Abstract
Background
Taxillus chinensis (DC.) Danser, the official species of parasitic loranthus that grows by parasitizing other plants, is used in various traditional Chinese medicine prescriptions. ABA-dependent and ABA-independent pathways are two major pathways in response to drought stress for plants and some genes have been reported to play a key role during the dehydration including dehydration-responsive protein RD22, late embryogenesis abundant (LEA) proteins, and various transcription factors (TFs) like MYB and WRKY. However, genes responding to dehydration are still unknown in loranthus.
Methods and Results
Initially, loranthus seeds were characterized as recalcitrant seeds. Then, biological replicates of fresh loranthus seeds (CK), and seeds after being dehydrated for 16 hours (Tac-16) and 36 hours (Tac-36) were sequenced by RNA-Seq, generating 386,542,846 high quality reads. A total of 164,546 transcripts corresponding to 114,971 genes were assembled by Trinity and annotated by mapping them to NCBI non-redundant (NR), UniProt, GO, KEGG pathway and COG databases. Transcriptome profiling identified 60,695, 56,027 and 66,389 transcripts (>1 FPKM) in CK, Tac-16 and Tac-36, respectively. Compared to CK, we obtained 2,102 up-regulated and 1,344 down-regulated transcripts in Tac-16 and 1,649 up-regulated and 2,135 down-regulated transcripts in Tac-36 by using edgeR. Among them some have been reported to function in dehydration process, such as RD22, heat shock proteins (HSP) and various TFs (MYB, WRKY and ethylene-responsive transcription factors). Interestingly, transcripts encoding ribosomal proteins peaked in Tac-16. It is indicated that HSPs and ribosomal proteins may function in early response to drought stress. Raw sequencing data can be accessed in NCBI SRA platform under the accession number SRA309567.
Conclusions
This is the first time to profile transcriptome globally in loranthus seeds. Our findings provide insights into the gene regulations of loranthus seeds in response to water loss and expand our current understanding of drought tolerance and germination of seeds.
Introduction
Taxillus chinensis (DC.) Danser, which is the official name of parasitic loranthus according to the Pharmacopoeia, is wildly used in various traditional Chinese medicine prescriptions such as the treatment of rheumatism, threatened abortion, hypertension, angina pectoris, stroke, and arrhythmia for many years in China [1–4]. There are a total of 51 species of parasitic loranthus, of which 23 are distributed in Guangxi of China [5]. It is also called “Sangjisheng” in China and grows by parasitizing other plants like Aceraceae, Anacardiaceae, Euphorbiaceae, Fabaceae, Fagaceae, Juglandaceae, Moraceae, Rosaceae, and Rutaceae [6]. In plants, developmental processes such as seed germination, seedling development, leaf development and flowering are always affected by various environmental stresses, such as drought, high salinity, and high or low temperatures [7–9]. However, it is still unknown about the effects of these environmental stresses on loranthaceous.
In plants, drought stress induces various biochemical and physiological responses, such as stomatal closure, repression of cell growth and photosynthesis, and activation of respiration [10]. At cellular and molecular levels, a large number of genes have been reported to respond to drought stress [11–13]. Large scale profiling methods like microarray and next-generation sequencing have been demonstrated to estimate the gene expression changes during dehydration process in several model species, such as Arabidopsis [14, 15], rice [16–18], soybean [19, 20] and other plants [21–23]. These studies have shown that the plant defense against drought stress starts with the perception of water loss, which can trigger the activation of abscisic acid (ABA)-dependent and ABA–independent regulatory systems [24]. According to their functions, the gene products induced by drought stress can be divided into two groups [12, 13, 24]. The first group includes proteins directly protecting against the drought stress, such as chaperones, late embryogenesis abundant (LEA) proteins, mRNA-binding proteins, water channel proteins, and lipid-transfer proteins. The second group contains various TFs that probably function in further regulation of signal transduction and gene expression, protein kinases, protein phosphatases, and other signaling molecules such as calmodulin-binding proteins.
RNA-Seq, a next generation sequencing technology, has become a useful tool for genome-wide gene expression analysis [25]. It enables the de novo assembly and gene expression analyses for those species, of which the genome sequences are not available currently [26]. To study drought stress induced genes, RNA-Seq has been used to assemble the transcriptome and profiled gene expression in Glycine max [20], Brassica rapa L. ssp. Pekinensis [21], Bryum argenteum [27], Brassica napus [28], and Gossypium arboretum [29]. In Jindou21 (drought-tolerant soybean genotype), 518 and 614 genes including genes ethylene-responsive factors, MYB TFs, and zinc finger proteins are differentially expressed under water deficit condition in leaves and roots, respectively [20]. Comparative transcriptome analysis of Brassica napus has shown that a total of 6,018 and 5,377 genes including AREB/ABF, NAC, WRKY and MYB/MYC TFs are induced in response to drought stress [28] in root and leaf, respectively. However, genes induced by drought stress in loranthus are still unknown.
Due to the importance of loranthus in medical use, it is necessary to identify drought responsive genes in loranthus seeds. According to the desiccation tolerance ability, seeds are mainly divided into two types: orthodox and recalcitrant. Recalcitrant seeds lack the mechanisms of metabolic “switch-off” and intracellular dedifferentiation, which contribute significantly to their desiccation sensitivity [30]. On molecular level, the abundance of LEA protein regulated by ABI3 (B3 domain-containing transcription factor) has been verified to link to the desiccation tolerance in recalcitrant and orthodox legume seeds [31]. In this study, we profiled the transcriptome of loranthus seeds during the dehydration using the Illumina HiSeq 2000 system. Differential gene expression analysis and annotation for these transcripts revealed that some gene products have been reported to be involved in drought tolerance, such as various transcription factors, dehydration-responsive protein RD22, ABI3, heat shock proteins and zinc finger proteins. It is interesting that transcripts encoding ribosomal proteins peaked in at loranthus seeds after 16 hours of dehydration. Down-regulation of auxin related proteins, RNA binding protiens, and dehydration-responsive element-binding proteins may be signals of lower germination rates and cell death. This is the first time to analyze transcriptome in loranthus species. Our findings will contribute to understand the drought tolerance mechanism in loranthus and contribute to the research field of loranthus in breeding programs.
Material and Methods
Ethics statement
No specific permits were required for the described field studies. The location is not privately-owned or protected in any way, and the field studies did not involve endangered or protected species.
Seed collection
The seeds of Taxillus chinensis (DC.) Danser were collected from 10 trees of Dracontomelon duperreanum Pierre in Guangxi Province of China in December of 2014. They were confirmed by senior botanists at Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences.
Seed water loss and water content assay
The dehydration and water content assay of mature loranthus seeds were performed according to the manufacturer’s protocol. In brief, the aluminum case was dried to constant weight, then 100 clean and fresh seeds were weighed (W1) within the aluminum case and weighed again after being dried at 100±2°C until constant weight (W2). We replicated four times to obtain the average moisture content (W0, shown in %) in fresh seeds using the formula below.
| (1) |
Whereas W1 means the average weight of fresh seeds and W2 means the average of dehydrated seeds. Next, another 200 clean and fresh seeds were divided into four groups (50 seeds in each group), incubated in sealed desiccants with silica gel after being weighted and weighted every 4 hours. So the moisture content in seeds after dehydration can be calculated by using this formula:
| (2) |
Here, and stand for the average weight of fresh seeds before and after dehydration, respectively.
Determination of seed viability by staining
The viability of loranthus seeds was assessed by immersing the seeds in a solution of 1% (w/v) 2,3,5-Triphenyl Tetrazolium Chloride (TTC, Sigma) according to the protocols [32, 33]. Briefly, using a sterile scalpel 25 seeds were cut for small incisions allowing the TTC to enter. After an eight-hour incubation in 1% TTC solution at 25°C, seeds were washed several times by sterile water. If viable, a redox reaction would change the embryo color from white to reddish brown during cellular respiration [34]. This experiment was replicated four times.
Germination experiment
Germination experiment was conducted exactly as described previously [35]. Briefly, at each dehydration time point 25 seeds were placed on wetted double layers of Fisher No. 1 filter papers in a dish and incubated at 25°C under 16 h photoperiod for two weeks, before germination rates were determined. Germination experiment was replicated four times for each time point.
RNA extraction
We selected fresh seeds (CK) and seeds after dehydration for 16 hours (Tac-16) and 36 hours (Tac-36) for deep sequencing. Total RNA was isolated from the seeds by using TRIzol® reagent (Invitrogen) according to the manufacturer’s protocol [36]. Briefly, 10 seeds (~ 3–4 g) was mixed with 1 ml of TRIzol® reagent, homogenized by power homogenizer and centrifuged at 12,000 ×g for 10 min at 4°C. Then, the fatty layer was discarded, cleared supernatant was transferred into a new tube. Next, 0.2 ml of chloroform was added into the tube, following by shaking the tube for 15 secs, centrifugation at 12,000 ×g for 15 min at 4°C and moving the aqueous phase into another new tube for RNA precipitation. We added 10 μg of RNase-free glycogen and 0.5 ml of 100% isopropanol into the aqueous phase, incubated the samples at room temperature for 10 min and centrifuged them at 12,000 ×g for 10 min at 4°C. Finally, the RNA pellet was washed by 1 ml of 75% ethanol, air-dried, suspended in RNase-free water and water-bathed at 60°C for 10 min. The quality of total RNA was evaluated and controlled by Agilent 2100 Bioanalyzer. For each sample we replicated total RNA isolation for three times and pooled them for cDNA library construction and sequencing.
cDNA library construction and sequencing
A total amount of 20 μg RNA was used for transcriptome cDNA library construction by using TruSeqTM RNA Sample Preparation Kit v2 (Illumina) and the cDNA library was sequenced on an Illumina HiSeq 2000 platform following the manufacturers’ protocols. In brief, poly(A) mRNAs were obtained by using Dynal Oligo(dT) beads (Invitrogen). mRNAs were then chemically fragmented into ~200 nt fragments. mRNA fragments were next copied into first strand cDNA by using reverse transcriptase and random primers, followed by the second strand cDNA synthesis using DNA Polymerase I (Invitrogen) and RNase H (Invitrogen) treatment. After end repaired by using End Repair Mix (Illumina) reagent, the cDNA fragments were purified and enriched to create the final cDNA library. A total of six libraries (each sample has two replicates) were sequenced by pair-end (2×90 bp) method on an Illumina HiSeqTM 2000 platform.
