Abstract
DNA methylation is thought to influence Quercus suber cork quality, which is the main constraint for its economic valorisation. However, a deep knowledge of the cytosine methylation patterns disclosing the epigenetic variability of trees with different cork quality types is totally missing. This study investigates the hypothesis that variations in DNA methylation contribute to differences in cork cellular characteristics directly related to original or traumatic phellogen activity. We used MSAPs (Methylation Sensitive Amplified Polymorphism) to assess DNA methylation patterns of cork and leaf tissues of Q. suber adult trees growing in three cork oak stands. The relationship between the detected polymorphisms and the diversity of cork quality traits was explored by a marker-trait analysis focusing on the most relevant quality characteristics. Populations differed widely in cork quality, but only slightly in degree of epigenetic differentiation. Four MSAP markers (1.3% of the total) were significantly associated with the most noteworthy quality traits: wood inclusions (nails) and porosity. This evidence supports the potential role of cytosine methylation in the modulation of differential phellogen activity either involved in localized cell death or in pore production, resulting in different cork qualities. Although, the underlying basis of the methylation polymorphism of loci affecting cork quality traits remain unclear, the disclosure of markers statistically associated with cork quality strengthens the potential role of DNA methylation in the regulation of these traits, namely at the phellogen level.
Introduction
Epigenetic variation can contribute to phenotypic plasticity and phenotype persistence in distinct environments, as has been recently suggested in long-lived forest trees (reviewed in [1]). Epigenetics refers to the mitotically and/or meiotically heritable changes in gene expression that do not imply modifications in DNA sequence [2–4]. Chromatin structure is affected by the interplay between DNA methylation, histone modifications and small RNAs, with cytosine methylation being the better-studied chromatin alteration. DNA methylation in plants is accomplished by a family of DNA methyltransferases comprising three specific functional classes [5]: the MET (Methyltransferase) class maintains the methylation in CG context found either within promoter regions [6,7] resulting in silencing [8] or within gene bodies [6,9,10] associated with gene expression [7]; the CMT (Chromomethylase) class maintains the methylation in CHG context occurring mainly in transposons and pseudogenes [10]; and the DRM (Domain-Rearranged-Methyltransferase) class associated with the de novo methylation in CHH sequences which are typically methylated at low levels in Arabidopsis [5].
Methylation-sensitive amplified polymorphisms (MSAP) is a practical method that has been widely used to investigate diversity of cytosine methylation and epigenetic structure in species lacking a sequenced genome for reference, as well as to investigate correlations between phenotypic functional traits [11–17]. Also, MSAP markers have been mapped to protein-coding genes in sorghum [18], maize [19,20] and rice [21], and also linked with stable epigenetic quantitative trait loci (QTLepi) of diverse agronomic traits [22]. Despite the limitations in acknowledging the exact genomic location for methylation and in whole genome coverage, MSAP still provides a significant number of anonymous loci randomly distributed throughout the genome in which the methylation state of restriction sites can be determined.
The cork oak (Quercus suber L.) is one of the most important forest species in the Mediterranean basin. Cork oak trees are currently the exclusive commercial source of cork, a thick periderm with insulating and protective role, made of dead cells with empty lumens and thin highly suberized walls [23,24]. Cork is formed by the division to the outside of the phellogen, a secondary meristem that is seasonally activated and that persists throughout the life of the tree [23,25]. Cork produced from the original phellogen is called ‘virgin’ cork and is usually harvested from 18 to 25 years-old trees, when stem perimeter reaches the legal size for extraction [26]. After cork removal, the phellogen dies and a new traumatic phellogen is formed by a process of meristematic activation in the underlying non-conducting phloem living cells [27]. Further cork extractions follow at a minimum of 9-years intervals, during which a cork thickness of 2–6 cm is reached. This capacity for self-regeneration of the phellogen determines cork oak uniqueness and makes cork the cornerstone of the economic sustainability of cork oak woodlands worldwide. Amadia cork, produced by a traumatic phellogen (3rd harvest onward) has singular characteristics derived from cellular structure and chemical composition. However, as a natural product, its quality criteria is determined by the cork tissue homogeneity and thickness, due to their industry applications, for some of them without viable substitutes [28].
Cork tissue homogeneity is affected by the incidence of discontinuities in the cork layer, such as lenticels (forming the lenticular channels) produced by the activity of the lenticular phellogen, and lignified phloem cells, known as ‘nail’ structures [29] and caused by an interrupted, abnormal phellogen activity which is highly detrimental to cork quality. In addition, cork thickness is mostly determined by the level of phellogen activity and corresponding cumulative annual cork growth (cork-ring width). A minimum thickness is needed in the raw cork planks in order to allow punching natural cork stoppers, the most valuable cork product. Despite these critical quality parameters of raw cork, the disclosure of factors regulating cork differentiation and quality at tree-level is in its infancy [30,31]. The high genetic variability found within cork oak populations [32,33], mixed with empirical assumptions of an effect of environmental conditions, hindered any causal association with the very high cork quality variability found within and between cork oak woodlands [29]. To better understand what affects cork quality, it is necessary to unravel the factors underlying such different phellogen activities giving rise to such variable phenotypes. Recent studies of the epigenetics of oak populations revealed patterns of epigenetic differentiation and single-methylation variants associated with climate variables [34,35], indicating that oak genomes exhibit phenotypic plasticity mediated by DNA methylation. Although the genus Quercus has already two sequenced genomes [36,37] no genomic data was available for cork oak. Some studies have been targeting cytosine methylation in this species [38–40] evidencing its association with cork quality, namely on cork tissue homogeneity [39].
Considering the role of DNA methylation in regulating phenotypic plasticity we hypothesised that DNA methylation variability could be related to original or traumatic phellogen activity, also contributing to cork cellular characteristics linked to quality. A deeper understanding of cytosine methylation variability at tree-level and its impact on relevant traits could lead to a better comprehension of its role in the modulation of cork tissue homogeneity. To test our hypothesis, we assessed the DNA methylation landscape of virgin and amadia cork tissues and leaves, focusing on CG and CCG context using MSAP analysis, in trees from three regions in Portugal with distinct edaphoclimatic conditions. Furthermore, to test for predicted relationships between the variation in DNA methylation and phenotypic diversity, the epigenetic polymorphisms detected were associated with the most relevant cork quality traits.
Materials and Methods
Plant material and DNA isolation
No specific permission was needed for the development of this study. Quercus suber is protected in Portugal against logging. The field studies did not involve endangered or other protected species.
