ABSTRACT
Mammalian cells express hundreds of intron-encoded box H/ACA RNAs which fold into a common hairpin-hinge-hairpin-tail structure, interact with 4 evolutionarily conserved proteins, dyskerin, Nop10, Nhp2 and Gar1, and function mainly in RNA pseudouridylation. The human telomerase H/ACA RNA (hTR) directs telomeric DNA synthesis and it carries a 5′-terminal domain encompassing the telomeric template sequence. The primary hTR transcript is synthesized from an independent gene by RNA polymerase II and undergoes 3′ end processing controlled by the 3′-terminal H/ACA domain. The apical stem-loop of the 3′ hairpin of hTR carries a unique biogenesis-promoting element, the BIO motif that promotes hTR processing and RNP assembly. AluACA RNAs represent a distinct class of human H/ACA RNAs; they are processed from intronic Alu repetitive sequences. As compared to canonical H/ACA RNAs, the AluACA RNAs carry unusually short or long 5′ hairpins and generally, they accumulate at low levels. Here, we demonstrate that the suboptimal 5′ hairpins are responsible for the weak expression of AluACA RNAs. We also show that AluACA RNAs frequently carry a processing/stabilization element that is structurally and functionally indistinguishable from the hTR BIO motif. Both hTR and AluACA biogenesis-promoting elements are located in the terminal stem-loop of the 3′-terminal H/ACA hairpin, they show perfect structural conservation and are functionally interchangeable in in vivo RNA processing reactions. Our results demonstrate that the BIO motif, instead of being confined to hTR, is a more general H/ACA RNP biogenesis-facilitating element that can also promote processing/assembly of intron-encoded AluACA RNPs.
KEYWORDS: AluACA RNAs, Box H/ACA RNAs, H/ACA RNP biogenesis, H/ACA RNA processing, intronic RNA processing, telomerase RNA
Introduction
The nuclei of human cells contain several hundreds of small nuclear RNAs (snRNAs) which, in the forms of small nuclear ribonucleoproteins (snRNPs), function in all steps of nuclear gene expression.1 Box H/ACA RNPs represent an abundant and evolutionarily conserved group of snRNPs.2-4 The major function of H/ACA RNPs is in site-specific pseudouridylation of rRNAs (rRNAs) and spliceosomal snRNAs, but they also participate in the nucleolytic processing of precursor rRNAs, telomeric DNA synthesis and probably, in other processes.4-9
Human H/ACA RNAs are typically composed of two 55- to 70-nucleotide- (nt)-long hairpins which are connected and followed by short single-stranded hinge and tail sequences carrying the conserved H (AgAnnA) and ACA boxes, respectively10-12 (Fig. 1A). Each H/ACA hairpin associates with 4 H/ACA proteins, Nhp2, Nop10, Gar1 and the pseudouridine synthase dyskerin.13-15 The 5′ and/or 3′ hairpins of pseudouridylation guide RNAs carry internal loops containing the bipartite target recognition sequences.5,16 The structure of archaeal pseudouridylation guide RNPs which are most frequently composed of a single-hairpin ACA RNA and 4 proteins, Cbf5 (dyskerin), L7Ae (Nhp2), Nop10 and Gar1 has been solved.17-22 The RNA hairpin is tightly docked to the surface of the Cbf5-Nop10-L7Ae ternary complex. Cbf5 binds to the ACA box, the basal hairpin stem and the 3′ side of the pseudouridylation loop as well as interacts with Nop10 that, in turn, provides a docking surface for L7Ae. Finally, L7Ae specifically interacts with a conserved RNA element, the kink-turn (K-turn or K-loop) that is composed of shared A-G and G-A base pairs and a flipped-out uridine in the terminal stem-loop of the RNA hairpin.23,24 The target recognition sequences base-pair with the target RNA to position the substrate uridine at the catalytic center of the pseudouridine synthase Cbf5.25
Figure 1.
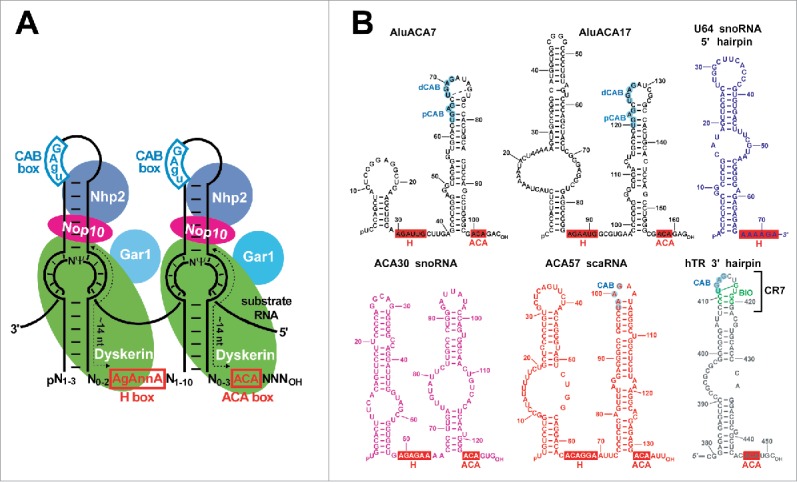
Structure of H/ACA RNAs and RNPs. (A) Schematic structure of eukaryotic double hairpin H/ACA RNPs. The consensus sequences of the conserved H and ACA boxes are highlighted in red boxes. The consensus CAB box sequences are in blue. Usually, the target uridine (Ψ) selected for pseudouridylation is separated by 13–15 nucleotides from the H and ACA box. Arrangement of the associated H/ACA RNP proteins dyskerin, Gar1, Nop10 and Nhp2 is indicated. (B) Computer-predicted secondary structures of human H/ACA RNAs used in this study. The AluACA7, AluACA17 (black), U64 (blue), ACA30 (pink), ACA57 (red) and telomerase RNA (hTR, gray) sequences are indicated in different colors. The conserved H and ACA motifs are in red boxes. The CAB boxes of ACA57 and hTR and the distal and proximal CAB boxes (dCAB and pCAB) of AluACA RNAs are highlighted in blue circles.
Eukaryotic double-hairpin H/ACA RNAs lack K-turns (K-loops) and the eukaryotic equivalent of L7Ae, Nhp2 lacks K-turn-binding specificity. It binds to the terminal loops of the H/ACA hairpins after formation of a complex with dyskerin, Nop10 and the Naf1 assembly factor.14,26-28 Gar1, that replaces Naf1 in the mature RNP, binds to the catalytic domain of both archaeal Cbf5 and eukaryotic dyskerin opposite to the RNA surface. Gar1 is essential for efficient pseudouridine synthesis, but dispensable for RNP accumulation.22,29-32 Eukaryotic H/ACA RNPs directing pseudouridylation of rRNAs and snRNAs accumulate in the nucleolus (small nucleolar RNPs, snoRNPs) and the Cajal body (CB) (small CB RNPs, scaRNPs), respectively.33 The terminal loops of H/ACA scaRNA hairpins carry the CB localization signal, the CAB box (consensus ugAG), which target scaRNPs into the CB through binding Wdr79 (also known as Wrap53).34-37
Human H/ACA RNAs are processed from spliced and debranched pre-mRNA introns by exonucleolytic activities.38 Assembly of precursor intronic H/ACA RNPs occurs in an RNA polymerase II- (RNAPII)-dependent manner already during pre-mRNA elongation and it is supported by several assembly factors.2,4,26,39-42 The associated RNP proteins protect the H/ACA RNA sequences from the processing exonucleases and thereby, delineate the termini of the mature RNA.
