Abstract
URI (unconventional prefoldin RPB5 interactor protein) is an unconventional prefoldin, RNA polymerase II interactor that functions as a transcriptional repressor and is part of a larger nuclear protein complex. The components of this complex and the mechanism of transcriptional repression have not been characterized. Here we show that KAP1 (KRAB-associated protein 1) and the protein phosphatase PP2A interact with URI. Mechanistically, we show that KAP1 phosphorylation is decreased following recruitment of PP2A by URI. We functionally characterize the novel URI-KAP1-PP2A complex, demonstrating a role of URI in retrotransposon repression, a key function previously demonstrated for the KAP1-SETDB1 complex. Microarray analysis of annotated transposons revealed a selective increase in the transcription of LINE-1 and L1PA2 retroelements upon knockdown of URI. These data unveil a new nuclear function of URI and identify a novel post-transcriptional regulation of KAP1 protein that may have important implications in reactivation of transposable elements in prostate cancer cells.
Keywords: phosphorylation, prostate cancer, protein phosphatase 2 (PP2A), transcription regulation, transcription repressor, unconventional prefoldin RPB5 interactor (URI)
Introduction
The URI (unconventional prefoldin RPB5 interactor protein) has structural homology to members of the prefoldin family of molecular chaperones. URI protein is distributed throughout the cell to coordinate cellular processes involving metabolism, proliferation, and gene expression. In the mitochondria, URI binds and represses PP1γ phosphatase, thus sustaining survival signaling through the mToR pathway (1, 2). Cytoplasmic URI is part of the Hsp90 and R2TP/prefoldin-like complex that assembles RNA polymerase II before nuclear translocation (3, 4). In the nucleus, URI functions as a transcriptional repressor that binds RPB5, the shared subunit of the RNA polymerases (5). Despite several reports demonstrating that URI binds assembling (3) and transcribing (6) RNA polymerase II, and key regulators of RNA polymerase II transcription (7, 8) and in conjunction with prefoldin-like protein ART-27, represses androgen receptor-dependent gene expression (9) in prostate cells, little is known about the mechanism of URI transcriptional repression. In Saccharomyces cerevisiae, URI binds the phosphorylated C-terminal domain (CTD)2 of RNA polymerase II and affects the recruitment of the chromatin remodeling complex RSC (10).
Here we show that in the nucleus URI interacts with KAP1 and the protein phosphatase PP2A. KAP1 is recruited on the DNA by zinc finger transcription factors containing a KRAB domain (KRAB-ZFP) (11, 12). KAP1 functions as a scaffold for the recruitment of a repression complex that includes histone deacetylases (13), HP1 (heterochromatin protein 1) (14), and the histone methyltransferase SETDB1 (15). The assembly of the KAP1 repression complex is tightly controlled. SUMOylation of KAP1 is necessary for recruitment of the repression machinery. ATM phosphorylation of KAP1 on Ser824 interferes with KAP1 SUMOylation and consequently, relieves the KAP1-mediated transcription repression through displacement of the repression complex (16). In embryonic stem cells, KAP1, acting through SETDB1 histone methyl transferase, is essential for silencing certain retroelements including human endogenous retroviruses, long interspersed element1 (LINE-1), and Alu elements (17–20). LINE-1 retrotransposon is the only active autonomous retrotransposon in the human genome (21, 22). LINE-1 transcription is driven by its 5′-UTR promoter, which mediates sense and antisense transcription (23, 24). Reactivation of retroelements has also been demonstrated in several types of cancers including prostate cancer, and it has been postulated to fuel genomic rearrangements (25–30).
The PPP family of serine/threonine phosphatases comprises type 1, 2A, and 2B protein phosphatases (PP1, PP2A, and PP2B) and others (31). The PP1 and PP2A catalytic subunits exist in different isoforms (PP1α, PP1β, PP1γ, PP2Aα, and PP2Aβ) that bind to regulatory proteins thought to confer substrate specificity. Serine and threonine phosphatases PP1α and PP1β were previously shown to dephosphorylate KAP1 Ser824, enabling repression of target genes (32).
Here we identify a novel multiprotein complex comprising URI, KAP1, and the phosphatase PP2A. In prostate cells, URI mediates dephosphorylation of KAP1 at Ser824 through recruitment of PP2A phosphatase. Misregulation of this newly identified complex leads to derepression of LINE-1 expression.
Results
URI Interacts with KAP1 in the Nucleus of Prostate Cells
URI/C19orf2 has been characterized as a transcriptional repressor, but little is known about the mechanisms by which URI inhibits gene transcription. We previously showed that URI, in complex with ART-27/UXT, represses androgen receptor-mediated gene transcription in prostate cells (9). Here we performed mass spectrometry analysis to identify URI interactors. We focused on nuclear proteins to investigate the largely unexplored role of URI in transcriptional regulation. Control LNCaP prostate cancer cells, stably overexpressing an empty vector (LNCaP-vect), or LNCaP cells constitutively expressing FLAG-tagged URI (LNCaP-URI) were used for the analysis. The number of peptides identified for each co-immunoprecipitated protein was compared between control cells and URI-overexpressing cells (Table 1). We identified a protein as a URI interactor if it immunoprecipitated from URI-overexpressing cells but not from control cells in replicate analyses. By these criteria, we identified 36 URI-interacting proteins from prostate cell nuclei. As expected, we identified 10 subunits of RNA polymerase I, II, or III (Table 1, asterisks) including RPB5/POLR2E, a previously identified URI interactor (4, 5, 10). We also confirmed URI interaction with ART-27/UXT and with all 10 components of the R2TP/prefoldin-like complex (Table 1, arrows) proposed to be important for the assembly of the RNA polymerase II complex (3). The two helicases RUVBL1 and RUVBL2, components of the R2TP/prefoldin-like complex, were also identified, although we noted a low number of the corresponding peptides also present in one of the two vector control samples. Among the novel URI interactors, we focused on the KAP1/TRIM28 protein, because of its previously characterized role in gene and transposon repression, and on the catalytic subunit of PP2A phosphatase, because of the role of URI in regulation of PP1γ phosphatase in the mitochondria (1) (Table 1). Interestingly, in at least one replicate experiment, peptides from the PP1α and PP1β phosphatases that interact with KAP1 (32) were also identified, as well as peptides from the regulatory subunit A of PP2A (although peptides from the latter were also present in the control immunoprecipitation), consistent with a URI-KAP1-phosphatase complex.
TABLE 1.
MS analysis of URI nuclear interactors
Shown is a complete list of confident nuclear URI interactors obtained from affinity purification followed by MudPIT LCLC-MS/MS analysis of FLAGURI from control cells (vector) and URI overexpressing cells (URI-FLAG). The analysis was repeated twice, and the number of peptides identified in the two analyses is reported with the international protein index (accession number) and the description for each protein. Additional proteins of interest that were also identified in control samples are reported in the bottom five rows. Arrows show the components of the R2TP/prefoldin-like complex, and asterisks show RNA polymerase subunits. The interaction of URI with the proteins in the rows with bold type is explored in this manuscript.