Cork oak adult trees were randomly chosen from three cork oak stands (montados) in different locations in Portugal: Barradas da Serra (BS), Herdade dos Leitões (HL) and Companhia das Lezírias (CL) (Table 1). Each stand comprised trees of controlled origin as well as trees resulting from natural regeneration. A detailed characterization and comparison of the three areas, in terms of climate and soil conditions, is described elsewhere [41]. Cork planks were harvested at breast height (at 1.30 m height from soil) during the more intense period of phellogen activity (the period of cork commercial harvesting), and phellogen with contiguous differentiating tissue (hereafter referred as cork tissues) were collected by scraping off the inner side of cork planks. Leaves were also collected from all trees. Cork planks were kept to evaluate cork quality and thickness. To avoid any developmental, and/or environmentally-related variation in DNA methylation, fully developed leaf blades and cork tissues were collected from each location on the same day. All living tissues were immediately stored in liquid nitrogen until further use. Total genomic DNA from leaves, and cork tissues was isolated according to Doyle and Doyle [42] with minor modifications: the isolation buffer contained 3% 2-mercaptoethanol and 2% PVP-40; proteinase K was added to a final concentration of 100 μg/mL before samples incubation at 65°C.
Table 1. Material collected in three cork oak populations in distinct edaphoclimatic conditions in Portugal.
| Populations | Geographical Coordinates | Treesb | ||
|---|---|---|---|---|
| Amadia cork | Virgin cork | Total | ||
| Grândola, Barradas da Serra (BS) | 38 11’ N, 8 37’ W 270 m a.s.l.a | 8 | 1 | 9 |
| Montargil, Herdade dos Leitões (HL) | 39 8’ N, 8 11’W 170 m a.s.l. a | 11 | 0 | 11 |
| Benavente, Companhia das Lezírias (CL) | 38 49’ N, 8 49’ W 20 m a.s.l. a | 10 | 11 | 21 |
a Above sea level
b Number of individuals sampled per population
MSAP procedure
MSAP is a modification of the AFLP method, which takes advantage of the differential behaviour of two isoschizomers, HpaII and MspI, in the presence of cytosine methylation in the CCGG context. These isoschizomers are coupled with EcoRI, which is thought to be negligibly influenced by DNA cytosine methylation. MspI can cleave non-methylated CCGG sequences and hemi- or fully-methylated CmCGG sequences but not hemi- and fully-methylated mCCGG and mCmCGG sequences. HpaII digests only non-methylated CCGG sequences and hemi-methylated mCCGG sequences from all possible methylated CCGG variants [43]. Therefore, distinct profiles obtained with EcoRI/HpaII and EcoRI/MspI reflect differences in the methylation status of these restriction sites.
For MSAP assays, genomic DNA (100 ng) isolated from cork tissues and leaves of the same genotype was first digested and ligated using 20 U of EcoRI and 5 U of MspI or HpaII (New England Biolabs, USA), 5 pmol Eco_adaptor, 50 pmol HM_adaptor (5 μM of EcoRI and 50 μM of MspI/HpaII adaptor pairs were previously prepared by mixing oligonucleotides and incubating at 98°C for 5 min followed by slow cooling in aluminium foil–S1 Table), 1 U of T4 DNA ligase (Invitrogen, USA) in 40 μL total volume supplemented with 50 mM NaCl and 100 ug/ml BSA for 6 h at 37°C. The enzymes were then inactivated by heating to 65°C for 15 min. A volume of 2 μL of the restriction/ligation products was used in 20 μL pre-selective amplification reactions containing 1X PCR buffer (Nzytech, Portugal), 30 ng of Pre_Eco (+A) primer and 30 ng of Pre_HM primer (S1 Table), 0.4 mM dNTPs, 1.5 mM MgCl2 and 2 U Taq polymerase (Nzytech, Portugal). PCR conditions were 2 min at 72°C followed by 25 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 2 min with a final extension step of 10 min at 72°C. Initially, 25 selective primer combinations were tested for fitness in identifying inter-specific variation and in generating reproducible MSAP profiles using two independent DNA extractions of each tissue from four representative genotypes (data not shown). From these, four primer combinations (S2 Table) were chosen for the comparative selective amplification. Selective PCRs were then performed using 5 μL of 1:10 dilution of pre-selective PCR products and the same reagents as the pre-selective amplification, but using 0.2 mM dNTPs and FAM labelled selective primers. The conditions of the touch-down selective PCR were as follows: 94°C for 30 s, 65°C for 30 s, 12 cycles of 72°C for 1 min (decreasing by 0.7°C each cycle), followed by 24 cycles of 94°C for 30 s, 56°C for 1 min, 72°C for 2 min, ending with 72°C for 5 min. Thereafter, 1 μL of selective amplification products was mixed with 0.3 μL of ROX dye and 12.2 μL of HiDi formamide (Applied Biosystems, USA), denatured by heating at 95°C for 5 min and snap cooled on ice for 5 min. Fragment separation and detection was made using an ABI PRISM 310 Genetic Analyser (Applied Biosystems, USA) and fragments were scored manually analysing the electropherograms with GENEMAPPER 3.7 software. For each primer combination, genotyping error rates were estimated through the comparison of two technical replicates profiles. MSAP genotyping error rates were estimated for each primer combination by running repeated EcoRI/HpaII and EcoRI/MspI analyses for all plants and both tissues and computed as described elsewhere [44]. Mean genotyping error rate (±SD) for the four MSAP primer combinations used was 0.004% ± 0.002. A combined digestion with EcoRI/HpaII + MspI was performed in parallel to improve the interpretation of the (absence, presence) pattern obtained for some epiloci in (EcoRI/MspI, EcoRI/HpaII) [43]. This approach helps to distinguish between the two situations that may generate that pattern: the cutting of hemimethylated sites mCCGG sites by HpaII and not by MspI; or the presence of internal CmCGG site(s) between the EcoRI site and the cleaved distal unmethylated CCGG [43].
MSAP data analysis
Reproducible peaks with lengths between 100 and 500 bp (to minimize the incidence of fragment size homoplasy [45]) were scored as presence (1) or absence (0) to form a raw data matrix. All epiloci showing a monomorphic pattern or any fragment present/absent in all but one individual were excluded from the data set, to prevent biased parameter estimates [46]. ‘Mixed Scoring 2’ scheme and the ‘Extract_MSAP_epigenotypes’ R function described by Schulz et al. [44] was used to convert the raw matrix (S3 Table) into three classes of markers corresponding to unmethylated (u-loci), HMeCG + MeCG methylation (internal methylation plus hemimethylation, m-loci) and HMeCCG methylation (external hemimethylation, h-loci). U-loci markers displaying the same profile either for EcoRI/MspI or EcoRI/HpaII in leaves and phellogen from the same genotype were considered as genetic variation and were removed from the epigenetic variability analysis (40 loci).
In order to measure the epigenetic diversity within leaves and cork tissues ‘MSAP_calc.R’ script [44] was used to calculate the number and percentage of polymorphic epiloci and Shannon’s information index for a dominant locus:
| (1) |
where pi is the frequency of the epigenetic marker presence per tissue. To test the null hypothesis that the distribution of Shannon´s information index is identical for both tissues, a Mann-Whitney-Wilcoxon test was performed in R environment. To infer patterns of individual and population epigenetic differentiation, a similarity matrix was assembled using the Soerensen and Dice distance and used to conduct the principal coordinate analysis (PCoA).
The epigenetic differentiation was estimated through Φst comparisons (an analogue of the fixation index Fst, measuring population differentiation) between leaves and cork tissues and among populations for both tissues using an analysis of molecular variance (AMOVA, [47]) and the same Soerensen and Dice similarity matrix. All these analyses were performed using R environment.
