ABSTRACT
Group II introns are large catalytic RNAs that form a ribonucleoprotein (RNP) complex by binding to an intron-encoded protein (IEP). The IEP, which facilitates both RNA splicing and intron mobility, has multiple activities including reverse transcriptase. Recent structures of a group II intron RNP complex and of IEPs from diverse bacteria fuel arguments that group II introns are ancestrally related to eukaryotic spliceosomes as well as to telomerase and viruses. Furthermore, recent structural studies of various functional states of the spliceosome allow us to draw parallels between the group II intron RNP and the spliceosome. Here we present an overview of these studies, with an emphasis on the structure of the IEPs in their isolated and RNA-bound states and on their evolutionary relatedness. In addition, we address the conundrum of the free, albeit truncated IEPs forming dimers, whereas the IEP bound to the intron ribozyme is a monomer in the mature RNP. Future studies needed to resolve some of the outstanding issues related to group II intron RNP function and dynamics are also discussed.
KEYWORDS: Cryo-EM, intron-encoded protein, intron RNP structure, Prp8, reverse transcriptase, telomerase
More than a quarter century ago, evolutionary connections started being inferred between the spliceosome and self-splicing RNAs, specifically the group II intron.1,2 The spliceosome's catalytic core was proposed to have arisen in an RNA world where nucleic acids are hypothesized to have been the prevailing macromolecular catalysts. Evidence in favor of the relatedness of group II intron RNA and the active center of the modern-day spliceosome mounted over the past decades, based on studies of splicing mechanism, RNA-metal ion coordination and RNA structure (reviewed in ref.3). This evidence has revolved around the nature of the RNA catalysts and seems incontrovertible. Now studies of group II intron-encoded proteins (IEPs) add an additional layer of similarity, not only very strikingly to the spliceosome, but also to other extant ribonucleoprotein (RNP) machines like viruses and telomerases.4,5
RNP and protein structures illuminate RNA-protein interactions
While previous efforts have revealed the structure of a group IIA intron at low resolution,6-8 the recent breakthrough to solve the group IIA intron RNA from Lactococcus lactis in complex with the intron encoded protein (IEP) provides a first near-atomic model of the overall architecture of a group II intron RNP (Fig. 1A).4 Also, we now have a close-up view of the interaction sites between excised RNA lariat and protein components, giving important insights into functions of the RNP. The IEP, which has multiple roles in splicing and mobility of the intron, has modules with reverse transcriptase (RT) and DNA-binding/endonuclease activities. The RT is subdivided into an RNA-binding region contained within an N-terminal extension (NTE, also referred to as RT0), a fingers-palm domain (RT1-RT7) and a thumb domain (Fig. 1B). The fingers-palm domain act together with the DNA endonuclease primarily to promote intron mobility, whereas the thumb domain functions as a maturase involved primarily in splicing, with some overlapping functionality. The RNA domains I through VI form the characteristic Y-shaped structure (Fig. 1A). DI is a folding scaffold that contains the exon-binding sites (EBS1 and EBS2), DII and DIII enhance catalytic activity, DIV is the major protein-binding domain, DV is the active site helix and DVI contains the nucleophile, a bulged adenosine (A) that initiates splicing and creates the branch-point (BP) of the intron lariat.9,10 The L. lactis IEP, called LtrA, makes several contacts with specific sites in DI and DIV. A remarkable feature of the RNP structure is that it contains the spliced mRNA, which pinpoints its interactions with EBS1 and EBS2 and its relationship to the IEP.4 This study not only represents a breakthrough in terms of a group II intron RNP structure, but it also reveals the first complete structure of a native IEP at near-atomic resolution (with an average resolution of 3.8 Å).
Figure 1.
