Abstract
Although the differentiation of oncogenically transformed basal progenitor cells is one of the key steps in prostate tumorigenesis, the mechanisms mediating this cellular process are still largely unknown. Here we demonstrate that an expanded p63+ and CK5+ basal/progenitor cell population, induced by the concomitant activation of oncogenic Kras(G12D) and androgen receptor (AR) signaling, underwent cell differentiation in vivo. The differentiation process led to suppression of p63-expressing cells with a decreased number of CK5+ basal cells but an increase of CK8+ luminal tumorigenic cells and revealed a hierarchal lineage pattern consisting of p63+/CK5+ progenitor, CK5+/CK8+ transitional progenitor, and CK8+ differentiated luminal cells. Further analysis of the phenotype showed that Kras-AR axis-induced tumorigenesis was mediated by Gli transcription factors. Combined blocking of the activators of this family of proteins (Gli1 and Gli2) inhibited the proliferation of p63+ and CK5+ basal/progenitor cells and development of tumors. Finally, we identified that Gli1 and Gli2 exhibited different functions in the regulation of p63 expression or proliferation of p63+ cells in Kras-AR driven tumors. Gli2, but not Gli1, transcriptionally regulated the expression levels of p63 and prostate sphere formation. Our study provides evidence of a novel mechanism mediating pathological dysregulation of basal/progenitor cells through the differential activation of the Gli transcription factors. Also, these findings define Gli proteins as new downstream mediators of the Kras-AR axis in prostate carcinogenesis and open a potential therapeutic avenue of targeting prostate cancer progression by inhibiting Gli signaling.
Keywords: androgen receptor, GTPase Kras (KRAS), Hedgehog signaling pathway, p63, prostate cancer
Introduction
Prostate cancer is the second most lethal cancer in men in North America and other Western countries (1). Prostate cancer progression has been characterized with multiple stages, including benign, prostatic intraepithelial neoplasia (PIN),2 invasive adenocarcinoma, and metastatic cancer (2). Numerous oncogenic driver genes, including loss of tumor suppressors, overexpression, and/or activation of oncogenes, have been identified based on genetic analysis of clinical prostate tumors. Dysregulation of ras signaling has been detected in 40% of prostate primary tumors and 90% of prostate metastatic disease (3). Gene fusion, genetic mutation, and prostate carcinogens have been reported in amplification or activation of Ras/Kras oncogenic signaling in prostate cancer (4–10). We have shown previously that the interplay of oncogenic Kras(G12D) and overexpression of AR signaling promotes prostate tumorigenesis through expansion of basal/progenitor cells (11). However, the molecular mechanisms underlying the transformation of these cells and the differentiation plasticity remain elusive.
The adult normal prostate gland consists of three types of epithelial cells: luminal, basal, and neuroendocrine cells. It has been reported that both basal and luminal cells serve as the cells of origin for the initiation of prostate cancer (12–15). Transformation of basal cells is one of the key steps in the initiation of prostate tumorigenesis. Experimental evidence has demonstrated that isolated naïve prostate basal cells are a target for oncogenic insults (12) and that transformation of basal cells induces differentiation to form luminal tumorigenic cells in vivo (13, 14).
While prostate luminal cells express cytokeratins (CKs) 8/18 with secretory function and mainly face the lumen in a tubule, basal cells usually express CK5/14 and p63, a member of the p53 protein family, and are aligned between the basal membranes of luminal cells (2). p63, a marker of prostate basal/progenitor cells, plays an essential role in the maintenance of prostate homeostasis and regeneration (16–18). Loss of p63 leads to severe developmental defects in mice, including lack of prostate, abnormal skin development, limb truncations, loss of hair follicles, and other defects (16, 19–21).
Although the homeostasis of p63 expression dictates the differentiation and renewal of prostate progenitor cells, dysregulation of p63 expression promotes tumor progression (22). However, the regulation of p63 expression and the role of p63-expressing cells in prostate tumor progression remain unclear under pathological conditions. Here we studied the differentiation potential and genetic regulation of p63-expressing cells in tumors mediated by oncogenic signaling of the Kras-AR axis. Basal/progenitor cells showed differentiation plasticity to a hierarchical pattern of multiple prostate lineages in vivo. Additionally, we provide experimental evidence that Gli transcription factors, effectors of multiple oncogenic pathways (23, 24), regulate the expression of p63 and an expansion of p63-expressing basal/progenitor cells. Our study helps to clarify the pathological role of p63 and provides a therapeutic strategy by targeting Gli signaling to inhibit prostate cancer progression.
Results
The Differentiation of p63 and/or CK5-expressing Cells to CK8+ Luminal Cells in Tumors
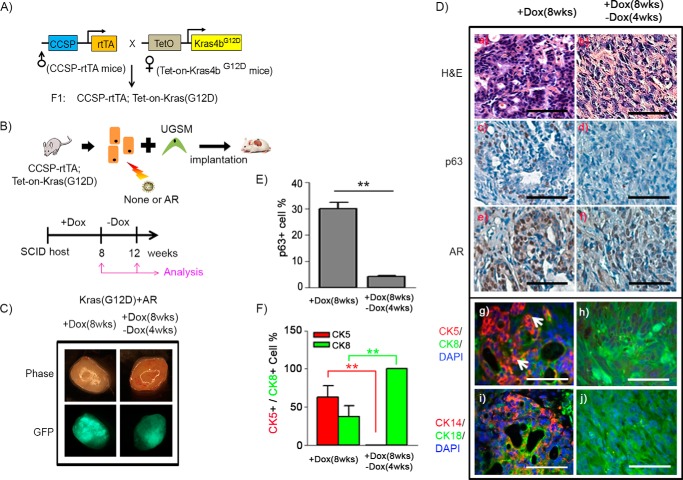
Kras and androgen receptor are two commonly dysregulated oncogenic signaling pathways in prostate cancer (3, 25). Previous studies have shown that the simultaneous activation of Kras and AR leads to an expansion of basal/progenitor cells (11). To monitor whether pathologically induced p63+ and CK5+ basal/progenitor cells have the potential to differentiate into luminal tumorigenic cells, we used a doxycycline-inducible Kras(G12D) model. CCSP-rtTA;Tet-on-Kras4bG12D mice were generated by mating CCSP-rtTA with Tet-on-Kras4bG12D transgenic mice (Fig. 1A). Clara cell secretory protein (CCSP), encoded by the SCGB1A1 gene, is primarily expressed in lung tissue (26), but its expression has also been shown in prostate or ovary tissue based on RNA sequencing analysis (GeneCards Database). Additionally, CCSP has been reported to be highly expressed in p63+ basal cells in murine tissue (27). After the genotype of the CCSP-rtTA;Tet-on-Kras4bG12D mice was confirmed by PCR analysis (supplemental Fig. S1A) (26), adult prostate tissue derived from CCSP-rtTA;Tet-on-Kras4bG12D mice was dissociated into single cells. Then, the prostate primary cells were transduced with/without AR carrying GFP as a marker (28) by lentiviral infection and combined with urogenital mesenchymal cells (Fig. 1B). The grafts were implanted subrenally in host SCID mice. Expression of Kras(G12D) in grafts was regulated by doxycycline supplied in the drinking water for 8 weeks, followed by an additional 4 weeks without doxycycline (Fig. 1B). The green fluorescence of the regenerated prostate grafts confirmed AR expression in tumors (Fig. 1C). Grafts derived from doxycycline-induced Kras(G12D) developed PIN lesions and exhibited epithelial proliferation surrounded by fibrous stroma (supplemental Fig. S1B). The proliferating cells had darkly stained nuclei, amphophilic cytoplasm, and a moderate nucleus:cytoplasm ratio. The cells largely formed a glandular structure with a well defined lumen. The glands were crowded and focally back-to-back, forming cribriform structures. In contrast, those from co-induction of doxycycline-induced Kras(G12D) and AR led to expansion of p63+ (Fig. 1D, c), CK5+ (Fig. 1D, g), or CK14+ cells (Fig. 1D, i) in prostate adenocarcinoma. The tumors showed epithelial proliferation in a haphazard pattern. In addition, the proliferating cells had darkly stained nuclei, amphophilic cytoplasm, and a high nucleus:cytoplasm ratio. The tumorigenic cells formed glandular structures, including well formed, moderately formed, and poorly formed glands. Tumor tissues contained 30%, 64%, and 37% of p63+, CK5+, and CK8+ cells, respectively (Fig. 1, E and F). Additionally, a fraction of cells was CK5+/CK8+ in Kras(G12D)+AR tumors (Fig. 1D, g).
