ABSTRACT
Nucleic acid aptamers are single-stranded DNA or RNA oligonucleotide sequences that bind to a specific target molecule with high affinity and specificity through their ability to adopt 3-dimensional structure in solution. Aptamers have huge potential as targeted therapeutics, diagnostics, delivery agents and as biosensors. However, aptamers composed of natural nucleotide monomers are quickly degraded in vivo and show poor pharmacodynamic properties. To overcome this, chemically-modified nucleic acid aptamers are developed by incorporating modified nucleotides after or during the selection process by Systematic Evolution of Ligands by EXponential enrichment (SELEX). This review will discuss the development of chemically-modified aptamers and provide the pros and cons, and new insights on in vitro aptamer selection strategies by using chemically-modified nucleic acid libraries.
KEYWORDS: Aptamers, chemical antibodies, chemically-modified aptamers, in vitro selection, modified nucleotides, nucleic acid ligands, SELEX
Abbreviations
- SELEX
Systematic Evolution of Ligands by EXponential enrichment
- US FDA
United States Food and Drug Administration
- AMD
Age-related macular degeneration
- VEGF
Vascular endothelial growth factor protein, 2′-NH2, 2′-Amino
- 2′-OH
2′-Hydroxyl
- Kd
Equilibrium dissociation constant
- 2′-OMe
2′-O-Methyl
- Bfgf
Basic fibroblast growth factor
- 2′-F
2′-Fluoro
- PSMA
Prostate specific membrane antigen
- IFN-γ
Interferon-gamma
- KGF
Keratinocyte growth factor
- 4′-S
4′-Thio
- 2′-FANA
2′-Fluroarabino nucleic acid
- HNA
1,5-Anhydro hexitol nucleic acid
- TAR
transactivation responsive element
- TNA
Threose nucleic acid
- LNA
Locked nucleic acid
- SOMAmers
Slow Off-rate Modified Aptamers
Introduction
Nucleic acid aptamer technology has attracted considerable attention in recent years in light of their widespread applications in therapeutic development, targeted drug delivery, bio-sensing and accurate molecular imaging. Aptamers are short single-stranded DNA or RNA oligonucleotides with unique 3-dimensional shape that can bind to their specific target with very high affinity and specificity.1-5 Aptamers are generally developed from a large pool of oligonucleotide libraries containing approximately 1014 members by a reiterative process referred to as SELEX which involves selection, separation and enrichment steps (Fig. 1).6,7 Till now, antibodies have been widely used for target specific molecular recognition.8 However, compared to antibody-based technologies, aptamers may possess a number of advantages including easy laboratory production in vitro effectively eliminating the use of live animals, no batch to batch variation, low or no immunogenicity, freedom to introduce multiple chemistries during synthesis without losing the affinity and specificity, small size that allows faster tissue penetration, ability to reverse target binding interactions using its complementary antidote sequence, significantly longer shelf-life and low cost. In 2004, an aptamer drug Macugen (Pegaptanib Sodium) was approved by United States Food and Drug Administration (US FDA) for the treatment of neovascular age-related macular degeneration (AMD) by targeting vascular endothelial growth factor protein 165 (VEGF165).9,10 Currently, a number of aptamer-based therapeutic candidates are in preclinical development and in different stages of clinical trials.11
Figure 1.
Schematic illustration of the SELEX and post-SELEX methods for developing aptamers.
Typically, aptamers are developed with naturally occurring nucleotides. However, aptamers composed of natural nucleotide monomers are not suitable for theranostic applications as they possess very poor resistance to enzymatic degradation and show decreased binding affinity, rendering poor pharmacokinetic properties. To circumvent these shortcomings, aptamers containing chemically-modified nucleotide analogs with high stability against nucleases are normally used. Early examples of modified aptamers were primarily produced by post-SELEX-based approach. In this process, the aptamers were first isolated using natural RNA or DNA random sequences by SELEX method and then modified as per demand on affinity, stability and functionality. For this purpose, appropriate chemically-modified nucleotides are systematically incorporated into an existing DNA/RNA aptamer during solid-phase oligonucleotide synthesis. Generally, a web-based secondary structure prediction algorithm (e.g. mfold,12 RNAfold13) is used as a tool to assist with the positioning of chemically-modified nucleotides and to truncate the overall size of the selected aptamers during chemical synthesis. Such chemically-fabricated aptamer variants are then tested for binding affinity, and the best candidates are used for further analysis and down-stream applications in vitro and in vivo. Evolution of chemically-modified aptamers would be a far more powerful approach to develop modified aptamers as this could generate aptamers with unique structures and with even shorter libraries.
Evolution of chemically-modified aptamers
A large repertoire of chemically-modified nucleotide analogs with remarkable biophysical properties have been developed in recent years and a few papers have reviewed the use of these chemically-modified nucleotides in the generation of aptamers and nucleic acids with enzymatic activity including DNAzymes and ribozymes.14-18 But, their applicability in de novo evolution of aptamers via SELEX methodologies are rather impeded by poor or lack of enzymatic recognition capabilities. Conventional selection methodologies involve multiple enzymatic steps that are required to amplify and regenerate chemically-modified nucleotide-containing libraries. Some sugar-modified nucleotides are reported to tolerate few commercially available DNA or RNA polymerases,19 making them promising candidates for aptamer selection. However, it is worth mentioning that the level of enzymatic recognition capabilities of the reported modified nucleotides varies depending on the specific chemical modification. Still, the substrate properties of a number of other promising analogs have not been reported which might be due to the lack of enzymatic recognition. One option is to evolve an enzyme specific to the modified nucleotide, and there are reports of successful aptamer selection using engineered enzymes for chemically-modified nucleotides.20,21 An overview of possible strategies that can be applied for selecting chemically-modified aptamers are outlined below (Fig. 2).
Figure 2.
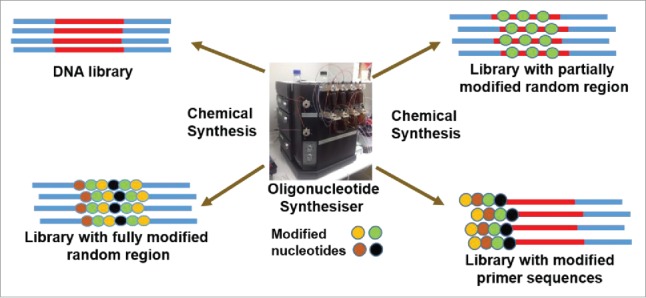
The construction of oligonucleotide libraries for developing chemically modified libraries.
Generation of chemically-modified libraries and selection strategies
Starting oligonucleotide libraries containing the desired chemically-modified nucleotide or a combination of different modifications can be chemically synthesized using an oligonucleotide synthesizer via standard phosphoramidite chemistry. Another approach could be to synthesize normal DNA library which subsequently can be converted to a chemically-modified library by following enzymatic protocols involving PCR and/or in vitro transcription reactions with modified nucleotide triphosphates using the specific polymerase which could also be used in subsequent amplification/regeneration steps of multiple selection rounds. Synthetic chemically-modified libraries can be constructed by incorporating the modified nucleotides in the randomized region or in the primer-binding region.
Library with modified random region
Modified nucleotides can be incorporated as mixmers together with natural nucleotide monomers or as fully modified in the random region to furnish a chemically-modified oligonucleotide library for aptamer selection. However, the libraries containing fully modified random region with all four modified nucleotide bases may be limited to use in one-step aptamer selection-based methodologies. This is mainly because the selected aptamer candidates are regenerated using triphosphate derivatives of the corresponding modified nucleotides, but during an enzymatic synthesis process, the primer-binding region will also be modified with the same modified nucleotides contrary to the starting library design. In SELEX to allow maximum base variations of the chemically-modified nucleotides (good substrates of specific enzymes) in the random region, the nucleotide bases can be mixed with one, two or three modified nucleotide bases of the same or different nucleotide analogs in combination with the natural counterparts. But, depending on the number of modified nucleotide bases in the randomized region, the primer-binding region needs to be adjusted with the base composition of unmodified nucleotides. In this approach, where nucleotide mixmers are randomized, it is difficult to locate the positioning of modified nucleotide analogs, and if the library is constructed with successive stretch of modified nucleotides it may be hard for the polymerases to regenerate these particular sequences for subsequent selection rounds.
Another design approach could be to fix the positioning of modified nucleotides in the random region, which will assist to keep track of the positioning of modified nucleotides after sequencing. But, this could limit the pool diversity to some extent, however, it is worthwhile to perform selection since some chemical modifications could generate unique shapes and enhance target binding affinity. It is best to limit up to two fixed modifications that could be incorporated individually at different positions or as a mixture at the same positions allowing two nucleotide variations. Again, the primer-binding region needs to be constructed only with the unmodified natural nucleotide bases (avoiding similar base counterparts of the chemically-modified nucleotides) to regenerate the selected aptamer candidates for use in the subsequent selection rounds. During the sequence alignment after sequencing, only those candidates that maintain the initial library design need to be considered for further analysis and chemical synthesis.
Library with modified primer-binding region
For some modified nucleotide analogs that offer potential biophysical properties, enzymatic recognition may be difficult limiting their application in aptamer selection using conventional SELEX method. But, such modifications can still be used in aptamer selection using a library with one of the primer-binding regions containing the desired chemically-modified nucleotides, and the other region with all four natural nucleotide monomers. This will essentially eliminate the need of an enzymatic recognition of modified nucleotides as a synthetic primer sequence with the chemical modifications can be used during the enrichment steps. But, the protocol may be limited to DNA-based oligonucleotides. This approach may not fit very well in line to the concept of aptamer selection where it is speculated that the randomized region would promote the formation of target-binding motifs due to high sequence variability. But, one cannot ignore the fact that a part of the primer-binding region also plays a role in the folding pattern of the binding aptamers in majority of the cases. On this ground, aptamer selection using a library with chemically-modified primer-binding regions can be well justified, and can be used as an alternate approach. This method has been successfully applied for generating LNA-modified aptamers.22,23
∞-aptamer library
Aptamer selections can also be performed using a library containing infinite range (∞) of modified nucleotides with non-natural structural chemistries of the nucleobase, sugar and phosphate backbone. Libraries containing infinite nucleotides could come with diverse range of chemistries bearing positive charges, hydrophobic groups, phosphorothioates, amino acids etc. that could improve and enhance the target binding interactions and nuclease resistance. Such libraries are mainly suitable for one-step selection protocols.24-27 ∞-nucleotides can be best positioned in the randomized regions. During synthesis, ∞-nucleotides of all four bases can also be mixed with their natural counterparts.
