Abstract
Nanos proteins repress the expression of target mRNAs by recruiting effector complexes through non‐conserved N‐terminal regions. In vertebrates, Nanos proteins interact with the NOT1 subunit of the CCR4–NOT effector complex through a NOT1 interacting motif (NIM), which is absent in Nanos orthologs from several invertebrate species. Therefore, it has remained unclear whether the Nanos repressive mechanism is conserved and whether it also involves direct interactions with the CCR4–NOT deadenylase complex in invertebrates. Here, we identify an effector domain (NED) that is necessary for the Drosophila melanogaster (Dm) Nanos to repress mRNA targets. The NED recruits the CCR4–NOT complex through multiple and redundant binding sites, including a central region that interacts with the NOT module, which comprises the C‐terminal domains of NOT1–3. The crystal structure of the NED central region bound to the NOT module reveals an unanticipated bipartite binding interface that contacts NOT1 and NOT3 and is distinct from the NIM of vertebrate Nanos. Thus, despite the absence of sequence conservation, the N‐terminal regions of Nanos proteins recruit CCR4–NOT to assemble analogous repressive complexes.
Keywords: deadenylation, decapping, mRNA decay, translational repression
Subject Categories: RNA Biology, Structural Biology
Introduction
Post‐transcriptional mRNA regulation plays an essential role in embryonic development. This regulation is mediated by RNA‐binding proteins that control the spatial and temporal expression of target mRNAs through the recruitment of effector complexes (Barckmann & Simonelig, 2013). The RNA‐binding proteins of the Nanos family are conserved post‐transcriptional mRNA regulators that play essential roles in embryonic germline specification, germline stem cell maintenance, and neuronal homeostasis in Drosophila melanogaster (Dm) and a wide range of other metazoans (Jaruzelska et al, 2003; Tsuda et al, 2003; Baines, 2005; Lai & King, 2013). The Dm Nanos protein is also required for posterior pattern formation in the embryo (Lehmann & Nüsslein‐Volhard, 1991).
Three Nanos paralogs (Nanos1–3) exist in vertebrates and various invertebrate species, whereas there is only one family member in Dm and other insects (Subramaniam & Seydoux, 1999; Mochizuki et al, 2000; Jaruzelska et al, 2003; Tsuda et al, 2003). This protein family is defined by a highly conserved CCHC‐type zinc‐finger (ZnF) domain (Curtis et al, 1997; Hashimoto et al, 2010) and divergent N‐ and C‐terminal unstructured regions of variable lengths (Fig 1A). The ZnF domain mediates binding to RNA and to Pumilio, a conserved Nanos partner that confers mRNA target specificity (Murata & Wharton, 1995; Curtis et al, 1997; Wreden et al, 1997; Asaoka‐Taguchi et al, 1999; Sonoda & Wharton, 1999; Jaruzelska et al, 2003). The unstructured regions are required for interaction with effector complexes (Verrotti & Wharton, 2000; Ginter‐Matuszewska et al, 2011), which include the CCR4–NOT deadenylase complex (Kadyrova et al, 2007; Suzuki et al, 2010, 2012; Joly et al, 2013; Bhandari et al, 2014).
Figure 1. Dm Nanos represses translation and promotes mRNA degradation.
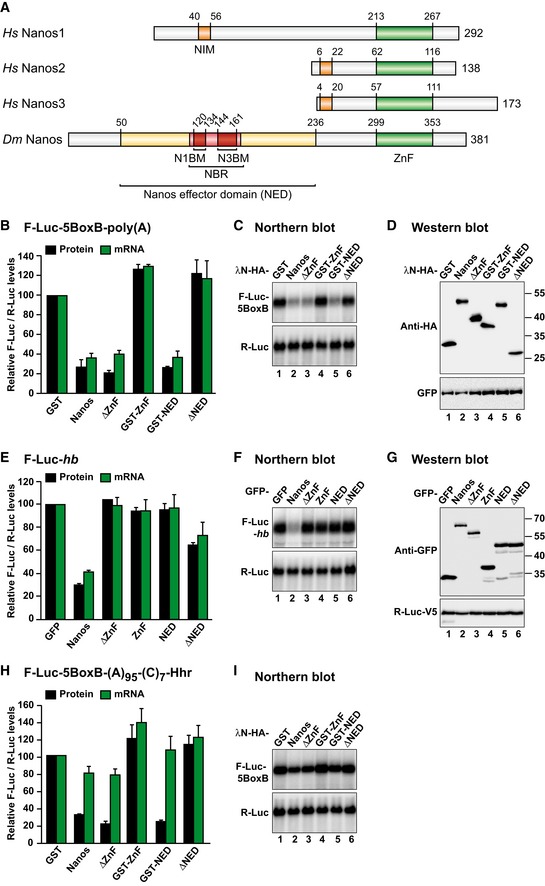
- Nanos comprises a highly conserved zinc‐finger RNA binding domain (ZnF) and non‐conserved N‐terminal and C‐terminal extensions (gray). NIM, NOT1‐interacting motif; NED, Nanos effector domain; NBR, NOT module binding region; N1BM and N3BM, NOT1 and NOT3 binding motifs, respectively. Numbers above the bars indicate residues at domain/motif boundaries.
- Tethering assay using the F‐Luc‐5BoxB reporter and the indicated λN‐HA‐tagged proteins in S2 cells. A plasmid expressing R‐Luc served as a transfection control. The F‐Luc activity (black bars) and mRNA levels (green bars) were normalized to those of the R‐Luc transfection control and set to 100 in the presence of the λN‐HA‐GST. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (B).
- Western blot analysis of the expression of the λN‐HA‐tagged proteins used in the experiments shown in (B) and (C). GFP served as a transfection control.
- GFP‐tagged Nanos fragments were tested for their ability to repress an F‐Luc reporter containing the hb 3′ UTR. A plasmid expressing GFP served as a negative control. F‐Luc activity (black bars) and mRNA levels (green bars) were normalized to those of an R‐Luc transfection control and analyzed as described in (B). The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (E).
- Western blot showing the expression of the GFP‐tagged proteins used in (E) and (F). R‐Luc‐V5 served as a transfection control.
- Tethering assay using the F‐Luc‐5BoxB‐A95‐C7‐Hhr reporter and the indicated λN‐HA‐tagged proteins in S2 cells. F‐Luc activity (black bars) and mRNA levels (green bars) were analyzed as described in (B). The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (H).
Source data are available online for this figure.
The CCR4–NOT complex catalyzes the removal of mRNA poly(A) tails and consequently represses translation. In addition, dead‐enylation by the CCR4–NOT complex is coupled to decapping and 5′‐to‐3′ exonucleolytic degradation by XRN1 and can therefore lead to full mRNA degradation in some cellular contexts (Temme et al, 2014). Furthermore, the CCR4–NOT complex can also repress translation independently of deadenylation (Cooke et al, 2010; Chekulaeva et al, 2011; Bawankar et al, 2013; Zekri et al, 2013).
The CCR4–NOT complex consists of several independent modules that dock with NOT1, a central scaffold subunit (Temme et al, 2014). NOT1 consists of independently folded α‐helical domains that provide binding sites for the individual modules (Fig EV1A). A central domain of NOT1, structurally related to the middle domain of eIF4G (the NOT1 MIF4G domain), provides a binding site for the catalytic module, which comprises two deadenylases, namely CAF1 (or its paralog POP2) and CCR4a (or its paralog CCR4b).
Figure EV1. Dm Nanos interacts with subunits of the deadenylase complexes.
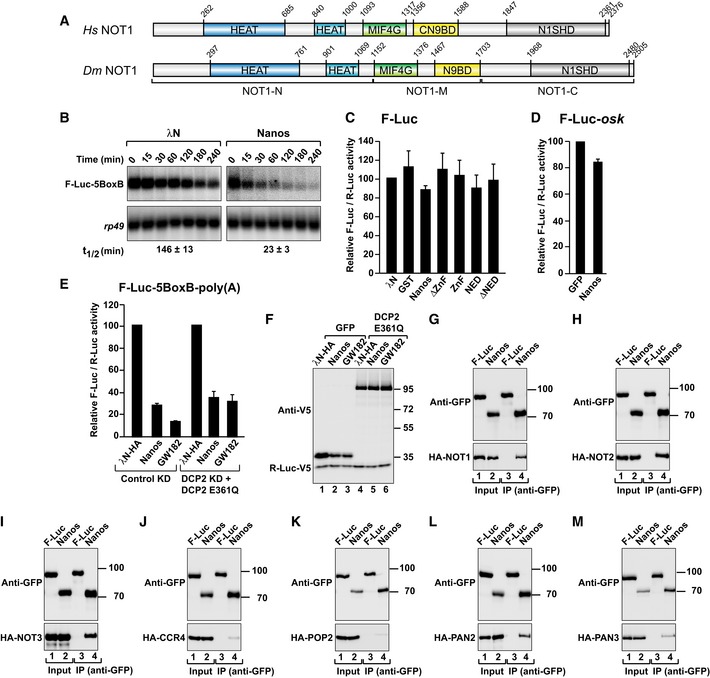
-
ADomain organization of Hs and Dm NOT1. NOT1 consists of N‐terminal (NOT1‐N), middle (NOT1‐M), and C‐terminal regions (NOT1‐C). NOT1‐N contains two HEAT repeat domains (dark and light blue); NOT1‐M contains a MIF4G domain that also consists of HEAT repeats and a three‐helix bundle domain (CN9BD). NOT1‐C contains another HEAT repeat domain, the NOT1 superfamily homology domain (SHD).
-
BNorthern blot analysis showing the decay of the F‐Luc‐5BoxB mRNA in S2 cells expressing the indicated proteins. The mRNA half‐lives (t1/2) ± standard deviations calculated from the decay curves of three independent experiments are indicated below the panels.
-
CTethering assay corresponding to the experiment described in Fig 1B but using an F‐Luc reporter that lacks the BoxB hairpins. F‐Luc activity was normalized to R‐Luc and set to 100 in cells expressing λN‐HA. The panel shows mean values ± standard deviations from three independent experiments.
-
DGFP‐tagged Nanos was coexpressed with an F‐Luc reporter containing the oskar 3′ UTR. R‐Luc served as a transfection control. F‐Luc activity was normalized to that of the R‐Luc transfection control and set to 100 in cells expressing GFP. The panel shows mean values ± standard deviations from three independent experiments.
-
ENormalized luciferase activities corresponding to the experiment shown in Fig 3A and B.
-
FWestern blot analysis showing the expression of the DCP2 mutant (DCP2 E361Q) in the experiment described in Fig 3A and B. R‐Luc‐V5 served as a transfection control.