Telomeric DNA is synthesized by the telomerase H/ACA scaRNP.43 The 5′ half of human telomerase RNA (hTR) carries the telomeric template sequence and interacts with the telomerase reverse transcriptase. The 3′ half of hTR folds into an H/ACA RNA structure and interacts with the 4 H/ACA proteins and the Wdr79 CB-localization protein.7,44,45 The H/ACA domain is required for accumulation, proper localization and function of telomerase and its malfunction can lead to improper telomere synthesis and various diseases.46 Instead of being processed from a pre-mRNA intron, hTR is transcribed from its independent gene by RNAPII. The 5′ end of hTR is defined by RNAPII transcription initiation and it is protected by the co-transcriptionally added 7-methylguanosine cap that is later converted into trimethylguanosine. The mature 3′ terminus of hTR is formed by post-transcriptional processing of the primary hTR transcript.47,48 The conserved terminal stem-loop region (CR7) of the 3′ hairpin of hTR encompasses 2 functionally distinct, but structurally overlapping elements, the CB localization signal (CAB box) that binds Wdr79 and the hTR-specific biogenesis-promoting element, the BIO motif, that is required for efficient 3′ end processing and H/ACA RNP assembly34,49-52 (Fig. 1B).
We have recently discovered a new, abundant group of human H/ACA RNAs, called the AluACA RNAs, which are processed from the right arms of intronic Alu repetitive sequences.8 Mature AluACA RNAs interact with the 4 H/ACA core proteins and the CB protein Wdr79. The function of AluACA RNPs remains unknown. As compared to canonical intron-encoded H/ACA sno/scaRNPs, the AluACA RNAs carry unusually short or long 5′ hairpins which apparently lack functional Nhp2 docking sites and therefore, are not expected to support efficient H/ACA RNP assembly. In great accordance with this, human AluACA RNPs, in general, accumulate at very low levels. However, in contrast to their aberrantly sized 5′ hairpins, some AluACA RNPs show unexpectedly efficient cellular accumulation. In this study, we have investigated the cis-acting RNA elements controlling the accumulation of AluACA RNPs. We demonstrate that human AluACA RNAs with reasonable expression carry an RNA processing/stabilization element that is structurally and functionally indistinguishable from the hTR BIO motif promoting hTR processing and telomerase assembly.
Results
Atypical 5′ hairpins account for weak accumulation of AluACA RNAs
Human cells express hundreds of AluACA snRNAs which belong to the family of box H/ACA RNAs, but are processed from intronic Alu repetitive sequences.8 In contrast to the efficiently expressed classical intron-encoded H/ACA snoRNAs and scaRNAs, the Alu-derived AluACA RNAs show weak accumulations, suggesting that they carry putative cis-acting elements modulating their processing efficiency and/or metabolic stability. To dissect the structural elements controlling the accumulation of AluACA RNAs, we investigated the in vivo expression of a series of mutant and chimeric AluACA RNAs containing classical H/ACA RNA sequences. The predicted 2-dimensional structures of human AluACA and traditional H/ACA RNAs used in this study are shown in Fig. 1B. The mutant and chimeric test AluACA RNA genes were inserted into the second intron of the human β-globin gene that had been placed under the control of the cytomegalovirus (CMV) promoter in the pCMV-globin expression plasmid (Fig. 2A).33,38 Upon transfection into HeLa cells, accumulation of the processed intronic RNAs and the spliced globin host mRNA was monitored by RNase A/T1 protection analysis with sequence-specific antisense RNA probes.
Figure 2.
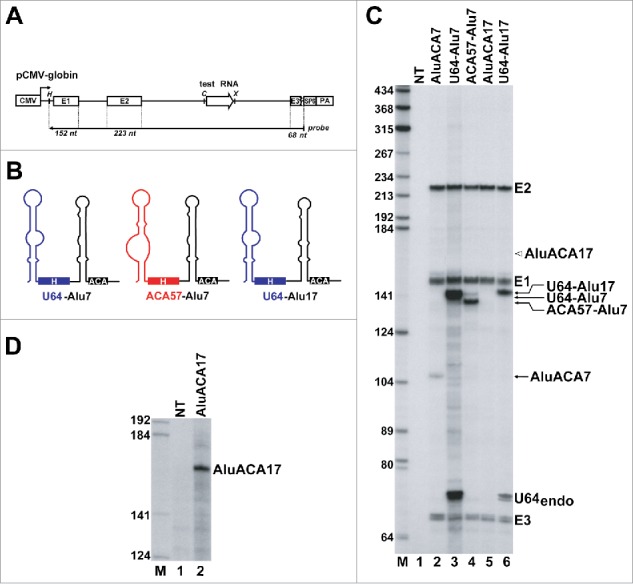
The atypical 5′ hairpins are responsible for inefficient expression of AluACA RNAs. (A) Schematic structure of the pCMV-globin expression vector. The cytomegalovirus (CMV) promoter and polyadenylation site (PA), the β-globin exons (E1, E2 and E3), the intronic test RNA (open arrow) and the SP6 RNA polymerase promoter are shown. The structure of the antisense RNA probe with the expected sizes of the protected fragments are shown. The relevant restriction sites (H, HindIII; C, ClaI; X, XhoI) are indicated. (B) Predicted schematic structures of chimeric H/ACA RNAs. The origin of RNA sequences is indicated by color code (blue, U64 snoRNA; black, AluACA7 and AluACA17; red, ACA57 scaRNA). (C) Processing of transiently expressed globin pre-mRNAs carrying intronic test RNAs. RNAs isolated from HeLa cells non-transfected (NT) or transfected with pCMV-globin expression plasmids carrying the indicated test RNA gene were RNase mapped with sequence-specific antisense RNA probes. The protected probe RNA fragments were separated on a 6% sequencing gel. Bands corresponding to the spliced globin exons (E1, E2 and E3) and the processed test RNAs are indicated. The fragment U64endo was protected by the HeLa endogenous U64 snoRNA and its identity was confirmed by control mappings (data not shown). Open arrow indicates the expected position of AluACA17. Lane M, DNA size markers in nucleotides. (D) Detection of transiently over-expressed AluACA17 RNA. Total RNA from HeLa cells transfected with the pCMV-globin-AluACA17 expression plasmid were mapped with a high specific activity antisense RNA probe complementary to AluACA17. Please note that under the applied mapping conditions, HeLa endogenous AluACA7 (panel C, lane 1) and AluACA17 (panel D, lane 1) remained indiscernible.
The most remarkable feature of AluACA RNAs is that they carry abnormally short or long 5′-terminal hairpin structures8 (Fig. 1B, see AluACA7 and AluACA17). It is noteworthy that AluACA RNAs with long 5′ hairpins are structurally reminiscent of the H/ACA domain of hTR that also features an exceptionally long 5′ hairpin (see below, Fig. 4A). Therefore, we first investigated whether the atypical 5′ hairpins are responsible for the weak accumulation of AluACA RNAs. The short and long 5′ hairpins of AluACA7 and AluACA17, respectively, were replaced for the 5′ hairpins of the efficiently accumulating U64 snoRNA and the ACA57 scaRNA (Fig. 2B). The chimeric test RNAs were transiently expressed within the globin pre-mRNA and their processing was monitored by RNase mapping (Fig. 2C). Although the globin host pre-mRNA was always efficiently synthesized and correctly spliced, see exons 1, 2 and 3 (E1, E2 and E3), accumulation of the processed intronic test RNAs showed a great variation. The expression level of the intronic RNAs was measured by PhosphorImager quantitation and normalized to the accumulation of the corresponding globin exons. Consistent with our previous observations, the wild-type AluACA7 and AluACA17 RNAs were processed with low efficiency from the globin pre-mRNA (lanes 2 and 5). The overexpressed AluACA17 RNA remained even undetectable under standard mapping conditions (lane 5); its accumulation was confirmed by mapping with a high specific activity probe (Fig. 2D, lane 2). In contrast, the U64-Alu7, ACA57-Alu7 and U64-Alu17 chimeric AluACA RNAs carrying the 5′ hairpins of U64 or ACA57 accumulated with about 50-, 20- and 100-times higher levels than their parental wild-type AluACA RNAs (lanes 3, 4 and 6). These results confirmed the idea that the unconventional 5′-terminal hairpins are responsible for the poor accumulation of human AluACA7 and AluACA17 RNAs.
