Abstract
Aim
The effect of interleukin 33 (IL-33) in the inflammatory process generates significant interest in the potential significance of IL-33 as a biomarker for coronary artery disease (CAD). Here, our objective was to analyze whether IL-33 gene polymorphisms are associated with premature CAD in a case-control association study.
Methods
Four IL-33 polymorphisms (rs7848215, rs16924144, rs16924159 and rs7044343) were genotyped by 5’ exonuclease TaqMan assays in 1095 patients with premature CAD and 1118 controls.
Results
The rs7044343 T allele was significantly associated with a diminished risk of premature CAD (OR = 0.81, 95% CI: 0.69–0.97, Pdom = 0.020; OR = 0.85, 95% CI: 0.75–0.96, Padd = 0.019) and central obesity (OR = 0.74, 95% CI: 0.58–0.93, Pdom = 0.0007), respectively. When patients were divided into groups with and without type 2 diabetes mellitus (T2DM), the rs7044343 T allele was associated with a reduced risk of premature CAD in patients without (OR = 0.85, 95% CI: 0.73–0.99, Padd = 0.038) and with T2DM (OR = 0.61, 95% CI: 0.38–0.97, Pdom = 0.039; OR = 0.69, 95% CI: 0.49–0.97, Padd = 0.035). In order to establish the functional effect of the rs7044343 polymorphism, the production of IL-33 was determined in monocytes of selected individuals. Monocytes from individuals with rs7044343 CC genotype produced higher levels of IL-33 than monocytes from individuals with other genotypes.
Conclusion
The results suggest that the IL-33 rs7044343 T allele could be a susceptibility marker for premature CAD and central obesity. The rs7044343 polymorphism could be involved in regulating the production of IL-33.
Introduction
Coronary artery disease (CAD) is a complex multifactorial disorder. This polygenic disease is caused by an inordinate inflammatory response to different forms of injuries to the arterial wall endothelium [1–3]. Admittedly, inflammation is as leading cause of atherogenesis since it disturbs lipoprotein metabolism and arterial wall biology. Infiltrates of T cells and activated macrophages are salient in atherosclerotic lesions of both humans and murines [4, 5]. The majority of T cells present in human atherosclerotic plaques belong to the CD4+ subset and produce predominantly cytokines of the Th1 subtype that have a critical pathogenic role in murine atherosclerosis models [6–10]. In contrast, it has been reported that the Th2 cells have an atheroprotective effect [11, 12].
IL-33 is a cytokine member of the IL-1 family, which includes IL-1 and IL-18 [13]. Unlike IL-1 and IL-18, which mainly promote Th1-associated responses, IL-33 predominantly induces the production of Th2 cytokines (IL-5 and IL-13) [14]. Miller et al. showed that IL-33 administration to ApoE-/- mice induced Th2 cytokines and protective ox-LDL antibodies, which significantly reduced atherosclerotic plaque development in the aortic sinus [15]. These data suggest that the gene that encodes IL-33 could be an important candidate gene for study in atherosclerosis. Recently, Tu et al. studied three IL-33 Tag SNPs (rs7025417, rs10975514, and rs10975519) in patients with CAD from the Chinese Han population [16]. In this study, the rs7025417 polymorphism was associated with CAD, with altered regulation of IL-33 gene expression and with high plasma IL-33 levels. Results of association studies may vary between populations due to genetic differences amongst them, including differences in allele frequencies and linkage disequilibrium (LD) structures. Therefore, it is important to examine multiple ethnic populations for the identification of ethnicity-specific loci as well as common susceptibility loci. The objective of our study was to evaluate whether IL-33 gene polymorphisms are associated with premature CAD in the Genetics of Atherosclerotic Disease (GEA) case-control association study. Also, the aim was to establish the possible effect of the associated polymorphism in the production of IL-33 in monocytes of individuals with different genotypes. After a functional prediction analysis, we selected four IL-33 gene polymorphisms (rs7848215, rs16924144, rs16924159, and rs7044343) with possible functional consequences and with minor allele frequency > 5% to be analyzed in the present study. The functional analysis showed that the rs7848215 produces a DNA binding site for the PBX1 transcription factor, the rs16924144 for SF/ASF, the rs16924159 for the SRp40 protein and the rs7044343 polymorphism produces binding site for the transcription factors SC35 and SF/ASF.
Material and Methods
Subjects
Every participant signed a written informed consent document. This protocol complies with the Declaration of Helsinki and was approved by the Ethics Committee of the Instituto Nacional de Cardiología Ignacio Chávez (INCICH). The GEA study focuses on the Mexican population and its main objective is to establish genetic factors linked with premature CAD and other coronary risk factors. All GEA study subjects are not blood related and are Mexican mestizos, who are defined as people born in Mexico, with an ancestry comprised of both indigenous inhabitants and individuals of African and/or Caucasian origin (mainly Spaniards), who had migrated to the Americas from the sixteenth century onward. A total of 2213 individuals were recruited, 1095 diagnosed with premature CAD and 1118 apparently healthy controls. History of myocardial infarction, angioplasty, revascularization surgery or coronary stenosis >50% on angiography (diagnosed before age 55 in men and before age 65 in women) was used to characterize premature CAD. Controls were seemingly healthy asymptomatic subjects without premature CAD family history, recruited from blood banks and Social Services centers. Congestive heart failure, liver, renal, thyroid or oncological disease were the exclusion criteria for controls. In an earlier report, we documented the selection of patients and controls of the GEA study [17]. Demographic, clinical, anthropometric, biochemical parameters and cardiovascular risk factors were assessed in all subjects. Qualified staff measured waist circumference, body mass index (BMI, kg/m2) and other anthropometric parameters. A sphygmomanometer was used to determine blood pressure (the average of the last two of three assessments). Patients with BMI ≥30 kg/m2 were classified as obese. Adult Treatment Panel III (ATP-III) criteria 2002 (Third report of the National Cholesterol Education Program) definitions were followed for central obesity, hypoalphalipoproteinemia, hypertriglyceridemia, and metabolic syndrome [18]. Total cholesterol (TC) levels ≥200 mg/dL defined hypercholesterolemia. Patients with systolic blood pressure ≥140 mmHg and/or diastolic blood pressure ≥90 mmHg, or the use of oral antihypertensive therapy were labeled as hypertense. And finally, we followed World Health Organization criteria to diagnose type 2 Diabetes mellitus (T2DM).
