ABSTRACT
Gene expression has been investigated in relation with growth rate in the yeast Saccharomyces cerevisiae, following different experimental strategies. The expression of some specific gene functional categories increases or decreases with growth rate. Our recently published results have unveiled that these changes in mRNA concentration with growth depend on the relative alteration of mRNA synthesis and decay, and that, in addition to this gene-specific transcriptomic signature of growth, global mRNA turnover increases with growth rate. We discuss here these results in relation with other previous and concurrent publications, and we add new evidence which indicates that growth rate controls mRNA turnover even under non-steady-state conditions.
KEYWORDS: Gene expression, growth rate, mRNA stability, Saccharomyces cerevisiae, transcription, yeast
Abbreviations
- GR
growth rate
- GRO
Genomic Run-On
- [mRNA]
mRNA concentration
- TR
transcription rate
- DR
degradation rate
- RP
ribosomal proteins
- RiBi
Ribosome Biogenesis
- ESR
environmental stress response
- iGR
instantaneous GR
- kd
DR constant
- TOR
target of rapamycin
- PKA
protein kinase A
Introduction
Growth is an inherent property of life beings.1 Since growth involves not only increase in volume, but also in mass, cells must synthesize new molecules to cope with this requirement. As proteins constitute a large part of the cell mass,2,3 most of this effort is devoted to protein synthesis, performed by ribosomes. Ribosomes are made of rRNA and ribosomal proteins (RP). In eukaryotes rRNAs are transcribed by RNA polymerases (RNA pol) I & III.4 According to these premises it seems logical that translation and RNA pol I & III transcription are unavoidably connected to cell growth rate (GR).5 Most genes, however, are transcribed by RNA pol II and encode proteins not directly related to ribosomes or translation. This raises the question as to how gene expression, considered either globally as the sum of all RNA pol II transcription or at the level of gene categories, is related or coordinated with GR.
Levels of mRNAs of specific gene functional categories correlate with growth rate
The intricate relationship between GR and gene expression has been intensively addressed by the D. Botstein's laboratory by performing chemostat experiments with the yeast Saccharomyces cerevisiae.6-8 The yeast cell adapts its GR to the availability of the limiting nutrient (a carbon, nitrogen or phosphate source). According to this experimental strategy GR is constant for long time periods and the physiology of cells is stably maintained. It can be assumed that gene expression is in a steady-state equilibrium in which mRNAs, proteins and all the other players are in constant concentrations.
Botstein's group found that about one tenth of the yeast transcriptome (628 genes) statistically and significantly correlates with GR. The mRNA level of about half of them increases with GR, and it decreases in the other half. The whole set clearly overlaps the environmental stress response (ESR)9 and fits with a slow growth signature observed in slow growth mutant strains.10 These authors and others have proposed that stress response and slow growth are intrinsically linked.10,11 By assuming that an optimal growth rate reflects the least stressful situation and vice versa, it seems that every GR is characterized by a transcriptome feature that reflects how stressed (how far from optimal) the cell is.
The mRNAs that decrease in this differential GR transcriptome are functionally connected to stress-response proteins and oxidative metabolism, whereas those that increase with GR are predominantly enriched in translation-related functions, such as ribosomal proteins and ribosome biogenesis (RiBi). These general correlations do not exclude more sophisticate regulations for specific gene subsets, like the differential response among core ribosomal proteins to GR.12
A problem with all these studies is that changes in specific mRNAs take place in relation to the population average which, in turn, is assumed to not change. Although in many instances this simplification could not be far from reality, it has not been proven that this is actually the case in all the experiments where GR varies, as pointed out by Athanasiadou et al.13 Moreover, the physiologically relevant parameter is not the amount of mRNA, but its concentration ([mRNA]), which depends on cell volume, a factor that can change with GR. This matter is normally disregarded in yeast studies.