Cork quality traits assessment
Amadia cork planks were processed according to a standard procedure [48] to assess cork quality traits but virgin corks were not scorable by the methods used. Amadia cork thickness was measured according to Ramos et al. [39]. Image acquisition of the two radial, two transversal and one tangential sections was made through scanning at a minimum resolution of 300 dpi. Images were uploaded and then analysed using ImageProPlus® (Media Cybernetics, USA) image-processing software.
In each cork sample, after a calibration based on an orthogonal position correction with an accuracy of 0.01 mm, the detection of cork tissue discontinuities was made based on threshold manipulation within a defined area of interest (AOI). Overall cork porosity was assessed at the tangential section, transverse and radial sections. However, only at transversal and radial sections, the overall porosity was discriminated by porosity (lenticels as lenticular channels, S1 Fig) and ‘nail’ (lignified phloem cells, S2 Fig). Cork tissue discontinuities datasets were obtained separately for each section, and based on the range variation of pore-variables. A set of four variables at cork sample/section level were selected for quality assessment: average pore area, average pore length, average pore roundness and average ‘nail’ area. Pore and ‘nail’ data were filtered out by area, and only cork tissue discontinuities with an area equal or superior to 0.8 mm2 were kept for analysis. Small porosity is functionally irrelevant and increases variance and variability of the sample [49]. Pore roundness is defined by the following formula:
| (2) |
where circular discontinuities will have roundness ≈ 1. When the pore is less circular the roundness value will be higher. The pore length is the length along the Y-axis (corresponding to the tree radial direction in the transverse and radial sections). At cork section level, the porosity coefficient and the nail coefficient were expressed as the percentage of the total pore and nail area of the AOI.
Assessment of cork ring widths was made in amadia cork samples according to a standard procedure [48], measuring the complete cork growth years in the transverse sections. The mean annual cork growth (mm yr-1) was then calculated per cork sample.
The population effect on the variability of cork quality traits was studied after testing for deviations from a normal distribution with a Shapiro-Wilk test. For traits that fitted a normal distribution, the effects of the population were evaluated using one way ANOVA tests, followed by Tukey´s multiple comparison test. For non-normally distributed data, Kruskal-Wallis non-parametric and Dunn´s Multiple Correction post-hoc test were used. To find if cork quality traits could be correlated, Pearson’s correlations between traits were computed. All these analyses were performed in R environment.
Association analysis
To assess whether observed differences in the considered cork quality traits within populations were related to DNA methylation polymorphisms, generalized linear models were fitted to higher cork quality presence data. For such approach, all cork quality traits measured were allocated into 2 classes—higher cork quality and lower cork quality—by applying a defined threshold according to trait distribution and taking into account their shape in each section (Table 2). For the response variable cork quality a logit link model was fitted assuming marker as a fixed-effects factor with two levels (presence/1 or absence/0). To test the null hypothesis of no effects of the marker, a likelihood ratio test was performed. The resulting p-values were used to identify significant associations (significance level = 0.05). False discovery rate adjusted p-value (q-value) were computed using the qvalue package [50]. We found the largest q-value leading to an expectation of less or equal to one false significant model [i.e. q-value x (number of models accepted as significant) ≤1]. For the interpretation of results, the estimated probability of success (higher cork quality, ) for the MSAP marker presence was computed from the inverse link function. Additionally, for the presence of MSAP marker, the estimated odds that higher cork quality is obtained, was computed as
| (3) |
All these analyses were performed using R environment.
Table 2. Thresholds of cork quality traits used for the association study with MSAP markers.
| Cork Quality Trait | Threshold a | Reference | Higher cork quality | Lower cork quality |
|---|---|---|---|---|
| Thickness | 27 mm | [49] | Equal or above threshold | Below threshold |
| Annual growth | 3 mm/year | [49] | ||
| % ‘nail’ _R | 1% | Outliers | Below threshold | Equal or above threshold |
| % ‘nail’ _Tr | 1% | Outliers | ||
| ‘nail’ Area_R | 3.98 mm2 | Median | ||
| ‘nail’ Area_Tr | 3.99 mm2 | Third Quartile | ||
| % Porosity_Ta | 6% | [49] | ||
| % Porosity_R | 6% | [49] | ||
| % Porosity_Tr | 6.8% | [49] | ||
| Pores Area_Ta | 2.57 | Median | ||
| Pores Area_R | 5 mm2 | [51] | ||
| Pores Area_Tr | 4.9 mm2 | [51] | ||
| Pores Roundness_Ta | 1.98 | Median | ||
| Pores Roundness_R | 4.03 | Median | ||
| Pores Roundness_Tr | 4.56 | Median | ||
| Pores Length_Ta | 2.5 | Median | ||
| Pores Length_R | 5 mm | [51] | ||
| Pores Length_Tr | 5 mm | [51] |
a Cork quality traits measured in all sections were allocated into 2 classes—higher cork quality and lower cork quality—by applying a defined threshold according to trait data distribution or previously reported values, and taking into account their shape in each section.
R—radial section; Tr—transverse section; Ta–tangential section.
Results
Epigenetic diversity and differentiation is higher in cork tissues
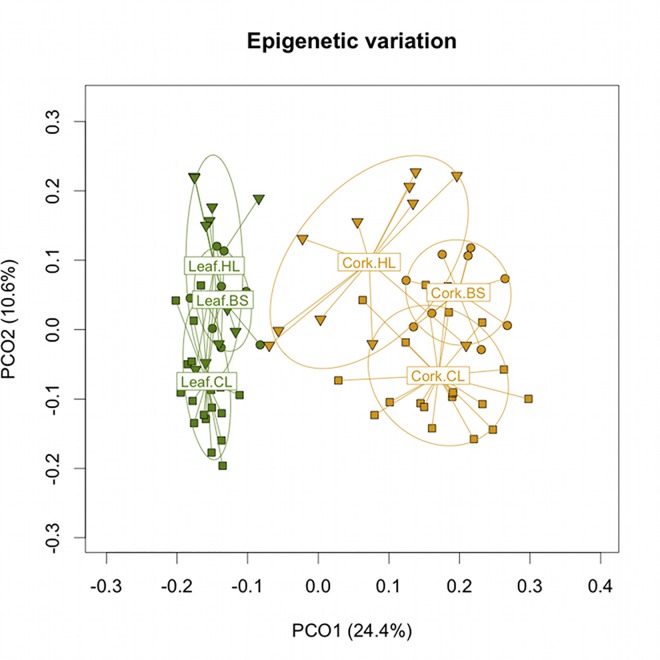
The methylation states of the scored loci were obtained after data transformation, yielding a total of 339 polymorphic epiloci for cork tissues (out of 340) and 303 polymorphic epiloci for leaves (out of 308) (Table 3). The Shannon’s information indexes for each marker were differentially distributed between the two tissues (W = 83102, p-value = 0.007749, Wilcoxon rank sum Test). Indeed, a higher number and frequency of u-loci was found for cork tissues among all individuals in contrast with a similar number but higher frequency of m-loci in leaves. The number of h-loci was higher in cork tissues but the frequency was higher in leaves. Comparing both tissues using total polymorphic loci, we found significant epigenetic differentiation (Fig 1, Φst = 0.3321, p-value < 0.0001), probably derived from their different nature i.e. meristematic vs. differentiated tissues. Comparing populations, epigenetic differentiation was found for cork tissues Φst = 0.168 (p-value < 0.0001) and for leaves Φst = 0.1608 (p-value < 0.0001).