Figure 4.
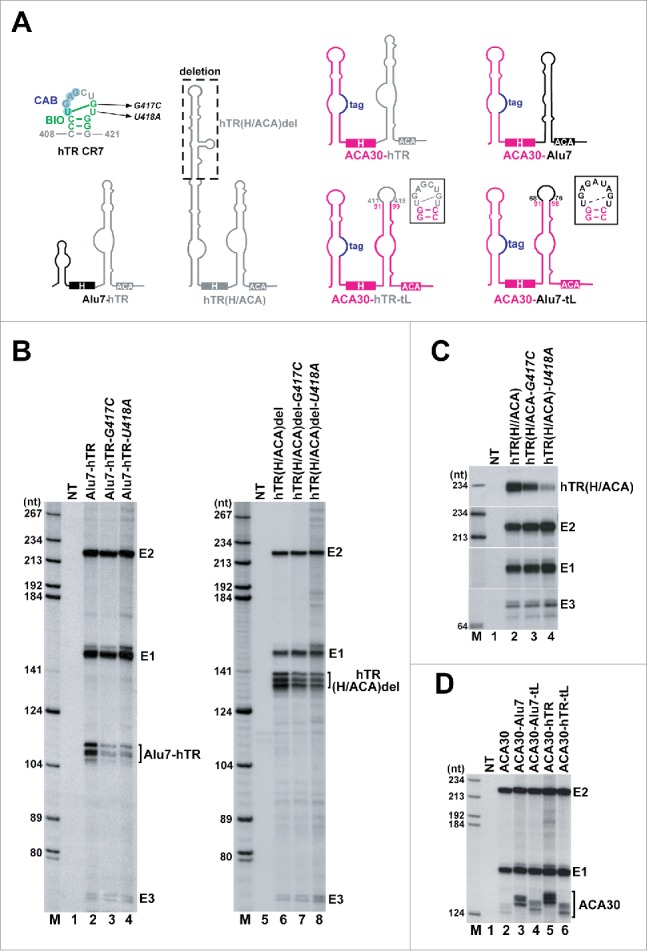
The BIO motif efficiently promotes accumulation of intronic H/ACA RNAs with suboptimal 5′ hairpins. (A) Schematic structures of artificial intronic H/ACA RNAs. The distal part of the unusually long 5′ hairpin of the H/ACA domain of hTR was deleted. Nucleotide alterations introduced into the hTR BIO motif are indicated. The predicted structures of the apical stem-loops of the 3′ hairpins of ACA30-hTR-tL and ACA30-Alu7-tL RNAs are shown. To distinguish between HeLa endogenous and transiently expressed ACA30 snoRNAs, a short, neutral sequence alteration (tag, indicated in blue) was introduced into the ectopically expressed ACA30 snoRNA. Other details are identical to the legends to Fig. 3. (B-D) Transient expression of artificial intronic H/ACA RNAs. The observed heterogeneity of the protected probe fragments represent RNase mapping artifacts. For other details, see the legends to Fig. 2.
The 3′ hairpins of AluACA7 and telomerase RNAs share a common element promoting in vivo RNA accumulation
Nhp2 binds to the terminal loop of the 5′ and 3′ hairpins located usually 24–28 nts upstream of the H and ACA boxes of H/ACA sno/scaRNAs (Fig. 1A, and also see Discussion). The unusually short and long 5′ hairpins of AluACA7 and AluACA17 lack terminal or internal loop structures located about 24–28 nts upstream of their H boxes and therefore, they are not expected to support assembly of the 5′ half of the dimeric H/ACA RNP. Consistent with this, processing of AluACA17 from the transiently expressed globin pre-mRNA was very inefficient (see Fig. 2C, lane 5). However, AluACA7 that carries a very short 5′ hairpin accumulated with unexpectedly high efficiency under the same expression conditions (lane 2). This prompted us to speculate that the 3′-terminal hairpin of AluACA7 might contain a putative stabilization element compensating for the weak accumulation implemented by the aberrant 5′ hairpin. To test this hypothesis, the short 5′ hairpin and the following H box of AluACA7 was fused to the 3′ hairpin-tail region of the ACA30 canonical snoRNA (Fig. 3A). In contrast to wild-type AluACA7, the Alu7-ACA30 chimeric RNA showed no detectable accumulation upon transient expression in HeLa cells, demonstrating that the 3′ hairpin of ACA30 cannot support AluACA7 accumulation (Fig. 3B, lanes 2 and 3). The same results were obtained when the 3′ hairpins of the U64 snoRNA and the ACA57 scaRNA were fused to the 5′ hairpin of AluACA7 (Fig. 3C, lane 3, and data not shown). These observations are consistent with the idea that the 3′ hairpin of AluACA7 carries a stabilization element that is missing from the 3′ hairpins of conventional intronic H/ACA sno/scaRNAs.
Figure 3.
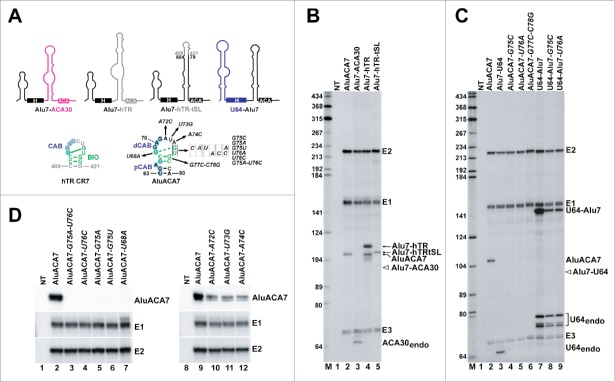
Identification and functional characterization of the BIO motif in the terminal stem-loop of the 3′ hairpin of AluACA7. (A) Schematic structures of chimeric Alu7-ACA30, Alu7-hTR, Alu7-hTR-tSL and U64-Alu7 RNAs. The origin of RNA sequences is indicated by color code (black, AluACA7 RNA; pink, ACA30 snoRNA; gray, hTR; blue U64 snoRNA). The two dimensional structures of the CR7 domain of hTR and the corresponding region of AluACA7 are shown. The BIO motif nucleotides are highlighted in green and the CAB box (CAB, pCAB and dCAB) nucleotides are in blue. The continuous and dashed green lines connecting the first U and penultimate G residues of the terminal loops indicate experimentally confirmed and predicted wobble base pairs, respectively.49 Nucleotide alterations introduced into AluACA7 are indicated. (B-D) In vivo accumulation of mutant and chimeric intron-encoded AluACA RNAs processed from transiently expressed globin pre-mRNAs. Total RNAs extracted from HeLa cells transfected with the indicated pCMV-globin expression constructs (see above the lanes) were analyzed by RNase A/T1 mappings with sequence-specific antisense probes. The protected RNA fragments were analyzed on 6% sequencing gels. Positions of the spliced globin exons (E1, E2 and E3) and the processed intronic RNAs are indicated on the right. The expected positions of Alu7-ACA30 and Alu7-U64 RNAs are indicated by open arrows. NT, control mapping with RNAs from non-transfected cells. Lanes M, size markers in nucleotides.
