Abstract
Medical errors remain a significant problem in healthcare. This paper investigates a data-driven outlier-based monitoring and alerting framework that uses data in the Electronic Medical Records (EMRs) repositories of past patient cases to identify any unusual clinical actions in the EMR of a current patient. Our conjecture is that these unusual clinical actions correspond to medical errors often enough to justify their detection and alerting. Our approach works by using EMR repositories to learn statistical models that relate patient states to patient-management actions. We evaluated this approach on the EMR data for 24,658 intensive care unit (ICU) patient cases. A total of 16,500 cases were used to train statistical models for ordering medications and laboratory tests given the patient state summarizing the patient’s clinical history. The models were applied to a separate test set of 8,158 ICU patient cases and used to generate alerts. A subset of 240 alerts generated by the models were evaluated and assessed by eighteen ICU clinicians. The overall true positive rates for the alerts (TPARs) ranged from 0.44 to 0.71. The TPAR for medication order alerts specifically ranged from 0.31 to 0.61 and for laboratory order alerts from 0.44 to 0.75. These results support outlier-based alerting as a promising new approach to data-driven clinical alerting that is generated automatically based on past EMR data.
Keywords: machine learning, outlier detection, medical errors, clinical monitoring and alerting, ICU care
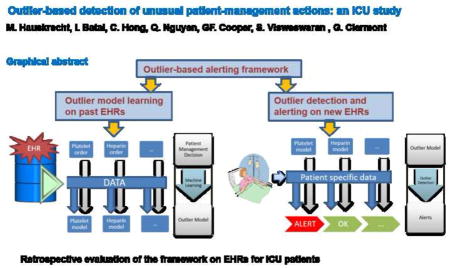
Graphical abstract
1 Introduction
Medical errors continue to be a significant problem in healthcare. In 2000, the Institute of Medicine published the report “To Err is Human – Building a Safer Health System”, which estimated that as many as 98,000 Americans were dying each year as a result of medical errors [1]. More recent studies suggest that the amount of harm due to such errors is even greater. A 2010 report by the U.S. Office of the Inspector General provides evidence to support a medical error rate of six percent among hospitalized Medicare beneficiaries (13.5% adverse event rate × 44% of adverse events that were clearly or likely preventable) [2]. A literature review by James analyzed four studies published between 2008 and 2011 and derived estimates of harm due to medical errors [3]. The number of patient deaths due to such errors was estimated to be more than 400,000 per year; serious harm was estimated to be at least 10-fold more common than lethal harm.
Studies support that some interventions are effective in reducing selected types of medical errors [4], including the use of computerized physician order entry [5]. Nonetheless, a recently published study showed no statistically significant decrease in overall hospital-based medical errors during the period from 2002 to 2007 in North Carolina, even though that state has had a high level of engagement in its efforts to improve patient safety [6]. The authors conclude that “our findings validate concern raised by patient-safety experts in the United States [4] and Europe [7] that harm resulting from medical care remains very common.” It is clear that there is considerable room for improvement in both the penetration of existing methods for reducing medical errors and the introduction of new, effective methods.
The focus of this paper is on the development and evaluation of a data-driven monitoring and alerting approach that relies on stored clinical information of past patient cases and on statistical methods for the identification of clinical outliers (anomalies) for a current patient. The key conjecture behind the approach is that the detection of anomalies corresponding to unusual patient management actions will help to identify medical errors. We have pioneered this new approach and reported its initial evaluation on post-cardiac surgical patient population in Hauskrecht et al [8–10]. In this paper, we describe further enhancements of the outlier detection methodology, including a new method for uniformly controlling the rate at which alerts are raised. We also present the results of a retrospective study of the methdology on patient cases from the intensive care unit (ICU). We report the true positive alert rate (TPAR) of the system for the different alert frequencies and demonstrate the improved TPAR for lower frequencies. Finally, we perform an indepth analysis of the reasons for the false positive alerts that occur.
The ICU has operating characteristics that predispose to medical errors, including that it is fast-paced, complex, and involves high-risk decision-making [11]. Multiple studies during the past 20+ years have documented that a significant number of medical errors occur in the ICU [12–14]. In a report published in 2005, Rothschild and colleagues at Harvard describe a year-long observational study that they conducted in a medical and a cardiac ICU, where they documented approximately one serious medical error per 5.4 patient days [15]. At least 65 percent of those errors were related to medication and laboratory orders, which are the focus of the investigation reported in the current paper. A recent systemic review and meta-analysis showed that on average those ICU patients with adverse events (whether or not they were medical errors) had significantly longer hospital stays (mean: 8.9 days; 95% CI: 3.3–14.7) and ICU stays (mean: 6.8 days; CI: 0.2–13.4) than did ICU patients without adverse events [16].
2 Background
Clinical alerting systems are designed to detect adverse events as early as possible. A common alerting approach uses rules that check EMR data of a patient for a specific clinical condition or a set of expert-defined physiologic criteria. If such patterns are identified in the data, an alert to the patient’s clinicians is raised [17]. The alert signal can take various forms, such as pop-up windows on an EMR interface or email/paging messages sent to the patient’s physician. Alerting systems have been extensively explored both academically [18–20] and commercially [21–23]. They have been applied in a variety of tasks, including detection of deviations from infectious disease treatment protocols [22], detection of adverse drug events [24, 25], detection of growth disorders [19], and detection of clinically important events in diabetes [18] and congestive heart failure management [26].
Knowledge-based alerting systems
The most common type of monitoring and alerting systems use rules that are manually constructed by clinical domain experts using their knowledge and personal experience. The alerts generated by these rules and their benefits are derived directly from the experts’ experience. Examples include rules that screen for drug allergies and interactions, as well as rules for syndromic diagnosis (e.g., suspected sepsis, acute lung injury) and disease severity estimation [27]. However, knowledge-based alerting systems based on rules (as currently being used) require manual construction, which can be time consuming and tedious. In addition, the expert-defined rules have, by their design, limited coverage of the large space of adverse events, particularly more complex adverse events. In other words, knowledge-based rules can only monitor and detect what they were explicitly built for. Finally, the rules are hard to tune in advance to achieve clinically acceptable performance in the environments in which they are deployed. It is not uncommon for alert rules that were built with significant expert effort to be retired (turned off) shortly after they are deployed due to high false alert rates [28, 29]. Even when such rules remain active, their alerts may be ignored due to false alert fatigue [28–30]. Thus, the performance of even carefully designed rules need to be adapted and optimized carefully to achieve positive clinical results, with minimization of false alarms being a key goal.
Outlier-based alerting
In our recent research work we have pioneered and developed a new approach for medical error detection [8, 9, 31] that is complementary to the knowledge-based alerting approach. Briefly, an alert is raised for clinical care decisions that are highly unusual (anomalous) with respect to past patients with the same or similar conditions. The rationale behind this approach is that the majority of past patient records stored in an EMR reflect the local standard of clinical care, and care that deviates from such standards (e.g., a medication decision) can be detected and will correspond to errors often enough to justify alerting. A major advantage of this anomaly-based alerting method is that unlike knowledge-based methods, it is data driven and does not depend on the monitoring and alerting rules being built by an expert. Instead, the outlier-based approach is driven by deviations from usual patterns of clinical care. These features make the approach applicable to a wide range of clinical environments and conditions, hence the clinical-alerting coverage is broad and deep (unlike for knowledge-based systems). Outlier-based monitoring and alerting has potential to complement the use of knowledge-based alerting systems that are currently deployed, thereby improving overall clinical coverage of current alerting systems.
