Abstract
Methods for genetic manipulation of Trypanosoma cruzi, the etiologic agent of Chagas disease, have been highly inefficient, and no endogenous tagging of genes has been reported to date. We report here the use of the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated gene 9) system for endogenously tagging genes in this parasite. The utility of the method was established by tagging genes encoding proteins of known localization such as TcFCaBP (flagellar calcium binding protein) and TcVP1 (vacuolar proton pyrophosphatase), and two proteins of undefined or disputed localization, the TcMCU (mitochondrial calcium uniporter) and TcIP3R (inositol 1,4,5-trisphosphate receptor). We confirmed the flagellar and acidocalcisome localization of TcFCaBP and TcVP1 by co-localization with antibodies to the flagellum and acidocalcisomes, respectively. As expected, TcMCU was co-localized with the voltage-dependent anion channel to the mitochondria. However, in contrast to previous reports and our own results using overexpressed TcIP3R, endogenously tagged TcIP3R showed co-localization with antibodies against VP1 to acidocalcisomes. These results are also in agreement with our previous reports on the localization of this channel to acidocalcisomes of Trypanosoma brucei and suggest that caution should be exercised when overexpression of tagged genes is done to localize proteins in T. cruzi.
Keywords: CRISPR/Cas; inositol 1,4,5-trisphosphate (IP3); mitochondria; Trypanosoma cruzi; vacuolar acidification; acidocalcisome; flagellum
Introduction
The application of the CRISPR/Cas9 technology to the study of protist parasites has dramatically increased the tools available for their genetic manipulation (1). Trypanosoma cruzi, the etiologic agent of Chagas disease, which is a significant cause of morbidity and mortality from the South of the United States to the South of Argentina and Chile, has been particularly refractory to genetic manipulation. However, the recent use of the CRISPR/Cas9 technology to knock down or knock out genes (2, 3) has revolutionized their study.
The localization of proteins is important to determine their cellular function, and previous studies in T. cruzi have used either antibodies or gene tagging methods with vectors that overexpressed the proteins (4). Although specific antibodies are useful to detect the endogenous proteins, it is not always possible to obtain them because either the proteins have low antigenicity or the antibodies cross-react with other proteins. Plasmids that enable the tagging of genes at their endogenous loci are not available for T. cruzi, and a major drawback of the overexpression of tagged proteins is that the proteins of interest sometimes are retained in the endoplasmic reticulum (ER)4 or localize to other compartments.
Here we have adapted the CRISPR/Cas9 system to tag genes of T. cruzi at their endogenous loci and tested this system with two genes encoding proteins of well recognized localization (flagellar calcium binding protein or TcFCaBP and the acidocalcisome vacuolar proton pyrophosphatase or TcVP1) and two encoding proteins of undefined or disputed localization (mitochondrial calcium uniporter or TcMCU and inositol 1,4,5-trisphosphate receptor or TcIP3R). TcMCU was functionally characterized more than 27 years ago as the calcium channel that transports Ca2+ into the mitochondria of the parasites (5, 6), and this finding was fundamental for the recent discovery of the gene encoding this channel in mammalian cells (7–9). The channel is localized to the inner mitochondrial membrane of a variety of cells, including Trypanosoma brucei (10). TcIP3R was reported to have ER localization in T. cruzi (11). However, the immunofluorescence evidence reported was disputed (12), because there was no clear reticular pattern or co-localization with a T. brucei ER marker, TbBiP, in the figures published (11). In addition, the T. brucei IP3R localized to the acidocalcisomes as demonstrated using antibodies against the endogenous tagged protein (13) and specific antibodies against the protein (14), as well as proteomic and functional analyses (13, 14). In this work, we report the acidocalcisome localization of TcIP3R.
The use of the CRISPR/Cas9 system for C-terminal tagging of genes was recently reported for three parasites: Toxoplasma gondii (15), Plasmodium yoelii (16), and Leishmania donovani (17), but has not been previously used in T. cruzi. The availability of this technique for T. cruzi has great potential for the functional analysis of proteins in this parasite.
Results
We first evaluated the endogenous C-terminal tagging method by introducing the epitope tag sequence into two different genes: the TcFCaBP gene and the TcVP1 gene. The proteins encoded by these genes are localized in well defined organelles in trypanosomes: flagellum (18) and acidocalcisomes (19), respectively. Monoclonal and polyclonal antibodies recognizing these proteins are available, as well as genetic information about the proteins. For 3×HA C-terminal tagging, we co-transfected a specific 3′ end-sgRNA/Cas9/pTREX construct with a specific DNA donor molecule for each gene amplified from the pMOTag-HX1–4H vector (Fig. 1A), whereas for 3×c-Myc C-terminal tagging, we co-transfected the same 3′ end-sgRNA/Cas9/pTREX constructs with a specific DNA donor molecule for each gene amplified from the pMOTag23M vector (Fig. 1B), as described under “Experimental Procedures.” We obtained G418/hygromycin- or G418/puromycin-resistant cell lines after 5 weeks under selective pressure. Transfectants were analyzed by PCR, using gDNA isolated from each one, and specific primer sets to distinguish between the wild type and the tagged cell lines.
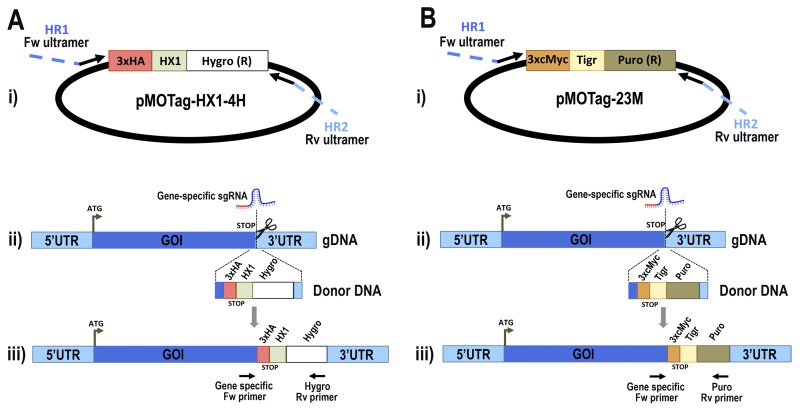
FIGURE 1.
Schematic representation of strategies used to generate endogenous C-terminal tagging in T. cruzi. A, panel i, pMOTag-HX1–4H vector map. The T. cruzi HX1 trans-splicing signal is located between the 3×HA tag sequence and the gene that confers resistance to hygromycin (Hygro (R)). HR1 Fw and HR2 Rv ultramers indicate oligonucleotides used to amplify DNA donor cassette. The annealing regions for ultramers to pMOTag-HX1–4H and genomic DNA (gDNA) are indicated in black and blue, respectively. Panel ii, a double-stranded gDNA break was produced by Cas9 targeted by the sgRNA both expressed from 3′ end-sgRNA/Cas9/pTREX plasmid downstream the STOP codon of the gene of interest (GOI) in the endogenous locus. Homologous directed repair was induced co-transfecting epimastigotes with the DNA donor cassette, containing homologous regions to the GOI 3′ end (blue) and to the GOI 3′UTR (light blue). Panel iii, integration of 3×HA and antibiotic resistance gene at 3′ end of GOI by homologous recombination. Arrows indicate primers used for checking integration of donor DNA. B, panel i, pMOTag23M vector map. The 3×c-Myc tag sequence and the puromycin resistance gene (Puro (R)) are separated by the T. brucei tubulin intergenic region (Tigr). The rest of the description of panels i–iii is similar to that for A. Hygro, hygromycin resistance gene; Puro, puromycin resistance gene; UTR, untranslated region; ATG, start codon.
