Abstract
By phosphorylating specific replication factors, cell cycle kinases ensure that eukaryotic DNA replication is initiated once and only once per mitotic cell division. New work in The EMBO Journal now reveals how DDK‐mediated phosphorylation of Mcm2‐7 helicase subunits is read out by Sld3, which provides further integration with CDK phosphorylation.
Subject Categories: DNA Replication, Repair & Recombination
Eukaryotic DNA replication is strictly coupled to cell cycle regulation, with the initiation step sensing and integrating signals from the cell cycle machinery. Replication initiates from so‐called origin sequences on chromosomal DNA, which in budding yeast are bound by origin recognition complex (ORC) proteins throughout the cell cycle. In the G1 phase, hexameric Mcm2‐7 helicase core complexes, which on their own do not yet display helicase activity, load onto ORC‐bound origins to form pre‐replicative complexes (pre‐RCs). After activation of the master cell cycle protein kinase CDK (cyclin‐dependent kinase) in late G1, budding yeast CDK phosphorylates two key replication factors, Sld2 and Sld3. This phosphorylation enhances interactions between Sld2, Sld3 and Dpb11, which in turn promotes the formation of active replicative DNA helicase, Cdc45‐Mcm2‐7‐GINS (CMG), on replication origins. A second protein kinase, Dbf4‐dependent kinase (DDK), is also activated at the G1/S transition and involved in assembly of the CMG helicase. DDK phosphorylates the Mcm2‐7 helicase core complex and promotes the loading of Sld3 and Cdc45 (Fig 1), but their mode of recruitment or how they recognize DDK‐mediated marks had remained unknown. A new study from John Diffley's laboratory (Deegan et al, 2016) now highlights the important role of Sld3 in this process.
Figure 1. Schematic of DDK‐dependent Sld3/7‐Cdc45 recruitment to Mcm2‐7 helicase double‐hexamers, followed by CDK‐dependent GINS recruitment for the formation of the active replicative helicase complex.
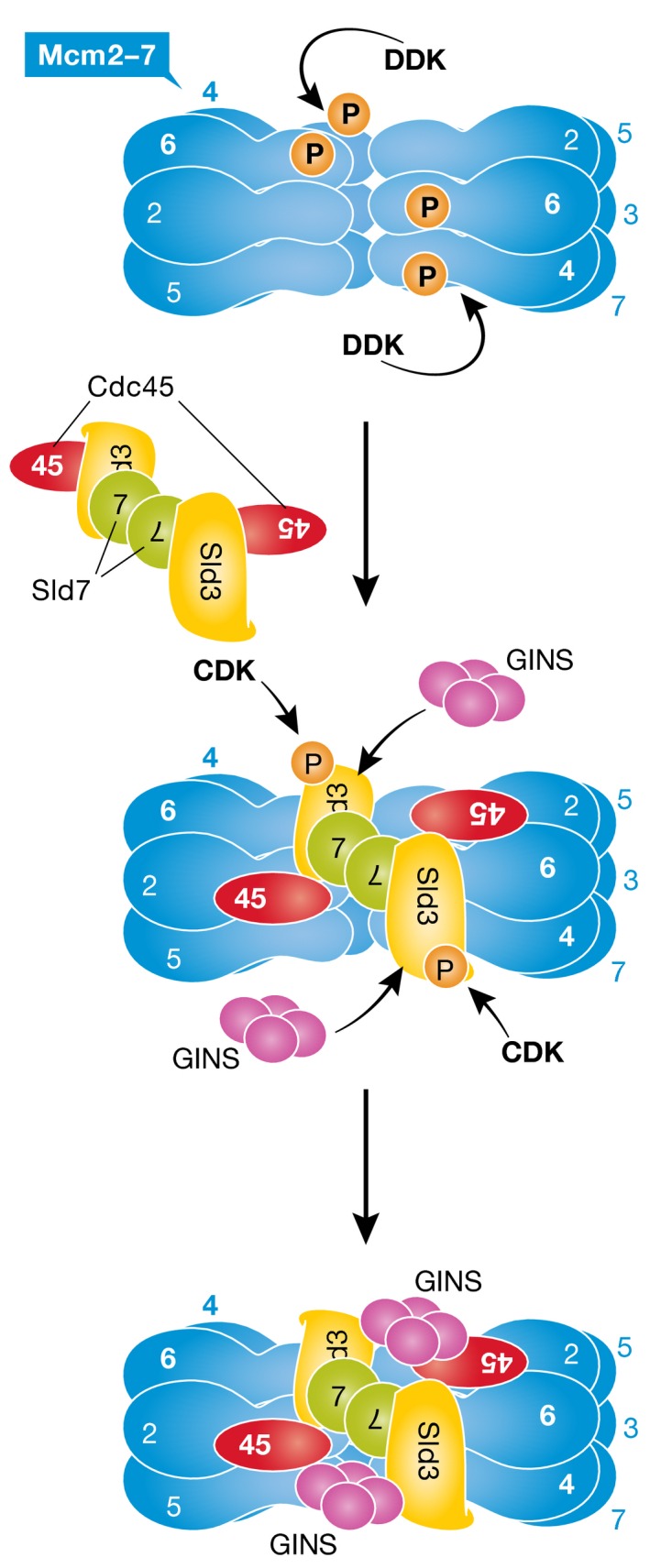
See text for details.