We noticed that the terminal stem-loop of the 3′ hairpin of AluACA7 is structurally highly reminiscent of the evolutionarily conserved CR7 domain of hTR that encompasses the CB localization signal sequence, the CAB box,34 and the hTR-specific processing/accumulation element, the BIO motif49,51 (Fig. 3A). In the AluACA7 terminal stem-loop, the left side of the loop contains the distal CAB box (dCAB, U68-G71). The first dCAB nucleotide, U68, has the potential to form a wobble base-pair with the penultimate G75 loop nucleotide, leaving the last loop nucleotide U76 unpaired, as is has been experimentally defined for the hTR BIO motif.49 Moreover, the terminal loop of AluACA7 is closed by C-G and G-C base-pairs that have been found to be important for efficient hTR accumulation.49 First we tested whether the 3′ hairpin of hTR is able to support accumulation of an H/ACA RNA with aberrantly short 5′ hairpin. To this end, the short 5′ hairpin and the following H box of AluACA7 was fused to the 3′-terminal hairpin region of hTR (Fig. 3A). The resulting Alu7-hTR composite RNA efficiently accumulated in transfected HeLa cells (Fig. 3B, lane 4). Please note that the multiple probe fragments protected by Alu7-hTR represent RNase A/T1 mapping artifacts which were reproducibly detected in hTR 3′ end mapping reactions. Moreover, the C408-G421 terminal stem-loop containing the CR7 domain of hTR was able to functionally replace the terminal stem-loop (G66-C78) of AluACA7 and to support accumulation of the chimeric Alu7-hTR-tSL RNA at the level of the wild-type AluACA7 (lane 5). These results strongly supported the idea that the terminal stem-loop regions of the 3′ hairpins of hTR and AluACA7 share a common RNA element, the BIO motif, that promotes processing and/or accumulation of these RNAs.
To further demonstrate its significance, we introduced nucleotide alterations into the predicted BIO motif of AluACA7 and tested the expression of the mutant RNAs (Fig. 3A). Replacement of the G75 residue that is predicted to form a wobble base-pair with U68 for a C (G75C), A (G75A) or U (G75U) abolished accumulation of the mutant AluACA7 RNAs (Fig. 3C, lane 4; Fig. 3D, lanes 5 and 6). Likewise, substitution of the bulged U76 with an A (U76A) or C (U76C) residue obliterated accumulation of the mutant AluACA7-G76A and AluACA7-U76C RNAs (Fig. 3C, lane 5 and Fig. 3D, lane 4). As expected, the double mutant AluACA7-G75A-U76C RNA also failed to accumulate upon transient expression in the globin pre-mRNA (Fig. 3D, lane 3). Likewise, swapping the first dCAB residue U68 for an A disrupted accumulation of AluACA7-U68A (lane 7). Finally, disruption of the G66C67-G77C78 stem closing the terminal loop also abolished accumulation of the AluACA7-G77C-C78G RNA (Fig. 3C, lane 6). In contrast, alteration of the loop nucleotides A72, U73 or A74 separating the distal CAB box from the BIO motif failed to abolish expression of the mutant AluACA-A72C, AluACA-U73G and AluACA-A74C RNAs, although reduced their accumulation by 50% (Fig. 3D, lanes 10, 11 and 12).
The U64-Alu7 artificial RNA composed of the 5′ hairpin of the U64 snoRNA and the 3′ hairpin of AluACA7 showed a very efficient accumulation in transfected HeLa cells (Fig. 2C, lane 3 and Fig. 3C, lane 7). Disruption of the BIO motif of U64-Alu7 through introduction of the G75C or U76A point mutations reduced expression of the mutant U64-Alu7-G75C and U64-Alu7-U76A RNAs by 90%, indicating that the BIO motif of AluACA7 can also promote accumulation of H/ACA RNAs carrying regular 5′ hairpins (Fig. 3C, lanes 7, 8 and 9).
BIO motif differentially contributes to the expression of H/ACA RNAs with standard and aberrantly sized 5′ hairpins
BIO motif mutations abolished expression of AluACA7 that carries an aberrantly short 5′ hairpin (Fig. 3C, lanes 4–6, Fig. 3D), but only reduced accumulation of the U64-Alu7 chimeric RNA encompassing the 5′ hairpin of the U64 canonical snoRNA (Fig. 3C, lanes 7–9). To further investigate the importance of the BIO motif for expression of structurally atypical and conventional H/ACA RNAs, we tested the impacts of BIO motif mutations on the in vivo accumulation of the Alu7-hTR chimeric RNA and the hTR(H/ACA)del RNA that represented only the 3′-terminal H/ACA domain of hTR except that its long natural 5′ hairpin was shortened to conform to the consensus size of H/ACA 5′ hairpins (Fig. 4A). The G417C and U418A BIO motif mutations were introduced into the CR7 domain of the 3′-terminal hTR hairpins of Alu7-hTR and hTR(H/ACA)del. Upon transient expression within the globin pre-mRNA, the processed mutant Alu7-hTR-G417C and Alu7-hTR-U418A RNAs carrying the aberrantly short 5′ hairpin of AluACA7 accumulated with 30% efficiency as compared to the control Alu7-hTR RNA (Fig. 4B, lanes 2–4). In contrast, the BIO motif mutations had less effect (only 30% reduction) on the accumulation of the mutant hTR(H/ACA)del-G417C and hTR(H/ACA)del-U418A RNAs with canonical H/ACA RNA structures (lanes 6–8). Next, we tested the effects of the G417C and U418A BIO motif mutations on the accumulation of the wild-type hTR(H/ACA) RNA that represented the full-length H/ACA domain of hTR with its long natural 5′ hairpin. When expressed in an intronic context, the mutant hTR(H/ACA)-G417C and hTR(H/ACA)-U418A RNAs accumulated with highly reduced efficiency (30% and 10%, respectively) compared to the wild-type hTR(H/ACA) RNA (Fig. 4C, lanes 2–4).
We next investigated whether the BIO motif can promote accumulation of canonical H/ACA RNAs. The natural terminal loop (G92-A98) of the 3′ hairpin of the ACA30 snoRNA was replaced with the U68-U76 and the U411-U418 terminal loop sequences of AluACA7 and hTR, respectively (Fig. 4A). Compared to the wild-type AluACA30 snoRNA, the resulting ACA30-Alu7-tL and ACA30-hTR-tL chimeric RNAs were 50% more efficiently processed from the transiently expressed globin pre-mRNA (Fig. 4D, lanes 2, 4 and 6). Moreover, replacement of the entire 3′ hairpin of the ACA30 with the 3′-terminal hairpin of AluACA7 or hTR largely improved expression of the ACA30-Alu7 and ACA30-hTR composite RNAs which accumulated 3 times more efficiently than the wild-type ACA30 snoRNA (lanes 3 and 5). These results, together with the above observation that BIO motif mutations abolish AluACA7 accumulation, but only partially inhibit U64-Alu7 expression (Fig. 3C), indicate that the BIO motif can promote accumulation of all types of H/ACA RNAs, but it plays a more critical role in the expression of H/ACA RNAs carrying structurally aberrant 5′ hairpins.