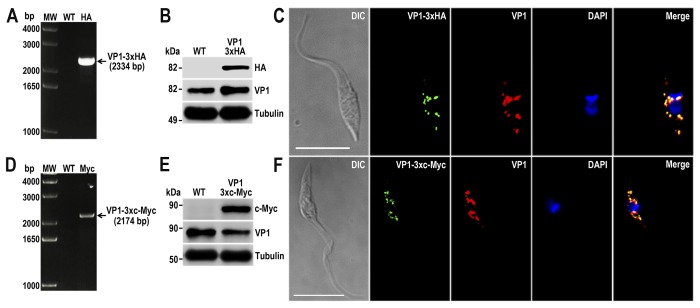
Fig. 2A shows that TcVP1–3×HA transfectants were efficiently tagged at the endogenous locus, because the corresponding band amplified with a reverse primer annealing on the hygromycin marker is only present in the resistant parasites (lane HA) but absent in the WT cells, which is the negative control of the reaction. We analyzed the TcVP1–3×HA transfectants by Western blotting, using commercial antibodies anti-HA tag, and a band of ∼85 kDa was clearly detected on the transfectant but absent in the wild type cells (Fig. 2B). A similar band appears in both wild type cells and TcVP1–3×HA transfectants when anti-TbVP1 antibodies were used (Fig. 2B). Immunofluorescence analysis (IFA) of the mutants verified the subcellular localization of the protein to the acidocalcisomes as it co-localizes with antibodies against TbVP1 (Fig. 2C), as expected (19). Similar results were obtained by 3×c-Myc C-terminal tagging using specific DNA donor molecules amplified from the pMOTag23M vector (Fig. 2, D–F). Site-specific insertion of DNA donor cassettes at the 3′ end of TcVP1 gene was verified by cloning and sequencing PCR products amplified from gDNA extracted from TcVP1–3×HA and TcVP1–3×c-Myc cell lines (Fig. 3A), confirming that the mechanism of homologous-directed DNA repair (HDR) took placed in almost the entire population, and a tagging efficiency of >95% was observed in both cell lines by IFA (data shown for TcVP1–3×c-Myc; Fig. 3B). These results also indicate that it is feasible to use the intergenic tubulin region of T. brucei as trans-splicing signal for T. cruzi.
FIGURE 2.
TcVP1 endogenous C-terminal tagging. A, PCR analysis using gDNA isolated from WT and TcVP1–3×HA cell lines. A DNA fragment was amplified in 3×HA-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT. B, Western blotting analysis of WT and TcVP1–3×HA cell lines. Anti-HA antibodies detect TcVP1–3×HA (expected size, 89 kDa), and anti-TbVP1 antibodies detect endogenous TcVP1 (85 kDa). Anti-α-tubulin antibody was used as a loading control. Antibodies are indicated on the right side of the blots, and molecular weights (MW) are on the left side. C, fluorescence microscopy of TcVP1–3×HA epimastigotes indicates localization of the endogenous tagged protein to acidocalcisomes. TcVP1–3×HA was detected with monoclonal anti-HA antibodies (green) or with polyclonal anti-TbVtc4 antibodies (red). D, PCR analysis of TcVP1–3×c-Myc epimastigotes. A DNA fragment was amplified in c-Myc-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT cells. E, Western blotting analysis of WT and TcVP1–3×c-Myc cell lines. Anti-c-Myc antibodies detect TcVP1–3×c-Myc (expected size, 91 kDa). Anti-TbVP1 antibodies detect endogenous TcVP1 (85 kDa). F, fluorescence microscopy of TcVP1–3×c-Myc epimastigotes indicates localization of the endogenous tagged protein to acidocalcisomes. TcVP1–3×c-Myc was detected with monoclonal anti-c-Myc antibodies (green) or with polyclonal anti-TbVP1 antibodies (red). The merge shows co-localization in yellow. Differential interference contrast (DIC) images are shown in the left panel. Nucleus and kinetoplast were labeled with DAPI (blue). Bars, 10 μm.
FIGURE 3.
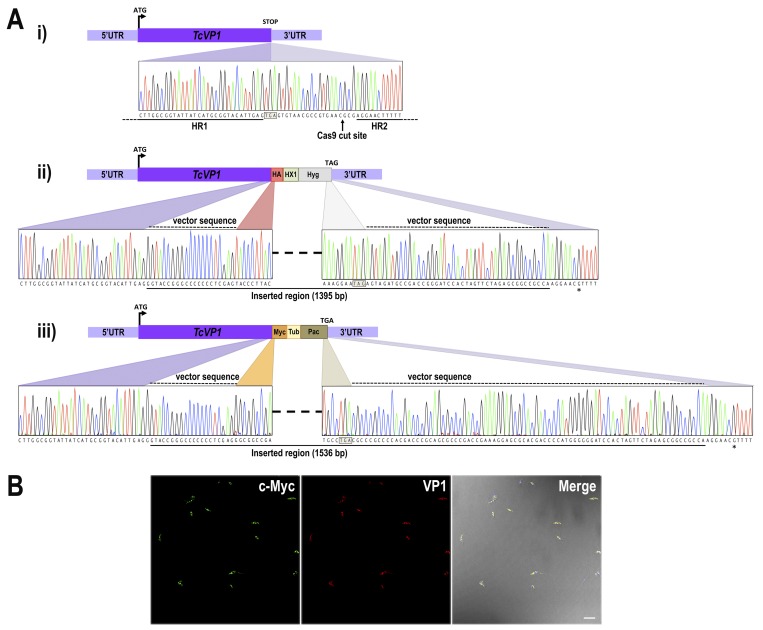
Sequence analysis of TcVP1 locus and tagging efficiency. A, panel i, nucleotide sequence of WT TcVP1 showing the Cas9-targeted cut site (vertical arrow) downstream the stop codon (boxed). Homologous template sequences included in ultramers to generate the DNA donor for HDR are shown as horizontal lines. A schematic representation of WT TcVP1 is shown at the top of the panel. Panels ii and iii, nucleotide sequences of 3×HA and 3×c-Myc tagged TcVP1 loci at the repaired region after Cas9-targeted double-stranded break in homogenously tagged populations. A schematic representation of tagged TcVP1 locus is shown on top of each panel. At the bottom of each panel the nucleotide sequence between the left and right arms of the homologous regions is shown. Colored regions indicate parental and tagged inserted sequences derived from pMOTag-HX1–4H (panel ii) and pMOTag-23M (panel iii). The dashed lines above the traces indicate the nucleotide sequence of each vector included in the donor DNA, located upstream and downstream the specific tag and the resistance marker, respectively. A continuous line under the nucleotide sequence indicates the inserted region in each tagged cell line. Stop codons of antibiotic resistance genes are shown in squared boxes. The asterisk indicates a nucleotide difference between WT (Y strain) and tagged cell lines, because the sequence of T. cruzi CL Brener Esmeraldo-like haplotype was used to design the ultramers for DNA donor amplification. B, immunofluorescence microscopy of TcVP1–3×c-Myc epimastigotes using monoclonal anti-c-Myc antibodies (green) and polyclonal anti-TbVP1 antibodies (red). DIC merged image is shown in the right panel. The image shows parasites observed in an entire field. Scale bar, 10 μm.