Sld3 (synthetically lethal with dpb11‐1) was first identified as a replication protein by yeast genetic screenings using a Dpb11 mutation (Kamimura et al, 2001). Its metazoan counterpart, Treslin/Ticrr, was later found as an interactor of the Dpb11 ortholog TopBP1 (Kumagai et al, 2010; Sansam et al, 2010). The Sld3 protein can be functionally divided into three parts: an N‐terminal domain interacting with Sld7 (Tanaka et al, 2011; Itou et al, 2015), a middle Cdc45 binding domain (CDB) (Itou et al, 2014) and a C‐terminal unstructured region containing CDK phosphorylation sites mediating CDK‐dependent binding to Dpb11. In Treslin/Ticrr, the N‐terminal moiety has similar characteristics as in Sld3 and mediates interaction with MTBP (MDM two binding protein), which has also been found to be essential for DNA replication (Boos et al, 2013); the middle region (called STD: Sld3‐Treslin Domain) is sequence‐related to the middle part of Sld3 and likely mediates Cdc45 binding (Sanchez‐Pulido et al, 2010); the C‐terminal portion again contains two well‐conserved CDK phosphorylation sites essential for binding to TopBP1 (Boos et al, 2011; Kumagai et al, 2011), with the remainder of it being dispensable for DNA replication.
In a recent breakthrough study, the Diffley group had succeeded in setting up an in vitro replication system reconstituted from purified yeast proteins (Yeeles et al, 2015). Here, they further extended this in vitro assay system for protein assembly on replication origins from purified yeast proteins and demonstrated that DDK‐dependent association of Sld3 and Cdc45 with pre‐RCs can be recapitulated even in vitro. Since DDK efficiently phosphorylated origin‐loaded Mcm2‐7 complexes, such complexes could now be disassembled and tested for Sld3 interaction in binding assays. These analyses revealed that phosphorylated Mcm4 and Mcm6 strongly bound to Sld3. Mcm2, Mcm4 and Mcm6 bear N‐terminal extension that has been shown to be extensively phosphorylated by DDK. Furthermore, Deegan et al (2016) found that several synthetic Mcm4 and Mcm6 phosphopeptides with possible DDK‐targeted sites also bind to Sld3 in vitro.
Deegan et al (2016) mapped the Mcm2‐7 binding site in Sld3 to its middle part, which also binds to Cdc45, and constructed specific mutations affecting either of these interactions. An unstructured loop in STD/CDB is known to be important for binding to Cdc45 (Itou et al, 2014), while basic amino acid residues after the STD/CDB were required for Mcm2‐7 binding. Substitution of conserved basic residues with acidic residues in either region selectively reduced the binding to Cdc45 and Mcm2‐7, respectively. This revealed that while Sld3 and Cdc45 are mutually dependent for loading onto phosphorylated Mcm2‐7 in vivo (Kamimura et al, 2001), Sld3 can be specifically recruited to Mcm2‐7 even in the absence of Cdc45 in vitro. Moreover, mutant Mcm4 and Mcm6 proteins with site‐specific substitutions mimicking DDK phosphorylation were able to bypass the requirement for Cdc7, the catalytic subunit of DDK. In fact, such mutants support Sld3 association with Mcm2‐7 in the absence of DDK phosphorylation in vitro. In an additional set of experiments, the authors could demonstrate that DDK‐dependent phosphorylation is only required at the step of replication initiation, but not for subsequent elongation. Stalling plasmid replication after initiation by omitting DNA topoisomerase from the in vitro replication system (Yeeles et al, 2015), they found that Mcm2‐7 dephosphorylation at this stage did not affect DNA replication efficiency once elongation was resumed by topoisomerase addition (Deegan et al, 2016). Finally, the Sld3 middle region is also phosphorylated by the replication checkpoint protein kinase Rad53, and the authors showed that this inhibits association with Mcm2‐7 in addition to Dpb11 and Cdc45 binding.
The new results by Deegan et al (2016) paper clearly demonstrate that Sld3 recognizes mainly DDK‐phosphorylated peptides found on Mcm4 and Mcm6. The three‐dimensional structure of the Sld3 phosphopeptide‐binding region remains to be determined, but it is likely that part of the STD/CDB contributes to the interaction as well, and we can probably expect interesting new structural inferences given the lack of resemblance to already known phosphopeptide‐binding domains. Moreover, because Sld3 binds to both Mcm4 and Mcm6, and two Sld3 molecules are connected by Sld7 in an anti‐parallel orientation (Itou et al, 2015), recruitment of Cdc45 and GINS to double‐hexameric Mcm2‐7 complexes may occur in trans (Fig 1). Such a putative mechanism may secure simultaneous activation of both Mcm2‐7 hexamers, to start DNA synthesis in both directions.
See also: TD Deegan et al (May 2016)
References
- Boos D, Sanchez‐Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF (2011) Regulation of DNA replication through Sld3‐Dpb11 interaction is conserved from yeast to humans. Curr Biol 21: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JFX (2013) Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340: 981–984 [DOI] [PubMed] [Google Scholar]
- Deegan TD, Yeeles JTP, Diffley JFX (2016) Phosphopeptide binding by Sld3 links Dbf4‐dependent kinase to MCM replicative helicase activation. EMBO J 35: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou H, Muramatsu S, Shirakihara Y, Araki H (2014) Crystal structure of the homology domain of the eukaryotic DNA replication proteins Sld3/Treslin. Structure 22: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Itou H, Shirakihara Y, Araki H (2015) The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr D Biol Crystallogr 71: 1649–1656 [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H (2001) Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae . EMBO J 20: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG (2010) Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2011) Direct regulation of Treslin by cyclin‐dependent kinase is essential for the onset of DNA replication. J Cell Biol 193: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Pulido L, Diffley JF, Ponting CP (2010) Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol 20: R509–R510 [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA (2010) A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 24: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Umemori T, Endo S, Muramatsu S, Kanemaki M, Kamimura Y, Obuse C, Araki H (2011) Sld7, an Sld3‐associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J 30: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]