FIGURE 1.
p63+ basal/progenitor cells possess differentiation potential to CK8+ luminal cells in Kras(G12D)+AR tumors. A, the CCSP-rtTA;Tet-on-Kras4bG12D mouse strain carrying doxycycline (Dox)-inducible Kras(G12D) was generated by crossing CCSP-rtTA mice with Tet-on-Kras4bG12D mice. B, prostate epithelial cells were isolated from prostate tissue of CCSP-rtTA;Tet-on-Kras4bG12D mice and transduced with AR along with GFP reporter by lentiviral infection. The infected cells were mixed with UGSM cells and implanted under the kidney capsule of CB.17SCID/SCID mice. The host mice were given drinking water containing Dox for 8 weeks to allow Kras(G12D) expression, followed by a Dox withdrawal period for an additional 4 weeks (a total of 12 weeks) to shut down Kras(G12D) expression. C, phase and fluorescence images of Kras(G12D)+AR grafts derived from 8 weeks of Dox induction (+Dox) or 8 weeks (wks) of Dox induction plus 4 weeks of Dox withdrawal (−Dox). Green fluorescence indicates that prostate cells were successfully infected by AR lentivirus. D, histological analysis of the regenerated grafts by H&E (a and b) and IHC staining of p63 (c and d), AR (e and f), CK5 (red)/CK8 (green)/DAPI (blue) (g and h), and CK14 (red)/CK18 (green)/DAPI (blue) (i and j). Scale bars = 50 μm. The white arrows indicate CK5+CK8+ cells. E and F, the number of p63+, CK5+, and CK8+ and total number of cells in tubules of regenerated tissue were counted. The percentage of p63+ (E) and CK5+ and CK8+ cells (F) per regenerated tubule was calculated. **, p < 0.01.
The regenerated prostate tissues derived from the Kras(G12D) group exhibited normal tubules after withdrawal of doxycycline (supplemental Fig. S1B), demonstrating a regression of Kras(G12D)-induced PIN lesions with the loss of Kras(G12D) activity. Interestingly, although the size of Kras(G12D)+AR tumors remained the same after withdrawal of doxycycline, the tumorigenic cells were mainly comprised of cells expressing CK8+ (Fig. 1D, h) or CK18+ (Fig. 1D, j) luminal markers but not p63+, CK5+, or CK14+ cells (Fig. 1D, d, h, and j). The data indicate that the increased number of CK8+ tumorigenic cells is associated with a decrease in p63+ and CK5+ cells, suggesting the differentiation of p63+ and/or CK5+ basal/progenitor cells to CK8+ luminal tumorigenic cells.
Withdrawal of Exogenous Androgen Reveals the Differentiation of p63 and/or CK5-expressing Cells to CK8+ Luminal Tumorigenic Cells
We have demonstrated previously that tumorigenic cells in tumors derived from constitutively active Kras(G12D) and AR maintain a similar pathological pattern in secondary recipients (11). To evaluate the importance of androgen levels in the oncogenic transformation of basal/progenitors cells, previously isolated Kras(G12D)+AR tumorigenic cells were implanted subcutaneously in a secondary recipient with or without an external testosterone pellet. Tumors grown with external testosterone were about 5-fold larger than those under physiological androgen levels (supplemental Fig. S2A). The secondary tumors showed predominantly CK5+ basal tumorigenic cells in both recipients (supplemental Fig. S2B). Additionally, Kras(G12D)+AR tumorigenic cells from our previous study developed tumors in host mice similar to those described previously; however, cells grown in castrated recipients did not develop tumors (supplemental Fig. S3B), suggesting that androgen levels in the host were required for the Kras(G12D)+AR engraftment.
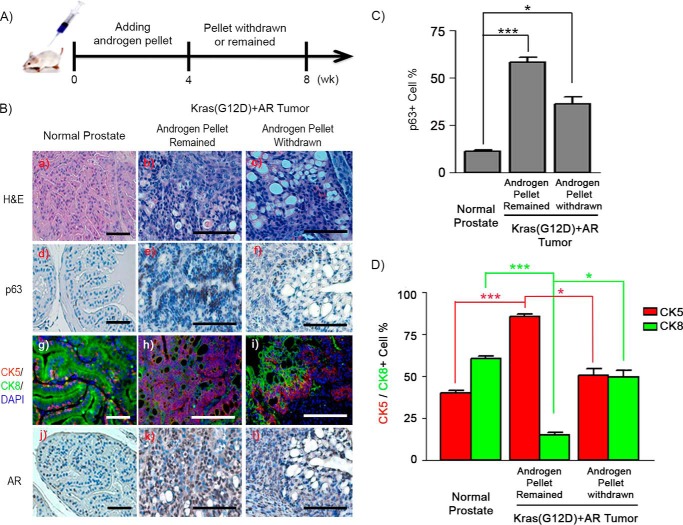
To further examine whether p63+ or CK5+ basal/progenitor cells had the potential to differentiate into luminal tumorigenic cells, the androgen levels of the secondary recipient were regulated by removal of the implanted testosterone pellet (Fig. 2A). As expected, tumor tissue in the group with a testosterone pellet showed an expansion of p63+ and/or CK5+ or CK14+ basal/progenitor cells in comparison with normal prostate tissues (Fig. 2, B–D, and supplemental Fig. S4C). CK5+ cells (Fig. 2, B, h, and D) or CK14+ (supplemental Fig. S4C) were predominant, and p63+ cells accounted for more than 50% within a tubule in the tumors (Fig. 2, B, e, and C). However, the percentage of p63+, CK5+, or CK14+ basal/progenitor cells significantly decreased with an increase in CK8+ or CK18+ luminal cells in tumors in the testosterone pellet removal group (Fig. 2, B–D, and supplemental Fig. S4C). After androgen pellet removal, p63+ cells regressed to become one to two layers in the basal layer (Fig. 2B, f, and supplemental Fig. S4A). CK5+ cells regressed and polarized at the basal layer, whereas multiple layers of CK8+ cells were polarized at the luminal region (Fig. 2B, i, and supplemental Fig. S4B). A layer of CK5+/CK8+ cells was localized between CK5+ and CK8+ layers (supplemental Fig. S4B). The hierarchal distribution pattern suggests that regression of pathologically expanded p63+ cells was associated with an decrease of CK5+ cells, leading to the occurrence of CK5+/CK8+ cells and an increase in CK8+ luminal tumorigenic cells in Kras(G12D)+AR tumors after removal of external androgen.
FIGURE 2.
p63+ and/or CK5+ basal/progenitor cells differentiate to CK8+ luminal cells after withdrawal of external androgen in Kras(G12D)+AR tumors. A, schematic of in vivo Kras(G12D)+AR tumor xenografts responding to withdrawal of external androgen in the host. Kras(G12D)+AR tumor xenografts were implanted subcutaneously into the flanks of SCID mice. Additionally, an androgen pellet was implanted subcutaneously on the back of host mice. After 4 weeks (wk), the androgen pellet was withdrawn from one group of host SCID mice, whereas it remained in the other group. Xenografts were harvested for pathology and IHC analysis. B, H&E (a–c) and IHC staining of p63 (d–f), CK5/CK8/DAPI (g–i), and AR (j–l) in Kras(G12D)+AR tumors carried by the host with an androgen pellet or withdrawal of the androgen pellet. Scale bars = 100 μm. C and D, the number of p63+ (C) and CK5+ and CK8+ (D) and total number of cells in tubules from Kras(G12D)+AR tumors or adult normal prostate tissue (control) was counted. The percentage of each type of the cells per tubule was calculated. *, p < 0.05; ***, p < 0.001.