-
G–MWestern blot analysis showing the interaction of GFP‐tagged Dm Nanos (full length) with HA‐tagged deadenylase subunits. GFP‐tagged firefly luciferase (F‐Luc) served as a negative control. Proteins were immunoprecipitated using a polyclonal anti‐GFP antibody. Inputs and immunoprecipitates were analyzed by Western blotting using anti‐GFP and anti‐HA antibodies. For the GFP‐tagged proteins, 3% of the inputs and 10% of the immunoprecipitates were loaded, whereas for the HA‐tagged proteins, 1% of the input and 30% of the immunoprecipitates were analyzed. In each panel, cell lysates were treated with RNase A prior to immunoprecipitation.
Source data are available online for this figure.
The C‐terminal region of NOT1 contains the NOT1 superfamily homology domain (SHD; Fig EV1A) and assembles with NOT2–NOT3 heterodimers to form the NOT module (Bhaskar et al, 2013; Boland et al, 2013). The NOT module provides binding sites for RNA‐binding proteins, such as vertebrate Nanos and Dm Bicaudal‐C, which recruit the CCR4–NOT complex to their mRNA targets (Chicoine et al, 2007; Suzuki et al, 2012; Bhandari et al, 2014).
The three vertebrate Nanos paralogs contain a 17‐amino acid NOT1‐interacting motif (NIM) that binds directly to the NOT1 SHD domain (Suzuki et al, 2012; Bhandari et al, 2014). Although the NIM is conserved in vertebrate Nanos, Nanos proteins of insects and worms do not have a detectable NIM (Lai et al, 2011; Suzuki et al, 2012; Bhandari et al, 2014). Nevertheless, Dm Nanos has been reported to interact with NOT4 through its unstructured N‐terminus (Kadyrova et al, 2007). However, because NOT4 is not stably associated with the CCR4–NOT complex in metazoans (Lau et al, 2009; Temme et al, 2010), it has remained unclear whether Dm Nanos recruits the CCR4–NOT complex to mRNA targets directly or rather relies on its interaction with additional partners, such as Pumilio (PUM) and Brain tumor (BRAT), to exert its repressive function (Wreden et al, 1997; Sonoda & Wharton, 1999, 2001).
In this study, we show that although Dm Nanos does not contain a NIM, it interacts directly with the CCR4–NOT complex using an extended region that we termed the Nanos effector domain (NED). The NED overlaps with a region previously shown to contribute to Nanos function in Dm embryos (Curtis et al, 1997; Arrizabalaga & Lehmann, 1999). The crystal structure of a central region of the NED (termed the NOT module binding region, NBR) bound to the NOT module revealed a bipartite interface that contacts both the NOT1 SHD and the NOT3 NOT‐box domains. The binding site for the Dm NBR on NOT1 does not overlap with the vertebrate NIM‐binding site. These results indicate that Nanos proteins have maintained the ability to interact with the CCR4–NOT complex using divergent motifs in disordered protein regions.
Results
Identification of the Dm Nanos effector domain (NED)
To investigate whether Dm Nanos possesses intrinsic mRNA repressive activity, we used a λN‐based tethering assay in Dm S2 cells (Behm‐Ansmant et al, 2006). This assay enabled the study of Nanos function independently of RNA‐binding activity. Tethered Dm Nanos caused fourfold repression of a firefly luciferase (F‐Luc) reporter containing five binding sites (Box B hairpins) for the λN‐tag that were inserted in the 3′ UTR (Fig 1B and C). The reduction in F‐Luc activity was predominantly explained by a corresponding decrease in the mRNA abundance (Fig 1B and C) and a shortening of the mRNA half‐life (Fig EV1B), indicating that Nanos induces mRNA degradation in S2 cells. Nanos did not affect the expression of an F‐Luc reporter lacking the BoxB hairpins (Fig EV1C); thus, Nanos must bind to the mRNA to induce degradation.
To define the sequences in Dm Nanos required for the repressive activity, we designed a series of deletion mutants. A Nanos protein lacking the ZnF domain retained the activity of the full‐length protein in the tethering assay, whereas the isolated ZnF domain was inactive (Fig 1A–C), as shown previously for vertebrate Nanos (Lai et al, 2011; Bhandari et al, 2014). Further analyses indicated that a region comprising residues 50–236 retained full repressive activity (Fig 1B and C) and was therefore termed the Nanos effector domain (NED). Conversely, deletion of residues 50–236 (ΔNED) abolished the ability of Nanos to repress the expression of the F‐Luc‐5BoxB reporter (Fig 1B and C). All protein fragments tested were expressed at comparable levels (Fig 1D), and none of them affected the expression of an F‐Luc reporter lacking the BoxB hairpins (Fig EV1C).
The results of the tethering assay were confirmed using a reporter containing the F‐Luc ORF fused to the 3′ UTR of the hunchback (hb) mRNA, a known Nanos target (Wang & Lehmann, 1991; Wreden et al, 1997; Sonoda & Wharton, 1999). Nanos degraded this mRNA (Fig 1E–G) but did not significantly affect the expression of an F‐Luc reporter containing the oskar 3′ UTR (Fig EV1D). Thus, Nanos causes mRNA degradation in S2 cells irrespective of whether it is artificially tethered or binds directly to an mRNA.
Importantly, deletion of the NED strongly impaired the ability of Nanos to repress the F‐Luc‐hb reporter (Fig 1E and F). However, the isolated NED did not repress the expression of this reporter (Fig 1E and F), most likely because it lacks RNA binding capacity. These results indicate that the Nanos NED confers repressive activity but requires the ZnF domain to bind to natural mRNA targets, as reported previously (Curtis et al, 1997; Hashimoto et al, 2010).
Nanos mediates translational repression in the absence of mRNA deadenylation
In addition to promoting mRNA target degradation, vertebrate Nanos proteins can repress translation in a deadenylation‐independent manner (Lai et al, 2011; Bhandari et al, 2014). Similarly, Dm Nanos promotes deadenylation and represses translation in the absence of mRNA degradation during oogenesis and early embryogenesis (Wharton & Struhl, 1991; Wreden et al, 1997; Chagnovich & Lehmann, 2001; Kadyrova et al, 2007). Therefore, we investigated whether Dm Nanos can repress translation in the absence of mRNA deadenylation in S2 cells. We used an mRNA with a 3′‐end generated by a self‐cleaving hammerhead ribozyme (HhR). This reporter is neither polyadenylated nor deadenylated (Zekri et al, 2013). Additionally, the reporter contains a DNA‐encoded poly(A) stretch of 95 nucleotides and a 3′ poly(C) stretch of seven nucleotides upstream of the ribozyme cleavage site (F‐Luc‐5BoxB‐A95C7‐HhR). This reporter is efficiently translated in S2 cells (Zekri et al, 2013). Tethered Nanos caused a threefold reduction in F‐Luc activity but only a 1.2‐fold reduction in mRNA levels, indicating that Nanos represses the expression of this reporter primarily at the translational level (Fig 1H and I). Furthermore, a Nanos protein lacking the NED had no repressive activity. Conversely, the NED was sufficient to repress translation of this reporter in the absence of mRNA degradation (Fig 1H and I). Thus, the NED is a major determinant for the repression mediated by Dm Nanos irrespective of whether the repression is caused by mRNA deadenylation and decay or by translational repression in the absence of deadenylation.
Nanos has intrinsic repressive activity independent of PUM and BRAT
Nanos functions together with PUM and BRAT to repress hb mRNA in Dm embryos (Sonoda & Wharton, 1999, 2001). The sequence in the hb 3′ UTR that binds PUM, BRAT, and Nanos consists of a duplicated Nanos response element (NRE). Each NRE is composed of one BoxA and one BoxB motif (Fig 2A; Wharton & Struhl, 1991; Murata & Wharton, 1995; Zamore et al, 1997; Wreden et al, 1997; Wharton et al, 1998; Sonoda & Wharton, 1999, 2001; Gupta et al, 2009 ; Loedige et al, 2014). BRAT binds to the BoxA motif of the NRE. PUM binds with high affinity to the BoxB motif but also exhibits low affinity for the BoxA motif (Zamore et al, 1997; Wang et al, 2002; Gupta et al, 2009; Loedige et al, 2014, 2015).
Figure 2. Dm Nanos exhibits intrinsic repressive activity.
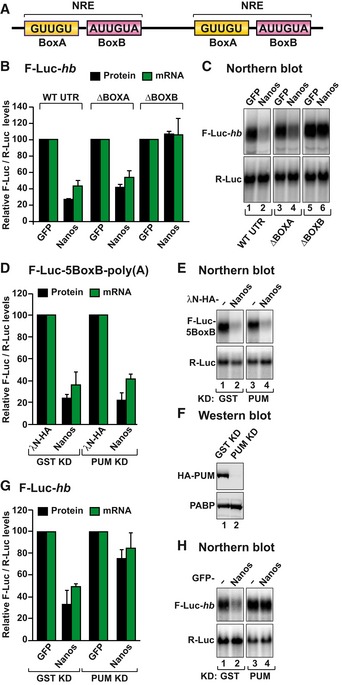
- Schematic representation of the Nanos response element in the 3′ UTR of hb mRNA.
- The activity of GFP‐tagged Dm Nanos was tested in S2 cells expressing an F‐Luc reporter containing the hb 3′ UTR (either wild type or mutants lacking the BoxA or BoxB sequences). A plasmid expressing GFP served as a negative control. F‐Luc activity (black bars) and mRNA levels (green bars) were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (B).
- Tethering assay using Dm Nanos and the F‐Luc‐5BoxB reporter in S2 cells depleted of PUM or control cells treated with a dsRNA targeting bacterial GST. A plasmid expressing R‐Luc mRNA served as a transfection control. The F‐Luc activity (black bars) and mRNA levels (green bars) were normalized to those of the R‐Luc transfection control and analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (D).
- Western blot analysis of S2 cells depleted of PUM and expressing HA‐PUM. Endogenous PABP served as a loading control.
- The ability of Nanos to repress the F‐Luc‐hb reporter was tested in S2 cells depleted of PUM as described in (D). The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (G).
Source data are available online for this figure.
To investigate whether Nanos requires BRAT or PUM to repress hb mRNA, we tested the effect of deleting the BoxA or BoxB motifs individually from the F‐Luc‐hb reporter. As shown in Fig 1E, transfected Nanos repressed and degraded the F‐Luc‐hb reporter in S2 cells (Fig 2B and C). These cells express both BRAT and PUM but not endogenous Nanos (Miles et al, 2012; Weidmann & Goldstrohm, 2012; Loedige et al, 2014). The deletion of the BoxA motif did not change the ability of Nanos to repress the reporter. By contrast, deletion of the BoxB sequences abrogated the ability of Nanos to degrade the reporter (Fig 2B), indicating that Nanos cooperates with endogenous PUM to repress the F‐Luc‐hb reporter, as expected (Murata & Wharton, 1995; Wreden et al, 1997; Sonoda & Wharton, 1999).
To determine whether Nanos activity in tethering assays was also dependent on its interaction with PUM, we performed the tethering assay in PUM‐depleted cells. PUM depletion did not affect Nanos activity in tethering assays (Fig 2D–F) but partially suppressed its ability to repress the F‐Luc‐hb reporter (Fig 2G and H).
