Abstract
We previously reported that delivery of a microRNA-138 mimic or siRNA against SIN3A to cultured cystic fibrosis (ΔF508/ΔF508) airway epithelia partially restored ΔF508-cystic fibrosis transmembrane conductance regulator (CFTR)-mediated cAMP-stimulated Cl− conductance. We hypothesized that dissecting this microRNA-138/SIN3A-regulated gene network would identify individual proteins contributing to the rescue of ΔF508-CFTR function. Among the genes in the network, we rigorously validated candidates using functional CFTR maturation and electrolyte transport assays in polarized airway epithelia. We found that depletion of the ubiquitin ligase SYVN1, the ubiquitin/proteasome system regulator NEDD8, or the F-box protein FBXO2 partially restored ΔF508-CFTR-mediated Cl− transport in primary cultures of human cystic fibrosis airway epithelia. Moreover, knockdown of SYVN1, NEDD8, or FBXO2 in combination with corrector compound 18 further potentiated rescue of ΔF508-CFTR-mediated Cl− conductance. This study provides new knowledge of the CFTR biosynthetic pathway. It suggests that SYVN1 and FBXO2 represent two distinct multiprotein complexes that may degrade ΔF508-CFTR in airway epithelia and identifies a new role for NEDD8 in regulating ΔF508-CFTR ubiquitination.
Keywords: cystic fibrosis, E3 ubiquitin ligase, endoplasmic-reticulum-associated protein degradation (ERAD), proteostasis, ubiquitylation (ubiquitination), airway epithelia, neddylation
Introduction
Cystic fibrosis (CF)3 is the most common lethal genetic disease among populations of Caucasian and northern European descent. CF is caused by autosomal recessive inheritance of mutations in the gene CF transmembrane conductance regulator (CFTR) (1), which encodes a phosphorylation- and nucleotide-gated anion channel (2, 3). The majority of CF-associated morbidity and mortality arise from progressive pulmonary infection and inflammation. Approximately 90% of people with CF have at least one mutant ΔF508-CFTR allele, making it the most common CFTR mutation (4, 5). The ΔF508 mutation results in CFTR protein misfolding, retention in the endoplasmic reticulum (ER), and degradation via the ER-associated degradation (ERAD) pathway (6–8).
A major focus of current CF research and therapeutics development is understanding factors regulating CFTR expression and biosynthesis and the cellular mechanisms underlying ΔF508-CFTR degradation and exploiting this knowledge to identify drugs or small molecules that might restore ΔF508-CFTR function. We recently identified a microRNA-regulated gene network that influences both CFTR transcription and processing (9). We discovered that miR-138 regulates wild-type CFTR and ΔF508-CFTR expression and biosynthesis through its interactions with the transcriptional regulatory protein SIN3A and a network of other gene products. miR-138 overexpression or SIN3A inhibition increased ΔF508-CFTR mRNA and protein levels in primary cultures of CF airway epithelia. Both interventions increased the abundance of fully glycosylated ΔF508-CFTR and partially restored ΔF508-CFTR-mediated Cl− transport in primary CF epithelia from multiple human donors (9). This novel finding identified a previously unrecognized mechanism regulating CFTR expression and biosynthesis.
We do not yet understand which miR-138/SIN3A-regulated genes contribute to the rescue of ΔF508-CFTR processing in airway epithelia. We hypothesize that only a subset of miR-138/SIN3A-regulated genes is sufficient and responsible for these effects. In the current study, we investigated candidate downstream target(s) of miR-138/SIN3A that may directly mediate the observed ΔF508-CFTR rescue. We identified synovial apoptosis inhibitor 1 (SYVN1) (Hrd1; E3 ubiquitin ligase), neural precursor cell expressed, developmentally down-regulated 8 (NEDD8; neddylation), and FBXO2 (Fbs1; E3 ubiquitin ligase) as members of this gene network that mediate partial restoration of ΔF508-CFTR trafficking and function. RNAi-mediated depletion of each of these factors increased ΔF508-CFTR protein maturation and significantly improved ΔF508-CFTR-mediated Cl− transport. These findings provide a new understanding of how CFTR expression is regulated in airway epithelia.
Results
MiR-138 and SIN3A Regulate Genes Influencing CFTR Biosynthesis
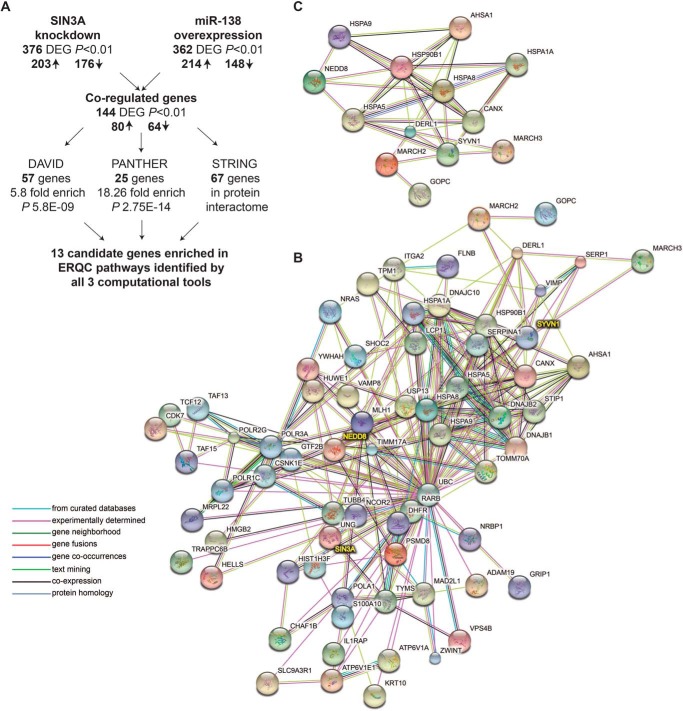
We previously reported that miR-138 overexpression or SIN3A knockdown improved ΔF508-CFTR trafficking and function in airway epithelia and altered transcript levels of target genes (9). Global mRNA transcript profiling after miR-138 overexpression and SIN3A inhibition were performed in the human airway Calu-3 cell line, a respiratory epithelial model (9–11). Using a p value cutoff of <0.01, we observed 80 genes up-regulated and 64 genes down-regulated in response to both interventions (Fig. 1A). We hypothesized that this miR-138/SIN3A-regulated gene network enhances CFTR biogenesis by influencing gene expression at multiple steps along its biosynthetic pathway.
FIGURE 1.
Prioritizing candidate genes by an integrated network analysis. A, flowchart describing identification of 13 candidate genes for the functional screen. DEG, differentially expressed gene; ↑, up-regulated; ↓, down-regulated. B, 67 of the 144 co-regulated differentially expressed genes demonstrate protein/protein interactions. STRING-based predicted protein/protein interactome for 67 genes is shown. C, STRING-based predicted protein/protein interactome for 13 candidate genes.
Here, we utilized three independent pathway and gene ontology analysis tools in parallel as an unbiased strategy to select genes from this list of 144 for further investigation. The Database for Annotation, Visualization, and Integrated Discovery (DAVID, version 6.7) (12) revealed a significant enrichment of 57 genes in pathways including chaperones (32 genes, 4.6-fold enrichment, p < 2.6E−09), unfolded protein response (25 genes, 5.8-fold enrichment, p < 1.1E−06), and ubiquitin proteasomal pathways (20 genes, 4.06-fold enrichment, p < 6.1E−04) (Fig. 1A). Classification of gene products using the tool Protein Analysis Through Evolutionary Relationships (PANTHER, version 10) (13) showed significant enrichment of the “protein folding” pathway (25 genes, 18.26-fold enrichment, p < 2.75E−14) (Fig. 1A). Finally, to account for physical and functional associations between gene products, we used the STRING database version 10 (14) to determine predicted protein/protein interactions within the 144 co-regulated genes. In all, 67 genes showed evidence of interactions (Fig. 1B).
Because many of these gene products are involved in protein trafficking or ER quality control (ERQC), we hypothesized that they may participate in the rescue phenotype observed with miR-138 overexpression and SIN3A inhibition. To identify candidates for further study, we intersected data outputs from all three computational tools. Thirteen candidate genes emerged from the intersection of DAVID, PANTHER, and STRING analyses (Fig. 1C and supplemental Table S1).