| Accession | Experiment 1 |
Experiment 2 |
Description | |||
|---|---|---|---|---|---|---|
| URI-FLAG | Vector | URI-FLAG | Vector | |||
| IPI00477619 | 70 | 0 | 55 | 0 | C19orf2 RPB5-mediating protein isoform a | ← |
| IPI00002408 | 37 | 0 | 27 | 0 | RPAP3 isoform 1 of RNA polymerase II-associated protein 3 | ← |
| IPI00024163 | 32 | 0 | 26 | 0 | POLR3A DNA-directed RNA polymerase III subunit RPC1 | * |
| IPI00291093 | 23 | 0 | 18 | 0 | POLR2E DNA-directed RNA polymerases I, II, and III subunit RPABC1 | * |
| IPI00465211 | 22 | 0 | 18 | 0 | WDR92 HZGJ | ← |
| IPI00021405 | 22 | 0 | 7 | 0 | LMNA isoform A of lamin A/C | |
| IPI00005657 | 19 | 0 | 15 | 0 | PFDN6 prefoldin subunit 6 | ← |
| IPI00006052 | 17 | 0 | 13 | 0 | PFDN2 prefoldin subunit 2 | ← |
| IPI00550995 | 15 | 0 | 13 | 0 | PIH1D1 PIH1 domain-containing protein 1 | ← |
| IPI00027887 | 14 | 0 | 13 | 0 | PDRG1 p53 and DNA damage-regulated protein 1 | ← |
| IPI00220045 | 13 | 0 | 3 | 0 | POLR3E isoform 1 of DNA-directed RNA polymerase III subunit RPC5 | * |
| IPI00170862 | 12 | 0 | 3 | 0 | UXT ubiquitously expressed transcript isoform 1 | ← |
| IPI00301346 | 10 | 0 | 13 | 0 | POLR3B DNA-directed RNA polymerase III subunit RPC2 | * |
| IPI00031960 | 9 | 0 | 9 | 0 | POLR1A DNA-directed RNA polymerase I subunit RPA1 | * |
| IPI00005179 | 8 | 0 | 9 | 0 | POLR1C isoform 1 of DNA-directed RNA polymerases I and III subunit RPAC1 | * |
| IPI00910997 | 6 | 0 | 4 | 0 | cDNA FLJ57486 | |
| IPI00026952 | 6 | 0 | 4 | 0 | PKP3 plakophilin-3 | |
| IPI00215978 | 6 | 0 | 3 | 0 | POLR3D DNA-directed RNA polymerase III subunit RPC4 | * |
| IPI00216318 | 6 | 0 | 2 | 0 | YWHAB isoform long of 14-3-3 protein β/α | |
| IPI00003309 | 5 | 0 | 4 | 0 | POLR2H DNA-directed RNA polymerases I, II, and III subunit RPABC3 | * |
| IPI00856080 | 4 | 0 | 4 | 0 | GPN3 isoform 2 of GPN loop GTPase 3 | |
| IPI00010157 | 4 | 0 | 2 | 0 | MAT2A S-adenosylmethionine synthetase isoform type-2 | |
| IPI00477279 | 4 | 0 | 2 | 0 | RPL12P2 similar to ribosomal protein L12 | |
| IPI00430770 | 3 | 0 | 5 | 0 | SUPT6H isoform 2 of transcription elongation factor SPT6 | |
| IPI00554788 | 3 | 0 | 5 | 0 | KRT18 keratin, type I cytoskeletal 18 | |
| IPI00796337 | 3 | 0 | 3 | 0 | PCBP2 poly(rC) binding protein 2 isoform a | |
| IPI00006379 | 3 | 0 | 3 | 0 | NOP58 nucleolar protein 5 | |
| IPI00429689 | 3 | 0 | 2 | 0 | PPP2CB serine/threonine-protein phosphatase 2A catalytic subunit β isoform | |
| IPI00438229 | 2 | 0 | 8 | 0 | TRIM28 isoform 1 of transcription intermediary factor 1β | |
| IPI00031627 | 2 | 0 | 5 | 0 | POLR2A DNA-directed RNA polymerase II subunit RPB1 | * |
| IPI00031661 | 2 | 0 | 4 | 0 | NOC4L nucleolar complex protein 4 homolog | |
| IPI00410067 | 2 | 0 | 3 | 0 | ZC3HAV1 isoform 1 of zinc finger CCCH-type antiviral protein 1 | |
| IPI00009659 | 2 | 0 | 3 | 0 | RPRD1B regulation of nuclear pre-mRNA domain-containing protein 1B | |
| IPI00032439 | 2 | 0 | 2 | 0 | POLR1D isoform 1 of DNA-directed RNA polymerases I and III subunit RPAC2 | * |
| IPI00032406 | 2 | 0 | 2 | 0 | DNAJA2 DnaJ homolog subfamily A member 2 | |
| IPI00015361 | 2 | 0 | 2 | 0 | PFDN5 prefoldin subunit 5 | |
| IPI00872177 | 0 | 0 | 4 | 0 | PPP1CB 41-kDa protein | |
| IPI00550451 | 3 | 0 | 0 | 0 | PPP1CA serine/threonine-protein phosphatase PP1α catalytic subunit | |
| IPI00554737 | 3 | 2 | 0 | 0 | PPP2R1A serine/threonine-protein phosphatase 2A 65-kDa regulatory subunit A α isoform | |
| IPI00021187 | 18 | 5 | 18 | 0 | RUVBL1 isoform 1 of RuvB-like 1 | ← |
| IPI00009104 | 18 | 2 | 21 | 0 | RUVBL2 RuvB-like 2 | ← |
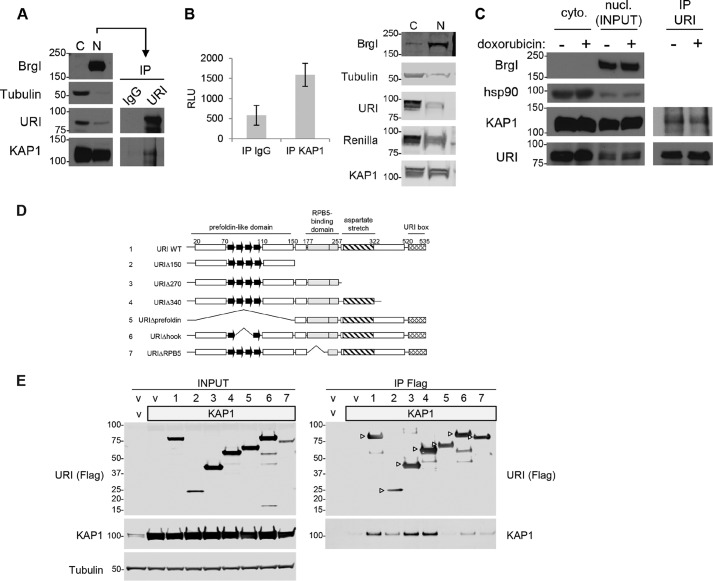
To validate URI interaction with KAP1, we performed co-immunoprecipitation experiments. Overexpressed and endogenous KAP1 protein co-immunoprecipitated with URI in HEK293 cells (Fig. 1E) and in LNCaP cell nuclear extracts (Fig. 1A). The reciprocal approach, detection of URI in KAP1 immunoprecipitated complexes, was much more challenging. We were able to immunoprecipitate KAP1 but were barely able to detect co-immunoprecipitated URI. The difficulty of the reciprocal immunoprecipitation is likely due to the abundance of KAP1 in LNCaP cells and that only a fraction of KAP1 interacts with URI. It is also possible that the polyclonal antibody used to immunoprecipitate (IP) KAP1 may interfere with the interaction with URI. Because we were only able to detect an extremely small amount of URI co-immunoprecipitated with KAP1 via Western blot (data not shown), we decided to utilize the LUMIER assay (33), which exploits the sensitivity of Renilla luciferase for the detection of protein-protein interactions. In Fig. 1B (left panel), URI N-terminally tagged with Renilla luciferase was expressed in LNCaP cells and endogenous KAP1 immunoprecipitated from the nuclear fraction. We show that Renilla activity is higher (∼2.7-fold) in the KAP1 IP compared with IgG control IP, demonstrating that KAP1 pulled down Renilla tagged URI. Isolation of the nuclear fraction is shown in Fig. 1B (right panel).
FIGURE 1.
URI interacts with KAP1. A, Western blot analysis of cytoplasmic (C) and nuclear (N) fractions isolated from LNCaP cells. Nuclear fractions were used for the immunoprecipitation of URI. A normal mouse IgG was used as control (IP IgG). BrgI and tubulin were used as nuclear and cytoplasmic markers, respectively (left panels). B, Renilla luciferase activity of URI tagged with Renilla luciferase co-immunoprecipitated with endogenous KAP1 from the nuclear fraction of LNCaP cells (left panel). The nuclear protein inputs are shown on the right panel. C, URI was immunoprecipitated as in A from LNCaP cells treated for 1 h with or without doxorubicin (1 μm). The data in A–C are representative of at least three independent experiments with standard deviation representing the error in B. nucl., nuclear; cyto., cytoplasmic. D, schematic of the seven URI constructs used to map domains of interaction between URI and KAP1. The known URI domains are indicated, and the numbers on top show the amino acid coordinates. E, an empty vector (v), KAP1 (gray box; 5 μg), and the various URI constructs were transfected in HEK293 cells (10 μg of total DNA was transfected in all transfections). 48 h after transfection cells were lysed. Inputs are shown in the left panel, and URI immunoprecipitated using FLAG antibodies is shown in the right panel. The immunocomplexes were analyzed by Western blot using the indicated antibodies. Arrowheads indicate the URI deletions/truncations. E is representative of three independent experiments.