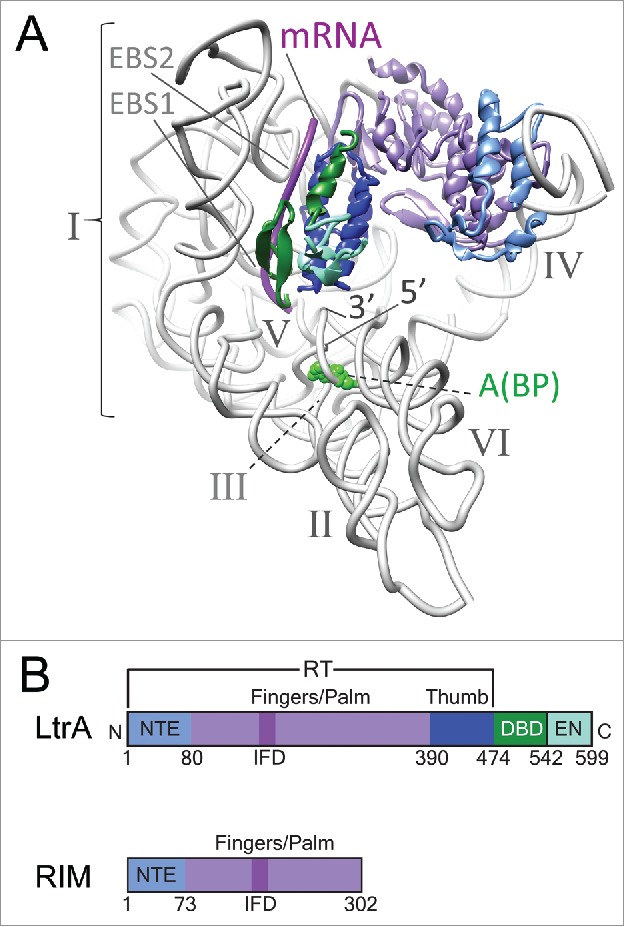
LtrA RNP with IEP consisting of different functional domains. (A) Structure of the group II intron-LrtA complex as established by cryo-EM. Six structural domains of the RNA (gray) are labeled I through VI. EBS1 and EBS2 point to the exon-binding sites within domain I of the intron RNA. The 5′ and 3′ ends of the intron RNA are marked, and the catalytic adenosine (A) at the branch point (BP) is highlighted green by space-filling. The bound fragment of mRNA captured in the cryo-EM study is magenta. Structural domains of LtrA are colored: NTE, light blue; RT fingers/palm domain, purple; thumb domain, dark blue; DNA-binding domain, green; and the endonuclease domain, cyan. (B) Comparison of domain organization of the 2 group II IEPs, LtrA and RIM, colored as in panel A. RIM is a truncated version of the full-length protein. Structures in this and subsequent figures were generated using Chimera 23.
Simultaneously, 2 structures of the fingers-palm domain alone of the RT from Roseburia intestinalis (Ri) and Eubacterium rectale (Er) bacteria were solved at 1.2 and 2.2 Å resolution, respectively.5 These two structures, referred to as maturases, encompass the RT0-RT7 motifs and include the NTE and fingers-palm domain, but not the thumb domain. The 2 structures are almost identical and the focus is therefore on Ri maturase (RIM), which is of exceptional resolution. Despite the absence of the thumb domain, RIM binds RNA from DIV with high affinity and specificity. Together the RIM and LtrA structures reinforce a common ancestry with the spliceosome, while they also manifest provocative differences.
RTs imply forks in the evolutionary tracks
In the almost 50 y since the discovery of RTs in retroviruses, these RNA-templated polymerases have presented a large and diverse population of enzymes that are common in selfish elements like retrotransposons, including group II introns, and viruses.11,12 RTs also occur in some cellular proteins like telomerase, which performs DNA synthesis at chromosome ends.11-13 Additionally, their similarity to a key spliceosomal protein Prp8, which, despite its lack of RT activity, provided the first suggestion of protein relatedness between group II introns and spliceosomes.14 The fingers-palm and thumb domains in all of these diverse RTs are assumed to have a common ancestor, and ancillary functions are thought to have been acquired as N- and C-terminal extensions and as insertions between the conserved RT1-RT7 motifs.13
The RT of the lactococcal LtrA bears close similarity to Prp8, most strikingly in the thumb domain. In addition to the structural parallel, the interactions of these 2 proteins with RNA are stunningly similar (Fig. 2).4 The thumb of LtrA and Prp8 both interact with RNA elements involved in 5′-splice site recognition, contacting the exon-binding EBS1 and U5 snRNP, respectively. These interactions both position the 5′ exon to the respective RNA active sites, which include a similar catalytic triad15 and the bulged adenosine nucleophile poised for catalysis. The most recently solved structures of the spliceosome in different functional states, the C complex state16,17 and the intron-lariat spliceosomal (ILS) complex state,18 show overall similarity but also distinct differences with the group IIA intron RNP complex at the splicing active site (Fig. 2). While the location of the spliced exon RNA in the group IIA intron is very similar to that of the equivalent exon in the C complex of spliceosome, the branch-point (BP) of the lariat adopts a similar position to the ILS complex, suggesting that the group II intron-LtrA complex represents an intermediate between these 2 splicesomal states. This underscores the close resemblance between the cryo-EM structure of the group IIA intron RNP complex and different functional states of the spliceosome. A common origin for these 2 RNPs seems irrefutable. The Ri and Er RT is also architecturally highly similar to the Prp8 protein, but the parallels reside in RT0, the NTE involved in RNA-binding, rather than in the thumb domain, which is not included in the crystal structures of the Ri and Er RTs. These observations suggest that whereas group II IEPs and the spliceosomal Prp8 evolved in parallel, the closely-related group II introns are somewhat divergent.