De novo assembly of the transcriptome
After adapter sequences and low quality reads were removed, raw sequencing reads were cleaned and quality controlled by FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, de novo assembly of high quality reads was carried out by Trinity software (release 2014-07-17) [37], according to the protocol [38].
Transcriptome annotation
Gene Ontology (GO) and biological pathway annotations for the assembled transcriptome were performed by mapping them to public databases, including NCBI non-redundant (NR), UniProt and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. First, BLAST software [39] was used to align all the transcripts to NR and UniProt databases. Matched transcripts were filtered by using a cut-off of e-value (1 × 10−5). Then, BLAST2GO software [40] was used to retrieve associated GO items describing biological processes (BPs), cellular components (CCs) and molecular functions (MFs) for the assembled transcripts. The enzyme commission numbers (EC) for each transcript were also annotated by BLAST2GO. Using a cut-off of e-value (≤1e-5), the transcripts with corresponding ECs were obtained and mapped to KEGG metabolic pathway database. Then, likely protein sequences were extracted from the assembled transcripts by TransDecoder, which is included in the Trinity software distribution. Potential signal peptides, transmembrane domains and rRNA transcripts were predicted by using SignalP [41], TMHMM Sever v2.0 [42] and RNAMMER [43], respectively. To identify the proteins distributed in EuKaryotic Orthologous Groups (KOG), Clusters of Orthologous Groups (COGs), and non-supervised orthologous groups (NOGs), the likely protein sequences were further used to search against EggNOG database (v4.1, http://eggnogdb.embl.de) [44]. Protein functional domains were identified by mapping the likely proteins to Pfam database [45] using HMMER [46] and filtered by using the cut-off of e-value (≤1e-5).
Reads alignment and transcriptome profile
To profile gene expression in loranthus seeds, Bowtie2 [47] and RSEM (RNA-Seq by Expectation-Maximization) [48] were used to map clean reads of CK (CK-1 and CK-2), Tac-16 (Tac-16-1 and Tac-16-2) and Tac-36 (Tac-36-1 and Tac-36-2) to the assembled transcriptome and evaluate the abundance of each transcript, respectively. To compare the expression levels of transcripts in different samples, we used FPKM (fragments per kilobase of transcript per million mapped reads) for normalization and to present the expression of transcripts.
Differentially expressed transcripts
Transcripts differentially expressed during the dehydration process were identified by using egdeR [49]. We used a strict criterial to select differentially expressed transcripts: normalized expression >1 FPKM, Log2FC (log2 fold change) >1 (up-regulated) or Log2FC <-1 (down-regulated), p-value <0.05 and FDR (false discovery rate) <0.05.
GO and KEGG pathway enrichment analysis
To identify significant GO terms and KEGG pathways enriched by candidate transcripts, we used p-value (Fisher’s exact test) to show the significance of enrichment for specific GO term. Then, q-value [50] was calculated to correct the p-value for each GO term or pathway and control the false discovery rate. Significant GO items and KEGG pathways should satisfy the critical: q-value<0.05. GO items and KEGG pathways not associated with plant bio activities were filtered.
Quantitative real-time PCR
To validate the expression of the assembled transcripts, quantitative real-time PCR (qRT-PCR) experiment was performed following the protocols. In brief, total RNA was extracted by using TRIzol® reagent (Invitrogen), as described. The quality of total RNA was evaluated and controlled by NanoDrop 1000 (Thermo Scientific). Forward and reverse primer sequences for candidate transcripts and control (actin-3) were predicted by Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and synthesized at BGI-Shenzhen. Then, FastQuant RT Kit (with gDNase, Tiangen) was used to synthesize cDNA from total RNA (1 μg). The cDNA samples (100 ng, 2 μl) were next mixed with 10 μl of SuperReal PreMix (SYBR Green, Tiangen), forward primer (0.8 μl), reverse primer (0.8 μl), 5 × gDNA Buffer (2 μl) and ddH2O (6.4 μl) to make the final PCR reaction mix. The final PCR reaction mix (SYBR Green) was amplified on a LightCycler480II (Roche) with three steps PCR, including 1 cycle initial denaturation (95°C for 3 min), 40 cycles of PCR reactions (95°C for 10 s, 60°C for 15 s and 72°C for 20 s) and a melting/dissociation curve stage (95°C for 10 s, 65°C for 1 min and a continuous temperature ramp (0.11°C/s) from 65 to 97°C). For each candidate we replicated three times of qRT-PCR in every sample. Average of the Ct (cycle threshold) for each candidate was calculated, ΔCt was used to evaluate the expression levels of candidate transcripts in each sample and ΔΔCt method was used to show the different expression of a particular transcript between two samples [51].
| (3) |
Whereas and stand for the average Ct values of a candidate transcript detected in dehydrated seeds and fresh seeds, respectively; and stand for the average Ct values of actin-3 detected in dehydrated seeds and fresh seeds, respectively. The relative normalized expression (RNE) was calculated by using 2-ΔΔCt method.
Availability of data and material
The raw sequencing files of these six samples (FASTQ formatted files) can be accessed in the NCBI Sequence Read Archive (SRA) database (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number of SRA309567 (CK-1: SRR2902061; CK-2: SRR4067162; Tac-16-1: SRR2902062; Tac-16-2: SRR4067163; Tac-36-1: SRR2902063; Tac-36-2 SRR4067164). The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GELW00000000. Scripts for key steps, such as transcriptome de novo assembly, annotation, expression profile and differential expression, can be seen in S1 File.
Results and Discussion
Seeds dehydration, viability test and germination experiment
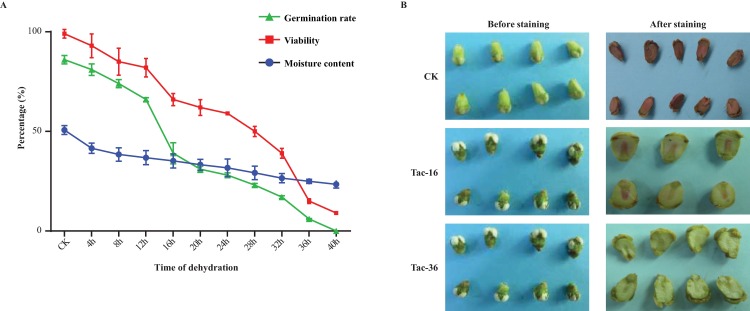
The loranthus seeds were collected from 10 trees of Dracontomelon duperreanum Pierre in Guangxi Province of China in December of 2014 and confirmed by senior botanists. Loranthus is wildly used in traditional Chinese medicine prescriptions and it is still unknown about the loranthus seed response to environmental stresses. Loranthus seeds always disperse as fresh seeds in wild by our long-term observations, so among the environmental stresses we first want to study the gene expression changes in loranthus seeds in response to water loss. Initially, we observed that loranthus seeds were recalcitrant because the viability and germination rate of loranthus seeds dropped quickly if they were stored in dry environment [52]. Using seed germination experiment we found after being stored for three days the germination rate decreased from 86% (germination rate of fresh seeds) to 40% and after six days the germination rate was only 5%. Then, we examined the moisture content of fresh loranthus seeds was 50.7% (w/w) on average (Fig 1A), which is similar to that in soybean [53]. Next, we performed the dehydration experiment and tested the viability of loranthus seeds (25 seeds × 4 replicates) using TTC staining. Fig 1A showed the moisture content, the viability and germination rate of loranthus seeds (25 seeds × 4 replicates) during the dehydration process. It is clear that the viability and germination rate of loranthus seeds were associated with the water content in seeds. The germination rate of loranthus fresh seeds was 86% (viability: 99%) and after being dehydrated for 16 hours it was dropped to 66% (viability: 66%, moisture content: 35.17%). If the seeds were dehydrated for 36 hours, the moisture content was decreased to 24.93%, the viability was decreased to 15% and the germination rate was 6%. After 40 hours’ dehydration the moisture content, viability and germination rate were examined as 23.47%, 9% and 0, respectively. Because both viability and germination rate were decreased most after being dehydrated for 16 hours (Tac-16) and 36 hours (Tac-36), we collected seeds from these two time points and CK to study the gene expression changes of loranthus seeds during the dehydration process. TTC staining of CK, Tac-16 and Tac-36 confirmed that the viability of loranthus seeds was affected by drought (Fig 1B). Loranthus seeds were abnormal after 16 hours’ dehydration and non-viable after 36 hours’ dehydration. In fact, moisture content has been experimented to play a key role in seed germination [54, 55], especially in early seed germination [56]. In Arabidopsis, water in seeds supplies several biochemical reactions during the seed germination, such as water up-taken from outside [53, 56]. At molecular level, drought stress induced genes have been characterized in different plant species including Arabidopsis [14, 15], rice [16–18], soybean [19, 20], and other plants [21–23].
Fig 1. Treatment of loranthus seeds.
(A) Moisture content assay, viability test and germination test of loranthus seeds. (B) TTC test of CK, Tac-16 and Tac-36 show fresh seeds (CK) are viable, seeds in Tac-16 group are abnormal and seeds in Tac-36 group are non-viable.