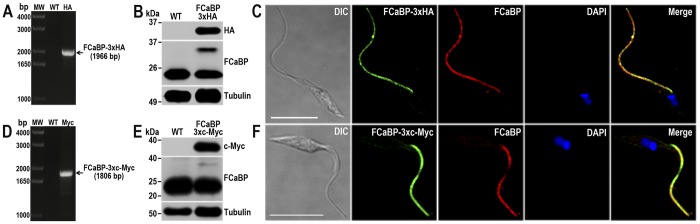
Using similar procedures we found that the TcFCaBP was efficiently tagged at the endogenous locus, as detected by PCR (Fig. 4, A and D), Western blotting analyses (Fig. 4, B and E) and IFA of epimastigotes generated using specific DNA donor molecules amplified from either the pMOTag-HX1–4H (Fig. 4C) or the pMOTag23M vector (Fig. 4F). TcFCaBP-3×HA and TcFCaBP-3×c-Myc exclusively localize in the flagellum, the expected localization of this protein (18), as shown by their co-localization with monoclonal antibodies against TcFCaBP (Fig. 4, C and F), which recognizes both the tagged and endogenous proteins by Western blotting analyses (Fig. 4, B and E). In both cases, detection of the endogenous non-tagged TcFCaBP was much stronger than the tagged version of the protein. We attribute this result to the fact that TcFCaBP is encoded by three identical copies of the gene arranged in tandem in the T. cruzi genome, and probably not all of them were tagged. The localization of C-terminal tagged TcVP1 and TcFCaBP at the expected compartments indicates that the method used is appropriate to detect the native localization of proteins in T. cruzi and that the two vectors employed, one of them designed for endogenous tagging of genes in T. brucei, are adequate for this purpose.
FIGURE 4.
TcFCaBP endogenous C-terminal tagging. A, PCR analysis using gDNA isolated from WT and TcFCaBP-3×HA cell lines. A DNA fragment was amplified in 3×HA-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT. B, Western blotting analysis of WT and TcFCaBP-3×HA cell lines. Anti-HA antibodies detect TcFCaBP-3×HA (predicted size, 28 kDa) and anti-FCaBP antibodies detect both endogenous (24 kDa) and tagged (28 kDa) TcFCaBP. Anti-α-tubulin antibody was used as a loading control. Antibodies are indicated on the right side of the blots, and molecular weights (MW) are on the left side. C, fluorescence microscopy of TcFCaBP-3×HA epimastigotes indicates localization of the endogenous tagged protein to flagellum. TcFCaBP-3×HA was detected with rat anti-HA antibodies (green) or with monoclonal anti-FCaBP antibodies (red). D, PCR analysis of TcFCaBP-3×c-Myc epimastigotes. A DNA fragment was amplified in c-Myc-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT cells. E, Western blotting analysis of WT and TcFCaBP-3×c-Myc cell lines. Anti-c-Myc antibodies detect TcFCaBP-3×c-Myc (predicted size, 30 kDa). Anti-FCaBP antibodies detect endogenous (24 kDa) and c-Myc-tagged (30 kDa) TcFCaBP. F, fluorescence microscopy of TcFCaBP-3×c-Myc epimastigotes indicates localization of the endogenous tagged protein to flagellum. TcFCaBP-3×c-Myc was detected with rabbit anti-c-Myc antibodies (green) or with monoclonal anti-TcFCaBP antibodies (red). The merge shows co-localization in yellow. DIC images are shown in the left panel. Nucleus and kinetoplast were labeled with DAPI (blue). Bars, 10 μm.
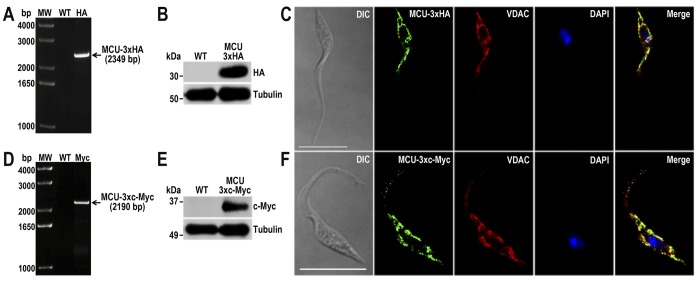
We next investigated the localization of two proteins for which either no previous localization studies have been reported (TcMCU) (20) or for which its localization has been disputed (TcIP3R) (12). TcMCU is the T. cruzi orthologue of the recently discovered MCU from vertebrate cells (8, 9) and of TbMCU (10). MCU localizes to the inner membrane of mitochondria in both vertebrate cells (8, 9) and T. brucei (10) and is solely responsible for mitochondrial Ca2+ uptake in T. brucei (10). Functional studies done in T. cruzi clearly established the presence of MCU in these cells (5, 6) and were important for the identification of the molecular nature of this channel in vertebrate cells (7). Using the same technique that we used to localize TcVP1 and TcFCaBP (see above), we found the TcMCU was tagged at the endogenous locus, as detected by PCR (Fig. 5, A and D), Western blotting analyses (Fig. 5, B and E), and IFA of cells obtained using specific DNA donor molecules amplified from either the pMOTag-HX1–4H (Fig. 5C) or the pMOTag23M vector (Fig. 5F). TcMCU co-localized with the mitochondrial voltage-dependent anion channel (VDAC; Fig. 5, C and F), as expected.
FIGURE 5.
TcMCU endogenous C-terminal tagging. A, PCR analysis using gDNA isolated from WT and TcMCU-3×HA cell lines. A DNA fragment was amplified in 3×HA-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT. B, Western blotting analysis of WT and TcMCU-3×HA cell lines. Anti-HA antibodies detect TcMCU-3×HA (predicted size, 35 kDa), whereas the band is absent in WT parasites. Anti-α-tubulin antibody was used as a loading control. Antibodies are indicated on the right side of the blots, and molecular weights are on the left side. C, fluorescence microscopy of TcMCU-3×HA epimastigotes indicates localization of the endogenous tagged protein to the mitochondria. TcMCU-3×HA was detected with monoclonal anti-HA antibodies (green). Polyclonal antibodies anti-TbVDAC were used to label mitochondria (red). The merge shows co-localization in yellow. D, PCR analysis of TcFCaBP-3×c-Myc epimastigotes. A DNA fragment was amplified in c-Myc-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT cells. E, Western blotting analysis of WT and TcMCU-3×c-Myc cell lines. Anti-c-Myc antibodies detect TcMCU-3×c-Myc (predicted size, 37 kDa), whereas the band is absent in WT epimastigotes. F, fluorescence microscopy of TcMCU-3×c-Myc epimastigotes indicates localization of the endogenous tagged protein to the mitochondria. TcMCU-3×c-Myc was detected with monoclonal anti-c-Myc antibody (green) or with polyclonal anti-TbVDAC antibodies (red). The merge shows co-localization in yellow. DIC images are shown in the left panel. Nucleus and kinetoplast were labeled with DAPI (blue). Bars, 10 μm.