Aptamer selection using sugar-modified nucleotides
Modifications of sugars will result in libraries with a greater functional diversity and can form stable aptamers with unique shapes. Sugar modifications could allow screening of ligands that bind with greater affinity to their targets than their unmodified counterparts. These unnatural modifications are less likely to be recognized by nucleases, making them more stable in serum. Most frequently, chemical modifications are introduced at the 2′-position of the nucleotide for increased nuclease resistance, and binding affinity. The SELEX evolution of various sugar- modified nucleotide (Fig. 3) containing aptamers against different targets, their binding affinities and stabilities are described below. Table 1 shows an overview of sugar-modified aptamer development in recent years.
Figure 3.
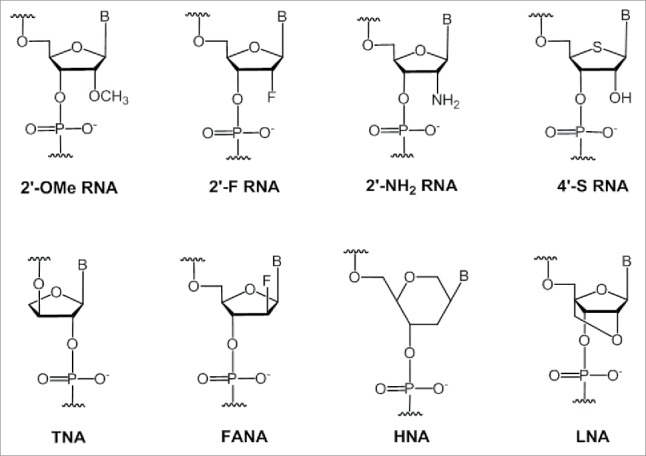
Structures of various sugar-modified nucleotides used in aptamer selection by SELEX methodologies.
Table 1.
Aptamers selected from sugar-modified DNA/RNA libraries and the binding affinity of aptamers to their targets.
| Modified nucleotide | Target | Selection Method | Polymerases Used | Binding Affinity (Kd) | Ref. |
|---|---|---|---|---|---|
| 2′-NH2 | Human neutrophil elastase | Nitrocellulose membrane filter binding | T7 RNA polymerase | 7–25 nM | 28 |
| VPF/VEGF | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 2.4 nM | 29 | |
| mRNAs | Affinity column | T7 RNA polymerase | No Kd (Kcat = 0.04 min−1) | 30 | |
| bFGF | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | Apparent Kd = 0.35 nM | 31 | |
| KGF | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 0.4 nM | 41 | |
| IFN-γ | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 1.8 nM | 40 | |
| MiniTAR | Magnetic bead immobilisation | DNA polymerase | 26–47 nM | 32 | |
| 2′-F | VEGF165 | Nitrocellulose membrane filter binding | T7 RNA Polymerase | 2 pM | 33 |
| PSMA | Magnetic bead immobilisation | Y639F T7 RNA Polymerase | No Kd (IC50 = 27 nM)Ki = 11.9 nM | 34, 35 | |
| Human complement C5 component | Nitrocellulose membrane filter binding | T7 RNA polymerase | 2–5 nM | 36 | |
| KGF | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 14F Kd = 0.3–3 pM | 41 | |
| Factor IXFactor IXa | Nitrocellulose membrane filter binding | AMV Reverse Transcriptase | Kd = 0.64 nM (9.3)Kd = 364 pM (Clone 9D-6) | 37,38 | |
| IFN-γ | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 106 nM | 40 | |
| IFN-γ | Nitrocellulose membrane filter binding | Y639F T7 RNA Polymerase | 6.8 nM | 40 | |
| Cancer Antigen 125 | Magnetic bead immobilisation | Y639F T7 RNA Polymerase | CA125.1 Kd = 4.13 nM | 39 | |
| 2′-OMe | VEGF | Electrophoretic mobility shift assay | Y639F/H784A/K378R T7 RNA Polymerase | 2 nM | 42 |
| Interleukin-23 and thrombin | Nitrocellulose membrane filter binding | Y639F/H784A/K378R T7 RNA Polymerase | Clone A5 Kd = 8.4 nMClone B4 Kd = 26 nM | 43 | |
| Tissue factor pathway inhibitor | Nitrocellulose membrane filter binding | Not Specified | ARC17480 Kd = 2.8 nMARC19499 IC50 = 17.9 nM | 44 | |
| 4′-S | Human thrombin | Nitrocellulose membrane filter binding | T7 RNA polymerase | 4.7 nM | 50 |
| 2′-FANA | HIV-1 reverse transcriptase | Electrophoretic mobility shift assay | Taq polymerase | 4 pM | 51 |
| HNA | TAR | Magnetic bead immobilisation | Pol6G12 | 28–67 nM | 52 |
| HIV Hen egg lysozyme | Magnetic bead immobilisation | Pol6G12 | HNA11 Kd = 107 nMHNA19 Kd = 141 nM | 52 | |
| TNA | Human thrombin | Capillary electrophoresis | Therminator DNA polymerase | 200–900 nM | 53 |
| BNA/LNA | Human thrombin | Capillary electrophoresis | an enzyme mix of KOD Dash and KOD mutant DNA polymerases | 0.26–27 nM | 23,70 |
| CD73 | Affinity column | KOD XL | 3.7 nM | 22 | |
| Spiegelmers | D-adenosine | Affinity column | T7 polymerase | 1.7 µM | 71 |
| L-arginine | Affinity column | T7 polymerase | Bound to L-arginine with Kd = 129 µM | 72 | |
| HIV-1 Tat protein | Bound to HIV-1 tat protein with Kd=26 µM | ||||
| D-vasopressin | Electrophoretic mobility shift assay | T7 polymerase | 0.9 µM | 73 | |
| D-staphylococcal enterotoxin B | Electrophoretic mobility shift assay | Taq DNA polymerase | 200 nM | 74 | |
| Nociceptin/ orphanin FQ | Affinity column | T7 polymerase | L-NOX2149 Kd = 0.3 µML-NOX2137 Kd = 0.7 µM | 77 | |
| D-gonadotropin-releasing hormone | Affinity column | T7 polymerase | L-S42 Kd = 45 nML-A10 Kd not specified (IC50 = 200 nM) | 78 | |
| Ghrelin | Electrophoretic mobility shift assay | T7 polymerase | 44.4 nM | 79,80 | |
| Calcitonin gene-related peptide | Affinity column | T7 polymerase | Apparent Kd = 2.5 nM | 75 | |
| Substance P | Affinity column | T7 polymerase | 40 nM | 76 |
2′-Amino (2′-NH2) modified aptamers
Aptamers have been selected from libraries where the 2′-hydroxyl (2′-OH) group of the pyrimidine is replaced by a NH2 group. A modified aptamer was screened from a 2′-NH2 -modified RNA library involving 2′-NH2-UTP and 2′-NH2-CTP replacing unmodified UTP and CTP during enrichment steps, specific to the human neutrophil elastase using nitrocellulose filter binding assay.28 The selected aptamers showed high affinity to the target with the equilibrium dissociation constant (Kd) in the range of 7–30 nM which is much lower than that of the unmodified RNA aptamer (Kd > 1 µM).28 These aptamers were found to be very selective to human neutrophil elastase.28 These modified RNA aptamers also improved nuclease resistance, with an extended half-life (20 h and 9 h in serum and urine respectively) than the unmodified aptamer (degraded in less than 8 and 5 mins in serum and urine respectively).28 Another 2′-NH2-pyrimidine-modified RNA aptamer was developed against VPF/VEGF by SELEX process.29 The affinity was measured through nitrocellulose filter binding assay and one aptamer NX-178 had a Kd of 2.4 nM, with an increased half-life in urine (17 h).29 Interestingly the introduction of a 2′-O-Methyl (2′-OMe) nucleotide post-SELEX further increased the binding affinity to Kd = 0.14 nM and half-life in urine to 31 h.29 This study suggests that the affinity and nuclease resistance of the RNA modified aptamers developed by SELEX methodology can be further improved by post-SELEX modification. In another study, Beaudry et al. developed a 2′-NH2 pyrimidine-modified ribozyme which was identified by affinity column-based SELEX.30 The selected aptamer had a half-life of 16 h in human serum, whereas the unmodified counterpart degraded in serum in just 5 mins.30 Jellinek et al. developed a different 2′-NH2 pyrimidine-modified RNA aptamer, selected through nitrocellulose membrane filter binding assay specific to basic fibroblast growth factor (bFGF).31 The identified aptamer m21A bound to bFGF with high binding affinity (Kd = 0.35 nM).31 The stability of this aptamer in serum is 100 fold higher than the natural RNA aptamer.31 In another study, Bugaut et al. used an interesting methodology whereby SELEX and dynamic combinatorial chemistry were combined to select conjugated RNA aptamers against MiniTAR by magnetic bead immobilisation using a 2′-NH2- modified RNA library. The resulting conjugated aptamers developed bound to MiniTAR with high affinity (Kd = 26–47 nM).32
2′-fluoro (2′-F) modified aptamers