mRNA concentrations result from 2 opposite rates: RNA pol II-dependent transcription and mRNA degradation.14 For this reason, we15 and others13 have studied the dependence of the yeast transcriptome on GR using previously published or new studies, in which different techniques have been used to quantify [mRNAs] and their turnover rates at the same time. These studies have found that cytosolic ribosome-related genes increase their [mRNA] both in relation to population average and in absolute terms.1,15 On the contrary, ESR-induced genes show a negative correlation with GR at the [mRNA] level.15 ESR-up and protein biosynthesis are functional groups that are always working in opposite directions.9 The TOR (target of rapamycin) and PKA (protein kinase A) regulatory pathways act on those regulons by means of specific protein kinases, which respond to nutrient availability and stress conditions.16 We have found that respiration-related genes also show a negative correlation with GR at the [mRNA] level.15 Interestingly the expression of some of those genes, especially those related to ethanol metabolism, have been found to positively correlate with GR in ethanol-based medium. This is clarifying because in that condition GR increase corresponds to higher respiration rate what means that is the GR, and not the carbon source, the cause of increased gene expression.8 This regulatory behavior of respiration-related genes is specific for Crabtree-positive yeasts,17 such as S. cerevisiae, because of the specialized evolved physiology of these organisms.16,18
It is noteworthy that the way to achieve the correlation with GR differs. On the one hand, the ribosome-related and ESR-up genes vary their mRNA levels by changing their synthesis rates. On the other hand, mitochondria and respiration-related mRNAs negatively adjust their mRNA concentrations to GR by controlling their stability (see Fig. 1). This differential strategy suggests the nature of the respective main regulators. Ribosome-related genes are controlled mostly by transcription factors, such as Rap1, Ifh1, Fhl1, Pbf1 and Pbf2 and Msn2/4.9,16,19 In contrast, nuclear-encoded mitochondrial genes are strongly regulated by the RNA-binding protein Puf3,20 which enables a post-transcriptional operon strategy.21
Figure 1.
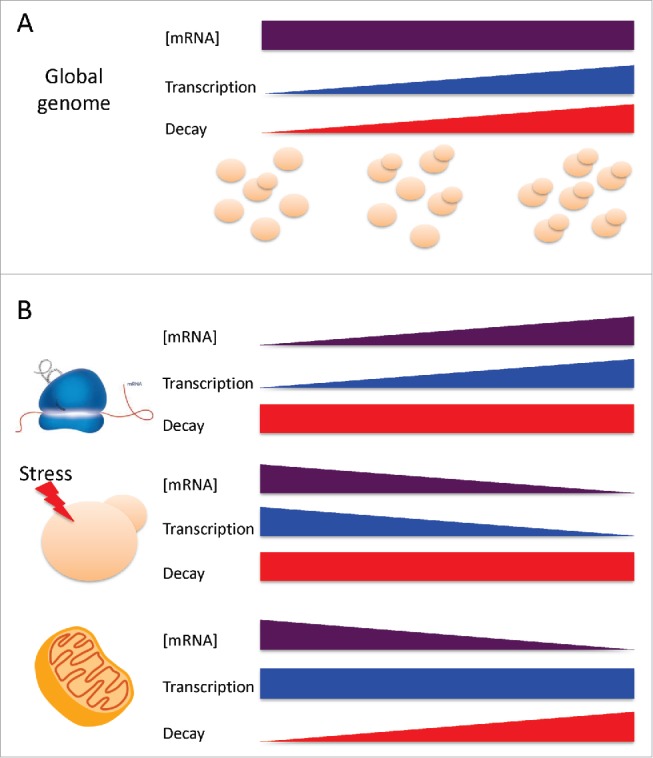
Changes in mRNA levels and turnover rates in growing yeast. (A) There is a constant mRNA concentration for most conditions with a change in GR (represented as a yeast cell population with variable budding index).15 Increasing mRNA turnover (resulting from parallel transcription and decay rates) accompanies an increase in GR (see26 for discussion). (B) Under steady-state conditions some gene functional groups change [mRNA] with GR by uncoupling the equilibrium between synthesis and decay rates. Three representative gene groups with different strategies are shown: protein biosynthesis, stress-induced and mitochondria-related genes (see text for discussion).