The proximal and distal CAB boxes of AluACA7 are not required for RNA accumulation
Finally, we investigated the importance of the proximal and distal CAB boxes (pCAB and dCAB) of AluACA7 for RNA accumulation. We had observed earlier that disruption of the pCAB and dCAB sequences inhibited Wdr79 binding and unexpectedly, also abolished the accumulation of AluACA7.8 This suggested that the 2 closely spaced CAB boxes support AluACA7 accumulation through Wdr79 recruitment. However, demonstration that the distal portion of the 3′ hairpin of AluACA7 encompassing the dCAB and pCAB boxes also accommodates an essential, structurally overlapping RNA processing/accumulation signal, the BIO motif, shed new lights on our previous observations. In the ugAG consensus CAB sequence of H/ACA scaRNAs and hTR, the highly conserved A3 and G4 residues are absolutely required for Wdr79 binding and CB localization.34,35 Consistent with our previous observations,8 simultaneous replacement of the A3 and G4 (dm1) or the G4 (dm3) residues for U and C residues in the pCAB and dCAB boxes of AluACA7 abolished accumulation of the mutant AluACA-dm1 and AluACA7-dm3 RNAs (Fig. 5A, lanes 3 and 5). However, besides disrupting Wdr79-binding,8 the dm1 and dm3 mutations are also predicted to destroy the functional integrity of the AluACA7 BIO motif through preventing formation of the essential G66-C67/G77-C78 terminal stem.49 In contrast, simultaneous alteration of the A3 residues in the dCAB and pCAB of AluACA7, while abolished Wdr79 binding as expected (Fig. 5B), failed to eliminate accumulation of the mutant AluACA-dm2 RNA (Fig 5A, lane 4). These results confirmed that the 2 closely spaced CAB boxes of AluACA7 are essential for Wdr79 binding, but in contrast to our previous conclusion, they demonstrated that recruitment of Wdr79 is not required for RNA processing and accumulation. As expected, disruption of the Wdr79-binding capacity and the BIO motif integrity of the U64-Alu7 and ACA57-Alu7 chimeric RNAs by replacement of the G4 residues in their pCAB and dCAB motifs with C residues slightly reduced (by 20%) the accumulations of the mutant U64-Alu7-dm3 and ACA57-Alu7-dm3 (compare lanes 7 and 8 and lanes 10 and 11). The same dm3 mutations had no detectable effect on the accumulation of the mutant U64-Alu17-dm3 RNA since wild-type AluACA17 lacks functional BIO motif (lane 12 and 13). Likewise, alteration of the A3 residues in the pCAB and dCAB of U64-Alu7 which abolished Wdr79 binding had no effect on the processing and accumulation of U64-Alu-dm2 (lane 9). These results confirmed the conclusion that the terminal stem-loop of the 3′ hairpin of AluACA7 encompasses 2 structurally overlapping but functionally distinct elements which direct Wdr79 binding and promote RNA processing and accumulation.
Figure 5.
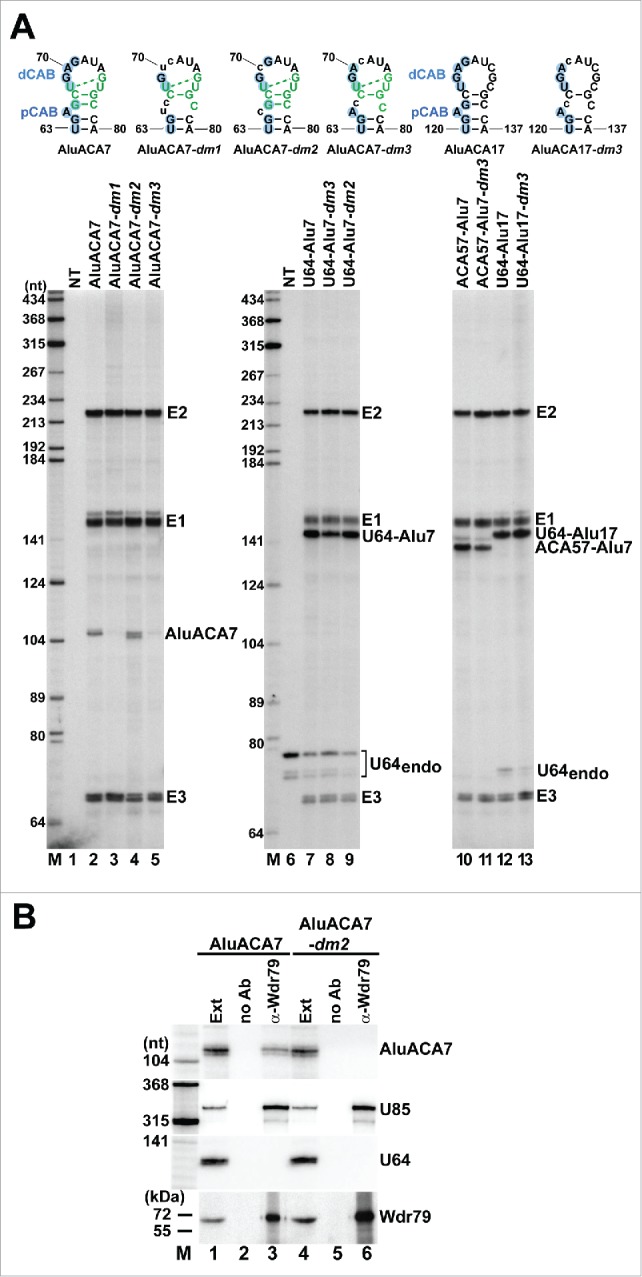
The CAB box function is not required for AluACA7 accumulation. (A) RNase mappings. The dm1, dm2 and dm3 CAB box mutations introduced into the proximal and distal CAB (pCAB and dCAB) sequences of AluACA7 and AluACA17 and their predicted consequences on the BIO motif structure are shown. For other details, see the legends to Fig. 3. (B) Wdr79 fails to interact with AluACA7-dm2. Wdr79 was immunoprecipitated with a specific antibody (α-Wdr79) from total extracts (Ext) prepared from HeLa cells transiently overexpressing AluACA7 or AluACA7-dm2 as indicated above the lanes. Co-IP of AluACA7 and AluACA7-dm2 was monitored by RNase mappings. Interaction of Wdr79 with the endogenous U85 H/ACA scaRNA (positive control) and the U64 snoRNA (negative control) was also tested. Recovery of Wdr79 was confirmed by western blot analysis. Lane no Ab, control IP performed without antibody.
Discussion
The apical stem-loop of the 3′-terminal hairpin of hTR carries the evolutionarily conserved CR7 domain that includes the CB localization signal, the CAB box,35 and the telomerase biogenesis-promoting element, the BIO motif.49-51 While the CAB box is present in all CB-specific H/ACA scaRNAs, the BIO motif has been considered as a telomerase-specific element that promotes 3′ end processing and assembly of the RNAPII-transcribed hTR precursor with H/ACA proteins. In this study, we have demonstrated that the 3′ hairpins of human intronic AluACA RNAs frequently carry a BIO motif that efficiently promotes accumulation of AluACA RNAs with aberrantly short or long 5′ hairpins.