In summary, Nanos requires the ZnF domain, PUM, and the BoxB motif to bind to the hb 3′ UTR and the NED to repress this mRNA. The requirement for the ZnF domain and PUM is bypassed when Nanos is directly tethered to the mRNA, indicating that Dm Nanos possesses intrinsic repressive activity, which is provided by the NED.
Nanos triggers deadenylation‐dependent decapping
The vertebrate Nanos proteins trigger deadenylation of mRNA targets by interacting with the CCR4–NOT complex (Suzuki et al, 2010, 2012; Bhandari et al, 2014). Deadenylation by CCR4–NOT is typically coupled to decapping and 5′‐to‐3′ exonucleolytic degradation by XRN1 (Temme et al, 2014). We therefore investigated whether Dm Nanos degrades bound mRNAs by promoting deadenylation‐dependent decapping.
If deadenylation precedes decapping and 5′‐to‐3′ mRNA degradation, then deadenylated mRNA decay intermediates are expected to accumulate in cells in which decapping is inhibited. To inhibit decapping, we overexpressed a catalytically inactive mutant of the decapping enzyme (DCP2*, E361Q) in cells depleted of endogenous DCP2 (Fig 3A and B). In these cells, degradation of the F‐Luc‐5BoxB reporter by Dm Nanos was inhibited and the F‐Luc‐5BoxB mRNA accumulated as a fast‐migrating form, corresponding to the deadenylated decay intermediate (A0; Fig 3B, lane 5). The luciferase activity is not restored despite restoration of mRNA levels (Fig EV1E), most likely because deadenylated transcripts are translated less efficiently. A similar fast‐migrating form accumulated when the GW182 protein was tethered (Fig 3B, lane 6). GW182 is known to cause deadenylation‐dependent decapping and thus served as a positive control (Behm‐Ansmant et al, 2006). The DCP2 mutant was expressed at comparable levels in each condition (Fig EV1F).
Figure 3. Dm Nanos induces deadenylation‐dependent decapping.
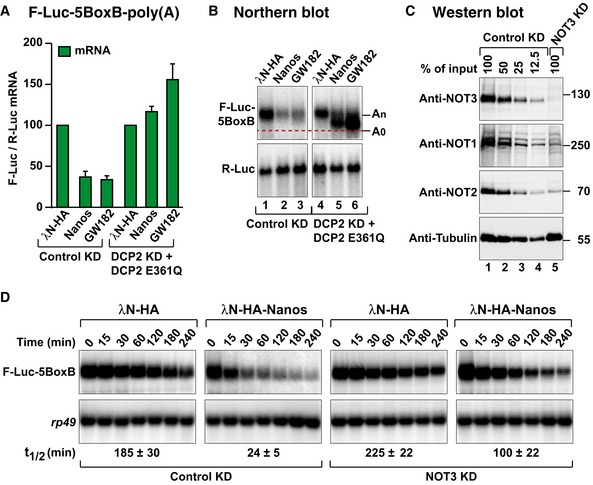
- Nanos tethering assay using the F‐Luc‐5BoxB reporter in control cells (treated with GST dsRNA) or in cells depleted of the decapping enzyme DCP2 (DCP2 KD) and expressing a catalytically inactive DCP2 mutant (E361Q). F‐Luc activity and mRNA levels were normalized to those of an R‐Luc transfection control and analyzed as described in Fig 1B. Normalized F‐Luc activities are shown in Fig EV1E. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (A). The positions of the polyadenylated (An) and deadenylated (A0) F‐Luc‐5BoxB mRNA are indicated on the right.
- Western blot analysis of S2 cells depleted of NOT3. Dilutions of control cell lysates were loaded in lanes 1–4 to estimate the efficacy of the depletion. α‐Tubulin served as a loading control. KD: knockdown. Endogenous NOT1 and NOT2 are co‐depleted with NOT3 as described previously (Boland et al, 2013).
- Northern blot analysis showing the decay of the F‐Luc‐5BoxB mRNA in control cells (treated with GST dsRNA) or in cells depleted of NOT3 expressing the indicated proteins. The mRNA half‐lives (t1/2) ± standard deviations calculated from the decay curves of three independent experiments are indicated below the panels.
Source data are available online for this figure.
To further investigate the dependence on the CCR4–NOT complex for Nanos‐mediated mRNA degradation, we depleted NOT3 in S2 cells. NOT3 depletion results in co‐depletion of NOT1 and NOT2 (Fig 3C; Temme et al, 2010; Boland et al, 2013). The ability of Nanos to elicit the degradation of F‐Luc‐5BoxB mRNA was partially suppressed in NOT3‐depleted cells (Fig 3D). Indeed, the half‐life of the F‐Luc‐5BoxB mRNA was increased fourfold in NOT3‐depleted cells expressing Nanos relative to that of control cells (Fig 3D).
The NED mediates binding to decay factors
Given that Nanos promotes deadenylation‐dependent decapping, we sought to determine whether it interacts with factors involved in the 5′‐to‐3′ decay pathway. We expressed Nanos with a GFP tag in S2 cells and tested for interactions with HA‐tagged subunits of the CCR4–NOT and PAN2–PAN3 deadenylase complexes as well as with decapping factors. Nanos interacted with NOT1, NOT2, NOT3, PAN2, and PAN3 (Figs 4A–D and EV1G–M). These interactions were observed in the presence of RNase A, suggesting that they are not mediated by RNA. In agreement with the tethering assays, the Nanos NED exhibited the same binding properties as full‐length Nanos, whereas a Nanos protein lacking the NED did not interact with the deadenylase subunits (Fig 4A–D).
Figure 4. The Nanos NED interacts with subunits of the deadenylase and decapping complexes.
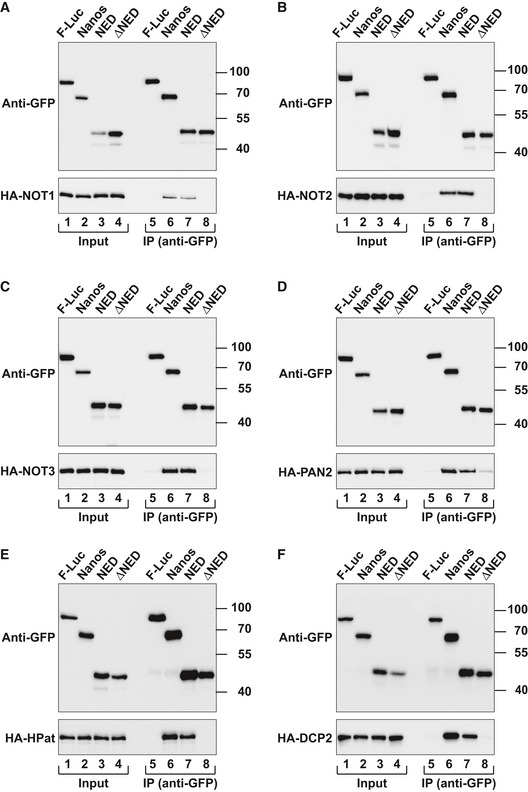
-
A–FWestern blots showing the interaction of GFP‐tagged Dm Nanos (either full length, NED, or ΔNED) and the indicated HA‐tagged proteins. The co‐immunoprecipitations were performed using a polyclonal anti‐GFP antibody. GFP‐tagged firefly luciferase (F‐Luc) served as a negative control. Inputs and immunoprecipitates were analyzed using anti‐GFP and anti‐HA antibodies. For the GFP‐tagged proteins, 3% of the inputs and 10% of the immunoprecipitates were loaded, whereas for the HA‐tagged proteins, 1% of the input and 30% of the immunoprecipitates were analyzed. In all panels, cell lysates were treated with RNase A prior to immunoprecipitation.
Source data are available online for this figure.
Nanos also interacted with the decapping enzyme DCP2 and the decapping factor HPat in an RNA independent manner but not with additional decapping factors (Figs 4E and F, and EV2A–F). Importantly, the NED was also necessary and sufficient for the interactions with DCP2 and HPat (Fig 4E and F).
Figure EV2. Dm Nanos interacts with decapping factors and the NOT module.
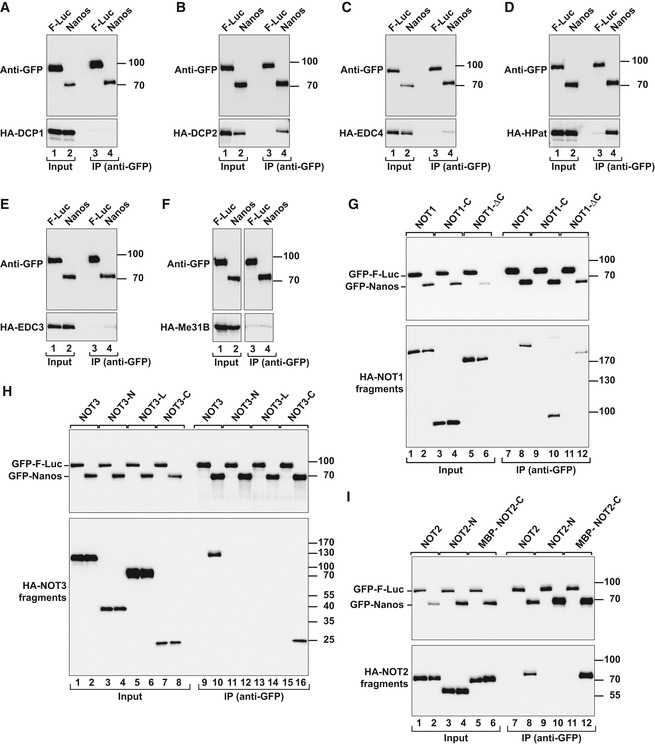
-
A–FCo‐immunoprecipitation assays using GFP‐tagged Dm Nanos (full length) and HA‐tagged decapping factors. Samples were analyzed as described in Fig EV1G–M.
-
G–IWestern blot analysis showing the interaction of GFP‐tagged Dm Nanos and HA‐tagged NOT1, NOT2, and NOT3 (either full length or the indicated fragments). Proteins were immunoprecipitated from RNase A‐treated cell lysates using anti‐GFP antibodies. GFP‐F‐Luc served as a negative control. For the detection of GFP‐tagged proteins, 3% of the input and 10% of the bound fractions were analyzed by Western blotting. For the detection of HA‐tagged NOT1, 1.5% of the input and 35% of the bound fractions were analyzed, whereas for HA–NOT2 and HA–NOT3 proteins, 1% of the input and 30% of the immunoprecipitates were analyzed.
Source data are available online for this figure.