Growth rate controls global mRNA turnover
The link between GR and gene expression has been analyzed to date in terms of mRNA levels. The studies referred to above6-8 assume that the vast majority of the genome produces mRNAs whose concentration does not change with growth. We checked that total [mRNA] does not have a dependency on GR by comparing a collection of yeast populations with different proliferation rates, these being either wild-type cells growing under optimal and suboptimal environmental conditions or mutant strains with limited proliferation speed in batch cultures.15 This constancy of [mRNA] was not found when GR was regulated in a chemostat by limiting nutrient supply. In this case, a direct relationship between [mRNA] and GR was detected.13 Under these conditions, cell volume also increased with growth,13 which clearly contrasted with the inverse correlation found in our collection of batch culture experiments.15 This observation suggests that [mRNA] homeostasis in chemostat experiments is affected by nutrient limitation, which perhaps induces a stress response, and also by the GR itself. In other experimental approaches, however, GR could be dependent on other issues such as temperature, carbon source or strain genotype. We conclude that the interdependence of GR with gene expression can have different outcomes depending on the cause that conditions the actual GR.
We wondered whether the [mRNA] homeostasis in rich medium, produced by the steady levels of most RNA pol II-dependent transcripts across the growth range, was the result of concomitantly constant rates of transcription and mRNA degradation. We have addressed this question and have found that global mRNA synthesis and decay rates rise with growth (Fig. 1A). As they change in parallel, [mRNA] does not increase, but mRNA turnover very significantly do.15
The parallel change of mRNA synthesis and decay rates with growth is likely facilitated by the mechanistic coupling of transcription and mRNA degradation machineries. We and others have demonstrated the existence of a feedback mechanism between mRNA decay and transcription, supported by the capacity of the mRNA degradation machinery to enter the nucleus and associate with the transcription machinery in transcribed genes.22,23 Another regulatory connection between transcription and mRNA decay, but in the direction from nucleus (transcription) to cytoplasm, has also been established since mRNAs can be co-transcriptionally bound by factors that imprint them and influence their stability and translatability.24,25
The disruption of the mechanistic coupling between transcription and mRNA degradation helps to explain the regulation of the mRNAs that do change with growth (Fig. 1B). Ribosome-related and ESR-up genes change mRNA synthesis with growth (in opposite directions), but maintain degradation rates (DR) constant. This uncoupling also happens in mitochondria and respiration-related genes in glucose-based growth. In this case however, mRNA degradation is regulated, while transcription rates (TR) remain unchanged. From this perspective, co-regulation of mRNA synthesis and decay with growth would be the general rule, and their uncoupling would allow specific mRNAs to accumulate or decrease according to GR. We have also proposed that reduced mRNA stability in the context of highly proliferating cells would weaken the phenotypic impact that inherited mRNAs would produce on daughter cells, to ensure their capacity to regulate gene expression in response to environmental changes.26
What happens under non-static conditions?
In chemostat cultures, in which all food supplies and external conditions remain constant, it is assumed that steady-state conditions apply for concentrations of macromolecules.6-8,13 For batch cultures in which the culture medium is not replaced, this is only true for short time periods. We proved during a 2-hour lapse around the middle exponential growth phase that the concentrations of most mRNAs remained essentially constant.27 A larger study in batch conditions28 has recently observed a slow, but continuous change in mRNA and protein concentrations in about 1000 genes that belong to those gene functional categories previously mentioned to change with GR: protein synthesis (decrease), respiration and ESR-up (increase). It is noteworthy that no GR alteration occurs during this change, which suggests an anticipation of the diauxic shift that occurs in the yeast culture when a mostly fermentative metabolism on glucose is replaced with a respiratory metabolism on ethanol. So changes in some growth-regulated mRNAs can occur, even in special constant-GR circumstances, to anticipate future metabolic alterations.