Transcriptome de novo assembly
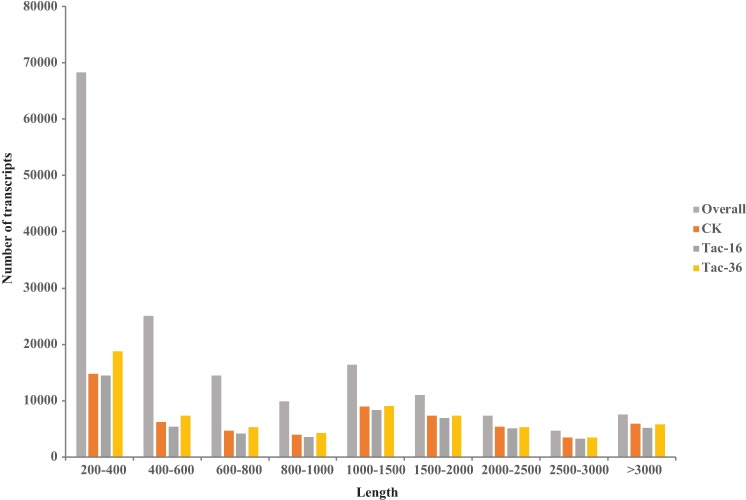
In this study, we conducted six cDNA libraries (in biological replicates) for CK, Tac-16 and Tac-36 and sequenced them using paired-end transcriptome sequencing. After the low quality reads were removed, 386,542,846 high quality reads were obtained and used for de novo assembly by using Trinity software [37]. A total of 164,546 transcripts corresponding to 114,971 genes (Table 1) were assembled. The assembly contains 149,031,959 bases (159 M in size), which was determined to be ‘strong’ and ‘fully consider’ for evaluating de novo transcriptome [57]. Next, other measures like GC content, N10, N20 and N50 were used to evaluate the transcriptome. The GC content, N10, N20 and N50 of the assembled transcriptome were calculated as 41.12%, 3,972, 3,040 and 1,610, respectively (Table 1). Of them, N50 is a statistical measure of average length of a set of sequences like genome and transcriptome sequences [58]. In addition, length distribution of the assembled transcripts (Fig 2) told us there were a total of 46,869 (28.48%) transcripts longer than 1,000 bp, of them 7,521 (4.57%) transcripts were longer than 3,000 bp. Except short transcripts (< 400 nt), the numbers of assembled transcripts, transcripts detected in CK, Tac-16 and Tac-36 were close to each other. This is the first time to study the loranthus seed transcriptome, it is hard to evaluate the numbers of transcripts and genes in loranthus due to the missing information of its genome sequence and annotation.
Table 1. Overview of the transcriptome de novo assembly.
| Type | Result |
|---|---|
| High quality reads | 386,542,846 |
| Total Trinity genes | 114,971 |
| Total Trinity transcripts | 164,546 |
| GC (%) | 41.12 |
| N10 | 3,972 |
| N20 | 3,040 |
| N50 | 1,610 |
| Total assembled bases | 149,031,959 |
Fig 2. Length distribution of the assembled transcripts and transcripts (> 1FPKM) detected in CK, Tac-16 and Tac-36.
Annotation of the assembled loranthus seed transcriptome
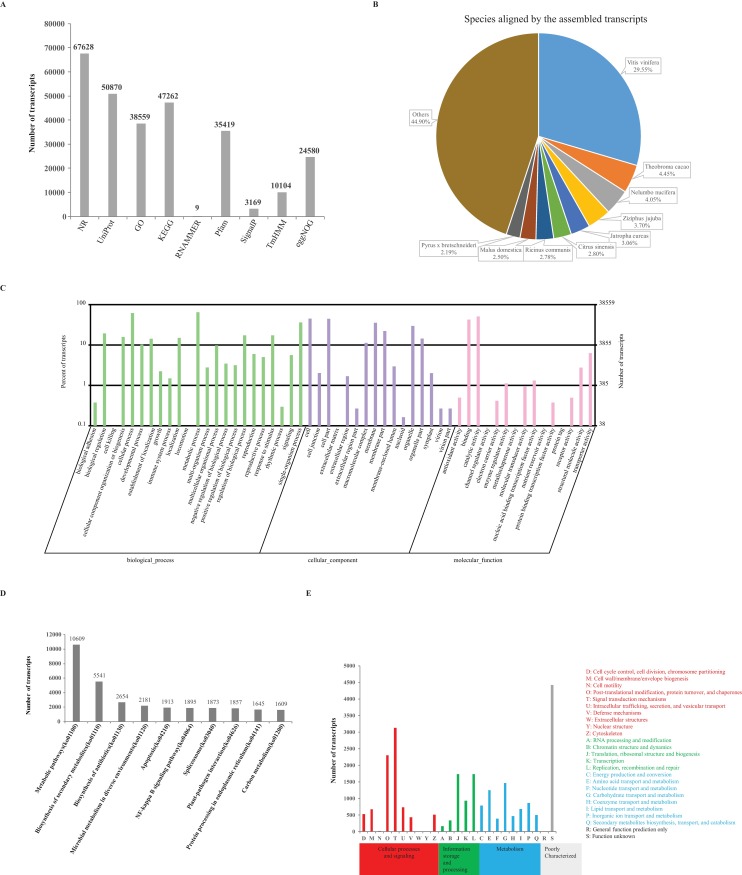
We next annotated the assembled transcriptome by mapping it to NCBI non-redundant (NR), UniProt, GO and KEGG databases and the numbers of transcripts matched to each database can be seen in Fig 3A. By using BLAST software [39] and a cut-off of e-value (< 1 × e-5), the largest number (67,628, 41.10%) of transcripts were aligned to NR database, followed by the UniProt database (50,870, 30.92%). We further explored the loranthus transcripts aligned to species in the NR mapping results (Fig 3B). There were 19,977 transcripts aligned to Vitis vinifera, taking 29.54% of all the transcripts aligned to NR database, followed by Theobroma cacao (3,010, 4.45%), Nelumbo nucifera (2,742, 7.05%) and Ziziphus jujuba (2,504, 3.70%). Using the NR and UniProt mapping results 38,559 (23.43%) transcripts (Fig 3A) whose orthologous sequences have GO annotations were divided into three categories: cellular component, biological process, and molecular function (S1 Table). GO analysis (Fig 3C) showed 19 GO items were enriched by more than 10% of the total assembled transcripts. Top 10 of them were “metabolic process” (25,062 transcripts), “cellular process” (23,847 transcripts), “catalytic activity” (19,620 transcripts), “cell” (17,340 transcripts), “cell part” (17,196 transcripts), “binding” (16,406 transcripts), “single-organism process” (13,988 transcripts), “membrane” (13,668 transcripts), “organelle” (11,457 transcripts) and “membrane part” (8,590 transcripts). In addition, 47,262 transcripts were identified to involve in 362 different KEGG pathways (Fig 3A). According to the numbers of transcripts, top 10 KEGG pathways can be seen in Fig 3D. The most significant KEGG pathway was “metabolic pathway” (ko01100), containing 10,609 transcripts. We also identified 1,857 and 1,645 transcripts playing a key role in the pathways of “plant-pathogen interaction” (ko04626) and “plant hormone signal transduction” (ko04075), respectively. The ontologies and pathways annotated for loranthus seed transcriptome showed their potential functions in seed development [59] and tolerance of environmental stresses [60]. Interestingly, 9 transcripts (Fig 3A) were predicted to be ribosomal RNAs by using RNAMMER [43].
Fig 3. Annotation of the assembled transcriptome.
(A) Number of transcripts aligned to different databases. (B) Species aligned by the assembled loranthus seed transcriptome. (C) Gene Ontology analysis for the assembled loranthus seed transcriptome. (D) Top 10 significant KEGG pathways. (E) COG annotation.
Next, we extracted likely proteins from the assembled transcripts using TransDecoder. In total, 49,004 transcripts (29.78% of the total assembled transcripts) were predicted to encode 61,610 proteins. Among the likely proteins, we identified 40,846 (24.82%) transcripts containing Pfam domain sequences, 3,169 (1.93%) with signal peptides and 10,104 (6.14%) transcripts encoding membrane related proteins (Fig 3A). Then, likely proteins encoded by the assembled transcripts were mapped to eggNOG database and proteins encoded by 24,580 (14.94%) transcripts were distributed in EuKaryotic Orthologous Groups (KOG), Clusters of Orthologous Groups (COGs), and non-supervised orthologous groups (NOGs), see S2 Table. As show in Fig 3E, 4,315 likely proteins were poorly characterized, 2,911 likely proteins were from COG of signal transduction mechanisms, and 2,138 likely proteins were from COG of post-translational modification, protein turnover, and chaperones. Annotations from different perspectives will give a better understanding of the functions of the assembled transcripts and help to identify transcripts involved in the dehydration process. In addition, the reasons of some transcripts annotated without encoding ability should be further explored [61].
Transcriptome profile and different expression
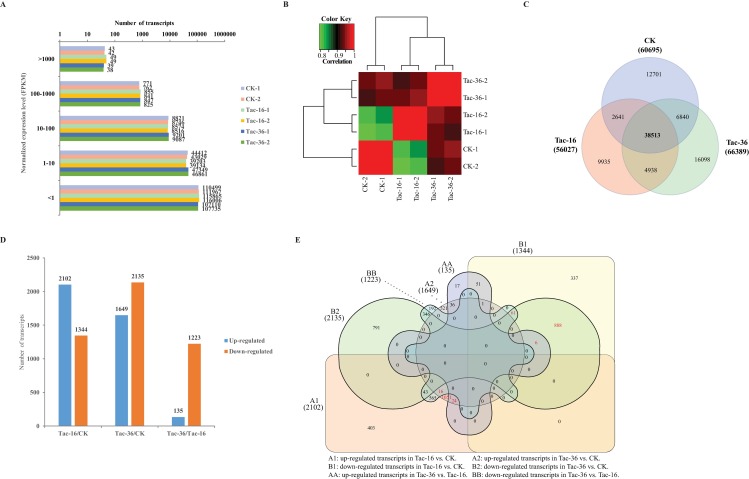
The viability and germination rate of loranthus seeds dropped quickly during dehydration (Fig 1A). In order to identify genes induced by drought stress and profile them in loranthus seeds, we performed two biological replicates for CK, Tac-16 and Tac-36 samples and the expression of transcripts was evaluated separately in each replicate. Bowtie2 [47] was used to align the high quality reads to the assembled transcriptome and RSEM [48] tool was used to profile gene expression in all samples. In total, we obtained 91,666 transcripts (>1 FPKM) and 54,047, 52,579, 48,681, 48,540, 57,436 and 56,811 transcripts (>1 FPKM) distributed in CK-1. CK-2, Tac-16-1, Tac-16-2, Tac-36-1 and Tac-36-2, respectively. Length distribution of transcripts detected in CK, Tac-16 and Tac-36 can be found in Fig 2and the distribution of their normalized expression (Fig 4A) showed 80.53% ~ 82.49% of the total detected transcripts (excluding transcripts < 1 FPKM) in each sample were less than 10 FPKM. Pearson correlations for the replicates were above 0.9 in CK, Tac-16 and Tac-36 well (Fig 4B), which indicated the replicates were performed very well. Venn diagram (Fig 4C) of detected transcripts in CK, Tac-16 and Tac-36 (> 1 FPKM in at least one of the two replicates) revealed 38,513 (42.01% of the total detected transcripts) were commonly detected and 12,701 (20.93% of transcripts detected in CK), 9,935 (17.73% of transcripts detected in Tac-16) and 16,098 (24.74% of transcripts detected in Tac-36) transcripts were detected exclusively in CK, Tac-16 and Tac-36, respectively.