Computed tomography of the chest and abdomen
Experienced radiologists interpreted the computed tomography of the chest and abdomen, performed using a 64-channel multi-detector helical computed tomography system (Somatom Sensation, Siemens). Coronary artery calcification (CAC) score was calculated using the Agatston method [19]. Total abdominal, subcutaneous and visceral adipose tissue areas (as described by Kvist et al.) were measured to assess the visceral to subcutaneous adipose tissue ratio (VAT/SAT) [20]. The hepatic to splenic attenuation ratio (LSAR) was estimated as described by Longo et al. [21]. CAC, VAT/SAT and LSAR were quantified using tomography scans. All patients and 1523 healthy controls underwent tomography. 405 controls were not considered for analysis, since their CAC score was positive, and they were thus considered as individuals with subclinical atherosclerosis (SA). The final control group included individuals (n = 1118) with only negative CAC scores.
Genetic analysis
We isolated genomic DNA from whole blood containing EDTA using standard techniques. The rs7848215, rs16924144, rs16924159, and rs7044343 IL-33 single nucleotide polymorphisms (SNPs) were genotyped using 5’ exonuclease TaqMan assays on an ABI Prism 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Genotyping call rate surpassed 95% for all SNPs tested, with no discordant genotypes in 10% of duplicate samples. We adhered to the manufacturer’s instructions to perform the assays.
Because the Mexican-Mestizo population is admixed, in order to assess the possible influence of population stratification, a panel of 265 ancestry informative markers (AIMs) distinguishing mainly Amerindian, European and African ancestry were selected [22] and genotyped on Illumina BeadStation using the GoldenGate assay. Duplicate control samples were genotyped on each chip, which also served as internal controls for quality of clustering and reproducibility. The primary analysis of the genotyping data with the Illumina Genome Studio software v.2011.1 was followed by visual inspection and assessment of data quality and clustering. Genotyping accuracy was also assessed by genotype clustering using the Illumina GeneTrain score, which is a measure of the clustering confidence of individual SNP alleles. Global Caucasian, Amerindian and African ancestry were determined in each individual using the ADMIXTURE software.
Functional prediction analysis
The effect of the IL-33 SNPs was predicted using the following bioinformatics software: FastSNP [23], SNP Function Prediction (http://snpinfo.niehs.nih.gov/snpfunc.htm), Human-transcriptome Database for Alternative Splicing (http://www.h-invitational.jp/h-dbas/), Splice Port: An Interactive Splice Site Analysis Tool (http://www.spliceport.cs.umd.edu/SplicingAnalyser2.html), ESE finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi), HSF (http://www.umd.be/HSF/), and SNPs3D (http://www.snps3d.org/).
Monocyte isolation
A sample of venous blood (40 mL) was obtained from 61 healthy controls selected according to the rs7044343 polymorphism (21 with CC, 21 with TC and 21 with TT genotypes). The peripheral blood mononuclear cell (PBMC) population was isolated by gradient centrifugation on Lymphoprep (Axis-Shield PoC AS, Oslo, Norway). Monocytes were isolated by positive selection with CD14-mAb-coated micro beads (Miltenyi Biotec, Bergisch Gladbach; Germany) following the manufacturer’s instructions (purity of 95–98%).
Monocyte cultures and IL-33 detection
Monocytes were counted in a Neubauer hemocytometer (Propper MFG Company, NY USA) chamber using 0.4% Trypan blue stain (Cambrex Bio Science, MD USA) to exclude dead cells. Monocyte density in culture was adjusted to 1 x106 per milliliter. Monocytes were cultured in RPMI-1640 medium (Sigma, Poole, UK), supplemented with 10% (volume/volume) heat-inactivated fetal bovine serum (Sigma), 0.1 mM L-glutamine, 100 U/ml penicillin and 100 U/ml streptomycin. Cells were stimulated with 100 ng/ml E coli lipopolysaccharide (from strain 0111:B4, Sigma) and 100 ng/ml of P gingivalis lipopolysaccharide (Invivogen, Calne, UK) for 6 hours in a humidified atmosphere with 5% CO2 at 37°C. Necrosis was induced by subjecting stimulated cells to five cycles of freezing to -70°C and thawing at 38°C. Necrotic cell preparations were centrifuged at 10,000 for 5 min and supernatants were kept at -70°C. The IL-33 levels were detected using specific ELISA kit for IL-33 (Biolegend, San Diego, CA); the sensitivity for the ELISA was 4.14 pg/ml.