RNA Interference Screen of 13 Candidates Reveals Role for SYVN1 and NEDD8 in Rescuing ΔF508-CFTR Biosynthesis
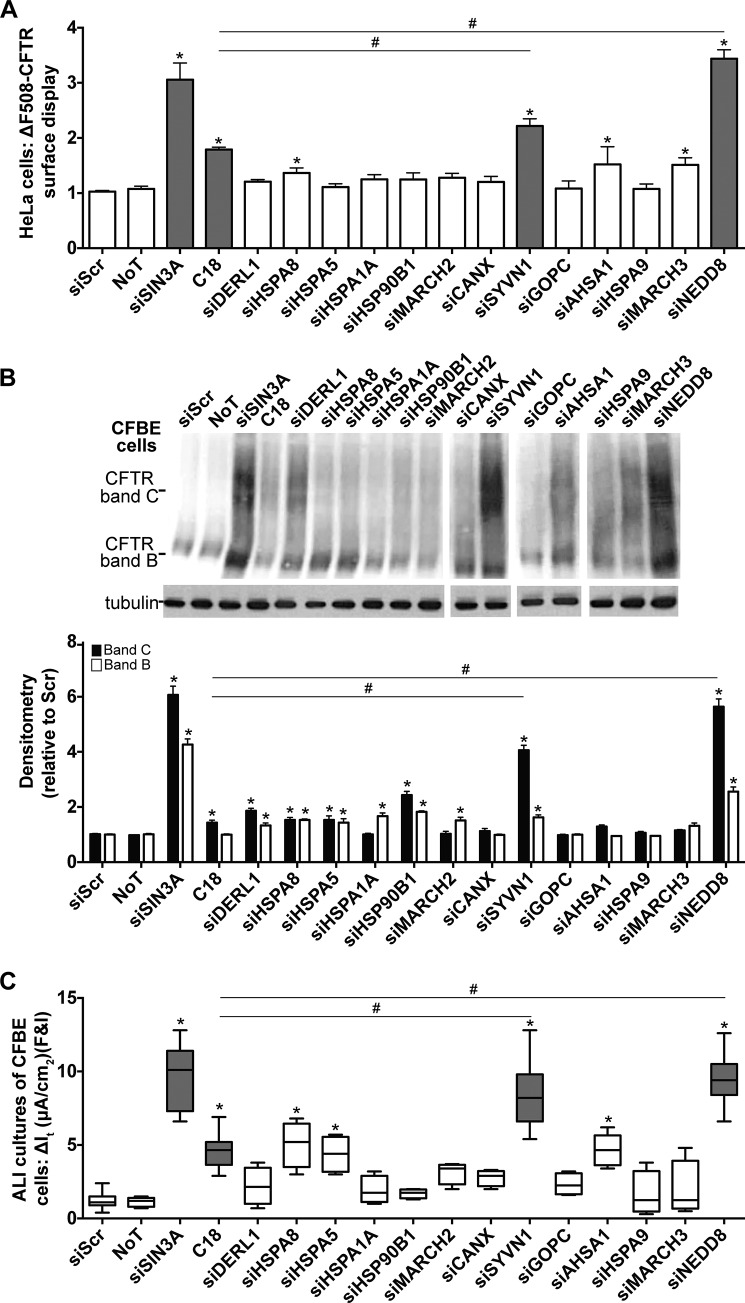
We used a loss of function screen and three assays to evaluate the effect of gene inhibition on ΔF508-CFTR biogenesis: 1) surface display of ΔF508-CFTR in HeLa cells stably expressing HA-tagged ΔF508-CFTR (HeLa-ΔF508-CFTR-HA) (Fig. 2A); 2) maturation of ΔF508-CFTR in cystic fibrosis bronchial epithelial (CFBE) cells, an immortalized cell line created from primary bronchial epithelia isolated from a patient with cystic fibrosis (15), as evidenced by fully glycosylated CFTR (band C) (Fig. 2B); and 3) functional rescue of ΔF508-CFTR in polarized CFBE cells (polarized cells grown at the air-liquid interface to model in vivo physiology) measured as cAMP agonist-induced Cl− transport (Fig. 2C). All assays were performed in parallel with two experimentally validated Dicer substrate siRNAs (DsiRNAs) for each candidate (supplemental Table S2).
FIGURE 2.
ΔF508-CFTR biosynthesis is regulated by SYVN1 and NEDD8. A, HeLa-ΔF508-CFTR-HA cells were transfected with the DsiRNAs targeting each candidate gene. 24 h post-transfection, ΔF508-CFTR surface display was measured using an anti-HA antibody. As negative controls, cells were transfected with siScr or left untreated (no treatment (NoT)). As positive controls, SIN3A expression was depleted with a DsiRNA, or cells were treated with the corrector compound C18 for 24 h (6 μm). -Fold increase and significance are plotted relative to siScr transfection. C18 (6 μm) was administered for 24 h. n = 48. B, representative immunoblot depicting ΔF508-CFTR expression in CFBE cells. CFBE cells were transfected with the DsiRNAs targeting each candidate gene. As negative controls, cells were transfected with siScr, or left untreated (no treatment (NoT)). As positive controls, SIN3A expression was depleted with a DsiRNA, or cells were treated with the corrector compound C18 for 24 h (6 μm). Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B in CFBE cells relative to siScr. n = 4. C18 (6 μm) was administered for 24 h. C, change in It in response to forskolin and IBMX (F&I) treatment in polarized ALI cultures of CFBE cells. n = 8 (minimum) cultures per treatment. C18 (6 μm) was administered basolaterally 24 h prior to electrophysiology study. A and B represent mean with error bars indicating S.E. C, box and whisker plot (minimum-maximum) represents median. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01.
High throughput screens have identified novel compounds, termed correctors, to rescue ΔF508-CFTR protein trafficking and function in vitro (16–22). The first of these to reach clinical trials is VX-809 (lumacaftor) (23), but it failed to be efficacious when administered alone (24). We used a VX-809 analog, the CFTR corrector corr-18 (C18) (20), as a positive control and a benchmark treatment. RNAi knockdown of several of the 13 candidate genes achieved significant improvement in one or more of the assays (Fig. 2, A–C). Two genes outperformed C18 on all three assays: SYVN1 and NEDD8. Inhibition of SYVN1 and NEDD8 expression rescued ΔF508-CFTR trafficking and maturation to levels significantly higher than C18 (Fig. 2, A and B) and improved cAMP-mediated Cl− transport significantly more than C18 treatment alone (Fig. 2C).
Loss of SYVN1 and NEDD8 Expression Alleviates ΔF508-CFTR Polyubiquitination
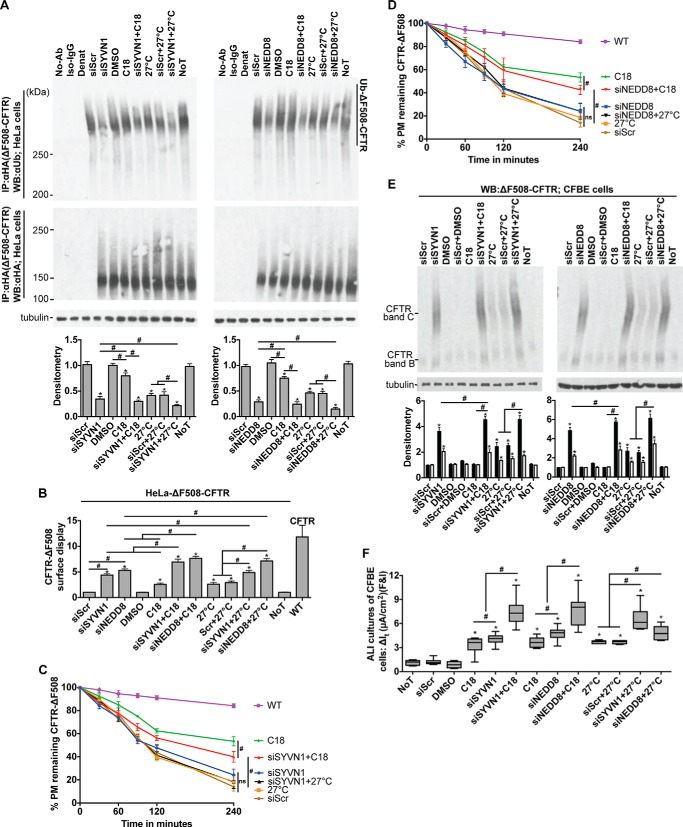
SYVN1 is an E3 ubiquitin-protein ligase and a component of the ERQC system called ERAD, which is involved in ubiquitin-dependent degradation of misfolded ER proteins (25–27). NEDD8 is a ubiquitin-like protein that is covalently attached to its substrates. Cullins form the largest class of NEDD8 substrates, and attachment of NEDD8 to cullins activates their associated E3 ubiquitin ligase activity and thus promotes polyubiquitination and proteasomal degradation of target proteins (28). The identification of these two genes whose functions converge on the polyubiquitination and proteasomal degradation pathway led us to hypothesize that SYVN1 and NEDD8 promote ΔF508-CFTR polyubiquitination and that depleting either protein would improve ΔF508-CFTR maturation in CF cells. To test this hypothesis, we determined the impact of depleting SYVN1 and NEDD8 on ΔF508-CFTR polyubiquitination. We transfected/treated HeLa-ΔF508-CFTR-HA cells with siSYVN1 and siNEDD8 to effect knockdown; 72 h later, we inhibited the proteasome with MG-132 (10 μm) for an hour, harvested protein, immunoprecipitated CFTR with an anti-HA antibody, and blotted for ubiquitin using an anti-ubiquitin antibody. SYVN1 and NEDD8 knockdown each significantly reduced ΔF508-CFTR ubiquitination compared with the scrambled oligonucleotide (siScr) control (Fig. 3A).
FIGURE 3.
SYVN1 or NEDD8 depletion in concert with C18 or low temperature enhance functional rescue of ΔF508-CFTR. A, ΔF508-CFTR ubiquitination measured 72 h after the indicated treatments. CFTR was immunoprecipitated with anti-HA antibody, and ubiquitin was detected with anti-ubiquitin antibody. CFTR protein levels were detected using the anti-HA antibody. C18 (6 μm) and 27 °C were administered 24 h prior to harvesting protein. Densitometry is relative to siScr. n = 4. B, surface display of ΔF508-CFTR in HeLa cells measured by cell surface ELISA 72 h after the indicated treatments. -Fold increase and significance relative to siScr transfection are shown. C18 (6 μm) was administered for 24 h; low temperature (27 °C) was administered for 24 h. n = 18. C and D, membrane stability of ΔF508-CFTR in HeLa cells measured by pulse-chase cell surface ELISA 72 h after the indicated treatments. Chase was performed at 37 °C. n = 18. E, representative immunoblot depicting ΔF508-CFTR expression in CFBE cells. Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B relative to siScr in CFBE cells. n = 4. F, change in It in response to forskolin and IBMX (F&I) treatment in polarized ALI cultures of CFBE cells. n = 10 (minimum) cultures per treatment. C18 (6 μm) and 27 °C treatment were for 24 h prior to electrophysiology study. A and B represent mean with error bars indicating S.E. F, box and whisker plot (minimum-maximum) represents median. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01. WB, Western blotting; Ab, antibody; NoT, no treatment; Denat, denatured; Ub, ubiquitin; PM, plasma membrane.