Because KAP1 was shown to be phosphorylated in response to DNA damage (16, 34–36) and URI is also modified upon a diverse range of treatments (9, 37), we tested whether doxorubicin treatment affected interaction between URI and KAP1. Equal levels of KAP1 protein co-immunoprecipitated with URI in LNCaP cells treated with or without doxorubicin for 1 h (Fig. 1C), suggesting that URI interacts with KAP1 independently of DNA damage and KAP1 phosphorylation. To identify the protein domains responsible for the interaction with KAP1, deletions and truncations of URI were generated (Fig. 1D). Immunoprecipitation assays showed that URI and KAP1 interaction likely involves multiple interaction domains (Fig. 1E): the prefoldin-like and the RPB5-binding domains. URI lacking the entire prefoldin-like domain (URI construct 5, bottom right panel) has a very weak interaction with KAP1 compared with the URIΔhook mutant (URI construct 6, bottom right panel), suggesting stronger interaction of KAP1 with the two α-helixes of the URI prefoldin-like domain. Altogether, these data show that URI binds KAP1 in prostate cells.
Depletion of URI Increases KAP1 Ser824 Phosphorylation in Response to DNA Damage
KAP1 was previously shown to be phosphorylated on serine 824 in response to DNA damage downstream of the ATM kinase (32). To explore the role of URI in KAP1 phosphorylation, LNCaP cells stably overexpressing an inducible shRNA control (LNCaP-shNS) or a shRNA against URI (LNCaP-shURI) were treated for 30 min with 1 μm doxorubicin, a strong inducer of the DNA damage response. Short exposure of LNCaP cells to 1 μm doxorubicin induced a negligible increase of nuclear KAP1 phosphorylation, but depletion of URI clearly increased nuclear KAP1 phosphorylation upon doxorubicin treatment (Fig. 2A). We note that the effect on KAP1 phosphorylation is clearly observed in nuclear extracts, but not in cytoplasmic extracts where we also detected abundant KAP1. Because URI interacts with the RPB5 subunit of RNA polymerase II (PolII), the phosphorylation status of the RNA polymerase CTD on serine 2 was also analyzed. PolII CTD serine 2 phosphorylation was not affected by URI depletion indicating that KAP1 is specifically hyperphosphorylated upon URI depletion. The same experiment shown above was also performed using the p53 and androgen receptor null prostate cell line PC3. The effect of URI depletion shown in LNCaP cells was recapitulated in the PC3 cells (Fig. 2B).
FIGURE 2.
URI depletion induces increased phosphorylation on serine 824 of nuclear KAP1. A, cytoplasmic and nuclear fractions were isolated from LNCaP-shNS and LNCaP-shURI cell lines. To induce expression of the shRNAs, cells were treated with doxycycline (1 μg/ml) for 3 days and then treated with or without doxorubicin (1 μm) for 30 min, prior to lysis. BrgI and tubulin were used as nuclear and cytoplasmic markers, respectively. B, PC3-shNS and PC3-shURI cell lines were treated as in A. BrgI and tubulin were used as nuclear and cytoplasmic markers, respectively. C, LNCaP-shNS and LNCaP-shURI were treated with or without 1 μm doxorubicin for 30 min, and ATM was immunoprecipitated (IP ATM). URI (input) depletion in the shURI cells is shown. Western blot analysis was performed using the indicated antibodies. The data are representative of at least three independent experiments.
We next asked whether the increase of KAP1 phosphorylation upon URI depletion was due to a direct role of URI on KAP1 or an indirect consequence of hyperactivation of ATM kinase by analyzing ATM phosphorylation on Ser1981, a site used as a marker for ATM activation (38). Treatment of LNCaP cells with doxorubicin induced activation of ATM kinase as measured by the increase in Ser1981 phosphorylation (Fig. 2C). Depletion of URI did not increase ATM Ser1981 phosphorylation (Fig. 2C). This indicates that the DNA damage response-dependent hyperphosphorylation of KAP1 upon URI depletion is likely a direct effect of URI on KAP1, presumably via reduced levels of bound phosphatase.
URI Binds Active PP2A Phosphatase
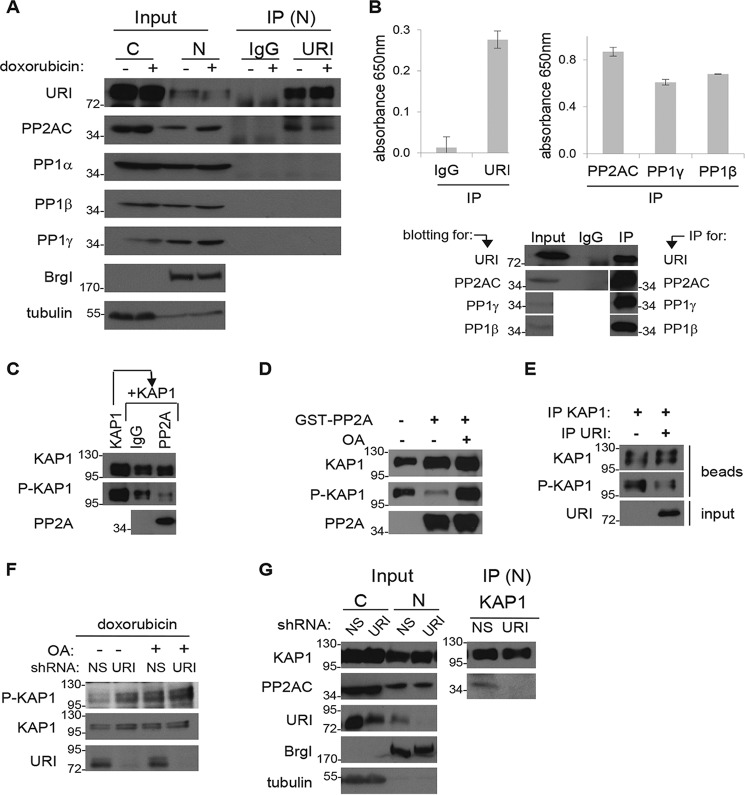
Our experiments suggest that URI binds and regulates the phosphorylation of KAP1. URI also binds to PP2A, suggesting that URI may recruit PP2A to dephosphorylate KAP1. We performed co-immunoprecipitations demonstrating that endogenous URI and PP2AC (PP2A catalytic subunit) interact in LNCaP nuclear extracts independent of DNA damage (Fig. 3A). We could not detect binding of URI to PP1α, β, or γ in the same immunoprecipitation experiments, suggesting that URI specifically binds PP2A phosphatase in the nucleus of prostate cells (Fig. 3A). Because previous studies showed that URI binds and inactivates PP1γ in the mitochondria (1), we reasoned that URI might also inactivate PP2A in prostate cells. We performed phosphatase activity assays measuring the dephosphorylation of a threonine phosphopeptide (KRpTIRR) that was used as a substrate for immunoprecipitated PP2A, URI, and control IgG from total lysates (Fig. 3B) or nuclear extracts (data not shown). Phosphatase assays demonstrated that PP2A co-immunoprecipitated with URI is active and has greater dephosphorylating activity compared with either PP1α or PP1β (Fig. 3B), probably because of its higher abundance in the nucleus of LNCaP cells. In contrast to URI-PP1γ binding in the mitochondria that suppresses phosphatase activity, these data suggest that URI is associated with active PP2A, and binding of URI to PP2A protein in the nucleus does not inhibit phosphatase activity.
FIGURE 3.
PP2A binding and dephosphorylation of KAP1 Ser824 is mediated by URI. A, nuclear URI was immunoprecipitated from LNCaP nuclear extracts treated with or without doxorubicin (1 μm) for 3 h. Proteins co-immunoprecipitated with URI were analyzed by Western blot using the indicated antibodies. C, cytoplasmic fraction; N, nuclear fraction. B, phosphatase activity assay was performed with immunoprecipitated URI, PP2A, PP1γ, and PP1β. IgG immunoprecipitation was used as a negative control. The histograms with standard deviation representing the error show the absorbance of a malachite solution proportional to free phosphates released by the phospho-substrate. Western blot of the immunoprecipitated proteins is shown. C–E, KAP1 immunoprecipitated from nuclear extracts of LNCaP cells treated with 5 μm doxorubicin for 1 h were mixed with PP2A phosphatase immunoprecipitated from nuclear extract of untreated LNCaP cells (C) or with GST-purified PP2A (D) in the presence or absence of 30 nm OA or URI immunoprecipitated from untreated LNCaP cells (E). The beads indicate immune complexes. P-KAP1 Ser824, total KAP1, and PP2AC were analyzed by Western blot. F, LNCaP-shNS and LNCaP-shURI were treated for 1 h with doxorubicin (1 μm) in the presence or absence of 30 nm okadaic acid. The nuclear and cytoplasmic fractions were analyzed by Western blot. G, PP2A association with KAP1 is URI-dependent. Western blot analysis of the nuclear fraction of LNCaP-shNS (non-silencing control) and LNCaP-shURI cell lines treated for 1 h with doxycycline and doxorubicin (1 μm). KAP1 was immunoprecipitated and blotted for KAP1 and PP2A. The data are representative of three independent experiments.