Statistical analysis
The SPSS version 18.0 statistical package (SPSS, Chicago, Il) was employed for the statistical estimation of means ± SD and frequencies of baseline characteristics. We compared frequencies using Chi-square tests, and means using the ANOVA and Students t-test. To determine the association between the polymorphisms and metabolic variables, we used ANCOVA and adjusted for age, gender, BMI, smoking history and alcohol consumption. The correlation of polymorphisms with premature CAD under dominant, recessive and additive inheritance models was analyzed with logistic regression analysis. Also, we utilized age, gender, BMI, smoking history, alcohol consumption and ancestry to adapt the models. They were constructed including one variable at time, and final models included variables with biological relevance or with statistical significance or both. Confounding bias was accepted when changes in estimated odds ratios (ORs) were equal or greater than 10%. When a principal effect model was reached, effect modification was also tested and interactions terms were constructed between the polymorphisms and different variables; the terms were included in the model when the significance of the p-value was greater or equal to 0.20. Hosmer–Lemeshow Goodness of Fit test was performed for each multiple logistic model. Bonferroni correction was used as appropriate. Statistical power to detect association with CAD was 0.80 as estimated with QUANTO software [http://hydra.usc.edu/GxE/]. The obtained genotype frequencies did not deviate from Hardy-Weinberg equilibrium (HWE, P > 0.05). Haploview version 4:1 (Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, USA) was used to calculate pairwise linkage disequilibrium (LD, D´) between polymorphisms and haplotype reconstruction.
Results
Tables 1 and 2 illustrate the general characteristics of the study population. Global ancestry was similar in patients and controls, showing 55.8% and 54.0% of Native American ancestry respectively; and 34.3% and 35.8% of Caucasian ancestry, respectively.
Table 1. Demographic characteristics of the population.
| Premature CAD (n = 1095) | Controls (n = 1118) | P value | ||||||
|---|---|---|---|---|---|---|---|---|
| P25 | Median | P75 | P25 | Median | P75 | |||
| Age (years) | 49 | 54 | 59 | 45.0 | 51.0 | 57.0 | <0.0001 | |
| Body Mass Index (Kg/m2) | 26.0 | 28.3 | 31.3 | 25.4 | 27.9 | 31.0 | 0.006 | |
| Waist circumference (cm) | 91.2 | 97.5 | 105.4 | 86.0 | 94.0 | 101.5 | <0.0001 | |
| Total Abdominal Fat (cm2) | 340 | 426 | 530.25 | 346 | 443 | 545 | 0.186 | |
| Subcutaneous Abdominal Fat (cm2) | 193 | 245.5 | 316 | 219.3 | 288.5 | 373.0 | <0.0001 | |
| Visceral Abdominal Fat (cm2) | 130 | 171 | 218.5 | 104 | 142 | 183 | <0.0001 | |
| Visceral/Subcutaneous adipose tissue ratio | 0.95 | 1.29 | 1.81 | 0.53 | 1.26 | 2.29 | 0.031 | |
| Blood Pressure (mmHg) | Systolic | 107 | 116.3 | 128 | 105.3 | 114.6 | 125.0 | 0.002 |
| Diastolic | 66 | 71.7 | 78.5 | 66.0 | 71.3 | 77.3 | 0.235 | |
| Heart Rate (bpm) | 57.5 | 64.3 | 72.7 | 59.5 | 65.3 | 71.0 | 0.158 | |
| Gender n (%) | Male | 905 (82.6) | 456 (40.8) | |||||
| Female | 192 (17.4) | 662 (59.2) | <0.0001 | |||||
| Weight n (%) | Normal weight | 190 (17.4) | 259 (23.2) | |||||
| Overweight | 515 (47) | 508 (45.4) | 0.005 | |||||
| Obesity | 390 (35.6) | 351 (31.4) | <0.000 | |||||
| Central obesity n (%) | 866 (79.2) | 868 (77.7) | 0.263 | |||||
| Tobacco smoking n (%) | Current | 135 (12.3) | 250 (22.4) | <0.0001 | ||||
| Former | 702 (65.4) | 346 (35.2) | <0.0001 | |||||
| Use of alcohol n (%) | 610 (55.7) | 823 (73.8) | <0.0001 | |||||
| Hypertension n (%) | 740 (67.6) | 302 (27.0) | <0.0001 | |||||
| Hypertensive Medication n (%) | 737 (71.3) | 170 (15.2) | <0.0001 | |||||
Data are expressed as median and percentiles 25 and 75.
*P values were estimated using Mann-Whitney U-test continuous variables and Chi-square or Fisher test for categorical values.
Table 2. Comparison of biochemical parameters in individuals with premature CAD and healthy controls.
| CAD premature | Controls | P value | |||||
|---|---|---|---|---|---|---|---|
| P25 | Median | P75 | P25 | Median | P75 | ||
| Total cholesterol (mg/dl) | 132.50 | 160.70 | 193.60 | 168.08 | 190.05 | 210.00 | <0.0001 |
| HDL-C (mg/dl) | 32.50 | 38.30 | 45.05 | 36.93 | 46.00 | 56.00 | <0.0001 |
| LDL-C (mg/dl) | 68.70 | 91.00 | 116.00 | 96.16 | 115.76 | 133.67 | <0.0001 |
| Triglycerides (mg/dl) | 119.00 | 162.80 | 221.60 | 108.10 | 143.20 | 199.50 | <0.0001 |
| ApoA1 (mg/dl) | 63 | 79 | 102 | 73 | 90 | 108 | <0.0001 |
| ApoB (mg/dl) | 102.00 | 120.00 | 136.90 | 114 | 134 | 157 | <0.0001 |
| Glucose (mg/dl) | 87 | 95 | 120 | 84 | 90 | 97 | <0.0001 |
| Insulin | 14.84 | 20.04 | 28.19 | 12.59 | 17.51 | 24.04 | <0.0001 |
| HOMA | 3.53 | 5.15 | 7.86 | 2.70 | 3.94 | 5.68 | <0.0001 |
| Alanine transaminase (IU/L) | 19.0 | 26.0 | 36.0 | 17.0 | 23.0 | 33.0 | 0.002 |
| Aspartate transaminase (IU/L) | 22.0 | 26.0 | 31.0 | 21.0 | 25.0 | 30.0 | 0.002 |
| Alkaline Phosphatase (IU/L) | 64.0 | 77.0 | 95.0 | 68.0 | 81.0 | 97.8 | 0.001 |
| Gamma-glutamyl transpeptidase (IU/L) | 23.0 | 33.0 | 50.0 | 17.0 | 25.0 | 41.0 | <0.0001 |
| TC > 200 mg/dL n (%) | 228 (20.8) | 401 (35.9) | <0.0001 | ||||
| Hypo-a-lipoproteinemia n (%) | 696 (63.6) | 565 (50.6) | <0.0001 | ||||
| Hypertriglyceridemia n (%) | 626 (57.2) | 520 (46.6) | <0.0001 | ||||
| Type 2 Diabetes Mellitus n (%) | 414 (37.8) | 109 (9.7) | <0.0001 | ||||
| Metabolic Syndrome n (%) | 551 (50.3) | 450 (40.3) | <0.0001 | ||||
Data are expressed as median and percentiles 25 and 75.