The NED interacts directly with the NOT module of the CCR4–NOT complex
To define more precisely how the CCR4–NOT complex interacts with Dm Nanos, we used truncated constructs of NOT1, NOT2, and NOT3 in co‐immunoprecipitation assays. Deletion of the NOT1 C‐terminal region that includes the SHD domain did not abolish the interaction of Nanos with NOT1 (Fig EV2G, lane 12; Fig EV1A and Appendix Table S1). Conversely, the NOT1 C‐terminal region was sufficient for Nanos binding (Fig EV2G, lane 10). These results suggest that NOT1 minimally provides two binding sites for Nanos. Additionally, the C‐terminal regions of NOT2 and NOT3 were sufficient for Nanos binding (Fig EV2H and I, and Appendix Table S1). The C‐terminal regions of NOT2 and NOT3 heterodimerize and interact with the NOT1 SHD to assemble the NOT module (Boland et al, 2013), suggesting that Nanos binds to the NOT module. Thus, Dm Nanos contains two or more regions that contact the CCR4–NOT complex, of which one binds to the NOT module and one contacts the NOT1 region N‐terminal to the SHD.
To determine whether the interaction of Nanos with the NOT module was direct, we performed pull‐down assays in vitro using purified recombinant proteins expressed in Escherichia coli. In this case, we used the human NOT module, which, in contrast to the Dm NOT module, is readily expressed in bacteria. The NOT module is highly conserved between the two species with 68, 55, and 72% identity for the NOT1 SHD and the NOT2 and NOT3 C‐terminal regions, respectively (Appendix Figs S1 and S2A and B).
The Nanos NED fused N‐terminally to glutathione S‐transferase (GST) pulled down the preassembled NOT module containing the NOT1 SHD together with the NOT2 and NOT3 C‐terminal regions (Fig 5A, lane 12), indicating that the interaction is direct.
Figure 5. The central region of the Nanos NED interacts directly with the NOT module.
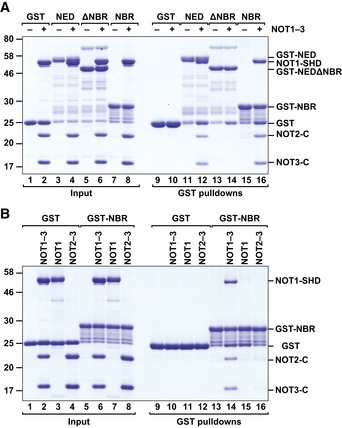
- GST pull‐down assay showing the interaction of the GST‐tagged Dm Nanos NED (wild type or lacking the NBR) and GST‐NBR with the recombinant Hs NOT module (NOT1–3; containing the NOT1 SHD and the NOT2 and NOT3 C‐terminal fragments).
- GST pull‐down assay showing the interaction of the GST‐tagged Dm Nanos NBR with the assembled NOT module (NOT1–3), the isolated NOT1 SHD (NOT1), or the NOT2–NOT3 dimers (NOT2–3).
Source data are available online for this figure.
The NED consists of several motifs that act redundantly to recruit the CCR4–NOT complex
To define the sequence requirements for binding of the Dm Nanos NED to the NOT module, we analyzed the alignment of NED sequences from various Drosophila species, which revealed three blocks of conserved residues (50–112, 116–163, and 207–236; Fig EV3A). Remarkably, each of these protein regions in isolation exhibited repressive activity in tethering assays, indicating functional redundancy (Fig EV3B–D). However, only the fragment comprising residues 116–163 was sufficient for binding to the purified NOT module complex in vitro (Fig 5A, lane 16, NBR), whereas deletion of residues 116–163 in the context of the NED abolished binding to the NOT module (Fig 5A, lane 14, ΔNBR). Thus, residues 116–163 within the NED represent a NOT module binding region (NBR).
Figure EV3. Sequence alignment of the Nanos NED regions of the indicated Drosophila species.
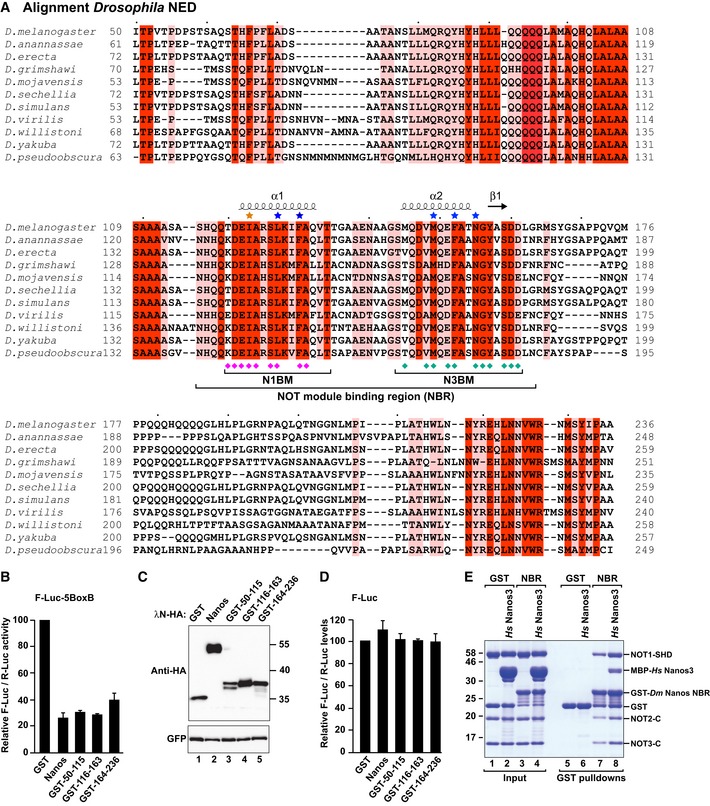
- The residues conserved in all of the aligned sequences are shown with a red background, and the residues with > 70% similarity are highlighted with a salmon background. The residues interacting with NOT1 and NOT3 are indicated by magenta and cyan diamonds, respectively, including main‐chain and minor side‐chain contacts. The residues mutated in this study are indicated by asterisks colored in blue (mutations that disrupt NOT module binding) and in orange (I123M mutation for the incorporation of selenomethionine as an anomalous scatterer).
- Tethering assay using the F‐Luc‐5BoxB reporter in S2 cells expressing the indicated λN‐HA‐tagged NED fragments. A plasmid‐expressing R‐Luc mRNA was used as a transfection control. The F‐Luc activities were normalized to those of the R‐Luc transfection control and set to 100 in the presence of the λN‐HA–GST. The panel shows mean values ± standard deviations from three independent experiments.
- Western blot analysis showing the expression of the λN‐HA‐tagged proteins used in the experiment described in (B). GFP served as a transfection control.
- An experiment similar to that described in (B) was performed using an F‐Luc reporter that lacks the BoxB hairpins. The panel shows mean values ± standard deviations from three independent experiments.
- GST pull‐down assay showing that the MBP‐tagged Nanos3 NIM peptide does not compete with the GST‐tagged Nanos NBR for binding to the purified NOT module.
Source data are available online for this figure.
The vertebrate Nanos NIM interacts directly with the NOT1 C‐terminal SHD domain and does not contact NOT2 or NOT3 (Bhandari et al, 2014). We therefore determined whether the Dm NBR interacted with the purified NOT1 SHD. However, the NBR interacted exclusively with the assembled NOT module but not with the isolated NOT1 SHD or the NOT2–NOT3 dimer (Fig 5B, lanes 14–16), indicating that the Dm NBR requires the entire NOT module for binding. Thus, the Dm NBR and the vertebrate NIM interact with the NOT module using different binding modes. Accordingly, the vertebrate Nanos3 NIM peptide (when added in excess to the NBR) did not compete with the NBR for binding to the NOT module; instead, both peptides bound simultaneously (Fig EV3E, lane 8).
Crystal structure of the Dm Nanos NBR bound to the NOT module
To explain the complexity of the interaction of the Dm NBR with the NOT module, we sought to obtain structural information. However, the failure to express soluble Dm NOT1 SHD in E. coli precluded the reconstitution of the Dm NOT module. Therefore, we turned to the human NOT module. On the basis of the available structure, we introduced mutations into the NOT1 SHD to improve crystallization. A triple substitution in the most C‐terminal loop of the NOT1 SHD domain (L19; Appendix Table S1 and Appendix Fig S1) resulted in highly reproducible crystals of the NOT module from which we obtained a structure at an improved resolution of 2.9 Å and in a new crystal packing environment with two copies of the complex per asymmetric unit (Fig EV4A and Table 1).
Figure EV4. Comparison of NOT module conformations and details of Dm Nanos NBR binding.
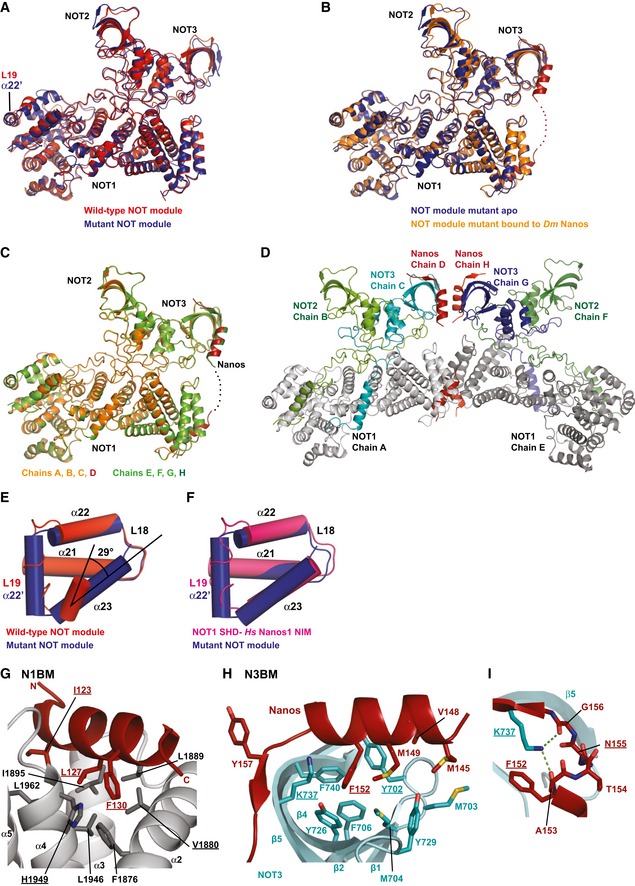
- Superposition of the mutant NOT module (apo form) crystallized in space group P21 (blue; chains A, B, C), and the wild‐type NOT module crystallized in space group P21212 (red; PDB code 4C0D; Boland et al, 2013).
- Superposition of the mutant NOT module structure obtained in the absence (blue, chains A, B, C) and presence of the Dm Nanos NBR (orange; chains A, B, C). The NBR peptide is shown in red.
- Superposition of the two NOT module complexes from the asymmetric unit of the crystals obtained in the presence of the Dm Nanos NBR peptide. Complex 1 (chains A, B, C) is colored in orange and complex 2 (chains E, F, G) in green. The NBR peptide is shown in red and dark green.
- Crystal packing of the NOT module mutant bound to the Dm Nanos NBR peptide.