KAP1 Ser824 Is a Substrate of URI-bound PP2A Phosphatase
To determine whether KAP1 Ser824 can be directly dephosphorylated by PP2A, we immunoprecipitated phospho-KAP1 Ser824 from the nucleus of LNCaP cells treated with doxorubicin. Immunoprecipitated phospho-KAP1 was mixed with PP2A immunoprecipitated from nuclear extracts of untreated LNCaP cells. Immunoblotting using a phospho-KAP1 Ser824 antibody confirmed robust phosphorylation of KAP1 upon doxorubicin treatment (Fig. 3C). Phosphorylation of KAP1 on Ser824 was substantially reduced in the presence of immunoprecipitated PP2A but not in a control (IgG) immunoprecipitation (Fig. 3C), suggesting that nuclear phospho-KAP1 Ser824 is a substrate of nuclear PP2A. To ensure that the dephosphorylation of KAP1 was mediated by PP2A, we also incubated nuclear phospho-KAP1 with purified GST-PP2AC protein. Immunoblotting showed a decrease in phosphorylated KAP1 in the presence of GST-PP2AC, and phosphorylation of KAP1 is unaffected when PP2A is inhibited by okadaic acid (Fig. 3D), indicating that KAP1 is a substrate of PP2A.
To demonstrate that phospho-KAP1 is dephosphorylated by URI-bound PP2A phosphatase, we immunoprecipitated FLAG-URI from nuclear extracts of LNCaP-URI cells, eluted the URI complex, and tested its ability to dephosphorylate KAP1. Western blot analysis showed that co-incubation of phospho-KAP1 with URI immunocomplexes (Fig. 3E) induced dephosphorylation of KAP1. These results suggest that URI-bound-PP2A dephosphorylates KAP1 at Ser824 in nuclear extracts of prostate cells. In addition pharmacological inhibition of PP2A via okadaic acid (OA) phenocopied the effect of URI depletion and enhanced KAP1 Ser824 phosphorylation. In LNCaP cells expressing URI (shRNA-NS) and treated with a concentration of OA (30 nm) that specifically inhibits PP2A, KAP1 was hyperphosphorylated to the same extent as cells depleted of URI (Fig. 3F, compare first and second lanes). These results support the hypothesis that in the nucleus URI modulates KAP1 Ser824 phosphorylation by recruiting PP2A phosphatase to KAP1.
KAP1 Binding to PP2A Is Mediated by URI
The data strongly suggest that URI mediates the binding of PP2A phosphatase to KAP1. To test this we performed co-immunoprecipitation experiments of KAP1 and PP2AC in LNCaP-shNS and LNCaP-shURI cells. Immunoprecipitation of KAP1 from the nuclear extract of LNCaP-shNS control cells treated with doxorubicin showed binding of PP2A phosphatase. This was abolished in LNCaP-shURI cells (Fig. 3G, right panel), indicating that in the presence of doxorubicin, PP2A recruitment to the KAP1 complex is URI-dependent. Based on these results we propose that URI targets PP2A to KAP1, and PP2A modulates the phosphorylation of KAP1 in response to environmental cues such as DNA damage insults (Fig. 4).
FIGURE 4.

URI modulates KAP1-mediated transcription repression through PP2A recruitment. Shown is a model of PP2A recruitment to the KAP1 complex through URI protein. Dephosphorylation of KAP1 on serine 824 by PP2A induces recruitment of the repression complex and repression of KAP1 regulated retrotransposons. Depletion of URI induces hyperphosphorylation of KAP1, release of the repression complex, and derepression of KAP1 regulated retrotransposons.
URI Depletion Induces Reactivation of Mobile Retroelements in Prostate Cancer Cell Lines
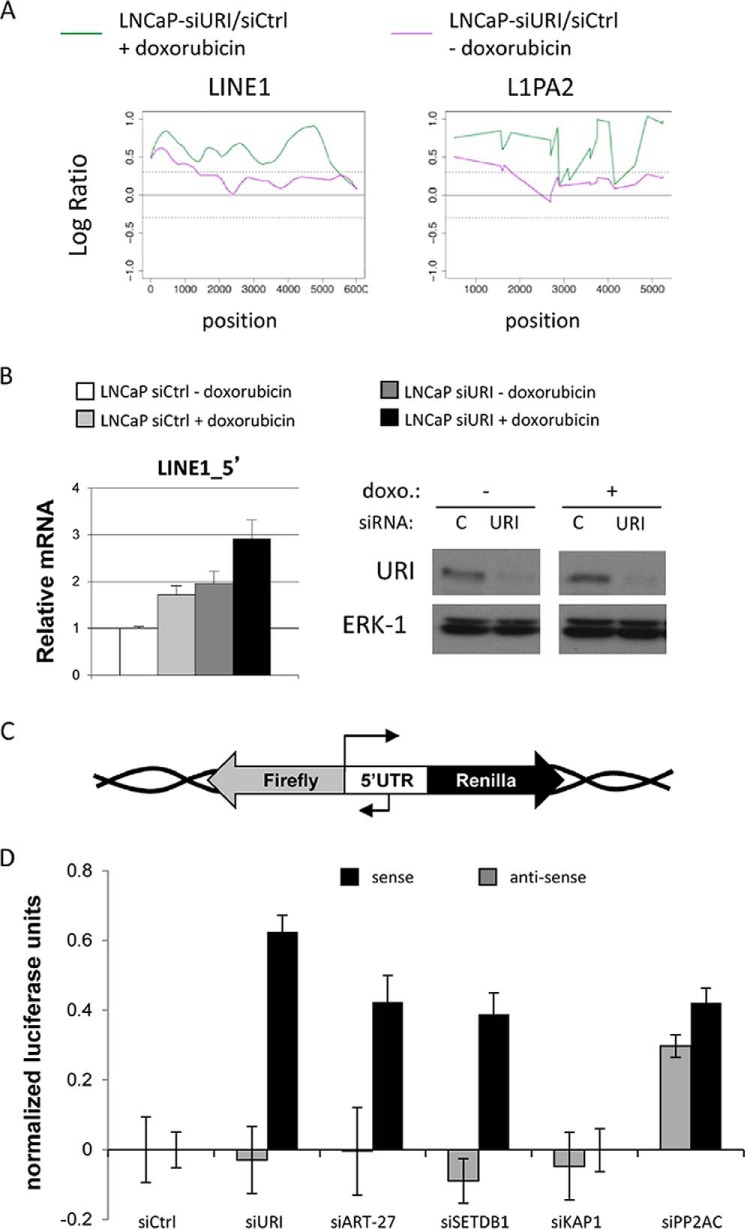
KAP1-mediated recruitment of SETDB1 methyltransferase plays a key role in the repression of some types of retroelements in embryonic stem cells (17–19). In somatic cells retrotransposons are tightly repressed; however, in conditions of misregulated cellular controls such as cancer, some classes of retrotransposons have been shown to evade suppression (27–30, 39). To show a functional relevance of the KAP1-URI-PP2A complex, we analyzed the effect of URI depletion on the expression of retroelements in prostate cancer cells. Because, to our knowledge, the complete profile of transposons reactivation in prostate cell lines is unknown, the effect of URI depletion on the expression of 511 annotated transposon transcripts in LNCaP cells was measured using a novel transposable element (TE) microarray platform (40). The effect of doxorubicin was also assessed to identify transposons potentially regulated by KAP1 phosphorylation. Transposable element expression in control cells was compared with the corresponding expression in URI-depleted cells (log ratio). Log ratio values bigger than 0 represent retroelement domains with increased expression upon URI depletion, whereas log ratio values smaller than 0 indicate regions repressed by URI depletion (Fig. 5A). We imposed a stringent threshold to focus only on expression of the most altered transposons. Transposable elements that showed altered expression with median absolute deviation (MAD) greater than 0.3 (Fig. 5A, horizontal dashed lines) in at least 500 bp were considered URI- or doxorubicin-regulated. The analysis revealed that LNCaP cells depleted of URI have increased expression of LINE-1 and L1PA2, an ancestral LINE-1 element. LINE-1 and L1PA2 were the only transposons affected by URI depletion and doxorubicin treatment. URI depletion induces a modest increase in expression of these retroelements (Fig. 5A, purple line), but concomitant doxorubicin treatment of URI depleted cells induced a considerable induction of LINE-1 and L1PA2 expression (Fig. 5A, green line). Increased LINE-1 expression was also validated by qPCR in Fig. 5B. Moreover we observed increased expression of LINE-1 in LNCaP cells depleted of ART-27, an interactor of URI that also regulates URI protein stability (4, 9) (data not shown).