Table 3. Epigenetic diversity of cork and leaf tissues within the three cork oak populations.
| Tissue | u-loci | m-loci | h-loci | Total PL |
|---|---|---|---|---|
| No./%PL | No./%PL | No./%PL | ||
| Cork tissues | 161/34.1 | 106/27.3 | 72/12.5 | 339 |
| Leaves | 133/25.1 | 108/37.5 | 62/10.2 | 303 |
u-loci–unmethylated loci; m-loci–methylated loci; h-loci–hemi-methylated loci; No.–number of polymorphic loci; %PL–Percentage of polymorphic loci; Total PL–Number of total polymorphic loci.
Fig 1. Principal Coordinate Analysis representing epigenetic differentiation between populations and tissues.
Graphical representation is based on the first two coordinates (PCO1 and PCO2) with the percentage of the variability shown between brackets. Inverted triangles represent trees from Herdade dos Leitões (HL), circles represent Barradas da Serra (BS) and squares represent Companhia das Lezírias (CL). The labels represent the centroid for the points cloud in each population and the ellipses represent the dispersion of those points around the centroid.
Epigenetic differentiation in cork tissues relates to phellogen origin
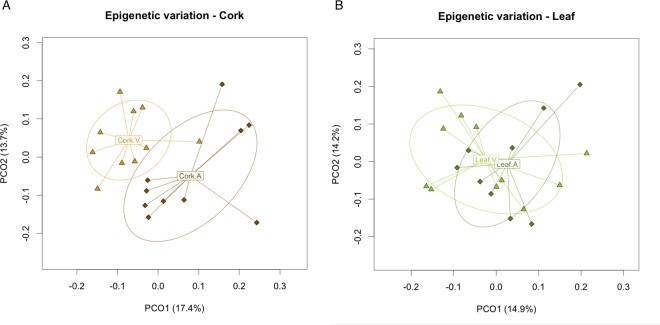
To investigate if phellogen origin (original or traumatic), and/or phellogen age could influence the trend of epigenetic differentiation, we analysed separately the amadia cork-producing trees (S3 Fig). Comparing with the whole sample results, a reduced epigenetic divergence between populations was found for cork tissues (Φst = 0.1432, p-value < 0.0001), indicating that virgin-producing trees could be contributing for higher population epigenetic diversity in these tissues. Considering these results, we further investigated whether virgin and amadia cork could be differentiated at the epigenetic level, using only leaf and cork tissues from CL population, where 11 trees produced virgin cork and 10 produced amadia cork. PCoA showed some epigenetic differentiation for cork samples, according to tissue type (Fig 2A) whereas no clear organization was found for leaves (Fig 2B). Significantly higher epigenetic differentiation was found for virgin cork tissues (Φst = 0.1196, p- value < 0.0001) when compared to leaves from the corresponding trees (Φst = 0.0271, p-value = 0.1598). These results suggest that the tissues generated from the original phellogen (originating virgin cork), although having similar frequencies of the three types of loci (u, m and h), may have different epigenetic marks as compared to the young traumatic phellogen formed after cork removal.
Fig 2.
Principal Coordinate Analysis representing epigenetic differentiation in cork (A) and leaf (B) tissues, between trees producing virgin cork (V) and amadia cork (A) from CL population. Graphical representation is based on the first two coordinates (PCO1 and PCO2) with the percentage of the variability explained shown between brackets. Triangles represent ‘virgin’ cork producing trees and diamonds amadia cork producing trees. The labels represent the centroid for the points cloud in each population and the ellipses represent the dispersion of those points around the centroid.
Cork quality traits are highly variable within populations
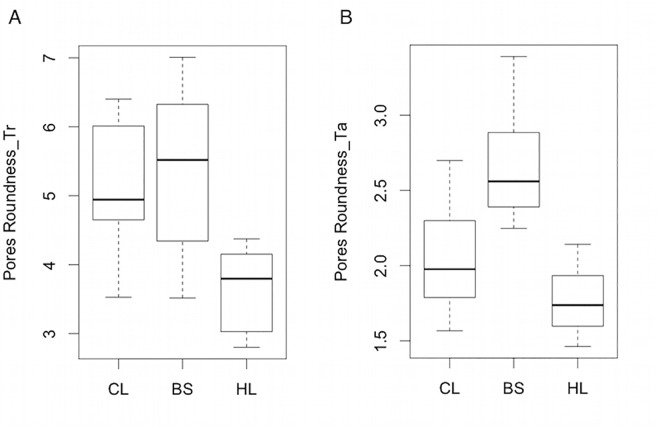
Comparing the cork quality traits assessed in the three populations, there was no statistically significant difference in the average values determined for almost all cork quality traits evaluated in each of the three different sections. In fact, most of the variability observed occurred within the populations rather than among them, as denoted by the larger coefficients of variation (S4 Table). One of the exceptions regards the pores roundness assessed in transverse sections (where pores should appear as rectangular channels). In fact, cork from HL population significantly differed from the other two populations (p-value = 0.0004, Fig 3A) due to its lower roundness values, which could be associated with shorter lenticular channels radially crossing the cork tissue. Interestingly, in radial sections this difference did not reach statistical significance. Moreover, in tangential sections (where pores should appear with an approximately circular form), corks from BS population displayed pores with a more irregular shape (higher values of roundness), as compared to corks from CL and HL populations (p-value < 0.0001, Fig 3B). Another exception concerns the pores’ length in corks from BS population, which was slightly higher than in HL population (p-value = 0.0209). This might be due to more elongated (elliptical) shape of lenticular channels in their tangential section, generally associated with lower cork porosity.
Fig 3. Boxplots representing the distribution of pores roundness.
(A) in transverse section (Tr) and (B) tangential section (Ta) in samples from amadia cork planks collected from the three populations: Companhia das Lezírias (CL), Barradas da Serra (BS) and Herdade dos Leitões (HL).
The occurrence of ‘nail’ is negatively correlated with cork growth
Considering the comprehensive amount of data collected for several cork traits determining quality, we have also assessed correlations between them (S4 Fig, S5 Table). In addition to the expected correlation between aspects of the same trait in transverse and radial sections, we also found a predictable strong positive correlation between cork thickness and annual growth (r = 0.875, p-value = 5.78e-07). Significant positive correlation was also found between cork thickness and pore area and pore length, in transverse sections, revealing that lenticular phellogen producing pores follow the same annual regularity as phellogen. Interestingly, we found a significant negative correlation between ‘nail’ parameters and cork thickness/ annual growth, suggesting that the localized death of phellogen (creating ‘nails’) has a negative impact on cork growth. This was particularly evident in transverse sections, in which the average area of ‘nail’ and cork thickness or annual growth showed a moderate correlation of -0.501 (p-value = 0.0065) or -0.434 (p-value = 0.0211), respectively. For ‘nail’ percentage, in the same section, the moderate correlation with cork thickness or annual growth was -0.4624957 (p-value = 0.01321) or -0.462 (p-value = 0.013), respectively.