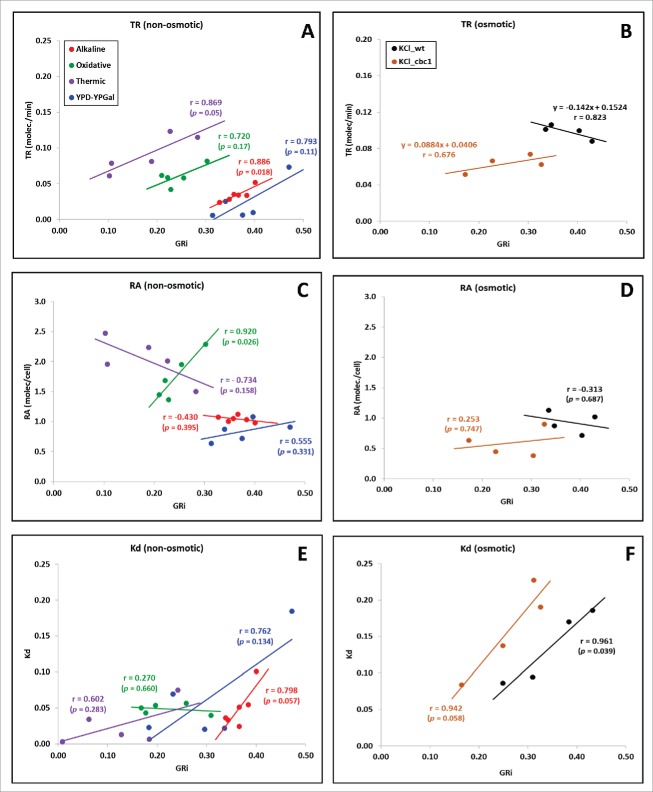
During sudden environmental changes, however, yeast cells cannot slowly adapt GR and physiology to the new situation. In this case, and depending on the intensity of stress, yeast cultures lower or stop GR and rapidly re-adapt their proteome to cope with the new conditions by developing both a general stress response (the ESR) and a specialized response to the particular stress (see9). As mentioned above, the ESR transcriptome signature is similar to the slow growth signature,10 so it would be interesting to analyze the behavior of the GR-dependent gene functional groups while a change in GR occurs due to stress. We analyzed a set of previously studied yeast responses to osmotic,29 oxidative,30 heat31 and alkaline32 stresses. Our Genomic Run-On (GRO) methodology,33 which simultaneously measures [mRNA], transcription (TR) and degradation (DR) rates, allowed us to describe previously that most mRNA changes were due to alterations in both synthesis and decay. In most cases the level of each mRNA was determined mainly by its TR, but DR usually plays an important role in fine-tuning the response and could even be a quantitatively important part of the response.32,34 However, the link between GR and gene expression has never been investigated because calculating GR in such a variable situation is not straightforward. Botstein's group created an algorithm based on a transcriptional signature to calculate an instantaneous growth rate (iGR) from the transcriptome at a particular time point.7 Now, we have used this algorithm to calculate iGR profiles from the transcriptomes ([mRNAs]) of our previous stress-response GRO studies.29-34 As expected, the common pattern was that iGR lowered immediately after applying stress and later recovered (Fig. S1). Then we analyzed the global tendency of [mRNA], TR and kd (degradation rate constant). Fig. 2 shows how analyzed stress responses can be classified into 2 groups according to their TR dependence on iGR: osmotic stresses have an almost flat tendency (Fig. 2B) and the other stresses show a positive correlation between TR and GR (Fig. 2A), rather like that observed under steady-state conditions (see18). On the contrary, the kd profiles show a positive correlation with iGR in all the stress responses (Fig. 2E–F). Thus the global mRNA level displays different tendencies, but tends to be a flat profile (Fig 2C–D). The general conclusion here is that, during stress responses, the direct correlation between mRNA turnover and GR, seen under steady-state conditions, is maintained, except for osmotic stresses. It has been seen that initial osmotic shock provokes shrinkage of cells and suddenly varies macromolecule concentrations and washes out RNA pol II and transcription factors from chromatin.35 This specific drop in transcription rates at the beginning of the stress response, when iGR is still high (Fig. S1), probably explains the distinctive profile of osmotic stress responses.
Figure 2.
Correlation between global mRNA turnover with the predicted yeast growth rate during stress responses. We plotted the predicted instantaneous growth rate (iGR in hr-1), calculated using the algorithm of Airoldi et al,7 for 5 different stress responses 29-33 (see Fig. S1) versus their median transcription rate (TR, A–B), total [mRNA] (RA, C–D) and the degradation rate constant (kd, E-F) of their transcriptomes (see15 for further details). Individual plots for each individual stress are shown, together with the linear regression, its Pearson correlation (r) and the associated p-value (p). Non-osmotic stresses (left panels) show clearly and positively correlate for both TR and kd with iGR, whereas osmotic stresses (to 0.4 M KCl in the wt and in the cbc1 mutant 29) show only a significant correlation with kd.