FIGURE 5.
Analysis of retrotransposon expression upon doxorubicin treatment and URI depletion. A, LNCaP cells were depleted of URI by siRNA treatment and subsequently treated with 1 μm doxorubicin for 9 h. The isolated mRNA was analyzed for the expression of all annotated transposons using microarray analysis. Duplicate microarrays were analyzed. The MAD is shown as a dotted line. B, the expression of LINE-1 retroelement was validated by qPCR. Representative of at least three independent experiments, and error is represented as standard deviation. The effective depletion of URI compared with non-targeting control (C) was verified by Western blot analysis; ERK1 is used as a loading control. C, schematic of the luciferase reporter cassette integrated in the 293T-REx reporter cell line. D, 293 LINE-1 5′-UTR reporter cells measuring firefly (antisense) and Renilla (sense) luciferase were assayed after 48 h following treatment with the indicated siRNAs. The values were normalized by protein content (Bradford) and by siCtrl sense values that were set to 0. Representative of three independent experiments and error is shown as standard deviation.
To demonstrate the role of the URI-ART-27-KAP1-PP2A complex in the suppression of LINE-1 retrotransposon transcription, we also measured the effect of their depletion on the activity of a LINE-1 promoter. LINE-1 transcription is mediated by a promoter present in its 5′-UTR that possesses forward and reverse activity (41). To interrogate LINE-1 promoter sense and antisense activity, we developed a reporter cassette with the 5′-UTR of LINE-1 (L1rp) driving Renilla luciferase expression in the sense orientation and firefly luciferase in the antisense orientation (Fig. 5C). The reporter cassette was integrated into the genome of 293T-REx cells using the Flp-In system. Depletion of URI, ART-27, and PP2A by siRNA shows a clear increase of the sense activity of LINE-1 promoter compared with cells treated with a control siRNA. Interestingly PP2A depletion also increased antisense transcription (Fig. 5D). Treatment with siRNA against KAP1 did not have any effect on firefly or Renilla transcription driven by the 5′-UTR of LINE-1. However, depletion of the methyltransferase SETDB1, recruited to the L1 promoter by KAP1, induced increased activity of the LINE-1 promoter. We therefore argue that KAP1 siRNA treatment is insufficient to deplete the highly expressed KAP1 protein, and the remaining levels of KAP1 are sufficient to mask the effect on LINE-1 transcription (Fig. 5D).
Overall, these results indicate that the newly identified complex comprising URI, its partner ART-27, and PP2A phosphatase affects one of the known KAP1 biological functions: retrotransposon repression. This finding may be relevant to the increased genomic instability resulting from URI deletion previously observed in the Drosophila (6) and Caenorhabditis elegans (42) germ lines and is consistent with the previously reported derepression of retroelements in prostate cancer cells (39). Future efforts will be needed to explore the relevance of retrotransposon activation in normal and cancerous prostate cells and the role of the novel URI complex here identified in the suppression of these dangerous mobile genomic elements.
Discussion
URI is a transcriptional repressor that interacts with RNA polymerase II through binding to the POLR2E/RPB5 subunit (4, 5). URI has been shown to repress transcription of reporter constructs controlled by the VP16 transactivator (5) and androgen receptor response elements (9). Despite the fact that the transcriptional repression function of URI was its first identified function, the mechanism of repression is still unknown. In an attempt to clarify the role of URI in transcription regulation, we performed mass spectrometry analysis of nuclear URI interactors using the LNCaP prostate cell line. This analysis confirmed several previously identified URI interactors such as RPB5, ART-27, and the R2TP/prefoldin-like complex. Among the novel interactors of URI, we found KAP1/TRIM28/TIF1β. KAP1 was demonstrated to mediate gene silencing through the recruitment of a repression complex comprising the methyltransferase SETDB1, HDAC1/2 histone deacetylases and HP1 proteins (43). Moreover KAP1 repression activity is regulated by phosphorylation and therefore by the activity of several phosphatases (32, 44). Here we show interaction of URI with KAP1 in the nucleus of prostate cells. This interaction modulates KAP1 phosphorylation on serine 824, a site that when phosphorylated interferes with KAP1 SUMOylation and consequently with recruitment of the repression complex by KAP1 protein. We show that URI affects KAP1 phosphorylation and activity through the recruitment of the PP2A phosphatase, another novel interactor of URI. We also demonstrate that PP2A is able to directly dephosphorylate KAP1 phosphoserine 824. Therefore, we propose a model whereby URI targets the PP2A phosphatase to the KAP1 complex to oppose the effect of KAP1 phosphorylation (Fig. 4). Interestingly, this function of URI resembles its function in the mitochondria where URI binds and represses PP1γ. Upon URI phosphorylation, PP1γ is released from URI binding, whereby it becomes active and dephosphorylates BAD (BCL2-associated agonist of cell death), triggering apoptosis (1, 2). In a similar manner we show that in the nucleus, URI binds PP2A phosphatase and targets it to the KAP1 complex. We show here that URI plays a role in modulating the response of KAP1 to the DNA damage response induced by doxorubicin. In the context of cancer cells, often characterized by the loss of control mechanisms, the PP2A-URI complex may play an important role in tumorigenesis and cancer development.
An interesting question open for future examination: what is the regulatory mechanism that recruits URI and PP2A activity to KAP1? We found that both URI interaction with KAP1 and URI interaction with PP2A are unaffected by doxorubicin treatment (Figs. 1C and 3A). It is possible that conformational changes triggered by unknown post-translational modifications of URI, KAP1, or PP2A downstream of stimuli such as doxorubicin may alter contact between phosphorylated KAP1 and the PP2A catalytic subunit. Further investigations are necessary to clearly elucidate this molecular process.
KAP1 and SETDB1 are essential for repression of retroelements in embryonic stem cells (17, 18, 20). Retroelements have been studied for possible deleterious effects on normal cell physiology caused by their intrinsic mobility throughout the genome (21, 25, 26). The majority of mobile elements are not active in somatic cells, but a growing body of evidence identified some subfamilies of retroelements that are still potentially “mobile” in the human genome, albeit only in germ line cells and specific other somatic cell types, notably early embryos and brain, as well as a variety of tumor types (26, 45–47). In prostate cancer in particular, LINE-1 retroelements have been suggested to be derepressed. The LINE-1 ORF2 encoded endonuclease was also proposed to be important for the formation of the TMPRSS2:ETS translocation found in more than 40% of prostate cancers (48). In this work we show that changes in URI expression can affect KAP1 phosphorylation previously linked to its repressive function. We show that the loss of URI, as well as some of its protein partners, increases expression of the LINE-1 retrotransposon. We utilized a transposon microarray platform to identify classes of transposons reactivated in LNCaP prostate cells. LINE-1 was the only retrotransposon with increased expression upon doxorubicin treatment and URI knockdown. We also confirmed LINE-1 activation with qPCR validation. Moreover, a luciferase reporter cell line showed that depletion of URI, ART-27, PP2A, and SETDB1 proteins by siRNA induces increased sense transcription from the LINE-1 promoter, supporting the idea that our newly identified complex plays a role in LINE-1 suppression. The small but reproducible changes in transposon expression that we observed by qPCR upon URI, or ART-27 depletion, are in line with low transposon expression in somatic cells and with a role of the URI-PP2A complex in reversing KAP1 transcriptional repression. Consistent with these observations, deletion of the yeast homolog of URI, Bud27, was found to induce a strong increase in Ty1 transposon mobility (49). It is intriguing to consider that alteration of URI levels can trigger misregulation of DNA structure through altered regulation of KAP1 function, therefore inducing aberrant regulation of retroelements usually kept dormant in heterochromatic regions. These observations can potentially lead to a deeper understanding of the role of retroelements in prostate cancer.