MSAP markers are associated with cork quality traits
The association study among the 315 MSAP polymorphic markers found for amadia cork tissues and the considered cork quality traits revealed 7 significant associations (Table 4). These significant marker-trait associations included 4 anonymous loci (1.3% of the total markers tested per trait), one of which in two different states of methylation and 3 of the studied traits that mostly contributes to cork quality: annual growth porosity and ‘nail’ presence. One MSAP marker (hemi-methylated C4_487) was associated with the ‘nail’ percentage in transverse sections, that when present there is an estimated probability that 95% of the individuals have a low percentage of ‘nail’ (high cork quality). Another interesting association was found between the C1_175 MSAP marker, that can be present either in methylated or unmethylated state, and the porosity coefficient and pores’ area. When this sequence is methylated, the estimated probability of corks showing lower pore area (high cork quality) is 94%.
Table 4. Statistically significant logistic regressions relating cork quality traits and MSAP marker presence.
| MSAP marker | Restriction site methylation state | Cork quality trait | Significance of MSAP marker effect a | q-value b | c | Odds d |
|---|---|---|---|---|---|---|
| p-value | ||||||
| C4_487 | Hemi-methylated | % ‘Nails’_Tr | 0.00094 | 0.0681 | 0.95 | 20.0 |
| C1_175 | Unmethylated | Pore area_Tr | 0.00020 | 0.0140 | 0.31 | 0.44 |
| % Porosity_R | 0.00054 | 0.0237 | 0.12 | 0.14 | ||
| Methylated | Pore area_Tr | 0.00020 | 0.0140 | 0.94 | 15.0 | |
| % Porosity_R | 0.00054 | 0.0237 | 0.50 | 1.00 | ||
| C2_101 | Unmethylated | Pore area_Tr | 0.00002 | 0.0040 | 0.27 | 0.36 |
| C1_296 | Unmethylated | Pore area_Tr | 0.00056 | 0.0293 | 0.18 | 0.22 |
Generalized linear models were fitted to higher cork quality presence data browsed for the N = 29 amadia cork samples (see Table 3 for details), using a logit link model and assuming fixed effects for MSAP marker.
a Likelihood ratio tests to verify the null hypothesis of no effects of the marker were performed.
b Largest q-value leading to an expectation of less or equal than one falsely significant model [i.e. q-value x (number of models accepted as significant) ≤1].
c Estimated probability of success (higher cork quality), when MSAP marker is present.
d The estimated odds to obtain higher cork quality (success) when the marker is present is .
R—radial section; Tr—transverse section
Discussion
In this study differences in DNA methylation patterns in virgin and amadia corks, and in leaf tissues of Q. suber adult trees were analysed. The comparison of DNA methylation profiles of cork and leaves, two different tissues from the same genotype, allowed differentiating genetic from epigenetic polymorphisms. The detected epigenetic polymorphisms were further associated with the most relevant cork quality traits: growth, porosity and presence of ‘nails’. MSAP was the method chosen for this study although the impossibility in identifying the sequence, location in the genome and the genes affected by the methylation. Regardless of these limitations we were able to assess the diversity of DNA methylation patterns in a significant number of CG and CCG loci, as well as to investigate associations with phenotypic traits, in a species without sequenced genome. Moreover, the MSAP scoring used in this study and previously suggested by Schulz and colleagues [44] allowed for scoring either methylated, unmethylated or external hemimethylated fragments, providing an extended view of genome methylation, that single scoring methods do not allow.
Epigenetic differentiation in cork stands
In order to differentiate methylated from mutated loci, cork tissues and leaves from the same individuals were used. However, this information could be achieved only for u-loci, when both HpaII and MspI were used for DNA digestion and equal MSAP profiles were found for both tissues in the same genotype. Thus, any difference in MSAP profiles between distinct genotypes should reflect mutated loci which were kept out of the analysis due to its reduced number, comparing with the number of epiloci found. To our knowledge, this is the first work making use of this approach in trees, to reduce some bias due to genetic variation.
Cork and leaf tissues showed a higher frequency of mCG methylation (m-loci) compared to mCCG (h-loci) in agreement with the overall methylation frequencies found in plant genomes [10,52]. Still, the frequency of all methylated loci (m-loci plus h-loci) was higher in leaves than in cork tissues and the opposite was found for unmethylated loci. However, it must be highlighted that MSAP does not detect all methylation contexts or fully methylated external sites associated with plant heterochromatin [44,52]. Nevertheless, these results may arise from the different nature of both tissues, as cells were fully differentiated in leaves while meristematic activity is found in the phellogen of cork tissues. Indeed, since fully differentiated cells in cork are devoid of cellular content [24], cork samples include mostly meristematic cells (in the phellogen most intense activity period) and contiguous young cells at early differentiation stage [53,54]. Also, the distinct cell differentiation stages present in these two tissues might contribute to the significantly higher cytosine methylation diversity found in cork tissues, as also observed in juvenile or undifferentiated Pinus tissues [55,56].
The three cork oak populations analysed showed a relatively high level of epigenetic differentiation (Φst = 0.16) for either leaf or cork samples. Previous work in Q. lobata, using reduced-representation bisulphite sequencing reported higher levels of population differentiation at CG contexts (FST = 0.28) than at CHG contexts (FST = 0.08) [34]. Differences in epigenetic differentiation between cork and valley oaks can be explained by the distinct methods used: for cork oak, CG and CCG contexts were evaluated together while for valley oaks these two contexts were used separately. Still, our result may reflect some degree of local adaptation related to polymorphic epiloci, as also suggested for valley oaks [34], probably due to stable epialleles appearing during these trees’ long life on the same environment. Nevertheless, most of the variability was found within populations which is consistent with the patterns observed also for the genetic variability of this oak populations [32,33] and for cork quality [29,57]. Heritable DNA methylation variants can arise spontaneously in the absence of genetic control [58,59] and potentially affect adaptation. However, it remains undetermined if the epigenetic variation found here is autonomous or an effect of underlying genetic variation, given the cork oak high genetic variability.