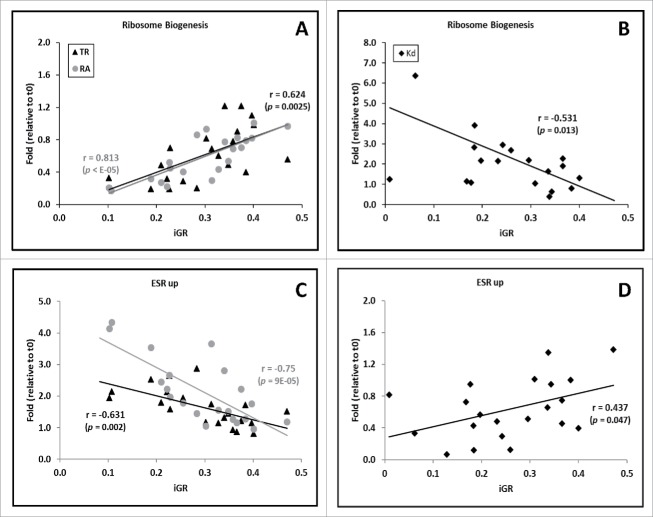
The analysis of particular gene categories shows that mRNA stability changes with iGR in all cases. In Fig. 3 we can see some gene functional groups analyzed for steady-state conditions in a previous work,15 in which mRNA stability did not play a role. During stress responses, mRNA stability actually does play a role in these functional groups. Interestingly, it works in the same direction as TR to raise (RiBi, see Fig. 3; RP not shown) or lower (ESR-up) mRNA levels. This means that mRNA decay control is more important in determining the mRNA profile during dynamic responses than in steady-state situations where it plays a more limited role.
Figure 3.
Correlation between mRNA turnover with the predicted yeast growth rate during stress responses for some selected gene groups. These analyses are identical to those in Fig. 2, but use only selected groups of functionally related genes. By way of example, we show here some groups analyzed in a previous work15 for non-osmotic stresses: Ribosome Biogenesis (A, B) and ESR-induced genes (C, D). The trends for those groups in [mRNA] (RA in graphs) are to increase (RiBi) or decrease (ESR-up). These trends were expected because the iGR calculation is, in part, based on part of their genes. Unlike previous work under steady-state conditions, these changes are due not only to parallel trends in TR, but also to inverse trends in kd profiles. p-values are shown below the r coefficient. A & C panels show TR and RA plots whereas B & D panels show kd plots.
Conclusions and future work
For most genes, the parallel change in synthesis and decay rates with growth involves no difference in [mRNA] and increased mRNA turnover. Therefore, the main difference in gene expression terms between fast- and slow-growing cells does not only consist in the differential levels of ribosome-related, respiration-related and ESR mRNAs, but also in the global change of mRNA turnover.15
In the non-steady-state, such as that caused by environmental stress, mRNA synthesis and DRs also correlate positively with the instantaneous GR deduced from transcriptomic patterns. So even when a steady state is absent, mRNA turnover seems to correlate with iGR (this article). Nutrient-limited conditions also bring about an increased TR in parallel to GR, but uncoupled synthesis and decay rates, which causes an increase in global [mRNA].15 It appears that global [mRNA] is less homeostatic under non-optimal growth conditions.
The results described and reviewed herein stem from experiments performed with budding yeast. We wondered if these phenomena were specific of this microorganism due to the specificity of its metabolic changes with growth, or whether they reflected a general paradigm that applies to other eukaryotes, including metazoan and human cells. The close similarity between yeast and cancer cells for the energetic changes associated with cell proliferation36 suggests that other fundamental processes linked to growth might be universally conserved, but experiments are needed to test this prediction. This is no easy task as measuring mRNA DRs is technically challenging in such experimental systems, particularly in living animals. New technologies need to be developed to achieve a fast, direct and unbiased mRNA turnover approach.
Finally, the mechanisms that connect GR with mRNA synthesis and degradation are unknown. They might be mediated by cell cycle regulation (discussed in15) or constitute an independent regulatory system. In both cases, the body of knowledge that accumulates on this topic in budding yeast should facilitate such unveiling.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. D. Gresham for helpful discussion and Dr. P Alepuz for critically reading the manuscript and all the members of the Valencia and Seville laboratories for their help and discussion.