Fig 4. Transcriptome profiling and differential expression.
(A) Distribution of normalized expression of transcripts detected in all samples. (B) Heat map of correlations between replicates. (C) Venn diagram of transcripts (>1 FPKM) detected in CK, Tac-16 and Tac-36. (D) Number of up- and down-regulated transcripts in Tac-16 and Tac-36 compared to CK. (E) Venn diagram of differentially expressed transcripts identified in all comparisons. Numbers in red represent commonly up- (1091) and down-regulated (955) transcripts in Tac16 and Tac-36 compared to CK.
To characterize drought induced genes in loranthus seeds, we employed edgeR [49] to identify transcripts differentially expressed in Tac-16 and Tac-36 compared to CK and used the criterial as follows: FPKM >5 in at least one sample, Log2FC >1 or Log2FC <-1, p-value <0.05 and FDR <0.05. Compared to CK, we obtained 2,102 up-regulated and 1,344 down-regulated transcripts in Tac-16 and 1,649 up-regulated and 2,135 down-regulated transcripts in Tac-36 (Fig 4D). We also compared Tac-16 and Tac-36 and found 1,358 transcripts differentially expressed including 135 transcripts up-regulated in Tac-36 and 1,223 transcripts up-regulated in Tac-16 (Fig 4D). In all three comparisons we obtained a total of 5,349 transcripts differentially expressed in loranthus seeds during the dehydration. Venn diagram of differentially expressed transcripts (Fig 4E) showed the numbers of transcripts commonly and specifically up-regulated or down-regulated in Tac-16 and Tac-36. It is revealed that 1,091 transcripts were commonly up-regulated and 955 transcripts were commonly down-regulated in Tac-16 and Tac-36 in comparison of CK. Interestingly, 24 transcripts such as c36451_g2_i1 (LEGB4_VICFA, legumin type B), c51416_g2_i1 (AB40G_ARATH, ABC transporter G family member 40) and c59053_g2_i1 (DIR23_ARATH, dirigent protein 23) kept increasing while 61 transcripts such as c60985_g1_i1 (12KD_FRAAN, auxin-repressed 12.5 kDa protein), c67927_g8_i3 (BH094_ARATH, transcription factor bHLH94), c49978_g1_i1 (DRE2D_ARATH, dehydration-responsive element-binding protein 2D) and c39272_g1_i1 (HS23C_OXYRB, small heat shock protein, chloroplastic) kept decreasing during the dehydration process. Detailed information of differentially expressed transcripts in Tac-16 and Tac-36 compared to CK can be accessed in S3 Table.
Functional analysis of differentially expressed transcripts
Next, we annotated the differentially expressed transcripts using GO and KEGG pathway databases. Overall, loranthus seed transcripts induced by drought were involved in metabolic pathways, such as “Metabolic pathways” (ko01100), “Riboflavin metabolism” (ko00740) and “Terpenoid backbone biosynthesis” (ko00900), signaling transduction pathway “Plant hormone signal transduction” (ko04075) and environmental adaption pathways such as “Plant-pathogen interaction” (ko04626), “Circadian entrainment” (ko04713) and “Circadian rhythm–plant” (ko04712) (Table 2).In addition, we found several pathways may be associated with loranthus seeds in response to dehydration at different stages. For example, there were 107 transcripts involved in “Ribosome” (ko03010) in Tac-16 (p-value = 2.2E-16, q-value = 6.8E-14) but only 40 in Tac-36 (p-value = 0.683, q-value = 1). And transcripts involved in “Neuroactive ligand-receptor interaction” (ko04080) decreased from 19 in Tac-16 (p-value = 1.37E-15, q-value = 4.24E-13) to 0 in Tac-36 (p-value = 1, q-value = 1). Although the numbers of transcripts annotated by KEGG pathway were decreased in Tac-36 compared to Tac-16, pathways of “Biosynthesis of secondary metabolites” (ko01110), “Biosynthesis of antibiotics” (ko01130) and “plant hormone signal transduction” (ko04075) had more differentially expressed transcripts in Tac-36 (Table 2).
Table 2. KEGG pathway analysis for differentially expressed transcripts.
| Group | Pathway | ID | Tac-16a | P-value | Q-value | Tac-36b | P-value | Q-value |
|---|---|---|---|---|---|---|---|---|
| Global and overview maps | Metabolic pathways | ko01100 | 426 | 2.2E-16 | 6.8E-14 | 572 | 2.2E-16 | 7.11E-14 |
| Global and overview maps | Biosynthesis of secondary metabolites | ko01110 | 232 | 1.07E-13 | 3.3E-11 | 391 | 2.2E-16 | 7.11E-14 |
| Signal transduction | Plant hormone signal transduction | ko04075 | 87 | 3.03E-12 | 9.35E-10 | 118 | 2.2E-16 | 7.11E-14 |
| Carbohydrate metabolism | Galactose metabolism | ko00052 | 44 | 4.19E-12 | 1.29E-09 | 48 | 2.16E-12 | 6.97E-10 |
| Biosynthesis of other secondary metabolites | Flavonoid biosynthesis | ko00941 | 11 | 0.005321 | 1 | 26 | 9.56E-12 | 3.09E-09 |
| Carbohydrate metabolism | Starch and sucrose metabolism | ko00500 | 68 | 1.42E-07 | 4.38E-05 | 86 | 2.21E-11 | 7.14E-09 |
| Biosynthesis of other secondary metabolites | Phenylpropanoid biosynthesis | ko00940 | 27 | 0.002195 | 0.678255 | 48 | 1.31E-10 | 4.23E-08 |
| Carbohydrate metabolism | Amino sugar and nucleotide sugar metabolism | ko00520 | 60 | 9E-12 | 2.78E-09 | 61 | 6.16E-10 | 1.99E-07 |
| Glycan biosynthesis and metabolism | Other glycan degradation | ko00511 | 27 | 1.29E-05 | 0.003974 | 37 | 3.12E-09 | 1.01E-06 |
| Metabolism of other amino acids | Cyanoamino acid metabolism | ko00460 | 17 | 0.009255 | 1 | 33 | 4.59E-09 | 1.48E-06 |
| Global and overview maps | Biosynthesis of antibiotics | ko01130 | 95 | 0.001081 | 0.334029 | 132 | 5.25E-09 | 1.7E-06 |
| Environmental adaptation | Plant-pathogen interaction | ko04626 | 86 | 2.19E-07 | 6.77E-05 | 99 | 1.85E-08 | 5.98E-06 |
| Biosynthesis of other secondary metabolites | Monobactam biosynthesis | ko00261 | 5 | 0.03746 | 1 | 14 | 1.92E-08 | 6.2E-06 |
| Global and overview maps | Microbial metabolism in diverse environments | ko01120 | 95 | 7.67E-07 | 0.000237 | 111 | 2.89E-08 | 9.34E-06 |
| Energy metabolism | Photosynthesis—antenna proteins | ko00196 | 0 | 1 | 1 | 9 | 3.46E-08 | 1.12E-05 |
| Amino acid metabolism | Lysine biosynthesis | ko00300 | 2 | 0.5836 | 1 | 14 | 4E-08 | 1.29E-05 |
| Metabolism of terpenoids and polyketides | Carotenoid biosynthesis | ko00906 | 9 | 0.003549 | 1 | 17 | 7.15E-08 | 2.31E-05 |
| Environmental adaptation | Circadian entrainment | ko04713 | 16 | 2.07E-06 | 0.000639 | 19 | 1.18E-07 | 3.8E-05 |
| Metabolism of cofactors and vitamins | Porphyrin and chlorophyll metabolism | ko00860 | 6 | 0.4873 | 1 | 23 | 1.37E-07 | 4.43E-05 |
| Xenobiotics biodegradation and metabolism | Naphthalene degradation | ko00626 | 3 | 0.02358 | 1 | 8 | 2.85E-07 | 9.19E-05 |
| Xenobiotics biodegradation and metabolism | Polycyclic aromatic hydrocarbon degradation | ko00624 | 15 | 1.32E-08 | 4.09E-06 | 14 | 4.91E-07 | 0.000158 |
| Biosynthesis of other secondary metabolites | Stilbenoid, diarylheptanoid and gingerol biosynthesis | ko00945 | 14 | 3.4E-06 | 0.00105 | 16 | 6.4E-07 | 0.000207 |
| Metabolism of other amino acids | Taurine and hypotaurine metabolism | ko00430 | 5 | 0.002268 | 0.700812 | 9 | 7.97E-07 | 0.000257 |
| Cell growth and death | Apoptosis | ko04210 | 66 | 0.01266 | 1 | 94 | 1.55E-06 | 0.000499 |
| Environmental adaptation | Circadian rhythm—plant | ko04712 | 20 | 0.000174 | 0.053828 | 26 | 1.94E-06 | 0.000627 |
| Biosynthesis of other secondary metabolites | Anthocyanin biosynthesis | ko00942 | 1 | 0.5807 | 1 | 8 | 4.24E-06 | 0.001368 |
| Excretory system | Aldosterone-regulated sodium reabsorption | ko04960 | 0 | 1 | 1 | 10 | 4.46E-06 | 0.001442 |
| Biosynthesis of other secondary metabolites | Isoflavonoid biosynthesis | ko00943 | 5 | 0.0185 | 1 | 10 | 8.61E-06 | 0.002782 |
| Metabolism of cofactors and vitamins | Riboflavin metabolism | ko00740 | 2 | 0.4556 | 1 | 10 | 8.61E-06 | 0.002782 |
| Global and overview maps | Degradation of aromatic compounds | ko01220 | 5 | 0.002567 | 0.793203 | 8 | 1.06E-05 | 0.003424 |
| Lipid metabolism | Glycerolipid metabolism | ko00561 | 16 | 0.09111 | 1 | 30 | 1.38E-05 | 0.00447 |
| Excretory system | Vasopressin-regulated water reabsorption | ko04962 | 8 | 0.001223 | 0.377907 | 11 | 2.74E-05 | 0.008863 |
| Carbohydrate metabolism | Glycolysis / Gluconeogenesis | ko00010 | 42 | 2.4E-05 | 0.00741 | 46 | 2.8E-05 | 0.00905 |
| Digestive system | Mineral absorption | ko04978 | 5 | 0.1158 | 1 | 12 | 3.83E-05 | 0.012368 |
| Lipid metabolism | Fatty acid biosynthesis | ko00061 | 10 | 0.04153 | 1 | 18 | 4.29E-05 | 0.013841 |
| Metabolism of terpenoids and polyketides | Terpenoid backbone biosynthesis | ko00900 | 15 | 0.003843 | 1 | 21 | 4.3E-05 | 0.013886 |
| Xenobiotics biodegradation and metabolism | Dioxin degradation | ko00621 | 3 | 0.000889 | 0.274577 | 4 | 4.92E-05 | 0.015888 |
| Metabolism of terpenoids and polyketides | Limonene and pinene degradation | ko00903 | 12 | 2.78E-05 | 0.008581 | 12 | 9.9E-05 | 0.031971 |
| Global and overview maps | 2-Oxocarboxylic acid metabolism | ko01210 | 10 | 0.3284 | 1 | 23 | 0.000112 | 0.036079 |
| Xenobiotics biodegradation and metabolism | Bisphenol degradation | ko00363 | 12 | 2.06E-06 | 0.000637 | 10 | 0.000214 | 0.06909 |
| Carbohydrate metabolism | Citrate cycle (TCA cycle) | ko00020 | 22 | 1.75E-05 | 0.00542 | 21 | 0.000326 | 0.105266 |
| Metabolism of other amino acids | Glutathione metabolism | ko00480 | 21 | 3.74E-06 | 0.001157 | 17 | 0.001681 | 0.542963 |
| Carbohydrate metabolism | Ascorbate and aldarate metabolism | ko00053 | 18 | 1.84E-05 | 0.00567 | 15 | 0.002348 | 0.758404 |
| Xenobiotics biodegradation and metabolism | Aminobenzoate degradation | ko00627 | 12 | 5.62E-05 | 0.017366 | 10 | 0.002609 | 0.842707 |
| Translation | Ribosome | ko03010 | 107 | 2.2E-16 | 6.8E-14 | 40 | 0.683 | 1 |
| Digestive system | Protein digestion and absorption | ko04974 | 25 | 2.2E-16 | 6.8E-14 | 4 | 0.3388 | 1 |
| Signaling molecules and interaction | Neuroactive ligand-receptor interaction | ko04080 | 19 | 1.37E-15 | 4.24E-13 | 0 | 1 | 1 |
| Sensory system | Phototransduction—fly | ko04745 | 20 | 3.63E-11 | 1.12E-08 | 9 | 0.009196 | 1 |
| Sensory system | Phototransduction | ko04744 | 12 | 5.86E-08 | 1.81E-05 | 6 | 0.01012 | 1 |
a. Number of differentially expressed transcripts in Tac-16 compared to CK.