This paper reports the next step of the development and testing of our outlier based alerting methodology. The paper describes a new way for precisely controlling the true alert rates that the method generates, and it reports a retrospective evaluation of the method on a set of ICU patient cases.
3 Methods
The outlier-based alerting framework we have developed works in two stages: a model-building stage and a model-application stage. In the model-building stage it uses cases from an EMR repository to train outlier models that summarize when certain patient management actions are typically made. Outlier models are built using statistical machine learning methods and represent probabilistic models of patient-care actions applied in response to various patient conditions in the past. There are multiple outlier models in the system covering different aspects of care, such as medication and laboratory (lab) orders. For example, a heparin model captures patient subpopulations for which heparin is typically prescribed, subpopulations for which it is not, and subpopulations for which it is typically discontinued. Similarly, a lab-order model summarizes patient conditions for which the lab is typically ordered and for which it is not. In general, a model, such as one predicting the ordering of a glucose level, will contain multiple predictors, such as functions of earlier medication orders, laboratory orders, and laboratory results. In the model-application stage, the outlier models are continuously applied to new patient data to identify those actions that are unusual and deviate from the prevalent pattern of care, as represented in the outlier models. An unusual action, which may correspond to an unusual omission or commission of a medication order or a lab order, is identified and an alert is raised. We next describe each of these stages in more depth.
3.1 Building (training) action-specific outlier models
Our framework first segments each patient case into multiple patient-state instances using fixed 24-hour time segmentation. It then uses feature construction methods proposed in [32] that take time series of measurements of labs, physiological parameters, and medication orders to create a wide assortment of temporal features representing a patient case and its history up to a specific segmentation time t. Examples of features generated for a lab time series (e.g., platelet count time series) are the last observed value of the lab, and the slope of the lab value that is derived from its two most recent values. Similar features are built for time series representing medication administration history, as for example a feature indicating that a certain medication (e.g., heparin) is currently given to the patient, or a feature reflecting the overall duration of the treatment. Appendix A gives a complete set of temporal features we generated for the different types of clinical variables (lab tests, physiological variables, and medications) and their time-series.
The features generated for the segmentation time t define a patient state instance s at that time and summarize what is known about the patient and her past up to that time. Every patient state instance s is then associated with clinical care actions (medication orders and lab orders) executed in next 24 hours, that is, actions observed in time interval (t, t + 24 hours]. The actions are encoded using a vector of binary values, where value 1 means the order was made in next 24 hours and value 0 that it was not. For example, if an INR lab test was ordered in next 24 hours, its value in the binary vector is 1. In the end, these steps allow us to produce a dataset of inputs s (representing individual patient states) and associated actions a that follow them.
Model learning
We used a Support Vector Machine (SVM) [33] with a linear kernel to learn a probabilistic model that predicts future clinical care actions from the patient-state features. We build models for each action a, that is, we build a separate model for predicting orders of heparin, aspirin, INR lab, etc. For a given action a, we determine how predictive is each clinical variable (and all its features) for that action individually. We assess the predictive ability of a clinical variable using a linear SVM method and internal train and test splits of the training data. The clinical variables are ranked in terms of the area under the ROC curve (AUROC). For each action a, we determine the 30 highest ranked clinical variables that predict it. We use the union of the full feature sets for these 30 variables to build the final model for predicting action a. Using these features and the training data, we apply the linear SVM method to learn a model that predicts the probability of action a given the features.
We note we chose the feature complexity of the current models by performing a limited experiment comparing the AUC performance of the models with features for the top 20 clinical variables (used in our previous study [8–10]), top 30 clinical variables (our current models), and for all clinical variables, respectively. The models with all features did not show any improvement over the models with 30 clinical variables which we attribute to a very large feature space the SVM algorithm had to optimize over and model overfitting. In addition, these optimizations were costly in terms of computational time, so the approach was clearly suboptimal. In terms of models with features based on the top 20 and the top 30 clinical variables, the models with 30 clinical variables were better than models with 20 clinical variables in 55% of medication order models, worse for 23%, and unchanged for 22% of models. Similarly, the models for lab order actions based on 30 clinical variables were better for 40% models, worse for 20%, and unchanged for 40%. All SVM models were built using the liblinear package [34] with hinge loss and the L2 regularization options.
Model calibration
The SVM model for action a defines a discriminative projection for a, reflecting whether it should be taken or not. We transform this projection into a calibrated probability [35] P(a = 1 | s) using a non-parametric approach. We chose a non-parametric calibration approach over monotonic ones because the distribution of positive and negative examples in the tails of the projection did not show monotonic improvements. In particular, we use a non-parametric calibration approach that relies on multiple histogram binning models, each defined by a different number of uniform size bins, and uses them to estimate P(a = 1 | s) by averaging over predictions of these models.
In the experiment section we use three binning models with 200, 400, and 600 bins, respectively, to calculate the estimates.
Model selection
Not all SVM models are good enogh and suitable for outlier calculations and alerting. Briefly, a good model should predict very well the action to be taken next. We used two criteria to select good predictive models: the area under the ROC (AUROC), and a special maxPPV10 statistic. The avgPPVtop10 statistic is based on identifying the ten highest values for P(a =1 | s) over all patient instances and using the average of those ten values. It is used to assess how strongly the model and the evidence in past data would support alerts for action a = 1. A higher value in both statistics indicates a better model. We used these statistics to define strong predictive models that satisfy AUROC > 0.7 and maxPPV10 > 0.5.
3.2 Monitoring and alerting
Outlier score
We used the probabilities derived as calibrated SVM model predictions for strong models to develop an outlier score which measures how unusual are patient-management actions. The score ranges from 0 to 1, with 1 (0) representing the most (least) unusual the action. In particular, an outlier for a binary action a (e.g., heparin is not ordered) is derived from the probability of the counterfactual action ¬a (heparin is ordered), that is, the action that was not taken. For example, suppose heparin was not ordered in 24 hours following a patient state s, and based on the trained model the probability of the counterfactual action -- heparin is ordered -- is P(heparin is ordered within 24 hours | s) = 0.98. Then the outlier score for heparin would be OutlierScore(s, heparin is not ordered within 24 hours following s) = 0.98. As mentioned, the closer the score of an action taken is to value 1, the more unusual (anomalous) the action is relative to the clinical context given by s.