Ruckman et al. developed an RNA aptamer specific to VEGF165 isoform by nitrocellulose filter binding assay using 2′-F modified RNA libraries.33 Most of the selected 2′-F RNA aptamers had affinities for VEGF165 in the pM range (aptamer with the lowest affinities had a Kd value of 2 pM).33 This candidate aptamer after further chemical modifications has been approved by the US FDA for the treatment of AMD. Lupold et al. developed another 2′-F modified RNA aptamer A10, targeting prostate specific membrane antigen (PSMA), cell surface receptors expressed on prostate cancer cells by SELEX using magnetic bead separation-based method.34 The aptamer A10 bound to PSMA with a Kd of 11.9 nM in LNCaP cells.34 This was later used to link siRNA to form aptamer-siRNA chimeras that allowed targeted delivery of siRNA to cells expressing PSMA wherein the siRNA reduced the expression of two survival genes, Polo-like kinase 1 and B-cell lymphoma 2.34,35 In another study, Biesecker et al. developed another aptamer against human complement C5 component using 2′-F pyrimidine-modified RNA libraries by nitrocellulose membrane filter binding assay.36 The aptamers generated had Kd of between 20–40 nM and this was further improved by a second biased SELEX experiment where the generated aptamers had a Kd of 2–5 nM.36 The aptamers generated were able to bind to human complement C5 component with high affinity and were able to inhibit its activity in human serum with high stability.36 Rusconi et al. developed 2′-F modified aptamers for Factor II, Factor VII, Factor IX and Factor X through nitrocellulose membrane filter binding assay.37,38 By this approach a RNA aptamer was identified against Factor IXa with the highest affinity (Kd of 0.64 nM), and was a potent anticoagulant.38 Another aptamer generated against Factor IX had a Kd of 364 pM.37 Lamberti et al. developed an aptamer against Cancer Antigen 125 by magnetic bead immobilisation using a 2′-F modified RNA library.39 Of the two RNA aptamers isolated, CA125.1 had a Kd of 4.15 nM and can be developed further for use as a diagnostic tool.39
Comparison of 2′-F and 2′-NH2 modifications
Various studies indicate that both 2′-F and 2′-NH2 modifications increase the nuclease resistance and binding affinity of aptamers toward their targets. There are a few studies comparing the two modifications to validate which modification was more powerful. Kubik et al. reported an aptamer that bound to interferon-gamma (IFN-γ) which was screened from RNA libraries modified at the 2′-position of pyrimidine nucleotides with F, NH2, or a mixture of F and NH2 (2′-F/NH2) groups using nitrocellulose filter binding assay.40 The binding affinity of the modified aptamers to IFN-γ differed depending on the sugar modifications.40 2′-NH2 modified aptamers had the highest binding affinity to IFN-γ with a Kd of 1.8 nM, whereas the 2′-F modified aptamers with the highest binding affinity had a Kd of 6.8 nM and the aptamers containing 2′-F/NH2 mixture had a Kd of 106 nM.40 The half-lives of these aptamers were measured through their stability in human serum, where 2′-NH2 modified aptamer had a half-life of 80 h, 2′-F modified aptamers had a half-life of 6 h and the mixed 2′F/NH2 modified aptamers had a half-life of 48 h.40 However, all three were more nuclease-resistant than the unmodified aptamer which had a half-life of 20 s in human serum.40 2′-NH2 modified aptamers seemed to confer the highest binding affinity to IFN-γ and nuclease resistance in human serum.40 2′-NH2 aptamer also inhibited IFN-γ binding to its receptor on A549 human lung carcinoma cells with an ID50 of 10 nM.40 Another study by Pagratis et al. suggested that 2′-F modified aptamers improved the binding affinity compared to the 2′-NH2 modified aptamers.41 In this study, aptamers have been selected successfully from libraries containing 2′-NH2 and 2′-F modified pyrimidines by nitrocellulose filter binding assay and compared the affinities of the two different aptamer variants to keratinocyte growth factor (KGF).41 The best 2′-F modified aptamer 14F bound to KGF with a higher affinity (Kd = 0.3–3 pM) than the best 2′-NH2 modified aptamer (Kd = 0.4 nM).41 2′-F modified aptamers (6F and 14F) inhibited KGF binding to their receptor in PC3 cells with a Ki of 100 and 200 pM respectively and had a much higher inhibitory activity than the 2′-NH2 modified variants, 14N and 29N (Ki = 1.4 nM).41 Although if 2′-F or 2′-NH2 modification is more powerful is arguable the use of 2′-NH2 modifications result in problems in solid-phase synthesis and the ribose sugar adopting an unfavorable conformation which has resulted in the reduced use of 2′-NH2.
2′-OMe modifications:
2′-OMe modification involves the replacement of the 2′-OH group of the nucleotide with a methoxy group. 2′-OMe is a common sugar modification that has been used post-SELEX to generate aptamers with high nuclease resistance. Burmeister et al. developed an aptamer against VEGF from oligonucleotide libraries modified with 2′-OMe adenine and guanines by electrophoretic mobility shift assay.42 The selected aptamer candidate, ARC245 had a Kd of 2 nM, however this is lower than Macugen, the FDA approved aptamer drug targeting VEGF (Kd = 50 pM).42 ARC245 showed inhibition of VEGF binding to VEGF receptor in 293 cells at 10 nM concentrations and was also highly stable in serum for up to 96 h.42 Burmeister et al. also screened aptamers comprising 2′-OMe pyrimidines by the SELEX process to multiple protein targets including thrombin and interleukin-23, using proteins immobilized on a 96-well plate and nitrocellulose filter binding assay.43 The identified aptamers targeting interleukin-23 had a Kd of 8.4 nM while the aptamer targeting thrombin had a Kd of 26 nM.43 Waters et al. developed 2′-OMe modified aptamers, ARC19499 and ARC17480 against tissue factor pathway inhibitor by nitrocellulose filter binding using a 2′-OMe modified library.44 ARC17480 bound to the tissue factor pathway inhibitor with a Kd of 2.8 nM, however as ARC19499 was not viable in experiments requiring radiolabelling a competition-binding experiment showed that ARC19499 competed with radiolabelled ARC17480 with an IC50 of 17.9 nM.44 2′-OMe incorporation into libraries and using these modified libraries for SELEX is hard due to lack of enzymes that are capable of recognizing these 2′-OMe modified bases. However, recently polymersases have been evolved that accept 2′-OMe modified triphosphates which would allow generation of additional 2′-OMe modified aptamers.21,45-49
4′-Thio (4′-S) modifications
4′-S modifications had a sulfur atom at the 4′-position of the sugar moiety. Kato et al. developed a 4′-S modified RNA aptamer that bound to human α-thrombin by nitrocellulose filter binding assay.50 In the presence of RNase, the stability of the aptamer candidate, thioRNA59, had a half-life of 1174 mins and was stable even after 12 h of incubation, which was 50 times greater than that of the corresponding natural RNA (completely degraded in the presence of RNase A in 3 h), and it also showed high binding affinity to thrombin with a Kd of 4.7 nM.50
2′-fluroarabino nucleic acid (2′-FANA) modified aptamers
Very recently, Alves Ferreira-Bravo et al. reported a 2′-FANA aptamer to human immunodeficiency virus-1 (HIV-1) reverse transcriptase.51 This 2′-FANA aptamer was isolated from a 2′-FANA modified DNA pool through SELEX using electrophoretic mobility shift assay.51 The developed 2′-FANA modified DNA aptamer, FA1 had a Kd value of 4 pM and showed greater resistance to nucleases.51
1,5-anhydro hexitol nucleic acid (HNA) modified aptamers
Pinheiro et al. developed HNA aptamers against HIV-1 transactivation responsive element (TAR) and hen egg lysozyme through SELEX using magnetic bead-based separation.52 HNA aptamer, T5-S8-7 bound to TAR with high specificity and had a Kd of 28–67 nM.52 HNA aptamers that bound specifically to hen egg lysozyme had a Kd of 107 to 141 nM as determined by surface plasmon resonance.52
Threose nucleic acid (TNA) modified aptamers
Yu et al. selected a TNA modified aptamer against thrombin using affinity column and capillary electrophoresis.53,54 TNA backbone is nuclease-resistant as TNA remained undigested even after 72 h of incubation with a pure nuclease unlike DNA and RNA which exhibit half-lives of 30 mins and less than 10s respectively.54 The developed TNA aptamers to thrombin had a Kd in the range of 200–900 nM.53 A new manganese-independent TNA polymerase was evolved using droplet-based optical polymerase sorting which can be used in TNA aptamer selection protocol.55
Locked nucleic acid (LNA) modified aptamers