The Expansion of Basal Cells Requires a Fully Functional AR
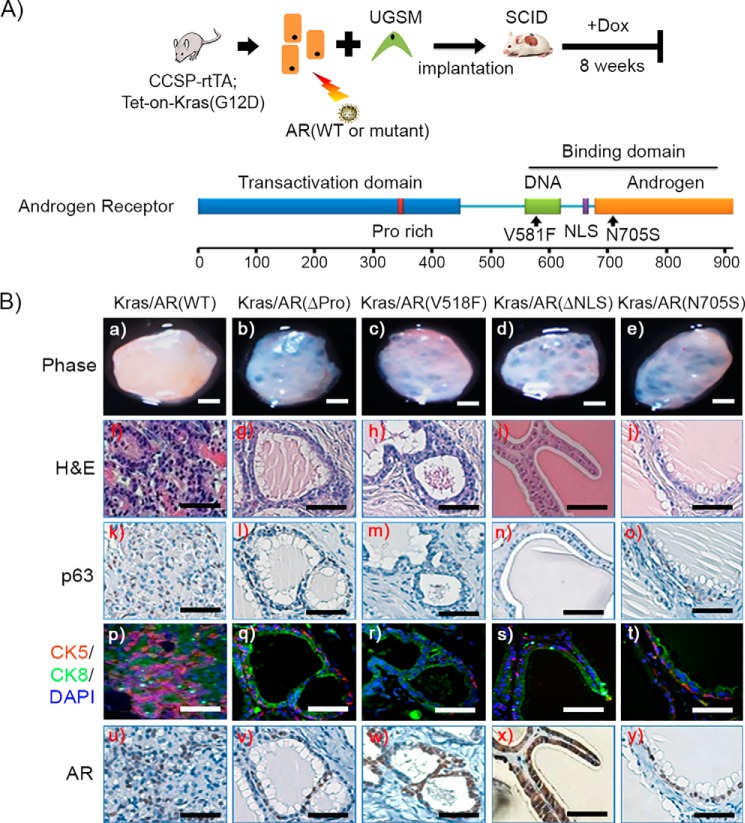
To examine whether the pathological expansion of p63+ and CK5+ cells required all AR functional domains, primary prostate cells derived from CCSP-rtTA;Tet-on-Kras4bG12D mice were transduced by lentiviral infection with wild-type AR or AR mutants (Fig. 3A). These mutants included AR(ΔPro), AR(V581F), AR(ΔNLS), and AR(N705S), which ablate the AR transactivation, DNA binding, nuclear localization, and androgen binding domains, respectively (29). Expression of the constructs was verified by Western blotting analysis (supplemental Fig. S6A). Prostate tissue regeneration was set up as described in Fig. 1, in which Kras(G12D) expression was regulated by the induction of doxycycline (Fig. 3A). As expected, overexpression of AR(WT) in combination with doxycycline-induced Kras(G12D) promoted tumorigenesis with an expansion of p63+ or CK5+ basal/progenitor cells (Fig. 3B, k and p). In contrast, regenerated tissues from Kras(G12D)+AR mutants exhibited normal tubule structure with a single layer of luminal and basal cells (Fig. 3B, g–j). Collectively, the data indicate that a fully functional AR is required for Kras signaling to pathologically regulate the transformation of basal/progenitor cells.
FIGURE 3.
The expansion of p63+ basal/progenitor cells requires a fully functional AR in Kras(G12D)+AR tumors. A, schematic of the in vivo prostate regeneration assay, including lentiviral infection and schedule for Dox induction. Prostate epithelial cells derived from CCSP-rtTA;Tet-on-Kras4bG12D transgenic mice were transduced with wild-type AR or AR mutants, mixed with UGSM, and implanted under the renal capsule. Host SCID mice were fed Dox in the drinking water for 8 weeks. A schematic of the AR functional domains is also shown. AR(ΔPro), AR(V518F), AR(ΔNLS) (a deletion mutant of AR nuclear localization signal), and AR(N705S) represent mutants that lost the functional domains of transcription activator, DNA binding, nuclear localization signal, and androgen binding, respectively. B, phase image of regenerated tissues (a–e) from the synergy of Kras(G12D) with AR (WT) or AR mutants (scale bars = 1 mm), H&E (f–j), and IHC staining of p63 (k–o), CK5 (red)/CK8 (green)/DAPI(blue) (p–t), and AR (u–y) of the generated grafts. AR expression is shown in the cytosol in x. Scale bars = 50 μm.
Blockade of Gli Transcription Factors Inhibits Kras(G12D)+AR-induced Tumors
Knowing that Kras-induced transformation requires active Gli signaling (30, 31), we examined whether the expression levels of Gli1 and Gli2, the two transcription factors known to cooperate with Kras induce transformation. Gli1 and Gli2 mRNAs (supplemental Fig. S5A) and protein (supplemental Fig. S5B) were significantly elevated in Kras+AR tumors in comparison with normal regenerated tissue.
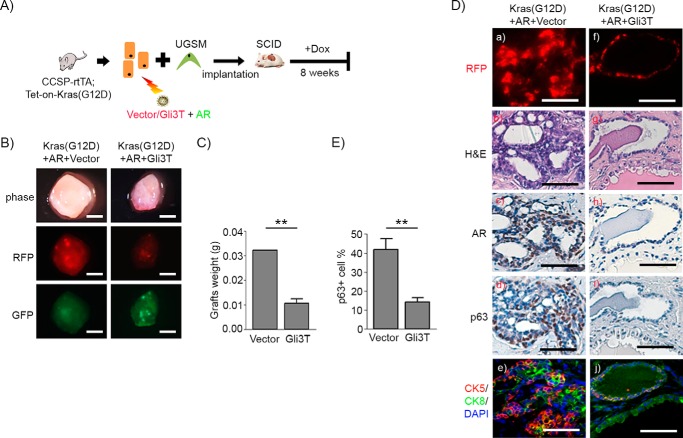
The reciprocal interaction of p63 and Gli transcription factors is essential for the maintenance of homeostasis of epithelial cells (32, 33). Thus, we further examined whether Gli signaling is essential for the expansion of p63 basal/progenitor cells in vivo. To this end, we used the N-terminal fragment of Gli3 (Gli3T) (supplemental Fig. S6B), a dominant negative repressor for Gli signaling (supplemental Fig. S6, C and D) (30). Primary prostate cells isolated from CCSP-rtTA;Tet-on-Kras4bG12D mice were co-transduced with or without AR together with control vector or Gli3T by lentiviral co-infection (Fig. 4A). The induction of Kras(G12D) expression was regulated by doxycycline in the drinking water (Fig. 4A). The regeneration tissue derived from the group without doxycycline in the drinking water led to normal tubules (supplemental Fig. S8). Overexpression of Gli3T showed a reduction in the size of the regenerated tissues in the Kras (supplemental Fig. S9) or normal regeneration group (supplemental Fig. S8), suggesting that blockage of Gli signaling with a dominant negative Gli3T lead to suppression of the regeneration process. However, regenerated prostate tubules (RFP+) in Gli3T groups expressed AR, p63, or CK5/CK8 (supplemental Figs. S8 and S9), suggesting that the regenerated prostate tubule structure was not affected by overexpression of Gli3T.
FIGURE 4.