Figure 2.
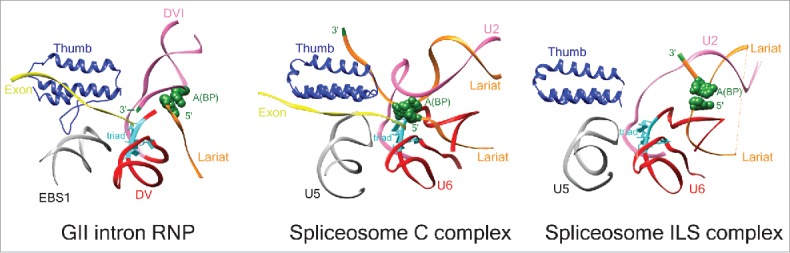
Configuration of active sites in group II intron and spliceosome. The important structural elements around the splicing active site are shown for the group II intron-LtrA complex (PDB ID: 5g2x, left), spliceosome complex C (PDB ID: 5gmk, middle), and spliceosome ILS complex (PDB ID: 3jb9, right), as revealed from their high resolution cryo-EM structures. The common functional features are depicted in the same color. The A(BP) is as in Fig. 1A, and other nucleotides in the catalytic triad are highlighted in cyan. The relative locations among the exon, the branch point, and the triad in the 3 structures suggest that the group II intron-LtrA complex represents an intermediate state between the C and ILS states of the spliceosome.
Intriguingly, a similarity of the fingers/palm of LtrA to the telomerase RT (TERT) also emerged.4 An artificial DNA-RNA heteroduplex from a co-crystal structure of the beetle Tribolium castaneum could be docked into the analogous site of LtrA without perturbing the main chain RNA structure and the heteroduplex contacts conserved amino acid residues at the active site. This similarity gives insight into the initial steps of intron retrohoming. A similarity of RIM to TERT also became apparent when the RT0 motif was removed from the analysis.5 Furthermore, whereas the fingers-palm RT1-RT7 of LtrA is related to TERT, the RIM structure of this domain shares substantial resemblance to the flaviviral hepatitis C virus RNA polymerase.
What might one make of these differences between the related bacterial group II intron maturases and their resemblances to extant eukaryotic RT-related proteins? Structure-based sequence of alignments of the RT fingers-palm domain indicate that the 3 IEPs have a very high degree of similarity, except that LtrA is less streamlined, with longer sequences between the RT1-RT7 motifs, including a lengthy amino acid insertion 4a between RT4 and RT5 (Figs. 3A and 4A). When a phylogenetic tree is constructed from the conserved motifs RT1-RT7, excluding the insertions, the 3 IEPs cluster together with very high statistical support (100% bootstrap value). These IEPs are also clearly related to Prp8, TERT and viral polymerases, but much more distantly so, with an unresolved branching order (Fig. 3B). From purely sequence-based comparisons, distinctive classes of group II introns, retroviruses, viral polymerases, TERT and other retroelements emerge,11-13 but the structure-based alignments give few clues as to ancestral origins, given the paucity of solved structures. Nevertheless, an unbiased comparison of the overall RMSD values for the structure of the RT domains (RT0-RT7) in RIM5 and LtrA4 with the structure of the RT domain in various related proteins finds HCV (PDB ID: 1C2P) with an RMSD of 2.2 Å and TERT (PDB ID: 3KYL) with an RMSD of 2.0 Å, respectively, as their closest matches.