Overall our data demonstrate a previously unexplored nuclear role of URI protein in the modulation of KAP1 phosphorylation and transcriptional repression. This modulation is mediated through URI binding to the PP2A phosphatase that we show reverses KAP1 serine 824 phosphorylation, thereby increasing retrotransposon expression.
Experimental Procedures
Subcloning of the URI Deletions
All the URI deletions and truncations were generated using a pcDNA3-FLAG-URI construct previously used in Ref. 9. Deletions were generated by PCR using specific primers with flanking XhoI restriction sequences. Therefore URI deletions have a unique XhoI restriction site at the point of deletion. Truncations were also generated by PCR using specific primers with XbaI restriction sequence. XbaI was chosen because it was a unique restriction site present just downstream of the URI STOP codon. The FLAG tag was maintained in all the deletions and truncations. The generated truncations of wild type URI were the following: URIΔ150 is a URI deleted of aa 151–535 and maintains only the prefoldin-like domain, URIΔ270 is a URI deleted of aa 271–535 and maintains the prefoldin-like and the RPB5 binding domains, and URIΔ340 is a URI deleted of aa 341–535 and maintains the prefoldin-like, the RPB5 binding, and the asparagine-rich domains. The generated deletions were the following: URIΔprefoldin is a URI deleted of aa 11–153 encompassing the entire prefoldin-like domain, URIΔhook is a URI deleted of aa 85–100 encoding two of the four β-strands of the prefoldin-like domain, and URIΔRPB5 is a URI deleted of aa 177–224 encompassing the essential domain for the binding of RPB5. All constructs were confirmed by sequencing.
Cell Culture and Stable Cell Lines
LNCaP (CRL-1740) and PC3 (CRL-1435) cell lines were purchased from ATCC (Manassas, VA). Cell lines are routinely tested for mycoplasma. The cells were cultured as previously reported in Ref. 9. LNCaP cells stably overexpressing the FLAG-URI construct or an empty vector as well as LNCaP stable cell lines overexpressing a non-silencing shRNA (shNS) or a shRNA against URI (shURI) were previously described (9). PC3 stable cell lines expressing shNS or shURI were generated with the same procedure used for LNCaP cells.
Luciferase 293T-REx Reporter Cell Lines
The 293T-REx-Flp-In cell line (R7807) was purchased from Life Technologies and was used to integrate a luciferase reporter construct into a modification of the pcDNA5/FRT/TO vector (Life Technologies). pcDNA5/FRT/TO vector was modified deleting the TO promoter and inserting a DNA fragment containing the 5′-UTR of L1rp human retrotransposon flanked by Renilla luciferase in the forward orientation and firefly luciferase in the antisense orientation. Renilla and firefly were generated through PCR using the pmirGLO dual luciferase vector (Promega) as a template. Stable cell lines were generated following the specification for Flp-In cell line generation.
Luciferase Assay
0.08 × 106 293T-REx reporter cells were plated in each well of a 24-well plate. The next day cells were transfected with siRNA control (siCtrl) or siRNA directed against the specified proteins (Life Technologies). RNAiMAX was used for transfection following the company specifications (Life Technologies). The following day a concentrated solution of doxorubicin was added to the medium to reach a final concentration of 50 nm. No appreciable cell death was observed at the end of the experiments. 48 h following transfection, the cells were lysed in reporter lysis buffer (Promega), and 30 μl of clarified cell lysate was used to measure firefly and Renilla luciferase with the Dual-Glo system (Promega). 1 μl of lysate was used to measure protein concentration by Bradford assay (Bio-Rad). Luciferase readings were normalized by protein concentration and then expressed as increase over control (siCtrl sample) set as 0.
Nuclear-Cytoplasmic Fractionation
For the isolation of cytoplasmic and nuclear fractions, the cells were scraped in PBS and resuspended in one packed cell volume of hypotonic 1× buffer A (10× buffer A: 100 mm Hepes, 15 mm MgCl2, 100 mm KCl; solution pH 7.9). The cells were left swelling in buffer A for 15 min on ice and then passed through a 23–26-gauge syringe needle. The solution was then centrifuged at 17,000 × g for 6 min at 4 °C. The supernatant (cytoplasmic fraction) was spun again at 17,000 × g for 15 min, whereas the pellet was resuspended in one pellet volume of buffer C (10 mm Hepes, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA; solution pH 7.9). Nuclei solution was incubated for 30 min on ice while stirring and then spun at 17,000 × g for 6 min. The supernatant (nuclear extract) was collected and the salt concentration adjusted to 300 mm using HEMG0 buffer (25 mm Hepes, 12.5 mm MgCl2, 10% glycerol, 1 mm EDTA). Triton X-100 was added to a final concentration of 1% and after 30 min of incubation on ice, the solution was centrifuged at 17,000 × g for 15 min. The supernatant was then used for immunoprecipitation experiments.
Antibodies and Immunoblotting
Protein lysates were prepared and immunoblotted as described in Ref. 9. The following antibodies were in used: BrgI (Abcam; ab4081), tubulin (AA13) (Covance; MMS-489P), URI (described in Ref. 1; generous gift from N. Djouder), KAP1 (Abcam; ab10483), P-KAP1 Ser824 (Abcam; ab70369), P-PolII S2 (Bethyl Laboratories; A300-654A), PolII (H-224) (Santa Cruz Biotechnology; sc-9001), Hsp90 (BD Transduction Lab; 610419), ATM (Abcam; ab17995), P-ATM (Abcam; ab36810), PP2AC (Santa Cruz 4B7; sc-13601), PP1α (EMD Biosciences;539517), PP1β (Bethyl Laboratories; A300-905A), PP1γ (C-19) (Santa Cruz Biotechnology; sc-6108), and ERK-1 (K-23) (Santa Cruz Biotechnology; sc-93).
The PAGE/Western blotting analyses presented in Fig. 1 (B, right panel, and E) were performed using NuPage 4–12% Bis-Tris gels run in MOPS buffer (Invitrogen). The proteins were transferred on PVDF-FL membranes (Bio-Rad) and processed for scanning on a LI-COR Odyssey CLx imaging system using IRDye secondary antibodies conjugated with infrared 680 and 800 IRDyes (LI-COR) against rabbit and mouse antibodies, respectively. Quantification was performed using Image Studio software (LI-COR).
Mass Spectrometry Analysis and Immunoprecipitation
15 × 108 LNCaP vector and LNCaP-FLAG-URI cells were used to isolate nuclear extracts. FLAG-URI was immunoprecipitated from the nuclear extracts using FLAG-conjugated agarose beads (Sigma). Immunoprecipitated and concentrated protein were then submitted to MS analysis.
Nuclear fractions of FLAG-URI and vector-only control were incubated with FLAG-conjugated agarose beads for 4 h. The beads were washed three times in HEMG300 (HEMG buffer with 300 mm KCl), three times in HEMG150 (HEMG buffer with 150 mm KCl), and twice in TBS. After the last wash, the beads were resuspended in 35 μl of TBS and 3 μl of 3× FLAG peptide (5 mg/ml) (Sigma; F4799). The solutions were incubated for 1 h at 4 °C. The supernatant containing the URI-interacting proteins was collected, and proteins were precipitated with TCA. TCA precipitate was resuspended in 8 m urea. Next the extracts were processed with ProteasMAX (Promega) per the manufacturer's instructions. The samples were subsequently reduced by 20 min of incubation with 5 mm tris-(2-carboxyethyl)phosphine at room temperature and alkylated in the dark by treatment with 10 mm iodoacetamide for 20 additional min. The proteins were digested overnight at 37 °C with sequencing grade modified trypsin (Promega), and the reaction was stopped by acidification with formic acid.