b. Number of differentially expressed transcripts in Tac-36 compared to CK.
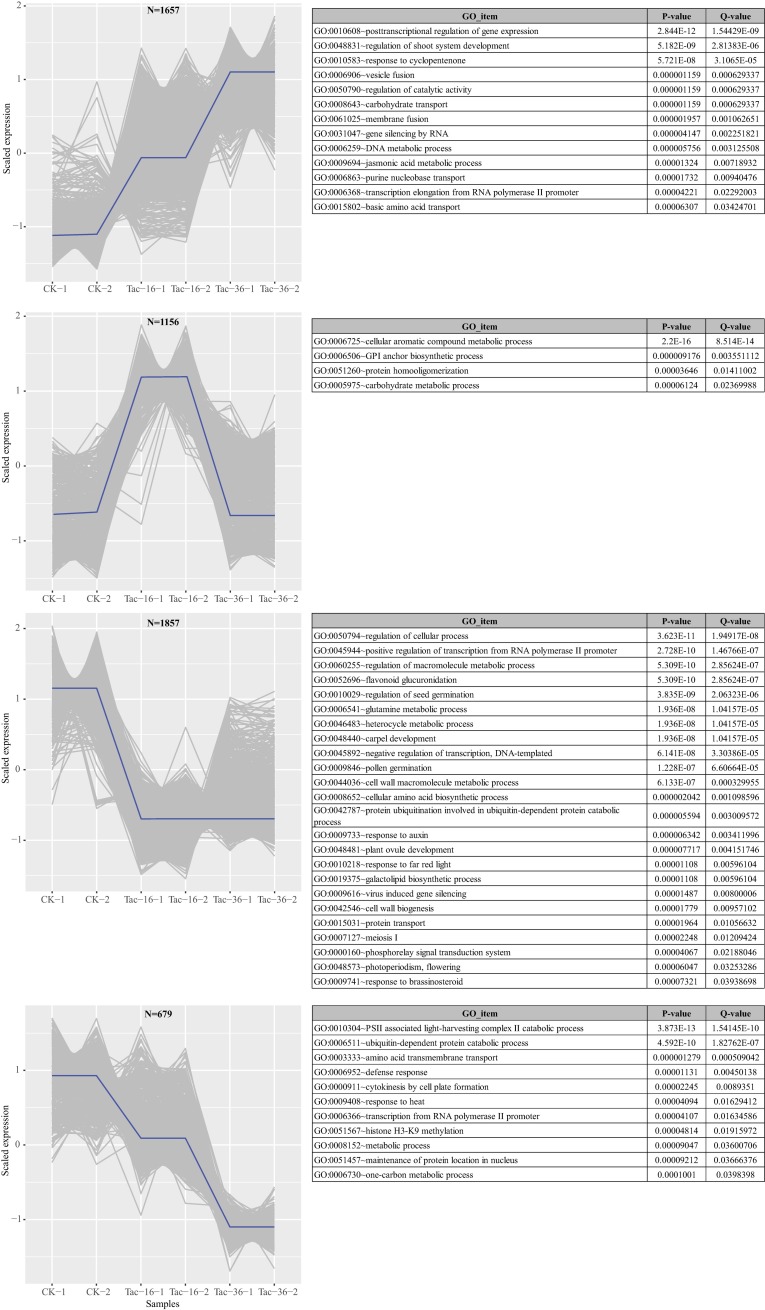
We performed K-means clustering analysis using MEV software (v4.9) and found four main groups of transcripts with different expression patterns in CK, Tac-16 and Tac-36 (Fig 5B). The biggest group containing 1,857 transcripts that were down-regulated in loranthus dehydrated seeds compared to CK. They were significantly enriched (p-value <0.05, q-value <0.05) in biological processes including “regulation of cellular process” (GO:0050794), “regulation of seed germination” (GO:0010029), “pollen germination” (GO:0009846), “response to auxin” (GO:0009733), “plant ovule development” (GO:0048481), “response to far red light” (GO:0010218), “photoperiodism, flowering” (GO:0048573) and “response to brassinosteroid” (GO:0009741). The decreased expression of transcripts involved in seed germination and cell development, especially the plant ovule development indicated the germination of loranthus seeds was affected due to the water loss, so we assume that loranthus seeds are recalcitrant. The second group contained 1657 transcripts that were up-regulated in loranthus seeds during the dehydration. GO analysis revealed these transcripts were involved in the biological processes such as “posttranscriptional regulation of gene expression” (GO:0010608), “regulation of shoot system development” (GO:0048831), “response to cyclopentenone” (GO:0010583), “vesicle fusion” (GO:0006906), “regulation of catalytic activity” (GO:0050790) and “jasmonic acid metabolic process” (GO:0009694). The third group contained 1,156 transcripts whose expression peaked in Tac-16. They were involved in “cellular aromatic compound metabolic process” (GO:0006725), “GPI anchor biosynthetic process” (GO:0006506), “protein homooligomerization” (GO:0051260) and “carbohydrate metabolic process” (GO:0005975). The last group contained 679 transcripts that were down-regulated in loranthus seeds during the dehydration process. Compared to the biggest group, the transcripts in this group were down-regulated in Tac-36 compared to Tac-16 and were involved in the biological processes like “defense response” (GO:0006952), “cytokinesis by cell plate formation” (GO:0000911), “response to heat” (GO:0009408), “transcription from RNA polymerase II promoter” (GO:0006366) and “metabolic process” (GO:0008152). Overall, in response to drought loranthus seeds actively or passively reduced the cell developmental activities, metabolism and transcripts involved in seed germination and plant defense system while transcripts associated with posttranscriptional regulation and gene silencing by RNA were up-regulated due to the water loss in loranthus seeds. In tobacco, reducing the metabolism is expected to play a major role at the early stage in programmed cell death [62]. Auxin and other hormones like abscisic acid (ABA), cytokinins and ethylene are well known to regulate the cell growth in multiple stages including cell development and cell death [63]. It is interesting that ABA also has the capacity of delaying the process of programmed cell death in aleurone cells although the mechanism is unknown [64, 65]. Another review says cell death is programmed when the metabolism is perturbed by abiotic stresses and the cells seem to be ready to die based on the integration of different signals including auxin, cytokinins, ethylene, and elicitors [66].
Fig 5. K-means clustering analysis of the differentially expressed transcripts in dehydrated loranthus seeds in comparison of CK and GO analysis.