Figure 3.
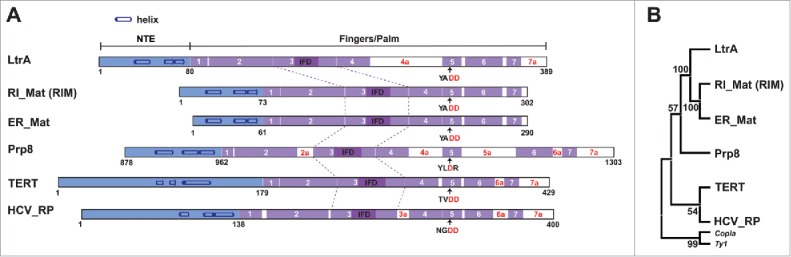
Structure-based sequence alignments and phylogenetic relatedness. (A) Organization of homologous RT domains in various IEP-related proteins. Conserved sequence motifs RT1–RT7 (purple) and IFDs (insertion in fingers domain, dark purple) are depicted. Insertions between RT1 and RT7 are designated as white spaces. The two catalytic aspartates are highlighted (red D's). (B) Phylogenetic tree showing evolutionary relatedness among proteins in panel A. The tree was constructed by the neighbor-joining method using structure-based sequence alignments. Copia and Ty1 RTs were added based on structure predictions as an out-group in the phylogenetic analysis.
Figure 4.
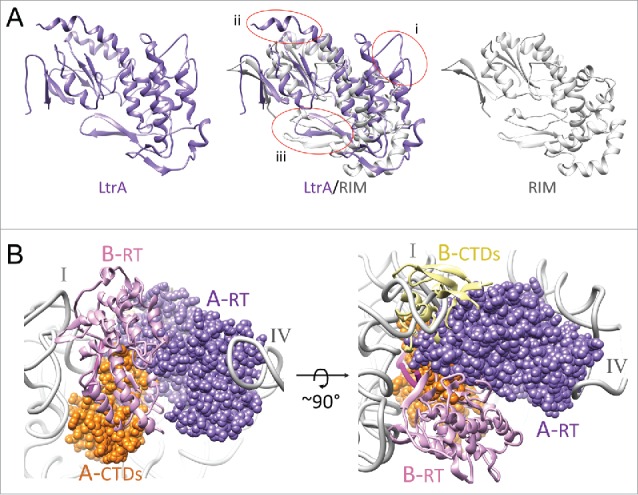
Differences between LtrA and RIM. (A) Comparison of LtrA and RIM structures. The RT domains of LtrA (PDB ID: 5g2x, purple) and RIM (PDB ID: 5hhj, gray) are shown as individual and superimposed ribbons. Three disparate regions between the 2 structures are circled (i – iii). LtrA is larger than RIM, with insertions in (i) the NTE and RT3 motif (encompassing the IFD), (ii) the 4a region (a significant portion of the 4a region that extended beyond the α-helix 5 was disordered in the RNP structure and could not be modeled), and (iii) the RT1/RT2 motif with a β-sheet hairpin, which is differently oriented between the structures. (B) Probing LtrA dimer formation. LtrA is shown in dimeric configuration of the RT domains RT0-RT7 as observed in the X-ray crystallographic study of RIM. The RNA-bound copy of the LtrA (chain A), from the cryo-EM study, is shown as a space-filled model, whereas the second superimposed copy of LtrA (chain B) is shown as ribbons. Labels: A-RT (dark purple) and B-RT (light purple) depict NTE and maturase domains of chains A and B, respectively; A-CTDs (orange) and B-CTDs (yellow) depict the thumb, DNA-binding, and endonuclease domains of chains A and B, respectively. I and IV, indicate the 2 LtrA-interacting domains of the RNA intron (gray).