Since AluACA RNAs are processed from intronic Alu repetitive sequences, they represent a distinct and structurally closely related group of H/ACA RNAs.8 A remarkable structural feature of mature AluACA RNAs is that compared to canonical intronic H/ACA sno/scaRNAs, they carry unusually short or long 5′-terminal hairpins. HeLa cells express several hundreds of different AluACA RNAs, but they accumulate at very low levels in general.8 Expression of artificial chimeric RNAs composed of AluACA and canonical H/ACA RNA elements demonstrated that the suboptimal 5′ hairpins account primarily for the poor accumulation of AluACA RNAs and that the BIO motif can significantly improve the inherently weak expression of AluACA RNAs. We detected perfectly conserved BIO motifs in more than 12% of the known 348 human AluACA RNAs. Usually, the BIO motif-containing RNAs were represented by high number of reads in our AluACA deep sequencing datasets,8 providing further support to the idea that the BIO motif promotes H/ACA RNA accumulation in vivo.
It has been proposed that the biogenesis-promoting element of hTR is composed of the C415, U416, G417 and U418 loop nucleotides following the CAB box.50,51 A detailed structural and functional analysis of the CR7 signal supporting hTR processing and CB-specific accumulation revealed that the hTR BIO motif comprises the terminal stem with 2 C-G bps at the top (C409-C410/G419-G420), a U.G wobble base-pair formed by the first U (U411) and the second-to-last G (G417) loop nucleotides and the unpaired U418 in the last position of the terminal loop49 (see Fig. 1B). The terminal stem-loop of the 3′ hairpin of 43 AluACA RNAs including AluACA7 carries a perfect copy of the experimentally defined hTR BIO motif. The C415 and U416 loop nucleotides proposed to be part of the hTR BIO box showed no conservation in AluACA RNAs. Consistently, alteration of the A72, U73 and A74 loop residues located between the dCAB and the BIO motif of AluACA7 only moderately affected AluACA7 accumulation (Fig. 3D).
The precise molecular role of the BIO motif played in stimulation of H/ACA RNP expression is uncertain. Disruption of the hTR BIO motif inhibited in vivo accumulation of mature-sized hTR, but increased the level of 3′-terminally extended primary hTR transcripts, suggesting that the BIO motif enhances hTR 3′ end processing.49 On the other hand, in in vitro RNP reconstitution experiments, the H/ACA core proteins dyskerin, Nop10 and Nhp2, while showed little affinity to the 3′ hairpins of canonical H/ACA snoRNAs, efficiently associated with the 3′-terminal hairpin of hTR in a BIO motif-dependent manner, leading to the conclusion that the BIO motif stimulates H/ACA RNP assembly.51,53 The BIO motif-promoted H/ACA RNP assembly might also stimulate hTR 3′ end processing. When incubated in nuclear extracts, H/ACA snoRNA transcripts carrying intronic flanking sequences are rapidly processed into mature-sized snoRNAs, but substrate snoRNAs with altered H or ACA boxes undergo only slow, non-specific degradation, suggesting that H/ACA RNP assembly promotes in vitro snoRNA processing38 (our unpublished data). This would also explain the observed overaccumulation of transiently expressed 3′-terminally extended hTR transcripts carrying mutant BIO motifs.49
During in vivo H/ACA RNP biogenesis, the preassembled H/ACA core complex containing dyskerin, Nop10, Nhp2 and the assembly factor Naf1 is co-transcriptionally and coordinately loaded onto both hairpins of the nascent H/ACA RNA.26,39,40,51 To promote concerted 2-hairpin H/ACA RNP assembly, the preassembled H/ACA core scaffold has been proposed to contain 2 sets of H/ACA core proteins.51 Specific recognition of nascent 2-hairpin H/ACA RNAs by the preassembled dimeric H/ACA core complex might have an important role in selection of bona-fide H/ACA RNA sequences in the complex RNAPII transcriptome. Systematic computer folding of experimentally validated human H/ACA RNA sequences available in the LBME SnoRNA Database (https://www-snorna.biotoul.fr) confirmed that H/ACA RNAs carry fairly uniformly sized 5′- and 3′-terminal hairpins. We found that the H and ACA boxes are usually separated from the terminal loops of the preceding hairpins by 24–28 nts (Fig. 6). The very few RNAs with extended hairpins, e.g. E2 RNA, carry internal loops positioned 24–26 nts upstream of the H or ACA box. The H/ACA hairpins are tightly docked to the dyskerin/Nop10/Nhp2 protein scaffold that may function as a molecular ruler measuring the distances between the dyskerin-bound H or ACA box and the Nhp2-bound terminal loop. Consistently, manipulation the 3′-terminal hairpin length greatly inhibited accumulation of hTR.51 Thus, we propose that the H/ACA hairpin length is an important structural hallmark that facilitates recognition of bone fide H/ACA RNAs in the complex eukaryotic transcriptomes.
Figure 6.
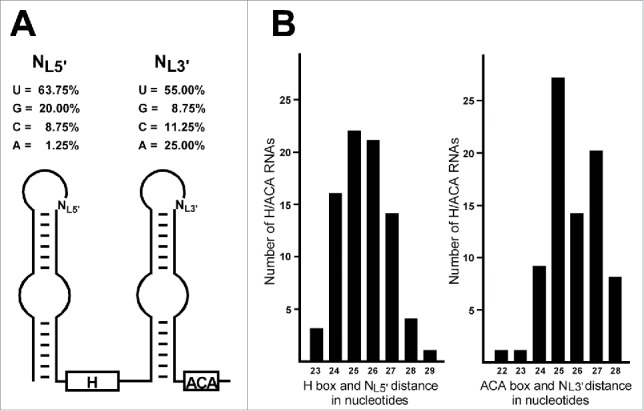
Structural properties of human intron-encoded H/ACA RNAs. (A) Uridine is overrepresented at the last positions of the terminal loops of the 5′ and 3′ hairpins of human H/ACA RNAs. Schematic structure of box H/ACA RNAs and the last unpaired terminal loop nucleotides (NL5′ and NL3′) are shown. Human H/ACA RNA sequences were collected from the LBME snoRNA database (https://www-snorna.biotoul.fr) and they were folded by the mfold web server (http://unafold.rna.albany.edu/?q=mfold/download-mfold). (B) Distance conservation between the terminal loops of 5′ and 3′ hairpins and the H/ACA boxes. Bar graphs indicate the frequencies of the number of nucleotides separating the last unpaired loop nucleotides (NL5′ and NL3′) from the H or ACA boxes.
It seems that the BIO motif specifically promotes assembly of H/ACA RNPs lacking functional 5′-terminal Nhp2 docking sites. Indeed, the BIO motif occurs in the 3′ hairpins of H/ACA RNAs which carry atypical 5′ hairpins and it has only a moderate stimulatory effect on the accumulation of canonical H/ACA sno/scaRNAs. Apart from Naf1, the hTR-associated H/ACA core complex lacks known H/ACA assembly factors, arguing against the possibility that the BIO motif recruits trans-acting H/ACA assembly factor(s) to hTR.51 We rather prefer the idea that the BIO motif is in fact an optimal Nhp2 binding site and its strong interaction with Nhp2 could stably anchor the dimeric dyskerin/Nop10/Nhp2 protein scaffold to the 3′-terminal H/ACA hairpin. Formation of an unusually stable 3′ hairpin H/ACA RNP could secure association of other part of the dimeric H/ACA core complex with the structurally aberrant 5′ hairpin even in the absence of a functional Nhp2 docking site. In the BIO motifs of AluACA7 and hTR, the unpaired U residues in the last position of the terminal loop are essential for in vivo RNA accumulation and they are reminiscent of the universally conserved bulged U in the K-loops/turns of archaeal H/ACA RNAs (Fig. 3).49 The Pro60 residue of L7Ae stacks over the flipped-out U of the archaeal H/ACA K-loops and it plays a central role in organizing the active center of L7Ae. This proline is universally conserved in yeast and vertebrate Nhp2 proteins and most probably, it has the same role in recognition of archaeal, yeast and human H/ACA RNAs.54 Lending support to this notion, the Feigon lab reported that 73% (27/37) yeast H/ACA snoRNAs have a U located 10 or 11 nts upstream from the top of the pseudouridylation loop.54 Closer inspection of the computer-predicted structures of human H/ACA RNAs revealed that U is highly overrepresented in the last position of the terminal loops of the 5′ (64%) and 3′ (55%) hairpins, indicating that human Nhp2 preferentially interacts with loop sequences terminated with an unpaired U (Fig. 6A). Thus, we propose that the molecular function of the BIO motif is in positioning the conserved terminal loop uridine into a structural context optimal for Nhp2 binding.
