ABSTRACT
Muscle development, or myogenesis, is a highly regulated, complex process. A subset of microRNAs (miRNAs) have been identified as critical regulators of myogenesis. Recently, miR-378a was found to be involved in myogenesis, but the mechanism of how miR-378a regulates the proliferation and differentiation of myoblasts has not been determined. We found that miR-378a-3p expression in muscle was significantly higher than in other tissues, suggesting an important effect on muscle development. Overexpression of miR-378a-3p increased the expression of MyoD and MHC in C2C12 myoblasts both at the level of mRNA and protein, confirming that miR-378a-3p promoted muscle cell differentiation. The forced expression of miR-378a-3p promoted apoptosis of C2C12 cells as evidenced by CCK-8 assay and Annexin V-FITC/PI staining results. Through TargetScan, histone acetylation enzyme 4 (HDAC4) was identified as a potential target of miR-378a-3p. We confirmed targeting of HDAC4 by miR-378a-3p using a dual luciferase assay and western blotting. Our RNAi analysis results also showed that HDAC4 significantly promoted differentiation of C2C12 cells and inhibited cell survival through Bcl-2. Therefore, we conclude that miR-378a-3p regulates skeletal muscle growth and promotes the differentiation of myoblasts through the post-transcriptional down-regulation of HDAC4.
KEYWORDS: Cell apoptosis, cell proliferation, HDAC4, miR-378a-3p, muscle differentiation
Introduction
Skeletal muscle growth and maintenance are essential for animal and human health, providing structural support that allows the control of motor movements and also allows energy storage.20 Therefore, muscle development plays a crucial role in overall body metabolism. Although many factors have been found to importantly contribute to the complex process of muscle development, the different mechanisms are not fully elucidated. MicroRNAs (miRNAs), 18–25 nucleotides, small single-stranded non-coding RNAs, negatively regulate gene expression and play crucial roles in many biological processes, especially muscle development.16 In animals, miRNAs are transcribed in the nucleus, are subsequently processed by 2 RNase III proteins (Drosha and Dicer), and are finally incorporated into RNA-induced silencing complexes that mediate translational inhibition or degradation of target mRNAs23 by base pairing with the 3′ untranslated region (UTR) of target mRNAs. By silencing the transcription of different target mRNAs, miRNAs are involved in nearly all developmental and pathological processes in animals including cell proliferation,6 cell differentiation,44 apoptosis,9,40,46 fat metabolism,12,28 and others.
These miRNAs also play roles in muscle development in mammals. Previous studies reported that several miRNAs, such as miR-1,47 miR-133,31,45,49 and miR-206,3,19,42 are muscle-specific miRNAs because they are key regulators in muscle development in human, pigs, and rats. Others miRNAs may also contribute to muscle development. For example, miR-1/206 may regulate prenatal skeletal muscle development by inhibiting SFRP1.48 Another miRNA, miR-27b, may affect bovine skeletal muscle growth and hypertrophy by targeting the muscle-specific gene MSTN.29 The overexpression of miR-29 in C2C12 myoblast represses proliferation but promotes myotube formation.39
Another miRNA, miR-378, affects cell survival, tumor growth, angiogenesis, and cell differentiation.7 Specifically, miR-378 can enhance tumor cell survival by repressing SuFu and Fus-1,24 increase the transcriptional activity of MyoD in part by repressing MyoR,13 promote BMP2-induced osteogenic differentiation,17 and also regulate osteoblast differentiation by targeting GalNT-7 in MC3T3-E1 cells.21 Previous studies also demonstrated that miR-378 suppressed cell migration and promoted cell apoptosis in prostate cancer8 and increased the size of lipid droplets and the incorporation of acetate into triacylglycerol.14 In mice, miR-378 participates in the regulation of mitochondrial metabolism and energy homeostasis.4 miR-378 suppress expression of Gli3 to inhibit activation of hepatic stellate cells and liver fibrosis.18 However, a role of miR-378 in the regulation of skeletal muscle development has not been described.
HDAC4, a member of the HDACs family, is as a crucial controller of cell growth,1 differentiation,43 and migration37 in various cell types. HDAC4 facilitates proliferation and migration of vascular smooth muscle cell during neointimal hyperplasia.37 HDAC4 in human glioblastoma leads to cell proliferation arrest and tumor growth impairment via inducing p21 expression.30,41 These reports demonstrate that HDAC4 plays a significant role in regulation of cell differentiation and proliferation. Nevertheless, the mechanism by which HDAC4 controls the differentiation and proliferation of myoblasts is unclear.
In this study, we investigated the role of miR-378a-3p in regulating muscle cell differentiation and proliferation. We found that miR-378a-3p showed a high expression level in skeletal muscle, promoted differentiation, and repressed proliferation of C2C12 cells. Additionally, we determined that miR-378a-3p overexpression can regulate the differentiation and proliferation of skeletal muscle myoblasts by targeting HDAC4.
Results
miRNAs expression profile in different tissues
To study the function of miRNAs in muscle development, 8 miRNAs including miR-378a-3p were selected as potential regulatory candidates based on our previous Solexa SBS technology sequencing results.36 The expression profile assay showed that miR-378a-3p expression was significantly higher than other miRNAs in the longissimus muscle (Fig. 1a), suggesting a potential role in muscle. We found that miR-378a-3p was expressed predominantly in muscle and weakly expressed in other tissues (Fig. 1b). Based on this, we speculated that miR-378a-3p could be a regulator in skeletal muscle development. Mature bta-miR-378 derives from the first intron of PPARG1B in chromosome 7 (Fig. 1c). A similarity analysis showed that miR-378a-3p was conserved among cattle, mouse, and humans (Fig. 1d). Thus, we chose mouse C2C12 cells as the experiment model.