SYVN1/NEDD8 Knockdown Enhances ΔF508-CFTR Biosynthesis by Proteasome Inhibition
Reduced ΔF508-CFTR ubiquitination in response to inhibiting SYVN1 or NEDD8 expression suggests either inactivation of the ΔF508-CFTR ubiquitination machinery or disruption of chaperone complexes that target the misfolded protein for ubiquitination. Either of these scenarios could explain the observed partial restoration of ΔF508-CFTR trafficking, maturation, and function. To elucidate the impact of SYVN1 and NEDD8 knockdown on CFTR, we measured the membrane stability of the rescued protein by pulse-chase live cell surface ELISA in HeLa-ΔF508-CFTR-HA cells. Twenty-four hours after transfecting cells with the noted reagents, we determined the residence time of ΔF508-CFTR at the membrane for five time points after beginning the chase (chase performed at 37 °C). Although SYVN1 or NEDD8 knockdown increased ΔF508-CFTR trafficking to the membrane (Fig. 3B), the treatments had little effect on ΔF508-CFTR membrane stability compared with the negative control (low temperature, 27 °C, treatment) (Fig. 3, C and D).
Because treatment with C18 and low temperature rescues ΔF508-CFTR processing and function (8, 29), we hypothesized that inhibiting SYVN1 or NEDD8 expression (for 72 h) in concert with C18 (6 μm for 24 h) or low temperature (27 °C for 24 h) would enhance functional rescue of ΔF508-CFTR. Combining SYVN1 or NEDD8 knockdown with C18 significantly increased ΔF508-CFTR trafficking (Fig. 3B) and maturation as measured by band C formation (Fig. 3E) compared with either treatment alone. Of note, C18 has little impact on ΔF508-CFTR maturation (29), and the absence of fully glycosylated CFTR (band C) is not an indication of drug inaction. A similar improvement in ΔF508-CFTR trafficking and maturation was observed upon combining SYVN1 or NEDD8 knockdown with low temperature treatment (Fig. 3, B and E). Combining SYVN1 or NEDD8 knockdown with C18 provided no further reduction in ΔF508-CFTR ubiquitination compared with either treatment alone (Fig. 3A) possibly because C18 treatment had little impact on ΔF508-CFTR ubiquitination levels. In contrast, a greater reduction in ΔF508-CFTR ubiquitination was observed upon combining low temperature treatment with either SYVN1 or NEDD8 knockdown (Fig. 3A). Of note, membrane residence of ΔF508-CFTR significantly improved only upon combining SYVN1 or NEDD8 inhibition with C18 treatment (Fig. 3, C and D). Finally, significantly more ΔF508-CFTR Cl− channel activity was observed in polarized CFBE cells upon combining C18 or low temperature with SYVN1 or NEDD8 knockdown compared with either treatment alone (Fig. 3F).
We selected C18 and low temperature for combinatorial treatments because, first, C18 interacts specifically with CFTR and has little impact on the ERQC/ubiquitination pathway (29), and second, ΔF508-CFTR processing is temperature-sensitive (8, 29–31), and the effect of low temperature on chaperones/co-chaperone expression in the ERQC/ubiquitination pathway is well characterized (19, 32, 33). The combination of SYVN1 and NEDD8 inhibition with either C18 or low temperature significantly enhanced rescue of ΔF508-CFTR maturation and function. Furthermore, inhibition of either gene reduced ubiquitination independently of C18 or low temperature, and SYVN1 and NEDD8 inhibition had no impact on ΔF508-CFTR membrane stability. Together, these findings suggest that both SYVN1 and NEDD8 modulate the ubiquitin proteasome pathway regulating ΔF508-CFTR processing and ubiquitin-mediated degradation.
Ubiquitin Ligase SYVN1 Regulates ΔF508-CFTR Polyubiquitination by the Ring Finger Protein 5 (RNF5)/Autocrine Motility Factor Receptor (AMFR) Pathway
Ubiquitination is a prerequisite for ΔF508-CFTR degradation by the ERAD pathway, which facilitates retrotranslocation and proteasomal targeting (27). Distinct E3 ubiquitin ligases have been identified that are involved in ΔF508-CFTR ubiquitination (34, 35). DNAJB12, HSPA8 (Hsc70), and RNF5 (RMA1) are involved in co-translational ubiquitination of ΔF508-CFTR (36, 37), whereas CHIP (STUB1) is involved with ubiquitination of full-length ΔF508-CFTR (36, 38). RNF5 (E3 ubiquitin-protein ligase) and AMFR (Gp78; E3 ubiquitin-protein ligase) are integral to the ΔF508-CFTR ubiquitination machinery in the ER (34, 37). AMFR complexes with RNF5 and specifically promotes polyubiquitination of ΔF508-CFTR (35). However, overexpression of wild-type AMFR has no effect on the degradation of ΔF508-CFTR, which suggests that an additional factor(s) is required for the degradation of polyubiquitinated ΔF508-CFTR (35).
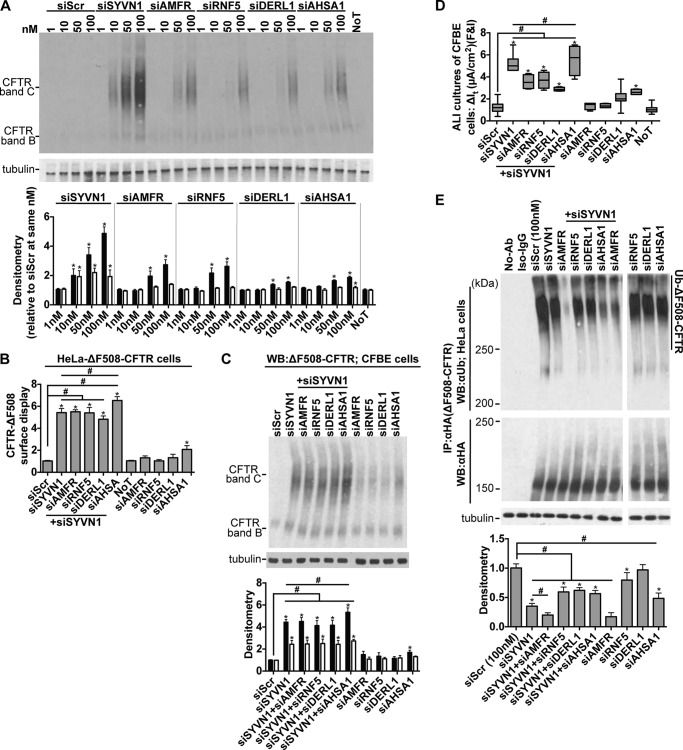
We hypothesized that SYVN1 might be the missing factor that regulates ΔF508-CFTR ubiquitination and degradation. We included AHSA1 in our studies as it stimulates Hsp90 ATPase activity, thereby regulating chaperone-mediated degradation of ΔF508-CFTR (21), a mechanism independent of the ER-based ubiquitination machinery. We also included DERL1 because it interacts with the ERAD machinery associated with ΔF508-CFTR degradation (39). We validated two different DsiRNAs against RNF5, AMFR, AHSA1, and DERL1 (supplemental Table S1) and demonstrated significant improvement in ΔF508-CFTR maturation upon target gene knockdown in a dose-dependent manner (Fig. 4A). Knockdown of AMFR, RNF5, DERL1, or AHSA1 improved ΔF508-CFTR trafficking (Fig. 4B), maturation (Fig. 4, A and C), and function in polarized cultures of CFBE cells (Fig. 4D) to varying degrees. These results suggest that multiple pathways may converge on and influence ubiquitin-mediated proteasomal degradation. Notably, SYVN1 knockdown achieved greater ΔF508-CFTR rescue (Fig. 4, B–D), suggesting that SYVN1 may participate in more than one of the aforementioned pathways for ΔF508-CFTR degradation. However, these results also raise the possibility that SYVN1 comprises an independent arm of the ERQC machinery that targets ΔF508-CFTR for ubiquitination and proteasomal degradation.
FIGURE 4.
SYVN1 regulates ΔF508-CFTR biosynthesis in part via the RNF5/AMFR pathway. A, representative immunoblots depicting rescue of ΔF508-CFTR maturation in CFBE cells. Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B relative to siScr in CFBE cells. n = 3. B, surface display of ΔF508-CFTR in HeLa cells measured by cell surface ELISA 72 h after the indicated treatments. -Fold increase and significance are relative to siScr transfection. n = 18. C, representative immunoblot depicting ΔF508-CFTR expression in CFBE cells. Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B relative to siScr in CFBE cells. n = 4. D, change in It in response to forskolin and IBMX (F&I) treatment in polarized ALI cultures of CFBE cells. n = 6. E, ΔF508-CFTR ubiquitination measured 72 h after the indicated treatments. CFTR was immunoprecipitated with anti-HA antibody, and ubiquitin was detected with anti-ubiquitin antibody. Densitometry is relative to siScr. n = 4. A, B, C, and E represent mean with error bars indicating S.E. D, box and whisker plot (minimum-maximum) represents median. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01. WB, Western blotting; Ab, antibody; NoT, no treatment; Ub, ubiquitin.
To investigate further how the ER membrane-associated SYVN1 influences ΔF508-CFTR ubiquitination, we hypothesized that a combinatorial gene silencing approach would help identify the pathways by which SYVN1 influences ΔF508-CFTR targeting to the proteasome. Using co-transfected DsiRNAs, we simultaneously reduced SYVN1 expression and either AMFR, RNF5, DERL1, or AHSA1. Although SYVN1 knockdown increased ΔF508-CFTR trafficking in HeLa cells (Fig. 4B), maturation in CFBE cells (Fig. 4C), and function in polarized cultures of CFBE cells (Fig. 4D), significantly greater rescue was observed only with the combined knockdown of SYVN1 and AHSA1 (Fig. 4, B–D). SYVN1 knockdown reduced ΔF508-CFTR ubiquitination (Fig. 4E), and only the dual inhibition of SYVN1 and AMFR yielded a greater reduction in ΔF508-CFTR ubiquitination (Fig. 4E) perhaps because of the role AMFR has in polyubiquitinating ΔF508-CFTR ubiquitin chains as an E4-like ligase (35) (Fig. 4E). These results suggest that SYVN1 augments the RNF5/AMFR ubiquitination machinery by acting either as a regulator or as a component in a larger complex.