In LNA, the sugar ring is locked by a 2′O, 4′C methylene linkage and is conformationally restricted adopting a C3′-endo conformation.56-61 Toward the development of LNA-modified aptamers, Veedu et al. and others have extensively investigated the enzymatic recognition capabilities of LNA-nucleotides.58-68 DNA aptamers containing LNA (BNA/LNA) nucleotides were developed against human thrombin using capillary electrophoresis SELEX. BNA/LNA-modified nucleotides were introduced in the primer-binding region of the library, and after selection the selected candidates were enriched by PCR using the LNA-modified primer sequence to regenerate the selected aptamer candidates using KOD polymerase.23,69 Many aptamers showed Kd values in the low nanomolar range.23,70 In another study, Elle et al. generated LNA-modified aptamers against CD73 immobilized on anti-His tag plates.22 A LNA-modified DNA aptamer NAC6772 showed high binding affinity to CD73 through surface plasmon resonance experiment with a Kd of 3.54 nM and inhibited the CD73 activity by 85%.22
Spiegelmers
Spiegelmers are mirror images of the natural aptamers in which the D-ribose (the natural ribose) are replaced with the unnatural L-ribose. Spiegelmers prevent enzymatic degradation as the chiral form of the nucleic acid (the L-form) is unnatural and is not recognized by nucleases. However, the L-form of the nucleic acid like their D-form natural counter parts can bind to the target with high affinity and will not trigger the immune response. Similar to the other sugar modifications described above, spiegelmers can be enriched by T7 RNA polymerases. Klussmann et al. generated one of the first spiegelmers through SELEX against the naturally occurring D-adenosine.71 The high affinity L-RNA aptamers against D-adenosine were isolated through affinity column and had a Kd of 1.7 µM which was comparable to the affinity displayed by the D-RNA aptamers.71 Nolte et al. reported another spiegelmer that bound to L-arginine that could also bind to a short peptide that contains an arginine-rich region in HIV-1 Tat protein.72 The aptamer, L-R16c was isolated through affinity column based SELEX.72 L-R16c bound to L-arginine with a Kd of 129 µM and to the Tat protein with a Kd of 26 µM.72 L-R16c was very stable in serum and did not degrade even after 60 h of incubation.72 In another study, spiegelmers were selected against D-vasopressin and D-staphylococcal enterotoxin B through SELEX using electrophoretic mobility shift assay.73 The L-DNA aptamers targeting D-vasopressin showed no degradation with exo- and endonucleases and showed high binding affinity for D-vasopressin with a Kd of 0.9 µM.73 The corresponding D-aptamer degraded in 10 s while the L-aptamer showed no degradation even after 10 days incubation with exo- and endonucleases and 7 days in serum.73 A DNA spiegelmer targeting D-staphylococcal enterotoxin B, B12b10 had a Kd of 200 nM.74 Vater et al. isolated a RNA spiegelmer against migraine associated calcitonin gene-related peptide through SELEX using affinity columns.75 The developed RNA spiegelmer STAR-F12 bound to calcitonin gene-related peptide with a Kd of 2.5 nM and inhibits its activity.75 In another study, a RNA spiegelmer targeting Substance P was generated through SELEX using affinity column.76 The developed L-RNA aptamer, SUP-A-004, which was further truncated and the resulting spiegelmer bound to Substance P with a Kd of 40 nM.76 SUP-A-004 inhibited L-substance P mediated Ca2+ release in AR42J pancreatic cells.76 Faulhammer et al., in another study reported the development of L-RNA aptamers NOX2149 and NOX2137 against Nociceptin/orphanin FQ through SELEX using affinity column.77 The L-NOX2149 and L-NOX2137 bound to the nociception/orphanin FQ with a Kd of 0.3 µM and 0.7 µM respectively which were similar to the binding affinity to their D-aptamer counterparts.77 However L-aptamers had the advantage of being more stable in serum.77 In another study, L-DNA/RNA aptamers that bind to D-gonadotropin-releasing hormone with high affinity and specificity were isolated out of L-RNA and DNA libraries through SELEX using affinity column.78 The L-DNA aptamer, S42 bound to gonadotropin-releasing hormone with a Kd of 45 nM which is similar to that of the D-DNA aptamer and the IC50 was 50 nM.78 The D-RNA aptamer, A10 bound with Kd of 55 nM, however the Kd of L-RNA aptamer could not be obtained.78 Both the DNA and RNA spiegelmers bound to gonadotropin-releasing hormones and inhibited their binding to their receptor in Chinese hamster ovary cells and there was no immune response produced in zimmermann rabbits.78 Helming et al. reported L-RNA aptamers targeting ghrelin through SELEX using electrophoretic mobility shift assay.79,80 L-NOX-B11, a L-RNA aptamer was isolated and bound to ghrelin with high affinity with a Kd of 44.4 nM.80 L-NOX-B11 inhibited ghrelin from binding to its receptor with an IC50 value of 5 nM in Chinese hamster ovary cells.79,80 NOXXON Pharma have developed many spiegelmers against various targets and a few of the spiegelmers are in various stages of the preclinical and clinical trials, two of which are described in a recent review by Sundaram et al. NOX-A12 is a spiegelmer that bound to and inhibits stroma cell-derived factor-1.81 After completion of Phase I and Phase IIa trials of NOX-A12, Phase 3 trials have been planned for multiple myeloma.81 Also, Phase 2b/3 trials have been planned for the same candidate against glioblastoma.81 Phase 2a clinical trials have been completed for another spiegelmer, NOX-E36, targeting monocyte chemoattractant- protein 1.81 Several other candidates are currently in different stages of clinical investigation.82 NOX-D20 is another spiegelmer that binds to anaphylatoxin C5a with picomolar affinity and is being considered for preclinical and clinical development.83 Wang et al. has described the generation of a chemically-synthesized D-amino acid polymerase that is capable of recognizing and catalyzing transcription and polymerisation of an L-DNA template.84 This polymerase could result in generation of new speigelmers in the future.84
Aptamer selection using modified nucleotide bases
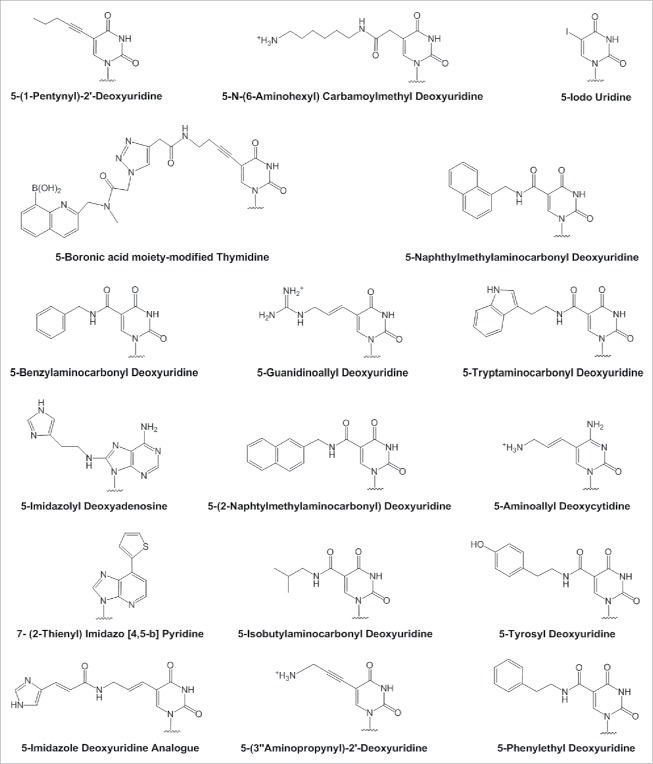
Incorporating chemical modification to the nucleotide bases could increase the stability and target-binding affinity of aptamers. A number of researchers have succeeded in generating aptamers with base-modified nucleotides (Fig. 4), some of which are in preclinical stages. Various base modifications are incorporated into the nucleic acid library and in many cases have been able to be amplified using family B-DNA polymerases.85 Table 2 provides a summary of base-modified aptamer development. There is also a new method called click-SELEX which is used to generate base-modified libraries by replacing the thymidines with C5-ethynyl-2′-deoxyuridine which is then further modified through click reaction with copper(I)-catalyzed alkyne-azide cycloaddition.86-91 The library is then used in selection experiments to evolve modified aptamers.86-91 This is a powerful method that allows generation of modified aptamers with new chemical modifications without problems caused by lack of enzymatic recognition as seen with the addition of a cluster of oligomannose glycans.86-91 This will allow targeting of many molecules and epitopes that is currently not possible with standard SELEX.86-91 Addition of new nucleotides and/or unnatural base pairs to the libraries and using these libraries for aptamer selection allows generation of modified aptamers with greater sequence and functional diversity.92-95 Zhang et al. has shown that addition of new nucleotides Z and P allowed generation of sequences that had better binding affinity to the target molecule indicating that new nucleotides are viable in the library and can show higher binding affinity than natural bases in some cases.92
Figure 4.
Structures of various base-modified nucleotides used in aptamer selection by SELEX methodologies.
Table 2.
Aptamers selected from base-modified DNA/RNA libraries and the binding affinity of aptamers to their targets.