Funding
This work has been supported by Spanish MiNECO and European Union funds (FEDER) to J.E.P-O. [BFU2013-48643-C3-3-P], and to S.C. [BFU2013-48643-C3-1-P], by the Regional Valencian Government [PROMETEO II 2015/006] to J.E.P-O, and by the Regional Andalusian Government [P12-BIO1938MO] to S.C.
References
- 1.Thompson DW. On Growth and Form. Cambridge University Press; Cambridge, UK: 1917; ISBN 0521 43776 8 [Google Scholar]
- 2.Schönheit P, Buckel W, Martin WF. On the Origin of Heterotrophy. Trends Microbiol 2016; 24:12-25; PMID:26578093; http://dx.doi.org/ 10.1016/j.tim.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 3.Caballero-Córdoba GM, Sgarbieri VC. Nutritional and toxicological evaluation of yeast (Saccharomyces cerevisiae) biomass and a yeast protein concentrate. J Sci Food Agric 2000; 80:341-51; http://dx.doi.org/ 10.1002/(SICI)1097-0010(200002)80:3%3c341::AID-JSFA533%3e3.0.CO;2-M [DOI] [Google Scholar]
- 4.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 1999; 24:437-440; PMID:10542411; http://dx.doi.org/ 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- 5.Mager WH, Planta RJ. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem 1991; 104:181-187; PMID:1921998; http://dx.doi.org/ 10.1007/BF00229818 [DOI] [PubMed] [Google Scholar]
- 6.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 2008; 19:352-67; PMID:17959824; http://dx.doi.org/ 10.1091/mbc.E07-08-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Airoldi EM, Huttenhower C, Gresham D, Lu C, Caudy AA, Dunham MJ, Broach JR, Botstein D, Troyanskaya OG. Predicting cellular growth from gene expression signatures. PLoS Comput Biol 2009; 5:e1000257; PMID:19119411; http://dx.doi.org/ 10.1371/journal.pcbi.1000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell 2011; 22:1997-2009; PMID:21525243; http://dx.doi.org/ 10.1091/mbc.E11-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000; 11:4241-57 ; PMID:11102521; http://dx.doi.org/ 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Duibhir E, Lijnzaad P, Benschop JJ, Lenstra TL, van Leenen D, Groot Koerkamp MJ, Margaritis T, Brok MO, Kemmeren P, Holstege FC. Cell cycle population effects in perturbation studies. Mol Syst Biol 2014; 10:732; PMID:24952590; http://dx.doi.org/ 10.15252/msb.20145172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slavov N, Airoldi EM, van Oudenaarden A, Botstein D. A conserved cell growth cycle can account for the environmental stress responses of divergent eukaryotes. Mol Biol Cell 2012; 23:1986-97; PMID:22456505; http://dx.doi.org/ 10.1091/mbc.E11-11-0961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A. Differential stoichiometry among core ribosomal proteins. Cell Rep. 2015; 13:865-73; PMID:26565899; http://dx.doi.org/ 10.1016/j.celrep.2015.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Athanasiadou R, Neymotin B, Brandt N, Miller D, Tranchina D, Gresham D. Growt rate-dependent global amplification of gene expression. BioRxiv 2016; http://dx.doi.org/ 10.1101/044735 [DOI] [Google Scholar]
- 14.Pérez-Ortín JE, Alepuz PM, Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet 2007; 23:250-7; PMID:17379352; http://dx.doi.org/21468232 10.1016/j.tig.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 15.García-Martínez J, Delgado-Ramos L, Ayala G, Pelechano V, Medina DA, Carrasco F, González R, Andrés-León E, Steinmetz L, Warringer J, et al.. The cellular growth rate controls overall mRNA turnover, and modulates either transcription or degradation rates of particular gene regulons. Nucleic Acids Res 2016; 44:3643-58; PMID:26717982; http://dx.doi.org/21468232 10.1093/nar/gkv1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosio MC, Negri R, Dieci G. Promoter architectures in the yeast ribosomal expression program. Transcription 2011; 2:71-7; PMID:21468232; http://dx.doi.org/ 10.4161/trns.2.2.14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman A, Piškur J. A study on the fundamental mechanism and the evolutionary driving forces behind aerobic fermentation in yeast. PLoS One 2015; 10:e0116942; PMID:25617754; http://dx.doi.org/ 10.1371/journal.pone.0116942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A 2005; 102:7203-8; PMID:15883364; http://dx.doi.org/ 10.1073/pnas.0502521102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 2000; 24:469-86; PMID:10978547; http://dx.doi.org/ 10.1111/j.1574-6976.2000.tb00551.x [DOI] [PubMed] [Google Scholar]