Differentially expressed transcripts in response to drought stress
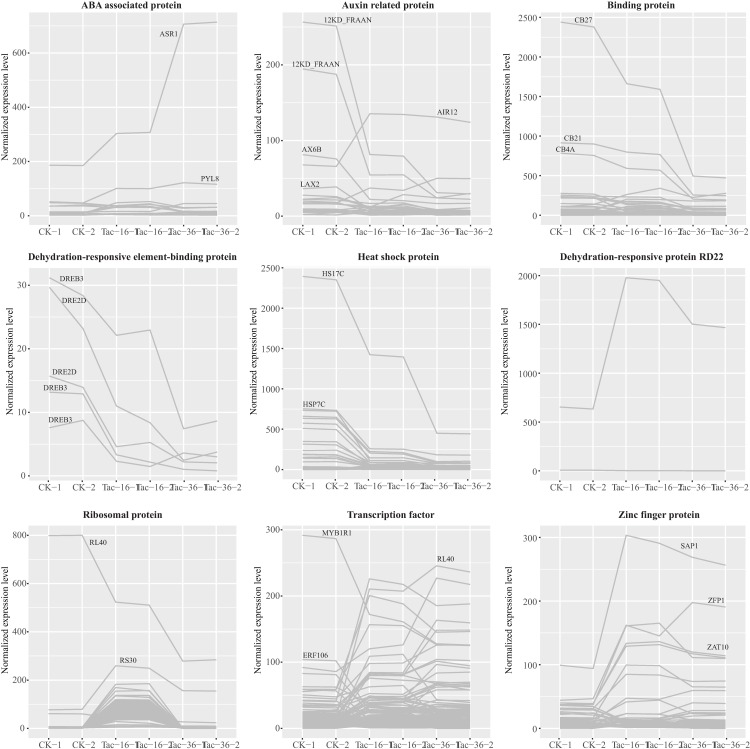
In order to understand the genes induced by drought in loranthus seeds, we further annotated the differentially expressed transcripts and found there were several genes whose products have been reported to be involved in cell development and cell death. It is well known that ABA-dependent and ABA-independent pathways are two main pathways for plants to respond the water loss [24, 67, 68], so we first examined the expression of ABA associated transcripts. In loranthus seed transcriptome we detected 49 ABA associated transcripts, of which 14 were differentially expressed in dehydrated seeds (Table 3, Fig 6, S3 Table). It is notable that ASR1 (Abscisic stress-ripening protein 1), which is induced by water and salt stress [69], was highly expressed (>200 FPKM) and up-regulated significantly in dehydrated loranthus seeds, compared to fresh loranthus seeds.
Table 3. Transcripts encoding different proteins are associated with dehydration tolerance.
| Gene family | Detected | Differentially expressed | Up-regulated (Tac-16/CK) | Down-regulated (Tac-16/CK) | Up-regulated (Tac-36/CK) | Down-regulated (Tac-36/CK) |
|---|---|---|---|---|---|---|
| ABA associated protein | 49 | 14 | 6 | 1 | 8 | 4 |
| Auxin related protein | 187 | 27 | 6 | 10 | 5 | 17 |
| Binding protein | 1544 | 94 | 37 | 28 | 22 | 34 |
| Dehydration-responsive element-binding protein | 12 | 5 | 0 | 3 | 0 | 5 |
| Dehydration-responsive protein RD22 | 11 | 2 | 1 | 0 | 1 | 1 |
| Heat shock protein | 163 | 48 | 18 | 28 | 0 | 25 |
| Late embryogenesis abundant protein | 5 | 0 | 0 | 0 | 0 | 0 |
| Ribosomal proteins | 1049 | 88 | 86 | 0 | 0 | 22 |
| Transcriptional factors | 1277 | 160 | 80 | 20 | 75 | 63 |
| Zinc finger proteins | 654 | 52 | 25 | 11 | 30 | 19 |
Fig 6. Differentially expressed transcripts involved in response to drought stress.
The functions of drought induced genes include protecting cells and regulating genes for signal transduction [70, 71]. According to their functions, these genes are classified into two groups. The first group of genes encoding proteins probably functions in stress tolerance, like LEA proteins, mRNA-binding proteins and aquaporins. In this study, we identified 5 and 55 transcripts encoding LEA proteins and aquaporins, respectively, but only three transcripts encoding aquaporins were differentially expressed significantly in dehydrated loranthus seeds. However, we found a number of binding proteins differentially expressed (Fig 6) and most of them were ATP-associated and RNA binding proteins (Table 3, S3 Table). The second group of gene products may be involved in further regulation of signal transduction and gene expression, like various transcription factors. In total, we detected 1,277 transcripts encoding transcription factors in all samples, of which 160 were differentially expressed during dehydration process. As shown in Table 3, 80 up-regulated and 20 down-regulated transcripts encoding TFs were identified in Tac-16 while 75 up-regulated and 63 down-regulated transcripts encoding TFs were identified in Tac-36, compared to CK. Differentially expressed transcripts encoding transcription factors include MYB, WRKY, and some ethylene-responsive transcription factors (S3 Table). These transcription factors have been reported to regulate ABA-dependent and ABA-independent pathways in plants to respond the dehydration stress and other stresses [19, 24, 72–75].
Among the ABA-dependent genes, RD22 (dehydration-responsive protein 22) is induced by drought stress because its promoter region contains a cis-acting element and can be recognized by MYB and MYC transcription factors [76]. We found RD22 mRNA was significantly up-regulated in dehydrated seeds compared to fresh seeds (Fig 6). Another group of ABA-dependent genes was heat shock protein (HSP). In response to dehydration, smHSPs (small HSPs) and HSPs are up-regulated in flesh fly, Sarcophaga crassipalpis [77, 78], the collembolan Folsomia candida [79], the eutardigrade Richtersius coronifer [80] and Belgica antarctica [81]. In contrast, we found smHSPs such as HS22C and HS23C and HSPs such as HS17C and HSP7C were down-regulated under drought stress in loranthus seeds (Table 3, Fig 6). It is hard to determine the expression patterns of HSPs under drought stress in plants. In Arabidopsis five transcripts encoding HSPs were up-regulated while one was down-regulated during rehydration process after dehydration [15]. In current study, we found 18 up-regulated and 28 down-regulated HSP transcripts in Tac-16 compared to CK, but they were down-regulated in loranthus seeds after being dehydrated over 36 hours. In addition, compared to the down-regulated HSPs, the expression levels of up-regulated HSPs in Tac-16 were much lower (Fig 6). Considering this we assume that the up-regulation of HSPs in orthodox seeds maybe one effective way to increase the tolerance to drought stress.
We also showed in Table 3and Fig 6five down-regulated transcripts encoding dehydration responsive element binding proteins (DRE2D and DREB3), which are ABA-independent genes. In Arabidopsis, Zea mays and Oryza sativa, DREB genes have been demonstrated to improve tolerance to drought, salt, cold and heat [82–84]. It is revealed in wheat and barley overexpression of DREB3 can elevate the expression of various stress responsive genes including CBF/DREB and LEA/COR/DHN genes, which means the DREB3 is inducible under drought stress and may help to increase the tolerance of water deficit [85]. However, both DRE2D and DREB3 were down-regulated in loranthus seeds during the dehydration process. In Table 3and Fig 6, we showed some other differentially expressed transcripts whose products might be associated with drought tolerance, including auxin related proteins, ribosomal proteins (RP) and zinc finger proteins (ZFNs). Of the 187 detected auxin related transcripts four (12KD_FRAAN, AX6B and LAX2) were down-regulated significantly in Tac-36 compared to CK. Although the relationship between 12KD_FRAAN (auxin-repressed 12.5 kDa protein) and drought is still unknown, 12KD_FRAAN has been reported to be correlated with fruit growth [86]. The decrease of auxin related proteins might explain the reduce of cell activities and viability of seeds during dehydration. In rice, overexpression of ZFNs can enhance drought and salt tolerance [87, 88]. But studies of ZFN gene expression changes in response of dehydration stresses are controversial [81, 89]. It is also hard to tell if ZFNs were up- or down-regulated in Tac-16 compared to CK (Table 3, Fig 6). We identified 25 up-regulated and 11 down-regulated ZFN transcripts in Tac-16, 30 up-regulated and 19 down-regulated ZFN transcripts in Tac-36, and it is implicated that ZFNs might function in drought tolerance at an early stage of dehydration in loranthus seeds. It is notable that the expression of 86 transcripts encoding ribosomal proteins was up-regulated in Tac-16 but down-regulated in Tac-36. Like ZFNs, ribosomal proteins function in early response of dehydration in Arabidopsis [90–92].
Transcripts involved in seed germination
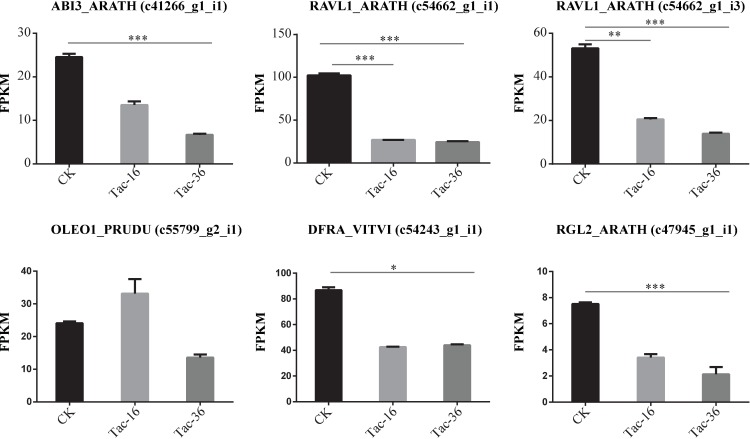
Previous studies have shown several genes are involved in the regulation of seed germination in Arabidopsis [93–96], Brassica oleracea [97] and Brassica napus [98]. Interestingly, some of them have been identified to respond to the drought stress as well. For example, ABI3, controls embryo degreening through SGR1 (Stay-green 1) and SGR2 (Stay-green 2) and participates in ABA-regulated gene expression during seed germination [96, 99]. In addition, COR47 (Cold-induced COR47 protein), OLEO1 (Oleosin), CHI (chalcone flavonone isomerase), CHS (chalcone synthase), DFR (dihydraflavonol-4-reductase), and RAB18 (Ras-related protein 18) have been characterized to regulate the process of Arabidopsis seed germination [96]. In rapeseeds, ZFN mRNAs were down regulated during the seed germination. In soybean seeds, LEA mRNAs are inducible by maturation or drying [100]. Some of them have been discussed according to their functions in response to drought stress previously. After removing lowly expressed transcripts strictly (<10 FRPKM), we found the transcripts encoding ABI3_ARATH, RAVL1_ARATH, DFRA_VITVI and RGL2_ARATH were down-regulated significantly in dehydrated loranthus seeds compared to fresh seeds (Fig 7). Because of water loss in loranthus seeds, four transcripts encoding CHS2_RUTGR, DFRA_VITVI and RAVL1_ARATH (encoded by two transcripts) were down-regulated while two transcripts encoding OLEO1_PRUDU and RAV2_ARATH increased more or less in Tac-16 and decreased quickly in Tac-36 (S3 Table). The down-regulation of seed germination key molecules might be related with the reduced seed viability and low rate of seed germination.