Figure 1.

Expression of miRNAs in Qinchuan cattle detected by QRT-PCR. (a) The expression of different miRNAs in skeletal muscle. (b) The expression of miR-378a-3p in different tissues of Qinchuan cattle. (c) pre-miR-378a-3p is located in the first intron of PPARGC1B gene in chromosome 7. (d) miR-378a-3p has high conservative in different species of mammal. The expression of miRNAs was normalized to U6. Values are mean ± SEM for 3 biological replicates, * P< 0.05, ** P < 0.01.
miR-378a-3p inhibited C2C12 cell proliferation
In order to explore an effect of miR-378a-3p on myoblast proliferation, C2C12 myoblasts were transfected with pcDNA3.1-miR-378a-3p, pcDNA3.1 (+) (control plasmid with no insert), and miR-378a-3p inhibitor. The transfection efficiency was detected by RT-qPCR and the results showed significantly different expression of miR-378a-3p (Fig. S1). Flow cytometry was used to analyze the cell cycle phase distribution, and the results showed that the number of cells at G0/G1 and G2 phases increased and there were reduced number of cells in S phase after transfection with pcDNA3.1-miR-378a-3p (Fig. 2a-c), which strongly suggests that miR-378a-3p arrests cells in the G0/G1 stage. This effect was reversed when the miR-378a-3p inhibitor was used (Fig. S2). To determine cell proliferation, we used the Cell Counting Kit-8 (CCK-8) assay and found that miR-378a-3p significantly inhibited cell proliferation (Fig. 2d). Cell proliferation was also detected using a 5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay. Similarly, C2C12 cells showed significantly less mitotic activity when miR-378a-3p was overexpressed (Fig. 2e, f). Taken together, miR-378a-3p was shown to inhibit the proliferation of C2C12 cells.
Figure 2.

miR-378a-3p represses the proliferation of C2C12 cell. (a, b, c) Cell were transfected by pcDNA3.1 (+) and pcDNA3.1-miR-378a-3p vector, and the cell cycle phase and proliferation index were analyzed by flow cytomctry. (d, e) Cell counting kit-8 (CCK-8) detected cell proliferation index. (e) 5-Ethynyl-2′-deoxyuridine (EdU) detected cell proliferation index, the scale bar stand 200μm. (f) Analysis results of EdU positive cells. Values are mean ± SEM for 3 biological replicates, * P< 0.05, ** P < 0.01.
miR-378a-3p promoted C2C12 cell apoptosis
To confirm the effect of miR-378a-3p on cell apoptosis, pcDNA3.1-miR-378a-3p was transfected into the C2C12 cells. The Annexin V- FITC/PtdIns staining assay showed the apoptosis index in the group with miR-378a-3p overexpression was significantly increased (Fig. 3a-c). We also detected the expression of cell survival related genes Bcl-2, Caspase 3, and BAX, and found that miR-378a-3p upregulated mRNA expression of these genes (Fig. 3d-g, S3), but showed the opposite effect when miR-378a-3p was inhibited (Fig. S2). These results demonstrate that miR-378a-3p promotes C2C12 cells apoptosis.
Figure 3.

miR378a-3p promotes C2C12 cell apoptosis. (a, b) C2C12 cells were transfected by pcDNA3.1 (+) and pcDNA3.1-miR-378a-3p, collected and stained by Annexin V-FITC/PtdIns. Cell apoptosis phase distribution were analyzed by flow cytomctry. (c) Cell apoptosis index were analyzed between the 2 groups. (d-g) Expression of cell apoptosis relative genes were detected by QRT-PCR and western blot. Values are mean ± SEM for 3 biological replicates, * P< 0.05, ** P < 0.01.
miR-378a-3p promoted C2C12 cell differentiation
To determine whether miR-378a-3p affects cell differentiation, the expression of miR-378a-3p was detected at different stages of differentiation. We found that the expression of miR-378a-3p generally increased in a time-dependent manner (Fig. 4a). The C2C12 cells transfected with pcDNA3.1-miR-378a-3p were induced to differentiate for 6 days and myoblast differentiation marker genes were detected by RT-qPCR and protein gel blot. We found that expression of MyoD and MHC was significantly upregulated in C2C12 cells that overexpressed miR-378a-3p, both at the mRNA and protein levels (Fig. 4b-d, S5). Taken together, these results revealed that miR-378a-3p promoted C2C12 cell differentiation.
Figure 4.
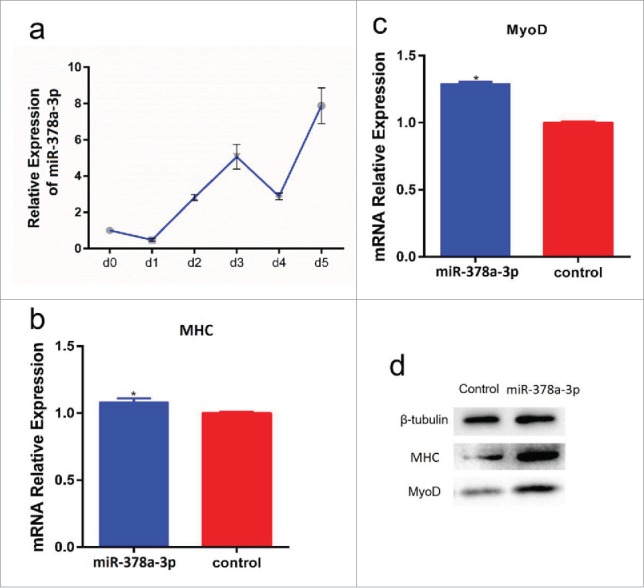
miR-378a-3p promotes C2C12 cell differentiation. (a) miR-378a-3p expression in different stages of differentiation. (b, c) Overexpression of miR-378a-3p in C2C12 cell and continues induced to undergo differentiation for 6 days, and subsequent detection the mRNA expression of MHC and MyoD by RT-PCR compared with control group. (d) The protein expression of MHC and MyoD were detected by protein gel blot. Days (d) indicate the time of cells in differentiation medium. Values are mean ± SEM for 3 biological replicates, * P < 0.05, ** P < 0.01.