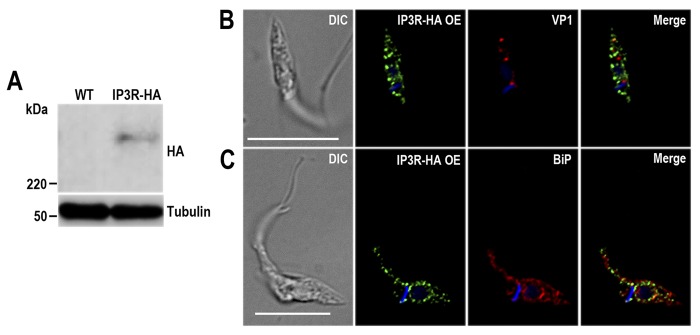
Before doing endogenous tagging of TcIP3R, we overexpressed the gene with an HA epitope tag (TcIP3R-HA-OE) to investigate the localization of the overexpressed protein. Fig. 6A shows the Western blotting analysis of lysates from WT and TcIP3R-HA-OE epimastigotes (IP3R-HA) incubated with anti-HA antibodies showing that transfected cells express the tagged protein of the expected size (∼340 kDa). Fig. 6B shows that the overexpressed protein does not co-localize with the acidocalcisome marker TcVP1, as detected with antibodies against HA and TbVP1, respectively. However, TcIP3R-HA-OE cells shows the same perinuclear and reticular localization pattern as BiP, an ER marker (21), as detected with antibodies against HA and TbBiP (Fig. 6C). Note that although the same distribution pattern is observed for both proteins, their localization is in general not superimposable. This is probably due to the membrane localization of TcIP3R-HA-OE, the intra-ER localization of the soluble BiP, and the fact that these images were deconvolved to eliminate background fluorescence.
FIGURE 6.
TcIP3R-HA overexpression exhibits an ER localization pattern. A, Western blotting analysis of WT and TcIP3R-HA-OE cell lines. Anti-HA antibodies detect TcIP3R-HA (predicted size, 340 kDa), whereas the band is absent in WT parasites. Anti-α-tubulin antibody was used as a loading control. Antibodies are indicated on the right side of the blots, and molecular masses are on the left side. B and C, fluorescence microscopy of TcIP3R-HA-OE epimastigotes indicates an ER localization pattern of the overexpressed tagged protein. TcIP3R-HA-OE was detected with rat anti-HA antibodies (1:100, green). Rabbit antibodies anti-TbVP1 (1:500) and anti-TbBiP (1:500) were used to label acidocalcisomes and ER, respectively (red). The merge images are shown in the right panel. DIC images are shown in the left panel. Nucleus and kinetoplast were labeled with DAPI (blue). Bars, 10 μm.
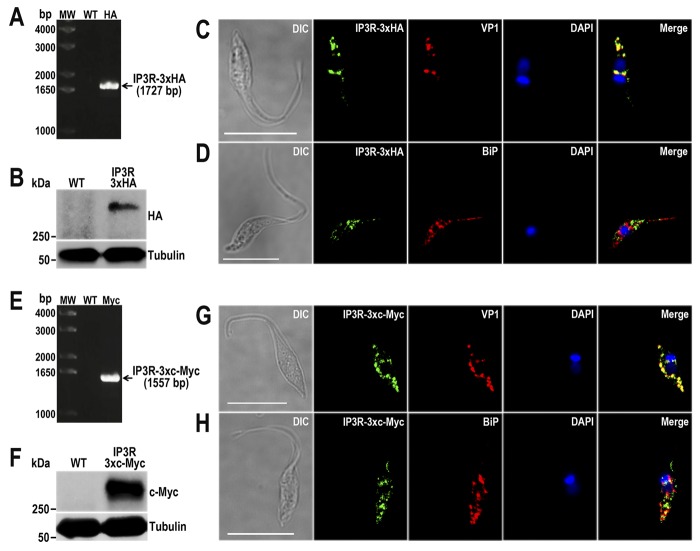
Fig. 7 shows the efficient tagging of TcIP3R at the endogenous locus, as detected by PCR (Fig. 7, A and E), Western blotting analyses (Fig. 7, B and F), and IFA of tagged cell lines generated using specific DNA donor molecules amplified from either the pMOTag-HX1–4H (Fig. 7, C and D) or the pMOTag23M vector (Fig. 7, G and H). TcIP3R-3×HA and TcIP3R-3×c-Myc localize to the acidocalcisomes, as previously described in T. brucei (13) and shown by the co-localization of anti-HA and anti-c-Myc antibodies with VP1 (Fig. 7, C and G). The anti-HA and anti-cMyc antibodies recognize the tagged proteins but not the endogenous IP3R in the WT by Western blotting analyses (Fig. 7, B and F). Fig. 7 (D and H) shows that there is no significant co-localization with the reticular distribution of TbBiP antibodies in the ER. Some co-localization with TbBiP antibodies, especially using TcIP3R-3×c-Mycs was also detected (Fig. 7H) and could correspond to the site of synthesis of the TcIP3R in the ER.
FIGURE 7.
Endogenous tagged TcIP3R localizes to acidocalcisomes. A, PCR analysis using gDNA isolated from WT and TcIP3R-3×HA cell lines. A DNA fragment was amplified in 3×HA-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT. B, Western blotting analysis of WT and TcIP3R-3×HA cell lines. Anti-HA antibodies detect TcIP3R-3×HA (predicted size, 342 kDa), whereas the band is absent in WT parasites. Anti-α-tubulin antibody was used as a loading control. Antibodies are indicated on the right side of the blots, and molecular weights (MW) are on the left side. C and D, fluorescence microscopy of TcIP3R-3×HA epimastigotes indicates localization of the endogenous tagged protein to aidocalcisomes. TcIP3R-3×HA was detected with monoclonal anti-HA antibodies (green). Polyclonal antibodies anti-TbVP1 (C) and anti-TbBiP (D) were used to label acidocalcisomes and ER, respectively (red). The merge shows co-localization with TcVP1 in yellow. E, PCR analysis of TcIP3R-3×c-Myc epimastigotes. A DNA fragment was amplified in c-Myc-tagged epimastigotes (indicated with arrow), whereas the band is absent in WT cells. F, Western blotting analysis of WT and TcIP3R-3×c-Myc cell lines. Anti-c-Myc antibodies detect TcIP3R-3×c-Myc (predicted size, 344 kDa), whereas the band is absent in WT epimastigotes. G and H, fluorescence microscopy of TcMCU-3×c-Myc epimastigotes indicates localization of the endogenous tagged protein to acidocalcisomes. TcMCU-3×c-Myc was detected with monoclonal anti-c-Myc antibody (green). Polyclonal antibodies anti-TbVP1 (G) and anti-TbBiP (H) were used to label acidocalcisomes and ER, respectively (red). The merge shows co-localization with TcVP1 in yellow. DIC images are shown in the left panel. Nucleus and kinetoplast were labeled with DAPI (blue). Bars, 10 μm.