- Orientation of NOT1 helix α23 in the wild type (PDB code 4C0D; Boland et al, 2013) and mutant NOT module apo complexes. Colors are as in (A). The black lines indicate the change in the relative orientation of the helix axes.
- Orientation of the NOT1 helix α23 in the mutant NOT module complex as compared to the orientation in the complex of the NOT1 SHD with the human Nanos1 NIM (PDB code 4CQO; Bhandari et al, 2014).
- Alternative view of the N1BM binding pocket centered on Dm Nanos F130. Selected residues of NOT1 and of the NBR peptide are shown as gray and red sticks, respectively. Residues mutated in this study are underlined.
- Alternative view of the N3BM binding pocket centered on Dm Nanos F152. Selected residues of NOT3 and of the NBR peptide are shown as cyan and red sticks, respectively. Residues mutated in this study are underlined.
- Additional close‐up of the N3BM binding site emphasizing the role of NOT3 K737 with hydrogen bonds as dashed green lines. Residues mutated in this study are underlined.
Table 1.
Data collection and refinement statistics
| NOT123–Nanos | NOT123–Nanos SeMet | NOT123 apo | |
|---|---|---|---|
| Space group | P 21 | P 21 | P 21 |
| Unit cell | |||
| dimensions a, b, c (Å) | 76.0, 135.7, 105.0 | 77.1, 135.8, 105.8 | 76.0, 135.3, 101.3 |
| angles α, β, γ (°) | 90, 108, 90 | 90, 108, 90 | 90, 108, 90 |
| Data collection a | |||
| Wavelength (Å) | 1.00005 | 0.97866 | 0.99982 |
| Resolution range (Å) | 50–3.1 (3.17–3.10) | 50–3.9 (4.00–3.90) | 50–2.9 (2.98–2.90) |
| R sym (%) | 7.5 (74.1) | 20.7 (81.3) | 7.5 (63.3) |
| Completeness (%) | 99.6 (98.2) | 99.7 (98.1) | 99.4 (99.7) |
| Mean I/σ(I) | 13.0 (1.7) | 8.6 (2.4) | 11.9 (2.1) |
| Unique reflections | 36,658 (2,706) | 18,985 (1,369) | 43,109 (3,191) |
| Multiplicity | 3.45 (3.45) | 6.92 (6.78) | 3.26 (3.27) |
| Refinement | |||
| R cryst (%) | 16.5 | 18.1 | |
| R free (%) | 22.7 | 22.1 | |
| Coordinate error (Å) | 0.42 | 0.35 | |
| Number of atoms | |||
| All atoms | 14,548 | 14,109 | |
| Protein | 14,548 | 14,109 | |
| Water | 0 | 0 | |
| Average B factor (Å2) | |||
| All atoms | 99.3 | 83.9 | |
| Ramachandran plot | |||
| Favored regions (%) | 95.3 | 95.8 | |
| Disallowed regions (%) | 0.3 | 0.4 | |
| RMSD from ideal geometry | |||
| Bond lengths (Å) | 0.010 | 0.010 | |
| Bond angles (°) | 1.10 | 1.10 | |
Values in parentheses are for highest resolution shell.
Furthermore, we also obtained the structure of the NOT module mutant in complex with the Dm Nanos NBR peptide at 3.1 Å resolution, which exhibited highly similar crystal packing and unit cell parameters closely related to the apo structure (Figs 6A and B, and EV4B and Table 1). The two copies of the complex in the asymmetric unit are structurally highly similar (Fig EV4C and D; RMSD 0.22 Å over 817 Cαs), and the Dm Nanos NBR peptide does not induce any major conformational changes in the NOT module (Fig EV4B; RMSD of 0.37 Å over 706 Cαs for the NOT module mutants in the bound and apo states).
Figure 6. Structure of the Dm Nanos NBR bound to the NOT module.
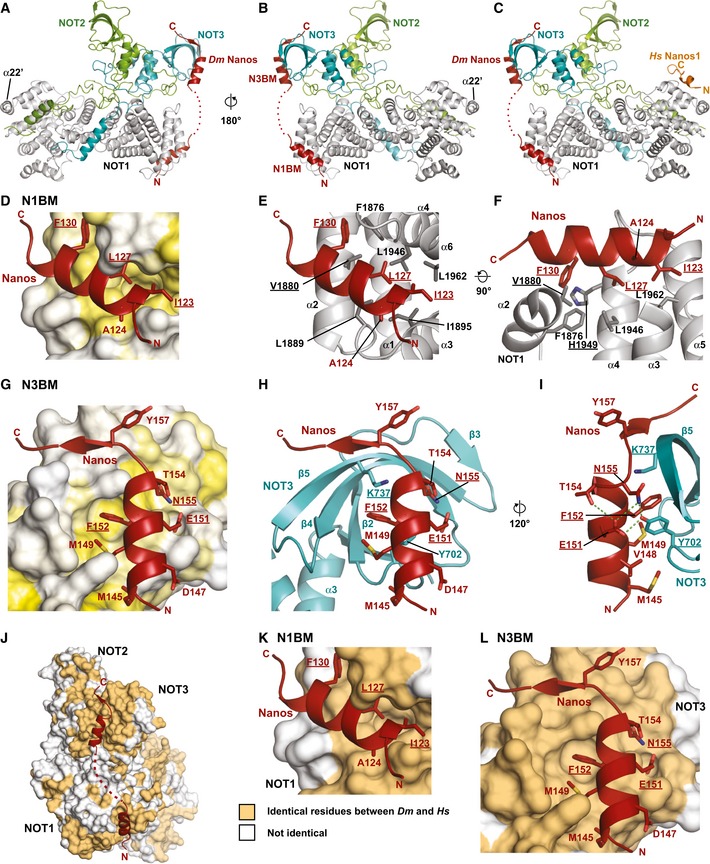
-
A, BCartoon representation of the Dm NBR peptide (red) bound to the NOT module in two orientations. The NOT1 SHD is colored in gray. NOT2 and NOT3 are shown in green and cyan, respectively.
-
CModel including the Nanos1 NIM peptide (from PDB entry 4CQO, Bhandari et al, 2014) obtained by superimposing the NOT1 SHD domains.
-
DSurface representation of the N1BM binding pocket of NOT1 with residues colored in a gradient from white to yellow with increasing hydrophobicity according to Kyte and Doolittle (1982).
-
ECartoon representation of the N1BM binding pocket. Selected residues of NOT1 and of the NBR peptide are shown as gray and red sticks, respectively. Residues mutated in this study are underlined.
-
FAlternative view of the N1BM binding pocket. NOT1 residues 1883–1894 of loop L2 have been omitted for clarity. Residues mutated in this study are underlined.
-
GSurface representation of the N3BM binding pocket of NOT3 colored as described in (D).
-
HCartoon representation of the N3BM binding pocket. Selected residues of NOT3 and of the NBR peptide are shown as cyan and red sticks, respectively. Residues mutated in this study are underlined.
-
IAlternative view of the N3BM binding pocket including selected hydrogen bonds as dashed green lines. Residues mutated in this study are underlined.
-
J–LConservation of the NBR binding sites on the NOT module surface. The NOT module is shown in surface representation. Surface residues that are identical between Hs and Dm are shown in orange, all other residues are shown in white. The views shown in (K) and (L) are in the same orientation as those shown in (D) and (G), respectively.
The most obvious difference to the previously reported structure of the human NOT module (Boland et al, 2013) is the formation of an additional α‐helix (α22') as part of the mutated loop L19 and the reorientation of the NOT1 C‐terminal helix α23 in both copies of the asymmetric unit (Fig EV4A and E). The rearrangement is probably due to the loss of crystal contacts by the mutated loop L19 sequence that can also explain the improved crystallization of the NOT module mutant compared with that of the wild type. Because helix α23 is now in a similar conformation as that in the complex of the NOT1 SHD domain with the Hs Nanos1 NIM peptide (Fig EV4F; Bhandari et al, 2014), we propose that this is the preferred conformation of helix α23.
Most strikingly, the Dm Nanos NBR peptide binds in a completely different location on the surface of the NOT module compared to the Hs Nanos1 NIM peptide (Fig 6C). Moreover, the Dm Nanos NBR peptide shows a very distinct bipartite binding mode, and it extends over the surfaces of both NOT1 and NOT3. The Dm Nanos NBR peptide folds into two α‐helices connected by a disordered linker region with lower sequence conservation (Figs 6A and B, and EV3A). The first α‐helix binds to the NOT1 SHD and represents the NOT1‐binding motif (N1BM, residues 120–134), whereas the second α‐helix is part of the NOT3‐binding motif (N3BM, residues 144–161) and binds to the NOT3–NOT‐box domain.
Interactions of the N1BM and the N3BM
As previously described, the NOT1 SHD consists of two perpendicular stacks of HEAT‐like repeats (Bhaskar et al, 2013; Boland et al, 2013). The NOT2 and NOT3 NOT‐box domains each adopt an SH3‐like fold comprising a five‐stranded half‐open β‐barrel as well as two and three α‐helices, respectively, which mediate heterodimerization. Furthermore, NOT2 and NOT3 have N‐terminal extensions that serve to embrace each other's NOT‐box domains and to tether the NOT2–NOT3 heterodimer to the surface of the NOT1 SHD (Fig 6A and B).
The N1BM folds into a three‐turn amphipathic α‐helix that docks into a groove on the surface of the NOT1 SHD (Figs 6D–F and EV4G). This groove forms between the termini of α‐helices 3 and 4 (HEAT repeat 2), and the flanking HEAT repeats 1 and 3 of the SHD. The hydrophobic surface of the N1BM α‐helix faces the groove with residues I123, A124, L127, and F130. These residues are matched by NOT1 SHD residues F1876, V1880, L1889, I1895, L1946, H1949, and L1962, leading to a predominantly hydrophobic interaction (Figs 6D–F, EV3A, EV4G, and Appendix Fig S1).
The N3BM also folds into an amphipathic helix. This helix docks onto the open side of the β‐barrel in the NOT3–NOT‐box domain, and it is additionally extended by a short β‐strand that augments the highly curved β‐sheet of the barrel (Figs 6G–I and EV4H and I). Again, the interaction is primarily hydrophobic with residues M145, V148, M149, and, in particular, F152 from the N3BM facing residues Y702, M703, M704, F706, Y726, Y729, K737, and F740 on the NOT3–NOT‐box (Figs 6G–I, EV3A, EV4H and I, and Appendix Fig S2B). Notably, the NOT3 residues Y702, F706, Y726, and F740 surround the aromatic ring of F152, and Y702 makes additional hydrogen bonds to E151 and N155 of N3BM (Figs 6I and EV4H). Furthermore, K737 contacts the F152 and G156 carbonyl oxygens and serves as an anchor for the following residues and stabilizes their main‐chain interactions with the outer edge of the NOT3 β‐sheet (Fig EV4I).