miR-378a-3p directly targeted the 3′UTR of the HDAC4 gene
To determine the mechanism by which miR-378a-3p promotes cell differentiation, we predicted potential target genes of miR-378a-3p using TargetScan6.2, miRanda, and the starBase database. HDAC4, one of the candidate genes, is related to muscle development and has a highly conserved binding site in the mRNA 3′UTR that is complementary to the miR-378a-3p seed. We found that this predicted binding site is highly conserved among vertebrates.
To establish the relationship between miR-378a-3p and HDAC4, a 500 bp fragment of HDAC4 3′ UTR containing the miR-378a-3p binding site was cloned into the psiCHECK-2 vector to yield psiCHECK-2-3′UTR-W. A separate fragment containing a 2-base mutation in the seed binding site of the HDAC4 3′ UTR was also moved into the psiCHECK-2 plasmid vector as psiCHECK-2-3′UTR-Mut (Fig.5a, b). The pcDNA3.1-miR-378a-3p was co-transfected with psiCHECK-2-3′UTR-W or psiCHECK-2-3′UTR-Mut into 293T cells. The Renilla luciferase activity was not reduced in cells containing the psiCHECK-2-3′UTR-Mut compared to the cells containing the psiCHECK-2-3′UTR-W (Fig. 5c). These results demonstrated the specific targeting relationship of miR-378a-3p and HDAC4 gene.
Figure 5.
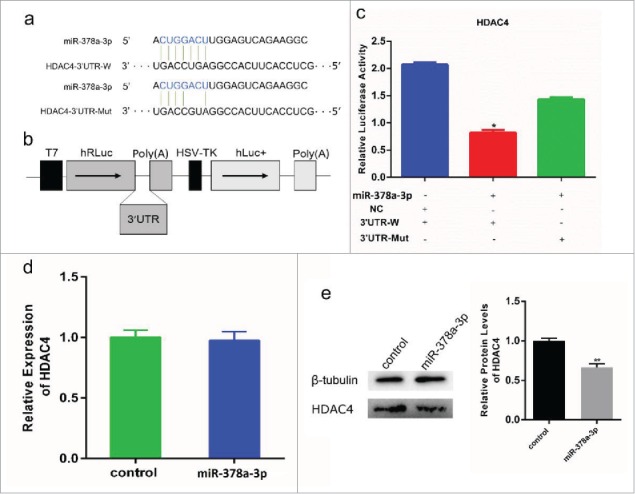
miR-378a-3p directly targets HDAC4 gene. (a) Sequence of miR-378a-3p and its predicted binding site in HDAC4 3′UTR and mutation 3′UTR. (b) The sketch map of psiCHECK2 vector in which HDAC4 3′UTR and mutation 3′UTR were inserted into the 3′ end of Renilla luciferase gene (hRluc). (c) miR-378a-3p/NC co-transfected with psiCHECK2-3′UTR-W and psiCHECK2-3′UTR-Mut into C2C12 cells individually, and Renilla luciferase activity was normalized to the firefly luciferase (hLuc+) activity. (d) HDAC4 mRNA expression in C2C12 myoblasts was detected by RT-PCR at 48 h post-transfection with miR-378a-3p and control. (e) HDAC4 protein expression was detected by western blot and gray value analysis. Values are mean ± SEM for 3 biological replicates, * P < 0.05, ** P < 0.01.
To confirm the effect of miR-378a-3p on HDAC4 in myoblasts, we examined the HDAC4 protein level following miR-378a-3p transfection. Western blot analysis indicated that miR-378a-3p overexpression suppressed HDAC4 expression (Fig. 5e); no effects were detected when miR-378a-3p interference was used to decrease activity (Fig. S6). Interestingly, we found the mRNA level of HDAC4 was not significantly changed between C2C12 cells that overexpressed miR-378a-3p or the control cells (Fig. 5d, S7). Therefore, we concluded that miR-378a-3p was directly targeting the 3′UTR of HDAC4 to inhibit its protein expression.
HDAC4 inhibited myogenic differentiation and promoted C2C12 cell proliferation
To determine the role of HDAC4 in myoblasts differentiation, C2C12 cells were transfected with siRNA against HDAC4. The results showed that HDAC4 mRNA expression was notably reduced in C2C12 cells transfected with siHDAC4 (Fig. 6a). The mRNA expression of MyoD, MHC, and MEF2C significantly increased in the C2C12 cells transfected with siHDAC4 (Fig. 6b), confirming the role of miR-378a-3p in muscle differentiation. To confirm the effects of HDCA4 on cell survival, cell proliferation and apoptosis were detected in C2C12 cells. Cell proliferation was detected by CCK-8 and EdU staining analysis. We found that siHDAC4 inhibited proliferation of C2C12 cells (Fig. 6d-f). Also, we analyzed the effect of siHDAC4 on cell apoptosis by flow cytometry, and found no significant difference in the distribution of cell apoptosis (Fig. 6g, h). In conclusion, these results demonstrated that HDAC4 promoted myogenic differentiation and suppressed myoblast proliferation.