Discussion
Our work demonstrates that the use of the CRISPR/Cas9 system in T. cruzi is not limited to loss of function studies (gene deletion/disruption/mutation) (2, 3) but could be used for C-terminal gene tagging. As proof of concept of the methodology employed, we confirmed the localization of TcVP1 and TcFCaBP to the acidocalcisomes and flagellum of the parasite, respectively. To our knowledge, this is the first report of endogenous tagging of proteins in T. cruzi. We also report the mitochondrial localization of TcMCU, the previously identified pore of the mitochondrial Ca2+ uniporter complex (22). In addition, we report the acidocalcisome localization of TcIP3R.
We previously reported in T. cruzi the HDR mechanism for double-stranded break repair in CRISPR/Cas9-induced PFR2 knock-out cell line (3). In that work we used a DNA donor molecule with 100-nt homology regions to induce DNA repair by homologous recombination in this organism, generating a homogeneous population where 100% cells exhibited gene disruption. Now, by providing a DNA donor template for CRISPR/Cas9-mediated gene tagging, we confirmed the high efficiency of this mechanism, because no other DNA repair mechanism was detected by sequencing in TcVP1–3×HA and TcVP1–3×c-Myc homogeneously tagged populations (Fig. 3A). High efficiency (>95%) gene tagging was observed in all tagged cell lines generated in this study, using DNA donor templates containing 100-nt homology arms, which makes this methodology a promising tool for cellular localization studies and immunoprecipitation assays.
Although most vertebrate IP3Rs reside in ER membranes, IP3 can stimulate Ca2+ release from the Golgi complex (23), the nucleus (24), and the secretory granules (25) of mammalian cells. IP3Rs can also be targeted to the plasma membrane, where they are important for Ca2+ entry (26). Secretory granules of a variety of cells (27–31) were reported to possess IP3Rs, although this was challenged (32). An IP3R localizes to the contractile vacuole in Paramecium tetraurelia, as detected with specific antibodies (33). The acidocalcisome localization of TbIP3R in T. brucei was demonstrated by endogenous gene tagging (13), by studies using specific antibodies (14), and by proteomic (14) and functional (13) studies. Interestingly, when TbIP3R (13) or the IP3Rs of Capsaspora owczarzaki (another protist in which biochemical characterization of the channel was done (34)) are expressed in DT40–3KO cells (chicken lymphocytes in which the three vertebrate IP3R have been knocked out), the proteins localize to the ER.
It was therefore puzzling that in the related trypanosomatid T. cruzi, the IP3R had an ER localization, as studied in epimastigotes overexpressing the channel tagged with GFP (11). This was also against our proteomic analysis of acidocalcisomes of T. cruzi,5 which supported the acidocalcisome localization. It has been reported that when overexpressed, membrane-targeted GFP fusion proteins have a propensity to form organelle aggregates that may lead to misinterpretations of sorting pathways of trafficked proteins (35). We found that this was indeed the case with the TcIP3R. Overexpression of TcIP3R resulted in the same pattern of perinuclear and reticular localization than that of the ER marker BiP. However, endogenous gene tagging of TcIP3R using CRISPR/Cas9 revealed the acidocalcisome localization of the channel. The presence of a Ca2+ uptake pump (Ca2+-ATPase) (36) and a Ca2+ release channel (TcIP3R, this work) in acidocalcisomes of T. cruzi suggests an important role of the organelles in Ca2+ signaling. The results also indicate that caution should be exercised when overexpression of tagged genes is done to localize proteins in T. cruzi.
In summary, the tools developed in this work will enable rapid endogenous gene tagging in T. cruzi, allowing us to establish the localization of proteins and to gain insight into their function. The molecular tools available in T. cruzi have lagged behind those developed for T. brucei (4). Our results indicate that it is possible to use the intergenic tubulin region of T. brucei as trans-splicing signal for T. cruzi, which expands the molecular toolbox available for T. cruzi, because pMOTag vectors (37) developed for C-terminal tagging in T. brucei could be also used for T. cruzi. The method developed in this work will facilitate the functional analysis of genes in T. cruzi, as well as physiological studies, allowing the identification of targets for drugs, diagnostics, and vaccines.
Experimental Procedures
Chemicals and Reagents
Hygromycin, blasticidin S HCl, BenchMark prestained protein ladder, BenchMark protein ladder, Alexa-conjugated secondary antibodies, and HRP-conjugated secondary antibodies were purchased from Life Technologies. Benzonase® nuclease was from Novagen (EMD Millipore, Billerica, MA). GoTaq DNA polymerase, pGEM-T easy Vector System and T4 DNA ligase were from Promega (Madison, WI). Antarctic phosphatase, restriction enzymes, and Q5® high fidelity DNA polymerase were from New England Biolabs (Ipswich, MA). Fluoromount-G® was from SouthernBiotech (Birmingham, AL). Anti-HA high affinity rat monoclonal antibody (clone 3F10) was purchased from Roche Applied Science. Rabbit antibody against T. brucei vacuolar H+-pyrophosphatase (TbVP1) (38) was from Dr. Norbert Bakalara (Ecole Nationale Supérieure de Chimie de Montpellier, Montpellier, France). Monoclonal antibody against FCaBP was from Dr. David Engman (Northwestern University, Evanston, IL). Polyclonal antibody against TbVDAC (39) was from Dr. Minu Chaudhuri (Meharry Medical College, Nashville, TN). Polyclonal antibody against TbBiP (21) was from Dr. Jay Bangs (State University of New York, Buffalo, NY). The pMOTag vectors (37) were from Dr. Thomas Seebeck (University of Bern, Bern, Switzerland). DNA oligonucleotides were purchased from Exxtend Biotecnologia Ltd. (Campinas, Brazil). Pierce BCA protein assay and HA epitope tag monoclonal antibody (clone 2-2.2.14) were from Thermo Fisher Scientific Inc. Rabbit anti-HA polyclonal antibody (Y-11), anti-c-Myc monoclonal antibody (clone 9E10), and rabbit anti-c-Myc polyclonal antibody (N-262) were from Santa Cruz Biotechnology (Dallas, TX). Anti-tubulin monoclonal antibody, puromycin, G418, mammalian cell protease inhibitor mixture (P8340), other protease inhibitors, and all other reagents of analytical grade were from Sigma.
Cell Culture
T. cruzi Y strain epimastigotes were cultured in liver infusion tryptose medium containing 10% heat-inactivated FBS at 28 °C (40). CRISPR/Cas9 mutant cell lines were maintained in medium containing 250 μg/ml G418 and 5 μg/ml puromycin or 350 μg/ml hygromycin. We determined the growth rate of epimastigotes by counting cells in a Neubauer chamber.