| Modified nucleotide | Target | Selection Method | Polymerases Used | Binding affinity (Kd) | Ref. |
|---|---|---|---|---|---|
| 5-pentynyl-dU | Human thrombin | Affinity column | Vent DNA Polymerase | 400 nM | 96 |
| 5-N-(6-aminohexyl) carbamoylmethyl-dU | Thalidomide (T5N and T5-IB) | Streptavidin-sepharose gel | KOD Dash DNA polymerase | T5N Kd= 113 µMT5-1B Kd= 133 µM | 100 |
| 5-boronic acid-dT | Fibrinogen | Magnetic bead immobilisation | Taq DNA Polymerase | 3–30 nM | 102 |
| 5-iodouridine | HIV-1 Rev protein. | Nitrocellulose membrane filter binding | T7 RNA polymerase | 0.8 nM | 97 |
| 5-(3′-aminopropynyl)-2′-dU | ATP (Sequence 409) | Capillary electrophoresis | Vent DNA polymerase | 6 nM | 98 |
| 5-benzylaminocarbonyl dU | Plasminogen Activator Inhibitor | Magnetic bead immobilisation | KOD DNA Polymerase | ≤30 nM | 107 |
| 5-isobutylaminocarbonyl –dU | Human mobility group -1 | ≤30 nM | |||
| 5-tryptaminocarbonyl -dU | Fractalkine | ≤30 nM | |||
| 5-naphthylmethylaminocarbonyl-dU | Human protein targets | ≤30 nM | |||
| 5-(3-aminoallyl)-dC5-guanidinoallyl-dU, 5-imidazolyl-dA | RNA | Magnetic bead immobilisation | Vent (exo -) DNA Polymerase | Kd not specified (DNAzyme12–91 Kobs = 0.06 min−1DNAzyme 16.2–11 Kcat= 1.4–1.5 min−1) | 106 |
| 7-(2-thienyl)imidazo[4,5-b]pyridine | VEGF165 | Magnetic bead immobilisation | Accu Prime Pfx DNA Polymerase | 0.65 pM | 105 |
| IFN-γ | 0.038 nM | ||||
| 5-tyrosyl-dU | Clostridium difficile binary toxins A and B | Magnetic bead immobilisation | KOD EX DNA polymerase | 1.4 nM (Toxin A)0.31 nM (Toxin B) | 109 |
| 5-(2-naphthylmethylaminocarbonyl)-dU | 9.2 nM (Toxin A)0.25 nM (Toxin B) | ||||
| 5-phenylethyl-dU | 13.1 nM (Toxin A)0.27 nM (Toxin B) | ||||
| 5-benzylaminocarbonyl-dU | 5 nM (Toxin B) | ||||
| 5-naphthylmethylaminocarbonyl-dU | 0.43 nM (Toxin B) | ||||
| 5-tryptaminocarbonyl-dU | 1.7 nM (Toxin A)0.45 nM (Toxin B) | ||||
| 5-carboxamide-modified-dU | Tumor necrosis factor receptor super family member 9 (6a and 6d) | Magnetic bead immobilisation | Deep Vent and KOD XL DNA Polymerases | 6a Kd= 6 nM 6d Kd= 4 nM | 103 |
| 5-imidazole-dU analog | RNA | Affinity column | DNA Polymerase | Kd not specified (DNAzyme 16.2–11 Kcat= 1.4–1.5 min−1 | 104 |
Uridine modifications
Latham et al. developed a modified aptamer containing 5-(1-pentynyl)-2′-deoxyuridine against thrombin by affinity column.96 The affinity of the selected aptamer pool was 400 nM.96 This was one of the first examples of using a base-modified nucleic acid library for screening aptamers. In another study, Jensen et al. developed RNA aptamers from a random RNA library containing photoreactive chromophore 5-iodouridine using crosslinking SELEX.97 The selected RNA aptamers bound and also cross-linked to the target with UV irradiation.97 These modified aptamers had high affinity with a Kd of 0.8 nM.97 Battersby et al. isolated another base-modified aptamer containing 5-aminopropynyl-deoxyuridine against ATP by electrophoretic mobility shift assay.98,99 One of the selected aptamer candidate, 409 was found to bind ATP with higher affinity (Kd = 6 µM) than their unmodified counterparts.98,99 Shoji et al. reported another base-modified aptamer with 5′-N-6-NH2-hexyl carbamoylmethyl dU.100,101 The modified library was used to screen aptamers against thalidomide using streptavidin-sepharose gel.100 Surface plasmon resonance showed that the selected base-modified aptamer T5N and T51B had a Kd of 113 µM and 133 µM respectively.100 Li et al. selected an aptamer containing boronic acid-modified thymidine against fibrinogen using magnetic bead immobilization-based SELEX.102 The resulting modified aptamers bound to fibrinogen with high affinity compared to that of the unmodified aptamers (Kd = 3–30 nM compared to Kd = 450 nM).102 Vaught et al. selected a DNA aptamer against tumor necrosis factor receptor super family 9 from a library possessing 5-amide-modified deoxyuridine via a magnetic bead-based SELEX method.103 Six 5-position modified 2′-dUTP derivatives were used.103 Two of the modified DNA aptamers developed showed high affinity (Kd = 6 nM and 4 nM respectively) compared to an unmodified RNA aptamer that has been previously reported (Kd = 40 nM).103
Imidazole modifications
Aptamers can also act as DNA enzymes which are catalytic nucleic acids and modified libraries have been used to select such aptamers. Sontoro et al. developed a modified DNAzyme aptamer via affinity column from a modified library containing C5-imidazole-functionalised dU.104 The isolated DNAzyme was one of the smallest nucleic acid enzymes, which showed multiple turnovers in the presence of millimolar concentrations of Zn2+, and a catalytic rate of 1.4–1.5 min−1 for cleavage.104 Kimoto et al. reported the selection of 7-(2-thienyl)imidazole[4,5-b]pyridine modified DNA aptamers against VEGF165 and IFN-γ by magnetic bead-based SELEX.105 The isolated DNA aptamers bound to VEGF165 and IFN-γ with 100 fold higher affinity than their natural counterparts with Kd values of 0.64 pM and 0.038 nM respectively.105 Hollenstein et al. reported the development of Dz12–91, a modified DNAzyme containing imidazole, ammonium and guanidinium groups. Dz12–91 was isolated through SELEX using magnetic streptavidin particles and cleaves all RNA sequence independently of M2+ with a Kobs of 0.06 min−1.106
Slow off-rate modified aptamers (SOMAmers)
Another class of aptamers with improved binding properties through base-modifications that result in slow dissociation rates called SOMAmers. There are many SOMAmer modifications that are used to generate aptamers to difficult protein targets.107 The affinities of the SOMAmers are consistently in the nanomolar range to their targets and show high nuclease resistance.107 Various proteins have been used as targets to generate SOMAmers.107,108 In many SOMAmers, the modifications are introduced at the 5-position of the uridine nucleotide.107 Gold et al. described the development of SOMAmers against more than 800 targets using multiplex SOMAmer affinity assay system from base-modified libraries including 5-benzylaminocarbonyl, 5-naphthylmethylaminocarbonyl, 5-tryptaminocarbonyl, 5-isobutylaminocarbonyl, 5-tyrosyl, 5-phenylethyl, and 5-guanidinoallyl.107,109 Ochsner et al. reported development of SOMAmers against toxins A, B and binary toxins using 5-benzylaminocarbonyl, 5-naphthylmethylaminocarbonyl, 5-tryptaminocarbonyl, 5-phenylethyl-1-aminocarbonyl, 5-tyrosylaminocarbonyl or 5-(2-naphthylmethyl)aminocarbonyl-modified uridine containing library using magnetic bead-based SELEX.109 Most of the modified aptamers bound to the toxin A and B with high binding affinity with the Kd in the nanomolar range.109 For example, the selected 5-tryptaminocarbonyl dU modified aptamer showed a Kd of 1.7 nM to Toxin A and 0.45 nM for Toxin B.109
Aptamer selection using phosphate modified nucleotides
A few phosphate-modified aptamers (Fig. 5) were reported as substrates of T7 RNA polymerase that could accept the triphosphate analogs of the modified phosphate moiety, mainly phosphorothioate and boranophosphates. Table 3 provides a summary of base-modified aptamer development.
Figure 5.
Structures of various phosphate-modified nucleotides used in aptamer selection by SELEX methodologies.
Table 3.
Aptamers selected from phosphate-modified DNA/RNA libraries and the binding affinity of aptamers to their targets.
| Modified nucleotide | Target | Selection Method | Polymerases Used | Binding affinity (Kd) | Ref. |
|---|---|---|---|---|---|
| Boranophosphate-5′-(α-P-borano)-G 5′-(α-P-borano)-U |
ATP | Affinity column | T7 RNA Polymerase | Not Specified | 113 |
| Phosphorothioate-modified DNA | bFGF | Nitrocellulose membrane filter binding | Taq polymerase | 1.8 nM | 110 |
| Phosphorothioate-dA | Nuclear factor for human Interleukin6 | Nitrocellulose membrane filter binding | Taq DNA Polymerase | Kd not specified (Kobs = 2 nM) | 112 |
| Phosphorothioate-dA | Human Nuclear factor-kappa B RelA (p65) | Nitrocellulose membrane filter binding | Taq DNA Polymerase | 4.8 nM | 111 |
| Human Nuclear Factor-kappa B RelA (p50) | 0.8 nM |
Phosphorothioate modifications
Through nitrocellulose membrane filter binding, phosphothioate-modifed RNA aptamer 11–20 was isolated that bound to bFGF with a Kd of 1.8 nM which was similar to the Kd of all RNA aptamers (Kd = 0.19 nM and 0.49 nM).110 Kd of another bFGF aptamer containing 2′-NH2 pyrimidines was 0.35 nM, which was also comparable to the unmodified RNA aptamers.110 These aptamers also bound to other bFGF members in the family, but not to the unrelated targets and were therefore “semi-specific” for the FGF family members.110 Another phosphorothioate DNA aptamer was selected against nuclear factor for human interleukin 6 which was nuclease-resistant to DNase 1 enzyme at the point of modifications.111,112 This modified DNA aptamer was selected through nitrocellulose filter binding from a library with thiophosphate backbone substitution at the thymidine positions and the modified aptamer was found to bind with stoichemistry of two protein dimers/duplex unlike the unmodified aptamer which bound with one protein dimer/duplex stoichemistry.112 The binding constant (Kobs) measured by fluorescence anisotropy was 2 nM for a 66-mer.112 Another phosphorothioate aptamer was selected against Nuclear Factor kappa B proteins Rel A (p65) and p50 and the selected aptamer had a Kd value of 4.8 nM and 0.8 nM respectively, which was comparable to the unmodified aptamer (Kd = 4.77 nM).111
Boranophosphate modification
Boronated nucleotide analog modifications of guanosine (5′(α-P-borano)triphosphate) for GTP and uridine (5′(α-P-borano)triphosphate) for UTP were also introduced into general libraries to select modified aptamers that bind ATP through affinity matrix containing C8-linked ATP agarose.113 Each of the 2 modifications were tolerated in the backbone alone, however both modifications in one backbone were not tolerated.113
Conclusions and future perspectives
Aptamers are nucleic acid ligands that are generally developed through SELEX and bind to their targets with high affinity and specificity. Aptamers can be used therapeutically, diagnostically, as molecular beacons or as DNAzymes in various diseases. Macugen is a therapeutic aptamer approved by the US FDA for the treatment of AMD. One of the major disadvantages of aptamers composed of natural nucleotides is their rapid degradation in vivo. To overcome this obstacle, chemically-modified aptamers have been developed through SELEX using modified-oligonucleotide libraries. There are many potential chemically-modified nucleotides with excellent properties, however many of them cannot be used in the SELEX processes to develop aptamers mainly because of their very limited enzymatic recognition capabilities which are required for enrichment steps. As reviewed here, it is very promising to see the development of chemically-modified aptamers using several sugar, base and phosphate-modified nucleotides. It is also very encouraging to note the development of novel engineered polymerases capable of recognizing a specific modified nucleotide. Recent developments in one-step-based selection methodologies further offer tremendous hope for developing aptamers containing modified aptamers with limited enzymatic recognition capabilities.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
RNV acknowledges the funding from McCusker Charitable Foundation, and Western Australian Neuroscience Research Institute. MC thanks the funding from Greg and Dale Higham.