Figure 6.
Knock down HDAC4 promotes differentiation and inhibits proliferation of C2C12 cells. (a) Transfection of siHDAC4 into C2C12 myoblasts to knock down the expression of HDAC4 mRNA and overexpression of miR-378a-3p to inhibited HDAC4 mRNA, detected by RT-PCR. (b) The mRNA expression of MHC, MyoD and MEF2C were detected by RT-PCR in siHDAC4 transfected and control group. (c) MHC and MyoD protein detected by protein gel blot. (d) Cell proliferation were detected with EdU stained in siHDAC4 and NC transfected of C2C12 cell. Nuclei were stained blue with DAPI, EdU were detected in red luciferase. Scale bar stand 200 μm. (e) EdU positive cells analysis. (f) CCK-8 were used to detected cell proliferation, and analyzed the absorbance value at 450 nm with Automatic microplate reader. (g, h) Cells were collected and stained by Annexin V-FITC/PI in siHDAC4 and NC groups, cell apoptosis distribution were analyzed by flow cytomctry. Values are mean ± SEM for 3 biological replicates, * P < 0.05, ** P < 0.01.
Discussion
Our previous high-throughput sequencing results on miRNAs in Qinchuan bovine skeletal muscle showed that miR-378a-3p was one of the highest expression miRNAs.36 We verified that the expression levels of miR-378a-3p was markedly higher in skeletal muscle than the levels in other tissues. The results suggested that miR-378a-3p might be a muscle-related miRNA, similar to other muscle-specific miRNAs that were reported previously.
Previous studies showed that miR-378 inhibits prostatic carcinoma cell proliferation8 and enhanced apoptosis of cardiomyocytes by reduced signaling in the Akt cascade.38 Additionally, miR-378-5p could suppress cell proliferation and promote cell apoptosis in CRC cells.22 In this research, we found that miR-378a-3p inhibited C2C12 cell proliferation and arrested cells in G0/G1 phase. The overexpression of miR-378a-3p promoted cell apoptosis. Cell proliferation and apoptosis are complicated processes, and our research showed that miR-378a-3p inhibits cell proliferation and promotes apoptosis. However, more studies are warranted to determine how miR-378a-3p regulates cell proliferation and apoptosis.
Our data demonstrated that miR-378a-3p reduced the expression of Bcl-2 and increased the expression of Bax and caspase-3. Bcl-2 family proteins regulate caspase activation and apoptosis by regulating mitochondrial outer membrane permeabilization, which releases mitochondrial proteins and promote apoptosis.15, 26 Caspase-3 is a crucial inducer of apoptosis, acting to induce apoptosis of human colon cancer cells.33,51 Downregulation of Bcl-2 expression and Caspase-3 activation could suppress proliferation and induce apoptosis in cancer cell lines.34 Usually, cell proliferation and apoptosis are regulated through complex interactions between caspase-3 and the Bcl-2/Bax pathway.26,34 Therefore, we concluded that miR-378 promotes apoptosis through regulating the expression of Bcl-2 and Bax.
Previous studies showed that miR-378 overexpression can increase MyoD expression which induces myoblast differentiation.13 Similarly, we demonstrated that miR-378a-3p overexpression significantly increased the expression of the mRNA and protein levels of MyoD and MHC in C2C12 cells, indicating that miR-378a-3p is an important regulator in myocyte differentiation. We also determined the effects of miR-378 to stimulate differentiation and apoptosis occurred by regulating the expression of HDAC4 at the post-transcriptional level. HDAC4 promotes proliferation in colon cancer cells41 and osteosarcoma cells.35 We showed that knocking down HDAC4 markedly inhibits C2C12 cell proliferation. HDAC4 is specifically involved in muscle development. HDAC4 reduced the ability of the myocyte enhancer factor 2 (MEF2) to access the regulatory regions of genes, resulting in repressed MEF2-dependent transcription.5,10,11,27 Moreover, members of the MEF2 family cooperate with the MyoD family to control the expression of muscle–specific genes.2,32 In the present study, we found that both HDAC4 knockdown and miR-378 overexpression promoted differentiation, inhibited the proliferation of C2C12 cells, and significantly increased the expression of MHC and MyoD. These findings may explain how miR-378a-3p promotes differentiation of C2C12 cells.
We also found that the knockdown of HDAC4 increased the mRNA level of MEF2C. MEF2C is a crucial transcriptional factors in regulating gene expression. A previous study reported that MEF2C expression is limited to the skeletal muscle25 and that the MEF2C protein was required in the p38 MAPK pathway that regulates the transcription of genes in skeletal muscle differentiation.50 Thus, miR-378a-3p regulated muscle differentiation by inhibiting HDAC4 expression, which could strengthen MEF2C expression of the activating p38 MAPK signal pathway.
In conclusion, our findings showed that miR-378a-3p may promote muscle differentiation by inhibiting HDAC4 at the post-transcriptional level. Moreover, miR-378a-3p suppressed cell proliferation and induced apoptosis in C2C12 cells (Fig. 7). However, this effect of miR-378a-3p on cell differentiation requires further studies.
Figure 7.