*P values were estimated using Mann-Whitney U-test continuous variables and Chi-square or Fisher test for categorical values.
Association of polymorphisms with premature CAD
Genotype frequencies in the polymorphic sites were in HWE. In all the evaluated models, the distribution of rs16924144, rs16924159, and rs7848215 polymorphisms was comparable in premature CAD patients and healthy controls. Conversely, the distribution of rs7044343 was not the same in the investigated groups. The rs7044343 T allele was associated with diminished risk of premature CAD when contrasted with to healthy controls (OR = 0.81, 95% CI = 0.69–0.97, Pdom = 0.020; OR = 0.85, 95% CI: 0.75–0.96, Padd = 0.019)) under dominant and additive models adjusted for age, gender, BMI, smoking history, alcohol consumption and ancestry (Table 3).
Table 3. Associations of IL33 polymorphisms with premature CAD.
| Polymorphism | Alleles | MAFa | MAFa | Genotypes | Genotypes | Phwe | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| CAD | Control | Premature CAD | Control | CAD/Control | Pdom value | ||
| rs16924144 | C/T | 0.48 | 0.48 | 281/564/250 | 294/569/255 | 0.33/0.55 | 0.85 (0.62–1.18); 0.34 |
| 0.25/0.52/0.23 | 0.26/0.51/0.23 | 1.00 (0.73–1.38); 0.97b 0.94 (0.77–1.15); 0.57c | |||||
| rs16924159 | A/G | 0.49 | 0.49 | 226/448/224 | 238/471/227 | 0.95/0.90 | 1.07 (0.74–1.55); 0.69 |
| 0.25/0.50/0.25 | 0.25/0.50/0.24 | 1.06 (0.72–1.56); 0.75b 1.05 (0.83–1.32); 0.66c | |||||
| rs7848215 | C/T | 0.14 | 0.14 | 827/238/30 | 832/263/23 | 0.01/0.71 | 0.91 (0.67–1.24); 0.58 |
| 0.75/0.22/0.03 | 0.74/0.24/0.02 | 1.60 (0.66–3.84); 0.29b 0.98 (0.75–1.27); 0.88c | |||||
| rs7044343 | C/T | 0.33 | 0.37 | 481/492/122 | 437/540/141 | 0.84/0.22 | 0.81 (0.69–0.97); 0.020 |
| 0.44/0.45/0.11 | 0.39/0.48/0.13 | 0.84 (0.56–1.27); 0.420 b | |||||
| 0.85 (0.75–0.96); 0.019 c |
Adjusted for age, gender, BMI, smoking history, alcohol consumption and ancestry.
a: MAF, minor allele frequency.
b: recessive model.
c: additive model.
CAD premature: Coronary artery disease premature.
Phwe: p value from Hardy-Weinberg equilibrium tests.
NS: Not significant.
*Underlined letter denotes the minor allele in the control samples.
Significant values are in bold.
The p values were corrected multiplying by 4, number of SNPs tested.
Association of the polymorphisms with cardiovascular risk factors
We considered the association of rs16924144, rs16924159, rs7848215 and rs7044343 polymorphisms with cardiovascular risk factors by comparing CAD patients and healthy controls. Under dominant model adjusted by age, gender, BMI, smoking history and alcohol consumption, the rs7044343 polymorphism was associated with reduced risk of central obesity (OR = 0.74, 95% CI = 0.58–0.93, Pdom = 0.0007) (Table 4).
Table 4. Association of the rs7044343 polymorphism with metabolic risk factors.
| Dominant model | Premature CAD | Control | OR (95% CI) | P value | |
|---|---|---|---|---|---|
| Obesity n (%) | C/C | 162 (0.15) | 124 (0.11) | ||
| C/T+TT | 221 (0.20) | 179 (0.16) | NS | - | |
| Central obesity | C/C | 372 (0.34) | 286 (0.26) | ||
| C/T+TT | 470 (0.43) | 475 (0.42) | 0.74 (0.58–0.93) | 0.0007 | |
| Hypo-α-lipoproteinemia n (%) | C/C | 287 (0.26) | 199 (0.18) | ||
| C/T+TT | 386 (0.35) | 287 (0.26) | NS | - | |
| Hypercholesterolemia n (%) | C/C | 102 (0.09) | 120 (0.11) | ||
| C/T+TT | 121 (0.11) | 225 (0.20) | NS | - | |
| Hypertriglyceridemia n (%) | C/C | 264 (0.24) | 173 (0.15) | ||
| C/T+TT | 346 (0.32) | 271 (0.24) | NS | - | |
| Metabolic syndrome n (%) | C/C | 229 (0.21) | 163 (0.15) | ||
| C/T+TT | 307 (0.28) | 228 (0.20) | NS | - | |
| Type 2 diabetes mellitus n (%) | C/C | 176 (0.16) | 51 (0.05) | ||
| C/T+TT | 226 (0.21) | 49 (0.04) | NS | - |
The model was adjusted by age, gender, BMI, smoking history and alcohol consumption.