References
- 1.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010; 9:537-50; PMID:20592747; http://dx.doi.org/ 10.1038/nrd3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med 2005; 56:555-83; PMID:15660527; http://dx.doi.org/ 10.1146/annurev.med.56.062904.144915 [DOI] [PubMed] [Google Scholar]
- 3.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 2007; 107:3715-43; PMID:17715981; http://dx.doi.org/ 10.1021/cr0306743 [DOI] [PubMed] [Google Scholar]
- 4.Veedu RN. Editorial (Thematic Issue: Medicinal Chemistry of Aptamers). Curr Top Med Chem 2015; 15:1065; PMID:25866280; http://dx.doi.org/ 10.2174/1568026615666150413161707 [DOI] [PubMed] [Google Scholar]
- 5.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 1999; 45:1628-50; PMID:10471678 [PubMed] [Google Scholar]
- 6.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 2007; 24:381-403; PMID:17627883; http://dx.doi.org/ 10.1016/j.bioeng.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 7.Gopinath SCB. Methods developed for SELEX. Anal Bioanal Chem 2007; 387:171-82; PMID:17072603; http://dx.doi.org/ 10.1007/s00216-006-0826-2 [DOI] [PubMed] [Google Scholar]
- 8.Demarest SJ, Glaser SM. Antibody therapeutics, antibody engineering, and the merits of protein stability. Curr Opin Drug Discov Devel 2008; 11:675-87; PMID:18729019 [PubMed] [Google Scholar]
- 9.Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 2006; 5:123-32; PMID:16518379; http://dx.doi.org/ 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- 10.Group MDRS. A phase II randomized double-masked trial of pegaptanib, an anti–vascular endothelial growth factor aptamer, for diabetic macular edema. Ophthalmology 2005; 112:1747-57; PMID:16154196; http://dx.doi.org/ 10.1016/j.ophtha.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 11.Kanwar JR, Roy K, Maremanda NG, Subramanian K, Veedu RN, Bawa R, Kanwar RK. Nucleic acid-based aptamers: applications, development and clinical trials. Curr Med Chem 2015; 22:2539-57; PMID:25723512; http://dx.doi.org/ 10.2174/0929867322666150227144909 [DOI] [PubMed] [Google Scholar]
- 12.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res 2003; 31:3429-31; PMID:12824340; http://dx.doi.org/ 10.1093/nar/gkg599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diafa S, Hollenstein M. Generation of aptamers with an expanded chemical repertoire. Molecules 2015; 20:16643-71; PMID:26389865; http://dx.doi.org/ 10.3390/molecules200916643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellafiore MA, Montserrat JM, Iribarren AM. Modified Nucleoside Triphosphates for In-vitro Selection Techniques. Front Chem 2016; 4:18; PMID:27200340; http://dx.doi.org/ 10.3389/fchem.2016.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong D, Yeung W, Hili R. Generation of Synthetic Copolymer Libraries by Combinatorial Assembly on Nucleic Acid Templates. ACS Comb Sci 2016; 18:355-70; PMID:27275512; http://dx.doi.org/ 10.1021/acscombsci.6b00059 [DOI] [PubMed] [Google Scholar]
- 17.Lapa SA, Chudinov AV, Timofeev EN. The toolbox for modified aptamers. Mol Biotechnol 2016; 58:79-92; PMID:26607475; http://dx.doi.org/ 10.1007/s12033-015-9907-9 [DOI] [PubMed] [Google Scholar]
- 18.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids 2014; 3:e201; PMID:25291143; http://dx.doi.org/ 10.1038/mtna.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauridsen LH, Rothnagel JA, Veedu RN. Enzymatic recognition of 2′‐modified ribonucleoside 5′‐triphosphates: Towards the evolution of versatile aptamers. ChemBioChem 2012; 13:19-25; PMID:22162282; http://dx.doi.org/ 10.1002/cbic.201100648 [DOI] [PubMed] [Google Scholar]
- 20.Loakes D, Holliger P. Polymerase engineering: towards the encoded synthesis of unnatural biopolymers. Chem Commun 2009:4619-31; PMID:19641798; http://dx.doi.org/27016740 10.1039/b903307f [DOI] [PubMed] [Google Scholar]
- 21.Aschenbrenner J, Marx A. Direct and site-specific quantification of RNA 2′-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res 2016; 44:3495-502; PMID:27016740; http://dx.doi.org/ 10.1093/nar/gkw200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elle IC, Karlsen KK, Terp MG, Larsen N, Nielsen R, Derbyshire N, Mandrup S, Ditzel HJ, Wengel J. Selection of LNA-containing DNA aptamers against recombinant human CD73. Mol Biosyst 2015; 11:1260-70; PMID:25720604; http://dx.doi.org/ 10.1039/C5MB00045A [DOI] [PubMed] [Google Scholar]
- 23.Kasahara Y, Irisawa Y, Ozaki H, Obika S, Kuwahara M. 2′, 4′-BNA/LNA aptamers: CE-SELEX using a DNA-based library of full-length 2′-O, 4′-C-methylene-bridged/linked bicyclic ribonucleotides. Bioorg Med Chem Lett 2013; 23:1288-92; PMID:23374873; http://dx.doi.org/ 10.1016/j.bmcl.2012.12.093 [DOI] [PubMed] [Google Scholar]
- 24.Lauridsen LH, Shamaileh HA, Edwards SL, Taran E, Veedu RN. Rapid one-step selection method for generating nucleic acid aptamers: Development of a DNA aptamer against α-bungarotoxin. PLoS One 2012; 7:e41702; PMID:22860007; http://dx.doi.org/ 10.1371/journal.pone.0041702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitsche A, Kurth A, Dunkhorst A, Pänke O, Sielaff H, Junge W, Muth D, Scheller F, Stöcklein W, Dahmen C et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol 2007; 7(1):48; PMID:17199888; http://dx.doi.org/ 10.1186/1472-6750-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng L, Stephens BJ, Bonin K, Cubicciotti R, Guthold M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Microsc Res Tech 2007; 70:372-81; PMID:17262788; http://dx.doi.org/ 10.1002/jemt.20421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan M, McBurnett SR, Andrews CJ, Allman AM, Bruno JG, Kiel JL. Aptamer selection express: a novel method for rapid single-step selection and sensing of aptamers. J Biomol Tech 2008; 19:311; PMID:19183794 [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res 1994; 22:5229-34; PMID:7529404; http://dx.doi.org/ 10.1093/nar/22.24.5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, Jucker FM, Janjić N. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem Biol 1995; 2:683-95; PMID:9383475; http://dx.doi.org/ 10.1016/1074-5521(95)90032-2 [DOI] [PubMed] [Google Scholar]
- 30.Beaudry A, DeFoe J, Zinnen S, Burgin A, Beigelman L. In vitro selection of a novel nuclease-resistant RNA phosphodiesterase. Chem Biol 2000; 7:323-34; PMID:10801472; http://dx.doi.org/ 10.1016/S1074-5521(00)00110-1 [DOI] [PubMed] [Google Scholar]
- 31.Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DP, Abesinghe P. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 1995; 34:11363-72; PMID:7547864; http://dx.doi.org/ 10.1021/bi00036a009 [DOI] [PubMed] [Google Scholar]
- 32.Bugaut A, Toulmé J-J, Rayner B. SELEX and dynamic combinatorial chemistry interplay for the selection of conjugated RNA aptamers. Org Biomol Chem 2006; 4:4082-8; PMID:17312962; http://dx.doi.org/ 10.1039/b610890c [DOI] [PubMed] [Google Scholar]
- 33.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 1998; 273:20556-67; PMID:9685413; http://dx.doi.org/ 10.1074/jbc.273.32.20556 [DOI] [PubMed] [Google Scholar]
- 34.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 2002; 62:4029-33; PMID:12124337; http://dx.doi.org/ 10.1158/0008-5472.can-12-2152 [DOI] [PubMed] [Google Scholar]
- 35.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 2006; 24:1005-15; PMID:16823371; http://dx.doi.org/ 10.1038/nbt1223 [DOI] [PubMed] [Google Scholar]
- 36.Biesecker G, Dihel L, Enney K, Bendele RA. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 1999; 42:219-30; PMID:10408383; http://dx.doi.org/ 10.1016/S0162-3109(99)00020-X [DOI] [PubMed] [Google Scholar]
- 37.Layzer JM, Sullenger BA. Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides 2007; 17:1-11; PMID:17461758; http://dx.doi.org/ 10.1089/oli.2006.0059 [DOI] [PubMed] [Google Scholar]
- 38.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 2002; 419:90-4; PMID:12214238; http://dx.doi.org/ 10.1038/nature00963 [DOI] [PubMed] [Google Scholar]
- 39.Lamberti I, Scarano S, Esposito CL, Antoccia A, Antonini G, Tanzarella C, De Franciscis V, Minunni M. In vitro selection of RNA aptamers against CA125 tumor marker in ovarian cancer and its study by optical biosensing. Methods 2016; 97:58-68; PMID:26542762; http://dx.doi.org/ 10.1016/j.ymeth.2015.10.022 [DOI] [PubMed] [Google Scholar]
- 40.Kubik MF, Bell C, Fitzwater T, Watson SR, Tasset DM. Isolation and characterization of 2′-fluoro-, 2′-amino-, and 2′-fluoro-/amino-modified RNA ligands to human IFN-gamma that inhibit receptor binding. J Immunol 1997; 159:259-67; PMID:9200462 [PubMed] [Google Scholar]
- 41.Pagratis NC, Bell C, Chang Y-F, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat Biotechnol 1997; 15:68-73; PMID:9035109; http://dx.doi.org/ 10.1038/nbt0197-68 [DOI] [PubMed] [Google Scholar]