Endogenous C-terminal Tagging by CRISPR/Cas9
To achieve the C-terminal tagging of endogenous proteins, we used the Cas9/pTREX-n vector we developed for T. cruzi (3) to clone a specific single guide RNA (sgRNA) sequence targeting the 3′ end of four different genes: TcVP1 (Gene ID TcCLB.510773.20), TcFCaBP (TcCLB.509391.20), TcMCU, (TcCLB.503893.120), and TcIP3R (T. cruzi Esmeraldo strain, contig KB205149.1, nt 21468–30512), encoding for the T. cruzi vacuolar proton pyrophosphatase, flagellar calcium binding protein, mitochondrial calcium uniporter, and inositol 1,4,5-trisphosphate receptor, respectively. Gene IDs are from the TritrypDB. Each one of these four constructs (3′ end-sgRNA/Cas9/pTREX-n), together with DNA donor cassettes to induce homology directed repair, were used to co-transfect T. cruzi epimastigotes and to insert a specific tag sequence (3×HA tag or 3×c-Myc tag) at the 3′ end of each gene.
sgRNA targeting the 3′ end of these genes were designed to induce the double-stranded break by Cas9 nuclease downstream their stop codons. Chimeric sgRNAs were obtained by PCR from plasmid pUC_sgRNA as previously described (3) using specific oligonucleotides (Table 1, primers 1–5), which include a BamHI restriction site, the 20-nt specific protospacer region, and a 20-nt sequence that anneals to the sgRNA backbone. Subsequently these sgRNAs were cloned into Cas9/pTREX-n vector through BamHI site. To avoid Cas9 off-targeting, protospacers were analyzed with ProtoMatch 1.0 script (15).
TABLE 1.
Oligonucleotides used in this study
Bold uppercase, specific protospacer; italic underlined uppercase, restriction site; lowercase, sgRNA annealing region; bold underlined uppercase, gene-specific homologous region; italic lowercase, pMOTag vector annealing region, italic bold uppercase, stop codon; bold double-underlined uppercase, HA tag sequence.
For the generation of a DNA donor cassette (DNA template to induce homologous-directed DNA repair) containing the tag sequence and a marker for antibiotic resistance, we used a modified version of the pMOTag4H vector (37), where the T. brucei tubulin intergenic region for trans-splicing was replaced by the HX1 T. cruzi trans-splicing signal present in the pTREX vector (41). The HX1 fragment was amplified using primers 6 and 7 from Table 1 and cloned into pMOTag4H vector by SalI/HindIII restriction sites. We named the resulting vector pMOTag-HX1–4H, and it allowed the generation of a DNA donor molecule containing a 3×HA tag and the hygromycin resistance marker (Fig. 1A). We also used the pMOTag23M vector (Fig. 1B) developed for C-terminal tagging in T. brucei (37). This vector contains a 3×c-Myc tag and the puromycin resistance gene. Templates for homologous recombination were amplified by PCR with 120-bp ultramers, of which 100 bp correspond to regions located right upstream of the stop codon (forward primer) and downstream of the Cas9 target site (reverse primer) of the TcVP1, TcFCaBP, TcMCU, and TcIP3R genes (Table 1, primers 8–15) and 20 bp for annealing on the pMOTag-HX1–4H and pMOTag23M vectors that were used as PCR templates (Fig. 1, A and B). PCRs were carried out using the following cycling conditions: initial denaturation for 2 min at 95 °C followed by 40 cycles of 20 s at 95 °C, 20 s at 63 °C, and 1 min 40 s at 72 °C and then a final extension for 10 min at 72 °C.
Epimastigotes co-transfected with specific combinations of 3′ end-sgRNA/Cas9/pTREX-n and DNA donor were cultured for 5 weeks with G418 and puromycin or hygromycin for selection of double resistant parasites. Endogenous gene tagging was verified by PCR from gDNA using gene-specific primer sets (Table 1, primers 16–21) and Western blotting analysis. All constructs were verified by sequencing.
Overexpression of TcIP3R
The TcIP3R gene (9044 nt) including the C-terminal human influenza HA tag was cloned into the pTREX-n vector following a three-step design and subsequently transfected to epimastigotes. Briefly, PCR was performed with primer set Fw_TcIP3R-XbaI/Rv_TcIP3R-BglII for N-terminal region (N), and primer set Fw_TcIP3R-BglII/Rv_TcIP3R-HA-XbaI_BglII for C-terminal region (C) (Table 1, primers 23–26), using T. cruzi Y strain gDNA as template. The amplified fragment N was cloned into pET-32 EK/LIC vector (Novagen) (N-TcIP3R/pET-32) by XbaI and BglII restriction sites. Then amplified fragment C was cloned by BglII into plasmid N-TcIP3R/pET-32, previously treated with Antarctic phosphatase, to obtain the TcIP3R/pET-32 plasmid. Next, the full sequence of TcIP3R-HA was excised with XbaI from TcIP3R/pET-32 plasmid and subcloned into dephosphorylated pTREX-n vector by XbaI to generate the TcIP3R-HA-OE/pTREX-n plasmid. Insert orientation was determined by PCR and sequencing.
Cell Transfections
Transfections were performed as previously described (3). Briefly, T. cruzi Y strain epimastigotes in early exponential phase (4 × 107 cells) were washed with PBS, pH 7.4, at room temperature and transfected in ice-cold CytoMix (25 mm Hepes, 120 mm KCl, 0.15 mm CaCl2, 10 mm K2HPO4, 2 mm EGTA, 5 mm MgCl2, 0.5% glucose, 100 μg/ml BSA, 1 mm hypoxanthine, pH 7.6) containing 25 μg of each plasmid construct and 25 μg of donor DNA in 4-mm electroporation cuvettes with three pulses (1,500 V, 25 microfarads) delivered by a Gene Pulser II (Bio-Rad). Stable cell lines were established and maintained under drug selection (250 μg/ml G418 and 350 μg/ml hygromycin or 5 μg/ml puromycin). Transfectant epimastigotes were cultured in liver infusion tryptose medium supplemented with 20% heat-inactivated FBS until stable cell lines were obtained.
PCR Analysis of Transfected Epimastigotes
Genomic DNA of double-resistant transfectants was used as template in PCRs to verify the integration of the DNA donor molecules into the 3′ end of the tagged genes. In each PCR was included a gene-specific forward primer (Table 1, primers 16–19) and a reverse primer annealing at 3′ end of the antibiotic marker present in the donor DNA (Table 1, primers 20 and 21). PCR conditions in a 25-μl reaction volume using GoTaq DNA polymerase with ∼20 ng of gDNA were as follows: 35 cycles of 95 °C for 20 s, 57 °C for 30 s, and 72 °C for 2 min 20 s followed by a final extension 72 °C for 10 min. TcVP1 site-directed tagging at nucleotide level was confirmed by sequencing several clones of PCR products obtained with primers 17 and 22 (Table 1) cloned into pGEM-T easy vector.