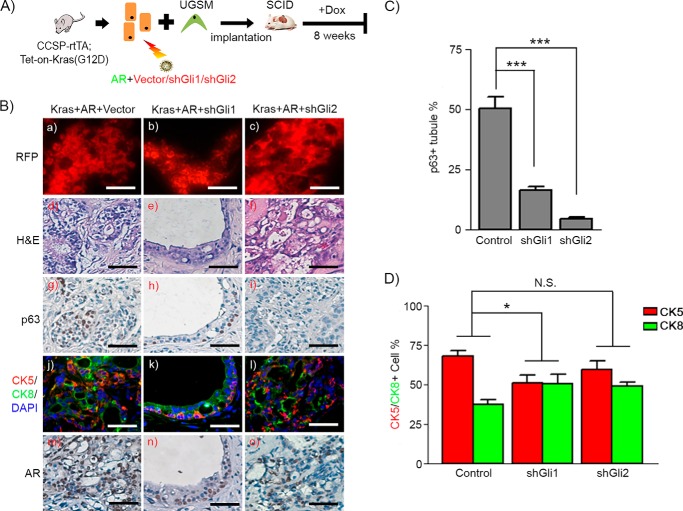
Gli signaling regulates the expansion of p63+ basal/progenitor cells and thus Kras(G12D)+AR tumors. A, schematic of the in vivo prostate regeneration assay, including lentiviral infection and the schedule for Dox induction. Prostate epithelial cells derived from CCSP-rtTA;Tet-on-Kras4bG12D transgenic mice were transduced with AR and control vector or Gli3T by lentiviral co-infection. The transduced cells were mixed with UGSM and implanted under the renal capsule. Host SCID mice were fed Dox in the drinking water for 8 weeks. B, phase and fluorescent images of regenerated prostate tissues of Kras(G12D)+AR+Vector and Kras(G12D)+Gli3T. Scale bars = 2 mm. C, the weight of the regenerated tissues was measured. D, RFP fluorescence (a and f), H&E (b and g), and IHC staining of AR (c and h), p63 (d and i), and CK5 (red)/CK8 (green)/DAPI (blue) (e and j) on regenerated tissues. Scale bars = 50 μm. E, the number of p63+ and total number cells in tubules from regenerated tissues was counted. The percentage of p63+ cells per regenerated tubule in the regenerated tissue was calculated. **, p < 0.01.
For Kras+AR mediated tumors, RFP/GFP signals in the regenerated prostate tissue suggests successful transduction of control vector or Gli3T and AR in the regenerated tissue (Fig. 4B). Grafts derived from those transduced with Gli3T weighed less in comparison with the control (Fig. 4C). Elevated AR expression was confirmed by immunohistochemistry of the regenerated tissues (Fig. 4D, c and h). Regenerated tissue in the Kras(G12D)+AR+vector group exhibited prostatic adenocarcinoma with an expansion of p63+ (Fig. 4D, d) and CK5+ (Fig. 4D, e) basal/progenitor cells, including CK5+/CK8+ cells. In contrast, tissues derived from the Kras(G12D)+AR+Gli3T group showed regenerated tubules that were histologically normal (Fig. 4D, i and j). These data strongly suggest that Gli signaling facilitates the pathological expansion of p63+ and CK5+ basal/progenitor cells and, thereby, Kras(G12D)+AR-induced tumorigenesis.
Gli1 and Gli2 Differentially Regulate Proliferation of p63-expressing Cells and Kras(G12D)+AR-induced Tumorigenesis in Vivo
Based on the above findings, we further examined the role of Gli1 and Gli2 in normal prostate regenerated tissues, Kras and Kras+AR transformed tissues. Primary prostate cells isolated from prostate tissue of CCSP-rtTA;Tet-on-Kras4bG12D mice were co-transduced with AR and shRNA-control, shRNA-Gli1, or shRNA-Gli2 by lentiviral co-infection, and then prostate regeneration grafts were established (Fig. 5A).
FIGURE 5.
Expression of Gli1 and Gli2 facilitates proliferation of p63+ basal/progenitor cells in Kras(G12D)+AR tumors. A, schematic of the in vivo prostate regeneration assay, including lentiviral infection and schedule for Dox induction. Prostate epithelial cells derived from CCSP-rtTA;Tet-on-Kras4bG12D transgenic mice were transduced with AR and control vector, shRNA-Gli1, or shRNA-Gli2. The transduced cells were mixed with UGSM and implanted under the renal capsule. Host SCID mice were fed Dox in the drinking water for 8 weeks. B, RFP fluorescence (a–c), H&E (d–f), and IHC staining of p63 (g–i), CK5 (red)/CK8 (green)/DAPI (blue) (j–l), and AR (m–o) in regenerated tissues of Kras(G12D)+AR+Vector, Kras(G12D)+AR+shGli1, and Kras(G12D)+AR+shGli2. Scale bars = 50 μm. C and D, the number of p63+, CK5+, and CK8+ and total number of cells in tubules from Kras(G12D)+AR+vector and Kras(G12D)+AR+shGli1/2 regenerated tissues was counted. The percentage of each type of cells was calculated. The ratio of CK5 to CK8 cell number was significantly reduced in Kras(G12D)+AR+shGli1 but not Kras(G12D)+AR+shGli2 in comparison with Kras(G12D)+AR+Vector. Two-way ANOVA analysis was applied. *, p < 0.05; ***, p < 0.001; N.S., not significant.
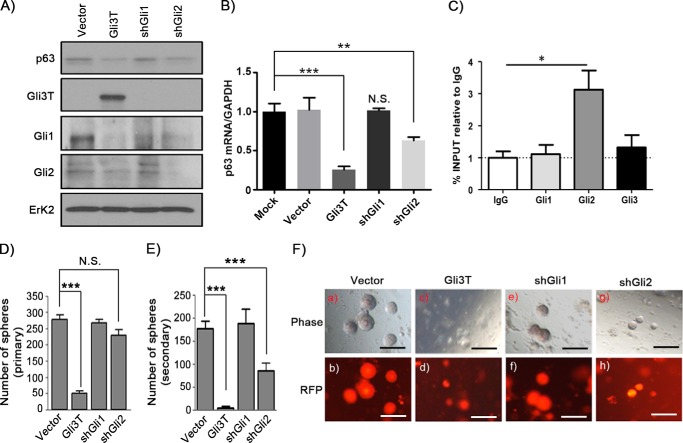
The knockdown of Gli1 or Gli2 by shRNA was confirmed at the mRNA (supplemental Fig. S7) and protein levels (Fig. 6A). The histology of shRNA-Gli1 or shRNA-Gli2 treatment was similar to the vector control in the normal regenerated prostate tissue (supplemental Fig. S8). AR, p63, or CK5/CK8 were expressed in regenerated prostate tubules (supplemental Fig. S8), suggesting that knockdown of Gli1 and Gli2 had no effect in the normal prostate regeneration process. However, PIN lesions induced by Kras were inhibited in the shRNA-Gli1 group and, to a lesser extent, in the shRNA-Gli2 group in comparison with the vector control (supplemental Fig. S9).
FIGURE 6.
Differential regulation of p63 expression by Gli1 and Gli2. A and B, mouse PEB cells were transduced with control vector, Gli3T, shRNA-Gli1 (shGli1), or shRNA-Gli2 (shGli2) by lentiviral infection. Cell lysates were collected and subjected to Western blotting analysis for expression of p63, Gli1, Gli2, Gli3T, and Erk2 (A) or quantitative RT-PCR for the expression of ΔNp63 and GAPDH (B). The vector group was normalized to GAPDH and set at 1. C, endogenous Gli1, 2, and 3 binding to the p63 promoter in PEB cells was examined by chromatin immunoprecipitation as described under “Experimental Procedures.” D–F, he primary prostate cells were transduced with control vector (Fu-CRW), Gli3T, shRNA-Gli1, or shRNA-Gli2 by lentiviral infection. After 5 days of growing in Matrigel, RFP was expressed in the transduced cells. RFP+ cells were sorted and remixed with Matrigel and subjected to the sphere formation assay. The number of primary (D) and secondary spheres (E) was recorded, and images of the spheres were taken (F). Scale bars = 200 μm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S., no significance (two-tailed Student t test).