- 42.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol 2005; 12:25-33; PMID:15664512; http://dx.doi.org/ 10.1016/j.chembiol.2004.10.017 [DOI] [PubMed] [Google Scholar]
- 43.Burmeister PE, Wang C, Killough JR, Lewis SD, Horwitz LR, Ferguson A, Thompson KM, Pendergrast PS, McCauley TG, Kurz M. 2-Deoxy purine, 2-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 2006; 16:337-51; PMID:17155909; http://dx.doi.org/ 10.1089/oli.2006.16.337 [DOI] [PubMed] [Google Scholar]
- 44.Waters EK, Genga RM, Schwartz MC, Nelson JA, Schaub RG, Olson KA, Kurz JC, McGinness KE. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood 2011; 117:5514-22; PMID:21389323; http://dx.doi.org/ 10.1182/blood-2010-10-311936 [DOI] [PubMed] [Google Scholar]
- 45.Chelliserrykattil J, Ellington A. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat Biotechnol 2004; 22:1155-60; PMID:15300257; http://dx.doi.org/ 10.1038/nbt1001 [DOI] [PubMed] [Google Scholar]
- 46.Fa M, Radeghieri A, Henry AA, Romesberg FE. Expanding the substrate repertoire of a DNA polymerase by directed evolution. J Am Chem Soc 2004; 126:1748-54; PMID:14871106; http://dx.doi.org/ 10.1021/ja038525p [DOI] [PubMed] [Google Scholar]
- 47.Siegmund V, Santner T, Micura R, Marx A. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2′-modified nucleotides. Chem Commun 2012; 48:9870-2; PMID:22932771; http://dx.doi.org/ 10.1039/c2cc35028a [DOI] [PubMed] [Google Scholar]
- 48.Padilla R, Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non‐canonical NTPs. Nucleic Acids Res 2002; 30:e138-e; PMID:12490729; http://dx.doi.org/ 10.1093/nar/gnf138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer–micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci 2010; 107:5-10; PMID:20080797; http://dx.doi.org/ 10.1073/pnas.0909611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, Matsuda A. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res 2005; 33:2942-51; PMID:15914669; http://dx.doi.org/ 10.1093/nar/gki578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreira-Bravo IA, Cozens C, Holliger P, DeStefano JJ. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res 2015; 43:9587-99; PMID:26476448; http://dx.doi.org/ 10.1093/nar/gkv1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew S-Y, McLaughlin SH. Synthetic genetic polymers capable of heredity and evolution. Science 2012; 336:341-4; PMID:22517858; http://dx.doi.org/ 10.1126/science.1217622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem 2012; 4:183-7; PMID:22354431; http://dx.doi.org/ 10.1038/nchem.1241 [DOI] [PubMed] [Google Scholar]
- 54.Yu H, Zhang S, Dunn MR, Chaput JC. An efficient and faithful in vitro replication system for threose nucleic acid. J Am Chem Soc 2013; 135:3583-91; PMID:23432469; http://dx.doi.org/ 10.1021/ja3118703 [DOI] [PubMed] [Google Scholar]
- 55.Larsen AC, Dunn MR, Hatch A, Sau SP, Youngbull C, Chaput JC. A general strategy for expanding polymerase function by droplet microfluidics. Nat Commun 2016; 7:11235; PMID:27044725; http://dx.doi.org/ 10.1038/ncomms11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veedu RN, Vester B, Wengel J. In vitro incorporation of LNA nucleotides. Nucleosides Nucleotides Nucleic Acids 2007; 26:1207-10; PMID:18058567; http://dx.doi.org/ 10.1080/15257770701527844 [DOI] [PubMed] [Google Scholar]
- 57.Veedu RN, Vester B, Wengel J. Novel applications of locked nucleic acids. Nucleic Acids Symp Ser: Oxford Univ Press, 2007; 51(1):29-30; PMID:18029570; http://dx.doi.org/17315250 10.1093/nass/nrm015 [DOI] [PubMed] [Google Scholar]
- 58.Veedu RN, Vester B, Wengel J. Enzymatic incorporation of LNA nucleotides into DNA strands. ChemBioChem 2007; 8:490-2; PMID:17315250; http://dx.doi.org/ 10.1002/cbic.200600501 [DOI] [PubMed] [Google Scholar]
- 59.Veedu RN, Vester B, Wengel J. Polymerase chain reaction and transcription using locked nucleic acid nucleotide triphosphates. J Am Chem Soc 2008; 130:8124-5; PMID:18533656; http://dx.doi.org/ 10.1021/ja801389n [DOI] [PubMed] [Google Scholar]
- 60.Veedu RN, Vester B, Wengel J. Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org Biomol Chem 2009; 7:1404-9; PMID:19300826; http://dx.doi.org/ 10.1039/b819946a [DOI] [PubMed] [Google Scholar]
- 61.Veedu RN, Wengel J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol Biosyst 2009; 5:787-92; PMID:19603111; http://dx.doi.org/ 10.1039/b905513b [DOI] [PubMed] [Google Scholar]
- 62.Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers 2010; 7:536-42; PMID:20232325; http://dx.doi.org/ 10.1002/cbdv.200900343 [DOI] [PubMed] [Google Scholar]
- 63.Veedu RN, Wengel J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol 2009; 6:321-3; PMID:19458498; http://dx.doi.org/ 10.4161/rna.6.3.8807 [DOI] [PubMed] [Google Scholar]
- 64.Crouzier L, Dubois C, Edwards SL, Lauridsen LH, Wengel J, Veedu RN. Efficient reverse transcription using locked nucleic acid nucleotides towards the evolution of nuclease resistant RNA aptamers. PLoS One 2012; 7:e35990; PMID:22558297; http://dx.doi.org/ 10.1371/journal.pone.0035990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Højland T, Veedu RN, Vester B, Wengel J. Enzymatic synthesis of DNA strands containing α-L-LNA (α-L-configured locked nucleic acid) thymine nucleotides. Artif DNA PNA XNA 2012; 3:14-21; PMID:22679529; http://dx.doi.org/ 10.4161/adna.19272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johannsen MW, Veedu RN, Madsen AS, Wengel J. Enzymatic polymerisation involving 2′-amino-LNA nucleotides. Bioorg Med Chem Lett 2012; 22:3522-6; PMID:22503454; http://dx.doi.org/ 10.1016/j.bmcl.2012.03.073 [DOI] [PubMed] [Google Scholar]
- 67.Veedu RN, Vester B, Wengel J. Polymerase directed incorporation studies of LNA-G nucleoside 5′-triphosphate and primer extension involving all four LNA nucleotides. New J Chem 2010; 34:877-9; http://dx.doi.org/ 10.1039/b9nj00628a [DOI] [Google Scholar]
- 68.Wheeler M, Chardon A, Goubet A, Morihiro K, Tsan SY, Edwards SL, Kodama T, Obika S, Veedu RN. Synthesis of selenomethylene-locked nucleic acid (SeLNA)-modified oligonucleotides by polymerases. Chem Commun 2012; 48:11020-2; PMID:23042489; http://dx.doi.org/ 10.1039/c2cc36464f [DOI] [PubMed] [Google Scholar]
- 69.Kuwahara M, Obika S, Nagashima J-i, Ohta Y, Suto Y, Ozaki H, Sawai H, Imanishi T. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′, 4′-bridged nucleosides. Nucleic Acids Res 2008; 36:4257-65; PMID:18583360; http://dx.doi.org/ 10.1093/nar/gkn404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasahara Y, Irisawa Y, Fujita H, Yahara A, Ozaki H, Obika S, Kuwahara M. Capillary electrophoresis–systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: target binding dominated by 2′-O, 4′-C-methylene-bridged/locked nucleic acid primer. Anal Chem 2013; 85:4961-7; PMID:23662585; http://dx.doi.org/ 10.1021/ac400058z [DOI] [PubMed] [Google Scholar]
- 71.Klussmann S, Nolte A, Bald R, Erdmann VA, Fürste JP. Mirror-image RNA that binds D-adenosine. Nat Biotechnol 1996; 14:1112-5; PMID:9631061; http://dx.doi.org/ 10.1038/nbt0996-1112 [DOI] [PubMed] [Google Scholar]
- 72.Nolte A, Klussmann S, Bald R, Erdmann VA, Fürste JP. Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat Biotechnol 1996; 14:1116-9; PMID:9631062; http://dx.doi.org/ 10.1038/nbt0996-1116 [DOI] [PubMed] [Google Scholar]
- 73.Williams KP, Liu X-H, Schumacher TNM, Lin HY, Ausiello DA, Kim PS, Bartel DP. Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc Natl Acad Sci 1997; 94:11285-90; PMID:9326601; http://dx.doi.org/ 10.1073/pnas.94.21.11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purschke WG, Radtke F, Kleinjung F, Klussmann S. A DNA Spiegelmer to staphylococcal enterotoxin B. Nucleic Acids Res 2003; 31:3027-32; PMID:12799428; http://dx.doi.org/ 10.1093/nar/gkg413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vater A, Jarosch F, Buchner K, Klussmann S. Short bioactive Spiegelmers to migraine‐associated calcitonin gene‐related peptide rapidly identified by a novel approach: Tailored‐SELEX. Nucleic Acids Res 2003; 31:e130-e; PMID:14576330; http://dx.doi.org/ 10.1093/nar/gng130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eulberg D, Buchner K, Maasch C, Klussmann S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res 2005; 33:e45-e; PMID:15745995; http://dx.doi.org/ 10.1093/nar/gni044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faulhammer D, Eschgaller B, Stark S, Burgstaller P, Englberger W, Erfurth J, Kleinjung F, Rupp J, Vulcu SD, Schroder W. Biostable aptamers with antagonistic properties to the neuropeptide nociceptin/orphanin FQ. RNA 2004; 10:516-27; PMID:14970396; http://dx.doi.org/ 10.1261/rna.5186504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S. GnRH binding RNA and DNA Spiegelmers: a novel approach toward GnRH antagonism. Chem Biol 2002; 9:351-9; PMID:11927260; http://dx.doi.org/ 10.1016/S1074-5521(02)00111-4 [DOI] [PubMed] [Google Scholar]