SYVN1 Catalytic Activity Regulates ΔF508-CFTR Polyubiquitination
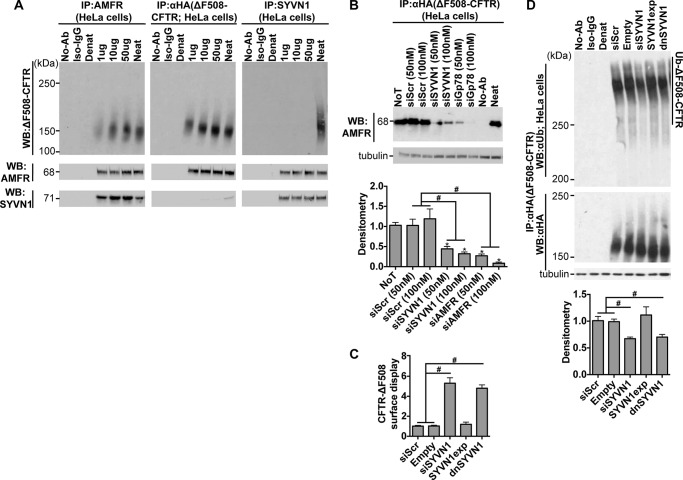
SYVN1 and AMFR form two distinct complexes: SYVN1-Sel1L-HERPUD1-DERL1-DERL2-OS9 and AMFR-UBAC2-UbxD8, which communicate with each other via DERL2/UbxD8 or SYVN1/AMFR interactions (40). To understand the interaction network existing among AMFR, SYVN1, and ΔF508-CFTR, we immunoprecipitated each of these proteins and blotted for all three proteins. AMFR physically interacts with both ΔF508-CFTR and SYVN1, but no physical interaction was observed between SYVN1 and ΔF508-CFTR (Fig. 5A). We next tested the impact of reducing SYVN1 levels on AMFR/ΔF508-CFTR interaction. We reduced SYVN1 levels using DsiRNAs in HeLa-ΔF508-CFTR-HA cells; 72 h later, we inhibited the proteasome with MG-132 (10 μm) for an hour, harvested protein, immunoprecipitated CFTR with an anti-HA antibody, and blotted for AMFR. SYVN1 inhibition significantly reduced AMFR/ΔF508-CFTR interaction (Fig. 5B) compared with control treatments. This result could explain why loss of SYVN1 expression resulted in reduced ΔF508-CFTR ubiquitination.
FIGURE 5.
Catalytic domain of SYVN1 is important to regulate ΔF508-CFTR biosynthesis. A, co-immunoprecipitation of AMFR, SYVN1, and ΔF508-CFTR after immunoprecipitation of AMFR (anti-AMFR antibody), SYVN1 (anti-SYVN1 antibody), or ΔF508-CFTR (anti-HA antibody). B, co-immunoprecipitation of AMFR with ΔF508-CFTR (immunoprecipitated with anti-HA antibody) measured 72 h after the indicated treatments. Densitometry is relative to siScr. n = 3. C, surface display of ΔF508-CFTR in HeLa cells measured by cell surface ELISA 72 h after the indicated treatments. -Fold increase and significance are relative to siScr transfection. n = 18. D, ΔF508-CFTR ubiquitination measured 72 h after the indicated treatments. CFTR was immunoprecipitated with anti-HA antibody, and ubiquitin was detected with anti-ubiquitin antibody. CFTR protein levels were detected using the anti-HA antibody. Densitometry is relative to siScr. n = 4. B–D represent mean with error bars indicating S.E. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01. WB, Western blotting; Ab, antibody; NoT, no treatment; Ub, ubiquitin; Denat, denatured; dnSYVN1, dominant-negative SYVN1; SYVN1exp, exogenously expressed wild-type SYVN1; Neat, total protein.
To test whether the regulatory role exerted by SYVN1 on ΔF508-CFTR ubiquitination is mediated by its catalytic activity, we exogenously expressed wild-type SYVN1 or a catalytically inactive dominant-negative SYVN1 by transfecting HeLa cells with the respective cDNAs. Exogenous expression of wild-type SYVN1 did not rescue ΔF508-CFTR trafficking to the cell surface (Fig. 5C) or reduce ubiquitinated ΔF508-CFTR levels (Fig. 5D). However, expression of the catalytically inactive dominant-negative SYVN1 improved ΔF508-CFTR trafficking (Fig. 5C) and reduced ΔF508-CFTR ubiquitination (Fig. 5D) to an extent similar to SYVN1 depletion by DsiRNAs. These results indicate that the rescue phenotype observed with SYVN1 knockdown is due to the loss of its catalytic activity.
NEDD8 Regulates ΔF508-CFTR Ubiquitination via the Skp, Cullin, F-box (SCF)-containing (SCFFBXO2) Complex
The SCFFBXO2 complex binds specifically to proteins with N-linked high mannose oligosaccharides and contributes to ubiquitination of N-glycosylated proteins (41). FBXO2 is an E3 ligase that directly interacts with ΔF508-CFTR; others include CHIP, RMA1, NEDD4-2, and AMFR (also an E4-like ligase) (34). Yoshida et al. (41) reported that ΔF508-CFTR is ubiquitinated by the SCFFBXO2 complex in HEK293T and COS7 cells and that loss of the F-box domain in FBXO2 significantly suppressed ΔF508-CFTR degradation. Their results and our findings suggest a possible mechanism for how NEDD8 inhibition might reduce ΔF508-CFTR degradation.
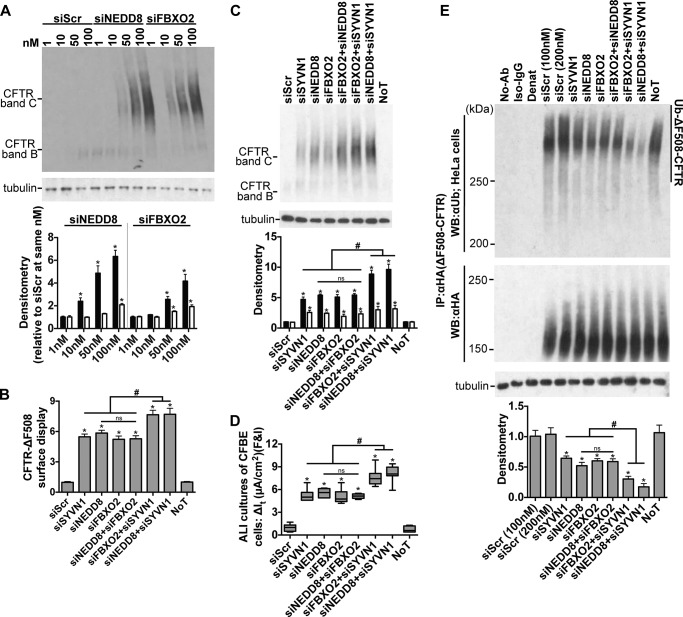
We validated two different DsiRNAs against FBXO2 (supplemental Table S1) and demonstrated significant improvement in ΔF508-CFTR maturation upon target gene knockdown in a dose-dependent manner (Fig. 6A). Inhibition of FBXO2 expression improved ΔF508-CFTR trafficking in HeLa cells (Fig. 6B), maturation in CFBE cells (Fig. 6, A and C), and function in polarized cultures of CFBE cells (Fig. 6D) to levels similar to that achieved with NEDD8 inhibition. FBXO2 knockdown also significantly reduced ΔF508-CFTR ubiquitination (Fig. 6E). As NEDD8 stimulates ubiquitination via the cullin-RING ubiquitin ligase (CRL) complexes, these results provide a direct link between NEDD8 and FBXO2-mediated ΔF508-CFTR ubiquitination.
FIGURE 6.
NEDD8 and FBXO2 exhibit overlapping action in regulating ΔF508-CFTR biosynthesis. A, representative immunoblots depicting rescue of ΔF508-CFTR maturation in CFBE cells. Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B relative to siScr in CFBE cells. n = 3. B, surface display of ΔF508-CFTR in HeLa cells measured by cell surface ELISA 72 h after the indicated treatments. -Fold increase and significance are relative to siScr transfection. n = 18. C, representative immunoblot depicting ΔF508-CFTR expression in CFBE cells. Protein was harvested 72 h post-treatment. Densitometry represents -fold increase of ΔF508-CFTR bands C and B relative to siScr in CFBE cells. n = 4. D, change in It in response to forskolin and IBMX (F&I) treatment in polarized ALI cultures of CFBE cells. n = 6. E, ΔF508-CFTR ubiquitination measured 72 h after the indicated treatments. CFTR was immunoprecipitated with anti-HA antibody, and ubiquitin was detected with anti-ubiquitin antibody. Densitometry is relative to siScr. n = 4. A, B, C, and E represent mean with error bars indicating S.E. D, box and whisker plot (minimum-maximum) represents median. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01; ns, not significant. WB, Western blotting; Ab, antibody; NoT, no treatment; Ub, ubiquitin; Denat, denatured.
We next used a combinatorial gene knockdown approach to assess the nature of interactions between NEDD8 and FBXO2 with respect to ΔF508-CFTR ubiquitination and trafficking. We co-transfected DsiRNAs to reduce NEDD8 and FBXO2 expression either alone or in combination. Remarkably, NEDD8 knockdown, FBXO2 knockdown, or the combined inhibition all yielded similar improvements in ΔF508-CFTR trafficking in HeLa cells (Fig. 6B), maturation in CFBE cells (Fig. 6, A and C), function in polarized cultures of CFBE cells (Fig. 6D), and reduction in ΔF508-CFTR ubiquitination (Fig. 6E), supporting their interdependence within the SCFFBXO2 complex for ΔF508-CFTR degradation. Interestingly, combining SYVN1 knockdown with either NEDD8 or FBXO2 inhibition further improved ΔF508-CFTR biosynthesis. Trafficking, maturation, and functional rescue of the mutant protein were significantly greater with the combinations of SYVN1/NEDD8 and SYVN1/FBXO2 compared with each knockdown alone (Fig. 6, B–D). Similarly, ΔF508-CFTR ubiquitination was maximally reduced by SYVN1 knockdown paired with either NEDD8 or FBXO2 inhibition (Fig. 6E). These results suggest that FBXO2 and NEDD8 act via the same pathway, which is distinct from the effects of SYVN1, and that the SCFFBXO2 complex is involved in ΔF508-CFTR ubiquitination and degradation in airway epithelia.