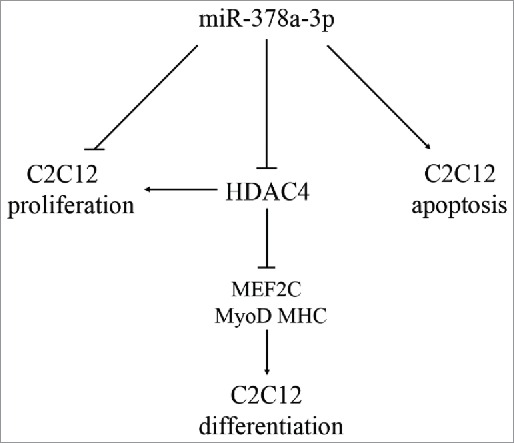
Model of miR-378a-3p functioning in C2C12 cells. miR-378a-3p promoted differentiation and repressed proliferation through targets HDAC4 in C2C12 cells, and promoted cell apoptosis.
Materials and methods
Tissues collected and RNA isolation
All animal samples used in this study were approved by the Animal Care and Use Committee of the College of Animal Science and Technology, Northwest A&F University. Bovine tissue samples included muscle, liver, heart, lung, spleen, kidney, brain, and fat. All samples were collected at a local slaughterhouse in xi'an. The bovines were slaughtered by exsanguination (n = 3), and all tissues were obtained under sterile conditions, washed with diethypyrocarbonate (DEPC) treated water, and immediately frozen in liquid nitrogen. The samples were stored at −80°C until RNA isolation. Total RNA was extracted from tissues or cells with Trizol reagent (TaKaRa, Japan).
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues or cells, and then 500 ng total RNA was converted to cDNA using the PrimeScript RT regent Kit (TaKaRa, Japan). Random primers, oligo (dT) or miRNA-specific stem-loop primers were designed by us and used for reverse-transcribed cDNA (Table S1). RT-qPCR was performed with the SYBR Green PCR Master Mix Reagent Kit (TaKaRa, Japan) and GAPDH and U6 were used for normalization of the data. The RT-qPCR procedure was as follows: cycle 1, 94°C for 3 min; cycle 2, 94°C for 5 s, 60°C for 25 s for 39 cycles, melt curve was generated using the cycle: 95°C for 10s, 65°C for 5s and 95°C for 5s. The fold-change of expression of the transcript mRNA or miRNA were analyzed using the 2−ΔΔCT method. The RT-qPCR primers listed in Table S2 were designed by Beacon Designer 8.
Vector construction
The overexpression vector pcDNA3.1-miR-378a-3p was obtained by PCR amplification of a fragment about 380 bp including the pre-miR-378a-3p complete sequence from bovine genomic DNA using primers containing restriction enzyme sites Hind III/Kpn I (TaKaRa, Japan); the forward primer was 5′-CCCAAGCTTTAGAAGGCTCCGAGAACCAG-3′ and reverse primer was 5′- GGGGTACCGAAGTTACAGGAAGGACCAGACA-3′. T4 DNA ligase was used to ligate the pre-miR-378a-3p fragment into the pcDNA3.1 (+) vector. miR-378a-3p inhibitor was compound by GenePharma, the sequence was GCCUUCUGACUCCAAGUCCAGU.
The HDAC4-3′UTR sequence including the miRNA binding site was amplified using a forward primer 5′-CCGCTCGAGCTGAACTTTGAAGCCTGTGG-3′ and reverse primer 5′-ATAAGAATGCGGCCGCAAGACCTTCCTGTCCTGCTC-3′. Separately, a 2-base mutagen in the miR-378a-3p-binding site of HDAC4 3′UTR (HDAC4-3′UTR-Mut) was generated with a pair of mutagenic primers: 5′-GACCAAAAGATGCCAGATTCTTGGACCG-3′ and 5′-CGGTCCAAGAATCTGGCATCTTTTGGTC-3′. The two fragments were ligated into the 3′-end of the Renilla gene in the psi-CHECK-2 dual-luciferase reporter vector (Promega, USA) using restriction enzymes Xho I and Not I (TaKaRa, Japan) and then ligated by T4 DNA ligase (TaKaRa, Japan).
Cell culture
HEK293T cells (ATCC, USA) and mouse C2C12 myoblasts were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Hyclone, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, USA) and 1% double antibiotics (penicillin and streptomycin) (GM) at 37°C with 5% CO2. To induce myogenic differentiation, culture medium was switched to differential medium (DMEM with 2% horse serum, DM) after cell growth had reached nearly 80% confluence.
Transfection
To detect the transfection efficiency of the recombinant vector, pcDNA3.1-miR-378a-3p was transfected into HEK293T cells. The cells at 90% confluence were plated at 5 × 105 cells/well in 6-well plates. When growth reached approximately 80% confluence, 2 μg of pcDNA3.1-miR-378a-3p was transfected into the HEK293T cells using Lipofectamine 2000 (Invitrogen, USA).
To confirm the effect of miR-378a-3p on muscle differentiation, pcDNA3.1-miR-378a-3p was transfected into C2C12 cells cultured in a 6-well plates. The cells were cultured in DM medium for 6 days, the cells were collected daily, and a group of cells transfected with pcDNA3.1 (+) vector were grown as a control. The siRNA of HDAC4 (siHDAC4) was transfected into C2C12 cells to explore the HDAC4 effect on cell differentiation; 50 nM siRNA was used for each well of the 6-well plates. The sequence of the siRNAs used in the transfection are 5′-GAGCAGCAGAGGAUCCACCAGUUAA-3′ and 5′-CACCGGAACCUGAACCACUGCAUUU-3′, and a negative control (NC) siRNA that did not target HDAC4was transfected into C2C12 cells as the control group; the sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′.