To confirm the sequence assignment of the relatively short N1BM and N3BM in the 3.1 Å electron density, we substituted I123 in the N1BM with methionine and crystallized the complex with a selenomethionine‐derivatized Dm Nanos mutant peptide. This allowed us to collect anomalous diffraction data using X‐ray energies at the selenium K‐edge (Table 1) and to calculate an anomalous difference Fourier map (Fig EV5A–D). The map unambiguously identified M123 in the N1BM as well as M145 and M149 in the N3BM, confirming that our initial sequence assignment was correct.
Figure EV5. Validation of the sequence assignment of the NBR peptide bound to the NOT module and activity of NBR mutants.
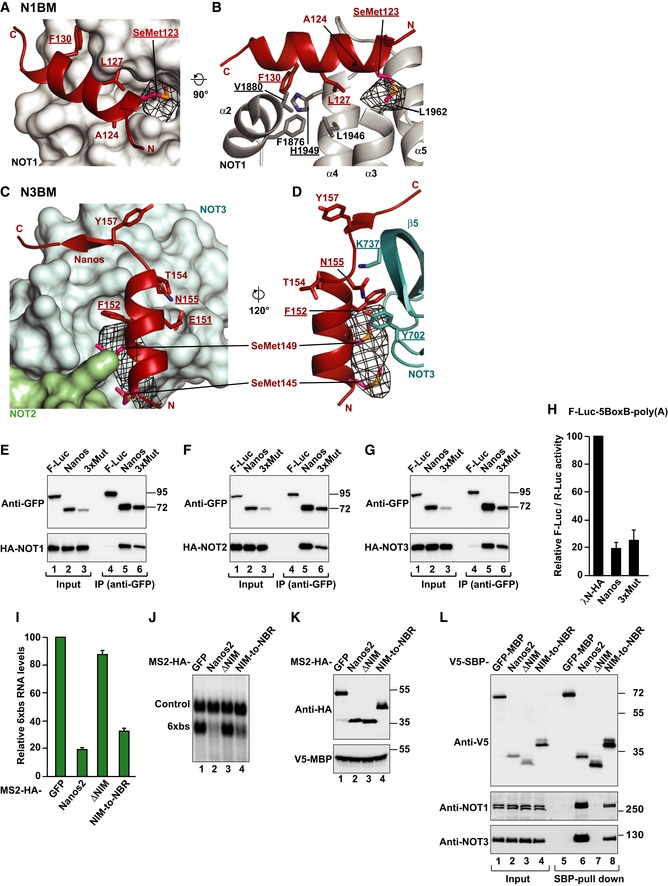
-
A–DAnomalous difference Fourier map (black mesh) calculated at a resolution of 7.5 Å and contoured at the 4.0 σ level. Data (available as source data) were collected at the Selenium K‐edge peak wavelength from a crystal containing selenomethionine‐substituted Dm Nanos NBR peptide (I123M mutant). The panels show close‐up views of the binding sites of the N1BM (A, B) and the N3BM (C, D) in the same orientations as in Fig 6. Residues mutated in this study are underlined.
-
E–GWestern blot analysis showing the interaction of GFP‐tagged Dm Nanos (wild type or 3xMut) with HA‐tagged NOT1, NOT2, and NOT3. GFP‐tagged firefly luciferase (F‐Luc) served as a negative control. Proteins were immunoprecipitated using a polyclonal anti‐GFP antibody. Inputs and immunoprecipitates were analyzed by Western blotting as described in Fig EV1G–M.
-
HA tethering assay using the F‐Luc‐5BoxB reporter and the indicated λN‐HA‐tagged proteins was performed in S2 as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
-
ITethering assays in human HEK293T cells, using a β‐globin reporter containing 6 binding sites (6xbs) for the MS2 protein and MS2‐HA‐tagged Hs Nanos2 (wild type or the indicated variants, Nanos2 ΔNIM and Nanos2 NIM‐to‐NBR). In the Nanos2 NIM‐to‐NBR protein, the NIM was replaced by the Dm Nanos NBR. A plasmid expressing an mRNA lacking MS2 binding sites (Control) served as a transfection control. The β‐globin‐6xbs mRNA levels were normalized to those of the control mRNA and set to 100 in the presence of MS2‐HA. The panel shows mean values ± standard deviations from three independent experiments.
-
JNorthern blot of representative RNA samples corresponding to the experiment shown in (I).
-
KWestern blot analysis showing the expression of the MS2‐tagged proteins used in the experiment shown in (I) and (J). V5‐MBP served as a transfection control.
-
LCo‐immunoprecipitation assay in human HEK293T cells showing the interaction of V5‐SBP‐tagged Nanos2 (wild type or the indicated variants) with endogenous NOT1 and NOT3. V5‐SBP‐tagged GFP‐MBP served as a negative control.
Source data are available online for this figure.
Validation of the binding interfaces
To confirm that the interfaces from the crystal structure are also relevant in solution, we introduced mutations in the Nanos peptide, NOT1 or NOT3, and tested binding of the peptide to the preassembled NOT modules using GST pull‐down assays in vitro. To validate the N1BM–NOT1 interface, we generated the NOT1 mutants V1880E and H1949D (corresponding to Dm NOT1 V2002E and H2067D, respectively). These mutations strongly reduced but did not abolish binding of the GST‐NBR fusion to the NOT module (Fig 7A, lanes 17 and 18 vs. 16). Conversely, the mutations in the Nanos N1BM (2xMut, L127D, F130D) strongly reduced binding to the NOT module (Fig 7A, lane 20).
Figure 7. Validation of the interaction interfaces.
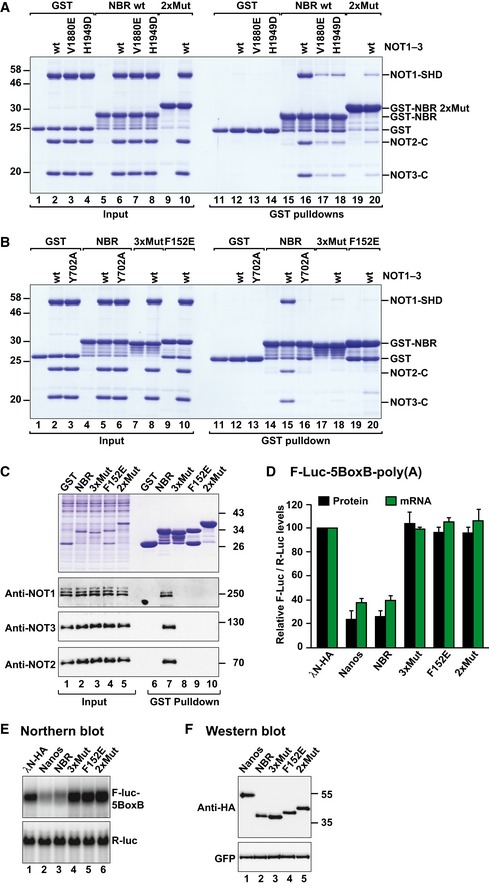
- GST pull‐down assay showing the interaction of the GST‐Dm Nanos NBR [wild type or 2xMut (L127D F130D)] with the recombinant NOT module (NOT1–3) containing the NOT1 SHD (either wild type or V1880E and H1949D). GST served as a negative control. Note that the GST‐Dm Nanos NBR 2xMut exhibits abnormal electrophoretic migration most likely due to the introduction of two negatively charged residues.
- GST pull‐down assay showing the interaction of GST‐Dm Nanos NBR [wild type, F152E and 3xMut (E151A, F152A, N155A)] with the recombinant NOT module containing wild‐type NOT3 or the Y702A mutant. GST served as a negative control.
- GST pull‐down assay showing the interaction of GST‐tagged Dm Nanos NBR (wild type, F152E, 2xMut, and 3xMut) with the Dm CCR4–NOT complex in S2 cell lysates. GST served as a negative control.
- Tethering assay using the Nanos NBR mutants and the F‐Luc‐5BoxB reporter in S2 cells. A plasmid expressing R‐Luc mRNA served as a transfection control. The F‐Luc activity (black bars) and mRNA levels (green bars) were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (D).
- Western blot showing the expression of the λN‐HA‐tagged proteins used in the experiments shown in (D) and (E). GFP served as a transfection and loading control.
Source data are available online for this figure.
To validate the N3BM–NOT3 interaction, we substituted the central NOT3 residue Y702 with alanine (Y702A). This mutation was sufficient to abolish binding of the NBR to the NOT module (Fig 7B, lane 16 vs. 15). Conversely, the F152E mutation in the Dm Nanos N3BM was sufficient to disrupt complex assembly (Fig 7B, lane 20), confirming the central role of this residue in the interaction. Because no residual binding was observed for the Nanos F152E mutant despite the presence of the N1BM, these results indicate that the N1BM is not sufficient for complex formation.
The residues in NOT1 and NOT3 interacting with the Nanos peptide are conserved in Dm (Fig 6J–L, and Appendix Figs S1 and S2B). Nevertheless, it was important to assess whether Dm Nanos binds to the Dm NOT module using a similar binding mode. To this end, we used the recombinant GST fusions of Dm Nanos NBR mutants described above and tested the interactions with the endogenous Dm CCR4–NOT complex in Dm S2 cell lysates. Mutations in the N1BM (2xMut) or in the N3BM (F152E or 3xMut (E151A F152A N155A); Appendix Table S1) were sufficient to disrupt the interaction (Fig 7C, lanes 8–10). In agreement with these results, the mutations abolished the activity of the NBR in tethering assays (Fig 7D–F). Notably, the mutations were ineffectual in the context of full‐length Nanos (Fig EV5E–H), in agreement with the notion that the NED contains multiple sequences that mediate the recruitment of the CCR4–NOT complex in a redundant manner.
The vertebrate Nanos NIM and the Dm NED perform analogous functions
Given that the Dm NED and the vertebrate NIM recruit the CCR4–NOT complex, we asked whether the NED could be replaced by the human Nanos NIM and elicit mRNA degradation in Dm cells. To test this hypothesis, we generated a chimeric protein containing the human Nanos2 NIM fused to the Dm Nanos ZnF domain (NIM–ZnF). This protein effectively repressed and degraded the F‐Luc‐5BoxB mRNA reporter in the tethering assays (Fig 8A and B). Importantly, the chimeric protein also degraded the F‐Luc‐hb reporter (Fig 8C and D) and interacted with the Dm CCR4–NOT complex (Fig 8E). Mutations of conserved aromatic residues in the NIM motif (NIM*: F6A and W9E; Appendix Table S1), which are essential for binding to the human NOT module (Bhandari et al, 2014), abolished the interaction of the chimeric protein with the Dm CCR4–NOT complex and the ability of the protein to degrade the F‐Luc‐5BoxB and F‐Luc‐hb mRNAs (Fig 8A–E). These results indicate that the vertebrate NIM interacts with the Dm CCR4–NOT complex using an interaction mode similar to that observed for the human complex. Accordingly, the NOT1 residues required for the interaction with the NIM are conserved in Dm (Appendix Fig S1; Bhandari et al, 2014).