Next, we examined the role of Gli1 and Gli2 in Kras (G12D)+AR-induced tumors in vivo. The RFP signal in the regenerated prostate tissues indicated transduction of shRNA-control or shRNA-Gli1/2 (Fig. 5B, a–c), and AR overexpression was confirmed by IHC (Fig. 5B, m–o). As expected, co-overexpression of Kras(G12D) and AR induced high-grade adenocarcinoma (Fig. 5B, d) with an expansion of p63+ and CK5+ basal/progenitor cells (Fig. 5, B, g and j, and C). In contrast, grafts derived from the Kras(G12D)+AR+shRNA-Gli1 group contained low-grade PIN lesions (Fig. 5B, e), inhibition of p63+ proliferation (Fig. 5, B, h, and C) and ratio of CK5:CK8 cells (Fig. 5, B, k, and D). However, grafts derived from the Kras(G12D)+AR+shRNA-Gli2 group displayed adenocarcinoma with high-grade lesions (Fig. 5B, f). Although tumorigenic tissue contained significant lower numbers of p63+-expressing basal/progenitor cells (Fig. 5, B, I, and C), it maintained an expansion of CK5+ cells with no significant difference in the ratio of CK5:CK8 cells in comparison with those in the Kras(G12D)+AR+vector group (Fig. 5, B, l, and D). The data suggest differential functional roles of Gli1 and Gli2 in the mediation of Kras(G12D)+AR tumors. Gli1 was essential for the proliferation of tumorigenic cells and tumor progression, and Gli2 largely regulated the expression of p63+-expressing cells.
Gli1 and Gli2 Differentially Regulate the Expression Levels of p63 and Sphere Formation
Next we examined whether Gli1 and Gli2 exhibited differential regulatory functions in p63 expression. Overexpression of Gli3T or knocking down Gli2, but not Gli1, suppressed the expression levels of p63 (Fig. 6, A and B). Additionally, Gli2, but not Gli1, showed direct binding to the p63 promoter (Fig. 6C).
p63 dictates the renewal potential of prostate progenitor and prostate sphere formation (34). We further examined whether Gli1 and Gli2 differentially regulated prostate sphere formation. Overexpression of Gli3T significantly suppressed prostate primary and secondary sphere formation (Fig. 6, D–F). Although knocking down Gli1 had no effect on sphere formation, knocking down Gli2 had no effect on primary sphere formation but significantly inhibited secondary sphere formation (Fig. 6, D–F). Collectively, the data suggest that Gli2 transcriptionally regulates the expression of p63 and, subsequently, the renewal potential of progenitor cells.
Discussion
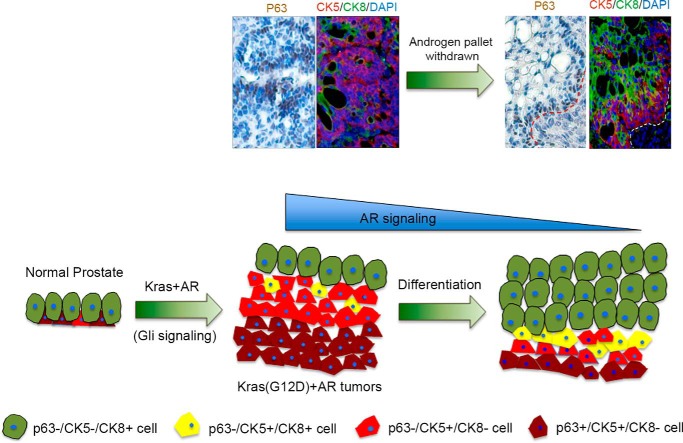
p63 serves as a marker of progenitor/basal cells in prostate tissue (18, 35). Our studies reveal a novel pathway regulating p63 expression and the proliferation of p63-expressing cells through synergy of doxycycline-inducible Kras and AR, two commonly activated oncogenes in prostate cancer (11). The expanded p63+CK5+ progenitor cells located at the basal membrane exhibited differentiation potential and correlated with the number of p63−CK5+ cells. By alteration of androgen levels or Kras activity in Kras(G12D)+AR tumors, the hierarchical lineage distribution pattern implies that p63+CK5+ cells located at the basal membrane differentiate to p63−CK5+ cells, further to CK5+/CK8+ transitional progenitor cells, and finally to CK5−/CK8+ luminal tumorigenic cells (Fig. 7). Our results illustrate a pathological differentiation process of p63+ or CK5+ progenitor/basal cells into CK8+ luminal tumorigenic cells. The data support the notion that basal cells serve as an oncogenic target for the cell of origin in prostate cancer (12, 15) and that oncogenic transformation of basal cells has the potential to differentiate into luminal cells and lead to proliferation of luminal tumorigenic cells during prostate cancer progression (13).
FIGURE 7.
Differentiation of pathological expanded progenitor cells reveals a hierarchical pattern of prostate progenitor and differentiated cells. Shown is a schematic for the response of Kras+AR tumors under reduced AR signaling. The alteration of tumorigenic cells expressing p63 and/or CK5/CK8 before and after withdrawal of exogenous androgen in Kras+AR-mediated tumors is highlighted. The represented figures are the highlight of Fig. 2B, e and h, for Kras+AR tumors grown under an external androgen pellet (before androgen pellet withdrawal) and Fig. 2B, f and i (for tumors after the withdrawal of the external androgen pellet). Synergy of Kras and AR signaling leads to an expansion of basal/progenitor cells mediated by Gli transcription factors. Multiple layers of p63+/CK5+/CK8− progenitor cells localizing at the basal membrane correlate with an elevated number of p63−/CK5+/CK8− cells in Kras+AR tumors. Upon down-regulation of Kras or AR signaling, p63+/CK5+/CK8− and p63−/CK5+/CK8− cells likely undergo differentiation, which leads to elevated numbers of p63−/CK5+/CK8+ transitional progenitor cells and completely differentiated p63−/CK5−/CK8+ tumorigenic cells.
Given the fact that Kras and AR signaling are commonly dysregulated in prostate cancer (3–5, 10, 25), a subset of tumorigenic cells could be maintained by the synergy of these oncogenic events. Currently, multiple drugs have been developed for androgen deprivation treatment (36). The suppression of androgen-AR signaling might promote the differentiation of tumorigenic basal cells in these tumors. As a result, the pathological lesion could be potentially maintained as a drug-resistant mechanism.
Our study demonstrates that Gli signaling facilitates Kras (G12D)+AR-mediated tumorigenesis. Gli signaling is usually activated by the hedgehog-dependent pathway; however, numerous oncogenic stimuli can bypass hedgehog-dependent signaling to induce Gli signaling (23, 37). For example, Kras(G12D)-mediated oncogenesis promotes the transcriptional activity of Gli family members in pancreatic ductal adenocarcinoma (30, 38). Loss of Gli1 interferes with the cross-talk between fibroblast and cancer cells and suppresses Kras-induced preneoplastic lesions (31).
Kras-induced Gli signaling might participate in the synergy with AR to promote p63 expression in Kras+AR tumors. This is further supported by evidence showing that Gli1 or Gli2 is associated with AR through direct protein-protein interactions and can regulate AR-controlled gene expression in prostate cancer (39, 40). Additionally, co-activation of AR with the C-terminal domain of Gli2 regulates androgen-responsive genes in prostate cancer cells (41). Given the fact that p63-expressing basal cells are essential for prostate sphere formation and prostate development (21, 35), future studies of whether and how p63 expression is transcriptionally regulated by the AR-Gli interaction will be helpful for an intervention of prostate cancer progression and relapse.
Gli signaling regulates prostate progenitor cell renewal, likely through modulation of p63 expression. Our study demonstrates that blocking Gli signaling (by overexpression of the dominant negative regulator Gli3T) inhibits p63 expression and pathological proliferation of p63-expressing cells in Kras+AR tumors. Our results support the reciprocal regulation of Gli signaling and p63 expression for maintenance of the epidermal homeostasis of multiple organs, including normal prostate development (32). Interference with the p63 and Gli signaling interaction network leads to dysregulation of differentiation and promotes initiation of carcinogenesis (22, 32, 33).