Of note, the SYVN1/NEDD8 combined knockdown gave consistently better results than the SYVN1/FBXO2 combination on all measures (Fig. 6, B–E), conferring the greatest overall improvement in ΔF508-CFTR biosynthesis. Together, these results suggest that SYVN1 and NEDD8, although acting via different pathways, are complementary in directing mutant CFTR to the proteasome and may be suitable therapeutic targets for optimal ΔF508-CFTR reconstitution.
Inhibiting SYVN1 and NEDD8 Expression Rescues cAMP-dependent Anion Transport in Cultured Primary CF Airway Epithelia
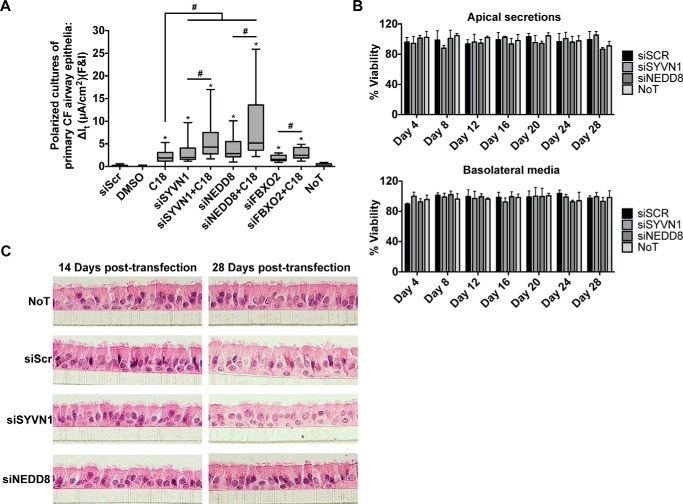
Encouraged by these results, we next reduced SYVN1, NEDD8, and FBXO2 expression in primary cultures of CF airway epithelia homozygous for the ΔF508 mutation. At 14 days post-transfection, we observed significantly improved cAMP-activated Cl− channel activity in cells from multiple human donors (Fig. 7A). Combining these individual treatments with C18 further improved CFTR-dependent Cl− transport (Fig. 7A).
FIGURE 7.
Inhibition of SYVN1, NEDD8, or FBXO2 partially restores ΔF508-CFTR function in primary CF airway epithelial cells. A, change in It in response to forskolin and IBMX (F&I) treatment in polarized primary airway epithelial cell cultures. n = 4 (minimum) cultures per donor; all treatments were replicated in six donors or more; FBXO2 treatment was replicated in three donors. B, LDH levels measured in the airway surface liquid and basolateral medium of primary air-liquid interface non-CF airway epithelial cultures every 4 days for a period of 28 days post-transfection with the noted reagents. n = 3 donors (three cultures per donor). Error bars indicate S.E. C, cell morphology was assessed by hematoxylin and eosin (H&E) staining and examination of sections of primary non-CF airway epithelial cell cultures at days 14 and 28 post-transfection with the noted interventions. A well differentiated morphology was maintained across all interventions. n = 3 donors (three cultures per donor). A, box and whisker plot (minimum-maximum) represents median. Statistical significance was determined by one-way ANOVA with Holm-Bonferroni correction: *, p < 0.01 (relative to siScr); #, p < 0.01. NoT, no treatment.
To evaluate the possibility that knockdown of either SYVN1 or NEDD8 was cytotoxic, we transfected primary airway epithelial cells from three non-CF donors and grew them at the air-liquid interface (ALI). We measured lactate dehydrogenase (LDH) release from the apical and basolateral compartments at 4-day intervals for 28 days (Fig. 7B) and performed hematoxylin and eosin staining on similar cultures at days 14 and 28 to assess changes in cell morphology (Fig. 7C). No differences were observed among untreated, siScr-, SYVN1 DsiRNA-, or NEDD8 DsiRNA-transfected cells. These results suggest that prolonged inhibition of SYVN1 or NEDD8 expression is well tolerated by airway epithelial cells.
Discussion
Our previous study revealed a gene expression network associated with improved ΔF508-CFTR biosynthesis and functional rescue (9). In the present study, we mined these data to identify individual gene targets with putative roles along the ERQC-ERAD pathways. Here, we show that inhibiting SYVN1, NEDD8, or FBXO2 expression restores intracellular transport and function of ΔF508-CFTR in primary CF airway epithelial cells. This rescue phenotype is most likely mediated by impairing the ERAD/ubiquitination machinery that targets ΔF508-CFTR for proteasomal degradation.
SYVN1 depletion decreased ΔF508-CFTR ubiquitination. This result suggested that SYVN1, an E3 ubiquitin ligase, was involved in regulating ΔF508-CFTR polyubiquitination. We also observed significantly less ubiquitinated ΔF508-CFTR upon combining SYVN1 knockdown with low temperature. In contrast, combining SYVN1 knockdown with C18 treatment failed to decrease ubiquitinated ΔF508-CFTR levels further. Upon measuring ΔF508-CFTR channel function, both C18 and low temperature treatment enhanced the level of rescue observed with SYVN1 knockdown alone. The results with low temperature treatment are particularly interesting. Although C18 specifically interacts with ΔF508-CFTR and modulates its folding (as observed with improved membrane residence times when combined with SYVN1 or NEDD8 knockdown), low temperature has a dramatic effect on chaperone and co-chaperone complexes that target ΔF508-CFTR to the proteasome. The observed cooperativity in maturation and functional rescue between SYVN1 knockdown and low temperature suggests a shared group of genes or protein complexes involved in ΔF508-CFTR ubiquitination and degradation. If such overlap exists it might provide insight into how, in the presence of low temperature or SYVN1 knockdown, ΔF508-CFTR escapes the Hsc70-CHIP E3 complex that monitors the conformation of different regions of nascent CFTR (36, 38).
Multiple pathways contribute to ΔF508-CFTR ubiquitination and delivery to the proteasome, a feature we exploited in determining whether SYVN1 was part of the RNF5-AMFR network. In addition to its known role in ubiquitinating multiple substrates, AMFR (gp78) also functions as an E4-like ligase and extends ubiquitin chains on ΔF508-CFTR (34, 35). Morito et al. (35) demonstrated that RNF5 (RMA1) worked upstream of AMFR by monoubiquitinating ΔF508-CFTR. AMFR recognizes the ubiquitin already conjugated to ΔF508-CFTR via the coupling of ubiquitin to ER degradation domain and further catalyzes ubiquitin chain formation in a manner similar to that of a multiubiquitin chain assembly factor (E4-like) (35). We noticed that ΔF508-CFTR rescue was further modestly increased when SYVN1 and AHSA1 knockdown were combined, whereas a similar combination of SYVN1 knockdown with depletion of RNF5 or AMFR failed to further enhance the rescue phenotype. This suggests that SYVN1 works together with RNF5-AMFR in the same pathway to mediate ubiquitination of ΔF508-CFTR. Furthermore, the rescue phenotype likely requires loss of SYVN1 catalytic activity as the expression of a catalytically inactive SYVN1 recapitulated results observed with SYVN1 knockdown.
Recent studies suggest that SYVN1 and AMFR act in sequence to promote misfolded protein turnover; SYVN1 is involved in retrotranslocation and ubiquitination of ERAD substrates, whereas AMFR provides an accessory function downstream of SYVN1 probably through interaction with the cytosolic chaperone BAG6 (40). This suggests that AMFR primarily functions to maintain the solubility of retrotranslocated polypeptides (42). Furthermore, SYVN1 is essential for ubiquitination of both luminal and membrane ERAD substrates, whereas AMFR does not share a similar role in retrotranslocation as transmembrane domains of AMFR are dispensable for ERAD (40, 43). These studies and others point to an increasingly important role for SYVN1 in ERAD and possibly an essential role in ΔF508-CFTR degradation.
Our results regarding the effects of SYVN1 inhibition and CFTR function must be interpreted within the confines of the cell system used. We examined CFTR within cultured primary airway epithelial cells, a model with many similarities to respiratory epithelia in vivo. Here, several complementary experimental approaches show that SYVN1 plays a significant role in degrading Δ508-CFTR. In other non-differentiated cell types and heterologous expression systems, the precise role of SYVN1 in ΔF508-CFTR degradation is controversial and may reflect cell type differences that are not applicable to CFTR biogenesis in pulmonary epithelia (35, 44). We also used experimentally validated, chemically modified DsiRNAs to inhibit gene expression, a method with high potency, reproducibility, and a low off-target profile (45). Moreover, AMFR is a highly unstable protein in contrast to the more stable SYVN1 (46, 47), suggesting that our approach captured the dynamic interaction between AMFR and SYVN1 and their influence on CFTR. Our results corroborate findings of Okiyoneda et al. (48) who showed that SYVN1 inhibition improved ΔF508-CFTR trafficking to the plasma membrane in HeLa cells. Additionally, Gnann et al. (49) demonstrated that SYVN1 knockdown in yeast stabilized ΔF508-CFTR.