Multidimensional Protein Identification Technology and LTQ Mass Spectrometry
The protein digest was pressure-loaded onto a 250-μm inner diameter capillary packed with 2.5 cm of 10-μm Jupiter C18 resin (Phenomenex, Torrance, CA) followed by an additional 2.5 cm of 5-μm Partisphere strong cation exchanger (Whatman, Clifton, NJ). The column was washed with buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid. After washing, a 100-μm inner diameter capillary with a 5-μm pulled tip packed with 15 cm of 4-μm Jupiter C18 resin (Phenomenex) was attached to the filter union, and the entire split column (desalting column filter union-analytical column) was placed in line with an Agilent 1100 quaternary HPLC (Palo Alto, CA) and analyzed using a modified five-step separation described previously (50). The buffer solutions used were 5% acetonitrile and 0.1% formic acid (buffer A), 80% acetonitrile and 0.1% formic acid (buffer B), and 500 mm ammonium acetate, 5% acetonitrile, and 0.1% formic acid (buffer C). Step 1 consisted of a 75-min gradient from 0–100% buffer B. Steps 2–5 had a similar profile except 3 min of 100% buffer A, 5 min of 10–100% buffer C, a 10-min gradient from 0–15% buffer B, and a 105-min gradient from 10–55% buffer B (except for step 5, which %B was increased from 10% to 100%). The 5 min of buffer C percentages were 10, 40, 60, and 100%, respectively, for the five-step analysis. Peptides eluted from the microcapillary column were electrosprayed directly into an LTQ mass spectrometer (ThermoFinnigan, Palo Alto, CA) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan mass spectrum (400–2000 m/z) followed by seven data-dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur datasystem.
Analysis of Tandem Mass Spectra
Protein identification and quantification analysis were performed with Integrated Proteomics Pipeline (Integrated Proteomics Applications, Inc., San Diego, CA) using ProLuCID and DTASelect2. Tandem mass spectra were extracted into ms1 and ms2 files (51) from raw files using RawExtract 1.9.9 and were searched against IPI human protein database (version 3_57_01, released on January 1, 2009; plus sequences of known contaminants such as keratin and porcine trypsin concatenated to a decoy database in which the sequence for each entry in the original database was reversed (52) using ProLuCID/Sequest (53). MS/MS data were searched with 3000.0 milli-amu precursor tolerance, and the fragment ions were restricted to a 600.0 ppm tolerance. All searches were parallelized (54) and performed on the Scripps Research Institute's Garibaldi 64-bit LINUX cluster with 2848 cores. Search space included all fully and half-tryptic peptide candidates with no missed cleavages restrictions. Carbamidomethylation (+57.02146) of cysteine was considered as a static modification; we required two peptides per protein and at least one tryptic terminus for each peptide identification. The ProLuCID search results were assembled and filtered using the DTASelect program (version 2.0) (55, 56) with a false discovery rate of 0.05; under such filtering conditions, the estimated false discovery rate was below ∼1% at the protein level in each individual analysis. The RAW files, parameter files, and full search results are available from the authors.
Phosphatase Activity Assay
PP2A phosphatase assays were performed using the PP2A immunoprecipitation phosphatase assay kit (Millipore; 17-313). More specifically, in the phosphatase assay presented in Fig. 3C, KAP1 was immunoprecipitated from LNCaP cells treated with doxorubicin and was divided into three fractions: one fraction was used as input IP and the other two fractions were co-incubated with separately immunoprecipitated PP2A or control IgG in phosphatase buffer and processed for the phosphatase assay. The immunoprecipitated KAP1 were co-incubated with immunoprecipitated PP2A or IgG and were incubated for 10 min at 30 °C, washed, and eluted using 1× SDS sample buffer with reducing agent. The input sample and the two phosphatase samples were then analyzed by Western blotting.
LUMIER Assay
A pcDNA3.1-based, Golden Gate (57) compatible plasmid was constructed to easily tag the N terminus of proteins with Renilla. URI DNA (9) used for the Golden Gate reaction was amplified by PCR with overhangs compatible with the Golden Gate sites of the constructed acceptor plasmid. The sequence of the obtained plasmid was confirmed by restriction digest and Sanger sequencing. LNCaP cells were transfected with URI-Renilla using Lipofectamine 3000 following the company's instructions (Thermo Fisher Scientific; L3000015). 48 h post-transfection, the cells were lysed, and the nuclear fraction was isolated. Endogenous KAP1 was immunoprecipitated using epoxy Dynabeads (Life Technologies; 14311D) conjugated to the KAP1 antibody (Abcam; ab10483). Beads conjugated to rabbit IgG were used as a control (Santa Cruz; sc-2027). After an overnight incubation, the beads were washed in HEMG300 and Triton buffer three times. The beads were then resuspended in 1× reporter buffer (Promega; E397A), and RenillaGlo substrate (Promega; E2710) was added. The mixture was incubated under agitation for 10 min, and the luminescence was measured on a Synergy H1 (BioTek) using Gen5 2.00 software (BioTek). A blank sample composed of 1× reporter buffer was also measured to establish the baseline.
siRNA Transient Transfections
siRNA transient transfection was performed as described in Ref. 9. 24 h post-transfection, doxorubicin (Sigma) was added.
qPCR
Total RNA was isolated using RNeasy Plus (Qiagen) according to the manufacturer's instructions. Total RNA was reverse transcribed using the Verso cDNA kit (Thermo Scientific), and genomic DNA contamination was digested using the Turbo DNA-free kit (Life Technologies). The absence of genomic DNA was confirmed by performing parallel cDNA synthesis reactions in the absence of reverse transcriptase. Cycle numbers greater than 35 were considered negative. Real time PCR was performed using gene-specific primers (32) and Fast SYBR Green Master Mix (Life Technologies). The data were analyzed by the ΔΔCt method using RPL19 as a control gene and were normalized to the values for control samples, which were arbitrarily set to 1. LINE-1 primers have been reported previously (39, 44).
Transposon Microarray
TE array is a novel microarray platform that probes all annotated transposon transcript sequences (40). LNCaP cells treated with siRNA control were compared with LNCaP cells treated with siRNA directed against URI. The samples were run on the array such that every siURI sample was compared with the siCtrl sample.
The raw microarray data were Loess normalized and processed using the R package Limma. To identify specific TEs that are differentially expressed, we used the MAD, which identifies the difference between a group of contiguous TE probes in a siRNA sample compared with the control sample. The expression maps of these differentially expressed (MAD > 0.1) TEs with M values (log2 ratios of the siURI sample compared with the respective control) on the y axis and probe position in each element consensus sequence on the x axis. The horizontal dotted lines mark log2 changes of 0.3 and −0.3 (see page for S. K. L. at New York University School of Medicine website for all annotated transcripts).
Author Contributions
Experiments were performed primarily by P. M. with help from E. M. B. and S. H. and important analysis by J. N. S. and V. G. P. M., J. N. S., J. R. Y., D. M. R., G. D., J. D. B., M. J. G., and S. K. L. planned and analyzed experiments. and P. M., S. H., J. D. B., M. J. G., and S. K. L. wrote the manuscript.
This work was supported by National Institutes of Health Grants R01 CA112226 (to S. K. L.) and P41 GM103533 and R01 MH067880 (to J. R. Y.), National Institute on Aging Fellowship F32 AG039127 (to J. N. S.), Department of Defense Fellowship W81XWH-10-1-0431 (to P. M.), and funds from the urology department at New York University School of Medicine (to S. K. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CTD
- C-terminal domain
- LINE-1
- long interspersed element 1
- PP
- protein phosphatase
- IP
- immunoprecipitated or immunoprecipitation
- Pol
- polymerase
- OA
- okadaic acid
- TE
- transposable element
- MAD
- median absolute deviation
- qPCR
- quantitative PCR
- aa
- amino acid(s)
- siCtrl
- siRNA control.