The monomer-dimer conundrum
A mystery raised by the IEP structures and by previous studies, is that the RIM RT forms a dimer,5 whereas LtrA clearly binds the intron RNA as a monomer.4 However, previous studies with RNPs assembled in vitro and in vivo also suggest that LtrA binds RNA as a dimer.7,19-21 Given this conundrum, further analysis is in order. One possibility is that the RIM structure represents a truncated protein from which a sequence that inhibits dimerization has been deleted and that full-length RIM also binds the intron RNA as a monomer. Another possibility is that LtrA associates with the intron as a dimer, where 2 copies of LtrA have different affinities for the intron, and the low-affinity LtrA molecule dissociates during flash freezing for cryo-EM grid preparation. Thus only the high-affinity LtrA copy is retained and visible in the 3D reconstruction. A labile association between the RNA is indeed suggested by the finding that about one-fourth of the particles imaged for cryo-EM are stripped of LtrA, which is the basis of the affinity purification.4 Such a possibility is also consistent with the observation that RNP particles from similar preparations examined by SEC-MALS (size exclusion chromatography-multi angle light scattering) had a calculated theoretical mass consistent with LtrA binding as a dimer.7
Then questions arise relating to functional and structural aspects of dimerization. Could dimerization possibly help bring the active sites of 2 functional domains of LtrA, the endonuclease and RT domains that are ∼45 Å apart in the cryo-EM structure of the RNP4 into functionally required proximity? We examined the possibility by modeling a RIM-inspired dimer of LrtA. The superimposition shows a significant steric clash between the C-terminal domains (CTDs), including the thumb, DNA-binding and endonuclease domains of the second copy of LtrA with the intron RNA (Fig. 4B). A significant portion of the thumb and endonuclease domains, which are absent from the RIM protein would be filling the space occupied by a portion of domain I of the intron RNA. Thus, a dimer of LtrA, in the configuration proposed in the X-ray crystallographic study of RIM is ruled out. Existence of alternative LtrA stoichiometry in different states of the RNP is a possibility to reconcile these differences,22 but our analysis suggests that 2 copies of LtrA would have to undergo very large inter-domain rearrangements. However, at the other end of the molecule, there is no steric concern with the N-terminal regions, including the NTE and α-helix 4 of the fingers and palm domain,4 so one could assume that the anchoring interactions between the LtrA and domain IVa of the intron RNA, as described in our cryo-EM structure, could be retained during dimerization of LtrA.
Looking to the past and the future
Sorting through different evolutionary connections between retroelements will certainly require more bioinformatics analyses and solved structures. A favored scenario is that the group II intron-like RNA is ancestral to the spliceosomal snRNA and that the IEP is the progenitor of Prp8, which lost RT activity.14 Similarities between the IEPs and TERT likewise suggest ancestral relationships and although some favor domestication of the group II intron as the origin of TERT, the order of events remains speculative (papers cited in ref.4). How viruses fit into the picture and whether they were derived from some of these retroelements or were their progenitors is even more ambiguous. What appears more certain is that the RT motifs and enzymatic machinery remained relatively fixed, while terminal and internal adaptations evolved to accommodate particular functions of the diverse RT families.
Comparing structures of these different group II IEPs and their RNPs will be mechanistically revealing while also solving the monomer-dimer conundrum. Then, examining the whole RNP as it engages in its various functions will begin to address intron dynamics. These different conformers will include not only the precursor RNP but also intermediates along the splicing pathway. There are also questions of how the excised intron RNP, captured by cryo-EM, contacts its DNA substrate as it initiates retromobility. Snapshots of such an RNP complex as it advances along its retromobility course, integrating into DNA, can be taken through the lens of crystallography, cryo-EM and single-molecule analysis. This combination of structure studies and kinetic analyses while interrogating the dynamics of conformational changes will undoubtedly illuminate the way.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are extremely thankful to 3 talented post-docs who helped with analyses and rendering of the figures: Prem Kaushal (Fig. 1A), Guosheng Qu (Fig. 3A) and Olga Novikova (Fig. 3B). We thank Drs. Irina Arkhipova and Alan Lambowitz for their insightful comments on the manuscript. Work in our labs is supported by grants from the NIH (GM061576 to RKA; GM39422 and GM44844 to MB) and the National Science Foundation of China (31270765 to H-WW).