NS: Not significant.
OR: Odds ratio.
CI: Confidence intervals.
Significant values are in bold.
The dominant model was analyzed considering 7 metabolic risk factors, so the p values were corrected multiplying by 7.
Association of the polymorphisms with metabolic parameters
We evaluated the effect of rs16924144, rs16924159, rs7848215 and rs7044343 polymorphisms on numerous metabolic parameters separately in controls (CAC score = 0), and premature CAD subjects. None of the studied polymorphisms was associated with metabolic parameters in the groups.
Association of the polymorphisms with premature CAD in patients with and without diabetes mellitus
Considering the high frequency of diabetes mellitus in our group of patients with CAD, we carried out an analysis in patients with and without this pathology in order to establish if the polymorphisms are associated with CAD or with T2DM. The rs7044343 T allele was associated with decreased risk of CAD in patients without T2DM (OR = 0.85, 95% CI = 0.73–0.99, Padd = 0.038) (Table 5) and with T2DM (OR = 0.61, 95% CI = 0.38–0.97, Pdom = 0.039; OR = 0.69, 95% CI = 0.49–0.97, Padd = 0.035) (Table 6). The models were adjusted for age, gender, BMI, smoking history, alcohol consumption and ancestry.
Table 5. Associations of IL33 polymorphisms in patients with premature CAD without diabetes mellitus.
| Polymorphism | Alleles | MAFa | MAFa | Genotypes | Genotypes | Phwe | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| CAD | Control | Premature CAD | Control | CAD/Control | Pdom value | ||
| rs16924144 | C/T | 0.49 | 0.48 | 168/355/159 | 240/460/211 | 0.45/0.38 | 0.86 (0.60–1.25); 0.45 |
| 0.24/0.52/0.23 | 0.26/0.51/0.23 | 0.99 (0.69–1.42); 0.96b 0.94 (0.75–1.18); 0.62c | |||||
| rs16924159 | A/G | 0.50 | 0.50 | 142/296/145 | 192/400/192 | 0.87/0.50 | 1.17 (0.77–1.78); 0.44 |
| 0.24/0.51/0.25 | 0.25/0.51/0.24 | 1.06 (0.68–1.65); 0.77b 1.09 (0.83–1.42); 0.51c | |||||
| rs7848215 | C/T | 0.15 | 0.14 | 500/162/20 | 678/213/20 | 0.09/0.52 | 0.97 (0.68–1.37); 0.88 |
| 0.73/0.24/0.03 | 0.74/0.23/0.02 | 1.30 (0.51–3.35); 0.57b1.01 (0.74–1.35); 0.95c | |||||
| rs7044343 | C/T | 0.34 | 0.38 | 300/306/76 | 343/447/121 | 0.87/0.29 | 1.03 (0.74–1.45); 0.826 |
| 0.44/0.45/0.11 | 0.38/0.49/0.13 | 0.81 (0.50–1.32); 0.404b | |||||
| 0.85 (0.73–0.99); 0.038c |
Adjusted for age, gender, BMI, smoking history, alcohol consumption and ancestry.
a: MAF, minor allele frequency.
b: recessive model.
c: additive model.
CAD premature: Coronary artery disease premature.
Phwe: p value from Hardy-Weinberg equilibrium tests.
NS: Not significant.
*Underlined letter denotes the minor allele in the control samples.
Significant values are in bold.
The p values were corrected multiplying by 4, number of SNPs tested.
Table 6. Associations of IL33 polymorphisms in patients with premature CAD and diabetes mellitus.
| Polymorphism | Alleles | MAFa | MAFa | Genotypes | Genotypes | Phwe | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| CAD | Control | Premature CAD | Control | CAD/Control | Pdom value | ||
| rs16924144 | C/T | 0.47 | 0.49 | 109/207/86 | 167/358/162 | 0.55/0.13 | 0.77 (0.46–1.31); 0.34 |
| 0.27/0.52/0.21 | 0.24/0.52/0.24 | 0.95 (0.56–1.61); 0.84b0.89 (0.64–1.23); 0.48c | |||||
| rs16924159 | A/G | 0.50 | 0.48 | 83/156/85 | 148/315/130 | 0.51/0.18 | 1.14 (0.64–2.04); 0.65 |
| 0.26/0.48/0.26 | 0.25/0.53/0.22 | 1.31 (0.71–2.42); 0.39b 1.16 (0.80–1.67); 0.42c | |||||
| rs7848215 | C/T | 0.12 | 0.15 | 316/75/11 | 500/172/15 | 0.03/1.00 | 0.68 (0.41–1.13);0.145 |
| 0.79/0.19/0.03 | 0.73/0.25/0.02 | 0.96 (0.24–3.72); 0.95b | |||||
| 0.75 (0.49–1.16); 0.202c | |||||||
| rs7044343 | C/T | 0.33 | 0.40 | 176/185/41 | 235/351/101 | 0.66/0.26 | 0.61 (0.38–0.97); 0.039 |
| 0.44/0.46/0.10 | 0.34/0.51/0.15 | 0.65 (0.34–1.27); 0.216b | |||||
| 0.69(0.49–0.97); 0.035c |
Adjusted for age, gender, BMI, smoking history, alcohol consumption and ancestry.
a: MAF, minor allele frequency.
b: recessive model.
c: additive model.
CAD premature: Coronary artery disease premature.
Phwe: p value from Hardy-Weinberg equilibrium tests.
NS: Not significant.
*Underlined letter denotes the minor allele in the control samples.
In this analysis the control group only included individuals without diabetes.
Significant values are in bold.