Alert score
The outlier score measures how anomalous is action a taken within 24 hours of patient state s, relative to the expected action. However, the anomaly with respect to s may not persist if the patient state has changed. We define an alert score to measure the persistence of the anomaly of action a in a new state s′ that is reached after 24 hours following s (call this time t′) during which a was taken. In essence, t′ denotes the current time at which the system is determining whether to raise an alert about action a. More specifically, we define the alert score for action a, as being AlertScore (s′, a) = min [OutlierScore (s, a), OutlierScore (s′, a)]. For AlertScore to be high, action a must be anomalous at both at the beginning and end of the 24-hour time period during which a was taken, which provides support that the action remains anomalous when an alert on it is raised at time t′. The alert score for each action in a monitored patient is recalculated every 24 hours and reflects the urgency to raise an alert with respect to that action at that time.
Controlling the alert rate
The alert scores for an action a when applied to many different patient states let us order (rank) these states in terms of the alert urgency for action a. A threshold defined relative to this order can be used to control the alert rate (the fraction of alerts sent) for action a. In principle one could attempt to combine outlier scores for different actions to control overall alert rates across all actions supported by strong models. However, this approach did not work well since alert scores across different actions varied widely contradicting the key assumption that outliers are generated by a random process. To address this issue and permit the uniform alert control, the alerts sent by our system and their frequency (rates) are controlled with the help of a shared alert parameter α ∈[0, 1] and action-specific alerting thresholds derived from this parameter. More specifically, let a be an observed clinical action (e.g., heparin not ordered) and Rate(¬a) be the average rate at which the counterfactual of the clinical action is observed in the training data (e.g., the number of heparin orders per patient per day). We define the alerting threshold θa for action a and alerting parameter α ∈ [0, 1] to be the product α * Rate(¬a). In other words, the threshold θa that determines the rate with which we wish to raise an alert for action a is a fraction of occurrence (as determined by α) of the counterfactual action in the data. For example, α might be 0.05, in which case we wish to raise an alert for the 5 percent of action a occurrences that are most anomalous. With this method, the expected alert rates for all models can be globally increased or decreased by changing an alerting parameter α that is used in setting θa for each a. Moreover, we can tune α so that the expected global alert rate is less than some specific limit (e.g., no more than one expected alert per 20 ICU patients per day).
We would like to note that the above alert control approach can be used for both retrospective and prospective selection of alerts. Briefly, using the past training data, for each supported action a we can estimate Rate(¬a). Then using the alert scores for past data we can find the action specific threshold θa that corresponds to the alert score for action a that would pass α * Rate(¬a) fraction of alerts. A set of action specific alert thresholds is then used to either pass or filter alert candidates.
4 Experiments on ICU dataset
The HIgh-DENsity Intensive Care (HIDENIC) dataset is a comprehensive database of 24,658 admissions (cases) to the ICUs between Dec 1999 and June 2004 that have been assembled from a variety of legacy services at the University of Pittsburgh Medical Center (UPMC). HIDENIC contains approximately 2,000 different variables are grouped in distinct domains of information that include extended demographics and hospital flow (hospital unit, admission/discharge/transfer date and time information); detailed physiology (vital signs, intake/output, severity of illness); prospective clinical diagnosis (APACHE scores); interventions (ventilation, dialysis, drugs with precise timing of administration, etc.); imaging study reports (radiology, echo, etc.); laboratory, pathology, microbiology orders and results; clinical notes; and administrative discharge abstracts (including ICD-9 diagnoses and time-stamped procedures). Among all the data in HIDENIC, we used the clinical information shown in Appendix A for the study reported in this paper.
4.1 Outlier model building and model selection
Model building
Each patient record was segmented into a time-series of increasing lengths of time (in 24 hour increments). Each time-series was summarized by a vector of over 14,000 different features that were constructed from clinical variables that included medication orders, laboratory orders, laboratory results, physiological variables, and volume measurements. These features constitute state s mentioned above, and this vector represents a patient instance. The data were split into a training dataset of 16,500 ICU patient cases (admitted from Dec 1999 till March 2003) and a testing dataset of 8,158 ICU patient cases (admitted from March 2003 till June 2004). The training and testing datasets are mutually exclusive, and the patient cases in the training dataset were admitted to the ICU prior to the patient cases in the test dataset, with a small portion of patients overlapping they stays. The training and testing patient cases were used to generate 225,894 training and 104,698 testing patient instances, respectively. We built a calibrated probabilistic SVM model for predicting each type of action (medication orders and laboratory orders) from the training set and applied the models to the test set for identifying omissions of medication orders and laboratory orders. In total, we built SVM models for 1075 different types medication orders and 222 different types of laboratory orders.
Model selection
As discussed above not all SVM models are suitable for outlier calculations and alerting. We proposed two statistics to select strong predictive models: the area under the ROC (AUROC), and a special avgPPV10 statistic. Appendix B summarizes the quality of predictive models built from the data and the distribution of their AUROC and avgPPV10 scores. Out of all the alert models, 99 laboratory omission models and 156 medication omission models were strong predictive models (AUROC > 0.7 and avgPPV10 > 0.5). These models were used to generate alerts for the study and were the basis of our analysis.
4.2 Study cases and their assessment
We applied strong alert models to test patient state instances and used them to calculate alert scores for the different lab and medication actions. For each patient state, action and their alert score we calculated minimum alert parameter value αmin, that is, the minimum α value that would lead to an alert. We used these αmin values to randomly select 420 alerts from all alert candidates and assess their quality. The number of alerts evaluated was constrained by practical considerations, including the number of critical care physicians we could recruit to assess them. The alert selection was weighted more heavily towards medication alerts for which we had a larger number of stronger models. In particular, out of 420 alerts selected 270 were medication omission alerts and 150 were lab omission alerts. We stratified the selection of alerts in order to represent the different degree of anomalousness as reflected by their αmin values. More specifically the alerts were stratified into subgroups that cover different αmin ranges (see Appendix D). The stratification helped us to evaluate and compare the performance of the alerting method at different operating points (alerting thresholds).
The alerts were evaluated by 18 physicians from the Departments of Critical Care Medicine and Surgery at the University of Pittsburgh. The physicians were divided into six groups of three physicians. Each physician evaluated 50 alerts, such that 40 alerts were shared and evaluated by all three members of the group, and 10 were unique and evaluated by only that individual reviewer. This led to 240 alerts evaluated by three physicians (shared alerts), and 180 alerts evaluated by just one physician for the total of 420 alerts. The analysis in this paper focuses on the shared alerts.
The 240 shared alerts that were generated included 165 medication-omission alerts (for 64 different types of medication orders) and 75 laboratory-omission alerts (for 22 different types of laboratory orders). Appendix C gives a list of medication and lab orders used to generate shared alerts. Appendix D summarizes the distribution of of shared alerts with respect to the different αmin alert threshold ranges.