- 79.Jarosch F, Buchner K, Klussmann S. In vitro selection using a dual RNA library that allows primerless selection. Nucleic Acids Res 2006; 34:e86-e; PMID:16855281; http://dx.doi.org/ 10.1093/nar/gkl463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helmling S, Maasch C, Eulberg D, Buchner K, Schröder W, Lange C, Vonhoff S, Wlotzka B, Tschöp MH, Rosewicz S. Inhibition of ghrelin action in vitro and in vivo by an RNA-Spiegelmer. Proc Natl Acad Sci U S A 2004; 101:13174-9; PMID:15329412; http://dx.doi.org/ 10.1073/pnas.0404175101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 2013; 48:259-71; PMID:23142634; http://dx.doi.org/ 10.1016/j.ejps.2012.10.014 [DOI] [PubMed] [Google Scholar]
- 82.2016 NPA NOXXON Pharma AG 2016. [Google Scholar]
- 83.Yatime L, Maasch C, Hoehlig K, Klussmann S, Andersen GR, Vater A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat Commun 2015; 6:6481; PMID:25901944; http://dx.doi.org/ 10.1038/ncomms7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, Xu W, Liu L, Zhu TF. A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat Chem 2016; 8:698-704; PMID:27325097; http://dx.doi.org/ 10.1038/nchem.2517 [DOI] [PubMed] [Google Scholar]
- 85.Kuwahara M, Nagashima J-i, Hasegawa M, Tamura T, Kitagata R, Hanawa K, Hososhima S-i, Kasamatsu T, Ozaki H, Sawai H. Systematic characterization of 2′-deoxynucleoside-5′-triphosphate analogs as substrates for DNA polymerases by polymerase chain reaction and kinetic studies on enzymatic production of modified DNA. Nucleic Acids Res 2006; 34:5383-94; PMID:17012278; http://dx.doi.org/ 10.1093/nar/gkl637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacPherson IS, Temme JS, Habeshian S, Felczak K, Pankiewicz K, Hedstrom L, Krauss IJ. Multivalent glycocluster design through directed evolution. Angew Chem Int Ed 2011; 50:11238-42; PMID:22191092; http://dx.doi.org/ 10.1002/anie.201105555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Temme JS, MacPherson IS, DeCourcey JF, Krauss IJ. High temperature SELMA: Evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnAb 2G12. J Am Chem Soc 2014; 136:1726-9; PMID:24446826; http://dx.doi.org/ 10.1021/ja411212q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tolle F, Brändle GM, Matzner D, Mayer G. A Versatile approach towards nucleobase‐modified aptamers. Angew Chem Int Ed 2015; 54:10971-4; PMID:26224087; http://dx.doi.org/ 10.1002/anie.201503652 [DOI] [PubMed] [Google Scholar]
- 89.Horiya S, MacPherson IS, Krauss IJ. Recent strategies targeting HIV glycans in vaccine design. Nat Chem Biol 2014; 10:990-9; PMID:25393493; http://dx.doi.org/ 10.1038/nchembio.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Temme JS, Drzyzga MG, MacPherson IS, Krauss IJ. Directed Evolution of 2G12‐Targeted Nonamannose Glycoclusters by SELMA. Chem Eur J 2013; 19:17291-5; PMID:24227340; http://dx.doi.org/ 10.1002/chem.201303848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Veedu R, Burri H, Kumar P, Sharma P, Hrdlicka P, Vester B, Wengel J. Polymerase-directed synthesis of C5-ethynyl locked nucleic acids. Bioorg Med Chem Lett 2010; 20:6565-8; PMID:20932755; http://dx.doi.org/ 10.1016/j.bmcl.2010.09.044 [DOI] [PubMed] [Google Scholar]
- 92.Zhang L, Yang Z, Sefah K, Bradley KM, Hoshika S, Kim M-J, Kim H-J, Zhu G, Jiménez E, Cansiz S. Evolution of functional six-nucleotide DNA. J Am Chem Soc 2015; 137:6734-7; PMID:25966323; http://dx.doi.org/ 10.1021/jacs.5b02251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thyer R, Ellefson J. Synthetic biology: New letters for life's alphabet. Nature 2014; 509:291-2; PMID:24805244; http://dx.doi.org/ 10.1038/nature13335 [DOI] [PubMed] [Google Scholar]
- 94.Sefah K, Yang Z, Bradley KM, Hoshika S, Jiménez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D. In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci 2014; 111:1449-54; PMID:24379378; http://dx.doi.org/ 10.1073/pnas.1311778111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Georgiadis MM, Singh I, Kellett WF, Hoshika S, Benner SA, Richards NG. Structural basis for a six nucleotide genetic alphabet. J Am Chem Soc 2015; 137:6947-55; PMID:25961938; http://dx.doi.org/ 10.1021/jacs.5b03482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Latham JA, Johnson R, Toole JJ. The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res 1994; 22:2817-22; PMID:7519769; http://dx.doi.org/ 10.1093/nar/22.14.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jensen KB, Atkinson BL, Willis MC, Koch TH, Gold L. Using in vitro selection to direct the covalent attachment of human immunodeficiency virus type 1 Rev protein to high-affinity RNA ligands. Proc Natl Acad Sci 1995; 92:12220-4; PMID:8618873; http://dx.doi.org/ 10.1073/pnas.92.26.12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Battersby TR, Ang DN, Burgstaller P, Jurczyk SC, Bowser MT, Buchanan DD, Kennedy RT, Benner SA. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J Am Chem Soc 1999; 121:9781-9; PMID:11543572; http://dx.doi.org/ 10.1021/ja9816436 [DOI] [PubMed] [Google Scholar]
- 99.Vaish NK, Larralde R, Fraley AW, Szostak JW, McLaughlin LW. A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 2003; 42:8842-51; PMID:12873145; http://dx.doi.org/ 10.1021/bi027354i [DOI] [PubMed] [Google Scholar]
- 100.Shoji A, Kuwahara M, Ozaki H, Sawai H. Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J Am Chem Soc 2007; 129:1456-64; PMID:17263432; http://dx.doi.org/ 10.1021/ja067098n [DOI] [PubMed] [Google Scholar]
- 101.Imaizumi Y, Kasahara Y, Fujita H, Kitadume S, Ozaki H, Endoh T, Kuwahara M, Sugimoto N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J Am Chem Soc 2013; 135:9412-9; PMID:23734784; http://dx.doi.org/ 10.1021/ja4012222 [DOI] [PubMed] [Google Scholar]
- 102.Li M, Lin N, Huang Z, Du L, Altier C, Fang H, Wang B. Selecting aptamers for a glycoprotein through the incorporation of the boronic acid moiety. J Am Chem Soc 2008; 130:12636-8; PMID:18763762; http://dx.doi.org/ 10.1021/ja801510d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton BE. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc 2010; 132:4141-51; PMID:20201573; http://dx.doi.org/ 10.1021/ja908035g [DOI] [PubMed] [Google Scholar]
- 104.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF. RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc 2000; 122:2433-9; PMID:11543272; http://dx.doi.org/ 10.1021/ja993688s [DOI] [PubMed] [Google Scholar]
- 105.Kimoto M, Yamashige R, Matsunaga K-i, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol 2013; 31:453-7; PMID:23563318; http://dx.doi.org/ 10.1038/nbt.2556 [DOI] [PubMed] [Google Scholar]
- 106.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Toward the combinatorial selection of chemically modified DNAzyme RNase A mimics active against all-RNA substrates. ACS Comb Sci 2013; 15:174-82; PMID:23485334; http://dx.doi.org/ 10.1021/co3001378 [DOI] [PubMed] [Google Scholar]
- 107.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010; 5:e15004; PMID:21165148; http://dx.doi.org/ 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O'Connell D, Waugh SM, Wolk SK, Mayfield WS, Burgin AB. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci 2012; 109:19971-6; PMID:23139410; http://dx.doi.org/ 10.1073/pnas.1213933109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ochsner UA, Katilius E, Janjic N. Detection of Clostridium difficile toxins A, B and binary toxin with slow off-rate modified aptamers. Diagn Microbiol Infect Dis 2013; 76:278-85; PMID:23680240; http://dx.doi.org/ 10.1016/j.diagmicrobio.2013.03.029 [DOI] [PubMed] [Google Scholar]
- 110.Jhaveri S, Olwin B, Ellington AD. In vitro selection of phosphorothiolated aptamers. Bioorg Med Chem Lett 1998; 8:2285-90; PMID:9873529; http://dx.doi.org/ 10.1016/S0960-894X(98)00414-4 [DOI] [PubMed] [Google Scholar]
- 111.King DJ, Bassett SE, Li X, Fennewald SA, Herzog NK, Luxon BA, Shope R, Gorenstein DG. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-κB RelA (p65) and p50. Biochemistry 2002; 41:9696-706; PMID:12135392; http://dx.doi.org/ 10.1021/bi020220k [DOI] [PubMed] [Google Scholar]
- 112.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry 1998; 37:16489-93; PMID:9843415; http://dx.doi.org/ 10.1021/bi981780f [DOI] [PubMed] [Google Scholar]
- 113.Lato SM, Ozerova NDS, He K, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamers to ATP. Nucleic Acids Res 2002; 30:1401-7; PMID:11884639; http://dx.doi.org/ 10.1093/nar/30.6.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]