The p values were corrected multiplying by 4, number of SNPs tested.
Haplotype analysis and SNP functional prediction
Even though the IL-33 polymorphisms were in high linkage disequilibrium (D’>0.8 and r2>0.9), the distribution of the haplotypes in premature CAD patients and healthy controls was comparable (data not included).
Interestingly, SNP functional prediction software results suggest that the rs7044343 polymorphism is functional. The variation in the rs7044343 polymorphism produces a DNA binding site for the transcription factors SC35 and SF/ASF with possible consequences in the expression of the IL-33.
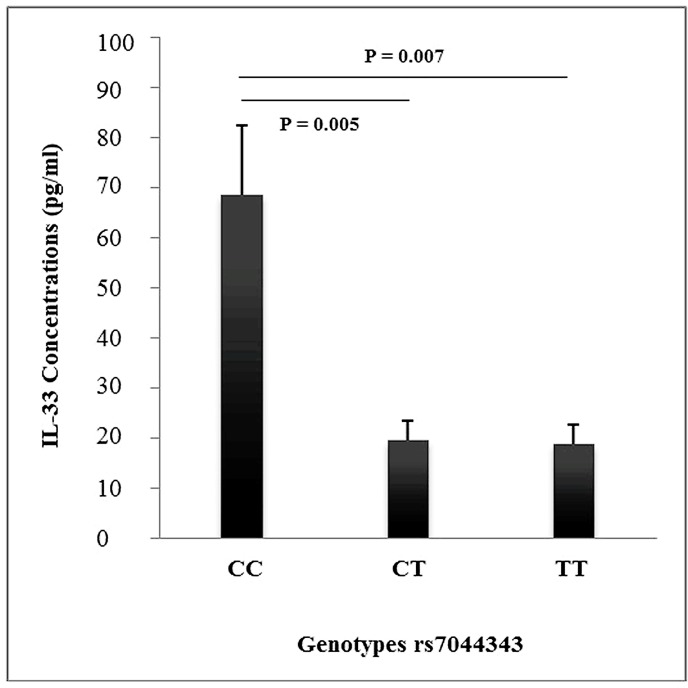
IL-33 levels in monocytes
In order to establish the functional effect of the rs7044343 polymorphism, the production of IL-33 was determined in monocytes of selected individuals (Fig 1). Monocytes from individuals with rs7044343 CC genotype produced higher levels of IL-33 (32.08+24.30) than those from patients with CT (16.32 ± 6.23) (P = 0.005) and TT (17.10 ± 5.48) (P = 0.007) genotypes.
Fig 1. Detection of IL-33 in monocytes from healthy individuals with rs7044343 CC, TC and TT genotypes (21 participants were included for each genotype).
Data are presented as mean ± SD. CC vs CT (P = 0.005) and CC vs TT (P = 0.007).
Discussion
IL-33 is a cytokine with an important role in the inflammatory process and in the pathogenesis of atherosclerosis [15]. Animal studies have indicated that IL-33 reduces macrophage foam cell formation [24] and inhibits the development of atherosclerosis in apolipoprotein E-deficient mice [15]. In spite of the important role of IL-33 in the development of atherosclerosis, too few studies have explored the possible role of the gene that encodes this cytokine in the genetic susceptibility to coronary artery disease. Tu et al. reported the association of the IL-33 rs7025417 polymorphism with the risk of developing CAD in a Chinese Han population, thus demonstrating an effect of this polymorphism in the IL-33 gene expression and plasma levels [16]. In another work, four IL-33 polymorphisms (rs1929992, rs10975520, rs11792633 and rs16924159) were studied in Chinese patients with CAD and none of them was associated with the disease [25]. Due to the effect of IL-33 in the inflammatory process, other polymorphisms in this gene have been associated with asthma, inflammatory bowel disease and Alzheimer’s disease [26–28]. In our study, four IL-33 gene polymorphisms (rs7848215, rs16924144, rs16924159, and rs7044343) were analyzed in order to establish their role as a susceptibility marker for premature CAD. We selected these polymorphisms considering their possible functional effect after an informatics analysis and considering that they have a minor allele frequency major to 5%. In our study, the rs7044343 was associated with reduced risk of developing CAD in patients with and without diabetes mellitus. This polymorphism was also associated with reduced risk of developing central obesity. The association of the rs7044343 genotypes in some diseases is contradictory. In the study of Li et al., the rs7044343 CC genotype was associated with decreased risk of developing rheumatoid arthritis (RA) and with low serum IL-33 levels [29]. Contrary to that detected in RA, in systemic sclerosis, the rs7044343 CC genotype was associated with increased risk of developing this disease [30]. Our result of rs7044343 polymorphism contrasts with that reported by Li et al., [29] in RA, however, are in line with that reported by Koca et al., [30] in systemic sclerosis, because in our study, the rs7044343 T allele was significantly associated with a diminished risk of premature CAD. Also, in the study of Li et al., [29] the rs7044343 CC genotype was associated with low serum IL-33 levels in AR patients. This result contrasts with our report, because, we detected a higher production of IL-33 in monocytes of individuals with the CC genotype, compared to those carrying CT and TT genotypes. Some methodological differences between the two studies could explain the apparent contradictory results. In the study by Li et al., [29] the measures were made in serum of RA patients, whereas in our study, the measures were made in monocytes cultures of healthy controls. The functional prediction software used here predicted that this polymorphism is functional. The presence of the C allele in this polymorphism produces a binding site for the transcription factors SC35 and SF/ASF proteins. These proteins belong to the family of SR proteins that regulate alternative splicing [31]. This polymorphism could have functional effects increasing the production of IL-33 isoforms with the consequent increase of the anti-atherogenic effect of this cytokine. In order to establish the functional effect of the rs7044343 polymorphism, the production of IL-33 was determined in monocytes of selected individuals. Monocytes from individuals with rs7044343 CC genotype produced higher levels of IL-33 than monocytes from individuals with other genotypes, suggesting a role of this polymorphism in the production of IL-33. It should be mentioned that monocytes used for this analysis were obtained from healthy individuals (without CAD or CAC). Therefore, the fact that individuals with the CC genotype produce more IL-33 does not necessarily mean the development of CAD. Alternatively, the inflammatory process in CAD includes the participation of several both pro- and anti-inflammatory cytokines. This is a complex phenomenon in which IL-33 may be playing a very important role. The functional analysis on monocytes only was made considering the rs7044343 polymorphism because the main objective of the study was to analyze whether IL-33 gene polymorphisms are associated with premature CAD. The analysis in the whole group of CAD patients and in the group of CAD patients with and without T2DM confirms the association of the rs7044343 polymorphism with CAD.