Figure 8. The NED and NIM are functionally equivalent.
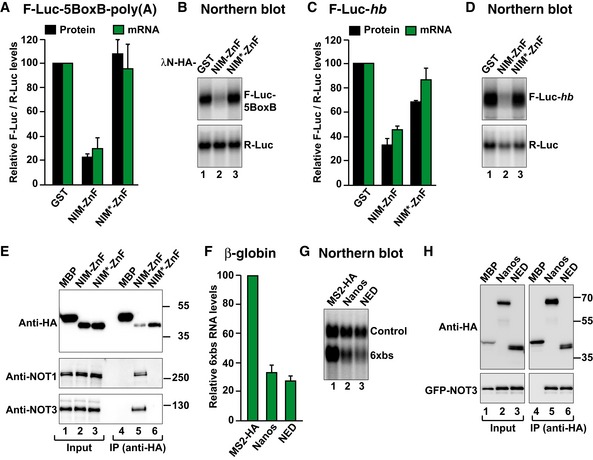
- Tethering assay using a chimeric Nanos protein and the F‐Luc‐5BoxB reporter in S2 cells. The chimeric Nanos protein contains the NIM of human Nanos2 (either wild type (NIM‐ZnF) or mutated (NIM*‐ZnF)), fused to the Dm ZnF domain. A plasmid expressing GST served as a negative control. F‐Luc activity and mRNA levels were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (A).
- The activity of GST‐HA‐tagged Nanos chimeric protein was tested in S2 cells expressing an F‐Luc‐hb reporter. A plasmid expressing GST served as a negative control. F‐Luc activity and mRNA levels were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (C).
- Western blot analysis showing the interaction of the HA‐tagged Nanos chimeric protein (NIM‐ZnF or NIM*‐ZnF) with endogenous Dm NOT1 and NOT3. HA‐MBP served as a negative control.
- Tethering assays in human HEK293T cells, using a β‐globin reporter containing 6 binding sites (6xbs) for the MS2 protein and MS2‐HA‐tagged Dm Nanos or NED fragment. A plasmid expressing an mRNA lacking MS2 binding sites (Control) served as a transfection control. The β‐globin‐6xbs mRNA levels were normalized to those of the control mRNA and set to 100 in the presence of MS2‐HA. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (F).
- Co‐immunoprecipitation assay in human HEK293T cells showing the interaction of the HA‐tagged Dm Nanos and the NED fragment with GFP‐tagged NOT3 in human cells. HA‐MBP served as a negative control. The inputs (1%) and bound fractions (10% of HA‐tagged proteins and 30% of GFP‐NOT3) were analyzed by Western blotting.
Source data are available online for this figure.
We also tested whether Dm Nanos repressed the expression of a reporter in tethering assays in human cells. We observed that both Dm Nanos and the NED caused mRNA degradation in tethering assays in human cells and interacted with human NOT3 (Fig 8F–H). Furthermore, when the NIM was replaced by the NBR in human Nanos2, the chimeric protein degraded the mRNA reporter and interacted with the endogenous CCR4–NOT complex in HEK293T cells (Fig EV5I–L).
Discussion
In this study, we showed that Dm Nanos recruits the CCR4–NOT complex directly through a Nanos effector domain (NED) that is conserved in the Drosophila species (Fig EV3). Similar to the vertebrate Nanos NIM, the Dm NED is necessary and sufficient to repress translation in the absence of mRNA degradation and to promote degradation of bound mRNAs. Thus, the NED and the NIM are the main determinants for the repressive activity of Nanos in Dm and vertebrates, respectively. Although the NED and the NIM are functionally analogous, they do not share sequence similarities, demonstrating that the absence of sequence conservation is not an indicator of functional irrelevance, in particular, when disordered, low complexity protein regions are involved. Such regions often mediate their function by interacting with binding partners using short linear motifs (SLiMs). SLiMs can evolve rapidly due to the lack of constraints to maintain a protein fold (Davey et al, 2012) and thus enable the evolution of distinct binding modes in orthologous proteins, especially in cases where these proteins are in a competitive scenario or even under positive selection. In this way, these proteins can maintain the ability to interact with the same partners using different binding modes.
The Dm NED uses multiple and redundant motifs to recruit the CCR4–NOT complex
The Dm NED and the vertebrate NIM use different modes to mediate the recruitment of the CCR4–NOT complex to Nanos mRNA targets. The NIM is a short 17‐residue motif present in the N‐terminal disordered region of vertebrate Nanos proteins, which binds to the NOT1 SHD domain (Bhandari et al, 2014). In contrast to vertebrates, flies have only a single Nanos protein but with an extended NED. The Dm NED is 187 amino acids in length and contains multiple and redundant binding sites for the CCR4–NOT complex. These multiple binding sites may increase the affinity of Dm Nanos for the CCR4–NOT complex through avidity effects. Redundancy may also confer a competitive advantage to Dm Nanos over other RNA‐binding proteins that compete for recruitment of the CCR4–NOT complex.
Interestingly, redundancy to recruit the CCR4–NOT complex is not only observed within the Nanos protein but also in the context of Nanos repressive complexes. Indeed, Nanos cooperates with PUM to bind and repress natural mRNA targets, and PUM also has the ability to recruit the CCR4–NOT complex independently of Nanos (Asaoka‐Taguchi et al, 1999; Sonoda & Wharton, 1999, 2001; Jaruzelska et al, 2003; Goldstrohm et al, 2006; Van Etten et al, 2012). This may partially obscure the effects of deletion or mutations in the Nanos NED. However, PUM does not act as a general substitute for Nanos because mutations in the Nanos ZnF domain that prevent mRNA binding caused strong developmental defects despite the presence of endogenous PUM (Arrizabalaga & Lehmann, 1999). Thus, PUM probably acts both additively and alternatively with the Nanos NED, resulting in distinct modes of engaging the CCR4–NOT complex. In their various combinations, the different binding modes can thus lead to a highly specific and tunable repression of mRNA targets in a cell context‐dependent manner.
RNA‐binding proteins recruit CCR4–NOT using a diversity of binding motifs
A large number of RNA‐associated proteins have been shown to recruit the CCR4–NOT complex to their mRNA targets to repress translation and/or to promote mRNA degradation. In addition to Nanos, these proteins include the GW182 proteins, which are involved in miRNA‐mediated gene silencing in animals, and the Dm proteins CUP, Bicaudal‐C, Smaug, and PUM (Semotok et al, 2005; Goldstrohm et al, 2006; Chicoine et al, 2007; Igreja & Izaurralde, 2011; Van Etten et al, 2012; Chen et al, 2014; Mathys et al, 2014). Additional examples from vertebrates are Roquin and tristetraprolin (TTP), a protein required for the degradation of mRNAs containing AU‐rich elements (ARE‐mediated mRNA decay) (Fabian et al, 2013; Leppek et al, 2013).
For the recruitment of the CCR4–NOT complex, most of these proteins rely on short linear motifs (SLiMs) embedded in peptide regions of predicted disorder. However, a detailed characterization of their interaction with the CCR4–NOT complex on a molecular level is only available for TTP, GW182, and vertebrate and Dm Nanos (Fabian et al, 2013; Bhandari et al, 2014; Chen et al, 2014; Mathys et al, 2014; this study). For TTP and vertebrate and Dm Nanos, the motifs adopt α‐helical conformations that possibly form only upon binding. Specificity results from aromatic and hydrophobic side chains that insert primarily into pockets on the surface of the NOT1 domains that consist of HEAT‐like repeats (Fabian et al, 2013; Bhandari et al, 2014; this study). By contrast, GW182 peptides likely bind to the CCR4–NOT complex in an extended conformation and insert tryptophan residues into tandem hydrophobic pockets exposed at the surface of the NOT9 subunit (also known as CAF40) of the CCR4–NOT complex and probably into additional pockets in NOT1 that remain to be identified (Chen et al, 2014; Mathys et al, 2014).
Similar to GW182 proteins, the Nanos NBR not only contacts NOT1 but also binds to NOT3, providing the first detailed insight into how an mRNA‐binding protein recruits the CCR4–NOT complex by contacting two of its subunits simultaneously. Interestingly, the surface on NOT1 that is contacted by Dm Nanos partially overlaps with the binding surface for NOT4 as observed in the Saccharomyces cerevisiae (Sc) complex (Bhaskar et al, 2015). This would suggest that Dm Nanos competes with NOT4 for binding to NOT1, providing additional opportunities for the regulation of gene expression. However, it is not known whether the NOT4 binding mode is conserved between Dm and Sc because NOT4 does not co‐purify with the CCR4–NOT complex in metazoans (Lau et al, 2009; Temme et al, 2010) and the Sc NOT4 sequences that bind NOT1 are not well conserved in metazoans (Bhaskar et al, 2015).
Together with previous studies (Fabian et al, 2013; Bhandari et al, 2014; Chen et al, 2014; Mathys et al, 2014), our results reveal that the recruitment of the CCR4–NOT complex is mediated by highly diverse sequence motifs and distinct binding modes. We speculate that these motifs represent a combinatorial code that is read by the CCR4–NOT complex to funnel the effects of diverse RNA‐binding proteins into a common repressive pathway, which results in the removal of the mRNA poly(A) tail, translational repression, and, depending on the cellular context, full degradation of the mRNA.
Materials and Methods
DNA constructs
The DNA constructs used in this study are provided in the Appendix Supplementary Materials and Methods and are listed in Appendix Table S1. All of the constructs and mutations were confirmed by sequencing.
Tethering assays and dsRNA interference
For the λN‐tethering assay, 2.5 × 106 S2 cells were seeded in 6‐well plates and co‐transfected with the following plasmids: 0.1 μg of reporter plasmid (F‐Luc‐5BoxB, F‐Luc‐5BoxB‐A95C7‐HhR or F‐Luc), 0.4 μg of R‐Luc‐A90‐HhR, and 25 ng of plasmid expressing λN‐HA or 2.5–25 ng of plasmid expressing λN‐HA–Nanos protein fusions. For the assay using F‐Luc‐hb, cells were co‐transfected with the following plasmids: 0.2 μg of F‐Luc‐hb reporter plasmid, 0.4 μg of R‐Luc‐A90‐HhR, and 20–30 ng of plasmids expressing GFP‐Nanos fusion proteins. Cells were harvested 3 days after transfection.
Knockdowns using dsRNA were performed as previously described (Behm‐Ansmant et al, 2006). To measure the mRNA half‐life, cells were treated with actinomycin D (5 μg/ml final concentration) three days post‐transfection and collected at the indicated time points. Firefly and Renilla luciferase activities were measured using a Dual‐Luciferase Reporter Assay system (Promega). Northern blotting was performed as previously described (Behm‐Ansmant et al, 2006).