References
- 1. Djouder N., Metzler S. C., Schmidt A., Wirbelauer C., Gstaiger M., Aebersold R., Hess D., and Krek W. (2007) S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol. Cell 28, 28–40 [DOI] [PubMed] [Google Scholar]
- 2. Guicciardi M. E., and Gores G. J. (2008) Cell stress gives a red light to the mitochondrial cell death pathway. Sci. Signal. 1, pe9. [DOI] [PubMed] [Google Scholar]
- 3. Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M. C., Ahmad Y., Neel H., Lamond A. I., and Bertrand E. (2010) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mita P., Savas J. N., Ha S., Djouder N., Yates J. R. 3rd, and Logan S. K. (2013) Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex. PLoS One 8, e63879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dorjsuren D., Lin Y., Wei W., Yamashita T., Nomura T., Hayashi N., and Murakami S. (1998) RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18, 7546–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirchner J., Vissi E., Gross S., Szoor B., Rudenko A., Alphey L., and White-Cooper H. (2008) Drosophila Uri, a PP1α binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC Mol. Biol. 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yart A., Gstaiger M., Wirbelauer C., Pecnik M., Anastasiou D., Hess D., and Krek W. (2005) The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 25, 5052–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei W., Gu J. X., Zhu C. Q., Sun F. Y., Dorjsuren D., Lin Y., and Murakami S. (2003) Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP). Cell Res. 13, 111–120 [DOI] [PubMed] [Google Scholar]
- 9. Mita P., Savas J. N., Djouder N., Yates J. R. 3rd, Ha S., Ruoff R., Schafler E. D., Nwachukwu J. C., Tanese N., Cowan N. J., Zavadil J., Garabedian M. J., and Logan S. K. (2011) Regulation of androgen receptor-mediated transcription by RPB5 binding protein URI/RMP. Mol. Cell. Biol. 31, 3639–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mirón-García M. C., Garrido-Godino A. I., Martínez-Fernández V., Fernández-Pevida A., Cuevas-Bermúdez A., Martín-Expósito M., Chávez S., de la Cruz J., and Navarro F. (2014) The yeast prefoldin-like URI-orthologue Bud27 associates with the RSC nucleosome remodeler and modulates transcription. Nucleic Acids Res. 42, 9666–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Urrutia R. (2003) KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolf G., Yang P., Füchtbauer A. C., Füchtbauer E. M., Silva A. M., Park C., Wu W., Nielsen A. L., Pedersen F. S., and Macfarlan T. S. (2015) The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 29, 538–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schultz D. C., Friedman J. R., and Rauscher F. J. 3rd (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2 α subunit of NuRD. Gene Dev. 15, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., and Losson R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18, 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., and Rauscher F. J. 3rd (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Gene Dev. 16, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee Y. K., Thomas S. N., Yang A. J., and Ann D. K. (2007) Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21(WAF1/CIP1) in breast cancer MCF-7 cells. J. Biol. Chem. 282, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 17. Karimi M. M., Goyal P., Maksakova I. A., Bilenky M., Leung D., Tang J. X., Shinkai Y., Mager D. L., Jones S., Hirst M., and Lorincz M. C. (2011) DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rowe H. M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P. V., Layard-Liesching H., Verp S., Marquis J., Spitz F., Constam D. B., and Trono D. (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 [DOI] [PubMed] [Google Scholar]
- 19. Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C., and Shinkai Y. (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 [DOI] [PubMed] [Google Scholar]
- 20. Castro-Diaz N., Ecco G., Coluccio A., Kapopoulou A., Yazdanpanah B., Friedli M., Duc J., Jang S. M., Turelli P., and Trono D. (2014) Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 28, 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burns K. H., and Boeke J. D. (2012) Human transposon tectonics. Cell 149, 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hancks D. C., and Kazazian H. H. Jr. (2012) Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nigumann P., Redik K., Mätlik K., and Speek M. (2002) Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79, 628–634 [DOI] [PubMed] [Google Scholar]
- 24. Alexandrova E. A., Olovnikov I. A., Malakhova G. V., Zabolotneva A. A., Suntsova M. V., Dmitriev S. E., and Buzdin A. A. (2012) Sense transcripts originated from an internal part of the human retrotransposon LINE-1 5′ UTR. Gene 511, 46–53 [DOI] [PubMed] [Google Scholar]
- 25. Belancio V. P., Roy-Engel A. M., and Deininger P. L. (2010) All y'all need to know 'bout retroelements in cancer. Semin. Cancer Biol. 20, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodić N., and Burns K. H. (2013) Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 9, e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doucet-O'Hare T. T., Rodić N., Sharma R., Darbari I., Abril G., Choi J. A., Young Ahn J., Cheng Y., Anders R. A., Burns K. H., Meltzer S. J., and Kazazian H. H. Jr. (2015) LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 112, E4894–E4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ewing A. D., Gacita A., Wood L. D., Ma F., Xing D., Kim M. S., Manda S. S., Abril G., Pereira G., Makohon-Moore A., Looijenga L. H., Gillis A. J., Hruban R. H., Anders R. A., Romans K. E., et al. (2015) Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 25, 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu Y., Feng F., Yang Y., Gao X., Cui J., Zhang C., Zhang F., Xu Z., Qv J., Wang C., Zeng Z., Zhu Y., and Yang Y. (2013) LINE-1 ORF-1p functions as a novel androgen receptor co-activator and promotes the growth of human prostatic carcinoma cells. Cell Signal. 25, 479–489 [DOI] [PubMed] [Google Scholar]
- 30. Rodić N., Steranka J. P., Makohon-Moore A., Moyer A., Shen P., Sharma R., Kohutek Z. A., Huang C. R., Ahn D., Mita P., Taylor M. S., Barker N. J., Hruban R. H., Iacobuzio-Donahue C. A., Boeke J. D., et al. (2015) Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ceulemans H., and Bollen M. (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 32. Li X., Lin H. H., Chen H., Xu X., Shih H. M., and Ann D. K. (2010) SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci. Signal. 3, ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blasche S., and Koegl M. (2013) Analysis of protein-protein interactions using LUMIER assays. Methods Mol. Biol. 1064, 17–27 [DOI] [PubMed] [Google Scholar]
- 34. Li X., Lee Y. K., Jeng J. C., Yen Y., Schultz D. C., Shih H. M., and Ann D. K. (2007) Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177–36189 [DOI] [PubMed] [Google Scholar]
- 35. Blasius M., Forment J. V., Thakkar N., Wagner S. A., Choudhary C., and Jackson S. P. (2011) A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 12, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goodarzi A. A., Kurka T., and Jeggo P. A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 [DOI] [PubMed] [Google Scholar]
- 37. Gstaiger M., Luke B., Hess D., Oakeley E. J., Wirbelauer C., Blondel M., Vigneron M., Peter M., and Krek W. (2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302, 1208–1212 [DOI] [PubMed] [Google Scholar]
- 38. Bakkenist C. J., and Kastan M. B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 39. Goering W., Ribarska T., and Schulz W. A. (2011) Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 32, 1484–1492 [DOI] [PubMed] [Google Scholar]
- 40. Gnanakkan V. P., Jaffe A. E., Dai L., Fu J., Wheelan S. J., Levitsky H. I., Boeke J. D., and Burns K. H. (2013) TE-array: a high throughput tool to study transposon transcription. BMC Genomics 14, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Speek M. (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parusel C. T., Kritikou E. A., Hengartner M. O., Krek W., and Gotta M. (2006) URI-1 is required for DNA stability in C. elegans. Development 133, 621–629 [DOI] [PubMed] [Google Scholar]
- 43. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., and Rauscher F. J. 3rd (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Gene Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 44. Liu J., Xu L., Zhong J., Liao J., Li J., and Xu X. (2012) Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle 11, 2643–2649 [DOI] [PubMed] [Google Scholar]
- 45. Belancio V. P., Roy-Engel A. M., Pochampally R. R., and Deininger P. (2010) Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 38, 3909–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kazazian H. H., Jr. (2011) Mobile DNA transposition in somatic cells. BMC Biol. 9, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rangwala S. H., Zhang L., and Kazazian H. H. Jr. (2009) Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 10, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin C., Yang L., Tanasa B., Hutt K., Ju B. G., Ohgi K., Zhang J., Rose D. W., Fu X. D., Glass C. K., and Rosenfeld M. G. (2009) Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139, 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nyswaner K. M., Checkley M. A., Yi M., Stephens R. M., and Garfinkel D. J. (2008) Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178, 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Washburn M. P., Wolters D., and Yates J. R. 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 51. McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., and Yates J. R. 3rd (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 52. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., and Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 53. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 54. Sadygov R. G., Eng J., Durr E., Saraf A., McDonald H., MacCoss M. J., and Yates J. R. 3rd. (2002) Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1, 211–215 [DOI] [PubMed] [Google Scholar]
- 55. Cociorva D., Tabb D. L., and Yates J. R. (2007) Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics, 16:13.4.1–13.4.14 [DOI] [PubMed] [Google Scholar]
- 56. Tabb D. L., McDonald W. H., and Yates J. R. 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Engler C., Kandzia R., and Marillonnet S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]