and Buschlen S. (1983) Incorporation of bacteriorhodopsin into large unilamellar liposomes by reverse phase evaporation. Biochem. Biophys. Res. Commun. 111, 373–382 [DOI] [PubMed] [Google Scholar]
- 13. Bogomolni R. A., Taylor M. E., and Stoeckenius W. (1984) Reconstitution of purified halorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 81, 5408–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dioumaev A. K., Wang J. M., Bálint Z., Váró G., and Lanyi J. K. (2003) Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry 42, 6582–6587 [DOI] [PubMed] [Google Scholar]
- 15. Rigaud J. L., Pitard B., and Levy D. (1995) Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223–246 [DOI] [PubMed] [Google Scholar]
- 16. Altenbach C., Greenhalgh D. A., Khorana H. G., and Hubbell W. L. (1994) A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 91, 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rink T., Riesle J., Oesterhelt D., Gerwert K., and Steinhoff H. J. (1997) Spin-labeling studies of the conformational changes in the vicinity of D36, D38, T46, and E161 of bacteriorhodopsin during the photocycle. Biophys. J. 73, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sineshchekov O. A., Govorunova E. G., Wang J., Li H., and Spudich J. L. (2013) Intramolecular proton transfer in channelrhodopsins. Biophys. J. 104, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindholm L., Ariöz C., Jawurek M., Liebau J., Mäler L., Wieslander Å., von Ballmoos C., and Barth A. (2015) Effect of lipid bilayer properties on the photocycle of green proteorhodopsin. Biochim. Biophys. Acta 1847, 698–708 [DOI] [PubMed] [Google Scholar]
- 20. Duschl A., McCloskey M. A., and Lanyi J. K. (1988) Functional reconstitution of halorhodopsin: properties of halorhodopsin-containing proteoliposomes. J. Biol. Chem. 263, 17016–17022 [PubMed] [Google Scholar]
- 21. Sineshchekov O. A., Govorunova E. G., Li H., and Spudich J. L. (2015) Gating mechanisms of a natural anion channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 112, 14236–14241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lórenz-Fonfría V. A., and Heberle J. (2014) Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta 1837, 626–642 [DOI] [PubMed] [Google Scholar]
- 23. Gulik-Krzywicki T., Seigneuret M., and Rigaud J. L. (1987) Monomer-oligomer equilibrium of bacteriorhodopsin in reconstituted proteoliposomes. A freeze-fracture electron microscope study. J. Biol. Chem. 262, 15580–15588 [PubMed] [Google Scholar]
- 24. Rigaud J. L., Paternostre M. T., and Bluzat A. (1988) Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry 27, 2677–2688 [DOI] [PubMed] [Google Scholar]
- 25. Leach S. J., and Scheraga H. A. (1960) Effect of light scattering on ultraviolet difference spectra1. J. Am. Chem. Soc. 82, 4790–4792 [Google Scholar]
- 26. Bruun S., Naumann H., Kuhlmann U., Schulz C., Stehfest K., Hegemann P., and Hildebrandt P. (2011) The chromophore structure of the long-lived intermediate of the C128T channelrhodopsin-2 variant. FEBS Lett. 585, 3998–4001 [DOI] [PubMed] [Google Scholar]
- 27. Ernst O. P., Sánchez Murcia P. A., Daldrop P., Tsunoda S. P., Kateriya S., and Hegemann P. (2008) Photoactivation of channelrhodopsin. J. Biol. Chem. 283, 1637–1643 [DOI] [PubMed] [Google Scholar]
- 28. Gan B. S., Krump E., Shrode L. D., and Grinstein S. (1998) Loading pyranine via purinergic receptors or hypotonic stress for measurement of cytosolic pH by imaging. Am. J. Physiol. 275, C1158–C1166 [DOI] [PubMed] [Google Scholar]
- 29. Govorunova E. G., Sineshchekov O. A., Li H., and Spudich J. L. (2017) Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu. Rev. Biochem. 86, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.