Our data illustrate different functional roles of Gli1 and Gli2 in mediation of the oncogenic synergy of Kras and AR. Although a decrease in Gli1 expression inhibits Kras (G12D)+AR-induced tumorigenesis, down-regulation of Gli2 suppresses the proliferation of p63+ basal/progenitor cells but not the Kras(G12D)+AR-induced prostatic lesion. This different functional role of Gli1 and Gli2 may be explained by the dissimilarity of the protein structures (37). Both Gli1 and 2 proteins contain the activator domain; however, Gli2, but not Gli1, has the inhibitory domain in the N terminus. This is in line with evidence showing that aberrant expression of Gli1 is involved in the formation of basal cell carcinoma (42, 43). In contrast, expression of Gli2 is regulated by p63, and p63-Gli2 cross-talk facilitates osteosarcoma progression (44). Additionally, mouse genetics show that Gli2 is essential in embryonic development, whereas Gli1 is dispensable (31). The redundant and distinctive functions among Gli family members should be further investigated in the pathological context. It remains to be determined whether Gli2, particularly the transcriptional repressor domain, could directly or indirectly regulate p63 expression in collaboration with AR signaling during tumor progression.
Dysregulation of hedgehog/Gli signaling is associated with numerous types of cancer, including prostate cancer (45). A variety of inhibitors have been developed to target different levels of hedgehog/Gli signaling, including suppression of SMO, interference with the hedgehog/patched interaction, and direct inhibition of Gli signaling. These compounds include SMO inhibitors such as vismodegib and sonidegib, two Food and Drug Administration-approved inhibitors, and others, such as BMS-833923, IPI-926, PF-04449913, and LY2940680, which are currently in clinical trials (46, 47). However, a variety of oncogenic pathways, including Kras, TGF-β, PI3K-AKT, PKC-α, and Kras+AR (this study), induce Gli signaling independent of hedgehog activation (23, 24). Inhibitors targeting SMO or hedgehog-dependent Gli signaling likely exhibit no efficacy in inhibition of non-canonical Gli signaling. Given that only very few Gli inhibitors have been developed, identifying inhibitors directly suppressing Gli signaling will provide an effective therapeutic approach in targeting tumor progression.
Experimental Procedures
Plasmid and Lentiviral Production
A lentiviral vector overexpressing Gli3T, designated FUCRW-Gli3T, was created for in vivo or in vitro experiments. The fragment corresponding to 1–743 amino acids of human Gli3 was PCR-amplified from template DNA, GLI-3 bs-2 (Addgene, catalog no. 16420), using hGli3-F and hGli3–743R primers (supplemental Table S1). The PCR product was inserted into the XbaI site of the FUCRW lentiviral vector. Additionally, multiple lentiviral vectors carrying shRNA were created to target Gli1 and Gli2 expression. Three pairs of shGli1 oligonucleotides and two pairs of shGli2 oligonucleotides were used that contain a target sequence of mouse Gli1 and Gli2, followed by the loop sequence (TTCAAGAGA) and the reverse complement of the targeting sequence (sequence shown in supplemental Table S1). One microgram of both forward and reverse oligonucleotides was annealed in buffer containing 10 mm Tris-HCl, 50 mm KCl, and 1.5 mm MgCl2. The product was purified and ligated into the BbsI site of the psiRNA H1.4 shuttle vector, which is immediately downstream of the H1 promoter and upstream of the heximer A termination sequence. The fragment of H1-shRNA was then digested from the shuttle vector by PacI and subcloned into the PacI site of the FUCRW vector. Because all lentiviral vectors were derived from the FUCRW parental vector, they carry an RFP marker under the CMV promoter. Lentivirus production and infection were performed as described previously (11). All procedures followed the safety guidelines and regulations of the University of Georgia.
Mouse Mating and Genotyping
CCSP-rtTA;Tet-on-Kras4bG12Dmice were generated by mating CCSP-rtTA mice (from the laboratory of D. Spyropoulos) with Tet-on-Kras4bG12D mice (from the Varmus laboratory). The genotype of the CCSP-rtTA;Tet-on-Kras4bG12D mice was genotyped by PCR analysis as described previously (supplemental Fig. S1A) (26). The genotyping primers can be found in supplemental Table 1. Expression of CCSP has been reported in lung, prostate, and ovary tissues based on RNA sequencing analysis (GeneCards Database).
Prostate Regeneration Assay and Secondary Implantation
For the prostate regeneration assay, primary prostate cells were isolated from 8- to 12-week-old CCSP-rtTA;Tet-on-Kras4bG12D male mice and infected with a lentivirus according to the experimental outline described in Figs. 1B, 3A, 4A, and 5A. Infected cells (2–3 × 105 cells/graft) were combined with urogenital sinus mesenchyme (UGSM) (2–3 × 105 cells/graft) together with 25 μl of collagen type I (adjusted to pH 7.0). After overnight incubation, grafts were implanted under the kidney capsule in CB.17SCID/SCID (SCID) mice by survival surgery.
In the experiment described in Fig. 2, Kras(G12D)+AR tumors were minced into small pieces, and 20–30 mg of tumor was implanted subcutaneously into the flanks of a SCID mouse. Additionally, an androgen pellet (testosterone) was also implanted under the back of SCID mice (in the experiment shown in supplemental Fig. S2 and Fig. 2), or no pellet was implanted (physiological level) (in the experiment shown in supplemental Fig. S2). After 4 weeks, testosterone was either removed in one group of mice or retained in the other group (in the experiment shown in Fig. 2). Alternatively, as described in the experiments shown in supplemental Fig. S3, Kras(G12D)+AR tumors were dissociated into single cells. 5 × 105 cells were injected subcutaneously into intact or 1-month-castrated SCID mice. All animals were maintained and used according to the surgical and experimental procedures of the protocol A2013 03-008, which was approved by the Institutional Animal Care and Use Committee of the University of Georgia.
Sphere Formation Assay for Primary Prostate Cells
Dissociated primary prostate cells from Bl6 mice as described above were counted and seeded at 5 × 105 cells/well in a 12-well plate. A lentivirus carrying the control vector or overexpression of Gli3T, shRNA-Gli1, or shRNA-Gli2 was added to dissociated primary prostate cells with a multiplicity of infection of 10–20, respectively. After 2 h of spin infection (at 1500 rpm), lentivirus was removed from each well. The transduced cells were resuspended from the well, washed twice, and finally resuspended in 50 μl of prostate epithelial cell growth medium (Lonza, catalog no. CC-3166). Fifty microliters of cell suspension was mixed with 50 μl of Matrigel and plated around the rims of the wells in a 12-well plate. After the cell-Matrigel mixture solidified at 37 °C for 20 min, 1 ml of PrEGM was added. Cells were cultured for 5 days to allow viral integration and RFP expression. After RFP expression was confirmed, dispase was added to digest the Matrigel matrix. Cell clusters were collected and treated sequentially with collagenase and trypsin as described above. Digested cells were passed through a 22-gauge syringe three times and filtered by a 40-μm cell strainer. Cells were then resuspended in 400 μl of PrEGM medium and sorted for RFP-positive cells by flow cytometry. Fifty microliters of 8 × 103 RFP+ cells in PrEGM were mixed with 50 μl of Matrigel and reseeded in one well of a 12-well plate. The number of spheres was counted after 10 days incubation. For the secondary spheres, primary spheres were dissociated by consecutive treatment of dispase, collagenase, and trypsin as described above, and 12,000 cells/well were seeded. The sphere growth procedures were the same as for the primary sphere assay (34).
Cell Culture, RNA Extraction, Real-time PCR, and Antibodies for Western Blotting Analysis and Immunohistochemistry
UGSM cells or mouse prostate epithelial basal (PEB) cells (a gift from the laboratory of Dr. E. Lynette Wilson) (48) were seeded into a 6-well plate and infected with the virus as indicated in supplemental Figs. S6D and S7B for UGSM cells and Fig. 6, A and B, for PEB cells. Three days after the cells were transduced with control vector or Gli3T, shRNA-Gli, or shRNA-Gli2 by lentiviral infection, the culture medium was removed, and cells were washed with PBS for RNA or protein extraction.