DNA methylation patterns differ according to the phellogen origin and age
Our results indicate that virgin and amadia corks show distinct DNA methylation profiles, leading to some degree of epigenetic differentiation, while no variation was found in leaves of the same trees. After formation, phellogen remains seasonally active throughout the life of the tree and produces virgin cork for several years. When cork is removed the original phellogen dies and a traumatic phellogen is formed in response to the wounding stimulus, by a process of meristematic activation of the non-conducting phloem living cells [28,60]. Therefore, virgin and amadia corks have distinct origins and develop from phellogens with different ages, since the traumatic phellogen has nine years old whereas the original phellogen has at least 25 years of age. Younger phellogens have been reported as having higher growth activity [48], and methylation profiles can be determined by underlying mechanistic differences linked to ageing, as seen in other plants [55,56,61]. Another hypothesis is related to the different origin and growth activity of original and traumatic phellogen. Plants have mechanisms to regenerate tissues via dedifferentiation or transdifferentiation, as seen in the xylem activation of meristematic activity during vascular repair (reviewed in [62]). Genes involved in chromatin modifications and remodelling, together with cell cycle genes were found to be up-regulated during the early regeneration process after bark girdling in Populus [63]. These results reveal that the capacity of xylem cells to change their fate is modulated by epigenetic regulation when cell cycle is activated [63]. Considering this, we may also hypothesize that after traumatic chromatin remodelling, some “memory” may have been imprinted in the newly formed phellogen, thus accounting for differences in methylation patterns between amadia and virgin cork tissues.
The presence of ‘nails’ is associated with DNA methylation
The extent of cork defects measured in this study, as well as the variability found among populations are within the values reported in previous publications [29,57]. Although the origin of these cork traits can be explained in the context of development, the genes controlling their frequency or shape are unknown. In this work we found a negative association between cork growth parameters (thickness and average annual growth) and the percentage of nail inclusions.
The width of the annual growth ring has been attributed to the number of cells produced by each phellogen mother-cell [64]. In turn, ‘nails’ are formed when small portions of the phellogen die and a new meristematic capacity arises in living cells inside the underlying phloem, isolating part of the later inside the suberized tissue [23]. Thus, both parameters are dependent on phellogen activity, and the negative correlation found between them can be explained by the temporary cessation of phellogen activity, even in small localized regions, having a drastic impact on cork growth and depreciating its quality. Interestingly, we found one DNA methylation polymorphism (C4_487) that strongly correlates with low percentage of ‘nails’, i.e. high cork quality, and more stable phellogens. This result seems to indicate a potential role of cytosine methylation in the regulation of phellogen-localized death, which is also corroborated by the higher expression of QsDMAP1 in low quality corks [39]. QsDMAP1 is a putative Methyltransferase 1 Associated Protein 1, homologous of the yeast chromatin remodelling complex SWR1 [65], shown to be involved in DNA methylation, DNA repair [66] and cell cycle control [67].
DNA methylation may be involved in lenticular phellogen activity
A significant positive correlation was found between cork thickness and the area and length of cork pores. Previous studies about the effect of growth rate on cork structure already describe this correlation [64]. Pores are naturally found in all cork planks, however their number, dimension and distribution vary widely among trees [23,24,27], with high quality corks showing fewer and small diameter pores and bad quality corks showing the opposite pattern. Pores have been regarded as gas trade structures that exchange gases across the trunk [68] resembling stomata in terms of function. Indeed, the lenticular channels in the young virgin cork are formed below stomata [24] suggesting a common initial development regulation. Moreover, a recent study shows that bad quality cork-producing trees have higher expression of putative stomatal/lenticular-associated genes than good quality corks [31]. Regardless of lenticels development below stomata in the cork oak first periderm, in amadia corks stomata do not occur, although lenticular channels are present. Actually, phellogen originates two different types of cells to the outside: cells that either differentiate as suberized cork cells or, in a very small proportion, cells forming a loosely filling tissue, whose disaggregation leads to the lenticular channels or pores that radially cross the cork [24]. Similarly to the phellogen, the vascular cambium contains two types of morphologically distinct cell types—fusiform initials and ray initials that give rise to the axial and radial components of xylem, respectively [60]. The identity of cambial cells is rather determined by positional cues (reviewed in [69]) but the determination of phellogen cells’ identity and the molecular mechanisms underlying those differences are intriguing issues still to be uncovered. Thus, it is not known how or when phellogen cells fate is established. However, DNA methylation is likely involved in such differentiation as we found a MSAP sequence that is unmethylated in almost all corks with high pore area. Another particular fragment appears in two states: unmethylated in almost all corks with high pore area, and methylated in three quarters of the corks with small pores. These MSAP sequences and their methylation status seem to be good candidates to be used as markers for the pore area, which is one of the most important cork quality traits.
Conclusions
This is the first work involving a genome-wide approach to correlate cork quality traits and DNA methylation in living cork cells. This study offers evidence that DNA methylation is associated with differences in cork cellular characteristics directly related to original or traumatic phellogen activity. The presence of cork quality trait-associated markers and their distribution across the three populations, support the hypothesis of a DNA methylation role in the regulation of cork quality traits. After confirmation of epigenetic status inheritance, further demonstration of a causative linkage between these methylation polymorphisms and cork quality traits will require additional investigation, namely by: (1) comparing with a reference genome to identify the sequence and genomic context of these markers, and their relationship with potential targets and gene expression, allowing to determine the methylation mechanisms underlying the phenotypic variation observed; (2) unravelling if these DNA methylation variants are independent of genetic variation by searching candidate genetic loci associated with them; (3) bisulphite-converted restriction site associated DNA sequencing (BsRADseq) [70] or whole epigenome analysis for a thorough genome DNA methylation study of the phellogen types producing contrasting cork qualities. This would allow mapping major DNA methylation differences, and finding other candidate epiloci associated with cork quality.
As final conclusion, our findings provide new tools to assess cork quality, and may still motivate further studies about the involvement of DNA methylation in altered forest trees phenotypes.
Supporting Information
(PNG)
(PNG)
(PNG)
Several parameters were measured for transversal (Tr), radial (R) and tangential (Ta) sections independently.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
(XLSX)
Acknowledgments
The authors are very indebted to Fundação João Lopes Fernandes for giving access to the Herdade dos Leitões trees and providing the best conditions for cork harvesting as well as to Companhia das Lezírias, S.A. and Herdade de Barradas da Serra. We also sincerely thank Helena Sapeta and Natacha Vieira (Instituto de Tecnologia Química e Biológica António Xavier, Oeiras, Portugal) for the help in sample collection in the field, Carmen Santos (Instituto Nacional de Investigação Agrária e Veterinária, I. P., Oeiras, Portugal) for support in the Genetic Analyser facilities and Inês Barbosa (NOVA University of Lisbon) for assistance in the cork quality image analysis. We also thank the two reviewers for the helpful suggestions and comments that significantly contributed to improve the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Portuguese Foundation for Science and Technology (FCT, http://www.fct.pt/) through the projects PTDC/AGR-CFL/104197/2008, PTDC/AGR-GPL 101785/2008, and PTDC/AGR-FOR/3356/2014. Also through funding to the Research unit GREEN-it "Bioresources for Sustainability" (UID/Multi/04551/2013), and LEAF Unit UID/AGR/04129/2013. VI—PhD grant SFRH/BD/85879/2012; PMB—Post-doc grant SFRH/BPD/86742/2012 and AC—Postdoctoral Fellow SFRH/BDP/97166/2013 were also supported by FCT. PMB also acknowledges the STSM financed by Cost Action FP1202 MaPFGR.