NEDD8 knockdown resulted in partial rescue of ΔF508-CFTR processing and function and decreased ΔF508-CFTR ubiquitination. NEDD8 stimulates ubiquitination via the CRL complexes upon covalent attachment to cullin. CRLs constitute the largest E3 ubiquitin ligase group, comprising >40% of all ubiquitin ligases (50). Our findings suggest a model wherein the positive effect of NEDD8 on ΔF508-CFTR rescue relates to its influence on the activity of the Cul1-based E3 ligase complex, SCFFBXO2. Therefore, the SCFFBXO2 complex may contribute significantly to the ubiquitination and degradation of ΔF508-CFTR in airway epithelia. Directly inhibiting FBXO2 expression also partially restored ΔF508-CFTR trafficking, maturation, and function while simultaneously reducing ΔF508-CFTR ubiquitination. The failure of the combined knockdown of NEDD8 and FBXO2 to exhibit additive effects on ΔF508-CFTR rescue supports the notion that FBXO2 acts downstream of NEDD8. Further studies are needed to understand whether the SCFFBXO2 complex is the only NEDD8-regulated complex ubiquitinating ΔF508-CFTR. It is worth noting that, although we achieved potent target gene inhibition and minimized off-target effects by using chemically modified DsiRNAs (45, 51), we did not assess whether restoration of ΔF508-CFTR trafficking, maturation, and/or function was a consequence of inhibiting two complementary pathways or severely impairing a single pathway. Such conclusions are beyond the scope of this work. Further studies will be required to understand how these pathways might interact and influence CFTR biosynthesis.
Although the biological significance of these findings on CFTR biology is evident, an important question is: are these pathways therapeutic targets for CF? The ubiquitin-proteasome system and SCF biology have been an active area of drug discovery for many years (52–54), and more broad spectrum inhibitors of the 26S proteosome such as bortezomib are in clinical trials to treat malignancies (55). MLN4924 targets the NEDD8-activating enzyme, thereby preventing the neddylation (activation) of cullin-RING proteins. This prevents ubiquitination by SCF complexes, promoting apoptosis and autophagy in targeted cells (56, 57). We tested MLN4924 for efficacy in rescuing ΔF508-CFTR-mediated Cl− transport in CFBE cells at six different doses ranging from 1 nm to 100 μm. Unfortunately, MLN4924 was toxic to epithelial cells and was only tolerated at doses that did not inhibit neddylation.4 Most F-box proteins are poorly understood, and thus inhibitor development has been slow. Recently, manipulating proteostasis has received attention as a means to rescue ΔF508-CFTR function (58–60). As more selective compounds are developed, targeting specific proteins in the ubiquitin-proteasome system or SCF complexes in airway epithelia may become feasible.
In summary, inhibition of SYVN1 (Hrd1; E3 ubiquitin ligase), FBXO2 (Fbs1; E3 ubiquitin ligase), or NEDD8 (neddylation) partially rescued ΔF508-CFTR protein maturation and significantly improved ΔF508-CFTR-mediated Cl− transport. Importantly, these effects were further amplified by co-treatment with a CFTR corrector compound (C18). These results suggest that SYVN1 and FBXO2 represent two distinct multiprotein complexes that may degrade ΔF508-CFTR in airway epithelia and identify a new role for NEDD8 in regulating ΔF508-CFTR ubiquitination. Our results provide new insights into CFTR biogenesis and confirm the validity of this discovery strategy. The gene products identified using this strategy may represent new targets for CF therapies.
Experimental Procedures
Primary Human Airway Epithelia
Airway epithelial cells from human trachea and primary bronchus were dissociated from native tissue by Pronase enzyme digestion. Permeable membrane inserts (0.6-cm2 Millipore polycarbonate filter and 0.33-cm2 Costar polyester) precoated with human placental collagen (type IV; Sigma) were seeded with freshly dissociated epithelia. Seeding culture medium used was DMEM/F-12 medium supplemented with 5% FBS, 50 units/ml penicillin, 50 μg/ml streptomycin, 50 μg/ml gentamicin, 2 μg/ml fluconazole, and 1.25 μg/ml amphotericin B. For epithelia from CF patients, the following additional antibiotics were used for the first 5 days: 77 μg/ml ceftazidime, 12.5 μg/ml imipenem and cilastatin, 80 μg/ml tobramycin, and 25 μg/ml piperacillin and tazobactam. After seeding, the cultures were maintained in DMEM/F-12 medium supplemented with 2% Ultroser G (Pall Biosepra) and the above listed antibiotics.
RNA Isolation
Total RNA from primary human airway epithelial cells, HeLa cells, and CFBE cells was isolated using the mirVanaTM microRNA isolation kit, TRIzol® reagent (Life Technologies) (61), or the SV96 Total RNA Isolation System (Promega, Madison, WI) according to the manufacturers' protocols. Total RNA was tested for quality on an Agilent Model 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only samples with an RNA integrity number over 7.0 were selected for downstream processing.
Oligonucleotide Transfections
These protocols were described in detail previously (62). Briefly, freshly dissociated human airway epithelial cells, CFBE cells, or HeLa cells were transfected in precoated 96-well plates (Costar) or TranswellTM permeable supports (0.33-cm2 0.4-μm polyester membrane, Costar 3470). LipofectamineTM RNAiMAX (Invitrogen) (manufacturer's recommended concentration) was used as a reverse transfection reagent. Precoated (with human placental collagen type IV) substrates were incubated with the transfection mixture comprising Opti-MEM (Invitrogen), oligonucleotide (Integrated DNA Technologies) and Lipofectamine RNAiMAX. 15–20 min later, 150,000 freshly dissociated cells suspended in DMEM/F-12 were added to each well/insert. Between 4 and 6 h later, all medium from the apical surface was aspirated, and complete medium was added to the basolateral surface. Medium on the basolateral surface was changed every 3–4 days. For human primary epithelial cultures, Ultroser G medium described above was used. For cultures from immortalized cell lines HeLa and CFBE41o− (termed CFBE throughout (15)), complete medium specific to each cell line was used (for HeLa, modified Eagle's medium (Gibco) + 10% FBS (Atlanta Biologicals) + 1% penicillin-streptomycin Strep (Gibco); for CFBE, advanced DMEM (Gibco) + 1% l-glutamine (Gibco) + 10% FBS + 1% penicillin-streptomycin).
Oligonucleotide Reagents
Ten DsiRNAs were designed and screened against each gene (data not shown), and the two best performing DsiRNAs were taken forward for additional studies (supplemental Table S2). The DsiRNAs were designed (64, 65), synthesized, and validated (45, 51) by Integrated DNA Technologies. All accompanying control sequences (Scr) were also generated by Integrated DNA Technologies.
Quantitative RT-PCR
First strand cDNA was synthesized using SuperScript® II (Invitrogen) with oligo(dT) and random hexamer priming. Sequence-specific PrimeTime® quantitative PCR assays were procured for the following human genes from Integrated DNA Technologies: SIN3A, DERL1, HSPA8, HSPA5, DNAJB12, BAG1, NHERF1, CAPNS1, HSPB1, HSPA1A, MARCH2, HSP90B1, RNF128, CANX, GRIP1, SYVN1, DAB2, RCN2, GOPC, HSPA9, MARCH3, PPP2R1B, RCN1, BAG2, STP6V1A, DNAJC3, CFTR, SIN3A, AMFR, RNF5, AHSA1, NEDD8, FBXO2, GAPDH, HPRT, and SFRS9. All reactions were set up using TaqMan® Fast Universal PCR Master Mix (Applied Biosystems) and run on the Applied Biosystems 7900 HT Real-Time PCR system. All experiments were performed in quadruplicate.
Electrophysiology Studies
Transepithelial Cl− current measurements were made in Ussing chambers ≥2 weeks postseeding as reported (66). Briefly, primary air-liquid interface cultures of airway epithelia or polarized CFBE cultures were mounted in Ussing chambers (EasyMount P2300 chamber system, Physiologic Instruments, San Diego, CA), voltage-clamped (model VCCMC8-4S, Physiologic Instruments), and connected to a computerized data acquisition system (Acquire & Analyze 2.3.181, Physiologic Instruments) to record short circuit currents and transepithelial resistance. Transepithelial Cl− current was measured under short circuit current conditions. After measuring baseline current, the transepithelial current (It) response to sequential apical addition of 100 μm amiloride, 100 μm 4,4′-diisothiocyanotostilbene-2,2′-disulfonic acid, 4.8 mm [Cl−], 10 μm forskolin, 100 μm 3-isobutyl-1-methylxanthine (IBMX), and 100 μm GlyH-101 was measured. Studies were conducted with a Cl− concentration gradient containing 135 mm NaCl, 1.2 mm MgCl2, 1.2 mm CaCl2, 2.4 mm K2PO4, 0.6 mm KH2PO4, 5 mm dextrose, and 5 mm Hepes, pH 7.4, on the basolateral surface and gluconate substituted for Cl− on the apical side.
SDS-PAGE and Immunoblotting
Cell lines were washed with PBS and lysed in freshly prepared lysis buffer (1% Triton, 25 mm Tris, pH 7.4, 150 mm NaCl, and protease inhibitors (cOmpleteTM, mini, EDTA-free, Roche Applied Science)) for 30 min at 4 °C. The lysates were centrifuged at 14,000 rpm for 20 min at 4 °C, and the supernatant was quantified by BCA Protein Assay kit (Pierce). CFTR was denatured in 6× sample SDS buffer (375 mm Tris-HCl, pH 6.8, 6% SDS, 48% glycerol, 9% 2-mercaptoethanol, and 0.03% bromphenol blue). 20 μg (HeLa and CFBE) of protein/lane was separated by 7% SDS-PAGE for Western blotting analysis. Protein abundance was quantified by densitometry using an Alpha Innotech Fluorochem imager. For CFTR, bands B and C were quantified separately. Western blots were probed, stripped, and reprobed as follows. PVDF membranes were first probed with the antibody against the gene of interest. After imaging, the PVDF membrane was stripped with Restore Western Blot Stripping Buffer (Thermo Scientific) for 15 min, washed in Tris-buffered saline-Tween (TBS-T) and blocked in 5% bovine serum albumin (BSA; Pierce) for 1 h. The membrane was washed in TBS-T and incubated with the goat anti-mouse secondary antibody (1:10,000; Sigma) for 1 h and imaged. If signal was detected, the stripping procedure was repeated until no signal was observed. The membrane was washed in TBS-T, blocked for 1 h in 5% BSA, and reprobed with the antibody against tubulin. Antibodies used were sourced as follows: CFTR (R-769, Cystic Fibrosis Foundation Therapeutics, Inc.), hemagglutinin (HA.11 clone 16B12 monoclonal antibody, Covance), α-tubulin (clone DM1A, Sigma), SYVN1 (ab38456, Abcam), NEDD8 (ab38634, Abcam), FBXO2 (ab96391, Abcam), AMFR (ab101284, Abcam), RNF5 (ab128200, Abcam), AHSA1 (ab56721, Abcam), and ubiquitin (ab140601, Abcam).