Cell proliferation assay
Cell counting kit-8 assay
C2C12 cells were transferred to 96-well plates at a density of 1×104 cells/well and then 100μL GM were added to each well. When the cells reached approximately 80% confluence, they were transfected with either the pcDNA3.1-miR-378a-3p or the pcDNA3.1 (+) vectors, respectively. After 24h of culture at 37°C, 10μL of CCK-8 reagent (Multisciences; China) was added to each well and incubation was continued for 4 h. The absorbance value of all samples were detected using an automatic microplate reader (Molecular Devices, USA) at 450 nm.
EdU proliferation assay
The cell proliferation was also assessed using the Cell-Light EdU DNA cell proliferation kit (RiboBio, China). C2C12 cells was seeded into 96-well culture plates containing 100μL GM. After 24 h, the cells were incubated into EdU medium. After 2 h, the test was performed according to the manufacturer's protocol.
Flow cytometry for the cell cycle assay
C2C12 cells were grown in 6-well plates (1×106 cells/well) with 2mL GM. The cells were treated with pcDNA3.1-miR-378a-3p and pcDNA3.1 (+). After 24 h, the Cell Cycle Assay Kit (Multisciences, China) was used to treat the cells. The cells were washed in PBS buffer and the supernatant was removed after centrifuging. Next, 1mL of DNA strain solution and 10 μL of permeabilization solution were added to the resuspended cells, and then blended by vortex shaking for 15 s. After incubating for 30 min in the dark at room temperature, the cell cycle was analyzed by Flow Cytometry (FACS CantoTM II, BD BioSciences, USA).
Cell apoptosis assay
Cell apoptosis was measured by Annexin V-FITC/PI staining assays. C2C12 cells were cultured in 6-well plates with 2 mL GM. When the cells reached a confluence of 80%-90%, the cells were transfected with pcDNA3.1-miR-378a-3p or pcDNA3.1 (+). After 24 h of incubation, the C2C12 cells were washed 3 times with PBS buffer. Cells were then harvested cells into 1.5 mL centrifuge tube and washed again, resuspending in 500 μL 1× binding buffer. Cells were then treated with Cell Apoptosis Assay Kit (Multisciences, China), incubated in the dark for 10 min at room temperature, then cell apoptosis was immediately analyzed using a Flow Cytometer.
Luciferase activity assay
Cells were cultured in 48-well plates when the cell growth reached about 80% confluence. The pcDNA3.1-miR-378a-3p and psiCHECK-2-HDAC4-3′UTR (HDAC4-UTR-W) or psiCHECK-2-HDAC4-mut-3′UTR (HDAC4-UTR-Mut) were co-transfected into cells by Lipofectamine2000. The transfection reagent was replaced with fresh growth medium (DMEM with 10% FBS) after transfection for 4∼6 h. Next, the cells were washed with PBS and harvested using 200μL Passive Lysis Buffer (PLB) and rocked for 30 min at room temperature. Dual-luciferase activity was measured by MPPC luminescence analyzer (HAMAMATSU; Beijing, China) and the Renilla Luciferase activity was normalized against Firefly Luciferase activity.
Western blot
The total proteins were extracted from cells using protein Lysis buffer RIPA containing 1mM PMSF (Solarbio; Beijing, China). The extracts were boiled with 4×SDS loading buffer (150 mM Tris-HCL (pH = 6.8), 12% SDS, 30% glycerol, 0.02% bromophenol blue, and 6% 2-mercaptoethanol) at 98°C for 10 min and then 20 μg total proteins were loaded and separated on 10% SDS-PAGE gels. After electrophoresis, the proteins were transferred to a 0.2 μm PVDF membrane that was soaked in formaldehyde, and then blocked with 5% skim milk in Tris Saline with Tween (TBST) buffer for about 2 h at room temperature. The membrane was then incubated overnight with primary antibodies specific for anti-HDAC4 (Dilution 1:1000; ab32534; Abcam, England), anti-MyoD (Dilution 1:1000; ab16148; Abcam, England), anti-MHC (Dilution 1:1000; ab24648; Abcam, England), anti-Bcl-2 (Dilution 1:1000; ab32124; Abcam, England), anti-Bax (Dilution 1:1000; ab32503; Abcam, England), anti-MEF2C (Dilution 1:1000; sc-365862; SANTA, USA) and anti-β-tubulin (Dilution 1:1000; KDM9003; Sungene Biotech, China) at 4°C. The PVDF membrane was washed 3 times with TBST buffer and then incubated with secondary antibody for the anti-immune rabbit IgG-HRP (Dilution 1:1000; LK2001; Sungene Biotech, China) 2 h at room temperature. β-tubulin was used as the internal control with a secondary antibody that was HRP-labeled anti-mouse IgG (Dilution 1:1000; LK2003; Sungene Biotech, China). Finally, antibody reacting bands were detected using ECL luminous fluid (Solarbio, China).