As for the limitations herein, we only included the study of four polymorphisms of IL-33, which seem to be functional based on the analysis of the prediction software used. In our study, we do not analyze the expression and neither plasma levels of IL-33 in CAD patients and healthy controls. However, we consider that the evaluation of IL-33 production in monocyte cultures of individuals with different genotypes could be a more direct approach of the effect of these genotypes in the production of IL-33. Since this is the first work that documents the correlation of the IL-33 polymorphisms with premature CAD and central obesity, further studies in an independent group of patients are required to validate the results. Indeed, a strength of our work is that the control group only included individuals without subclinical atherosclerosis (individuals without coronary artery calcification).
The IL-33 polymorphisms were in strong linkage disequilibrium in the present work; and still, the haplotypes were not associated with premature CAD. Crawford et al. described that the haplotype architecture of candidate genes across the human genome is convoluted. Also, they mentioned that a considerable the amount of sequence variation has not been documented yet [32]. Consequently, the absence of association of IL-33 haplotypes in our study is not definitive, owing to the incomplete knowledge of both the genetic variation within the IL-33 gene and the structure of linkage disequilibrium in the analyzed region.
Conclusions
In conclusion, the association of the IL-33 rs7044343 polymorphism with both premature CAD and central obesity is established here. This polymorphism had functional effects, based on an in silico prediction analysis. In this study, we demonstrate that the rs7044343 polymorphism has an effect in the production of IL-33 in monocytes stimulated by lipopolysaccharide. Notably, Mexican people form a population with a distinctive genetic background and important differences [33–36]. Thus, owing to these genetic characteristics, the associations of the IL-33 polymorphisms shown here are not definitive and should be tested in other independent populations.
Acknowledgments
This work was submitted in partial fulfillment of the doctoral degree requirements of Javier Angeles-Martínez at the Graduate Studies in Biomedical Sciences of the Universidad Nacional Autónoma de México. The authors are grateful to the study participants.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the Consejo Nacional de Ciencia y Tecnología (projects 156911) and Fundación Gonzalo Rio Arronte, Mexico City, Mexico. Javier Angeles-Martínez was supported by a fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACyT). All of the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ross R (1999) Atherosclerosis an inflammatory disease. N Engl J Med 340:115–126. 10.1056/NEJM199901143400207 [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Moll X (2005) Inflammatory and Anti-Inflammatory Markers in Acute Coronary Syndromes. Ready for Use in the Clinical Setting?. Rev Esp Cardiol 58:615–617. [PubMed] [Google Scholar]
- 3.Lusis AJ (2000) Atherosclerosis. Nature 407:233–241. 10.1038/35025203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westerterp M, Murphy AJ, Wang M, Pagler TA, Vengrenyuk Y, Kappus MS, et al. (2013) Deficiency of ATP-binding cassette transporters A1 and G1 in macrophages increases inflammation and accelerates atherosclerosis in mice. Circ Res 112:1456–1465. 10.1161/CIRCRESAHA.113.301086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinetti-Gbaguidi G, Staels B (2011) Macrophage polarization in metabolic disorders: functions and regulation. Curr Opin Lipidol 22:365–372. 10.1097/MOL.0b013e32834a77b4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer OJ, van der Wal AC, Verhagen CE, Becker AE (1999) Cytokine secretion profiles of cloned T cells from human aortic atherosclerotic plaques. J Pathol 188:174–179. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X, Nicoletti A, Elhage R, Hansson GK (2000) Transfer of CD4(+) T cells aggravates atherosclerosis in immunodeficient apolipoprotein E knockout mice. Circulation 102:2919–2922. [DOI] [PubMed] [Google Scholar]
- 8.Davenport P, Tipping PG (2003) The role of interleukin-4 and interleukin-12 in the progression of atherosclerosis in apolipoprotein E-deficient mice. Am J Pathol 163:1117–1125. 10.1016/S0002-9440(10)63471-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitman SC, Ravisankar P, Daugherty A (2002) Interleukin-18 enhances atherosclerosis in apolipoprotein E(-/-) mice through release of interferon-gamma. Circ Res 90:E34–E38. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Pablo AM, Jiang X, Wang N, Tall AR, Schindler C (1997) IFN-gamma potentiates atherosclerosis in ApoE knock-out mice. J Clin Invest 99:2752–2761. 10.1172/JCI119465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansson GK, Libby P (2006) The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol 6:508–519. 10.1038/nri1882 [DOI] [PubMed] [Google Scholar]
- 12.Buono C, Binder CJ, Stavrakis G, Witztum JL, Glimcher LH, Lichtman AH (2005) T-bet deficiency reduces atherosclerosis and alters plaque antigen-specific immune responses. Proc Natl Acad Sci USA 102:1596–1601. 10.1073/pnas.0409015102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arend WP, Palmer G, Gabay C (2008) IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev 223:20–38. 10.1111/j.1600-065X.2008.00624.x [DOI] [PubMed] [Google Scholar]