CFTR Ubiquitination Measurements
Cells were treated with 10 μm MG-132 in the last 1 h of incubation and then lysed in lysis buffer (1% Triton, 25 mm Tris, pH 7.4, 150 mm NaCl, protease inhibitors (cOmpleteTM, mini, EDTA-free), 5 mm N-ethylmaleimide, and 20 μm MG-132) for 30 min at 4 °C. The lysates were centrifuged at 14,000 rpm for 20 min at 4 °C, and the supernatant was quantified by BCA Protein Assay kit. CFTR was precipitated with the anti-HA antibody. The immunoprecipitates were analyzed by immunoblotting with anti-ubiquitin and anti-HA antibodies. CFTR ubiquitination level with molecular masses >180 kDa was measured by densitometry and normalized for the CFTR level in the precipitate.
Immunoprecipitation
Immunoprecipitation (IP) experiments were performed in HeLa cells stably expressing ΔF508-CFTR-HA. For IP of ΔF508-CFTR, cells were lysed (as described above), and supernatant (20–50 μg of protein) was incubated with either anti-HA (for IP of CFTR) or anti-AMFR for 1 h at 4 °C followed by incubation with protein G-agarose (Invitrogen) for 1 h at 4 °C. Immunoprecipitates were washed four times with lysis buffer and eluted in 6× sample SDS buffer. Samples were analyzed by immunoblotting as described above.
Measuring Cell Surface Display of CFTR
HeLa cells stably expressing wild-type CFTR or CFTR-ΔF508 were kindly provided by Dr. G. Lukacs (31, 63). Cell surface ELISA was performed on these cells (48) after noted treatments. HeLa cells were transfected/treated in 96-well plates (Costar). Briefly, the plate containing the cells was moved to a cold room (4 °C), and the medium used was ice-cold. Cells were washed with PBS and blocked for 30 min with PBS containing 5% BSA. Anti-HA primary antibody (Covance) was added in 5% BSA in PBS at a 1:1000 concentration for 1 h. Cells were washed with PBS, and anti-mouse secondary antibody (HRP-conjugated; Amersham Biosciences) was added to cells at 1:1000 concentration in 5% BSA in PBS for 1 h. Cells were washed thoroughly, and signal was developed using SureBlue ReserveTM 3,3′,5,5′-tetramethylbenzidine microwell substrate (Kirkegaard & Perry Laboratories, Inc.). The reaction was stopped and read on a VersaMaxTM microplate reader (Molecular Devices) at 540 nm using the SoftMax® Pro Software (Molecular Devices). For normalization, cells were lysed, and total protein was quantitated using the BCA Protein Assay kit. The experiment was performed in quadruplicate, and the data are presented as a mean ± S.D. of individual data points.
Measuring Membrane Residence of ΔF508-CFTR in HeLa Cells
The methods for measuring membrane residence time are similar to that of measuring cell surface display with slight modifications. 72 h post-treatment, cells were moved to 4 °C. The cells were incubated in primary anti-HA antibody for 1 h and washed in ice-cold PBS. The cells were then exposed to the secondary antibody for 1 h, washed with ice-cold PBS, and incubated at 37 °C for the chase. Surface ΔF508-CFTR was detected using Amplex Ultra Red reagent. The readings were made at 37 °C at the following time points: 0, 30, 60, 90, 120, and 240 min.
LDH Cytotoxicity Assay
Primary airway epithelial cell cultures from three non-CF human donors were untreated or transfected with the following reagents: siScr, SYVN1 DsiRNA, and NEDD8 DsiRNA. The apical surface was washed, and the basolateral medium was collected on days 4, 8, 12, 16, 20, 24, and 28 post-transfection. An LDH cytotoxicity assay kit (Cayman Chemical) was used to measure the levels of lactate dehydrogenase in the washes and basolateral medium. Percentage of toxicity and viability were computed based on LDH levels. Data were normalized to untransfected cells.
Histochemistry
Epithelial sheets on filters were fixed with zinc formalin, embedded in paraffin, sectioned at 5-μm thickness, and stained with hematoxylin (Leica Biosystems) and eosin (Sigma) stain. Sections were visualized by light microscopy.
Statistical Analysis
All statistical analysis was done using GraphPad Prism version 6.0h. Normality of data was tested using the D'Agostino and Pearson omnibus normality test. One-way ANOVA with Holm-Bonferroni multiple comparison test was used to determine significance. Statistics used in individual experiments along with the method of presenting data and error bars are described in the figure legends.
Author Contributions
S. R. designed the study, performed experiments, interpreted results, and prepared the manuscript. S. R. O and A. M. J performed experiments. K. R. P. contributed new reagents/analytic tools. M. A. B interpreted results and prepared the manuscript. P. B. M. designed the study, interpreted results, and prepared the manuscript.
Supplementary Material
Acknowledgments
We thank Rob Piper, Chris Adams, Lynda Ostedgaard, Patrick Sinn, Colleen Stein, and Carolyn Yrigollen for critical comments on the manuscript. We also thank Philip Karp, Sarah Ernst, Lynda Ostedgaard, and Christine Wohlford-Lenane for technical assistance. We also acknowledge the support of the In Vitro Models and Cell Culture Core, which is partly supported by National Institutes of Health Grants P01 HL091842 and P30 DK54759 and the Cystic Fibrosis Foundation.
This work was supported by National Institutes of Health Grants R21 HL104337 and RO1 HL118000 (to P. B. M.) and the Roy J. Carver Charitable Trust. P. B. M. is named as inventor on submitted patent application (United States Patent 62061500, held by the University of Iowa Research Foundation) describing targets for cystic fibrosis therapies. S. R. is named as inventor on submitted patent application (United States Patent 62061500, held by the University of Iowa Research Foundation) describing targets for cystic fibrosis therapies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1 and S2.
S. Ramachandran, S. R. Osterhaus, K. R. Parekh, and P. B. McCray, Jr., unpublished observations.
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- miR
- microRNA
- ERQC
- ER quality control
- CFBE
- cystic fibrosis bronchial epithelial
- DsiRNA
- Dicer substrate siRNA
- C18
- corr-18
- SYVN1
- synovial apoptosis inhibitor 1
- NEDD8
- neural precursor cell expressed, developmentally down-regulated 8
- siScr
- scrambled oligonucleotide
- CHIP (also known as STUB1)
- STIP1 homology and U-box containing protein 1
- RNF5
- ring finger protein 5
- AMFR
- autocrine motility factor receptor
- SCF
- Skp, cullin, F-box
- CRL
- cullin-RING ubiquitin ligase
- ALI
- air-liquid interface
- LDH
- lactate dehydrogenase
- IP
- immunoprecipitation
- ANOVA
- analysis of variance
- IBMX
- 3-isobutyl-1-methylxanthine
- It
- transepithelial current
- HERPUD1
- homocystein inducible ER protein with ubiquitin like domain 1
- CANX
- calnexin
- GOPC
- golgi-associated PDZ and coiled-coil motif containing
- HPRT
- hypoxanthine phosphoribosyltransferase 1.