Statistical analysis
The quantitative results are presented as mean ± standard error of the mean (SEM) based on at least 3 independent experiments. All data in this study were analyzed by one-way analysis of variance (ANOVA) for P-value calculations using SPSS v17.0 software. P < 0.05 was considered statistically significant differences among means. The software Image J was utilized for gels image gray value analysis.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Investig 2006; 116(7):1853-64; PMID:16767219; http://dx.doi.org/ 10.1172/JCI27438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 2005; 16(4–5):585-95; PMID:16099183; http://dx.doi.org/25530839 10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 3.Boettger T, Wüst S, Nolte H, Braun T. The miR-206/133b cluster is dispensable for development, survival and regeneration of skeletal muscle. Skeletal Muscle 2014; 4(1):1; PMID:25530839; http://dx.doi.org/ 10.1186/s13395-014-0023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, Bassel-Duby R, Olson EN. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci 2012; 109(38):15330-5; PMID:22949648; http://dx.doi.org/12709441 10.1073/pnas.1207605109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JK, Sun L, Yang XJ, Zhu G, Wu Z. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J Biol Chem 2003; 278(26):23515-21; PMID:12709441; http://dx.doi.org/ 10.1074/jbc.M301922200 [DOI] [PubMed] [Google Scholar]
- 6.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38(2):228-33; PMID:16380711; http://dx.doi.org/ 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, Wang SF. MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol 2012; 29(3):1673-80; PMID:22052152; http://dx.doi.org/ 10.1007/s12032-011-0083-x [DOI] [PubMed] [Google Scholar]
- 8.Chen QG, Zhou W, Han T, Du SQ, Li ZH, Zhang Z, Shan GY, Kong CZ. MiR-378 suppresses prostate cancer cell growth through downregulation of MAPK1 in vitro and in vivo. Tumor Biol 2015; 37(1):1-9; PMID:26432328; http://dx.doi.org/ 10.1007/s13277-015-3996-8 [DOI] [PubMed] [Google Scholar]
- 9.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005; 33(4):1290-7; PMID:15741182; http://dx.doi.org/ 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi MC, Cohen TJ, Barrientos T, Wang B, Li M, Simmons BJ, Yang JS, Cox GA, Zhao Y, Yao TP. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell 2012; 47(1):122-32; PMID:22658415; http://dx.doi.org/ 10.1016/j.molcel.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen TJ, Barrientos T, Hartman ZC, Garvey SM, Cox GA, Yao TP. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J 2009; 23(1):99-106; PMID:18780762; http://dx.doi.org/ 10.1096/fj.08-115931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci 2011; 108(22):9232-7; PMID:Can't; http://dx.doi.org/ 10.1073/pnas.1102281108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem 2011; 286(22):19431-8; PMID:21471220; http://dx.doi.org/ 10.1074/jbc.M111.219006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol-Endocrinol Metab 2010; 299(2):E198-206; PMID:20484008; http://dx.doi.org/ 10.1152/ajpendo.00179.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol 2015; 7(12):a006080; PMID:26626938; http://dx.doi.org/ 10.1101/cshperspect.a006080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15(8):509-524; PMID:25027649; http://dx.doi.org/ 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 17.Hupkes M, Sotoca AM, Hendriks JM, van Zoelen EJ, Dechering KJ. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells. BMC Mol Biol 2014; 15(1):1; PMID:24467925; http://dx.doi.org/ 10.1186/1471-2199-15-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha C-S, Kim SW, Yun YH, Jung Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 2016; 7:10993; PMID:27001906; http://dx.doi.org/23071643 10.1038/ncomms10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jalali S, Ramanathan GK, Parthasarathy PT, Aljubran S, Galam L, Yunus A, Garcia S, Cox RR Jr, Lockey RF, Kolliputi N. Mir-206 regulates pulmonary artery smooth muscle cell proliferation and differentiation. PloS one 2012; 7(10):e46808; PMID:23071643; http://dx.doi.org/ 10.1371/journal.pone.0046808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javed R, Jing L, Yang J, Li X, Cao J, Zhao S. miRNA transcriptome of hypertrophic skeletal muscle with overexpressed myostatin propeptide. Bio Med Res Int 2014; 2014:328935; PMID:25147795; http://dx.doi.org/ 10.1155/2014/328935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One 2009; 4(10):e7535; PMID:19844573; http://dx.doi.org/ 10.1371/journal.pone.0007535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, Gupta M. A novel cardiomyocyte-enriched MicroRNA, miR-378, targets insulin-like growth factor 1 receptor implications in postnatal cardiac remodeling and cell survival. J Biol Chem 2012; 287(16):12913-26; PMID:22367207; http://dx.doi.org/ 10.1074/jbc.M111.331751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11(9):597-610; PMID:20661255; http://dx.doi.org/ 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 24.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci 2007; 104(51):20350-5; PMID:18077375; http://dx.doi.org/23652597 10.1073/pnas.0706901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Schneider MF. Opposing HDAC4 nuclear fluxes due to phosphorylation by β-adrenergic activated protein kinase A or by activity or Epac activated CaMKII in skeletal muscle fibres. J Physiol 2013; 591(14):3605-23; PMID:23652597; http://dx.doi.org/ 10.1113/jphysiol.2013.256263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S. BOK Is a Non-canonical BCL-2 Family Effector of Apoptosis Regulated by ER-Associated Degradation. Cell 2016; 165(2):421-33; PMID:26949185; http://dx.doi.org/ 10.1016/j.cell.2016.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000; 408(6808):106-11; PMID:11081517; http://dx.doi.org/ 10.1038/35040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meerson A, Traurig M, Ossowski V, Fleming J, Mullins M, Baier L. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia 2013; 56(9):1971-9; PMID:23756832; http://dx.doi.org/ 10.1007/s00125-013-2950-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miretti S, Martignani E, Accornero P, Baratta M. Functional effect of mir-27b on myostatin expression: a relationship in piedmontese cattle with double-muscled phenotype. BMC genomics 2013; 14(1):1; PMID:23323973; http://dx.doi.org/ 10.1186/1471-2164-14-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, Bellahcene A, Verdin E, Castronovo V. HDAC4 represses p21WAF1/Cip1 expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene 2009; 28(2):243-56; PMID:18850004; http://dx.doi.org/ 10.1038/onc.2008.371 [DOI] [PubMed] [Google Scholar]