Fig 7. Transcripts associated with seed germination.
Different symbols are used show the significance of different expression: < 0.05 (*), <0.01 (**) and <0.001 (***). Error bar represents the standard deviation.
Validation by quantitative real-time PCR
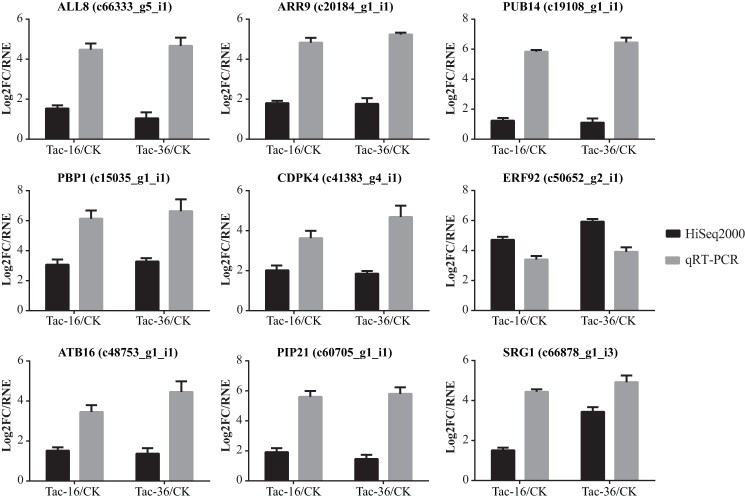
To validate the expression of differentially expressed transcripts in dehydrated and fresh loranthus seeds, quantitative real-time PCR (qRT-PCR) experiment was performed due to its high throughput, sensitivity and accuracy. It is widely used to determine the accuracy of transcripts and their expression identified by RNA-Seq [101, 102]. In view of this, 9 transcripts were randomly selected for qRT-PCR and actin-3 was used as control. The expected size of target transcripts ranged from 96 to 200 bp (S4 Table). For each transcript we performed three times in every sample. After qRT-PCR amplification ΔCt was calculated. Then, to compare a transcript in different samples, we used ΔΔCt method [51], shown as RNE. Fig 8showed the Log2FC identified by RNA-Seq (HiSeq2500) and RNE detected by qRT-PCR. It is clear that qRT-PCR confirmed the up-regulation of these transcripts.
Fig 8. qRT-PCR validation for 9 candidate transcripts.
Log2FC means log2 fold change in RNA-Seq experiment while RNE means relative normalized expression (2-ΔΔCt) in qRT-PCR experiment. Error bar represents the standard deviation.
Conclusions
In conclusion, we assembled the transcriptome of seeds during dehydration process in Taxillus chinensis (DC.) Danser using Illumina RNA-Seq system. A total of 164,546 transcripts corresponding to 114,971 genes were assembled from three libraries–fresh seeds (CK), seeds after 16 hours (Tac-16) and 36 hours (Tac-36) dehydration. Gene expression profiles showed 38,513 transcripts were commonly detected in all samples and 12,701, 9,935 and 16,098 transcripts were detected exclusively in CK, Tac-16 and Tac-36, respectively. Compared to CK, differential expression analysis demonstrated by edgeR characterized 2,102 up-regulated and 1,344 down-regulated transcripts in Tac-16 and 1,649 up-regulated and 2,135 down-regulated transcripts in Tac-36. K-means clustering analysis divided them into four groups with different expression patters and functions. Annotation of differentially expressed transcripts revealed transcripts encoding ABA associated proteins (ASR1 and ABAH4), dehydration-responsive protein RD22, zinc finger proteins and some TFs were up-regulated in Tac-16 and Tac-36. We also found controversial dysregulations of heat shock proteins in response to drought stress in loranthus seeds. Interestingly, transcripts encoding ribosomal proteins peaked in Tac-16, indicating they might be functional in early dehydration process. This is the first time to report loranthus transcriptome. The output of this study will contribute understanding the mechanism of gene regulation in loranthus seeds during dehydration and give new insights of genes induced by drought stress.
Supporting Information
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This study was funded by National Youth Science Foundation of China (81403045), Guangxi Youth Science Foundation (2014GXNSFBA118204) and National Science and Technology Major Project of China (2012ZX09304006).
Data Availability
The raw sequencing files of these six samples (FASTQ formatted files) can be accessed in the NCBI Sequence Read Archive (SRA) database (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number of SRA309567 (CK-1: SRR2902061; CK-2: SRR4067162; Tac-16-1: SRR2902062; Tac-16-2: SRR4067163; Tac-36-1: SRR2902063; Tac-36-2 SRR4067164). The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GELW00000000.
Funding Statement
This study was funded by National Youth Science Foundation of China (81403045), Guangxi Youth Science Foundation (2014GXNSFBA118204) and National Science and Technology Major Project of China (2012ZX09304006).
References
- 1.Commission CP. Chinese pharmacopoeia. Beijing: Chemical Industry Publishing House; 2010. p. p280. [Google Scholar]
- 2.Wang Y, Deng M, Zhang SY, Zhou ZK, Tian WX. Parasitic loranthus from Loranthaceae rather than Viscaceae potently inhibits fatty acid synthase and reduces body weight in mice. J Ethnopharmacol. 2008;118(3):473–8. 10.1016/j.jep.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 3.Xueying S, Min H, Quan Z. Inhibition effect of estradiol on proliferation of splenocytes and intervention of Sangjisheng (桑寄生)[J]. Pharmacology and Clinics of Chinese Materia Medica. 2005;6:022. [Google Scholar]
- 4.Ding B, Dai Y, Hou YL, Wu XM, Chen X, Yao XS. Four new hemiterpenoid derivatives from Taxillus chinensis. Fitoterapia. 2013;86:1–5. 10.1016/j.fitote.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 5.Li YH RJ, Chen SL, Lu D, Zhu KX, Zhao MH. Studies on medicinal plants of Loranthaceae resources in China. World Sci Technol Mod Tradit Chin Med. 2009;5:5. [Google Scholar]
- 6.Liu C-Y, Lin Y-C, Deng J-S, Liao J-C, Peng W-H, Huang G-J. Antioxidant, anti-inflammatory, and antiproliferative activities of Taxillus sutchuenensis. The American journal of Chinese medicine. 2012;40(02):335–48. [DOI] [PubMed] [Google Scholar]
- 7.Wang WB, Kim YH, Lee HS, Kim KY, Deng XP, Kwak SS. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol Biochem. 2009;47(7):570–7. 10.1016/j.plaphy.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 8.Farooq M, Wahid A, Kobayashi N, Fujita D, Basra S. Plant drought stress: effects, mechanisms and management Sustainable agriculture: Springer; 2009. p. 153–88. [Google Scholar]
- 9.Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18(10):575–83. 10.1016/j.tplants.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58(2):221–7. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- 11.Ingram J, Bartels D. The Molecular Basis of Dehydration Tolerance in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. 10.1146/annurev.arplant.47.1.377 [DOI] [PubMed] [Google Scholar]
- 12.Shinozaki K, Yamaguchi-Shinozaki K. Gene Expression and Signal Transduction in Water-Stress Response. Plant Physiol. 1997;115(2):327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray EA. Plant responses to water deficit. Trends in plant science. 1997;2(2):48–54. [Google Scholar]
- 14.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant Journal. 2002;31(3):279–92. [DOI] [PubMed] [Google Scholar]
- 15.Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, et al. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca 7000 full-length cDNA microarray. Plant J. 2003;34(6):868–87. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13(4):889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133(4):1755–67. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139(2):822–35. 10.1104/pp.105.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabara RC, Tripathi P, Lin J, Rushton PJ. Dehydration-induced WRKY genes from tobacco and soybean respond to jasmonic acid treatments in BY-2 cell culture. Biochem Biophys Res Commun. 2013;431(3):409–14. 10.1016/j.bbrc.2012.12.156 [DOI] [PubMed] [Google Scholar]
- 20.Chen LM, Zhou XA, Li WB, Chang W, Zhou R, Wang C, et al. Genome-wide transcriptional analysis of two soybean genotypes under dehydration and rehydration conditions. BMC Genomics. 2013;14:687 10.1186/1471-2164-14-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S, Zhang F, Yu Y, Zhang D, Zhao X, Wang W. Transcriptome profiling of dehydration stress in the Chinese cabbage (Brassica rapa L. ssp. pekinensis) by tag sequencing. Plant Molecular Biology Reporter. 2012;30(1):17–28. [Google Scholar]
- 22.Kido EA, Ferreira Neto JR, Silva RL, Pandolfi V, Guimaraes AC, Veiga DT, et al. New insights in the sugarcane transcriptome responding to drought stress as revealed by superSAGE. ScientificWorldJournal. 2012;2012:821062 10.1100/2012/821062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, et al. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot. 2009;60(12):3531–44. 10.1093/jxb/erp194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- 25.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17. 10.1101/gr.079558.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao B, Zhang D, Li X, Yang H, Zhang Y, Wood AJ. De novo transcriptome characterization and gene expression profiling of the desiccation tolerant moss Bryum argenteum following rehydration. BMC Genomics. 2015;16:416 10.1186/s12864-015-1633-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Zhang X, Zhang K, An H, Hu K, Wen J, et al. Comparative Analysis of the Brassica napus Root and Leaf Transcript Profiling in Response to Drought Stress. Int J Mol Sci. 2015;16(8):18752–77. 10.3390/ijms160818752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Yao D, Wang Q, Xu W, Wei Q, Wang C, et al. mRNA-seq analysis of the Gossypium arboreum transcriptome reveals tissue selective signaling in response to water stress during seedling stage. PLoS One. 2013;8(1):e54762 10.1371/journal.pone.0054762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berjak P, Pammenter NW. Implications of the lack of desiccation tolerance in recalcitrant seeds. Front Plant Sci. 2013;4:478 10.3389/fpls.2013.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delahaie J, Hundertmark M, Bove J, Leprince O, Rogniaux H, Buitink J. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J Exp Bot. 2013;64(14):4559–73. 10.1093/jxb/ert274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conacher C, Poiner I, Butler J, Pun S, Tree D. Germination, storage and viability testing of seeds of Zostera capricorni Aschers. from a tropical bay in Australia. Aquatic Botany. 1994;49(1):47–58. [Google Scholar]
- 33.Cabaço S, Santos R. Reproduction of the eelgrass Zostera marina at the species southern distributional limit in the Eastern Atlantic. Marine Ecology. 2010;31(2):300–8. [Google Scholar]
- 34.Smith FE. Tetrazolium salt. Science. 1951;113(2948):751–4. [DOI] [PubMed] [Google Scholar]
- 35.Gutterman Y, Gendler T, Rachmilevitch S. Survival of Schismus arabicus seedlings exposed to desiccation depends on annual periodicity. Planta. 2010;231(6):1475–82. 10.1007/s00425-010-1124-y [DOI] [PubMed] [Google Scholar]
- 36.Ji H, Chen M, Greening DW, He W, Rai A, Zhang W, et al. Deep Sequencing of RNA from Three Different Extracellular Vesicle (EV) Subtypes Released from the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct Mirna-Enrichment Signatures. 2014;9(10). 10.1371/journal.pone.0110314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 41.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods. 2011;8(10):785–6. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 42.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305(3):567–80. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 43.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42(Database issue):D231–9. 10.1093/nar/gkt1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 52.King M, Roberts E. The storage of recalcitrant seeds: achievements and possible approaches1979.