Western Blot Analysis
Western blotting analyses were performed using standard procedures used in our laboratory (42, 43). Parental and mutant cell lines were harvested separately. Parasites were washed twice in PBS and resuspended in radio immunoprecipitation assay buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% SDS, 0.1% Triton X-100) plus a mammalian cell protease inhibitor mixture (diluted 1:250), 1 mm phenylmethylsulfonyl fluoride, 2.5 mm tosylphenylalanyl chloromethyl ketone, 100 μm N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E64), and benzonase nuclease (25 units/ml of culture). The cells were then incubated for 1 h on ice. Cell lysis was verified under a light microscope, and protein concentration was determined by a bicinchoninic acid protein assay. 30 μg of protein from each cell lysate were mixed with 6× Laemmli sample buffer (125 mm Tris-HCl, pH 7, 10% (w/v) β-mercaptoethanol, 20% (v/v) glycerol, 4.0% (w/v) SDS, 4.0% (w/v) bromphenol blue) before application to 10%, 12% or 6–12% (gradient) SDS-polyacrylamide gels (depending on antigen size) without previous boiling. Separated proteins were transferred onto nitrocellulose membranes (Bio-Rad) with a Bio-Rad Trans-blot apparatus. The membranes were blocked with 5% nonfat dried skim milk in PBS-T (PBS containing 0.1% (v/v) Tween 20) overnight at 4 °C. Next blots were incubated for 1 h at room temperature with a primary antibody, i.e. monoclonal anti-TcFCaBP antibody (1:100 dilution), polyclonal rabbit anti-TbVP1 antibody (1:2,000), monoclonal anti-HA-Tag (1:5,000), monoclonal anti-c-Myc tag (1:100), and monoclonal anti-tubulin (1:40,000). After three washes with PBS-T, blots were incubated with the secondary antibody (goat anti-mouse IgG or goat anti-rabbit IgG, HRP-conjugated antibody, diluted 1:10,000). The membranes were washed three times with PBS-T, and Western blotting images were obtained and processed with a C-DiGit Blot Scanner (LI-COR Biosciences).
Immunofluorescence Analysis
Epimastigotes were washed with PBS and fixed with 4% paraformaldehyde in PBS for 1 h at room temperature. The cells were allowed to adhere to poly-l-lysine-coated coverslips and then permeabilized for 5 min with 0.1% Triton X-100. Permeabilized cells were blocked with PBS containing 3% BSA, 1% fish gelatin, 50 mm NH4Cl, and 5% goat serum overnight at 4 °C. Then cells were incubated with a primary antibody (monoclonal anti-TbFCaBP, 1:10; polyclonal rabbit anti-TbVP1, 1:250; monoclonal anti-HA tag, 1:500; rat anti-HA tag, 1:10; monoclonal anti-c-Myc tag, 1:10; rabbit anti-c-Myc tag, 1:50; rabbit anti-TbBiP, 1:50; rabbit anti-TbVDAC, 1:200) diluted in 1% BSA in PBS (pH 8.0) for 1 h at room temperature. The cells were washed three times with 1% BSA in PBS (pH 8.0) and then incubated for 1 h at room temperature in the dark with Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 546-conjugated goat anti-rabbit or Alexa Fluor 546-conjugated goat anti-mouse and Alexa Fluor 488-conjugated goat anti-rabbit secondary antibodies (1:1,000). Following incubation with the secondary antibody, the cells were washed and mounted on slides. DAPI (5 μg/ml) was included in the Fluoromount-G mounting medium to stain DNA. Controls were performed as described above but in the absence of a primary antibody. Differential interference contrast and fluorescence optical images were captured with a 100× objective (1.35 aperture) under nonsaturating conditions with an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ charge-coupled device camera driven by DeltaVision software (Applied Precision, Issaquah, WA) and deconvolved for 15 cycles using Softwarx deconvolution software (Fig. 6) or with a confocal microscope Leica TCS SP5 II, with a 100× objective (1.44 aperture) under nonsaturating conditions, that uses photomultiplier tubes for detection of emission, and LAS AF software (Leica, Wetzlar, Germany) for acquisition and processing of digital images (Figs. 2–5 and 7).
Author Contributions
N. L. and M. A. C. designed and conducted the experiments and analyzed the data. R. D. wrote the majority of the manuscript, with specific sections contributed by N. L. and M. A. C. R. D. and A. E. V. supervised the work and contributed to the analysis of experiments. M. S. performed the IFA experiments with TcIP3R-HA-OE cells.
Acknowledgments
We thank David Engman, Minu Chadhuri, Jay Bangs, and Norbert Bakalara for antibodies against TcFCaBP, TbVDAC, TbBiP, and TbVP1, respectively; Thomas Seebeck for pMOTag vectors; and the staff of the Life Sciences Core Facility at the State University of Campinas for the acquisition of the confocal microscopy images.
This work was funded by São Paulo Research Foundation Grant 2013/50624-0, and National Institutes of Health Grant AI107663, and São Paulo Research Foundation Postdoctoral Fellowships 2014/08995-4 (to N. L.) and 2014/13148-9 (to M. A. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
R. Docampo, unpublished results.
- ER
- endoplasmic reticulum
- IFA
- immunofluorescence analysis
- VDAC
- voltage-dependent anion channel
- IP3R
- inositol 1,4,5-trisphosphate receptor
- nt
- nucleotide(s)
- DIC
- differential interference contrast.