- 14.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, et al. (2005) IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor- related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23:479–490. 10.1016/j.immuni.2005.09.015 [DOI] [PubMed] [Google Scholar]
- 15.Miller AM, Xu D, Asquith DL, Denby L, Li Y, Sattar N, et al. (2008) IL-33 reduces the development of atherosclerosis. J Exp Med 205:339–346. 10.1084/jem.20071868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu X, Nie S, Liao Y, Zhang H, Fan Q, Xu C, et al. (2013) The IL-33-ST2L Pathway Is Associated with Coronary Artery Disease in a Chinese Han Population. Am J Hum Genet 93:652–660. 10.1016/j.ajhg.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villarreal-Molina T, Posadas-Romero C, Romero-Hidalgo S, Antúnez-Argüelles E, Bautista-Grande A, Vargas-Alarcón G, et al. (2012) The ABCA1 gene R230C variant is associated with decreased risk of premature coronary artery disease: the genetics of atherosclerotic disease (GEA) study. PLoS One 7:e49285 10.1371/journal.pone.0049285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation 106:3134–3421. [PubMed] [Google Scholar]
- 19.Mautner GC, Mautner SL, Froehlich J, Feuerstein IM, Proschan MA, Roberts WC, et al. (1994) Coronary artery calcification: assessment with electron beam CT and histomorphometric correlation. Radiology 192:619–623. 10.1148/radiology.192.3.8058924 [DOI] [PubMed] [Google Scholar]
- 20.Kvist H, Chowdhury B, Grangard U, Tylén U, Sjöström L (1988) Total and visceral adipose-tissue volumes derived from measurements with computed tomography in adult men and women: predictive equations. Am J Clin Nutr 48:1351–1361. [DOI] [PubMed] [Google Scholar]
- 21.Longo R, Ricci C, Masutti F, Vidimari R, Crocé LS, Bercich L, et al. (1993) Fatty infiltration of the liver. Quantification by 1H localized magnetic resonance spectroscopy and comparison with computed tomography. Invest Radiol 28:297–302. [PubMed] [Google Scholar]
- 22.Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, et al. (2009) Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A 106:8611–8616. 10.1073/pnas.0903045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. (2006) FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34 (Web Server issue):W635–641 [accessed on 2012]. 10.1093/nar/gkl236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren JE, Michael DR, Salter RC, Ashlin TG, Calder CJ, Miller AM, et al. (2010) IL-33 reduces macrophage foam cell formation. J Immunol 185:1222–1229. 10.4049/jimmunol.1000520 [DOI] [PubMed] [Google Scholar]
- 25.Wu F, He M, Wen Q, Zhang W, Yang J, Zhang X, et al. (2014) Associations between variants in IL-33/ST2 signaling pathway genes and coronary heart disease risk. Int J Mol Sci 15:23227–23239. 10.3390/ijms151223227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savenije OE, Mahachie John JM, Granell R, Kerkhof M, Dijk FN, de Jongste JC, et al. (2014) Association of IL-33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with Wheeling phenotype and asthma in childhood. J Allergy Clin Immunol 134:170–177. 10.1016/j.jaci.2013.12.1080 [DOI] [PubMed] [Google Scholar]
- 27.Latiano A, Palmieri O, Pastorelli L, Vecchi M, Pizarro TT, Bossa F, et al. (2013) Association between genetic polymorphisms in IL-33, IL1R1 and risk for inflammatory bowel disease. Plos One 8:e62144 10.1371/journal.pone.0062144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JT, Song JH, Wang ND, Wu ZC, Zhang Q, Zhang N, et al. (2012) Implication of IL-33 gene polymorphism in Chinese patients with Alzheimer´s disease. Neurobiol Aging 33:1014. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Mu R, Guo J, Wu X, Tu X, Liu X, et al. (2014) Genetic variant in IL33 is associated with susceptibility to rheumatoid arthritis. Arthritis Res Ther 16:R105 10.1186/ar4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koca SS, Pehlivan Y, Kara M, Alibaz-Oner F, Oztuzcu S, Yilmaz N, et al. (2016) The IL-33 gene is related to increased susceptibility to systemic sclerosis. Rheumatol Int 36:579–584. 10.1007/s00296-015-3417-8 [DOI] [PubMed] [Google Scholar]
- 31.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J (2001) SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20:1785–2001. 10.1093/emboj/20.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, et al. (2004) Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet 74:610–622. 10.1086/382227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lisker R, Perez-Briceño R, Granados J, Babinsky V, de Rubens J, Armendares S, et al. (1986) Gene frequencies and admixture estimates in a Mexico City population. Am J Phys Anthropol 71:203–207. 10.1002/ajpa.1330710207 [DOI] [PubMed] [Google Scholar]
- 34.Lisker R, Pérez-Briceno R, Granados J, Babinsky V (1988) Gene frequencies and admixture estimates in the state of Puebla, Mexico. Am J Phys Anthropol 76:331–335. 10.1002/ajpa.1330760307 [DOI] [PubMed] [Google Scholar]
- 35.Lisker R, Ramirez E, Briceño RP, Granados J, Babinsky V (1988) Gene frequencies and admixture estimates in four Mexican urban centers. Hum Biol 62:791–801. [PubMed] [Google Scholar]
- 36.Juárez-Cedillo T, Zuñiga J, Acuña-Alonzo V, Pérez-Hernández N, Rodríguez-Pérez JM, Barquera R, et al. (2008) Genetic admixture and diversity estimations in the Mexican Mestizo population from Mexico City using 15 STR polymorphic markers. Forensic Sci Int Genet 2:e37–39. 10.1016/j.fsigen.2007.08.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.