Assessments of alerts
The alerts were presented to the reviewers using a case review interface we developed called PATRIA that graphically displays the information in a patient’s EMR up to the time of the alert. The interface lets the reviewer see the alert raised and peruse the EMR data of the patient that are known prior to the alert, including all text reports (progress notes, operative and procedure notes, radiology reports, EEG and EKG reports), lab results, medications, physiological parameters, volumes, and procedures performed. After perusing the case, the reviewer completed an electronic questionnaire to specify: (1) the appropriateness of the alert raised, (2) a free text comment section asking the reviewer to justify agreement/disagreement with the alert, (3) whether the reviewer would follow up on the alert with a clinical action, if the reviewer were managing the case prospectively, and (4) the clinical importance of any such action for patient management. The main study question: Will you take a clinical action based on receiving this alert? was used to assess each alert. We used the majority vote to define the reviewers’ consensus. That is, if at least 2 out of 3 of the reviewers answered “yes” to the question, then the alert was assessed to be a true positive and labeled as “appropriate.” Appendix E shows the pairwise Cohen’s kappa agreements of the reviewers in each of the six groups. Somewhat lower kappa statistics observed in some of the groups are discussed in Section 6.
True positive alert rate
The number of true positive alerts divided by the total number of alerts, which we call the true positive alert rate (TPAR), is the key statistic that we used to evaluate the alerting system. We evaluated TPAR at different values of the alert parameter α, which represents different operating points of the system. The secondary statistic that we calculated and analyzed was the alert rate, which reflects the average frequency of alerts raised by the system per unit time.
4.3 Analysis of incorrect alerts
The TPAR results demonstrate the potential of our approach in raising clinically important alerts. However, an equally important issue is to understand when the framework makes mistakes and raises incorrect alerts. To obtain an understanding of such alerts, we analyzed free-text answers provided by the evaluators.
We applied the following procedure to conduct the free-text analysis. First, from all shared alerts (240) and their reviews (720 = 240 × 3) we selected the alerts the evaluators deemed as inappropriate. The alerts marked as an ‘inappropriate alert’ by one of the 18 evaluators included 81 lab alerts (out 225 evaluated lab alerts) and 261 medication alerts (out of 495 medication alerts). Two of the authors of this paper (Clermont and Visweswaran) then analyzed the answers and developed a set of qualitative categories representing the different reasons that reviewers disagreed with the alerts. Both authors are clinicians and neither were reviewers in the alerting study.
Next, the two clinicians individually reviewed each alert assessment marked as an ‘inappropriate alert’ by one of the 18 clinicians evaluating alerts and assigned it to one of the categories explaining the reasons for the disagreement. After the initial assignment, the two clinicians met and reconciled the differences among them through discussion and consensus. This process led to a unique assignment of all ‘inappropriate alerts’ to one of the qualitative categories.
5 Results
This section first reviews the alert performance of the system and then describes the types of errors that it made.
5.1 Analysis of alert performance
Table 1 summarizes results based on the assessments of the 240 alerts that were each reviewed by three reviewers. The table lists the TPARs for the different thresholds on the selected models and the alert rate, which is measured as the number of alerts per patient per day for those models. The results for medication omission and lab order omission alerts are tabulated both separately and combined. Consider, for example, an entry for alert parameter α = 0.025. If our system was operated at this parameter value, the estimated TPAR for the medication omission alerts is 0.4396, or just under 1 correct alert per two alerts raised, and the estimated average alert rate is 0.0318, which is a little over 3 such alerts raised for every 100 patient days.
Table 1.
TPARs for the outlier-based alerting framework and the different values of alert parameter α that were estimated based on the reviewers’ assessments and their standard errors (Std err). Also included is the alert rate for each corresponding alert threshold and TPAR. The alert rate estimates are based on 104,698 test patient instances leading to standard errors very close to zero for all alert rate entries; hence they are excluded from the table.
| alert parameter | lab orders (22 models) | med orders (64 models) | combined (86 models) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| TPAR | Std err | Alert rate* | TPAR | Std err | Alert rate* | TPAR | Std err | Alert rate* | |
|
| |||||||||
| 0.001 | 0.750 | 0.129 | 0.004 | 0.583 | 0.142 | 0.001 | 0.712 | 0.105 | 0.005 |
| 0.0025 | 0.643 | 0.121 | 0.011 | 0.603 | 0.090 | 0.003 | 0.634 | 0.096 | 0.014 |
| 0.005 | 0.657 | 0.110 | 0.022 | 0.523 | 0.085 | 0.006 | 0.627 | 0.087 | 0.028 |
| 0.01 | 0.482 | 0.103 | 0.044 | 0.437 | 0.068 | 0.013 | 0.472 | 0.081 | 0.056 |
| 0.015 | 0.445 | 0.124 | 0.066 | 0.416 | 0.059 | 0.019 | 0.439 | 0.096 | 0.085 |
| 0.025 | 0.469 | 0.119 | 0.109 | 0.440 | 0.063 | 0.032 | 0.462 | 0.093 | 0.141 |
| 0.04 | 0.603 | 0.089 | 0.175 | 0.464 | 0.062 | 0.051 | 0.572 | 0.070 | 0.226 |
| 0.06 | 0.612 | 0.089 | 0.263 | 0.424 | 0.063 | 0.077 | 0.569 | 0.070 | 0.339 |
| 0.08 | 0.608 | 0.101 | 0.350 | 0.319 | 0.056 | 0.102 | 0.542 | 0.079 | 0.453 |
| 0.1 | 0.608 | 0.101 | 0.438 | 0.309 | 0.054 | 0.128 | 0.520 | 0.073 | 0.566 |
Number of alerts per patient per day.
Table 1 estimates TPARs based on the reviewers’ answers. Since the analysis was done by retrospective review of past patient cases in the test set, we also had access to data and actions taken by physicians after the alerts would have been raised. In particular, we could see and analyze if the actions our system alerted on were taken by physicians in the next 24 hours after the alerts were raised. (of course without them seeing the alert). Table 2 shows the TPAR results derived from the observed action for all alerts raised for the models in the study at varying thresholds.
Table 2.
TPARs for the outlier-based alerting framework estimated based on follow-up actions observed in the EMR data and standard errors of these estimates. Also included are alert rates for each threshold and corresponding TPARs.
| alert parameter | lab orders (22 models) | med orders (64 models) | combined (86 models) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| TPAR | Std err | Alert rate | TPAR | Std err | Alert rate | TPAR | Std err | Alert rate | |
|
| |||||||||
| | 0.001 | 0.557 | 0.024 | 0.004 | 0.289 | 0.045 | 0.001 | 0.507 | 0.021 | 0.005 |
| | 0.0025 | 0.529 | 0.015 | 0.011 | 0.267 | 0.025 | 0.003 | 0.474 | 0.013 | 0.014 |
| | 0.005 | 0.519 | 0.010 | 0.022 | 0.247 | 0.017 | 0.006 | 0.459 | 0.009 | 0.028 |
| | 0.01 | 0.520 | 0.007 | 0.044 | 0.230 | 0.012 | 0.013 | 0.456 | 0.006 | 0.056 |
| | 0.015 | 0.519 | 0.006 | 0.066 | 0.211 | 0.009 | 0.019 | 0.450 | 0.005 | 0.085 |
| | 0.025 | 0.512 | 0.005 | 0.109 | 0.216 | 0.007 | 0.032 | 0.445 | 0.004 | 0.141 |
| | 0.04 | 0.494 | 0.004 | 0.175 | 0.210 | 0.006 | 0.051 | 0.430 | 0.003 | 0.226 |
| | 0.06 | 0.478 | 0.003 | 0.263 | 0.202 | 0.004 | 0.077 | 0.415 | 0.003 | 0.339 |
| | 0.08 | 0.451 | 0.003 | 0.350 | 0.197 | 0.004 | 0.102 | 0.394 | 0.002 | 0.453 |
| | 0.1 | 0.419 | 0.002 | 0.438 | 0.191 | 0.003 | 0.128 | 0.367 | 0.002 | 0.566 |
The results in Table 1 show that our alerting approach yields good TPARs across a wide range of alert thresholds explored in the study. In particular, TPARs for medication omission alerts vary from 0.3 to 0.58 and TPARs for lab omission alerts from 0.45 to 0.75. The results also indicate that by tightening the alerting threshold one is able to control the TPAR, that is, the TPAR tends to be higher for the tighter threshold and decreases by gradually relaxing the threshold.