References
- 1. Rowe S. M., Miller S., and Sorscher E. J. (2005) Cystic fibrosis. N. Engl. J. Med. 352, 1992–2001 [DOI] [PubMed] [Google Scholar]
- 2. Anderson M. P., Gregory R. J., Thompson S., Souza D. W., Paul S., Mulligan R. C., Smith A. E., and Welsh M. J. (1991) Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253, 202–205 [DOI] [PubMed] [Google Scholar]
- 3. Anderson M. P., Rich D. P., Gregory R. J., Smith A. E., and Welsh M. J. (1991) Generation of cAMP-activated chloride currents by expression of CFTR. Science 251, 679–682 [DOI] [PubMed] [Google Scholar]
- 4. Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., and Tsui L. C. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 5. Tsui L. C. (1992) Mutations and sequence variations detected in the cystic fibrosis transmembrane conductance regulator (CFTR) gene: a report from the Cystic Fibrosis Genetic Analysis Consortium. Hum. Mutat. 1, 197–203 [DOI] [PubMed] [Google Scholar]
- 6. Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., and Smith A. E. (1990) Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 7. Ward C. L., Omura S., and Kopito R. R. (1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 8. Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., and Welsh M. J. (1992) Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 9. Ramachandran S., Karp P. H., Jiang P., Ostedgaard L. S., Walz A. E., Fisher J. T., Keshavjee S., Lennox K. A., Jacobi A. M., Rose S. D., Behlke M. A., Welsh M. J., Xing Y., and McCray P. B. Jr. (2012) A microRNA network regulates expression and biosynthesis of wild-type and ΔF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A. 109, 13362–13367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grainger C. I., Greenwell L. L., Lockley D. J., Martin G. P., and Forbes B. (2006) Culture of Calu-3 cells at the air interface provides a representative model of the airway epithelial barrier. Pharm. Res. 23, 1482–1490 [DOI] [PubMed] [Google Scholar]
- 11. Haws C., Finkbeiner W. E., Widdicombe J. H., and Wine J. J. (1994) CFTR in Calu-3 human airway cells: channel properties and role in cAMP-activated Cl− conductance. Am. J. Physiol. Lung Cell. Mol. Physiol. 266, L502–L512 [DOI] [PubMed] [Google Scholar]
- 12. Dennis G. Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., and Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 13. Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., and Thomas P. D. (2010) PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., and von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunzelmann K., Schwiebert E. M., Zeitlin P. L., Kuo W. L., Stanton B. A., and Gruenert D. C. (1993) An immortalized cystic fibrosis tracheal epithelial cell line homozygous for the ΔF508 CFTR mutation. Am. J. Respir. Cell Mol. Biol. 8, 522–529 [DOI] [PubMed] [Google Scholar]
- 16. Hutt D. M., Herman D., Rodrigues A. P., Noel S., Pilewski J. M., Matteson J., Hoch B., Kellner W., Kelly J. W., Schmidt A., Thomas P. J., Matsumura Y., Skach W. R., Gentzsch M., Riordan J. R., et al. (2010) Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat. Chem. Biol. 6, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalid O., Mense M., Fischman S., Shitrit A., Bihler H., Ben-Zeev E., Schutz N., Pedemonte N., Thomas P. J., Bridges R. J., Wetmore D. R., Marantz Y., and Senderowitz H. (2010) Small molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J. Comput.-Aided Mol. Des. 24, 971–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pedemonte N., Lukacs G. L., Du K., Caci E., Zegarra-Moran O., Galietta L. J., and Verkman A. S. (2005) Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Investig. 115, 2564–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sondo E., Tomati V., Caci E., Esposito A. I., Pfeffer U., Pedemonte N., and Galietta L. J. (2011) Rescue of the mutant CFTR chloride channel by pharmacological correctors and low temperature analyzed by gene expression profiling. Am. J. Physiol. Cell Physiol. 301, C872–C885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Goor F., Hadida S., Grootenhuis P. D., Burton B., Stack J. H., Straley K. S., Decker C. J., Miller M., McCartney J., Olson E. R., Wine J. J., Frizzell R. A., Ashlock M., and Negulescu P. A. (2011) Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. U.S.A. 108, 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R. 3rd, and Balch W. E. (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 22. Holleran J. P., Glover M. L., Peters K. W., Bertrand C. A., Watkins S. C., Jarvik J. W., and Frizzell R. A. (2012) Pharmacological rescue of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) detected by use of a novel fluorescence platform. Mol. Med. 18, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Accurso F. J., Rowe S. M., Clancy J. P., Boyle M. P., Dunitz J. M., Durie P. R., Sagel S. D., Hornick D. B., Konstan M. W., Donaldson S. H., Moss R. B., Pilewski J. M., Rubenstein R. C., Uluer A. Z., Aitken M. L., et al. (2010) Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N. Engl. J. Med. 363, 1991–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clancy J. P., Rowe S. M., Accurso F. J., Aitken M. L., Amin R. S., Ashlock M. A., Ballmann M., Boyle M. P., Bronsveld I., Campbell P. W., De Boeck K., Donaldson S. H., Dorkin H. L., Dunitz J. M., Durie P. R., et al. (2012) Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 67, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., Fujii R., Miura N., Komiya S., Nishioka K., Maruyama I., et al. (2003) Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 17, 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaneko M., Ishiguro M., Niinuma Y., Uesugi M., and Nomura Y. (2002) Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532, 147–152 [DOI] [PubMed] [Google Scholar]
- 27. Metzger M. B., Hristova V. A., and Weissman A. M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bosu D. R., and Kipreos E. T. (2008) Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okiyoneda T., Veit G., Dekkers J. F., Bagdany M., Soya N., Xu H., Roldan A., Verkman A. S., Kurth M., Simon A., Hegedus T., Beekman J. M., and Lukacs G. L. (2013) Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat. Chem. Biol. 9, 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X., Koulov A. V., Kellner W. A., Riordan J. R., and Balch W. E. (2008) Chemical and biological folding contribute to temperature-sensitive ΔF508 CFTR trafficking. Traffic 9, 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma M., Benharouga M., Hu W., and Lukacs G. L. (2001) Conformational and temperature-sensitive stability defects of the ΔF508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J. Biol. Chem. 276, 8942–8950 [DOI] [PubMed] [Google Scholar]
- 32. Zhang D., Ciciriello F., Anjos S. M., Carissimo A., Liao J., Carlile G. W., Balghi H., Robert R., Luini A., Hanrahan J. W., and Thomas D. Y. (2012) Ouabain mimics low temperature rescue of F508del-CFTR in cystic fibrosis epithelial cells. Front. Pharmacol. 3, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gomes-Alves P., Neves S., Coelho A. V., and Penque D. (2009) Low temperature restoring effect on F508del-CFTR misprocessing: a proteomic approach. J. Proteomics 73, 218–230 [DOI] [PubMed] [Google Scholar]
- 34. Lukacs G. L., and Verkman A. S. (2012) CFTR: folding, misfolding and correcting the ΔF508 conformational defect. Trends Mol. Med. 18, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., and Nagata K. (2008) Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meacham G. C., Patterson C., Zhang W., Younger J. M., and Cyr D. M. (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 37. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., and Cyr D. M. (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 38. Caohuy H., Jozwik C., and Pollard H. B. (2009) Rescue of ΔF508-CFTR by the SGK1/Nedd4–2 signaling pathway. J. Biol. Chem. 284, 25241–25253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun F., Zhang R., Gong X., Geng X., Drain P. F., and Frizzell R. A. (2006) Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J. Biol. Chem. 281, 36856–36863 [DOI] [PubMed] [Google Scholar]
- 40. Zhang T., Xu Y., Liu Y., and Ye Y. (2015) gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Mol. Biol. Cell 26, 4438–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoshida Y., Chiba T., Tokunaga F., Kawasaki H., Iwai K., Suzuki T., Ito Y., Matsuoka K., Yoshida M., Tanaka K., and Tai T. (2002) E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 [DOI] [PubMed] [Google Scholar]
- 42. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., and Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., and Weissman A. M. (2007) The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 13, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 44. Ballar P., Ors A. U., Yang H., and Fang S. (2010) Differential regulation of CFTRΔF508 degradation by ubiquitin ligases gp78 and Hrd1. Int. J. Biochem. Cell Biol. 42, 167–173 [DOI] [PubMed] [Google Scholar]
- 45. Collingwood M. A., Rose S. D., Huang L., Hillier C., Amarzguioui M., Wiiger M. T., Soifer H. S., Rossi J. J., and Behlke M. A. (2008) Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides 18, 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., and Weissman A. M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., and Wiertz E. (2004) Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 48. Okiyoneda T., Barrière H., Bagdány M., Rabeh W. M., Du K., Höhfeld J., Young J. C., and Lukacs G. L. (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gnann A., Riordan J. R., and Wolf D. H. (2004) Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol. Biol. Cell 15, 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., et al. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 51. Behlke M. A. (2008) Chemical modification of siRNAs for in vivo use. Oligonucleotides 18, 305–319 [DOI] [PubMed] [Google Scholar]
- 52. Bulatov E., and Ciulli A. (2015) Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem. J. 467, 365–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang X., and Dixit V. M. (2016) Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hussain M., Lu Y., Liu Y. Q., Su K., Zhang J., Liu J., and Zhou G. B. (2016) Skp1: implications in cancer and SCF-oriented anti-cancer drug discovery. Pharmacol. Res. 111, 34–42 [DOI] [PubMed] [Google Scholar]
- 55. Adams J., and Kauffman M. (2004) Development of the proteasome inhibitor Velcade (bortezomib). Cancer Invest. 22, 304–311 [DOI] [PubMed] [Google Scholar]
- 56. Godbersen J. C., Humphries L. A., Danilova O. V., Kebbekus P. E., Brown J. R., Eastman A., and Danilov A. V. (2014) The Nedd8-activating enzyme inhibitor MLN4924 thwarts microenvironment-driven NF-κB activation and induces apoptosis in chronic lymphocytic leukemia B cells. Clin. Cancer Res. 20, 1576–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nawrocki S. T., Griffin P., Kelly K. R., and Carew J. S. (2012) MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin. Investig. Drugs 21, 1563–1573 [DOI] [PubMed] [Google Scholar]
- 58. Esposito S., Tosco A., Villella V. R., Raia V., Kroemer G., and Maiuri L. (2016) Manipulating proteostasis to repair the F508del-CFTR defect in cystic fibrosis. Mol. Cell. Pediatr. 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pesce E., Gorrieri G., Sirci F., Napolitano F., Carrella D., Caci E., Tomati V., Zegarra-Moran O., di Bernardo D., and Galietta L. J. (2016) Evaluation of a systems biology approach to identify pharmacological correctors of the mutant CFTR chloride channel. J. Cyst. Fibros. 15, 425–435 [DOI] [PubMed] [Google Scholar]
- 60. Roth D. M., Hutt D. M., Tong J., Bouchecareilh M., Wang N., Seeley T., Dekkers J. F., Beekman J. M., Garza D., Drew L., Masliah E., Morimoto R. I., and Balch W. E. (2014) Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 12, e1001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramachandran S., Clarke L. A., Scheetz T. E., Amaral M. D., and McCray P. B. Jr. (2011) Microarray mRNA expression profiling to study cystic fibrosis. Methods Mol. Biol. 742, 193–212 [DOI] [PubMed] [Google Scholar]
- 62. Ramachandran S., Krishnamurthy S., Jacobi A. M., Wohlford-Lenane C., Behlke M. A., Davidson B. L., and McCray P. B. Jr. (2013) Efficient delivery of RNA interference oligonucleotides to polarized airway epithelia in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L23–L32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K. G., Papsin B., Zerangue N., Stenmark H., and Lukacs G. L. (2004) Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J. Cell Biol. 164, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim D. H., Behlke M. A., Rose S. D., Chang M. S., Choi S., and Rossi J. J. (2005) Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 [DOI] [PubMed] [Google Scholar]
- 65. Rose S. D., Kim D. H., Amarzguioui M., Heidel J. D., Collingwood M. A., Davis M. E., Rossi J. J., and Behlke M. A. (2005) Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 33, 4140–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Itani O. A., Chen J. H., Karp P. H., Ernst S., Keshavjee S., Parekh K., Klesney-Tait J., Zabner J., and Welsh M. J. (2011) Human cystic fibrosis airway epithelia have reduced Cl− conductance but not increased Na+ conductance. Proc. Natl. Acad. Sci. U.S.A. 108, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.