References
- 1.Bräutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Marcos JG, et al. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol. 2013;3: 399–415. 10.1002/ece3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riggs AD, Martienssen RA RV. Introduction Epigenetic mechanisms of gene regulation (ed Russo VEA, et al). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1996. pp. 1–4. [Google Scholar]
- 3.Eichten SR, Schmitz RJ, Springer NM. Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiol. 2014;165: 933–947. 10.1104/pp.113.234211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenfeld G. A brief history of epigenetics. Cold Spring Harb Perspect Biol. 2014;6:a018200 10.1101/cshperspect.a018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. Nature Publishing Group; 2010;11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, et al. Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell. 2006;126: 1189–1201. 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 7.Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, et al. Patterns of population epigenomic diversity. Nature. 2013;495: 193–198. 10.1038/nature11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar a. H, et al. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell. 2008;133: 523–536. 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robert K. Tran, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff and S. DNA Methylation Profiling Identifies CG Methylation Clusters in Arabidopsis Genes. Curr Biol. 2005;15: 154–159. 10.1016/j.cub.2005.01.008 [DOI] [PubMed] [Google Scholar]
- 10.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452: 215–219.4 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyna-López GE, Simpson J, Ruiz-Herrera J. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet. 1997;253: 703–710. [DOI] [PubMed] [Google Scholar]
- 12.Herrera CM, Bazaga P. Epigenetic correlates of plant phenotypic plasticity: DNA methylation differs between prickly and nonprickly leaves in heterophyllous Ilex aquifolium (Aquifoliaceae) trees. Bot J Linn Soc. 2013;171: 441–452. [Google Scholar]
- 13.Peraza-Echeverria S, Herrera-Valencia VA, Kay A-J. Detection of DNA methylation changes in micropropagated banana plants using methylation-sensitive amplification polymorphism (MSAP). Plant Sci. 2001;161: 359–367. [DOI] [PubMed] [Google Scholar]
- 14.Paun O, Bateman RM, Fay MF, Hedren M, Civeyrel L, Chase MW. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol Biol Evol. 2010;27: 2465–2473. 10.1093/molbev/msq150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera CM, Bazaga P. Untangling individual variation in natural populations: ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol Ecol. 2011;20: 1675–1688. 10.1111/j.1365-294X.2011.05026.x [DOI] [PubMed] [Google Scholar]
- 16.Medrano M, Herrera CM, Bazaga P. Epigenetic variation predicts regional and local intraspecific functional diversity in a perennial herb. Mol Ecol. 2014;23: 4926–38. 10.1111/mec.12911 [DOI] [PubMed] [Google Scholar]
- 17.Wilschut R, Oplaat C, Snoek B, Kirschner J, Verhoeven K. Natural epigenetic variation contributes to heritable flowering divergence in a widespread asexual dandelion lineage. Mol Ecol. 2016;25: 1759–1768. 10.1111/mec.13502 [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Xu C, von Wettstein D, Liu B. Tissue-specific differences in cytosine methylation and their association with differential gene expression in sorghum. Plant Physiol. 2011;156: 1955–1966. 10.1104/pp.111.176842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candaele J, Demuynck K, Mosoti DR, Mosoti DR, Beemster GTS, Inze D, et al. Differential methylation during maize leaf growth targets developmentally regulated genes. Plant Physiol. 2014;164: 1350–64. 10.1104/pp.113.233312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A, et al. Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell. 2004;16: 510–522. 10.1105/tpc.017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W-S, Pan Y-J, Zhao X-Q, Dwivedi D, Zhu L-H, Ali J, et al. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J Exp Bot. 2011;62: 1951–1960. 10.1093/jxb/erq391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long Y, Xia W, Li R, Wang J, Shao M, Feng J, et al. Epigenetic QTL mapping in Brassica napus. Genetics. 2011;189: 1093–1102. 10.1534/genetics.111.131615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira H. Cork biology, production and uses—Elsevier Publications Elsevier Publications; 2007. [Google Scholar]
- 24.Graça J, Pereira H. The periderm development in Quercus suber. Iawa J. 2004;25: 325–335. [Google Scholar]
- 25.Natividade J V. Subericultura. Second Edi. Moeda IN da C da, editor. Lisboa: Lisboa: Ministério da Agricultura Pescas e Alimentação, Direcção Geral das Florestas; 1950.
- 26.Oliveira G, Costa A. How resilient is Quercus suber L. to cork harvesting? A review and identification of knowledge gaps. For Ecol Manage. 2012;270: 257–272. [Google Scholar]
- 27.Fortes MA, Rosa ME, Pereira H. A cortiça. Lisboa: ISTPress; 2004. [Google Scholar]
- 28.Silva SP, Sabino MA, Fernandes EM, Correlo VM, Boesel LF, Reis RL. Cork: properties, capabilities and applications. Int Mater Rev. 2005;50: 345–365. [Google Scholar]
- 29.Pereira H, Lopes F, Graça J. The evaluation of the quality of cork planks by image analysis. Holzforsch—Int J Biol Chem Phys Technol Wood. 1996;50: 111–115. [Google Scholar]
- 30.Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M. A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiol. 2007;144: 419–431. 10.1104/pp.106.094227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teixeira RT, Fortes AM, Pinheiro C, Pereira H. Comparison of good- and bad-quality cork: application of high-throughput sequencing of phellogenic tissue. J Exp Bot. 2014;65: 4887–4905. 10.1093/jxb/eru252 [DOI] [PubMed] [Google Scholar]
- 32.Simeone, Cosimo M, Papini A, Vessella F, Bellarosa R, Spada F, Schirone B. Multiple genome relationships and a complex biogeographic history in the eastern range of Quercus suber L. (Fagaceae) implied by nuclear and chloroplast DNA variation. Caryologia. 2009;62: 236–252. [Google Scholar]
- 33.Coelho AC, Lima MB, Neves D, Cravador A. Genetic diversity of two evergreen oaks [Quercus suber (L.) and Quercus ilex subsp. rotundifolia (Lam.)] in Portugal using AFLP markers. Silvae Genet. 2006;55: 105–118. [Google Scholar]
- 34.Platt A, Gugger PF, Pellegrini M, Sork VL. Genome-wide signature of local adaptation linked to variable CpG methylation in oak populations. Mol Ecol. 2015;24: 3823–3830. 10.1111/mec.13230 [DOI] [PubMed] [Google Scholar]
- 35.Gugger PF, Fitz-Gibbon S, Pellegrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol. 2016;25: 1665–1680. 10.1111/mec.13563 [DOI] [PubMed] [Google Scholar]