References
- 1.Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 1986; 44:207-10; PMID:2417724; http://dx.doi.org/ 10.1016/0092-8674(86)90751-8 [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. Five easy pieces. Science 1991; 254:663; PMID:1948046; http://dx.doi.org/ 10.1126/science.1948046 [DOI] [PubMed] [Google Scholar]
- 3.Lambowitz AM, Belfort M. Mobile bacterial group II introns at the crux of eukaryotic evolution. Mobile DNA III: Microbiol Spectr./ASM Press 2015; 3:1209-36; PMID:26104554; http://dx.doi.org/27136327 10.1128/microbiolspec.MDNA3-0050-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang HW. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol 2016; 23:549-59; PMID:27136327; http://dx.doi.org/ 10.1038/nsmb.3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C, Pyle AM. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol 2016; 23:558-65; PMID:27136328; http://dx.doi.org/ 10.1038/nsmb.3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slagter-Jäger JG, Allen GS, Smith D, Hahn IA, Frank J, Belfort M. Visualization of a group II intron in the 23S rRNA of a stable ribosome. Proc Nat Acad Sci USA 2006; 103:9838-43; PMID:16785426; http://dx.doi.org/ 10.1073/pnas.0603956103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta K, Contreras LM, Smith D, Qu G, Huang T, Spruce LA, Seeholzer SH, Belfort M, Van Duyne GD. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res 2014; 42:5347-60; PMID:24567547; http://dx.doi.org/ 10.1093/nar/gku140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang T, Shaikh TR, Gupta K, Contreras-Martin LM, Grassucci RA, Van Duyne GD, Frank J, Belfort M. The group II intron ribonucleoprotein precursor is a large, loosely packed structure. Nucleic Acids Res 2011; 39:2845-54; PMID:21131279; http://dx.doi.org/ 10.1093/nar/gkq1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harbor Perspectives in Biol 2011; 3:a003616; PMID:20463000; http://dx.doi.org/ 10.1101/cshperspect.a003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pyle AM. Group II Intron Self-Splicing. Ann Rev Biophys 2016; 45:183-205; PMID:27391926; http://dx.doi.org/ 10.1146/annurev-biophys-062215-011149 [DOI] [PubMed] [Google Scholar]
- 11.Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell 1998; 92:587-90; PMID:9506510; http://dx.doi.org/ 10.1016/S0092-8674(00)81123-X [DOI] [PubMed] [Google Scholar]
- 12.Eickbush TH. Telomerase and retrotransposons: which came first? Science 1997; 277:911-2; PMID:9281073; http://dx.doi.org/ 10.1126/science.277.5328.911 [DOI] [PubMed] [Google Scholar]
- 13.Gladyshev EA, Arkhipova IR. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A 2011; 108:20311-6; PMID:21876125; http://dx.doi.org/ 10.1073/pnas.1100266108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 2013; 493:638-43; PMID:23354046; http://dx.doi.org/ 10.1038/nature11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol 2014; 21:464-71; PMID:24747940; http://dx.doi.org/ 10.1038/nsmb.2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 2016; 353:895-904; PMID:27445308; http://dx.doi.org/ 10.1126/science.aag2235 [DOI] [PubMed] [Google Scholar]
- 17.Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016; 537:196-201; PMID:27459055; http://dx.doi.org/ 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015; 349:1182-91; PMID:26292707; http://dx.doi.org/ 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- 19.Rambo RP, Doudna JA. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 2004; 43:6486-97; PMID:15157082; http://dx.doi.org/ 10.1021/bi049912u [DOI] [PubMed] [Google Scholar]
- 20.Blocker FH, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 2005; 11:14-28; PMID:15574519; http://dx.doi.org/ 10.1261/rna.7181105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 1999; 38:9069-83; PMID:10413481; http://dx.doi.org/ 10.1021/bi982799l [DOI] [PubMed] [Google Scholar]
- 22.Piccirilli JA, Staley JP. Reverse transcriptases lend a hand in splicing catalysis. Nat Struct Mol Biol 2016; 23:507-9; PMID:27273636; http://dx.doi.org/ 10.1038/nsmb.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605-12; PMID:15264254; http://dx.doi.org/ 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]