We note that the certainty of the estimates in Table 1 (estimates based on reviewers’ feedback) is influenced by the limited number of expert assessments of alerts that were practical to obtain, and the standard errors are rather high, especially for lab omission results. Thus, an iregularity in the expected drop in TPARs for higher threshold values is likely the result of estimation error due to the limited sample size. The estimates in Table 2 are based on a much larger sample size and hence the estimates are more certain. These TPAR values are monotonically decreasing, as we would expect. The results clearly show this TPAR can be effectively controlled by changing alert parameter α. The differences between the TPARs in the two tables are expected. The TPARs derived in Table 2 can be viewed as approximating a lower bound on the performance of the system. Briefly, this TPAR is based on the exact match in actions we (hypothetically) alerted on. However, in real patient management (without alerts) the need to order a medication or a lab may have been overlooked for more than 24 hours, or a comparable action may have been taken instead of the action recommended by our system.
5.2 Analysis of incorrect alerts
For each alert that a clinician-rater judged rated as “inappropriate”, we assigned it to a category that captures the reason the alert was judged as inappropriate. Tables 3 and 4 summarize the results for lab and medication alerts, respectively.
Table 3.
Categories of inappropriate lab-order alerts and their counts.
| Category | # alerts |
|---|---|
| Lab test results recently obtained | 9 |
| A different lab test might provide more information | 3 |
| A different lab test was obtained with same information | 1 |
| No clinical indication to order lab (lab would not help medical decision making, condition being alerted upon was resolved/resolving/stable, comfort care, patient death) | 68 |
| 81 (total) |
Table 4.
Categories of inappropriate medication-order alerts and their counts.
| Category | # alerts |
|---|---|
| Already receiving the alerted medication or an alternative, equivalent medication | 38 |
| Contraindication | 29 |
| Old indication | 8 |
| No indication (no clinical justification, medical justification has elapsed, comfort care, patient death) | 142 |
| Insufficient data to make determination | 44 |
| 261 (total) |
For lab order omissions, the reviewers thought that in 13 cases the lab test either correctly ordered or that there was a more informative lab other than the alerted on that was either ordered or should have been ordered, such as INR lab instead of PTT. The largest number of incorrect lab order alerts (68) was associated with alerts that did not seem to have any indication or the clinicians thought they were not needed. Examples included instances of comfort care, or when previously high values of a lab were trending down and the physician believed further frequent monitoring of values was not needed.
For medication order alerts, the reviewers believed that in 38 cases the patient was receiving the medication the system alerted on or was receiving an equivalent medication. One example is alerting on the absence of an order for nizatidine when the patient was already on famotidine. In 29 cases the reviewers thought the order alerted on was contraindicated. An example is an alert on the absence of an order for amlodipine while the patient was on vasopressors. Another example is an alert on the omission of furosemide when the patient was allergic to that drug. In 8 cases the reviewers believed the agent that was recommended by the system would not be used today; note the training data in this study spanned the years 2000–2004. The majority of inappropriate medication order instances (142) were categorized into a “no indication” category. One example of this category is an alert on the omission of a potassium supplement for a patient with acute kidney injury that will likely need hemodialysis. Another example is alerting on a hematopoetic agent for the patient who was low on hemoglobin due to a known GI bleed. Finally, in 44 cases the physicians believed there were insufficient data in the EMR to make the determination of the alert, such as a recommendation of Percocet with no information on patient’s level of pain.
6 Discussion
The results of this study show that outlier models built from a subset of lab and medication orders can raise clinically useful alerts with true alerts rates of 30 to 60% for medication omissions and 45 to 75% for lab order omissions. These rates compare favorably to the performance of knowledge-based alerting systems reported in the literature. Briefly, knowledge-based alerting systems in the literature are usually evaluated in terms of alert override rates [28, 29, 34, 36, 37]. The override rates may be influenced by multiple factors, such as the frequency of alerts and their quality [36, 38]. In general, high frequency and low quality alerts can lead to alert fatigue and subsequent high override rates [28, 29, 36, 38, 39]. The override alert rates that have been reported for a variety of drug safety systems in the above literature are in the 0.49 to 0.96 range. If override rates approximate false alert rates, then the TPARs corresponding to the override alert rates just quoted are in the 0.04 to 0.51 range, and thus, the TPARs in our study compare very favorably to them. Similarly, our results compare favorably to TPARs of 0.01 to 0.14 for clinical monitoring systems reported by Graham and Cvach [40]. The experimental data are consistent with higher true positive alert rates occurring when we restrict the alerts to those with higher alerting scores. This finding suggests that we may be able to adjust true positive alert rates of such a monitoring system to achieve performance that is clinically acceptable. Such adjustments are not supported by knowledge-based alerting systems.
6.1 Limitations
Our methodology relies on a probability estimate of an action to be taken. To calculate this estimate, we use a linear SVM model, combined with a calibration approach that is based on averaging over multiple binnings models. Obtaining high-quality probability estimates from limited sample sizes in general is a hard task. Our approach is limited by the particular methods we use for modeling (linear models) and calibration (averaging of binning models).
Another open issue is the methodology for controlling the alert rates. An ideal solution would be to control the alerts for all actions by thresholding a single alert quantity that would reflect the deviation from the standard care and hence the desirability of raising an alert on that patient state. One possible approach would be to control alerts directly based on the alert score derived from the probabilistic models in Section 3.2. However, the quality of our models for the different actions vary considerably, due to various model assumptions (see above) and the sample sizes used to estimate them. This in turn would bias alert selection based on the alert score only to models capable of predicting high probabilities of actions, other models would not be able to reach high probability scores and hence would not be alerted on. Our current methodology overcomes this problem by controlling alerts using a shared alert parameter α, and the alert score is used only to rank alerts for each action individually. Doing so permits a wider range of alerts to be included and raised, but, at the same time opens up a possibility that alerts for bad (unpredictive or weakly predictive) models will be generated, leading to random or close to random alerts. The model selection step assures that only models of certain minimal quality are used to generate the alerts which in the end prevents a generation of such alerts.