Materials and methods
General procedures
Unless described otherwise, all DNA, RNA and oligonucleotide manipulations, protein IP experiments were performed according to standard laboratory protocols. Oligodeoxynucleotides were purchased from Eurofins MWG. The identity of all expression constructs was verified by sequence analysis. Human HeLa cells were grown as monolayer cultures in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), 50 U/ml penicillin and 50 µg/ml streptomycin (Invitrogen). Transient transfection of HeLa cells was performed by using jetPRIME (Polyplus-transfection SA) transfection reagent according to the instructions of the manufacturer. Anti-Wdr79 antibody (WRAP53 C2) was purchased from Innovagen AB. RNA secondary structure predictions were performed by using the mfold web server (http://unafold.rna.albany.edu/?q=mfold/download-mfold).
Expression constructs
The pCMV-AluACA7, pCMV-AluACA17, pCMV-U64-ACA7 and pCMV-hTR(H/ACA) expression constructs have been described.8,49 To generate pCMV-ACA30, the human ACA30 snoRNA gene together with its 50 and 48 nt-long flanking sequences was PCR-amplified and inserted into the ClaI and XhoI sites in the second intron of the β-globin gene placed under the control of the cytomegalovirus promoter in the pCMV-globin expression vector.33 To distinguish between HeLa endogenous ACA30 and the ectopically expressed ACA30 snoRNAs, the G47-U52 region of the cloned ACA30 RNA was replaced with complementary sequences. For construction of pCMV-ACA57-Alu7, the 5′ half of the ACA57 gene (T1-A69) was fused to the 3′ part of the AluACA7 gene (C36-C105) by using the PCR megaprimer approach.55 The amplified DNA was inserted into the ClaI and XhoI sites of the pCMV-globin expression plasmid. The same approach was used for construction of the pCMV-U64-Alu17, pCMV-Alu7-ACA30, pCMV-Alu7-hTR, pCMV-ACA30-Alu7 and pCMV-ACA30-hTR plasmids; the 5′ halves of the U64 (A1-A72), AluACA7 (T1-G35) and the ACA30 (T1-A64) genes and the 3′ halves of the AluACA17 (G92-G162), ACA30 (A65-G128) and hTR (C379-C451) genes were fused and inserted into the pCMV-globin plasmid. To generate pCMV-Alu7-hTR-tSL, the G66-C78 internal region of AluACA7 was replaced for the C408-G421 fragment of hTR in the pCMV-AluACA7 expression plasmid. To obtain the pCMV-ACA30-Alu7-tL and pCMV-ACA30-hTR-tL expression constructs, in the pCMV-ACA30 plasmid, the G92-A98 internal region of the ACA30 gene was replaced for the T68-T76 and T411-G418 internal sequences of the AluACA7 and hTR genes, respectively. The pCMV-hTR(H/ACA)del plasmid was generated through deletion of the C234-G337 region of the hTR(H/ACA) gene from the pCMV-hTR(H/ACA) plasmid. All expression vectors carrying full-length or chimeric mutant AluACA RNA genes were generated by PCR mutagenesis using appropriately designed PCR primers and the parental “wild-type” expression plasmids as templates.
RNA extraction
About 3 × 106 HeLa cells were washed with phosphate-buffered saline (pH 7.4) and resuspended in 0.7 ml of lysis solution (4M guanidium thiocyanate, 25 mM sodium-citrate, 0.5% N-lauroylsarcosine, 0.1M β-mercaptoethanol, pH 7.4). The cell lysate was supplemented with 40 µl of 3M sodium-acetate (pH 5.0) and extracted twice with 0.6 ml water-saturated phenol and 0.3 ml chloroform/isoamyl alcohol (24:1). RNA was precipitated from the aqueous phase with 0.7 ml isopropanol, collected by centrifugation and precipitated again from 0.3 ml of 0.3 M sodium-acetate (pH 5.0) with an equal volume of isopropanol. RNA was collected and subjected to DNase digestion for 30 min in 0.1 ml of 1x DNase I buffer containing 25U of RNase-free DNase I (Thermo Scientific) and 40 U of RNasin (Promega). After extraction with equal volume of phenol/chloroform/isoamyl alcohol (25:24:1), RNA was precipitated with ethanol, dissolved in water and used for RNase protection analysis.
RNase protection assay
Sequence-specific antisense RNA probes were synthesized in the presence of [α-P32]CTP (specific activity 220 Ci/mmol) with SP6 RNA polymerase using the appropriate HindIII-digested pCMV-globin expression plasmids as templates. For synthesis of high specific activity antisense AluACA17 RNA probe, the pCMV-globin-AluACA17 expression plasmid was linearized by ClaI and used as a template for SP6 transcription in the presence of [α-P32]CTP (specific activity 2700 Ci/mmol). Each probe was purified on a 6% sequencing gel and the purified antisense RNA corresponding to 50,000 cpm was hybridized with 10 μg test RNA in 10 µl of hybridization buffer containing 40 mM PIPES, pH 6.7, 400 mM NaCl, 1 mM EDTA, 80% formamide at 50°C for 6–12 hours. Single-stranded RNA was digested in 100 µl RNase solution containing 10 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5 mM EDTA, 100 mM LiCl, 40 µg/ml RNase A and 100 U/ml RNase T1 for 1h at room temperature. RNase digestion was stopped by addition of 2 µl of 10% SDS and 20 µg of proteinase K. After incubation at 37°C for 30 min, RNA was purified by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and recovered by ethanol precipitation. The protected RNAs were analyzed on 6% sequencing gels and visualized by autoradiography.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Our work was supported by grants from la Fondation pour la Recherche Médicale and Agence Nationale de la Recherche.