- 36.Sork VL, Fitz-Gibbon ST, Puiu D, Crepeau M, Gugger PF, Sherman R, et al. First Draft Assembly and Annotation of the Genome of a California Endemic Oak Quercus lobata Née (Fagaceae). G3 (Bethesda). 2016;6: g3.116.030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plomion C, Aury JM, Amselem J, Alaeitabar T, Barbe V, Belser C, et al. Decoding the oak genome: Public release of sequence data, assembly, annotation and publication strategies. Mol Ecol Resour. 2016;16: 254–265. 10.1111/1755-0998.12425 [DOI] [PubMed] [Google Scholar]
- 38.Ribeiro T, Viegas W, Morais-Cecílio L. Epigenetic marks in the mature pollen of Quercus suber L. (Fagaceae). Sex Plant Reprod. 2009;22: 1–7. 10.1007/s00497-008-0083-y [DOI] [PubMed] [Google Scholar]
- 39.Ramos M, Rocheta M, Carvalho L, Inácio V, Graça J, Morais-Cecilio L. Expression of DNA methyltransferases is involved in Quercus suber cork quality. Tree Genet Genomes. 2013;9: 1481–1492. [Google Scholar]
- 40.Correia B, Valledor L, Meijón M, Rodriguez JL, Dias MC, Santos C, et al. Is the interplay between epigenetic markers related to the acclimation of cork oak plants to high temperatures? PLoS One. 2013;8: e53543 10.1371/journal.pone.0053543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa A, Barbosa I, Roussado C, Graça J, Spiecker H. Climate response of cork growth in the Mediterranean oak (Quercus suber L.) woodlands of southwestern Portugal. Dendrochronologia. 2016;38: 72–81. [Google Scholar]
- 42.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus (Madison). 1990;12: 13–15. [Google Scholar]
- 43.Fulneček J, Kovařík A. How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet. 2014;15: 2 10.1186/1471-2156-15-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz B, Eckstein RL, Durka W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol Ecol Resour. 2013;13: 642–653. 10.1111/1755-0998.12100 [DOI] [PubMed] [Google Scholar]
- 45.Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol. 2002;11: 139–151. [DOI] [PubMed] [Google Scholar]
- 46.Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004;13: 3261–3273. 10.1111/j.1365-294X.2004.02346.x [DOI] [PubMed] [Google Scholar]
- 47.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa A, Nunes LC, Spiecker H, Graça J. Insights into the responsiveness of cork oak (Quercus suber L.) to bark harvesting. Econ Bot. 2015;69: 171–184. [Google Scholar]
- 49.Costa A, Pereira H. Influence of vision systems, black and white, colored and visual digitalization, in natural cork stopper quality estimation. J Sci Food Agric. 2007;87: 2222–2228. [Google Scholar]
- 50.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100: 9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa A, Pereira H. Decision rules for computer-vision quality classification of wine natural cork stoppers. Am J Enol Vitic. 2006;57: 210–219. [Google Scholar]
- 52.Fulnecek J, Matyásek R, Kovarík A. Distribution of 5-methylcytosine residues in 5S rRNA genes in Arabidopsis thaliana and Secale cereale. Mol Genet Genomics. 2002;268: 510–517. 10.1007/s00438-002-0761-7 [DOI] [PubMed] [Google Scholar]
- 53.Şen A, Quilhó T, Pereira H. The cellular structure of cork from Quercus cerris var. cerris bark in a materials’ perspective. Ind Crops Prod. 2011;34: 929–936. [Google Scholar]
- 54.Soler M, Serra O, Molinas M, García-Berthou E, Caritat A, Figueras M. Seasonal variation in transcript abundance in cork tissue analyzed by real time RT-PCR. Tree Physiol. 2008;28: 743–751. [DOI] [PubMed] [Google Scholar]
- 55.Fraga MF, Cañal MJ, Rodríguez R. Phase-change related epigenetic and physiological changes in Pinus radiata D. Don. Planta. 2002;215: 672–678. 10.1007/s00425-002-0795-4 [DOI] [PubMed] [Google Scholar]
- 56.Fraga MF, Rodriguez R, Canal MJ. Genomic DNA methylation-demethylation during aging and reinvigoration of Pinus radiata. Tree Physiol. 2002;22: 813–816. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez-Adrados JR, Lopes F, Pereira H. Quality grading of cork planks with classification models based on defect characterisation. Holz als Roh- und Werkst. 2000;58: 39–45. [Google Scholar]
- 58.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401: 157–161. 10.1038/43657 [DOI] [PubMed] [Google Scholar]
- 59.Graaf A Van Der, Wardenaar R, Neumann DA, Taudt A, Shaw RG, Jansen RC. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc Natl Acad Sci U S A. 2015;112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evert RF. Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: their Structure, Function, and Development. 3rd edn. John Wiley & Sons, Inc.; 2006. [Google Scholar]
- 61.Santamaría M, Hasbún R, Valera MJ, Meijón M, Valledor L, Rodríguez JL, et al. Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J Plant Physiol. 2009;166: 1360–1369. 10.1016/j.jplph.2009.02.014 [DOI] [PubMed] [Google Scholar]
- 62.Birnbaum KD, Alvarado AS. Slicing across Kingdoms: Regeneration in Plants and Animals. Cell. 2008;132: 697–710. 10.1016/j.cell.2008.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Gao G, Chen J, Taylor G, Cui K, He X. Molecular features of secondary vascular tissue regeneration after bark girdling in Populus. New Phytol. 2011;192: 869–884. 10.1111/j.1469-8137.2011.03855.x [DOI] [PubMed] [Google Scholar]
- 64.Pereira H, Graça J, Baptista C. The effect of growth rate on the structure and compressive properties of cork. IAWA Bull. 1992;13: 389–396. [Google Scholar]
- 65.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14: 147–154. 10.1016/j.gde.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 66.Negishi M, Chiba T, Saraya A, Miyagi S, Iwama A. Dmap1 plays an essential role in the maintenance of genome integrity through the DNA repair process. Genes to Cells. 2009;14: 1347–1357. 10.1111/j.1365-2443.2009.01352.x [DOI] [PubMed] [Google Scholar]
- 67.Shin JH, Kang HC, Park YY, Ha DH, Choi YH, Eum HY, et al. Corepressor MMTR/DMAP1 is an intrinsic negative regulator of CAK kinase to regulate cell cycle progression. Biochem Biophys Res Commun. Elsevier Inc.; 2010;402: 110–115. 10.1016/j.bbrc.2010.09.126 [DOI] [PubMed] [Google Scholar]
- 68.Lendzian KJ. Survival strategies of plants during secondary growth: Barrier properties of phellems and lenticels towards water, oxygen, and carbon dioxide. J Exp Bot. 2006;57: 2535–2546. 10.1093/jxb/erl014 [DOI] [PubMed] [Google Scholar]
- 69.Prislan P, Čufar K, Koch G, Schmitt U, Gričar J. Review of cellular and subcellular changes in the cambium. IAWA J. 2013;34: 391–407. [Google Scholar]
- 70.Trucchi E, Mazzarella AB, Gilfillan GD, Lorenzo MT, Schönswetter P, Paun O. BsRADseq: screening DNA methylation in natural populations of non-model species. Mol Ecol. 2016;25: 1697–1713. 10.1111/mec.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG)
(PNG)
(PNG)
Several parameters were measured for transversal (Tr), radial (R) and tangential (Ta) sections independently.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.