The study results were calculated based on the majority consensus of the raters evaluating the alerts. The analysis of raters’ agreements (Appendix E) showed lower kappa statistics among the raters in some of the reviewers’ groups than others. One reason for variation may be that the raters where not highly familiar with our case-review inteface and different reviewers found and used different sources of information to make their assessments. We believe, however, that in the majority of cases the disagreements show natural variability and diversity of opinion among the clinician raters about the appropriateness and utility of the raised alerts. If we had used an alert threshold that was highly lenient and admitted many alerts, even those that are not very anomalous, we would expect high rater agreement that most alerts were inappropriate. If the alert threshold were highly restrictive and admitted alerts only on very anomalous actions, we would expect relatively high rater agreement that the alerts are appropriate. When working within a range of alerting thresholds that are between those two extremes, as we did, it is not surprising that there may be a wider range of opinion about the appropriateness of the alerts, and thus, lower kappa statistics.
6.2 Advantages
The advantages of outlier-based alerting include that it provides broad coverage of clinical care, and it can be learned automatically from an archive of EMR data, updated automatically over time, and adapted to a local healthcare setting. The outlier-based approach we developed is also probabilistic, which provides a clear semantics for alerting and for explaining alerts to users. A clinical action that is an outlier is not necessarily an error. Nonetheless, we conjecture that a system can be developed in which outliers will be errors often enough such that it is worthwhile to raise them. The alert that is raised states that “this is an outlier” and not “this is an error.” It is up to the clinician to determine if the outlier is an error that needs attention. There are several reasons why an outlier might not be a medical error. Inadequate training data or a machine-learning model that is not sufficiently sophisticated might result in an appropriate clinical action being labeled as an outlier. Also, in highly unusual or complex situations, there may be little or no precedence for the clinical action that is taken, which would make it an outlier, even though the action may be appropriate. It is also possible that for some clinical actions the usual practice is not the best approach; in that case, the best clinical action could be flagged as an outlier. It is an empirical question whether our conjecture above is valid in particular clinical environments. The results of this study provide support for it, although additional study is still needed.
Knowledge-based alerting and outlier-based alerting could be used together. The knowledge-based alerts might encode relatively rare or complex clinical situations that would be difficult for an outlier-based system to learn. The outlier-based system could provide broad coverage of many clinical situations for which it would be impractical to write all the rules manually. With such a dual system, it is possible that they may sometimes contradict each other (or at least appear to do so). For example, suppose a clinician ordered medication med1 for a given patient. The knowledge-base system might raise an alert indicating that medication med2 is more appropriate. The clinician then switches the order to med2, but receives an alert from the outlier-based system saying that med2 is unusal in the patient’s current clinical context. This sequence of alerts is not neecssarily contradictory, because both alerts might be valid for what they each represent. Nevertheless, in such a situation, the system could provide the clinician with both alerts and indicate the need for the clinician to resolve them.
6.3 Future work
The analysis of incorrect alerts revealed the limitations of our current method and suggests possible avenues for model improvements. First, while the models were able to capture the main patterns of care, they made mistakes in cases where a special condition made the order inappropriate or when one of many alternative treatments was already given to treat the problem instead. The reasons for not capturing these special conditions were either because the data recording them were not used in the model (e.g., we did not use allergy data) or the relations in between these special conditions and the orders were not represented abundantly enough in the training data to capture these associations. One way to improve the methodology is to automatically learn when actions do and do not tend to co-occur. Such a capability will generalize the current system, which only alerts on one action at a time, to a system that considers joint actions in raising alerts.
Our current plans are to develop and evaluate a real-time version of an outlier-based alerting system. This system will use an archive of ICU EMR data for training that is much larger than used in the study reported here. We plan to investigate several improvements in model training, including increasing the temporal granularity of alerting and modeling medication dosages. As the data capture infrastructure is constantly updated, models can be recomputed and we expect the set of active models to evolve in time, as medical practive evolves. The system will monitor current ICU patients in (almost) real time and raise outlier-based alerts. The alerts will be received and assessed by Critical Care physicians who are off service for a month. These physicians will be able to access the real-time EMR system to inform their evaluation of the alerts. If this evaluation obtains positive results, as we anticipate, then we plan to carry out a clinical trial to examine whether an outlier-based alerting system impacts outcomes such as length of stay and patient morbidity and mortality.
Paper highlights.
A framework for detecting unusual patient-management decisions from EHR data
An unusual (or outlier) patient management action may correspond to a medical error
Evaluates the outlier based alerting approach using expert reviews on EHR data for ICU patients
The overall true positive rates for the alerts (TPARs) ranged from 0.44 to 0.71
A promising new approach to data-driven clinical alerting and medical error detection
Acknowledgments
This research was supported by grants R01-GM088224 from the NIH. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Appendices
Appendix A
Clinical information (and related features) used for constructing predictive models from EMR data. The features were extracted from time series data in EMRs.
| Clinical variable type | features | |
|---|---|---|
| Basic patient info | age sex height result indicator height |
weight result indicator weight BMI indicator BMI |
| ICU admission info | Admission diagnosis code Time since admission Time since diagnosis change Apache3 score |
Admission categories (19 categories such as trauma, infectious, surgical, etc) |
| Lab test t | last value measurement for lab t time elapsed since last measurement for t pending result for lab t known value indicator for t known trend indicator for t 2nd last value measurement for t difference for last two measurements for t slope for last two measurements of t % drop for last two measurements of t % drop for last two measurements per day nadir value for lab t difference for last and nadir values for t slope for last and nadir values for t % drop for last and nadir values of t |
% drop for last and nadir values per day apex value for t difference for last and apex values for t slope for last and apex values of t % drop for last and apex values of t % drop for last and apex values per day baseline (1st) value for lab t difference for last and baseline values for t slope for last and first values for t % drop for last and first values of t % drop for last and first values per day overall slope for lab t difference for last and mean value of t time difference between two last lab values |
| Physiological parameter s | last value of parameter s time elapsed since last reading of s s value known indicator average value of s over last 2 hours average value of s over last 4 hours average value of s over last 6 hours average value of s over last 24 hours difference of average over last 2 hours from the mean value of s difference of average over last 4 hours from the mean value of s difference of average over last 6 hours from the mean value of s |
difference of average over last 24 hours from the mean value of s difference of last value from average over last 2 hours difference of last value from average over last 4 hours difference of last value from average over last 6 hours difference of last value from average over last 24 hours nadir value of s over last 24 hours apex value of s over last 24 hours difference of last value from 24 hour nadir difference of 24hour apex from last value difference of 24hour apex and 24hour nadir |
| I/O volumes | Known I/O volumes indicator Total intake over last 24 hours Total intake via IV over 24 hours Total intake oral over last 24 hours Total intake other over last 24 hours |
Total output over last 24 hours Total output urine over last 24 hours Total output other over last 24 hours I/O balance over last 24 hours |
| Medication m | Patient on medication m Time elapsed since last administration of medication m Time elapsed since first administration of medication m Time elapsed since last change in medication administration m |
|
| Procedure p | Patient had procedure p in past 24 hours Patient had procedure p during the stay Time elapsed since last procedure p Time elapsed since first procedure p |
|
Appendix B
Area under the ROC curves.
ranges for all models built from data for predicting lab-order and medication-order omissions.
| AUROC range | # of lab-order omission models | # of medication-order omission models | total # of models |
|---|---|---|---|
| (0.95, 1.00] | 24 | 15 | 39 |
| (0.90, 0.95] | 18 | 79 | 97 |
| (0.85, 0.9] | 54 | 151 | 205 |
| (0.80, 0.85] | 44 | 187 | 231 |
| (0.75, 0.80] | 14 | 154 | 168 |
| (0.70, 0.75] | 23 | 58 | 81 |
| (0.65, 0.70] | 0 | 28 | 28 |
| (0.60, 0.65] | 0 | 18 | 18 |
| (0.55, 0.60] | 0 | 14 | 14 |
| (0.50, 0.55] | 0 | 6 | 6 |
| 0.50 | 45 | 365 | 410 |
| all | 222 | 1075 | 1297 |
avgPPVtop10 statistics.