Tethering assays in human cells using the β‐globin reporter containing six MS2 binding sites (6xbs; Lykke‐Andersen et al, 2000) were performed as previously described (Bhandari et al, 2014). The transfection mixtures contained 0.1 or 1 μg of plasmids expressing MS2‐HA‐tagged Nanos or the NED fragment, respectively.
Co‐immunoprecipitation assays and Western blotting
For co‐immunoprecipitation assays in S2 cells, 2.5 × 106 cells were seeded per well in 6‐well plates and transfected using Effectene transfection reagent (Qiagen). The transfection mixtures contained 1 μg of plasmid expressing HA‐tagged deadenylase or decapping factors and 1–2 μg of GFP‐tagged Nanos (either full length or fragments). Cells were harvested 3 days after transfection, and co‐immunoprecipitation assays were performed as previously described (Braun et al, 2011). Co‐immunoprecipitation assays in human cells were performed as previously described (Bhandari et al, 2014). All Western blots were developed with the ECL Western Blotting Detection System (GE Healthcare) according to the manufacturer's recommendations. Antibodies used in this study are listed in Appendix Table S2.
Protein expression and purification
All proteins for crystallization and in vitro pull‐down assays were expressed in E. coli BL21 (DE3) Star cells (Invitrogen) in ZY medium at 20°C overnight. A detailed description of the purification procedures is included in the Appendix Supplementary Materials and Methods.
Crystallization, data collection, and structure determination
A detailed description of the crystallization conditions and the structure determination process is included in the Appendix Supplementary Materials and Methods. All diffraction data sets were recorded on a PILATUS 6M detector at the PXII beamline of the Swiss Light Source at a temperature of 100 K. The diffraction data and refinement statistics are summarized in Table 1.
In vitro GST pull‐down assays
In vitro pull‐down assays were performed as previously described (Bhandari et al, 2014). Briefly, purified GST‐tagged Nanos (full length or fragments) was incubated with untagged NOT module and Protino glutathione agarose 4B beads (Macherey‐Nagel) in pull‐down buffer containing 50 mM HEPES (pH 7.5), 200 mM NaCl, and 2 mM DTT. After a 1 h incubation, the beads were washed four times with pull‐down buffer. The proteins were eluted using pull‐down buffer supplemented with 25 mM glutathione, precipitated with trichloroacetic acid, and analyzed by SDS–PAGE.
For the experiment shown in Fig 7C, 25 × 106 S2 cells were suspended in 1 ml of hypotonic buffer (10 mM HEPES (pH 7.6), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and 1× Roche‐EDTA free protease inhibitor cocktail) for 15 min on ice. Cells were lysed by sonication (3 times for 30 s followed by 30 s on ice) followed by applying 10 strokes of a Dounce homogenizer. Cell lysates were supplemented with 200 mM NaCl. Cell debris was removed by 15‐min centrifugation at 16,000 × g at 4°C. The supernatant was collected, treated with RNase A, and supplemented with 40 μg of purified GST‐tagged Nanos NBR peptide (wild type or mutant) and 40 μl Protino glutathione agarose 4B beads (Macherey‐Nagel). After 1.5 h incubation on a rotating wheel at 4°C, the beads were washed three times with binding buffer (hypotonic buffer supplemented with 200 mM NaCl). Bound proteins were eluted in 50 μl of binding buffer containing 25 mM reduced glutathione solution and analyzed by SDS–PAGE and Western blotting.
Accession numbers
The structures were deposited in the Protein Data Bank under accession codes 5FU6 for the NOT module and 5FU7 for the NOT module with the NBR peptide.
Author contributions
TR and KS cloned, expressed, and purified proteins for crystallization and pull‐down assays. TR and KS crystallized the NOT module mutant and the NOT module–Nanos complex. TR solved the structures and performed in vitro pull‐down assays. DB and SH cloned proteins for expression in human and S2 cells. SH carried out co‐immunoprecipitation assays. DB designed, performed, and analyzed functional assays in human and S2 cells. EV and OW supervised the structural studies and assisted with refinement. EV designed the NOT module crystallization mutant. OW conceived and analyzed the selenomethionine experiment. EI designed and supervised the project. EI and OW wrote the manuscript with help from TR, DB, and EV.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We are grateful to Heike Budde and Catrin Weiler for excellent technical support. We thank the staff at the PX beamlines of the Swiss Light Source, Villigen, for assistance with data collection. This work was supported by the Max Planck Society and the Gottfried Wilhelm Leibniz Program awarded to E.I.
The EMBO Journal (2016) 35: 974–990
References
- Arrizabalaga G, Lehmann R (1999) A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics 153: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka‐Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S (1999) Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol 1: 431–437 [DOI] [PubMed] [Google Scholar]
- Baines RA (2005) Neuronal homeostasis through translational control. Mol Neurobiol 32: 113–121 [DOI] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M (2013) Control of maternal mRNA stability in germ cells and early embryos. Biochim Biophys Acta 1829: 714–724 [DOI] [PubMed] [Google Scholar]
- Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E (2013) NOT10 and C2orf29/NOT11 form a conserved module of the CCR4‐NOT complex that docks onto the NOT1 N‐terminal domain. RNA Biol 10: 228–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm‐Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E (2014) Structural basis for the Nanos‐mediated recruitment of the CCR4‐NOT complex and translational repression. Genes Dev 28: 888–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Basquin J, Conti E (2015) Architecture of the ubiquitylation module of the yeast Ccr4‐Not complex. Structure 23: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Seraphin B, Conti E (2013) Structure and RNA‐binding properties of the Not1‐Not2‐Not5 module of the yeast Ccr4‐Not complex. Nat Struct Mol Biol 20: 1281–1288 [DOI] [PubMed] [Google Scholar]
- Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu‐Öztürk D, Wohlbold L, Weichenrieder O, Izaurralde E (2013) Structure and assembly of the NOT module of the human CCR4‐NOT complex. Nat Struct Mol Biol 20: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E (2011) GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Chagnovich D, Lehmann R (2001) Poly(A)‐independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci USA 98: 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W (2011) miRNA repression involves GW182‐mediated recruitment of CCR4‐NOT through conserved W‐containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoğlu‐Öztürk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E (2014) A DDX6‐CNOT1 complex and W‐binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54: 737–750 [DOI] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P (2007) Bicaudal‐C recruits CCR4‐NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704 [DOI] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M (2010) Translational repression by deadenylases. J Biol Chem 285: 28506–28513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R (1997) A CCHC metal‐binding domain in Nanos is essential for translational regulation. EMBO J 16: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ (2012) Attributes of short linear motifs. Mol BioSyst 8: 268–281 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N (2013) Structural basis for the recruitment of the human CCR4‐NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginter‐Matuszewska B, Kusz K, Spik A, Grzeszkowiak D, Rembiszewska A, Kupryjanczyk J, Jaruzelska J (2011) NANOS1 and PUMILIO2 bind microRNA biogenesis factor GEMIN3, within chromatoid body in human germ cells. Histochem Cell Biol 136: 279–287 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M (2006) PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK (2009) Co‐occupancy of two Pumilio molecules on a single hunchback NRE. RNA 15: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Hara K, Hishiki A, Kawaguchi S, Shichijo N, Nakamura K, Unzai S, Tamaru Y, Shimizu T, Sato M (2010) Crystal structure of zinc‐finger domain of Nanos and its functional implications. EMBO Rep 11: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Izaurralde E (2011) CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 25: 1955–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA (2003) Conservation of a Pumilio‐Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol 213: 120–126 [DOI] [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas‐Rios P, Busseau I, Simonelig M (2013) The CCR4 Deadenylase Acts with Nanos and Pumilio in the Fine‐Tuning of Mei‐P26 Expression to Promote Germline Stem Cell Self‐Renewal. Stem Cell Reports 1: 411–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP (2007) Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134: 1519–1527 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lai F, King ML (2013) Repressive translational control in germ cells. Mol Reprod Dev 80: 665–676 [DOI] [PubMed] [Google Scholar]
- Lai F, Zhou Y, Luo X, Fox J, King ML (2011) Nanos1 functions as a translational repressor in the Xenopus germline. Mech Dev 128: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT (2009) Human Ccr4‐Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453 [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nüsslein‐Volhard C (1991) The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112: 679–691 [DOI] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G (2013) Roquin promotes constitutive mRNA decay via a conserved class of stem‐loop recognition motifs. Cell 153: 869–881 [DOI] [PubMed] [Google Scholar]
- Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, Engelmann JC, Krahn MP, Meister G (2015) The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain‐Tumor‐Mediated Gene Regulation. Cell Rep 13: 1206–1220 [DOI] [PubMed] [Google Scholar]
- Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Löffler P, Längst G, Merkl R, Urlaub H, Meister G (2014) The NHL domain of BRAT is an RNA‐binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev 28: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke‐Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense‐mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki‐Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W (2014) Structural and Biochemical Insights to the Role of the CCR4‐NOT Complex and DDX6 ATPase in MicroRNA Repression. Mol Cell 54: 751–765 [DOI] [PubMed] [Google Scholar]
- Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ (2012) Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev 26: 356–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Sano H, Kobayashi S, Nishimiya‐Fujisawa C, Fujisawa T (2000) Expression and evolutionary conservation of nanos‐related genes in Hydra. Dev Genes Evol 210: 591–602 [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP (1995) Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756 [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA (2005) Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Current Biol 15: 284–294 [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13: 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP (2001) Drosophila Brain Tumor is a translational repressor. Genes Dev 15: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G (1999) nos‐1 and nos‐2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans . Development 126: 4861–4871 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y (2010) NANOS2 interacts with the CCR4‐NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci USA 107: 3594–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y (2012) Interaction between NANOS2 and the CCR4‐NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE 7: e33558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Simonelig M, Wahle E (2014) Deadenylation of mRNA by the CCR4‐NOT complex in Drosophila: molecular and developmental aspects. Front Genet 5: 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E (2010) Subunits of the Drosophila CCR4‐NOT complex and their roles in mRNA deadenylation. RNA 16: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y (2003) Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann C, Brumbaugh J, Coon JJ, Goldstrohm AC (2012) Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287: 36370–36383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrotti AC, Wharton RP (2000) Nanos interacts with cup in the female germline of Drosophila . Development 127: 5225–5232 [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R (1991) Nanos is the localized posterior determinant in Drosophila . Cell 66: 637–647 [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM (2002) Modular recognition of RNA by a human pumilio‐homology domain. Cell 110: 501–512 [DOI] [PubMed] [Google Scholar]
- Weidmann CA, Goldstrohm AC (2012) Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol 32: 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y (1998) The Pumilio RNA‐binding domain is also a translational regulator. Mol Cell 1: 863–872 [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967 [DOI] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S (1997) Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 124: 3015–3023 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R (1997) The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA‐binding proteins. RNA 3: 1421–1433 [PMC free article] [PubMed] [Google Scholar]
- Zekri L, Kuzuoğlu‐Öztürk D, Izaurralde E (2013) GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 32: 1052–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7
Source Data for Figure 8