Total RNA was isolated by TRIzol reagent (Life Technologies), following the protocol of the manufacturer. In brief, cells were lysed in the plate by adding 1 ml of TRIzol reagent per well. The cell lysate was resuspended several times and transferred to a 1.5-ml tube. 0.2 ml of chloroform was added to each tube and vortexed for 10 s. After centrifugation at 12,000 rpm for 1 min, the supernatant was transferred into a new 1.5-ml tube, and 600 μl of chloroform was added and vortexed for 10 s. The mixture was centrifuged at 12,000 rpm for 1 min, and the upper aqueous phase was collected. After repeating the chloroform cleaning step, RNA from the aqueous phase was precipitated by adding 600 μl of isopropanol and incubation at −20 °C for 30 min. The mixture was further centrifuged at 12,000 rpm for 15 min. The supernatant was removed, and the RNA pellet was washed with cold 75% ethanol. Total RNA was obtained by removing ethanol, air-dried, and redissolved in 10 μl of nuclease-free water.
Complementary DNA was reverse-transcribed from 1 μg of total RNA in a 20-μl reaction with a high-capacity cDNA reverse transcription kit (Life Technologies). The RT products were diluted with 80 μl of distilled H2O (making the total volume 100 μl), and 2 μl was used for each real-time PCR reaction. The relative quantification in -fold changes in gene expression was obtained by 2-ΔΔCt method with GAPDH as the internal reference gene. Real-time PCR reactions were performed using the PerfeCTa SYBR Green FastMix (Quanta Biosciences). Each reaction contained 10 μl of 2× SYBR Green FastMix, 1 μl of primer pairs (20 ng/μl), 2 μl of cDNA, and 7 μl of distilled water. The thermal cycling conditions were composed of an initial denaturation step at 95 °C for 1 min, 40 cycles at 95 °C for 10 s, and 60 °C for 50 s. The experiments were carried out in triplicate.
For Western blotting analysis, cells were lysed with radioimmune precipitation assay buffer containing standard protease inhibitors. For immunohistochemistry, formalin-fixed/paraffin-embedded specimens were sectioned at 4-μm thickness and mounted on positively charged slides. Sections were stained with H&E, and IHC analysis as described previously (11, 29, 49). The following antibodies were used for Western blotting analysis and immunohistochemistry: Gli3 (Abcam, Ab69838, 1:1000), Actin (Abcam, Ab49900, 1:5000), Erk (Santa Cruz Biotechnology, SC-154, 1:5000), p63 (Santa Cruz Biotechnology, 8431, 1:250), AR (Santa Cruz Biotechnology, SC-816, 1:200), CK5 (BioLegend, 905501, 1:500), CK8 (BioLegend, 904801, 1:1000), Gli1 (Cell Signaling Technology, 2534, 1:500), Gli2 (Abcam, ab26056, 1:500), CK14 (Abcam, ab7800, 1:250), and CK18 (Abcam, ab181597, 1:800).
ChIP Assay
Briefly, 5 × 106 PEB cells were cross-linked with 1% formaldehyde, followed by cell lysis. DNA was sheared using a Bioruptor 300 (Diagenode, Denville, NJ) to fragment DNA to ∼600 bp. Aliquots of the sheared chromatin were then immunoprecipitated using magnetic beads and Gli1 (Novus Biologicals, NB600-600), Gli2 (Novus Biologicals, NB600-874), or Gli3 (Santa Cruz Biotechnology, sc-20688 X) antibodies or a normal rabbit IgG separately. Following immunoprecipitation, cross-links were removed, and immunoprecipitated DNA was purified using spin columns and subsequently amplified by quantitative PCR. The following quantitative PCR primers were designed to amplify regions of the ΔNp63 promoter containing a potential Gli binding site: sense, GCCTTTGAGATGCCTCTGT; antisense, TCCAGCTACACAGCAGAAAC. Quantitative SYBR PCR was performed in triplicate using the C1000 thermal cycler (Bio-Rad). Results are represented as percentage of input.
Statistical Analysis
Prism software was used to carry out a statistical analysis. The data were presented as mean ± S.E. and analyzed by Student's t test. All t tests were performed at the two-sided 0.05 level for significance.
Author Contributions
M. W. and H. C. designed the experiments and wrote the manuscript. M. W. performed the majority of the experiments. L. I., Q.L., and S. K. helped with immunohistochemistry, lentiviral vectors, and preparation of some lentiviruses in some experiments. D. D. S. initiated and provided prostate tissue of the doxycycline-regulated Kras mouse colony. Z. B. participated in writing the manuscript. E. J. T., R. E. V., and M. E. F. Z. conducted the ChIP assay, provided prostate tissue of transgenic mice, and participated in manuscript preparation. Y. M. performed real time PCR and J. H. performed pathological analysis.
Supplementary Material
This work was supported by National Institutes of Health Grants R01CA172495 (to H. C.) and CA136526 (to M. E. F. Z.) and DOD Grant W81XWH-15-1-0507 (to H. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S9 and Table S1.
- PIN
- prostatic intraepithelial neoplasia
- CK
- cytokeratin
- AR
- androgen receptor
- CCSP
- Clara cell secretory protein
- SCID
- severe combined immunodeficiency
- IHC
- immunohistochemistry
- UGSM
- urogenital sinus mesenchyme
- PEB
- prostate epithelial basal
- Dox
- doxycycline
- RFP
- red fluorescent protein
- SMO
- smoothened.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2015) Cancer statistics, 2015. CA-Cancer J. Clin. 65, 5–29 [DOI] [PubMed] [Google Scholar]
- 2. Abate-Shen C., and Shen M. M. (2000) Molecular genetics of prostate cancer. Genes Dev. 14, 2410–2434 [DOI] [PubMed] [Google Scholar]
- 3. Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., et al. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X. S., Shankar S., Dhanasekaran S. M., Ateeq B., Sasaki A. T., Jing X., Robinson D., Cao Q., Prensner J. R., Yocum A. K., Wang R., Fries D. F., Han B., Asangani I. A., Cao X., et al. (2011) Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 1, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palanisamy N., Ateeq B., Kalyana-Sundaram S., Pflueger D., Ramnarayanan K., Shankar S., Han B., Cao Q., Cao X., Suleman K., Kumar-Sinha C., Dhanasekaran S. M., Chen Y. B., Esgueva R., Banerjee S., et al. (2010) Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat. Med. 16, 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Konishi N., Enomoto T., Buzard G., Ohshima M., Ward J. M., and Rice J. M. (1992) K-ras activation and ras p21 expression in latent prostatic carcinoma in Japanese men. Cancer 69, 2293–2299 [DOI] [PubMed] [Google Scholar]
- 7. Salmaninejad A., Ghadami S., Dizaji M. Z., Golchehre Z., Estiar M. A., Zamani M. R., Ebrahimzadeh-Vesal R., Nowroozi M. R., and Shakoori A. (2015) Molecular characterization of KRAS, BRAF, and EGFR genes in cases with prostatic adenocarcinoma: reporting bioinformatics description and recurrent mutations. Clin. Lab. 61, 749–759 [DOI] [PubMed] [Google Scholar]
- 8. Ngalame N. N., Tokar E. J., Person R. J., and Waalkes M. P. (2014) Silencing KRAS overexpression in arsenic-transformed prostate epithelial and stem cells partially mitigates malignant phenotype. Toxicol. Sci. 142, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ngalame N. N., Waalkes M. P., and Tokar E. J. (2016) Silencing KRAS overexpression in cadmium-transformed prostate epithelial cells mitigates malignant phenotype. Chem. Res. Toxicol. 29, 1458–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weber M. J., and Gioeli D. (2004) Ras signaling in prostate cancer progression. J. Cell. Biochem. 91, 13–25 [DOI] [PubMed] [Google Scholar]