The quality of predictive models built from data in terms of avgPPVtop10 statistics and their distribution. Only models that passed the AUROC > 0.70 threshold are included. Higher avgPPVtop10 values reflect models that are stronger for predicting outliers and alerts. The N/A entry records the number of models for which less than 10 patient instances had action a =1.
| avgPPVtop10 range | # of lab-order omission models | # of medication-order omission models | total # of models |
|---|---|---|---|
|
| |||
| (0.90, 1.00] | 77 | 62 | 139 |
| (0.80, 0.90] | 12 | 22 | 34 |
| (0.70, 0.80] | 4 | 24 | 28 |
| (0.60, 0.70] | 4 | 18 | 22 |
| (0.50, 0.60] | 2 | 30 | 32 |
| (0.40, 0.50] | 5 | 28 | 33 |
| (0.30, 0.40] | 4 | 17 | 21 |
| (0.20, 0.30] | 6 | 33 | 39 |
| (0.10, 0.20] | 17 | 72 | 89 |
| [0, 0.10] | 38 | 304 | 342 |
| N/A | 8 | 80 | 88 |
|
| |||
| all | 177 | 690 | 867 |
Appendix C
Medication order alerts.
64 medication order alerts analyzed during the evaluation study. C prefix denotes classes of medications.
| fentanyl | lansoprazole | C_SkeletalMuscleRelaxants |
| norepinephrine | haloperidol | C_HematopoieticAgents |
| metronidazole | Bactrim | C_LoopDiuretics |
| metoprolol | amlodipine | C_DirectVasodilators |
| albuterol | multivitamin | C_MiscellaneousAntibacterials |
| dobutamine | casanthranol | C_Anticonvulsants, Miscellaneous |
| furosemide | simvastatin | C_AntiarrhythmicAgents |
| potassium | lisinopril | C_NitratesandNitrites |
| famotidine | epoetinalfa | C_NonsteroidalAnti-inflammatoryAgents |
| morphine | bisacodyl | |
| nizatidine | sodiumphosphate | C_IronPreparations |
| nitroprusside | dextrose | C_Dihydropyridines |
| magnesium | cytomegalovirusimmuneglobulin | C_Antidepressants |
| diltiazem | C_AntiulcerAgents, AcidSuppressants | C_Antipsychotics |
| pantoprazole | C_Anticonvulsants | C_Platelet-aggregationInhibitors |
| acyclovir | C_Anxiolytics, Sedatives, Hypnotics | C_5-HT3ReceptorAntagonists |
| iron | C_Diuretics | C_Biguanides |
| aspirin | C_ImmunosuppressiveAgents | C_AntiheparinAgents |
| calcium | C_Mydriatics | C_SelectiveBeta1AdrenergicAgonists |
| percocet | C_Antivirals | C_ClassIIIAntiarrhythmics |
| sodiumbicarbonate | C_ThyroidandAntithyroidAgents | C_FourthGenerationCephalosporins |
| vasopressin | C_CoumarinDerivatives |
Lab order alerts.
22 lab order alerts analyzed during the evaluation study.
| Base Deficit | PTT | CPK, Total |
| HCO3 | RDW | CPK-MB |
| ABS Lymphs | WBC | Creatinine & GFR |
| Basophils | ALT_B_SGPT | Ionized Ca |
| INR | AST_B_SGOT | LDH |
| Lymphs | Bili, Delta | FM_Monocytes (Fluid) |
| MPV PT |
Bili, Total | Bilirubin (Urine) |
Appendix D
The number of shared alerts (and their subtypes) that were evaluated by the reviewers for the different αmin alert parameter ranges. The total number of alerts is 240.
| alert parameter (αmin) | Number of lab alerts | Number of med alerts | Number of all alerts |
|---|---|---|---|
|
| |||
| (0, 0.001] | 12 | 12 | 24 |
| (0.001, 0.0025] | 10 | 18 | 28 |
| (0.0025, 0.005] | 9 | 15 | 24 |
| (0.005, 0.01] | 9 | 22 | 31 |
| (0.01, 0.015] | 5 | 23 | 28 |
| (0.015, 0.025] | 6 | 17 | 23 |
| (0.025, 0.04] | 10 | 18 | 28 |
| (0.04, 0.06] | 8 | 14 | 22 |
| (0.06, 0.08] | 5 | 9 | 14 |
| (0.08, 0.1] | 1 | 14 | 15 |
| > 0.1 | 0 | 3 | 3 |
Appendix E
Pairwise Cohen’s kappa statistics for the reviewers in the six groups.
Briefly, with three reviewers in each group we can calculate three different kappa scores. The scores in the table are ordered with the minimum and maximum kappa scores per group shown on the left and righ respectively.