References
- 1. Yu Y-T, Scharl EC, Smith CM, Steitz JA The growing world of small nuclear ribonucleoproteins In: Gesteland R.F., Cech T.R., Atkins J.F. , eds. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1999; 37:487-524; http://dx.doi.org/ 10.1101/087969589.37.487 [DOI] [Google Scholar]
- 2.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell 2010; 37:597-606; PMID:20227365; http://dx.doi.org 10.1016/j.molcel.2010.01.032 [DOI] [PubMed] [Google Scholar]
- 3.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; http://dx.doi.org 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 4.Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine–pseudouridylation. RNA Biol 2014; 11:1483-94; PMID:25590339; http://dx.doi.org 10.4161/15476286.2014.972855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; http://dx.doi.org 10.1016/S0092-8674(00)80263-9 [DOI] [PubMed] [Google Scholar]
- 6.Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J 2009; 28:1260-70; PMID:19322192; http://dx.doi.org 10.1038/emboj.2009.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 1999; 19:567-76; PMID:9858580; http://dx.doi.org 10.1128/MCB.19.1.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jády BE, Ketele A, Kiss T. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes Dev 2012; 26:1897-910; PMID:22892240; http://dx.doi.org9160748 10.1101/gad.197467.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997; 89:565-73; PMID:9160748; http://dx.doi.org 10.1016/S0092-8674(00)80238-X [DOI] [PubMed] [Google Scholar]
- 10.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 1997; 11:941-56; PMID:9106664; http://dx.doi.org 10.1101/gad.11.7.941 [DOI] [PubMed] [Google Scholar]
- 11.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 1996; 86:823-34; PMID:8797828; http://dx.doi.org 10.1016/S0092-8674(00)80156-7 [DOI] [PubMed] [Google Scholar]
- 12.Kiss T, Bortolin ML, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol 1996; 16:1391-400; PMID:8657112; http://dx.doi.org 10.1128/MCB.16.4.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 2010; 30:2775-86; PMID:20351177; http://dx.doi.org 10.1128/MCB.00151-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Duan J, Li D, Yang B, Dong M, Ye K. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 2011; 25:2409-21; PMID:22085967; http://dx.doi.org 10.1101/gad.175299.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Duan J, Li D, Ma S, Ye K. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J 2011; 30:5010-20; PMID:22117216; http://dx.doi.org 10.1038/emboj.2011.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, Loria JP, Moore PB. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Mol Cell 2007; 26:205-15; PMID:17466623; http://dx.doi.org 10.1016/j.molcel.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 17.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 2006; 443:302-7; PMID:16943774; http://dx.doi.org 10.1038/nature05151 [DOI] [PubMed] [Google Scholar]
- 18.Hamma T, Reichow SL, Varani G, Ferre-D'Amare AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol 2005; 12:1101-7; PMID:16286935; http://dx.doi.org 10.1038/nsmb1036 [DOI] [PubMed] [Google Scholar]
- 19.Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 2009; 16:740-6; PMID:19478803; http://dx.doi.org 10.1038/nsmb.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res 2006; 34:826-39; PMID:16456033; http://dx.doi.org 10.1093/nar/gkj482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 2006; 21:249-60; PMID:16427014; http://dx.doi.org 10.1016/j.molcel.2005.11.017 [DOI] [PubMed] [Google Scholar]
- 22.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 2009; 34:427-39; PMID:19481523; http://dx.doi.org 10.1016/j.molcel.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 23.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 2003; 31:869-77; PMID:12560482; http://dx.doi.org 10.1093/nar/gkg175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daldrop P, Lilley DM. The plasticity of a structural motif in RNA: structural polymorphism of a kink turn as a function of its environment. RNA 2013; 19:357-64; PMID:23325110; http://dx.doi.org18059286 10.1261/rna.036657.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang B, Xue S, Terns RM, Terns MP, Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 2007; 14:1189-95; PMID:18059286; http://dx.doi.org 10.1038/nsmb1336 [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 2004; 23:1857-67; PMID:15044956; http://dx.doi.org 10.1038/sj.emboj.7600181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs RNA New York, NY: 2004; 10:1704-12; PMID:15388873; http://dx.doi.org/11433018 10.1261/rna.7770604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 2001; 29:2733-46; PMID:11433018; http://dx.doi.org 10.1093/nar/29.13.2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev 2005; 19:1238-48; PMID:15870259; http://dx.doi.org 10.1101/gad.1309605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bousquet-Antonelli C, Henry Y, G'Elugne JP, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J 1997; 16:4770-6; PMID:9303321; http://dx.doi.org 10.1093/emboj/16.15.4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 2005; 33:3133-44; PMID:15933208; http://dx.doi.org 10.1093/nar/gki630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Yang L, Gao YQ, Zhao XS. Accurate placement of substrate RNA by Gar1 in H/ACA RNA-guided pseudouridylation. Nucleic Acids Res 2015; 43:7207-16; PMID:26206671; http://dx.doi.org 10.1093/nar/gkv757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 2002; 21:2746-56; PMID:12032087; http://dx.doi.org 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647-52; PMID:14981093; http://dx.doi.org12912925 10.1083/jcb.200310138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J 2003; 22:4283-93; PMID:12912925; http://dx.doi.org 10.1093/emboj/cdg394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47-57; PMID:19285445; http://dx.doi.org 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009; 323:644-8; PMID:19179534; http://dx.doi.org 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev 1995; 9:1411-24; PMID:7797080; http://dx.doi.org 10.1101/gad.9.11.1411 [DOI] [PubMed] [Google Scholar]
- 39.Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 2006; 26:630-42; PMID:16382153; http://dx.doi.org 10.1128/MCB.26.7.2540-2549.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 2006; 173:207-18; PMID:16618814; http://dx.doi.org 10.1083/jcb.200601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 2005; 25:3295-304; PMID:15798213; http://dx.doi.org 10.1128/MCB.25.8.3295-3304.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 2005; 25:5396-403; PMID:15964797; http://dx.doi.org 10.1128/MCB.25.13.5396-5403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev 2015; 29:1095-105; PMID:26063571; http://dx.doi.org 10.1101/gad.263863.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci U S A 2011; 108:20325-32; PMID:21844345; http://dx.doi.org 10.1073/pnas.1100279108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell 2000; 100:503-14; PMID:10721988; http://dx.doi.org 10.1016/S0092-8674(00)80687-X [DOI] [PubMed] [Google Scholar]
- 46.Batista LF, Artandi SE. Understanding telomere diseases through analysis of patient-derived iPS cells. Curr Opin Genet Dev 2013; 23:526-33; PMID:23993228; http://dx.doi.org 10.1016/j.gde.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng CK, Wang HF, Burns AM, Schroeder MR, Gaspari M, Baumann P. Human telomerase RNA processing and quality control. Cell Rep 2015; 13:2232-43; PMID:26628367; http://dx.doi.org 10.1016/j.celrep.2015.10.075 [DOI] [PubMed] [Google Scholar]
- 48.Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. A Polyadenylation-Dependent 3′ End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA. Cell Rep 2015; 13:2244-57; PMID:26628368; http://dx.doi.org 10.1016/j.celrep.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 49.Theimer CA, Jady BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol Cell 2007; 27:869-81; PMID:17889661; http://dx.doi.org 10.1016/j.molcel.2007.07.017 [DOI] [PubMed] [Google Scholar]
- 50.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 2003; 11:1361-72; PMID:12769858; http://dx.doi.org 10.1016/S1097-2765(03)00196-5 [DOI] [PubMed] [Google Scholar]
- 51.Egan ED, Collins K. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol 2012; 32:2428-39; PMID:22527283; http://dx.doi.org 10.1128/MCB.00286-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin-Rivera L, Blasco MA. Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J Biol Chem 2001; 276:5856-65; PMID:11056167; http://dx.doi.org 10.1074/jbc.M008419200 [DOI] [PubMed] [Google Scholar]
- 53.Dragon F, Pogacic V, Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol 2000; 20:3037-48; PMID:10757788; http://dx.doi.org 10.1128/MCB.20.9.3037-3048.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koo BK, Park CJ, Fernandez CF, Chim N, Ding Y, Chanfreau G, Feigon J. Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol 2011; 411:927-42; PMID:21708174; http://dx.doi.org 10.1016/j.jmb.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta AK. Efficient amplification using [megaprimer] by asymmetric polymerase chain reaction. Nucleic Acids Res 1995; 23:4530-1; PMID:7501483; http://dx.doi.org 10.1093/nar/23.21.4530 [DOI] [PMC free article] [PubMed] [Google Scholar]