- 20.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 2000; 19:6602-11; PMID:11101532; http://dx.doi.org/ 10.1093/emboj/19.23.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007; 8:533-43; PMID:17572691; http://dx.doi.org/ 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- 22.Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 2013; 153:1000-11; PMID:23706738; http://dx.doi.org/ 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 23.Medina DA, Jordán-Pla A, Millán-Zambrano G, Chávez S, Choder M, Pérez-Ortín JE. Cytoplasmic 5'-3' exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front Genet 2014; 5:1; PMID:24567736; http://dx.doi.org/ 10.3389/fgene.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 2010; 143:552-63; PMID:21074047; http://dx.doi.org/ 10.1016/j.cell.2010.10.033 [DOI] [PubMed] [Google Scholar]
- 25.Gupta I, Villanyi Z, Kassem S, Hughes C, Panasenko OO, Steinmetz LM, Collart MA. Translational capacity of a cell is determined during transcription elongation via the Ccr4-Not complex. Cell Rep 2016; 15:1782-94; PMID:27184853; http://dx.doi.org/ 10.1016/j.celrep.2016.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chávez S, García-Martínez J, Delgado-Ramos L, Pérez-Ortín JE. The importance of controlling mRNA turnover during cell proliferation. Curr Genet 2016; PMID:27007479; http://dx.doi.org/20301094 10.1007/s00294-016-0594-2 [DOI] [PubMed] [Google Scholar]
- 27.Pelechano V, Pérez-Ortín JE. There is a steady-state transcriptome in exponentially growing yeast cells. Yeast 2010; 27:413-22; PMID:20301094; http://dx.doi.org/ 10.1002/yea.1768 [DOI] [PubMed] [Google Scholar]
- 28.Slavov N, Budnik BA, Schwab D, Airoldi EM, van Oudenaarden A. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep 2014; 7:705-14; PMID:24767987; http://dx.doi.org/ 10.1016/j.celrep.2014.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, De Clercq N, Medina DA, Garre E, Sunnerhagen P, Pérez-Ortín JE, Alepuz P. The mRNA cap-binding protein Cbc1 is required for high and timely expression of genes by promoting the accumulation of gene-specific activators at promoters. Biochim Biophys Acta 2016; 1859:405-19; PMID:26775127; http://dx.doi.org/ 10.1016/j.bbagrm.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 30.Molina-Navarro MM, Castells-Roca L, Belli G, García-Martínez J, Marín-Navarro J, Moreno J, Pérez-Ortín JE, Herrero E. Comprehensive transcriptional analysis of the oxidative response in yeast. J Biol Chem 2008; 283:17908-18; PMID:18424442; http://dx.doi.org/ 10.1074/jbc.M800295200 [DOI] [PubMed] [Google Scholar]
- 31.Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí G, Pérez-Ortín JE. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One 2011; 6:e17272; PMID:21364882; http://dx.doi.org/ 10.1371/journal.pone.0017272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canadell D, García-Martínez J, Alepuz P, Pérez-Ortín JE, Ariño J. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim Biophys Acta 2015; 1849:653-64; PMID:25900709; http://dx.doi.org/ 10.1016/j.bbagrm.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 33.García-Martínez J, Aranda A, Pérez-Ortín JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 2004; 15:303-13; PMID:15260981; http://dx.doi.org/19369426 10.1016/j.molcel.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 34.Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 2009; 15:1110-20; PMID:19369426; http://dx.doi.org/ 10.1261/rna.1435709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 2004; 118:351-61; PMID:15294160; http://dx.doi.org/ 10.1016/j.cell.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Ruiz R, Uribe-Carvajal S, Devin A, Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim Biophys Acta 2009; 1796:252-65; PMID:19682552 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.