| Group id | minimum | maximum | |
|---|---|---|---|
|
| |||
| 1 | −0.0417 | 0.1071 | 0.3204 |
| 2 | 0.3407 | 0.3407 | 0.5604 |
| 3 | 0.0500 | 0.1688 | 0.2500 |
| 4 | 0.2947 | 0.4074 | 0.4217 |
| 5 | −0.2276 | −0.2178 | 0.0074 |
| 6 | 0.2147 | 0.2947 | 0.3401 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohn LT, Corrigan JM, Donaldson MS. To err is human: Building a safer health system. 2000. [PubMed] [Google Scholar]
- 2.Levinson DR. Adverse events in hospitals: National incidence among Medicare beneficiaries. 2010. [Google Scholar]
- 3.James J. A new, evidence-based estimate of patient harms associated with hospital care. Journal of Patient Safety. 2013;9(3):122–128. doi: 10.1097/PTS.0b013e3182948a69. [DOI] [PubMed] [Google Scholar]
- 4.Leape LL, Berwick DM. Five years after To Err Is Human. Journal of the American Medical Association. 2005;293:2384–2390. doi: 10.1001/jama.293.19.2384. [DOI] [PubMed] [Google Scholar]
- 5.Bates DW, et al. The impact of computerized physician order entry on medication error prevention. Journal of the American Medical Informatics Association. 1999;6:313–321. doi: 10.1136/jamia.1999.00660313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landrigan CP, et al. Temporal trends in rates of patient harm resulting from medical care. New England Journal of Medicine. 2010;363:2124–2134. doi: 10.1056/NEJMsa1004404. [DOI] [PubMed] [Google Scholar]
- 7.Vincent C, Aylin P, Franklin BD. Is health care getting safer? British Medical Journal. 2008;337:2426. doi: 10.1136/bmj.a2426. [DOI] [PubMed] [Google Scholar]
- 8.Hauskrecht M, et al. Outlier detection for patient monitoring and alerting. Journal of biomedical informatics. 2013;46(1):47–55. doi: 10.1016/j.jbi.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauskrecht M, et al. Conditional outlier detection for clinical alerting. AMIA Annu Symp Proc. 2010;2010:286–90. [PMC free article] [PubMed] [Google Scholar]
- 10.Valko M, et al. Conditional anomaly detection with soft harmonic functions. Data Mining (ICDM), 2011 IEEE 11th International Conference on; IEEE; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubins HB, Moskowitz MA. Complications of care in a medical intensive care unit. Journal of General Internal Medicine. 1990;5:104–109. doi: 10.1007/BF02600508. [DOI] [PubMed] [Google Scholar]
- 12.Donchin Y, et al. A look into the nature and causes of human errors in the intensive care unit. Critical Care Medicine. 1995;23:294–300. doi: 10.1097/00003246-199502000-00015. [DOI] [PubMed] [Google Scholar]
- 13.Ferraris VA, Propp ME. Outcome in critical care patients: A multivariate study. Critical Care Medicine. 1992;20:967–976. doi: 10.1097/00003246-199207000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Giraud T, et al. Iatrogenic complications in adult intensive care units: A prospective two-center study. Critical Care Medicine. 1993;21:40–51. doi: 10.1097/00003246-199301000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Rothschild JM, et al. The critical care safety study: The incidence and nature of adverse events and serious medical errors in intensive care. Critical Care Medicine. 2005;33:1694–1700. doi: 10.1097/01.ccm.0000171609.91035.bd. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed A, et al. Outcome of adverse events and medical errors in the intensive care unit: A systematic review and meta-analysis. Journal of American Medical Quality. 2015;30(1):23–30. doi: 10.1177/1062860613514770. [DOI] [PubMed] [Google Scholar]
- 17.Classen DC, et al. Computerized surveillance of adverse drug events in hospital patients. Journal of the American Medical Association. 1991;266(20):2847–51. [PubMed] [Google Scholar]
- 18.Bellazzi R, Larizza C, Riva A. Temporal Abstractions for Interpreting Diabetic Patients Monitoring Data. Intelligent Data Analysis. 1998;2(1):97–122. [Google Scholar]
- 19.Haimowitz IJ, I, Kohane S. Managing temporal worlds for medical trend diagnosis. Artificial Intelligence in Medicine. 1996;8(3):299–321. doi: 10.1016/0933-3657(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 20.Haimowitz IJ, Le PP, Kohane IS. Clinical monitoring using regression-based trend templates. Artificial Intelligence in Medicine. 1995;7(6):473–96. doi: 10.1016/0933-3657(95)00023-6. [DOI] [PubMed] [Google Scholar]
- 21.Cerner. Cerner’s Discern Expert. 2011 Available from: http://www.cerner.com/public/MillenniumSolution.asp?id=3579/
- 22.Evans RS, et al. A computer-assisted management program for antibiotics and other anti-infective agents. New England Journal of Medicine. 1998;338:232–238. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 23.Misys. Misys Insight Alert Systems. 2011 Available from: http://www.misyshealthcare.com/Products/Product+Portfolio/misys+insight/
- 24.Jha AK, et al. Identifying adverse drug events: development of a computer-based monitor and comparison with chart review and stimulated voluntary report. Journal of the American Medical Informatics Association. 1998;5(3):305–14. doi: 10.1136/jamia.1998.0050305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozich JD, Haraden CR, Resar RK. Adverse drug event trigger tool: a practical methodology for measuring medication related harm. Quality & Safety in Health Care. 2003;12(3):194–200. doi: 10.1136/qhc.12.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadhwa R, et al. Analysis of a failed clinical decision support system for management of congestive heart failure. Proceedings of the Symposium of the American Medical Informatics Association. 2008:773–777. [PMC free article] [PubMed] [Google Scholar]
- 27.Schedlbauer A, et al. What evidence supports the use of computerized alerts and prompts to improve clinicians’ prescribing behavior? Journal of the American Medical Informatics Association. 2009;16:531–538. doi: 10.1197/jamia.M2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh TC, et al. Characteristics and consequences of drug allergy alert overrides in a computerized physician order entry system. Journal of the American Medical Informatics Association. 2004;11(6):482–491. doi: 10.1197/jamia.M1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weingart SN, et al. Physicians’ decisions to override computerized drug alerts in primary care. Arch Intern Med. 2003;163:2625–2631. doi: 10.1001/archinte.163.21.2625. [DOI] [PubMed] [Google Scholar]
- 30.Bitan Y, et al. Nurses; reactions to alarms in a neonatal intensive care unit. Cognitive Technology Work. 2004;6(4):239–246. [Google Scholar]
- 31.Hauskrecht M, et al. Evidence-based anomaly detection in clinical domains. AMIA Annu Symp Proc. 2007:319–23. [PMC free article] [PubMed] [Google Scholar]
- 32.Valko M, Hauskrecht M. Feature importance analysis for patient management decisions. Stud Health Technol Inform. 2010;160(Pt 2):861–5. [PMC free article] [PubMed] [Google Scholar]
- 33.Vapnik V. The Nature of Statistical Learning Theory. New York: Springer-Verlag; 1995. [Google Scholar]
- 34.Fan R-E, et al. LIBLINEAR: A Library for Large Linear Classification. J Mach Learn Res. 2008;9:1871–1874. [Google Scholar]
- 35.DeGroot M, Fienberg S. The comparison and evaluation of forecasters. The Statistician. 1983;32:12–22. [Google Scholar]
- 36.Baker DE. Medication alert fatigue: The potential for compromised patient safety. Hospital Pharmacy. 2009;44:460–462. [Google Scholar]
- 37.van der Sijs H, et al. Overriding of drug safety alerts in computerized physician order entry. Journal of the American Medical Informatics Association. 2006;13:138–147. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah NR, et al. Improving acceptance of computerized prescribing alerts in ambulatory care. Journal of the American Medical Informatics Association. 2006;13:5–11. doi: 10.1197/jamia.M1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidling HM, et al. Factors influencing alert acceptance: a novel approach for predicting the success of clinical decision support. J Am Med Inform Assoc. 2011;18(4):479–84. doi: 10.1136/amiajnl-2010-000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham KC, Cvach M. Monitor alarm fatigue: standardizing use of physiological monitors and decreasing nuisance alarms. Am J Crit Care. 2010;19(1):28–34. doi: 10.4037/ajcc2010651. [DOI] [PubMed] [Google Scholar]