- 53.Adams C, Rinne R. Moisture content as a controlling factor in seed development and germination. International Review of Cytology. 1980;68:1–8. [Google Scholar]
- 54.Senaratna T, McKersie BD. Dehydration Injury in Germinating Soybean (Glycine max L. Merr.) Seeds. Plant Physiol. 1983;72(3):620–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, et al. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 2013;163(1):205–15. 10.1104/pp.113.223511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weitbrecht K, Muller K, Leubner-Metzger G. First off the mark: early seed germination. J Exp Bot. 2011;62(10):3289–309. 10.1093/jxb/err030 [DOI] [PubMed] [Google Scholar]
- 57.O'Neil ST, Emrich SJ. Assessing De Novo transcriptome assembly metrics for consistency and utility. BMC Genomics. 2013;14:465 10.1186/1471-2164-14-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95(6):315–27. 10.1016/j.ygeno.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mauch F, Mauch-Mani B, Boller T. Antifungal Hydrolases in Pea Tissue: II. Inhibition of Fungal Growth by Combinations of Chitinase and beta-1,3-Glucanase. Plant Physiol. 1988;88(3):936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bewley JD. Seed Germination and Dormancy. Plant Cell. 1997;9(7):1055–66. 10.1105/tpc.9.7.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, et al. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12:131 10.1186/1471-2164-12-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiology. 2004;134(3):1100–12. 10.1104/pp.103.035956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu HM, Cheun AY. Programmed cell death in plant reproduction. Plant Mol Biol. 2000;44(3):267–81. [DOI] [PubMed] [Google Scholar]
- 64.Fath A, Bethke P, Lonsdale J, Meza-Romero R, Jones R. Programmed cell death in cereal aleurone. Plant Mol Biol. 2000;44(3):255–66. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z. Small GTPases: versatile signaling switches in plants. Plant Cell. 2002;14 Suppl:S375–88. 10.1105/tpc.001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones AM. Programmed cell death in development and defense. Plant Physiol. 2001;125(1):94–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94. 10.1016/j.tplants.2004.12.012 [DOI] [PubMed] [Google Scholar]
- 68.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):410–7. [DOI] [PubMed] [Google Scholar]
- 69.Kalifa Y, Perlson E, Gilad A, Konrad Z, Scolnik P, BAR‐ZVI D. Over‐expression of the water and salt stress‐regulated Asr1 gene confers an increased salt tolerance. Plant, Cell & Environment. 2004;27(12):1459–68. [Google Scholar]
- 70.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675–90. 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130(4):2129–41. 10.1104/pp.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of plant research. 2011;124(4):509–25. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- 73.Channeliere S, Riviere S, Scalliet G, Szecsi J, Jullien F, Dolle C, et al. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Lett. 2002;515(1–3):35–8. [DOI] [PubMed] [Google Scholar]
- 74.Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, et al. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnol J. 2004;2(2):155–68. 10.1111/j.1467-7652.2004.00059.x [DOI] [PubMed] [Google Scholar]
- 75.Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot. 2007;58(11):2873–85. 10.1093/jxb/erm076 [DOI] [PubMed] [Google Scholar]
- 76.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9(10):1859–68. 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tammariello SP, Rinehart JP, Denlinger DL. Desiccation elicits heat shock protein transcription in the flesh fly, Sarcophaga crassipalpis, but does not enhance tolerance to high or low temperatures. J Insect Physiol. 1999;45(10):933–8. [DOI] [PubMed] [Google Scholar]
- 78.Hayward SA, Rinehart JP, Denlinger DL. Desiccation and rehydration elicit distinct heat shock protein transcript responses in flesh fly pupae. J Exp Biol. 2004;207(Pt 6):963–71. [DOI] [PubMed] [Google Scholar]
- 79.Bayley M, Petersen SO, Knigge T, Kohler H, Holmstrup M. Drought acclimation confers cold tolerance in the soil collembolan Folsomia candida. J Insect Physiol. 2001;47(10):1197–204. [DOI] [PubMed] [Google Scholar]
- 80.Jonsson KI, Schill RO. Induction of Hsp70 by desiccation, ionising radiation and heat-shock in the eutardigrade Richtersius coronifer. Comp Biochem Physiol B Biochem Mol Biol. 2007;146(4):456–60. 10.1016/j.cbpb.2006.10.111 [DOI] [PubMed] [Google Scholar]
- 81.Lopez-Martinez G, Benoit JB, Rinehart JP, Elnitsky MA, Lee RE Jr., Denlinger DL. Dehydration, rehydration, and overhydration alter patterns of gene expression in the Antarctic midge, Belgica antarctica. J Comp Physiol B. 2009;179(4):481–91. 10.1007/s00360-008-0334-0 [DOI] [PubMed] [Google Scholar]
- 82.Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007;50(1):54–69. 10.1111/j.1365-313X.2007.03034.x [DOI] [PubMed] [Google Scholar]
- 83.Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47(1):141–53. 10.1093/pcp/pci230 [DOI] [PubMed] [Google Scholar]
- 84.Lin RC, Park HJ, Wang HY. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant. 2008;1(1):42–57. 10.1093/mp/ssm004 [DOI] [PubMed] [Google Scholar]
- 85.Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J. 2011;9(2):230–49. 10.1111/j.1467-7652.2010.00547.x [DOI] [PubMed] [Google Scholar]
- 86.Reddy AS, Poovaiah BW. Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxin-regulated gene. Plant Mol Biol. 1990;14(2):127–36. [DOI] [PubMed] [Google Scholar]
- 87.Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, et al. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett. 2008;582(7):1037–43. 10.1016/j.febslet.2008.02.052 [DOI] [PubMed] [Google Scholar]
- 88.Liu K, Wang L, Xu Y, Chen N, Ma Q, Li F, et al. Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta. 2007;226(4):1007–16. 10.1007/s00425-007-0548-5 [DOI] [PubMed] [Google Scholar]
- 89.Mukhopadhyay A, Vij S, Tyagi AK. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci U S A. 2004;101(16):6309–14. 10.1073/pnas.0401572101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiyosue T, Abe H, Yamaguchi-Shinozaki K, Shinozaki K. ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim Biophys Acta. 1998;1370(2):187–91. [DOI] [PubMed] [Google Scholar]
- 91.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol. 1994;25(5):791–8. [DOI] [PubMed] [Google Scholar]
- 92.Kawaguchi R, Girke T, Bray EA, Bailey-Serres J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 2004;38(5):823–39. 10.1111/j.1365-313X.2004.02090.x [DOI] [PubMed] [Google Scholar]
- 93.Barrero JM, Millar AA, Griffiths J, Czechowski T, Scheible WR, Udvardi M, et al. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010;61(4):611–22. 10.1111/j.1365-313X.2009.04088.x [DOI] [PubMed] [Google Scholar]
- 94.Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16(5):646–58. 10.1101/gad.969002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leeggangers HA, Folta A, Muras A, Nap JP, Mlynarova L. Reduced seed germination in Arabidopsis over-expressing SWI/SNF2 ATPase genes. Physiol Plant. 2015;153(2):318–26. 10.1111/ppl.12231 [DOI] [PubMed] [Google Scholar]
- 96.Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6(11):1567–82. 10.1105/tpc.6.11.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Soeda Y, Konings MC, Vorst O, van Houwelingen AM, Stoopen GM, Maliepaard CA, et al. Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiol. 2005;137(1):354–68. 10.1104/pp.104.051664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fei H, Tsang E, Cutler AJ. Gene expression during seed maturation in Brassica napus in relation to the induction of secondary dormancy. Genomics. 2007;89(3):419–28. 10.1016/j.ygeno.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 99.Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, et al. ABI3 controls embryo degreening through Mendel's I locus. Proc Natl Acad Sci U S A. 2013;110(40):E3888–94. 10.1073/pnas.1308114110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsing YC, Chen ZY, Shih MD, Hsieh JS, Chow TY. Unusual sequences of group 3 LEA mRNA inducible by maturation or drying in soybean seeds. Plant Mol Biol. 1995;29(4):863–8. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–47. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang H, Liu J, Huang S, Guo T, Deng L, Hua W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene. 2014;538(1):113–22. 10.1016/j.gene.2013.12.057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The raw sequencing files of these six samples (FASTQ formatted files) can be accessed in the NCBI Sequence Read Archive (SRA) database (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number of SRA309567 (CK-1: SRR2902061; CK-2: SRR4067162; Tac-16-1: SRR2902062; Tac-16-2: SRR4067163; Tac-36-1: SRR2902063; Tac-36-2 SRR4067164). The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GELW00000000.
The raw sequencing files of these six samples (FASTQ formatted files) can be accessed in the NCBI Sequence Read Archive (SRA) database (http://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession number of SRA309567 (CK-1: SRR2902061; CK-2: SRR4067162; Tac-16-1: SRR2902062; Tac-16-2: SRR4067163; Tac-36-1: SRR2902063; Tac-36-2 SRR4067164). The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GELW00000000. Scripts for key steps, such as transcriptome de novo assembly, annotation, expression profile and differential expression, can be seen in S1 File.