- 11. Cai H., Memarzadeh S., Stoyanova T., Beharry Z., Kraft A. S., and Witte O. N. (2012) Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer. Cancer Res. 72, 4672–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawson D. A., Zong Y., Memarzadeh S., Xin L., Huang J., and Witte O. N. (2010) Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 2610–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi N., Zhang B., Zhang L., Ittmann M., and Xin L. (2012) Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z. A., Mitrofanova A., Bergren S. K., Abate-Shen C., Cardiff R. D., Califano A., and Shen M. M. (2013) Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., Kruithof-de Julio M., Economides K. D., Walker D., Yu H., Halili M. V., Hu Y. P., Price S. M., Abate-Shen C., and Shen M. M. (2009) A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Signoretti S., Waltregny D., Dilks J., Isaac B., Lin D., Garraway L., Yang A., Montironi R., McKeon F., and Loda M. (2000) p63 is a prostate basal cell marker and is required for prostate development. Am. J. Pathol. 157, 1769–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Signoretti S., Pires M. M., Lindauer M., Horner J. W., Grisanzio C., Dhar S., Majumder P., McKeon F., Kantoff P. W., Sellers W. R., and Loda M. (2005) p63 regulates commitment to the prostate cell lineage. Proc. Natl. Acad. Sci. U.S.A. 102, 11355–11360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurita T., Medina R. T., Mills A. A., and Cunha G. R. (2004) Role of p63 and basal cells in the prostate. Development 131, 4955–4964 [DOI] [PubMed] [Google Scholar]
- 19. Romano R. A., Smalley K., Magraw C., Serna V. A., Kurita T., Raghavan S., and Sinha S. (2012) ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139, 772–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suh E. K., Yang A., Kettenbach A., Bamberger C., Michaelis A. H., Zhu Z., Elvin J. A., Bronson R. T., Crum C. P., and McKeon F. (2006) p63 protects the female germ line during meiotic arrest. Nature 444, 624–628 [DOI] [PubMed] [Google Scholar]
- 21. Pignon J. C., Grisanzio C., Geng Y., Song J., Shivdasani R. A., and Signoretti S. (2013) p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. U.S.A. 110, 8105–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caserta T. M., Kommagani R., Yuan Z., Robbins D. J., Mercer C. A., and Kadakia M. P. (2006) p63 overexpression induces the expression of Sonic Hedgehog. Mol. Cancer Res. 4, 759–768 [DOI] [PubMed] [Google Scholar]
- 23. Aberger F., Kern D., Greil R., and Hartmann T. N. (2012) Canonical and noncanonical Hedgehog/GLI signaling in hematological malignancies. Vitam. Horm. 88, 25–54 [DOI] [PubMed] [Google Scholar]
- 24. Fernández-Zapico M. E. (2008) Primers on molecular pathways GLI: more than just Hedgehog? Pancreatology 8, 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y., Sawyers C. L., and Scher H. I. (2008) Targeting the androgen receptor pathway in prostate cancer. Curr. Opin. Pharmacol. 8, 440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fisher G. H., Wellen S. L., Klimstra D., Lenczowski J. M., Tichelaar J. W., Lizak M. J., Whitsett J. A., Koretsky A., and Varmus H. E. (2001) Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15, 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng D., Yin L., and Chen J. (2014) Evidence for Scgb1a1+ cells in the generation of p63+ cells in the damaged lung parenchyma. Am. J. Respir. Cell Mol. Biol. 50, 595–604 [DOI] [PubMed] [Google Scholar]
- 28. Cai H., Babic I., Wei X., Huang J., and Witte O. N. (2011) Invasive prostate carcinoma driven by c-Src and androgen receptor synergy. Cancer Res. 71, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xin L., Teitell M. A., Lawson D. A., Kwon A., Mellinghoff I. K., and Witte O. N. (2006) Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 7789–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajurkar M., De Jesus-Monge W. E., Driscoll D. R., Appleman V. A., Huang H., Cotton J. L., Klimstra D. S., Zhu L. J., Simin K., Xu L., McMahon A. P., Lewis B. C., and Mao J. (2012) The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 109, E1038–E1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mills L. D., Zhang Y., Marler R. J., Herreros-Villanueva M., Zhang L., Almada L. L., Couch F., Wetmore C., Pasca di Magliano M., and Fernandez-Zapico M. E. (2013) Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J. Biol. Chem. 288, 11786–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chari N. S., Romano R. A., Koster M. I., Jaks V., Roop D., Flores E. R., Teglund S., Sinha S., Gruber W., Aberger F., Medeiros L. J., Toftgard R., and McDonnell T. J. (2013) Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ. 20, 1080–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Memmi E. M., Sanarico A. G., Giacobbe A., Peschiaroli A., Frezza V., Cicalese A., Pisati F., Tosoni D., Zhou H., Tonon G., Antonov A., Melino G., Pelicci P. G., and Bernassola F. (2015) p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 3499–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lukacs R. U., Goldstein A. S., Lawson D. A., Cheng D., and Witte O. N. (2010) Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 5, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Y., Hamana T., Liu J., Wang C., An L., You P., Chang J. Y., Xu J., McKeehan W. L., and Wang F. (2015) Prostate sphere-forming stem cells are derived from the P63-expressing basal compartment. J. Biol. Chem. 290, 17745–17752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Graham L., and Schweizer M. T. (2016) Targeting persistent androgen receptor signaling in castration-resistant prostate cancer. Med. Oncol. 33, 44. [DOI] [PubMed] [Google Scholar]
- 37. Hui C. C., and Angers S. (2011) Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 [DOI] [PubMed] [Google Scholar]
- 38. Ji Z., Mei F. C., Xie J., and Cheng X. (2007) Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 282, 14048–14055 [DOI] [PubMed] [Google Scholar]
- 39. Chen G., Goto Y., Sakamoto R., Tanaka K., Matsubara E., Nakamura M., Zheng H., Lu J., Takayanagi R., and Nomura M. (2011) GLI1, a crucial mediator of sonic hedgehog signaling in prostate cancer, functions as a negative modulator for androgen receptor. Biochem. Biophys. Res. Commun. 404, 809–815 [DOI] [PubMed] [Google Scholar]
- 40. Chen M., Feuerstein M. A., Levina E., Baghel P. S., Carkner R. D., Tanner M. J., Shtutman M., Vacherot F., Terry S., de la Taille A., and Buttyan R. (2010) Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Mol. Cancer 9, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li N., Chen M., Truong S., Yan C., and Buttyan R. (2014) Determinants of Gli2 co-activation of wildtype and naturally truncated androgen receptors. Prostate 74, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 42. Dahmane N., Lee J., Robins P., Heller P., and Ruiz i and Altaba A. (1997) Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 389, 876–881 [DOI] [PubMed] [Google Scholar]
- 43. Wang X. Q., and Rothnagel J. A. (2001) Post-transcriptional regulation of the gli1 oncogene by the expression of alternative 5′ untranslated regions. J. Biol. Chem. 276, 1311–1316 [DOI] [PubMed] [Google Scholar]
- 44. Ram Kumar R. M., Betz M. M., Robl B., Born W., and Fuchs B. (2014) ΔNp63α enhances the oncogenic phenotype of osteosarcoma cells by inducing the expression of GLI2. BMC Cancer 14, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shigemura K., and Fujisawa M. (2015) Hedgehog signaling and urological cancers. Curr. Drug Targets 16, 258–271 [DOI] [PubMed] [Google Scholar]
- 46. Pandolfi S., and Stecca B. (2015) Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways: implications for cancer therapy. Expert Rev. Mol. Med. 17, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rimkus T. K., Carpenter R. L., Qasem S., Chan M., and Lo H. W. (2016) Targeting the Sonic Hedgehog signaling pathway: review of Smoothened and GLI inhibitors. Cancers 8, 22–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salm S. N., Takao T., Tsujimura A., Coetzee S., Moscatelli D., and Wilson E. L. (2002) Differentiation and stromal-induced growth promotion of murine prostatic tumors. Prostate 51, 175–188 [DOI] [PubMed] [Google Scholar]
- 49. Xin L., Ide H., Kim Y., Dubey P., and Witte O. N. (2003) In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc. Natl. Acad. Sci. U.S.A. 100, 11896–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.