References
- 1. Lander N., Chiurillo M. A., and Docampo R. (2016) Genome editing by CRISPR/Cas9: a game change in the genetic manipulation of protists. J. Eukaryot. Microbiol. 63, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peng D., Kurup S. P., Yao P. Y., Minning T. A., and Tarleton R. L. (2014) CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio 6, e02097–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lander N., Li Z. H., Niyogi S., and Docampo R. (2015) CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. MBio 6, e01012–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Docampo R. (2011) Molecular parasitology in the 21st century. Essays Biochem. 51, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Docampo R., and Vercesi A. E. (1989) Characteristics of Ca2+ transport by Trypanosoma cruzi mitochondria in situ. Arch. Biochem. Biophys. 272, 122–129 [DOI] [PubMed] [Google Scholar]
- 6. Docampo R., and Vercesi A. E. (1989) Ca2+ transport by coupled Trypanosoma cruzi mitochondria in situ. J. Biol. Chem. 264, 108–111 [PubMed] [Google Scholar]
- 7. Docampo R., and Lukes J. (2012) Trypanosomes and the solution to a 50-year mitochondrial calcium mystery. Trends Parasitol. 28, 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baughman J. M., Perocchi F., Girgis H. S., Plovanich M., Belcher-Timme C. A., Sancak Y., Bao X. R., Strittmatter L., Goldberger O., Bogorad R. L., Koteliansky V., and Mootha V. K. (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Stefani D., Raffaello A., Teardo E., Szabò I., and Rizzuto R. (2011) A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang G., Vercesi A. E., and Docampo R. (2013) Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat. Commun. 4, 2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimoto M., Enomoto M., Morales J., Kurebayashi N., Sakurai T., Hashimoto T., Nara T., and Mikoshiba K. (2013) Inositol 1,4,5-trisphosphate receptor regulates replication, differentiation, infectivity and virulence of the parasitic protist Trypanosoma cruzi. Mol. Microbiol. 87, 1133–1150 [DOI] [PubMed] [Google Scholar]
- 12. Docampo R., and Huang G. (2015) Calcium signaling in trypanosomatid parasites. Cell Calcium 57, 194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang G., Bartlett P. J., Thomas A. P., Moreno S. N., and Docampo R. (2013) Acidocalcisomes of Trypanosoma brucei have an inositol 1,4,5-trisphosphate receptor that is required for growth and infectivity. Proc. Natl. Acad. Sci. U.S.A. 110, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang G., Ulrich P. N., Storey M., Johnson D., Tischer J., Tovar J. A., Moreno S. N., Orlando R., and Docampo R. (2014) Proteomic analysis of the acidocalcisome, an organelle conserved from bacteria to human cells. PLoS Pathog. 10, e1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sidik S. M., Hackett C. G., Tran F., Westwood N. J., and Lourido S. (2014) Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9, e100450, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C., Xiao B., Jiang Y., Zhao Y., Li Z., Gao H., Ling Y., Wei J., Li S., Lu M., Su X. Z., Cui H., and Yuan J. (2014) Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5, e01414–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang W. W., and Matlashewski G. (2015) CRISPR-Cas9-mediated genome editing in Leishmania donovani. MBio 6, e00861–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Engman D. M., Krause K. H., Blumin J. H., Kim K. S., Kirchhoff L. V., and Donelson J. E. (1989) A novel flagellar Ca2+-binding protein in trypanosomes. J. Biol. Chem. 264, 18627–18631 [PubMed] [Google Scholar]
- 19. Scott D. A., de Souza W., Benchimol M., Zhong L., Lu H. G., Moreno S. N., and Docampo R. (1998) Presence of a plant-like proton-pumping pyrophosphatase in acidocalcisomes of Trypanosoma cruzi. J. Biol. Chem. 273, 22151–22158 [DOI] [PubMed] [Google Scholar]
- 20. Docampo R., Vercesi A. E., and Huang G. (2014) Mitochondrial calcium transport in trypanosomes. Mol. Biochem. Parasitol. 196, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bangs J. D., Uyetake L., Brickman M. J., Balber A. E., and Boothroyd J. C. (1993) Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei: divergent ER retention signals in a lower eukaryote. J. Cell Sci. 105, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 22. Chaudhuri D., Sancak Y., Mootha V. K., and Clapham D. E. (2013) MCU encodes the pore conducting mitochondrial calcium currents. Elife 2, e00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinton P., Pozzan T., and Rizzuto R. (1998) The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 17, 5298–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Echevarría W., Leite M. F., Guerra M. T., Zipfel W. R., and Nathanson M. H. (2003) Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 5, 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoo S. H. (2010) Secretory granules in inositol 1,4,5-trisphosphate-dependent Ca2+ signaling in the cytoplasm of neuroendocrine cells. FASEB J. 24, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dellis O., Dedos S. G., Tovey S. C., Taufiq-Ur-Rahman, Dubel S. J., and Taylor C. W. (2006) Ca2+ entry through plasma membrane IP3 receptors. Science 313, 229–233 [DOI] [PubMed] [Google Scholar]
- 27. Yoo S. H. (1994) pH-dependent interaction of chromogranin A with integral membrane proteins of secretory vesicle including 260-kDa protein reactive to inositol 1,4,5-triphosphate receptor antibody. J. Biol. Chem. 269, 12001–12006 [PubMed] [Google Scholar]
- 28. Blondel O., Moody M. M., Depaoli A. M., Sharp A. H., Ross C. A., Swift H., and Bell G. I. (1994) Localization of inositol trisphosphate receptor subtype 3 to insulin and somatostatin secretory granules and regulation of expression in islets and insulinoma cells. Proc. Natl. Acad. Sci. U.S.A. 91, 7777–7781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerasimenko O. V., Gerasimenko J. V., Belan P. V., and Petersen O. H. (1996) Inositol trisphosphate and cyclic ADP-ribose-mediated release of Ca2+ from single isolated pancreatic zymogen granules. Cell 84, 473–480 [DOI] [PubMed] [Google Scholar]
- 30. Nguyen T., Chin W. C., and Verdugo P. (1998) Role of Ca2+/K+ ion exchange in intracellular storage and release of Ca2+. Nature 395, 908–912 [DOI] [PubMed] [Google Scholar]
- 31. Quesada I., Chin W. C., Steed J., Campos-Bedolla P., and Verdugo P. (2001) Mouse mast cell secretory granules can function as intracellular ionic oscillators. Biophys. J. 80, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alvarez J. (2012) Calcium dynamics in the secretory granules of neuroendocrine cells. Cell Calcium 51, 331–337 [DOI] [PubMed] [Google Scholar]
- 33. Ladenburger E. M., Korn I., Kasielke N., Wassmer T., and Plattner H. (2006) An Ins(1,4,5)P3 receptor in paramecium is associated with the osmoregulatory system. J. Cell Sci. 119, 3705–3717 [DOI] [PubMed] [Google Scholar]
- 34. Alzayady K. J., Sebé-Pedrós A., Chandrasekhar R., Wang L., Ruiz-Trillo I., and Yule D. I. (2015) Tracing the evolutionary history of inositol,1,4,5-trisphosphate receptor: insights from analyses of Capsaspora owczarzaki Ca2+ release channel orthologs. Mol. Biol. Evol. 32, 2236–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lisenbee C. S., Karnik S. K., and Trelease R. N. (2003) Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4, 491–501 [DOI] [PubMed] [Google Scholar]
- 36. Lu H. G., Zhong L., de Souza W., Benchimol M., Moreno S., and Docampo R. (1998) Ca2+ content and expression of an acidocalcisomal calcium pump are elevated in intracellular forms of Trypanosoma cruzi. Mol. Cell. Biol. 18, 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oberholzer M., Morand S., Kunz S., and Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 38. Lemercier G., Dutoya S., Luo S., Ruiz F. A., Rodrigues C. O., Baltz T., Docampo R., and Bakalara N. (2002) A vacuolar-type H+-pyrophosphatase governs maintenance of functional acidocalcisomes and growth of the insect and mammalian forms of Trypanosoma brucei. J. Biol. Chem. 277, 37369–37376 [DOI] [PubMed] [Google Scholar]
- 39. Singha U. K., Sharma S., and Chaudhuri M. (2009) Downregulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell 8, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bone G. J., and Steinert M. (1956) Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178, 308–309 [DOI] [PubMed] [Google Scholar]
- 41. Vazquez M. P., and Levin M. J. (1999) Functional analysis of the intergenic regions of TcP2beta gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene 239, 217–225 [DOI] [PubMed] [Google Scholar]
- 42. Lander N., Bernal C., Diez N., Añez N., Docampo R., and Ramírez J. L. (2010) Localization and developmental regulation of a dispersed gene family 1 protein in Trypanosoma cruzi. Infect. Immun. 78, 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lander N., Ulrich P. N., and Docampo R. (2013) Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J. Biol. Chem. 288, 34205–34216 [DOI] [PMC free article] [PubMed] [Google Scholar]