- 31.Nariyama M, Mori M, Shimazaki E, Ando H, Ohnuki Y, Abo T, Yamane A, Asada Y. Functions of miR-1 and miR-133a during the postnatal development of masseter and gastrocnemius muscles. Mol Cell Biochem 2015; 407(1–2):17-27; PMID:25981536; http://dx.doi.org/ 10.1007/s11010-015-2450-y [DOI] [PubMed] [Google Scholar]
- 32.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol 2007; 27(23):8143-51; PMID:17875930; http://dx.doi.org/ 10.1128/MCB.01187-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito M, Yaguchi T, Yasuda Y, Nakano T, Nishizaki T. Adenosine suppresses CW2 human colonic cancer growth by inducing apoptosis via A 1 adenosine receptors. Cancer Lett 2010; 290(2):211-5; PMID:19822392; http://dx.doi.org/ 10.1016/j.canlet.2009.09.011 [DOI] [PubMed] [Google Scholar]
- 34.Shirali S, Aghaei M, Shabani M, Fathi M, Sohrabi M, Moeinifard M. Adenosine induces cell cycle arrest and apoptosis via cyclinD1/Cdk4 and Bcl-2/Bax pathways in human ovarian cancer cell line OVCAR-3. Tumor Biology 2013; 34(2):1085-95; PMID:23345014; http://dx.doi.org/ 10.1007/s13277-013-0650-1 [DOI] [PubMed] [Google Scholar]
- 35.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 2009; 28(46):4065-74; PMID:19734943; http://dx.doi.org/ 10.1038/onc.2009.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J, Li M, Li Z, Xue J, Lan X, Zhang C, Lei C, Chen H. Identification and profiling of conserved and novel microRNAs from Chinese Qinchuan bovine longissimus thoracis. BMC Genomics 2013; 14(1):42; PMID:23332031; http://dx.doi.org/ 10.1186/1471-2164-14-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usui T, Morita T, Okada M, Yamawaki H. Histone deacetylase 4 controls neointimal hyperplasia via stimulating proliferation and migration of vascular smooth muscle cells. Hypertension 2014; 63(2):397-403; PMID:24166750; http://dx.doi.org/ 10.1161/HYPERTENSIONAHA.113.01843 [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Ma B, Ji X, Deng Y, Zhang T, Zhang X, Gao H, Sun H, Wu H, Chen X. MicroRNA-378-5p suppresses cell proliferation and induces apoptosis in colorectal cancer cells by targeting BRAF. Cancer Cell Int 2015; 15(1):1; PMID:25678856; http://dx.doi.org/ 10.1186/s12935-015-0156-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei W, He H, Zhang W, Zhang H, Bai J, Liu H, Cao J, Chang K, Li X, Zhao S. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis 2013; 4(6):e668; PMID:23764849; http://dx.doi.org/ 10.1038/cddis.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch C, Chen Y, Stallings R. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 2007; 26(34):5017-22; PMID:17297439; http://dx.doi.org/ 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- 41.Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, Figueroa M, Melnick A, Kao GD, Augenlicht LH. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell 2008; 19(10):4062-75; PMID:18632985; http://dx.doi.org/ 10.1091/mbc.E08-02-0139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winbanks CE, Beyer C, Hagg A, Qian H, Sepulveda PV, Gregorevic P. miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS One 2013; 8(9):e73589; PMID:24023888; http://dx.doi.org/ 10.1371/journal.pone.0073589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, Gregorevic P. TGF-β regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem 2011; 286(16):13805-14; PMID:21324893; http://dx.doi.org/ 10.1074/jbc.M110.192625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007; 131(1):146-59; PMID:17923094; http://dx.doi.org/ 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 45.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 2007; 120(17):3045-52; PMID:17715156; http://dx.doi.org/ 10.1242/jcs.010728 [DOI] [PubMed] [Google Scholar]
- 46.Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, Wu F. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int 2012; 32(5):752-760; PMID:22276989; http://dx.doi.org/ 10.1111/j.1478-3231.2011.02750.x [DOI] [PubMed] [Google Scholar]
- 47.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 2007; 13(4):486-91; PMID:17401374; http://dx.doi.org/ 10.1038/nm1569 [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Sun W, Wang R, Lei C, Zhou R, Tang Z, Li K. Wnt antagonist, secreted frizzled-related protein 1, is involved in prenatal skeletal muscle development and is a target of miRNA-1/206 in pigs. BMC Mol Biol 2015; 16(1):4; PMID:25888412; http://dx.doi.org/ 10.1186/s12867-015-0035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van IJcken W, Grosveld F. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 2013; 17(2):210-24; PMID:23395168; http://dx.doi.org/ 10.1016/j.cmet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation participation of the MEF2C transcription factor. J Biol Chem 1999; 274(8):5193-200; PMID:9988769; http://dx.doi.org/ 10.1074/jbc.274.8.5193 [DOI] [PubMed] [Google Scholar]
- 51.Zhang YX, Xia D, Wang M. Effect of protein kinase C alpha, caspase-3, and survivin on apoptosis of oral cancer cells induced by staurosporine1. Acta Pharmacologica Sinica 2005; 26(11):1365-72; PMID:16225760; http://dx.doi.org/ 10.1111/j.1745-7254.2005.00205.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



